|
1
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
La Vecchia C: Ovarian cancer: Epidemiology
and risk factors. Eur J Cancer Prev. 26:55–62. 2017. View Article : Google Scholar
|
|
3
|
Kuroki L and Guntupalli SR: Treatment of
epithelial ovarian cancer. BMJ. 371:m37732020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Armstrong DK, Alvarez RD, Backes FJ,
Bakkum-Gamez JN, Barroilhet L, Behbakht K, Berchuck A, Chen LM,
Chitiyo VC, Cristea M, et al: NCCN guidelines® insights:
Ovarian cancer, version 3.2022. J Natl Compr Canc Netw. 20:972–980.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P
and Wu M: Circrna: Functions and properties of a novel potential
biomarker for cancer. Mol Cancer. 16:942017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Franco-Zorrilla JM, Valli A, Todesco M,
Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA and
Paz-Ares J: Target mimicry provides a new mechanism for regulation
of microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tang X, Ren H, Guo M, Qian J, Yang Y and
Gu C: Review on circular RNAs and new insights into their roles in
cancer. Comput Struct Biotechnol J. 19:910–928. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding J, Wang Q, Guo N, Wang H, Chen H, Ni
G and Li P: CircRNA circ_0072995 promotes the progression of
epithelial ovarian cancer by modulating miR-147a/CDK6 axis. Aging
(Albany NY). 12:17209–17223. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guan X, Zong ZH, Liu Y, Chen S, Wang LL
and Zhao Y: circPUM1 promotes tumorigenesis and progression of
ovarian cancer by sponging miR-615-5p and miR-6753-5p. Mol Ther
Nucleic Acids. 18:882–892. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet. 15:R17–R29. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jeck WR and Sharpless NE: Detecting and
characterizing circular RNAs. Nat Biotechnol. 32:453–461. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
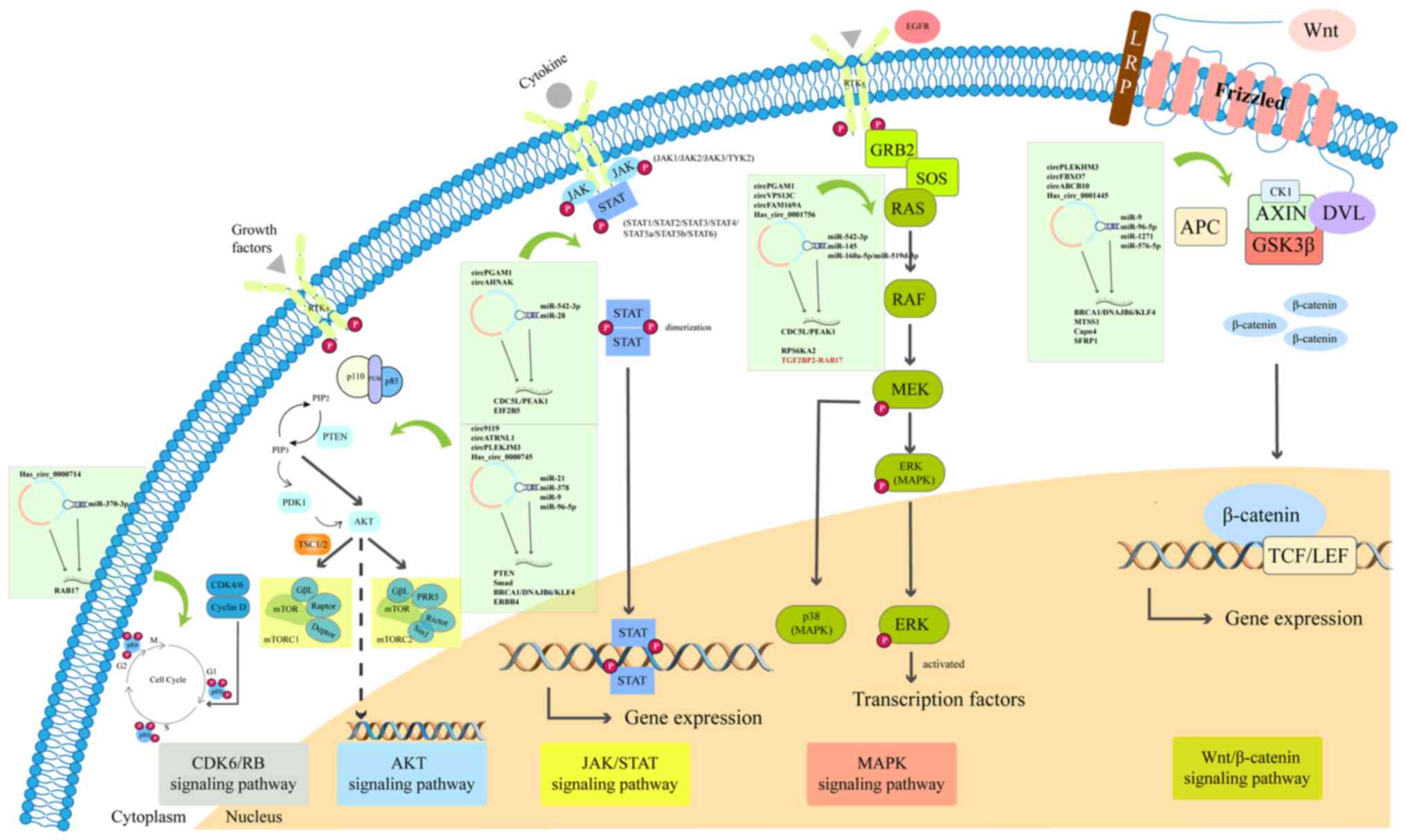
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang S, Cheng J, Quan C, Wen H, Feng Z,
