Introduction
Recent global cancer statistics have shown that the
incidence of lung cancer ranks second after breast cancer, and that
it has the highest mortality rate among malignant tumors (1). Non-small cell lung cancer (NSCLC)
accounts for >80% of all types of lung cancer, and >70% of
patients pathologically diagnosed with NSCLC are in the advanced
stages, often making them unsuitable for surgery (2). Compared with the stronger toxic
side effects of chemotherapy, first-generation EGFR-tyrosine kinase
inhibitors (TKIs), such as gefitinib and erlotinib, can
significantly improve the efficacy and quality of life of patients
with EGFR-sensitive mutations. Resistance is a common problem that
affects the efficacy of clinical drugs, and studies on acquired
resistance of first-and second-generation EGFR-TKIs have found that
the T790M mutation in exon 20 of EGFR is the main mechanism of
resistance (3). Based on these
results, osimertinib, a third-generation EGFR-TKI was developed,
which has been shown to significantly prolong the median
progression-free survival and median overall survival (18.9 and
38.6 months, respectively) of patients with advanced NSCLC with an
EGFR mutation (4), and has
become one of the most effective and safe standard treatment
regimens for advanced patients with NSCLC and the EGFR T790M
mutation (5). Furthermore, its
high blood-brain barrier penetration can control central nervous
system metastasis (6). However,
long-term use of osimertinib often results in diminished drug
efficacy, rendering patients less sensitive to the drug and leading
to treatment failure. Possible mechanisms underlying osimertinib
resistance have been reported to include EGFR-dependent and
EGFR-independent mechanisms, and also involve alterations in
signaling pathways, oncogenic gene fusions and autophagy (7-12); however, the underlying mechanisms
remain to be explored. Therefore, constructing
osimertinib-resistant NSCLC cell lines in vitro, initially
exploring the mechanism of osimertinib resistance and identifying
methods to reverse drug resistance are important areas in the field
of lung cancer research.
Circular RNA (circRNA) is a type of non-coding RNA
characterized by a covalently closed loop, which is different from
traditional linear RNA. circRNA has the characteristics of tissue
specificity and evolutionary conservation, and is not degraded by
common exonucleases. It can stably exist in complex intracellular
and extracellular environments, and can bind microRNA (miRNA) or
protein (13). In addition,
circRNA can regulate the expression of downstream genes and is
involved in a variety of tumor signaling pathways, where it
participates in the regulation of gene transcription, protein
translation and other functions, providing a novel direction for
NSCLC drug resistance research (14-17). A previous study reported that
circFGFR1 serves a role as an oncogene in NSCLC (18). In addition, hsa_circ_0000567 is
upregulated in NSCLC cells with acquired gefitinib resistance
(19). The expression levels of
hsa_circ_0043632 in osimertinib-resistant NSCLC cells are also
higher than those in sensitive NSCLC cells; however, the mechanism
remains unclear (20).
circRNA serves an important regulatory role in the
occurrence and development of lung cancer and drug resistance;
however, there are few reports on circRNA and osimertinib
resistance in NSCLC (21,22).
The present study assessed the abnormal expression of circRNAs in
osimertinib-resistant NSCLC and the molecular mechanism underlying
the regulation of targeted drug resistance. It will further enrich
the current theory of EGFR-TKI resistance and create a new area of
study for the investigation of therapeutic methods to overcome
EGFR-TKI resistance.
Materials and methods
Cell culture and reagents
NCI-H1975 (EGFR, L858R and T790M mutations) and
HCC827 (EGFR, E746-A750 deletion) NSCLC cells were obtained from
The Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. Immortalized human umbilical vein endothelial cells
(HUVECs) were purchased from Zhongqiao Xinzhou Biotechnology Co.,
Ltd. (cat. no. ZQY004). H1975 and HCC827 cells were cultured in
RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc.), whereas
HUVECs were cultured in Endothelial Cell Medium (ScienCell Research
Laboratories, Inc.). Media were supplemented with 10% fetal bovine
serum (Shanghai Yeasen Biotechnology Co., Ltd.), 100 U/ml
penicillin and 100 µg/ml streptomycin. All cells were
cultured in a humidified incubator at 37°C with 5% CO2.
Osimertinib, rapamycin (RAPA; cat. no. HY-10219) and
3-methyladenine (3-MA; cat. no. HY-19312) were purchased from
MedChemExpress. HCC827, HCC827-OR, H1975 and H1975-OR cells were
treated with 10 µM RAPA or 1 nM 3-MA for 24 h at 37°C, to
detect the expression levels of autophagy-related proteins.
Establishment of osimertinib-resistant
H1975 and HCC827 cells lines
The osimertinib-resistant H1975 and HCC827 cells
were established by continuous exposure to stepwise-increasing
concentrations of osimertinib. Osimertinib was dissolved in
dimethyl sulfoxide before being added to the cell culture medium.
The initial concentrations of osimertinib were equal to the
half-maximal inhibitory concentration (IC50) in H1975 (2
nM) and HCC827 (10 nM) cells (23). Cells were then incubated in
drug-free medium until the surviving cells had recovered and
exhibited a normal exponential growth rate. The dosing was
continued at the original concentration until the cells could grow
normally at this concentration. Subsequently, the drug
concentration was increased until the cells could grow stably. When
the drug screening concentration was 5 times the IC50
value of parental cells, the concentration difference was increased
by 20 nM until the cells could grow stably. When the drug screening
concentration was 10 times the IC50 value of parental
cells, the concentration difference was increased by 50 nM until
the cells could grow stably. When the drug screening concentration
was 20 times the IC50 value of the parental cells, the
concentration was increased by 100 nM until the cells could grow
stably. After 10 months, the newly established
osimertinib-resistant cell lines, which were indicated as H1975-OR
and HCC827-OR cells, were maintained in medium containing 1.5 and
2.5 µM osimertinib, respectively. The parental H1975 and
HCC827 cells were cultured in drug-free medium in parallel. The
drug resistance index (RI) is equal to the IC50 value of
the resistant cells divided by the IC50 value of the
parental cells.
Morphological analysis
H1975, H1975-OR, HCC827 and HCC827-OR cells were
cultured in a 6-well plate at a density of 1×105
cells/well. After incubation at 37°C for 24 h, the cellular
morphology was observed using an inverted light microscope (Olympus
Corporation).
Cell transfection
Plasmids were purified using DNA Midiprep Kits
(Qiagen, Inc.) and transfected into cells using Polyjet
transfection reagent (SignaGen Laboratories). Small interfering
RNAs (siRNAs) against circRNAs and a siRNA negative control
obtained from Shanghai Genepharma Co., Ltd. were transfected into
cells (HCC827-OR, H1975, H1975-OR) using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Cells and siRNAs were incubated with
Lipofectamine 2000 at 37°C for 6 h. Subsequently, the transfection
reagents were replaced with complete RPMI 1640 medium. A total of
48 h post-transfection the cells were harvested for subsequent
experiments. A total of 100 pmol siRNA was used for transfection in
one well of a six-well plate. The siRNA sequences are listed in
Table I. circPDLIM5
overexpression plasmid was generated by chemical gene synthesis,
and PLVX-puro vector was used as the plasmid backbone (TsingKe
Biological Technology). HCC827-OR cells were chosen for plasmid
transfection when cell density reached the optimal 70-80%
confluence at the time of transfection. A total of 1 µg
plasmid was used for transfection in one well of a six-well plate.
Cells and plasmids were incubated with Polyjet at room temperature
for 15 min. Cells were harvested for subsequent experiments 48 h
post-transfection. An empty PLVX-puro vector was used as the
negative control for plasmid transfection.
 | Table IPrimer and siRNA sequences. |
Table I
Primer and siRNA sequences.
| Name | Sequence,
5′-3′ |
|---|
| circPLOD2-F |
AATGGAAATGGACCCACCAAG |
| circPLOD2-R |
ATCACCACCTCTCCATTCTTCTC |
| circTNFRSF21-F |
GCCATTGTGGAAAAGGCAGG |
| circTNFRSF21-R |
GGCTTGTGTTGGTACAATGCT |
| circPPP4R1-F |
GGGAGTCCGAAAGGCTTGTG |
| circPPP4R1-R |
AGTCATCCACACCAACCAAC |
| circMUC16-F |
GGACCCCATAGTCTCCCCTCT |
| circMUC16-R |
TGCAGGTTGGTGATGGTGAAG |
| circANKRD11-F |
TATACAGCCCTGCTCTGGGA |
| circANKRD11-R |
CTGAAAGTCAGTGCTGACGAG |
| circGLS-F |
AAGGCACAGACATGGTTGGTA |
| circGLS-R |
AACCTTTCCTCCAGACTGCT |
| circRSF1-F |
AAGCTACAGATTCTTCTGTGTGA |
| circRSF1-R |
AGGTGGGCAGAACCATTCTC |
| circPDLIM5-F |
GGAACAACTCAGTCTCGCTCT |
| circPDLIM5-R |
ACACAGCAGGAGATGTAATGGG |
| circNAV3-F |
GACAGTGGCACAAAGCAGTG |
| circNAV3-R |
GGTTGGCCCAGTCAGTGTAA |
| circSNX13-F |
ATGCGATACTTTGTCAGGGAAAT |
| circSNX13-R |
AAGGACAATGCCAAGGCTTCC |
| GAPDH-F |
AGAAGGCTGGGGCTCATTTG |
| GAPDH-R |
AGGGGCCATCCACAGTCTTC |
| GPC3-F |
CCTTTGAAATTGTTGTTCGCCA |
| GPC3-R |
CCTGGGTTCATTAGCTGGGTA |
| SOX2-F |
GCCGAGTGGAAACTTTTGTCG |
| SOX2-R |
GGCAGCGTGTACTTATCCTTCT |
| AUTS2-F |
GGGCTCCGACAAGGAAGAC |
| AUTS2-R |
TGGCGTTTCTCCACACGTTC |
| CFH-F |
GTGAAGTGTTTACCAGTGACAGC |
| CFH-R |
AACCGTACTGCTTGTCCAAAA |
| SESN3-F |
ACCTGCTCTGTACCAACTGC |
| SESN3-R |
GACGACCGGATGTAGAGTATTCT |
| COL1A2-F |
GTTGCTGCTTGCAGTAACCTT |
| COL1A2-R |
AGGGCCAAGTCCAACTCCTT |
| LOX-F |
CGGCGGAGGAAAACTGTCT |
| LOX-R |
TCGGCTGGGTAAGAAATCTGA |
| VCAN-F |
GTAACCCATGCGCTACATAAAGT |
| VCAN-R |
GGCAAAGTAGGCATCGTTGAAA |
| NNMT-F |
ATATTCTGCCTAGACGGTGTGA |
| NNMT-R |
TCAGTGACGACGATCTCCTTAAA |
| C3-F |
GGGGAGTCCCATGTACTCTATC |
| C3-R |
GGAAGTCGTGGACAGTAACAG |
| ALDH1A1-F |
GCACGCCAGACTTACCTGTC |
| ALDH1A1-R |
CCTCCTCAGTTGCAGGATTAAAG |
| PTPRD-F |
CTCCAAGGTTTACACGAACACC |
| PTPRD-R |
AGTCCGTAAGGGTTGTATTCTGA |
| SPP1-F |
CTCCATTGACTCGAACGACTC |
| SPP1-R |
CAGGTCTGCGAAACTTCTTAGAT |
| SEMA6A-F |
AATCAGTATTTCGCATGGCAACT |
| SEMA6A-R |
GCAATGTAGAGGGTTCCGTTCA |
| UGT3A1-F |
AGTCCATCTTCCATCCCCGA |
| UGT3A1-R |
AACAGCCCTGAATGGGCTAC |
| TMPRSS11E-F |
CCTGGCAGTGTGCATTGGA |
| TMPRSS11E-R |
CAAGTCTCTGGCTCATTTCTGT |
| ALDH1L2-F |
GCTGAAGTTGGCACTAATTGGC |
| ALDH1L2-R |
TGAACACCCCTACTACTCGGT |
| SPTB-F |
CCAGCCACCTTACAGCAGG |
| SPTB-R |
TGCGAGTTCACCCATTTCGT |
| SPARC-F |
TGAGGTATCTGTGGGAGCTAATC |
| SPARC-R |
CCTTGCCGTGTTTGCAGTG |
| NRK-F |
AGGGAGGTCACGGATCTGG |
| NRK-R |
CATAAGTACCAAGGCCAATGGTT |
|
si-circPPP4R1-1#-sense |
CGUUGGUUGGUGUGGAUGATT |
|
si-circPPP4R1-1#-antisense |
UCAUCCACACCAACCAACGTT |
|
si-circPPP4R1-2#-sense |
UUGGUUGGUGUGGAUGACUTT |
|
si-circPPP4R1-2#-antisense |
AGUCAUCCACACCAACCAATT |
|
si-circPDLIM5-1#-sense |
AGAAGGCAAAAGCAUCUGCTT |
|
si-circPDLIM5-1#-antisense |
GCAGAUGCUUUUGCCUUCUTT |
|
si-circPDLIM5-2#-sense |
AAAGAAGGCAAAAGCAUCUTT |
|
si-circPDLIM5-2#-antisense |
AGAUGCUUUUGCCUUCUUUTT |
| si-NC-F |
UUCUCCGAACGUGUCACGUTT |
| si-NC-R |
ACGUGACACGUUCGGAGAATT |
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) analyses
Total RNA was extracted from H1975, H1975-OR, HCC827
and HCC827-OR cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. RNA was digested with RNase R (Epicentre; Illumina, Inc.)