Hu Q, Zhu J, Huang Y and Wu X: circCELSR1 (hsa_circ_0063809)
contributes to paclitaxel resistance of ovarian cancer cells by
regulating FOXR2 expression via miR-1252. Mol Ther Nucleic Acids.
19:718–730. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang F, Nazarali AJ and Ji S: Circular
RNAs as potential biomarkers for cancer diagnosis and therapy. Am J
Cancer Res. 6:1167–1176. 2016.PubMed/NCBI
|
|
18
|
Yang X, Mei J, Wang H, Gu D, Ding J and
Liu C: The emerging roles of circular RNAs in ovarian cancer.
Cancer Cell Int. 20:2652020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: miRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang A, Zheng H, Wu Z, Chen M and Huang
Y: Circular RNA-protein interactions: Functions, mechanisms, and
identification. Theranostics. 10:3503–3517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Qu S, Yang X, Li X, Wang J, Gao Y, Shang
R, Sun W, Dou K and Li H: Circular RNA: A new star of noncoding
RNAs. Cancer Lett. 365:141–148. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Panda AC, Grammatikakis I, Munk R, Gorospe
M and Abdelmohsen K: Emerging roles and context of circular RNAs.
Wiley Interdiscip Rev RNA. 8: View Article : Google Scholar : 2017.
|
|
23
|
Su Q and Lv X: Revealing new landscape of
cardiovascular disease through circular RNA-miRNA-mRNA axis.
Genomics. 112:1680–1685. 2020. View Article : Google Scholar
|
|
24
|
Su L, Li R, Zhang Z, Liu J, Du J and Wei
H: Identification of altered exosomal microRNAs and mRNAs in
Alzheimer's disease. Ageing Res Rev. 73:1014972022. View Article : Google Scholar
|
|
25
|
Zhang J, Luo Q, Li X, Guo J, Zhu Q, Lu X,
Wei L, Xiang Z, Peng M, Ou C and Zou Y: Novel role of
immune-related non-coding RNAs as potential biomarkers regulating
tumour immunoresponse via MICA/NKG2D pathway. Biomark Res.
11:862023. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lu S, Zhu N, Guo W, Wang X, Li K, Yan J,
Jiang C, Han S, Xiang H, Wu X, et al: RNA-Seq revealed a circular
RNA-microRNA-mRNA regulatory network in hantaan virus infection.