to eliminate linear RNAs and enrich circRNAs. RNA was not digested
with RNase R when detecting linear RNAs. Subsequently, total RNA
(1.0 µg) was reverse transcribed to cDNA using a Reverse
Transcription kit (Takara Biotechnology Co., Ltd.) according to
manufacturer's protocol, and qPCR analyses were performed using
SYBR Green (Takara Biotechnology Co., Ltd.) to determine the
relative expression levels of the specific genes. qPCR was
performed as follows: Initial denaturation at 95°C for 30 sec; 40
cycles at 95°C for 5 sec and 60°C for 34 sec; followed by melting
curve at 95°C for 5 sec and 60°C for 60 sec. The primer sequences
are listed in Table I. Relative
mRNA expression levels were analyzed using the 2−ΔΔCq
method with GAPDH as the control (24).
Next-generation sequencing
DNA was extracted from the cell samples according to
the instructions of the DNA extraction kit (cat. no. DC102-01;
Vazyme Biotech Co., Ltd.). DNA quantification was performed on the
Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Inc.) with the
dsDNA HS Assay kit (Thermo Fisher Scientific, Inc.). The DNA was
purified using VAHTS DNA Clean Beads (cat. no. N411-02; Vazyme
Biotech Co., Ltd.), and was then fragmented and DNA libraries were
constructed using the KAPA Hyper Prep kit (KAPA Biosystems; Roche
Diagnostics) according to the manufacturer's instructions. The
loading concentration of the final library was 1.75 nM. The
concentration was measured by qPCR analysis as aforementioned with
the standard curve (25). A
panel targeting 139 cancer-related genes (GeneseeqPrime™; Geneseeq
Technology, Inc.) using customized xGen lockdown probes (Integrated
DNA Technologies, Inc.) was used for hybridization capture. The
hybridization reaction was performed with Dynabeads M-279 (Thermo
Fisher Scientific, Inc.), and xGen lockdown hybridization and wash
kit (Integrated DNA Technologies, Inc.). The target-enriched
libraries were sequenced on the Illumina Hiseq4000 NGS platform
(HiSeq 3000/4000 SBS kit, cat. no. FC-410-1001; HiSeq 3000/4000 SR
Cluster kit, cat. no. GD-410-1001; HiSeq 3000/4000 PE Cluster kit,
cat. no. PE-410-1001; HiSeq 4000 Sequencing System, cat. no.
SY-401-4001; all from Illumina, Inc.) using the 150 bp paired-end
reading according to the manufacturer's protocol. Somatic single
nucleotide variants and small insertion/deletions were called by
VarScan2 (26). Copy number
variation (CNV) was screened using CNVkit (27). A fold-change of >1.50 was
considered to be a copy number gain and <0.65 was defined as
copy number loss.
High-throughput RNA sequencing
(RNA-seq)
The preparation of whole transcriptome libraries and
deep sequencing was performed by Shanghai BIO Biotech. VAHTS Total
RNA-Seq(H/M/R) Library PrepKit for Illumina (cat. no. NR605-02;
Vazyme Biotech Co., Ltd.) was used to prepare RNA samples for
sequencing. The RNA integrity number of the extracted RNA was
assessed using an Agilent Bioanalyzer 2100 (Agilent Technologies,
Inc.). Qubit®2.0 Fluorometer (Thermo Fisher Scientific,
Inc.) was used to detect the concentration of the library and
Agilent 4200 (Agilent Technologies, Inc. was used to detect the
size of the library; the loading concentration and volume of the
final library was 35 ng/µl and 15 µl. High-throughput
RNA-seq was then performed after sequencing reagents were prepared
according to the Illumina user guide. Resulting libraries were
sequenced initially on an Illumina nova-seq instrument (HiSeq X Ten
Reagent kit v2.5, cat. no. FC-501-2501; HiSeq X Ten System, cat.
no. SY-412-1001; both from Illumina, Inc.) that generated
paired-end reads of 150 nucleotides. Quality control standards for
sequencing results were as follows: The amount of data was ~15
G/sample, and Q20 ratio (%) was not less than 90%. Finally, the
clean reads were compared with the reference genome using an
algorithm for aligning sequence reads against the reference genome
(BWA-MEM; https://github.com/lh3/bwa) with
default parameters. The reference genome used was GRCh38. For mRNA
analyses, the RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq) and Ensemble
(https://www.ensembl.org/index.html)
transcript databases were chosen as the annotation references.
CircRNA distribution in the genome was explored by comparing the
circRNA sequence with the genome. The host linear transcripts were
annotated according to the location of the circRNA sequence in the
chromosome. The expression of circRNAs was calculated using
junction reads at the back-splicing sites, and SRPBM was used to
normalize the reads. SRPBM value was calculated using the following
formula: SRPBM=(SR x109)/N, in which SR is the number of
spliced reads, and N is the total number of mapped reads in
samples. The differential analysis of circRNAs between the two
groups (HCC827 vs. HCC827-OR, H1975 vs. H1975-OR) was performed
using the negative binomial distribution test based on the DESeq
package (https://www.rdocumentation.org/packages/DESeq/versions/1.24.0).
Hierarchical clustering analysis between the two groups (HCC827 vs.
HCC827-OR, H1975 vs. H1975-OR) was conducted using the R stats
package (https://www.rdocumentation.org/packages/stats/versions/3.6.2).
After the P-value was obtained via Fisher's exact test, the P-value
threshold was determined by false discovery rate. The differential
expression multiple (fold-change) was also calculated based on the
SRPBM values. There were two criteria used to screen for
differentially expressed genes, as follows: One was the fold-change
of the same circRNA between two groups, indicating how much the
expression value of this gene differs between the two groups, and
the other was P-value (P<0.05) or false discovery rate, showing
whether the results can be trusted.
Functional and pathway enrichment
analysis of the host genes of differentially expressed
circRNAs
According to the position information of circRNAs,
the protein-coding gene corresponding to the genomic position of
circRNA can be obtained using CIRI software (https://sourceforge.net/projects/ciri/files/CIRI-full/CIRI_Full_v2.1.1.jar/download),
which is known as the host gene. Gene Ontology (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were
performed on the host genes of the differentially expressed
circRNAs via the clusterProfiler package. clusterProfiler
(https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
is a Bioconductor-dependent R software package that automates the
biological term classification process and gene cluster enrichment
analysis.
Sanger sequencing
circPDLIM5 and circPPP4R1 sequences were obtained
using divergent primers sent to TsingKe Biological Technology for
Sanger sequencing analysis. Primers are listed in Table I.
Calculation of IC50
The IC50 values were determined using the
Cell Counting Kit 8 (CCK8) cell viability assay (cat. no.
40203ES80; Shanghai Yeasen Biotechnology Co., Ltd.). H1975,
H1975-OR, HCC827 and HCC827-OR cells were seeded
(3.0×103 cells/well) in 96-well plates. Subsequently,
0.25, 0.5, 1, 2, 4, 8, 16 and 32 nM osimertinib were added to H1975
cells; 5, 10, 20, 40, 80, 160, 320 and 640 nM osimertinib were
added to HCC827 cells; 0.25, 0.5, 1, 2, 4, 8, 16 and 24 µM
osimertinib were added to H1975-OR cells; 0.5, 1, 2, 4, 8, 12, 16
and 32 µM osimertinib were added to HCC827-OR cells. After
treatment with osimertinib for 72 h at 37°C, 100 µl CCK8
solution was added to each well (1:10 dilution) for 2 h at 37°C,
after which, the absorbance was measured at 450 nm. According to
drug concentration and corresponding OD value, the drug
concentration-cell inhibition rate curve was drawn to calculate
IC50 values.
Cell proliferation assays
Cells were seeded at a density of 2.0×103
cells/well in 96-well plates. Six replicate wells were used for
each analysis. The untransfected cells were cultured in different
concentrations of osimertinib (HCC827 and HCC827-OR cells: 0, 15 nM
and 6.5 µM osimertinib; H1975 and H1975-OR cells: 0, 2.5 nM
and 2.5 µM osimertinib). The transfected cells were cultured
in 0 and 1 µM osimertinib. The CCK8 solution was added to
each well prior to the endpoint of incubation at a 1:10 (v/v)
dilution per 100 µl, and was incubated for 2 h at 37°C. Cell
viability was determined by measuring the absorbance of the
converted dye at 450 nm.
EdU assay
The cellular proliferative capability was assessed
using the EdU assay kit (Guangzhou RiboBio Co., Ltd.). Cells were
cultured in 24-well plates at 2×104 cells/well.
Subsequently, 50 µM EdU labeling medium was added to the
24-well plates and they were incubated for 2 h at 37°C. The
following experimental procedures were performed at room
temperature. After treatment with 4% paraformaldehyde for 0.5 h and
0.5% Triton X-100 for 1 h, cells were stained with EdU reaction
solution for 30 min. DAPI was used to label cell nuclei.
EdU-positive cells were observed and imaged under a fluorescence
microscope (Olympus Corporation).
Colony formation assay
The untransfected cells were seeded in a 6-well
plate at a density of 1,000 cells/well, and cultured in different
concentrations of osimertinib (HCC827 and HCC827-OR cells: 0, 15 nM
and 6.5 µM osimertinib; H1975 and H1975-OR cells: 0, 2.5 nM
and 2.5 µM osimertinib). The transfected cells were cultured
in 0 and 1 µM osimertinib. They were incubated in a cell
culture incubator until colony formation occurred for 10 days at
37°C. The colonies were washed with PBS and fixed with 4%
polyformaldehyde for 30 min at room temperature. Subsequently, the
colonies were stained with 0.1% crystal violet at room temperature
for 30 min. Images of the colonies (>50 cells) were captured
using digital camera (Panasonic) and calculated by ImageJ software
(version 1.5.3; National Institutes of Health).
Transwell assay
Transwell chambers (MilliporeSigma; pore size, 8.0
µm) were used to assess migration. The transfected cells
(5×104) were directly added to the insert membranes.
Serum-free medium containing 0 and 1 µM osimertinib was
placed into the upper chamber and medium containing 10% FBS was
added to the lower chamber. Following incubation for 48 h at 37°C,
the remaining cells on the upper membrane were removed with cotton
wool. Invasive cells were fixed with methanol for 30 min at room
temperature and stained with 0.1% crystal violet for 30 min at room
temperature. Stained cells were counted under a light microscope
(Olympus Corporation) and images were captured.