Front Cell Infect Microbiol. 10:972020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang X, Ye T, Liu H, Lv P, Duan C, Wu X,
Jiang K, Lu H, Xia D, Peng E, et al: Expression profiles,
biological functions and clinical significance of circRNAs in
bladder cancer. Mol Cancer. 20:42021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Najafi S: Circular RNAs as emerging
players in cervical cancer tumorigenesis; A review to roles and
biomarker potentials. Int J Biol Macromol. 206:939–953. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Najafi S: The emerging roles and potential
applications of circular RNAs in ovarian cancer: A comprehensive
review. J Cancer Res Clin Oncol. 149:2211–2234. 2023. View Article : Google Scholar
|
|
30
|
Fattahi M, Shahrabi S, Saadatpour F,
Rezaee D, Beyglu Z, Delavari S, Amrolahi A, Ahmadi S,
Bagheri-Mohammadi S, Noori E, et al: microRNA-382 as a tumor
suppressor? Roles in tumorigenesis and clinical significance. Int J
Biol Macromol. 250:1258632023. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Pordel S, Khorrami M, Saadatpour F, Rezaee
D, Cho WC, Jahani S, Aghaei-Zarch SM, Hashemi E and Najafi S: The
role of microRNA-185 in the pathogenesis of human diseases: A focus
on cancer. Pathol Res Pract. 249:1547292023. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Najafi S, Aghaei Zarch SM, Majidpoor J,
Pordel S, Aghamiri S, Fatih Rasul M, Asemani Y, Vakili O, Mohammadi
V, Movahedpour A and Arghiani N: Recent Insights into the roles of
circular rnas in human brain development and neurologic diseases.
Int J Biol Macromol. 225:1038–1048. 2023. View Article : Google Scholar
|
|
33
|
Xu YX, Pu SD, Li X, Yu ZW, Zhang YT, Tong
XW, Shan YY and Gao XY: Exosomal ncRNAs: Novel therapeutic target
and biomarker for diabetic complications. Pharmacol Res.
178:1061352022. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang
W, Wang G, Wu P, Wang H, Jiang L, et al: Exosomal circRNAs:
Biogenesis, effect and application in human diseases. Mol Cancer.
18:1162019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Karreth FA and Pandolfi PP: ceRNA
cross-talk in cancer: When ce-bling rivalries go awry. Cancer
Discov. 3:1113–1121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: the Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lan C, Peng H, Hutvagner G and Li J:
Construction of competing endogenous RNA networks from paired
RNA-seq data sets by pointwise mutual information. BMC Genomics.
20(Suppl 9): S9432019. View Article : Google Scholar
|
|
39
|
Kristensen LS, Andersen MS, Stagsted LVW,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Peng Z, Fang S, Jiang M, Zhao X, Zhou C
and Gong Z: Circular RNAs: Regulatory functions in respiratory
tract cancers. Clin Chim Acta. 510:264–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li R, Jiang J, Shi H, Qian H, Zhang X and
Xu W: CircRNA: A rising star in gastric cancer. Cell Mol Life Sci.
77:1661–1680. 2020. View Article : Google Scholar
|
|
42
|
Rong Z, Xu J, Shi S, Tan Z, Meng Q, Hua J,
Liu J, Zhang B, Wang W, Yu X and Liang C: Circular RNA in
pancreatic cancer: A novel avenue for the roles of diagnosis and
treatment. Theranostics. 11:2755–2769. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chaichian S, Shafabakhsh R, Mirhashemi SM,
Moazzami B and Asemi Z: Circular RNAs: A novel biomarker for
cervical cancer. J Cell Physiol. 235:718–274. 2020. View Article : Google Scholar
|
|
44
|
Razavi ZS, Tajiknia V, Majidi S, Ghandali
M, Mirzaei HR, Rahimian N, Hamblin MR and Mirzaei H: Gynecologic
cancers and non-coding RNAs: Epigenetic regulators with emerging
roles. Crit Rev Oncol Hematol. 157:1031922021. View Article : Google Scholar
|
|
45
|
Sang Y, Chen B, Song X, Li Y, Liang Y, Han
D, Zhang N, Zhang H, Liu Y, Chen T, et al: circRNA_0025202
regulates tamoxifen sensitivity and tumor progression via
regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther.
27:1638–1652. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan
Y, Kong X, Bu J, Liu M and Xu S: circRNA-002178 act as a ceRNA to
promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis.