Flow cytometric analysis
The Annexin V-FITC Apoptosis Detection Kit (cat. no.
556547; BD Biosciences) was used to assess apoptosis. Cells were
cultured in a 6-well plate at a density of 1×105
cells/well and were treated with different concentrations of
osimertinib (HCC827, HCC827-OR: 0, 15 nM and 6.5 µM; H1975,
H1975-OR: 0, 2.5 nM and 2.5 µM) or were transfected with 100
pmol siRNA. After 48 h, the cells in each well were collected and
dissolved in 1 ml 1X Annexin V Binding Buffer (cat. no. 51-66121E;
BD Biosciences). Subsequently, 10 µl Annexin V-FITC (cat.
no. 51-65874X; BD Biosciences) was added and mixed for Annexin
V-FITC labeling. The cells were incubated at room temperature in
the dark for 15 min. A total of 5 min before analysis, 10 µl
PI Staining Solution (cat. no. 51-66211E; BD Biosciences) was added
for staining at room temperature in the dark for 15 min. The
cellular DNA content in the treated cells was quantified using a
FACScan flow cytometer (BD Biosciences) and the results were
analyzed using FlowJo software v10.6.2 (FlowJo, LLC,).
For cell cycle distribution analysis,
1×105 cells were treated with different concentrations
of osimertinib (HCC827, HCC827-OR: 0, 15 nM and 6.5 µM;
H1975, H1975-OR: 0, 2.5 nM and 2.5 µM) or transfected with
100 pmol siRNA. After 48 h, the cells were fixed in ice-cold 70%
ethanol. The cells were resuspended in 0.5 ml PI/RNase staining
solution (cat. no. 550825; BD Biosciences) and incubated at room
temperature in the dark for 15 min. Prior to analysis, the samples
can be temporarily stored at 4°C in the dark. Within 1 h, the
samples were quantitatively detected using a FACScan flow
cytometer. The results were analyzed using FlowJo v10.6.2
software.
Tube formation assay
A Matrigel-based tube formation assay was performed
to assess tumor cell vasculogenesis. Briefly, 2×104/well
HUVECs were seeded onto 96-well plates coated with 50 µl
Matrigel (BD Biosciences) and allowed to polymerize for 10 min at
37°C. H1975-OR or HCC827-OR cells transfected with siRNAs were
centrifuged at room temperature for 5 min at 300 × g. The
conditioned medium containing 0 or 1 µM osimertinib was
collected from the supernatant and added to the well. After
incubation for 4-6 h at 37°C, the formation of tube structures was
observed under an inverted light microscope, and representative
images were captured. Total tubule length and number of tubule
branches in each well was calculated by ImageJ software (version
1.5.3). All experiments were repeated independently in
triplicate.
Western blot analysis
After treatment with different concentrations (0,
10, 100, 1,000 and 10,000 nM) of osimertinib, cells were lysed with
RIPA extraction reagent (Beyotime Institute of Biotechnology)
supplemented with a protease and phosphatase inhibitor cocktail
(MedChemExpress). Protein concentrations were measured using the
BCA Protein Assay Kit (Beyotime Institute of Biotechnology).
Proteins (20 µg/lane) were separated by SDS-PAGE on 10% gels
and were transferred to 0.22-µm polyvinylidene difluoride
membranes. Membranes was blocked with 1X TBS-0.1% Tween-20 (TBST)
containing 5% BSA (Beyotime Institute of Biotechnology) with
constant agitation for 1 h at room temperature. Membranes were then
incubated with primary antibodies (1:1,000) against EGFR (cat. no.
4267), phosphorylated (p)-EGFR (cat. no. 3777), AKT (cat. no.
4685), p-AKT (cat. no. 4060), ERK (cat. no. 4695), p-ERK (cat. no.
4370) (all from Cell Signaling Technology, Inc.), P62 (cat. no.
66184-1-Ig), LC3-I/II (cat. no. 14600-1-AP) and GAPDH (cat. no.
60004-1-Ig) (all from Proteintech Group, Inc.) overnight at 4°C.
HRP-conjugated goat anti-rabbit or anti-mouse IgG H&L
pre-adsorbed secondary antibodies (1:10,000; cat. nos. SA00001-2
and SA00001-1; Proteintech Group, Inc.) were then used to incubate
the membranes for 1 h at room temperature. Subsequently, the
membranes were incubated with Super ECL Detection Reagent (cat. no.
36208ES60; Shanghai Yeasen Biotechnology Co., Ltd.) for 1 min and a
chemiluminescence imaging system (cat. no. 5200; Tanon Science and
Technology Co., Ltd.) was used for visualization of the blots.
Statistical analysis
Statistical analyses were performed using SPSS 20.0
(IBM Corp.) and GraphPad Prism 9 software (Dotmatics). All
experiments were performed at least three times and data are
presented as the mean ± standard deviation. When the normality of
data distribution and equal variance between groups was met,
unpaired Student's t-test was applied in two-group comparisons. For
multiple comparisons, one-way ANOVA or two-way ANOVA and Bonferroni
post hoc test were used, or Kruskal-Wallis test and Dunn post hoc
test were used if the normality of data distribution and equal
variance between groups was not met. P<0.05 was considered to
indicate a statistically significant difference.
Results
Establishment and identification of
osimertinib-resistant cell lines
Among the NSCLC cell lines, H1975 and HCC827 are
sensitive to osimertinib, although they have different mutations;
H1975 cells harbor the L858R/T790M double mutation, and HCC827
cells harbor the E746-A750 deletion. These two cell lines with
different gene mutation backgrounds were selected to construct
osimertinib-resistant cell lines in the present study, with the aim
of understanding the mechanism of osimertinib resistance more
comprehensively. Osimertinib-resistant cell lines were induced by
increasing the drug concentration. Initially, the parental cells
HCC827 and H1975 were cultured to a stable state, and were then
treated with osimertinib, the proliferation inhibition rate was
detected and the IC50 value was calculated, which was
used as the initial concentration. The initial concentrations of
osimertinib used to treat HCC827 and H1975 cells were 10 and 2 nM,
respectively. The dosing was continued at the original
concentration until the cells could grow normally at this
concentration, after which the drug concentration was increased, as
aforementioned. Finally, after 10 months, osimertinib-resistant
cell lines were successfully established, named HCC827-OR and
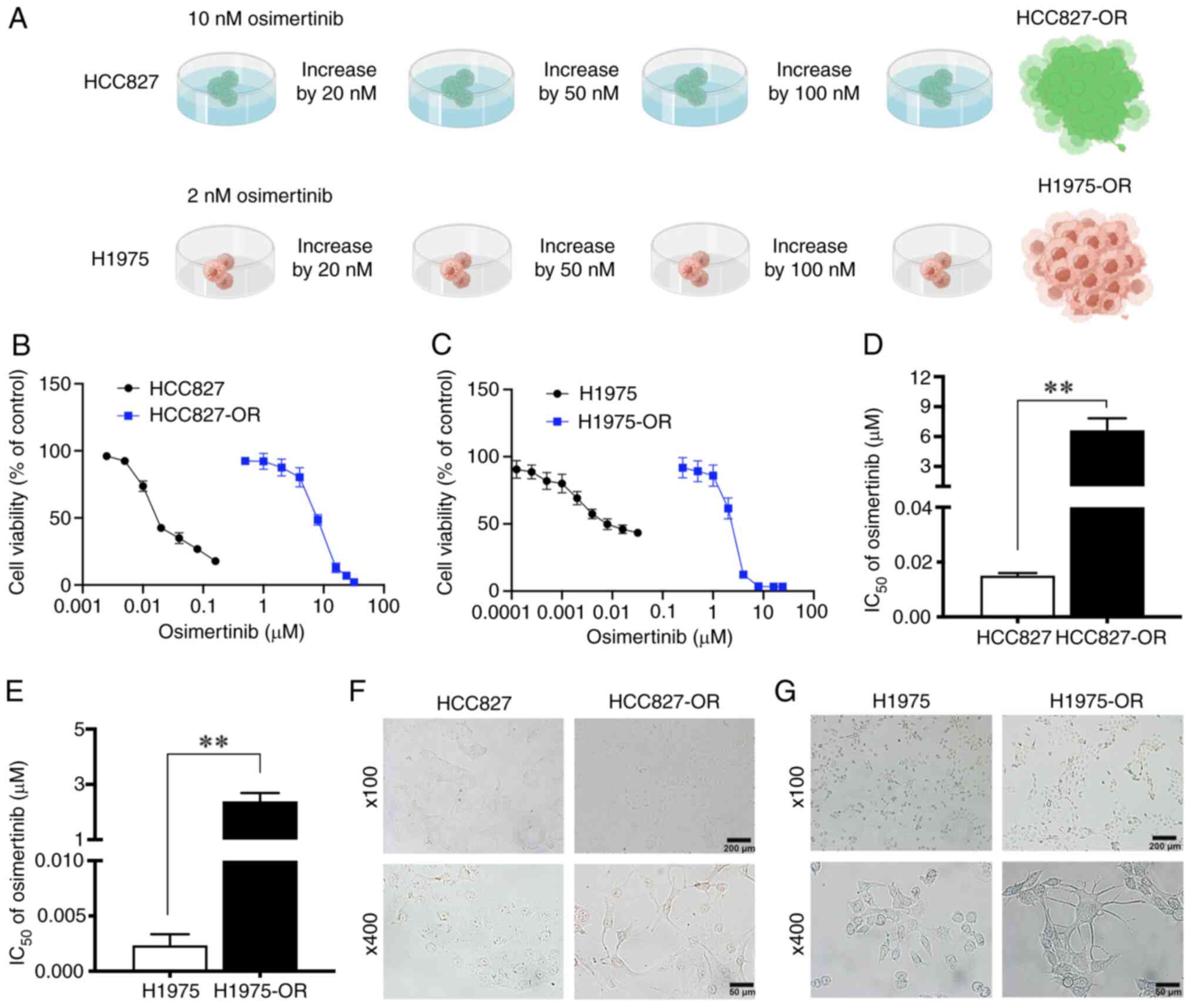
H1975-OR (Fig. 1A).
CCK8 assays were then performed to detect the cell
viability of the four cell lines in response to treatment with
different concentrations of osimertinib. The viability of HCC827-OR
and H1975-OR cells did not decrease as markedly as that of HCC827
and H1975 cells (Fig. 1B and C).
The IC50 values of osimertinib for HCC827 and HCC827-OR
cells were 15.04 nM and 6.64 µM, and the IC50
values of osimertinib for H1975 and H1975-OR cells were 2.34 nM and
2.38 µM respectively (Fig. 1D
and E). The drug RI of HCC827-OR and H1975-OR cells was 441.49
and 1,017.09, respectively.
During the 10 months of exposure to osimertinib, the
morphology of cells was markedly changed. HCC827 cells appeared
closely linked and clumped, with irregular and unclear cell
contours, and a few cells had short pseudofoot protrusions.
HCC827-OR cells were not closely connected, with obvious cell
contours and a mesenchymal phenotype, including a fusiform
appearance. In addition, some cells exhibited long pseudopodia. At
a high magnification, the cytoplasmic proportion of HCC827-OR cells
was increased compared with that of HCC827 cells (Fig. 1F). At low magnification, H1975
cells were loosely arranged with irregular contours. H1975-OR cells
were closely arranged in a spindle shape and grew in clumps. At
high magnification, compared with H1975 cells, some H1975-OR cells
exhibited pseudopodia and presented a dendritic shape (Fig. 1G).
The present study also performed next-generation
sequencing to evaluate the gene mutation status of
osimertinib-resistant and -sensitive cell lines. The mutant
abundance of T790M was much higher in H1975-OR cells than in H1975
cells. Copy number amplification of AKT1, CDK6 and MYC was also
detected in H1975-OR cells (Table
SI). The abundance of EGFR and CDK4 mutations were lower in
HCC827-OR cells than in HCC827 cells (Table SII). NGS results not only
demonstrated the presence of EGFR mutations in the
osimertinib-resistant cells constructed for the present study, but
also revealed potential drug resistance mechanisms, such as copy
number amplification of CDK6, MYC and AKT1, and CDK4 gene mutation.
In summary, osimertinib-resistant cell lines were successfully
established and the details of these resistant cell lines were
identified.
Characterization of the proliferation and
drug resistance of osimertinib-resistant and -sensitive cells
After long-term treatment with osimertinib, the cell
viability of sensitive and resistant cells was affected. Cell
viability was detected by CCK8 assay, and cell proliferation
ability and proliferation rate could also be detected.
Drug-resistant cells and parental cells were monitored using CCK8
reagent for 5 consecutive days (days 0, 1, 2, 3 and 4). The
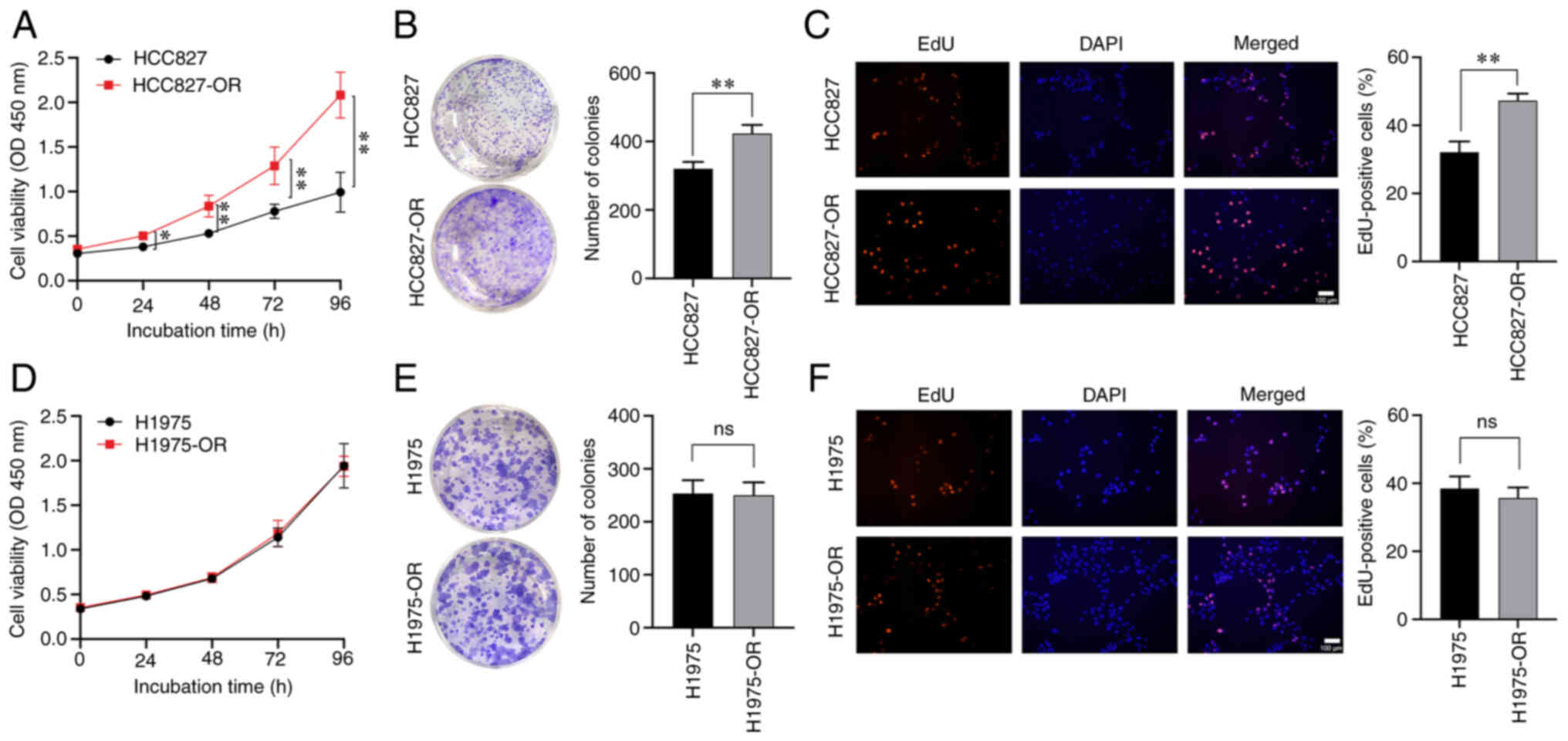
viability of HCC827-OR cells was higher than that of HCC827 cells
on day 1, whereas the difference between the two cell lines on days
2, 3 and 4 was more significant (Fig. 2A). Notably, there was no
significant difference in viability between the H1975 and H1975-OR
cells (Fig. 2D). Colony
formation assays showed that HCC827-OR cells exhibited stronger
proliferative ability than HCC827 cells (Fig. 2B), whereas there was no
significant difference between H1975-OR and H1975 cells regarding
proliferative ability (Fig. 2E).
The EdU assay was also used to evaluate the proliferation of these
four cell lines, and the results were consistent with those of CCK8
and colony formation assays (Fig. 2C
and F).
To further compare the effects of osimertinib on
proliferation of drug-resistant cell lines and parental cell lines,
the cells were treated with different concentrations of
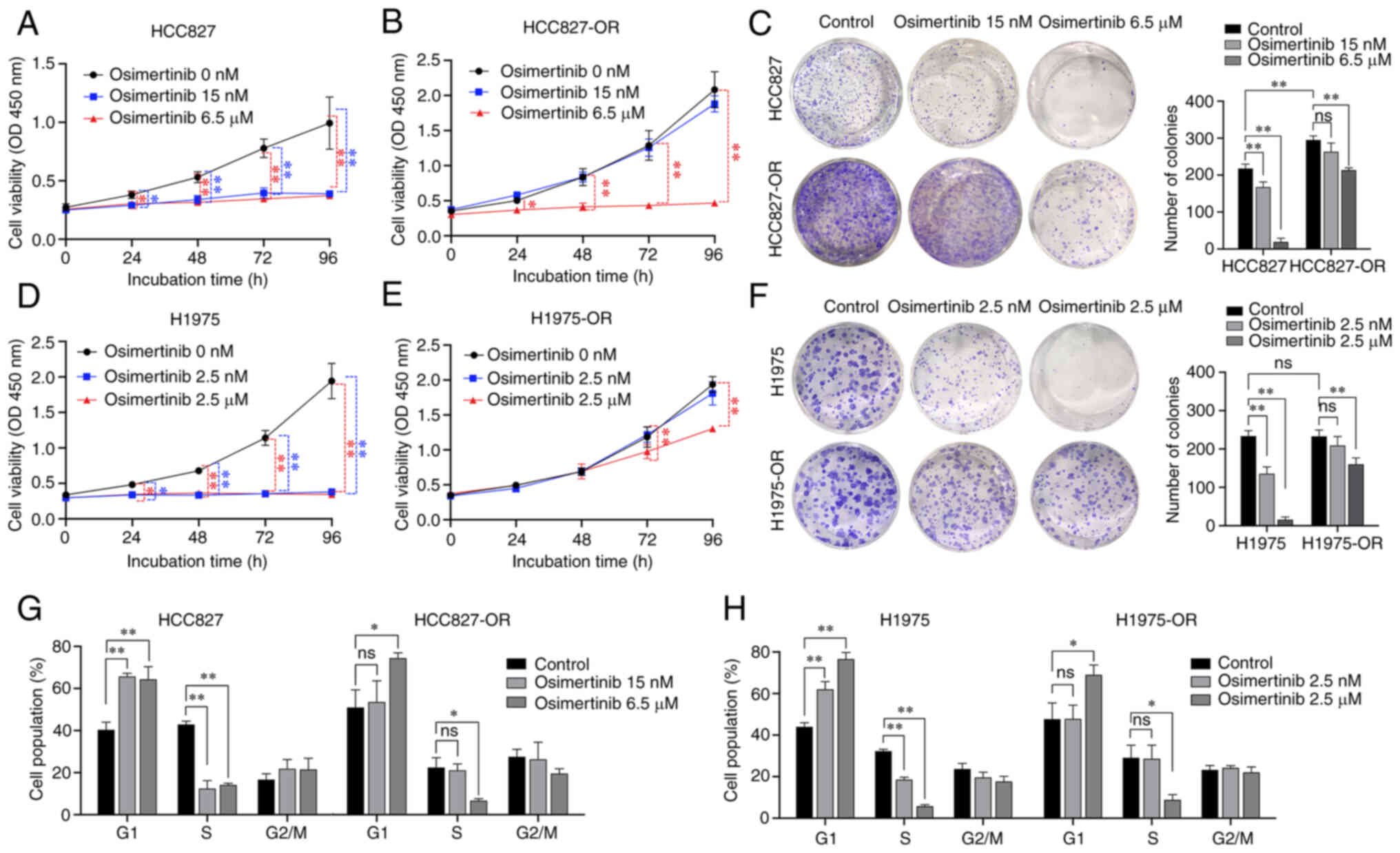
osimertinib. For HCC827 and HCC827-OR cells, the three treatment
groups were 0, 15 nM osimertinib (low concentration) and 6.5
µM osimertinib (high concentration) (Fig. 3A and B). H1975 and H1975-OR cells
were treated with 0, 2.5 nM osimertinib (low concentration) and 2.5
µM osimertinib (high concentration) (Fig. 3D and E). The low concentrations
of osimertinib significantly inhibited HCC827 and H1975 cell
viability, but did not inhibit HCC827-OR and H1975-OR cell
viability. The high concentrations of osimertinib significantly
inhibited all of the four cell lines. Notably, colony formation of
HCC827-OR cells was stronger than that of HCC827 cells, but there
was no significant difference between H1975-OR and H1975 cells
(Fig. 3C and F). Both low and
high concentrations of osimertinib inhibited the colony formation
of the parental cells, HCC827 and H1975. The low concentrations of
osimertinib did not inhibit the colony formation of drug-resistant
cells; however, the high concentrations of osimertinib
significantly inhibited the colony formation of HCC827-OR and
H1975-OR cells. These results confirmed the effects of osimertinib
on the proliferation and drug resistance of osimertinib-resistant
and -sensitive cells.
Osimertinib-resistant cells display
decreased sensitivity to osimertinib-induced apoptosis and cell
cycle arrest
Flow cytometry was used to examine cell cycle
distribution and apoptosis in an effort to better understand how
osimertinib affects resistant and sensitive cells. Osimertinib was
applied to sensitive and resistant cells at various concentrations,
as aforementioned. Compared with that in the control group,
following treatment with low and high concentrations of
osimertinib, the number of HCC827 cells was increased in
G1 phase and decreased in S phase (Figs. 3G and S1A). Compared with that in the control
group, following treatment with the high concentration of
osimertinib, the number of HCC827-OR cells was increased in
G1 phase and decreased in S phase. Compared with that in
the control group, following treatment with low and high
concentrations of osimertinib, the number of H1975 cells was
increased in G1 phase and decreased in S phase (Figs. 3H and S1B). Compared with that in the control
group, following treatment with the high concentration of
osimertinib, the number of H1975-OR cells was increased in
G1 phase and decreased in S phase. These findings
indicated that the low concentration of osimertinib exerted
considerable influence on the cell cycle distribution of HCC827 and
H1975 cells, whereas it had no significant effect on HCC827-OR and
H1975-OR cells. However, high-concentration osimertinib could
affect the cell cycle distribution of osimertinib-resistant cells
and parental cells.