11:322020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Huang G, Liang M, Liu H, Huang J, Li P,
Wang C, Zhang Y, Lin Y and Jiang X: CircRNA hsa_circRNA_104348
promotes hepatocellular carcinoma progression through modulating
miR-187-3p/RTKN2 axis and activating Wnt/β-catenin pathway. Cell
Death Dis. 11:10652020. View Article : Google Scholar
|
|
48
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Liu T, Yuan L and Zou X: Circular RNA
circ-BNC2 (hsa_ circ_0008732) inhibits the progression of ovarian
cancer through microRNA-223-3p/FBXW7 axis. J Ovarian Res.
15:952022. View Article : Google Scholar
|
|
50
|
Xu Q, Deng B, Li M, Chen Y and Zhuan L:
circRNA-UBAP2 promotes the proliferation and inhibits apoptosis of
ovarian cancer though miR-382-5p/PRPF8 axis. J Ovarian Res.
13:812020. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Li M, Chi C, Zhou L, Chen Y and Tang X:
Circular PVT1 regulates cell proliferation and invasion via
miR-149-5p/FOXM1 axis in ovarian cancer. J Cancer. 12:611–621.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Liang Y, Meng K and Qiu R: Circular RNA
Circ_0013958 functions as a tumor promoter in ovarian cancer by
regulating miR-637/PLXNB2 axis. Front Genet. 12:6444512021.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Yu W, Goncalves KA, Li S, Kishikawa H, Sun
G, Yang H, Vanli N, Wu Y, Jiang Y, Hu MG, et al: Plexin-B2 mediates
physiologic and pathologic functions of angiogenin. Cell.
171:849–864.e25. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Li X, He S and Ma B: Autophagy and
autophagy-related proteins in cancer. Mol Cancer. 19:122020.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wang Y, Mo Y, Peng M, Zhang S, Gong Z, Yan
Q, Tang Y, He Y, Liao Q, Li X, et al: The influence of circular
RNAs on autophagy and disease progression. Autophagy. 18:240–253.
2022. View Article : Google Scholar :
|
|
56
|
Zhang Z, Zhu H and Hu J: CircRAB11FIP1
promoted autophagy flux of ovarian cancer through DSC1 and miR-129.
Cell Death Dis. 12:2192021. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Gan X, Zhu H, Jiang X, Obiegbusi SC, Yong
M, Long X and Hu J: CircMUC16 promotes autophagy of epithelial
ovarian cancer via interaction with ATG13 and miR-199a. Mol Cancer.
19:452020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Song W, Zeng Z, Zhang Y, Li H, Cheng H,
Wang J and Wu F: CircRNF144B/miR-342-3p/FBXL11 axis reduced
autophagy and promoted the progression of ovarian cancer by
increasing the ubiquitination of Beclin-1. Cell Death Dis.
13:8572022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Claesson-Welsh L and Welsh M: Vegfa and
tumour angiogenesis. J Intern Med. 273:114–127. 2013. View Article : Google Scholar
|
|
60
|
Wang J, Li Y, Zhou JH, Shen FR, Shi X and
Chen YG: CircATRNL1 activates Smad4 signaling to inhibit
angiogenesis and ovarian cancer metastasis via miR-378. Mol Oncol.
15:1217–1233. 2021. View Article : Google Scholar :
|
|
61
|
Schwarte-Waldhoff I and Schmiegel W: Smad4
transcriptional pathways and angiogenesis. Int J Gastrointest
Cancer. 31:47–59. 2002. View Article : Google Scholar
|
|
62
|
Chen J, Li X, Yang L, Li M, Zhang Y and
Zhang J: CircASH2L promotes ovarian cancer tumorigenesis,
angiogenesis, and lymphangiogenesis by regulating the miR-665/VEGFA
axis as a competing endogenous RNA. Front Cell Dev Biol.
8:5955852020. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ma L, Liu W and Li M: Circ_0061140
contributes to ovarian cancer progression by targeting
miR-761/LETM1 signaling. Biochem Genet. 61:628–650. 2023.
View Article : Google Scholar
|
|
64
|
Pastushenko I and Blanpain C: EMT
transition states during tumor progression and metastasis. Trends
Cell Biol. 29:212–226. 2019. View Article : Google Scholar
|
|
65
|
Shang BQ, Li ML, Quan HY, Hou PF, Li ZW,
Chu SF, Zheng JN and Bai J: Functional roles of circular RNAs
during epithelial-to-mesenchymal transition. Mol Cancer.
18:1382019. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang L, Zhou Q, Qiu Q, Hou L, Wu M, Li J,
Li X, Lu B, Cheng X, Liu P, et al: CircPLEKHM3 acts as a tumor
suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1
axis in ovarian cancer. Mol Cancer. 18:1442019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Bai F, Zhang LH, Liu X, Wang C, Zheng C,
Sun J, Li M, Zhu WG and Pei XH: GATA3 functions downstream of BRCA1
to suppress EMT in breast cancer. Theranostics. 11:8218–8233. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Menezes ME, Mitra A, Shevde LA and Samant
RS: DNAJB6 governs a novel regulatory loop determining
Wnt/β-catenin signalling activity. Biochem J. 444:573–580. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Zeng XY, Yuan J, Wang C, Zeng D, Yong JH,
Jiang XY, Lan H and Xiao SS: circCELSR1 facilitates ovarian cancer
proliferation and metastasis by sponging miR-598 to activate BRD4
signals. Mol Med. 26:702020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Li X, Lin S, Mo Z, Jiang J, Tang H, Wu C
and Song J: CircRNA_100395 inhibits cell proliferation and
metastasis in ovarian cancer via regulating
miR-1228/p53/epithelial-mesenchymal transition (EMT) axis. J
Cancer. 11:599–609. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhou J, Dong ZN, Qiu BQ, Hu M, Liang XQ,
Dai X, Hong D and Sun YF: CircRNA FGFR3 induces
epithelial-mesenchymal transition of ovarian cancer by regulating
miR-29a-3p/E2F1 axis. Aging (Albany NY). 12:14080–14091. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wu SG, Zhou P, Chen JX, Lei J, Hua L, Dong
Y, Hu M, Lian CL, Yang LC and Zhou J: circ-PTK2 (hsa_circ_0008305)
regulates the pathogenic processes of ovarian cancer via miR-639
and FOXC1 regulatory cascade. Cancer Cell Int. 21:2772021.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhang F, Xu Y, Ye W, Jiang J and Wu C:
Circular RNA S-7 promotes ovarian cancer EMT via sponging miR-641
to up-regulate ZEB1 and MDM2. Biosci Rep. 40:BSR202008252020.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Chen Q, Zhang J, He Y and Wang Y:
hsa_circ_0061140 knockdown reverses FOXM1-mediated cell growth and
metastasis in ovarian cancer through miR-370 sponge activity. Mol
Ther Nucleic Acids. 13:55–63. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wang T, Chen X, Qiao W, Kong L, Sun D and
Li Z: Transcription factor E2F1 promotes EMT by regulating ZEB2 in
small cell lung cancer. BMC Cancer. 17:7192017. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Li Y and Chen X: miR-4792 inhibits
epithelial-mesenchymal transition and invasion in nasopharyngeal
carcinoma by targeting FOXC1. Biochem Biophys Res Commun.
468:863–869. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Caramel J, Ligier M and Puisieux A:
Pleiotropic roles for ZEB1 in cancer. Cancer Res. 78:30–35. 2018.
View Article : Google Scholar
|
|
79
|
Katoh M, Igarashi M, Fukuda H, Nakagama H
and Katoh M: Cancer genetics and genomics of human FOX family
genes. Cancer Lett. 328:198–206. 2013. View Article : Google Scholar
|
|
80
|
Wang Y and Patti GJ: The Warburg effect: A
signature of mitochondrial overload. Trends Cell Boil.
33:1014–1020. 2023. View Article : Google Scholar
|
|
81
|
Yu G, Yang Z, Peng T and Lv Y: Circular
RNAs: Rising stars in lipid metabolism and lipid disorders. J Cell
Physiol. 236:4797–4806. 2021. View Article : Google Scholar
|
|
82
|
Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T
and Shu Y: CircRNAs in cancer metabolism: A review. J Hematol
Oncol. 12:902019. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Lin C, Xu X, Yang Q, Liang L and Qiao S:
Circular RNA ITCH suppresses proliferation, invasion, and
glycolysis of ovarian cancer cells by up-regulating CDH1 via
sponging miR-106a. Cancer Cell Int. 20:3362020. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Xie W, Liu LU, He C, Zhao M, Ni R, Zhang Z
and Shui C: Circ_0002711 knockdown suppresses cell growth and
aerobic glycolysis by modulating miR-1244/ROCK1 axis in ovarian
cancer. J Biosci. 46:212021. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hou W and Zhang Y: Circ_0025033 promotes
the progression of ovarian cancer by activating the expression of
LSM4 via targeting miR-184. Pathol Res Pract. 217:1532752021.