In addition to cell cycle analysis, the effect of
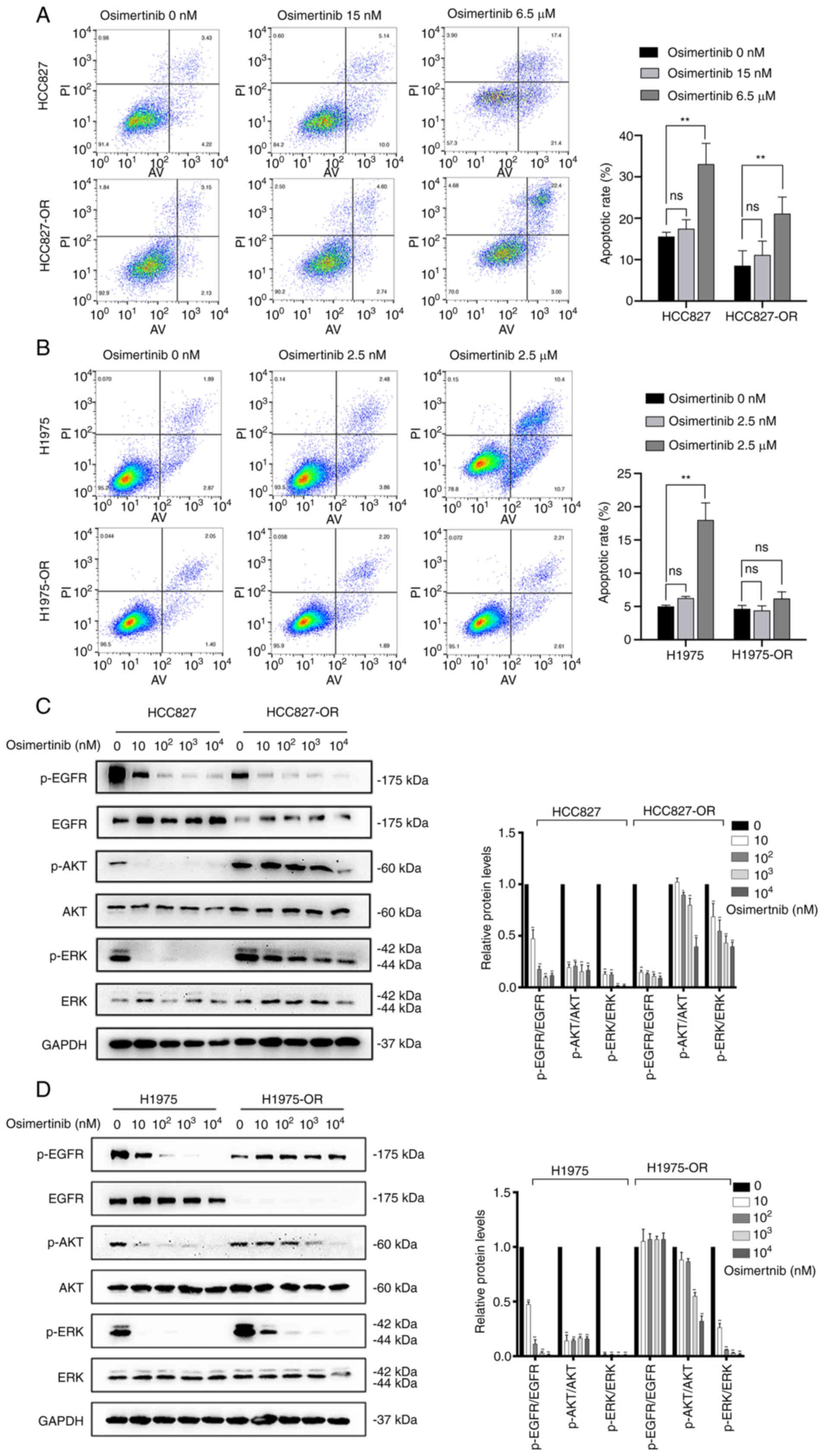
osimertinib on the apoptosis of resistant cells and sensitive cells
was examined. The low concentration of osimertinib had no
significant effects on cell apoptosis in both sensitive and
resistant cells. Similar results were observed in H1975 and
H1975-OR cells (Fig. 4A and B).
By contrast, the high concentration of osimertinib significantly
promoted the apoptosis of parental cells. In resistant cells, the
high concentration of osimertinib promoted the apoptosis of
HCC827-OR cells, but had no significant effect on the apoptosis of
H1975-OR cells. Collectively, parental cells appeared to be more
sensitive to osimertinib-induced apoptosis and cell cycle
arrest.
Characterization of EGFR and downstream
proteins in osimertinib-sensitive and -resistant cells
During the development of osimertinib resistance,
the levels of some key proteins are changed, including AKT, ERK and
EGFR (28). Compared with those
in HCC827 cells, the expression levels of EGFR in HCC827-OR cells
were decreased, whereas the expression levels of AKT and ERK were
not affected. Notably, the expression levels of p-proteins, such as
p-EGFR, p-ERK and p-AKT, in HCC827-OR and HCC827 cells were
associated with the concentration of osimertinib. The expression
levels of p-EGFR, p-ERK and p-AKT in HCC827-OR and HCC827 cells
were decreased with the increasing osimertinib concentration
(Fig. 4C). The expression levels
of p-EGFR in HCC827-OR cells were lower than those in HCC827 cells
either in the control group or osimertinib treatment groups. By
contrast, the expression levels of p-AKT and p-ERK were higher in
HCC827-OR cells than those in HCC827 cells in the same
concentration group. In addition, the inhibitory effect of
osimertinib treatment on p-AKT and p-ERK was weaker than that of
p-EGFR.
After 4 h of osimertinib treatment, the expression
levels of p-EGFR, p-AKT and p-ERK were significantly downregulated
in H1975 cells (Fig. 4D).
Similarly, the expression levels of p-AKT and p-ERK in H1975-OR
cells were decreased with the increase in osimertinib
concentration. In contrast to H1975 cells, p-EGFR expression in
H1975-OR cells was not affected by osimertinib.
Taken together, the expression of EGFR in HCC827-OR
and H1975-OR cells was decreased, whereas the expression of AKT and
ERK was not markedly affected. In addition, p-AKT and p-ERK of
HCC827-OR and H1975-OR were markedly activated compared with those
in HCC827 and H1975 cells, and the inhibitory effect of osimertinib
on p-AKT and p-ERK was weaker in resistant cells than that in
parental cells. Furthermore, the expression levels of p-EGFR were
inhibited in HCC827-OR cells, whereas they were not affected in
H1975-OR cells treated with various osimertinib concentrations.
These results indicated that the AKT and ERK
signaling pathways may be involved in osimertinib resistance, and
that the activation of p-AKT and p-ERK may serve an important role
in the development of osimertinib resistance.
circRNA expression profiles and pathway
analysis in osimertinib-sensitive and -resistant cells
To identify the differentially expressed circRNAs in
osimertinib-resistant cell lines and sensitive cell lines, unbiased
RNA-seq analysis of HCC827, HCC827-OR, H1975 and H1975-OR cells was
performed. The results of hierarchical clustering suggested that
the circRNA expression patterns were distinguishable between
resistant and sensitive cells (Fig.
5A and B). A total of 67 upregulated and 142 downregulated
circRNAs were identified. Notably, 65 circRNAs were considered new
circRNAs with no annotations. GO and KEGG pathway analyses were
performed with circRNA host genes to evaluate the roles of
differentially expressed circRNAs. The results of KEGG pathway
analysis are shown in Fig. 5C and
D, while the results of GO analysis are not shown. The host
genes of differentially expressed circRNAs in H1975-OR and H1975
cells were mainly associated with 'regulation of autophagy'.
Notably, the host genes of differentially expressed circRNAs in
HCC827-OR and HCC827 cells were associated with 'endocytosis'.
Autophagy can affect endocytosis and cell signaling, and
endocytosis can also regulate the different steps of autophagy
(29-31). Autophagy and endocytosis serve an
important role in drug resistance of NSCLC. The present study
subsequently performed western blot analysis to detect the
expression levels of autophagy-related proteins in
osimertinib-sensitive and -resistant cell lines. Cells were treated
with the autophagy agonist RAPA and the autophagy inhibitor 3-MA.
Western blot analysis showed that RAPA enhanced the expression
levels of autophagy-related proteins, whereas 3-MA inhibited the
expression levels of autophagy-related proteins (Fig. 5E and F). Compared with in the
parental cells, HCC827-OR and H1975-OR cells exhibited reduced
protein expression levels of P62 and an increase in LC3-II
conversion, indicating that autophagy was activated in
osimertinib-resistant cells and autophagy may be one of the
underlying mechanisms of osimertinib resistance.
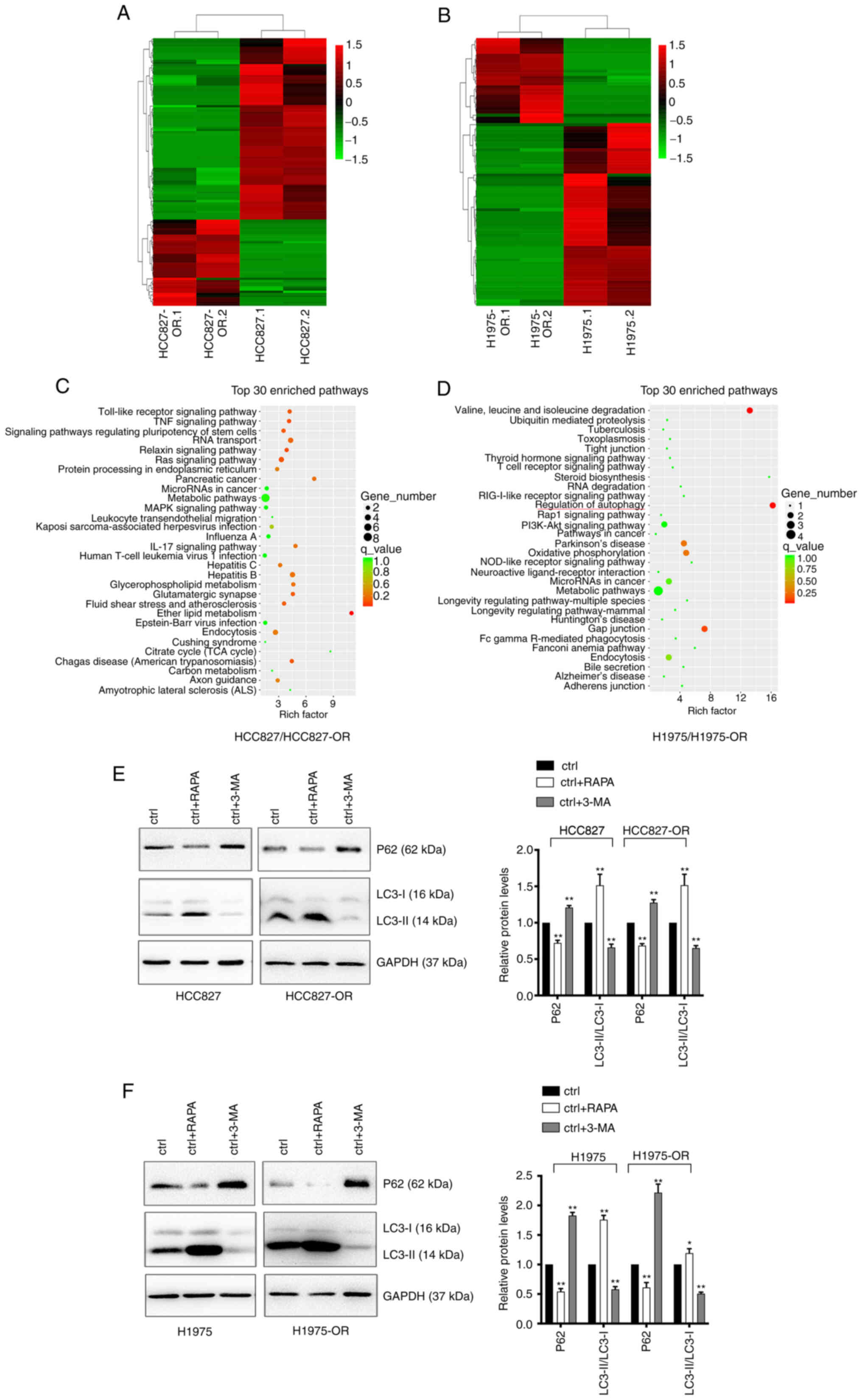 | Figure 5Profiles of differentially expressed
circRNAs in HCC827, HCC827-OR, H1975 and H1975-OR cells.