View Article : Google Scholar
|
|
86
|
Chen L, Lin YH, Liu GQ, Huang JE, Wei W,
Yang ZH, Hu YM, Xie JH and Yu HZ: Clinical significance and
potential role of LSM4 overexpression in hepatocellular carcinoma:
An integrated analysis based on multiple databases. Front Genet.
12:8049162022. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Ma H, Qu S, Zhai Y and Yang X:
circ_0025033 promotes ovarian cancer development via regulating the
hsa_miR-370-3p/SLC1A5 axis. Cell Mol Biol Lett. 27:942022.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Chen Y, Ye X, Xia X and Lin X: Circular
RNA ABCB10 correlates with advanced clinicopathological features
and unfavorable survival, and promotes cell proliferation while
reduces cell apoptosis in epithelial ovarian cancer. Cancer
Biomark. 26:151–161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Hu Y, Zhu Y, Zhang W, Lang J and Ning L:
Utility of plasma circBNC2 as a diagnostic biomarker in epithelial
ovarian cancer. Onco Targets Ther. 12:9715–9723. 2019. View Article : Google Scholar
|
|
90
|
Ning L, Lang J and Wu L: Plasma
circN4BP2L2 is a promising novel diagnostic biomarker for
epithelial ovarian cancer. BMC Cancer. 22:62022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Wang S, Zhang K, Tan S, Xin J, Yuan Q, Xu
H, Xu X, Liang Q, Christiani DC, Wang M, et al: Circular RNAs in
body fluids as cancer biomarkers: the new frontier of liquid
biopsies. Mol Cancer. 20:132021. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Zheng Y, Li Z, Yang S, Wang Y and Luan Z:
CircEXOC6B suppresses the proliferation and motility and sensitizes
ovarian cancer cells to paclitaxel through miR-376c-3p/FOXO3 axis.
Cancer Biother Radiopharm. 37:802–814. 2022.
|
|
93
|
Zhu J, Luo JE, Chen Y and Wu Q:
Circ_0061140 knockdown inhibits tumorigenesis and improves PTX
sensitivity by regulating miR-136/CBX2 axis in ovarian cancer. J
Ovarian Res. 14:1362021. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Huang H, Yan L, Zhong J, Hong L, Zhang N
and Luo X: Circ_0025033 deficiency suppresses paclitaxel resistance
and malignant development of paclitaxel-resistant ovarian cancer
cells by modulating the miR-532-3p/FOXM1 network. Immunopharmacol
Immunotoxicol. 44:275–286. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Xia B, Zhao Z, Wu Y, Wang Y, Zhao Y and
Wang J: Circular RNA circTNPO3 regulates paclitaxel resistance of
ovarian cancer cells by miR-1299/NEK2 signaling pathway. Mol Ther
Nucleic Acids. 21:780–791. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Yuan D, Guo T, Qian H, Ge H, Zhao Y, Huang
A, Wang X, Cao X, Zhu D, He C and Yu H: Icariside II suppresses the
tumorigenesis and development of ovarian cancer by regulating
miR-144-3p/IGF2R axis. Drug Dev Res. 83:1383–1393. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Luo Y and Gui R: Circulating exosomal
circFoxp1 confers cisplatin resistance in epithelial ovarian cancer
cells. J Gynecol Oncol. 31:e752020. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Liu X, Yin Z, Wu Y, Zhan Q, Huang H and
Fan J: Circular RNA lysophosphatidic acid receptor 3 (circ-LPAR3)
enhances the cisplatin resistance of ovarian cancer. Bioengineered.