Unsupervised hierarchical clustering analysis of the differentially
expressed circRNAs between (A) HCC827 and HCC827-OR cells, and (B)
H1975 and H1975-OR cells. Kyoto Encyclopedia of Genes and Genomes
analysis for the host genes of differentially expressed circRNAs in
(C) HCC827 and HCC827-OR cells, and (D) H1975 and H1975-OR cells.
Rich factor represents the enrichment degree of differentially
expressed genes. The y-axis shows the name of the enriched
pathways. The area of each node represents the number of enriched
host genes of differentially expressed circRNAs. The P-value is
represented by a color scale. (E) HCC827 and HCC827-OR cells, and
(F) H1975 and H1975-OR cells were treated with 10 µM RAPA or
1 nM 3-MA for 24 h at 37°C, and the expression levels of
autophagy-related proteins were detected by western blot analysis.
Western blots were semi-quantified. *P<0.05 and
**P<0.01 vs. ctrl. 3-MA, 3-methyladenine; circRNA,
circular RNA; ctrl, control; RAPA, rapamycin. |
Validation of the expression level and
biological role of circP-DLIM5 and circPPP4R1 in
osimertinib-resistant and -sensitive cells
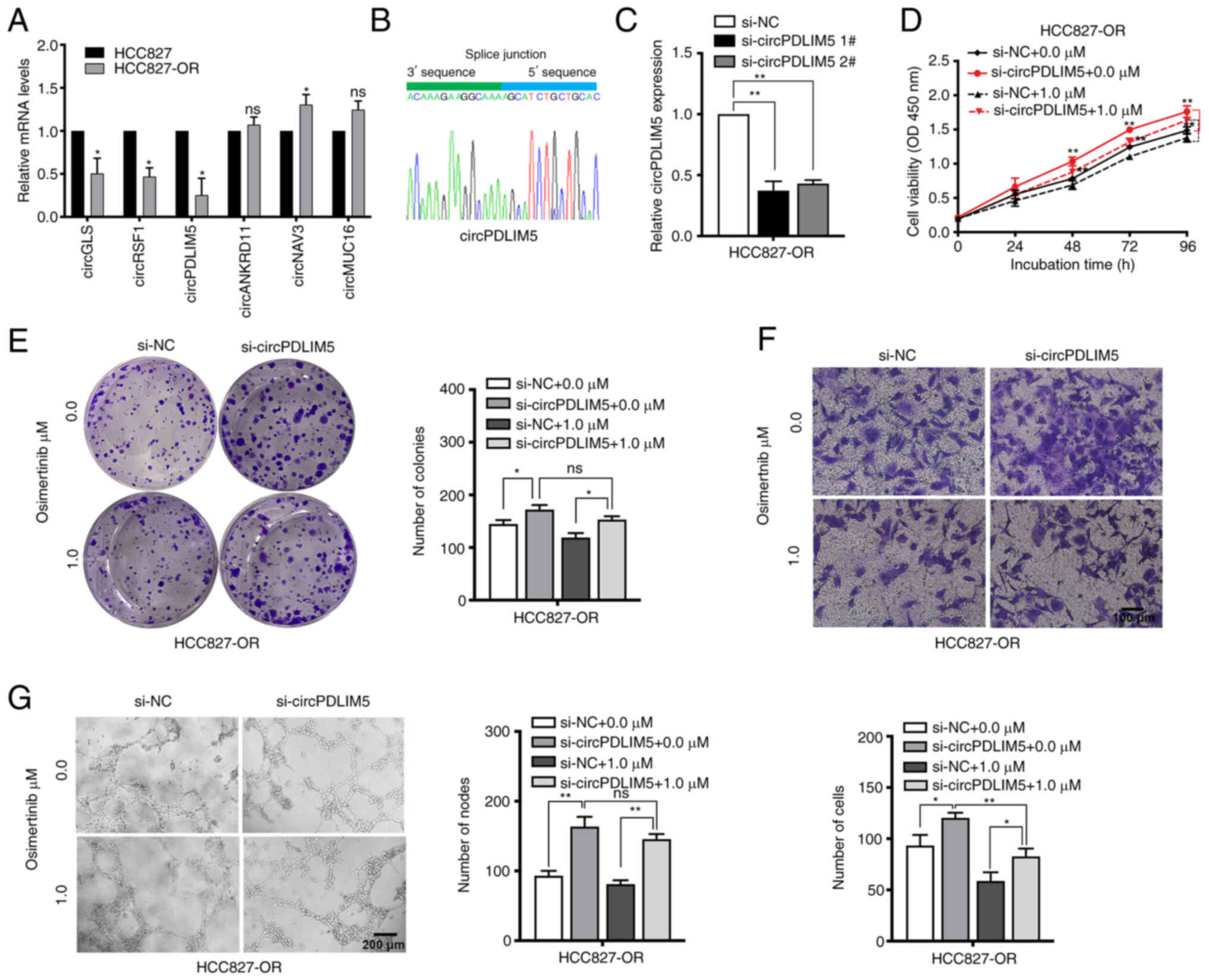
The top 10 dysregulated circRNAs are summarized in
Tables II and III. RT-qPCR was used to validate the
RNA-seq data. The expression levels of those dysregulated circRNAs
and mRNAs (including ALDH1A1, PTPRD, TMPRSS11E and SPTB) were
measured in HCC827 and HCC827-OR cells (Figs. 6A, and S2A and B). The most dysregulated
circRNA (circPDLIM5) was selected for further study. Sanger
sequencing was used to confirm the back-splice junction sites of
circPDLIM5 (Fig. 6B). To explore
the biological role of circP-DLIM5 in osimertinib-resistant cells,
specific siRNAs targeting the back-splice junction sites of
circPDLIM5 were designed and synthesized to silence circPDLIM5
expression (Fig. 6C).
si-circPDLIM5 1# was chosen for subsequent experiments due to its
high interference efficiency. Cell proliferation was measured by
the CCK8 assay, and silencing of circPDLIM5 significantly promoted
cell proliferation in HCC827-OR cells (Fig. 6D). The colony formation assay
showed that knockdown of circPDLIM5 significantly increased the
colony-forming ability of HCC827-OR cells (Fig. 6E). In cell cycle and apoptosis
analyses, knockdown of circPDLIM5 markedly reduced the apoptotic
rate and G0/G1 distribution in HCC827-OR
cells (Fig. S3A and B).
Epithelial-mesenchymal transition and angiogenesis are closely
related to the development of osimertinib resistance (32,33); therefore, Transwell migration and
angiogenesis assays were performed. Silencing of circPDLIM5
promoted the migration and angiogenesis of HCC827-OR cells
(Fig. 6F and G). In addition,
circPDLIM5 was overexpressed in HCC827-OR cells; overexpression of
circPDLIM5 inhibited cell proliferation and migration, and promoted
cell apoptosis (Fig. S3C-G).
These findings indicated that inhibition of circP-DLIM5
significantly promoted the proliferation, migration and
angiogenesis of HCC827-OR cells.
 | Table IITop 10 upregulated and downregulated
circular RNAs in HCC827-OR cells compared with in HCC827 cells. |
Table II
Top 10 upregulated and downregulated
circular RNAs in HCC827-OR cells compared with in HCC827 cells.
| circRNA ID | circBase_ID | Chromosome | Gene symbol | log2FC | P-value | Regulation |
|---|
| circANKRD11 |
hsa_circ_0004566 | 16 | ANKRD11 | 2.277435 | 0.001953 | Up |
| circMUC16 | - | 19 | MUC16 | 1.534303 | 0.037548 | Up |
| circRYK |
hsa_circ_0003633 | 3 | RYK | 1.089538 | 0.026517 | Up |
| circNAV3 |
hsa_circ_0005450 | 12 | NAV3 | 1.402228 | 0.046951 | Up |
| circMKLN1 |
hsa_circ_0001746 | 7 | MKLN1 | 1.401088 | 0.008609 | Up |
| circRSF1 |
hsa_circ_0000344 | 11 | RSF1 | -2.83892 | 0.039224 | Down |
| circPDLIM5 |
hsa_circ_0070467 | 4 | PDLIM5 | -2.72013 | 0.00267 | Down |
| circGLS |
hsa_circ_0001085 | 2 | GLS | -2.69737 | 0.000753 | Down |
| circMIB1 |
hsa_circ_0000836 | 18 | MIB1 | -2.51699 | 0.025407 | Down |
| circOMA1 |
hsa_circ_0000072 | 1 | OMA1 | -2.47581 | 0.02635 | Down |
 | Table IIITop 10 upregulated and downregulated
circular RNAs in H1975-OR cells compared with in H1975 cells. |
Table III
Top 10 upregulated and downregulated
circular RNAs in H1975-OR cells compared with in H1975 cells.
| circRNA ID | circBase_ID | Chromosome | Gene symbol | log2FC | P-value | Regulation |
|---|
| circSNX13 |
hsa_circ_0004671 | 7 | SNX13 | -3.81645 | 0.023204 | Down |
| circPLOD2 |
hsa_circ_0067682 | 3 | PLOD2 | -2.98033 | 0.018598 | Down |
| circTNFRSF21 |
hsa_circ_0001610 | 6 | TNFRSF21 | -2.86512 | 0.013664 | Down |
| circDCBLD2 |
hsa_circ_0066631 | 3 | DCBLD2 | -2.66131 | 0.014213 | Down |
| circMPP6 |
hsa_circ_0001686 | 7 | MPP6 | -2.22047 | 0.020604 | Down |
| circSATB2 |
hsa_circ_0003915 | 2 | SATB2 | -3.71277 | 0.026945 | Down |
| circMUC16 | - | 19 | MUC16 | 3.958709 | 0.00559 | Up |
| circPPP4R1 |
hsa_circ_0007509 | 18 | PPP4R1 | 3.193046 | 0.043673 | Up |
| circSPAG16 |
hsa_circ_0058040 | 2 | SPAG16 | 1.857802 | 0.028686 | Up |
| circMYH9 | - | 22 | MYH9 | 1.454296 | 0.015441 | Up |
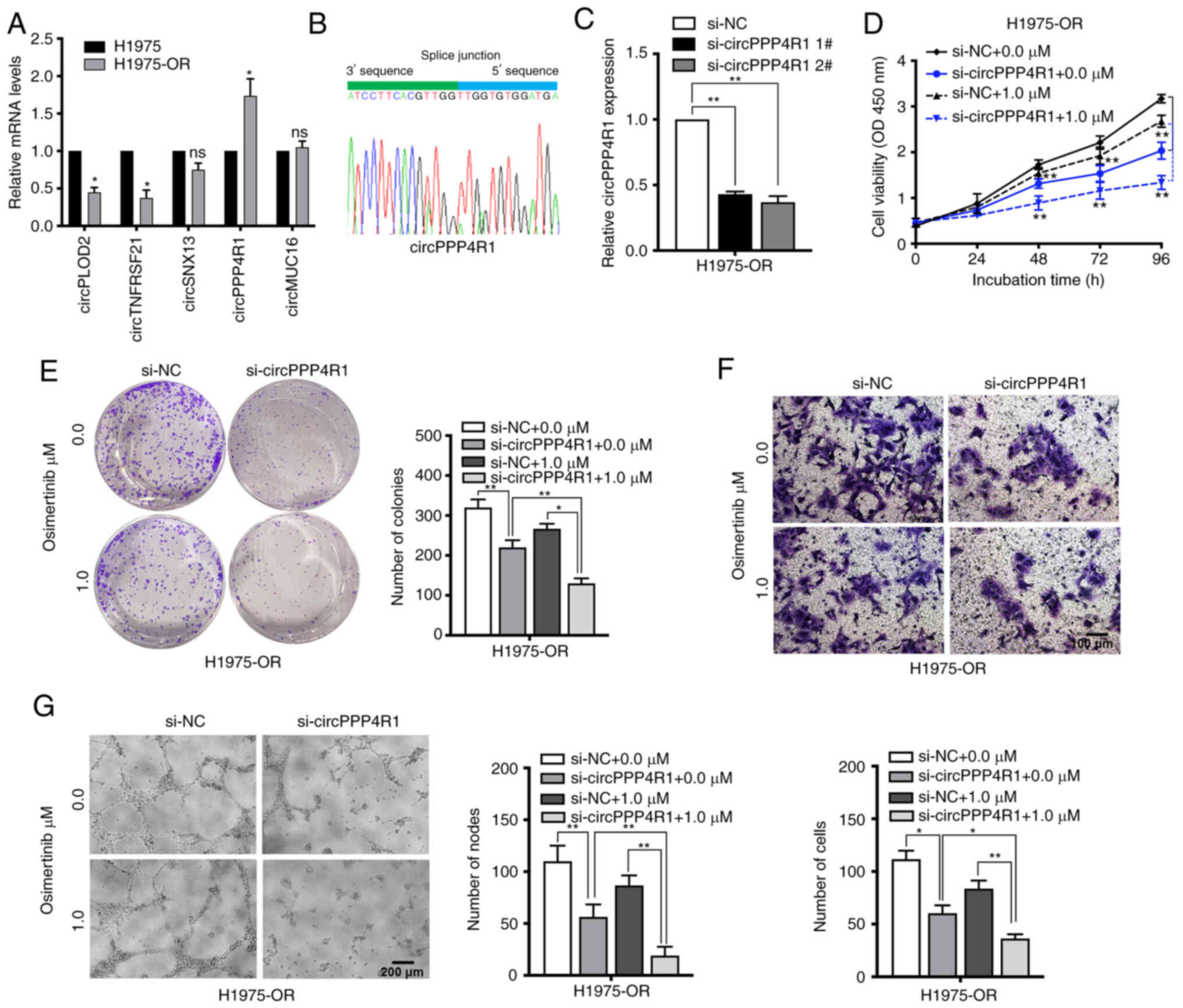
The expression levels of dysregulated circRNAs and
mRNAs were also measured in H1975 and H1975-OR cells (Figs. 7A, and S2C and D). circPPP4R1 was chosen as
the most dysregulated circRNA in H1975 and H1975-OR cells. Sanger
sequencing was used to confirm the back-splice junction sites of
circPPP4R1, confirming the head-to-tail splicing in the RT-qPCR
product of circPPP4R1 (Fig. 7B).
Specific siRNAs targeting the back-splice junction sites of
circPPP4R1 were designed and synthesized to silence circPPP4R1
expression (Fig. 7C).
si-circPPP4R1 2# was chosen for subsequent experiments due to its
high interference efficiency. CCK8 and colony formation assay
showed that silencing of circPPP4R1 significantly inhibited cell
proliferation in H1975-OR cells (Fig. 7D and E). Knockdown of circPPP4R1
also markedly increased the apoptotic rate and
G0/G1 distribution in H1975-OR cells
(Fig. S4A and B). Inhibition of
circPPP4R1 also significantly reduced the migration and
angiogenesis of H1975-OR cells, and the inhibitory impact was more
pronounced following osimertinib treatment, whereas it had no
significant effect on parental H1975 cells (Figs. 7F and G and S4C-G). These results suggested that
dysregulated circRNAs may have an important role in NSCLC drug
resistance.
Discussion
EGFR is one of the driver genes closely related to
the occurrence and development of NSCLC. EGFR-TKIs targeting EGFR
have improved the survival of patients with lung cancer, bringing
marked efficacy to patients with EGFR mutations. In particular,
osimertinib, a third-generation irreversible EGFR-TKI, has achieved
significant clinical benefits in both first-line and second-line
treatment. Notably, the effect of osimertinib has brought great
hope for patients with NSCLC; however, most of the patients still
have inevitable disease progression, which is usually caused by the
generation of acquired resistance to osimertinib (34). Osimertinib resistance in tumors
is caused by the regulation of a multi-factor network, and there is
still a lack of clinically applicable efficacy prediction markers
and effective intervention strategies based on drug resistance
molecular mechanisms. With the occurrence of osimertinib
resistance, it is necessary to discover the possible mechanisms of
osimertinib resistance in advance and to formulate effective
strategies (12).
In a previous study, a gefitinib-resistant PC9 cell
line was established by stepwise escalation method: Parental PC9
cells were cultured with a stepwise escalation in gefitinib
concentration from 5 nM to 5 µM over 6 months (35). In order to explore the mechanism
of osimertinib resistance, the present study established
osimertinib-resistant cell lines. The method of establishment in
the present study is similar to the clinical low-dose
administration method of osimertinib. After 10 months,
osimertinib-resistant cell lines HCC827-OR and H1975-OR were
successfully induced. Verusingam et al (36) previously generated an
osimertinib-resistant cell line from H1975, whereas the present
study developed two resistant cell lines. Sensitive and resistant
cell lines were then treated with different concentrations of
osimertinib, and the specific differences between sensitive and
resistant cell lines were analyzed. Tian et al (9) established three resistant cell
lines, PC9, HCC827 and H1975. Cao et al (11) also used osimertinib-resistant
cell lines to explore the potential mechanism of osimertinib
resistance; however, details of the method of establishment were
not provided.
The present study demonstrated that the
drug-resistant cells HCC827-OR and H1975-OR had considerably
elevated levels of p-AKT and p-ERK compared with in parental cells.
In addition, the inhibitory effect of osimertinib on p-AKT and
p-ERK was less pronounced in the resistant cells than in the
parental cells. By contrast, HCC827-OR cells exhibited a clearer
inhibitory impact of osimertinib on p-EGFR. The preliminary
findings of Tian et al (9) are in agreement with the present
study. In PC9 osimertinib-resistant cells, ERK was significantly
activated and AKT/GSK3β were partially activated in PC9-osimertinib
resistant cells. AKT was the dominant activated signaling factor,
not ERK, in HCC827-osimertinib resistant and H1975-osimertinib
resistant cells. These findings indicated that ERK and AKT
activation may be contributors to osimertinib resistance (9). Booth et al (10) also found activated AKT in
osimertinib-resistant cells.
circRNAs have been identified as a novel class of
non-coding RNA, which are different from traditional linear RNAs.
It has become known that circRNAs function in multiple biological
processes and are involved in several types of cancer (37-40). circ-0003418 exerts an
antitumorigenic role in hepatocellular carcinoma (HCC) and advances
the sensitivity of HCC cells to cisplatin by restraining the
Wnt/β-catenin pathway (41).
circPVT1 is related with the cisplatin and pemetrexed insensitivity
of patients with lung adenocarcinoma; notably, circPVT1 contributes
to cisplatin and pemetrexed chemotherapy resistance through the
miR-145-5p/ABCC1 axis (42).
Hsa_circ_0005576 promotes osimertinib resistance through the
miR-512-5p/IGF1R axis in lung adenocarcinoma cells (43). Inhibition of circ7312 reduces
osimertinib resistance by promoting pyroptosis and apoptosis
through the miR-764/MAPK1 axis (44). METTL3 initiates the m6A
modification of circKRT17, thereby enhancing the stability of YAP1
by recruiting EIF4A4 and promoting osimertinib resistance in lung
adenocarcinoma (45). Hsa_
circRNA_0002130 is upregulated in the serum exosomes of patients
with osimertinib-resistant NSCLC and is closely associated with
glycolysis (21). These previous
findings indicated that circRNA may have a key regulatory role in
tumorigenesis and drug resistance. Although there are some reports
on circRNA and osimertinib resistance in lung cancer, the complex
mechanism underlying circRNA dysregulation and their biological
effects has not been clearly elucidated.
The present study screened the differentially
expressed circRNAs between HCC827, HCC827-OR, H1975 and H1975-OR
cells by high-throughput sequencing. KEGG pathway analysis revealed
that the host genes of differentially expressed circRNAs in
resistant and sensitive cells were mainly associated with
regulation of autophagy. Compared with in the parental cells,
HCC827-OR and H1975-OR cells showed reduced protein levels of P62
and an increase in LC3-II conversion, indicating that the resistant
cell lines may have increased autophagy levels, which is consistent
with previous results (11).
Current studies have shown that inhibition of autophagy can
increase the sensitivity of tumor cells to drugs, and the
activation of autophagy can lead to increased drug resistance in
tumor cells (46-48). Osimertinib has also been shown to
induce autophagy in NSCLC cell lines (11). Drug-resistant cells have
increased autophagy activity when compared with their sensitive
parental cells. A novel anticancer drug GZ17-6.02 that interacts
with osimertinib can increase autophagosome formation (10). Increased protective autophagy may
be one of the mechanisms underlying NSCLC resistance to EGFR-TKIs
(11).
The present study validated the RNA-seq results and
chose the most dysregulated circRNAs (circPDLIM5 and circPPP4R1)
for further study. In vitro studies showed that knockdown of
circPDLIM5 could promote HCC827-OR cell proliferation, migration
and angiogenesis, and inhibit apoptosis. By contrast, inhibition of
circPPP4R1 could reduce H1975-OR cell proliferation, migration and
angiogenesis, and promote apoptosis. Through the establishment and
analysis of drug-resistant cell lines, novel mechanisms and
pathways in drug resistance may be identified, which can lay a
foundation for further research and provide a scientific basis for
clinical drug use.
In conclusion, the present study established
osimertinib-resistant cell lines and revealed the aberrant
expression of circRNAs. The results indicated that circRNAs may
have important roles in NSCLC osimertinib resistance, providing
potential diagnostic or therapeutic biomarkers for EGFR-TKI therapy
of NSCLC. Further studies should be conducted to further the
investigation of the mechanism underlying osimertinib
resistance.
Supplementary Data
Availability of data and materials
The high-throughput RNA sequencing and NGS data in
the present study have been deposited in the BioProject (https://www.ncbi.nlm.nih.gov/bioproject/) under
accession number PRJNA1000087 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1000087/)
and PRJNA1000497 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1000497/).
The other datasets used and/or analyzed during the current study
are available from the corresponding author on reasonable
request.
Authors' contributions
ZXW and ZYC designed the study. XC, JYG and JLH
designed the main experiments, detected the cell biological
function, conducted the RT-qPCR analysis, performed the statistical
analysis, and wrote the manuscript. KW and GZ participated in the
design of the experiments and statistical analysis. JLH and KW
carried out the western blot analyses. All authors read and
approved the final manuscript. XC and ZXW confirm the authenticity
of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
This work was supported by grants from the National Natural
Science Foundation of China (grant nos. 82072591 and 82203058), the
Key Research and Development Plan (Social Development) of Science
And Technology Department of Jiangsu Province (grant no.
BE2019760), the Jiangsu Province Capability Improvement Project
through Science, Technology and Education, Jiangsu Provincial
Medical Key Discipline Cultivation Unit (grant no. JSDW202235), the
'789' Excellent Talent Training Plan of the Second Affiliated
Hospital of Nanjing Medical University (grant no.
789ZYRC202090146), and the China Postdoctoral Science Foundation
(grant nos. 2022M710753 and 2023T160120).
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Testa U, Castelli G and Pelosi E: Lung
cancers: Molecular characterization, clonal heterogeneity and
evolution, and cancer stem cells. Cancers (Basel). 10:2482018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Normanno N, Maiello MR, Chicchinelli N,
Iannaccone A, Esposito C, De Cecio R, D'alessio A and De Luca A:
Targeting the EGFR T790M mutation in non-small-cell lung cancer.
Expert Opin Ther Targets. 21:159–165. 2017. View Article : Google Scholar
|
|
4
|
Ramalingam SS, Vansteenkiste J, Planchard
D, Cho BC, Gray JE, Ohe Y, Zhou C, Reungwetwattana T, Cheng Y,
Chewaskulyong B, et al: Overall survival with osimertinib in
untreated, EGFR-mutated advanced NSCLC. N Engl J Med. 382:41–50.
2020. View Article : Google Scholar
|
|
5
|
Nagasaka M, Zhu VW, Lim SM, Greco M, Wu F
and Ou SI: Beyond osimertinib: the development of third-generation
EGFR tyrosine kinase inhibitors for advanced EGFR+ NSCLC. J Thorac
Oncol. 16:740–763. 2021. View Article : Google Scholar
|
|
6
|
Park S and Ahn MJ: Osimertinib in central
nervous system progressive EGFR-mutant lung cancer: Do we need to
detect T790M? Ann Oncol. 31:15822020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nakatani K, Yamaoka T, Ohba M, Fujita KI,
Arata S, Kusumoto S, Taki-Takemoto I, Kamei D, Iwai S, Tsurutani J
and Ohmori T: KRAS and EGFR amplifications mediate resistance to
rociletinib and osimertinib in acquired afatinib-resistant NSCLC
harboring exon 19 deletion/T790M in EGFR. Mol Cancer Ther.
18:112–126. 2019. View Article : Google Scholar
|
|
8
|
Nishiyama A, Takeuchi S, Adachi Y, Otani
S, Tanimoto A, Sasaki M, Matsumoto S, Goto K and Yano S: MET
amplification results in heterogeneous responses to osimertinib in
EGFR-mutant lung cancer treated with erlotinib. Cancer Sci.
111:3813–3823. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tian X, Wang R, Gu T, Ma F, Laster KV, Li
X, Liu K, Lee MH and Dong Z: Costunolide is a dual inhibitor of
MEK1 and AKT1/2 that overcomes osimertinib resistance in lung
cancer. Mol Cancer. 21:1932022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Booth L, West C, Moore RP, Von Hoff D and
Dent P: GZ17-6.02 and pemetrexed interact to kill
osimertinib-resistant NSCLC cells that express mutant ERBB1
proteins. Front Oncol. 11:7110432021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao P, Li Y, Shi R, Yuan Y, Gong H, Zhu G,
Zhang Z, Chen C, Zhang H, Liu M, et al: Combining EGFR-TKI with
SAHA overcomes EGFR-TKI-acquired resistance by reducing the
protective autophagy in non-small cell lung cancer. Front Chem.
10:8379872022. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu K, Xie F, Wang F and Fu L: Therapeutic
strategies for EGFR-mutated non-small cell lung cancer patients
with osimertinib resistance. J Hematol Oncol. 15:1732022.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ebbesen KK, Hansen TB and Kjems J:
Insights into circular RNA biology. RNA Biol. 14:1035–1045. 2017.
View Article : Google Scholar :
|
|
14
|
Huang W, Yang Y, Wu J, Niu Y, Yao Y, Zhang
J, Huang X, Liang S, Chen R, Chen S and Guo L: Circular RNA cESRP1
sensitises small cell lung cancer cells to chemotherapy by sponging
miR-93-5p to inhibit TGF-β signalling. Cell Death Differ.
27:1709–1727. 2020. View Article : Google Scholar
|
|
15
|
Wang C, Tan S, Li J, Liu WR, Peng Y and Li
W: CircRNAs in lung cancer-biogenesis, function and clinical
implication. Cancer Lett. 492:106–115. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang T, Liu Z, She Y, Deng J, Zhong Y,
Zhao M, Li S, Xie D, Sun X, Hu X and Chen C: A novel protein
encoded by circASK1 ameliorates gefitinib resistance in lung
adenocarcinoma by competitively activating ASK1-dependent
apoptosis. Cancer Lett. 520:321–331. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang LX, Gao J, Long X, Zhang PF, Yang X,
Zhu SQ, Pei X, Qiu BQ, Chen SW, Lu F, et al: The circular RNA
circHMGB2 drives immunosuppression and anti-PD-1 resistance in lung
adenocarcinomas and squamous cell carcinomas via the
miR-181a-5p/CARM1 axis. Mol Cancer. 21:1102022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang PF, Pei X, Li KS, Jin LN, Wang F, Wu
J and Zhang XM: Circular RNA circFGFR1 promotes progression and
anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung
cancer cells. Mol Cancer. 18:1792019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wen C, Xu G, He S, Huang Y, Shi J, Wu L
and Zhou H: Screening circular RNAs related to acquired gefitinib
resistance in non-small cell lung cancer cell lines. J Cancer.
11:3816–3826. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen T, Luo J, Gu Y, Huang J, Luo Q and
Yang Y: Comprehensive analysis of circular RNA profiling in
AZD9291-resistant non-small cell lung cancer cell lines. Thorac
Cancer. 10:930–941. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ma J, Qi G and Li L: A novel serum
exosomes-based biomarker hsa_circ_0002130 facilitates
osimertinib-resistance in non-small cell lung cancer by sponging
miR-498. Onco Targets Ther. 13:5293–5307. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pan J, Xing J, Yu H, Wang Z, Wang W and
Pan Y: CircRBM33 promotes migration, invasion and mediates
osimertinib resistance in non-small cell lung cancer cell line. Ann
Transl Med. 11:2522023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang ZH, Jiang XM, Guo X, Fong CM, Chen X
and Lu JJ: Characterization of osimertinib (AZD9291)-resistant
non-small cell lung cancer NCI-H1975/OSIR cell line. Oncotarget.
7:81598–81610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Damgaard MV and Treebak JT: Protocol for
qPCR analysis that corrects for cDNA amplification efficiency. STAR
Protoc. 3:1015152022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koboldt DC, Zhang Q, Larson DE, Shen D,
McLellan MD, Lin L, Miller CA, Mardis ER, Ding L and Wilson RK:
VarScan 2: Somatic mutation and copy number alteration discovery in
cancer by exome sequencing. Genome Res. 22:568–576. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Talevich E, Shain AH, Botton T and Bastian
BC: CNVkit: Genome-wide copy number detection and visualization
from targeted DNA sequencing. PLoS Comput Biol. 12:e10048732016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang X, Maity TK, Ross KE, Qi Y, Cultraro
CM, Bahta M, Pitts S, Keswani M, Gao S, Nguyen KDP, et al:
Alterations in the global proteome and phosphoproteome in third
generation EGFR TKI resistance reveal drug targets to circumvent
resistance. Cancer Res. 81:3051–3066. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Birgisdottir ÅB and Johansen T: Autophagy
and endocytosis-interconnections and interdependencies. J Cell Sci.
133:jcs2281142020. View Article : Google Scholar
|
|
30
|
Ganapathy AS, Saha K, Suchanec E, Singh V,
Verma A, Yochum G, Koltun W, Nighot M, Ma T and Nighot P: AP2M1
mediates autophagy-induced CLDN2 (claudin 2) degradation through
endocytosis and interaction with LC3 and reduces intestinal
epithelial tight junction permeability. Autophagy. 18:2086–2103.
2022. View Article : Google Scholar :
|
|
31
|
Nnah IC, Wang B, Saqcena C, Weber GF,
Bonder EM, Bagley D, De Cegli R, Napolitano G, Medina DL, Ballabio
A and Dobrowolski R: TFEB-driven endocytosis coordinates MTORC1
signaling and autophagy. Autophagy. 15:151–164. 2019. View Article : Google Scholar
|
|
32
|
Yochum ZA, Cades J, Wang H, Chatterjee S,
Simons BW, O'Brien JP, Khetarpal SK, Lemtiri-Chlieh G, Myers KV,
Huang EH, et al: Targeting the EMT transcription factor TWIST1
overcomes resistance to EGFR inhibitors in EGFR-mutant
non-small-cell lung cancer. Oncogene. 38:656–670. 2019. View Article : Google Scholar
|
|
33
|
Yu HA, Schoenfeld AJ, Makhnin A, Kim R,
Rizvi H, Tsui D, Falcon C, Houck‑Loomis B, Meng F, Yang JL, et al:
Effect of osimertinib and bevacizumab on progression-free survival
for patients with metastatic EGFR-mutant lung cancers: A phase 1/2
single-group open-label trial. JAMA Oncol. 6:1048–1054. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tang ZH and Lu JJ: Osimertinib resistance
in non-small cell lung cancer: Mechanisms and therapeutic
strategies. Cancer Lett. 420:242–246. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Chen Z, Chen Q, Cheng Z, Gu J, Feng W, Lei
T, Huang J, Pu J, Chen X and Wang Z: Long non-coding RNA CASC9
promotes gefitinib resistance in NSCLC by epigenetic repression of
DUSP1. Cell Death Dis. 11:8582020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Verusingam ND, Chen YC, Lin HF, Liu CY,
Lee MC, Lu KH, Cheong SK, Han-Kiat Ong A, Chiou SH and Wang ML:
Generation of osimertinib-resistant cells from epidermal growth
factor receptor L858R/T790M mutant non-small cell lung carcinoma
cell line. J Chin Med Assoc. 84:248–254. 2021. View Article : Google Scholar
|
|
37
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kristensen LS, Jakobsen T, Hager H and
Kjems J: The emerging roles of circRNAs in cancer and oncology. Nat
Rev Clin Oncol. 19:188–206. 2022. View Article : Google Scholar
|
|
39
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar
|
|
40
|
Zhang M, Bai X, Zeng X, Liu J, Liu F and
Zhang Z: circRNA-miRNA-mRNA in breast cancer. Clin Chim Acta.
523:120–130. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen H, Liu S, Li M, Huang P and Li X:
circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance
through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco
Targets Ther. 12:9539–9549. 2019. View Article : Google Scholar :
|
|
42
|
Zheng F and Xu R: CircPVT1 contributes to
chemotherapy resistance of lung adenocarcinoma through
miR-145-5p/ABCC1 axis. Biomed Pharmacother. 124:1098282020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu S, Jiang Z, Xiao P, Li X, Chen Y, Tang
H, Chai Y, Liu Y, Zhu Z, Xie Q, et al: Hsa_circ_0005576 promotes
osimertinib resistance through the miR‑512‑5p/IGF1R axis in lung
adenocarcinoma cells. Cancer Sci. 113:79–90. 2022. View Article : Google Scholar
|
|
44
|
Dai C, Ma Z, Si J, An G, Zhang W, Li S and
Ma Y: Hsa_circ_0007312 promotes third‑generation epidermal growth
factor receptor‑tyrosine kinase inhibitor resistance through
pyroptosis and apoptosis via the MiR‑764/MAPK1 axis in lung
adenocarcinoma cells. J Cancer. 13:2798–2809. 2022. View Article : Google Scholar :
|
|
45
|
Ji Y, Zhao Q, Feng W, Peng Y, Hu B and
Chen Q: N6-methyladenosine modification of CIRCKRT17 initiated by
METTL3 promotes osimertinib resistance of lung adenocarcinoma by
EIF4A3 to enhance YAP1 stability. Cancers (Basel). 14:55822022.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lin Z, Niu Y, Wan A, Chen D, Liang H, Chen
X, Sun L, Zhan S, Chen L, Cheng C, et al: RNA m6A
methylation regulates sorafenib resistance in liver cancer through
FOXO3-mediated autophagy. EMBO J. 39:e1031812020. View Article : Google Scholar
|
|
47
|
Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang
C, Wu Z, Rong X, Huang N, Sun L, et al: Long noncoding RNA (lncRNA)
EIF3J-DT induces chemoresistance of gastric cancer via autophagy
activation. Autophagy. 17:4083–4101. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Paquette M, El-Houjeiri L, C Zirden L,
Puustinen P, Blanchette P, Jeong H, Dejgaard K, Siegel PM and Pause
A: AMPK-dependent phosphorylation is required for transcriptional
activation of TFEB and TFE3. Autophagy. 17:3957–3975. 2021.
View Article : Google Scholar : PubMed/NCBI
|