13:3739–3750. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Cheng Y, Ban R, Liu W, Wang H, Li S, Yue
Z, Zhu G, Zhuan Y and Wang C: MiRNA-409-3p enhances
cisplatin-sensitivity of ovarian cancer cells by blocking the
autophagy mediated by Fip200. Oncol Res. Jan 2–2018.Epub ahead of
print. View Article : Google Scholar
|
|
100
|
Ghafouri-Fard S, Khoshbakht T, Bahranian
A, Taheri M and Hallajnejad M: CircMTO1: A circular RNA with roles
in the carcinogenesis. Biomed Pharmacother. 142:1120252021.
View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Wang J, Wu A, Yang B, Zhu X, Teng Y and Ai
Z: Profiling and bioinformatics analyses reveal differential
circular RNA expression in ovarian cancer. Gene. 724:1441502020.
View Article : Google Scholar
|
|
102
|
Zhang PF, Gao C, Huang XY, Lu JC, Guo XJ,
Shi GM, Cai JB and Ke AW: Cancer cell-derived exosomal circUHRF1
induces natural killer cell exhaustion and may cause resistance to
anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer.
19:1102020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Gong J, Xu X, Zhang X and Zhou Y: Circular
RNA-9119 suppresses in ovarian cancer cell viability via targeting
the microRNA-21-5p-PTEN-Akt pathway. Aging (Albany NY).
12:14314–14328. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Guo M, Li S, Zhao X, Yuan Y, Zhang B and
Guan Y: Knockdown of circular RNA Hsa_circ_0000714 can regulate
RAB17 by sponging miR-370-3p to reduce paclitaxel resistance of
ovarian cancer through CDK6/RB pathway. Onco Targets Ther.
13:13211–13224. 2020. View Article : Google Scholar :
|
|
105
|
Ji J, Li C, Wang J, Wang L, Huang H, Li Y
and Fang J: Hsa_circ_0001756 promotes ovarian cancer progression
through regulating IGF2BP2-mediated RAB5A expression and the
EGFR/MAPK signaling pathway. Cell Cycle. 21:685–696. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang C, Li Y, Zhao W, Liu G and Yang Q:
Circ-PGAM1 promotes malignant progression of epithelial ovarian
cancer through regulation of the miR-542-3p/CDC5L/PEAK1 pathway.
Cancer Med. 9:3500–3521. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
He SL, Zhao X and Yi SJ: CircAHNAK
upregulates EIF2B5 expression to inhibit the progression of ovarian
cancer by modulating the JAK2/STAT3 signaling pathway.
Carcinogenesis. 43:941–955. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Lu H, Zheng G, Gao X, Chen C, Zhou M and
Zhang L: Propofol suppresses cell viability, cell cycle progression
and motility and induces cell apoptosis of ovarian cancer cells
through suppressing MEK/ERK signaling via targeting
circVPS13C/miR-145 axis. J Ovarian Res. 14:302021. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Fu Z, Ding C, Gong W and Lu C: ncRNAs
mediated RPS6KA2 inhibits ovarian cancer proliferation via p38/MAPK
signaling pathway. Front Oncol. 13:10283012023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Wang S, Li Z, Zhu G, Hong L, Hu C, Wang K,
Cui K and Hao C: RNA-binding protein IGF2BP2 enhances circ_0000745
abundancy and promotes aggressiveness and stemness of ovarian
cancer cells via the microRNA-3187-3p/ERBB4/PI3K/AKT axis. J
Ovarian Res. 14:1542021. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Wu M, Qiu Q, Zhou Q, Li J, Yang J, Zheng
C, Luo A, Li X, Zhang H, Cheng X, et al: circFBXO7/miR-96-5p/MTSS1
axis is an important regulator in the Wnt signaling pathway in
ovarian cancer. Mol Cancer. 21:1372022. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Lin X, Chen Y, Ye X and Xia X: Circular
RNA ABCB10 promotes cell proliferation and invasion, but inhibits
apoptosis via regulating the microRNA-1271-mediated
Capn4/Wnt/β-catenin signaling pathway in epithelial ovarian cancer.
Mol Med Rep. 23:3872021. View Article : Google Scholar
|
|
113
|
Wu Y, Zhou J, Li Y, Shi X, Shen F, Chen M,
Chen Y and Wang J: Hsa_circ_0001445 works as a cancer suppressor
via miR-576-5p/SFRP1 axis regulation in ovarian cancer. Cancer Med.
12:5736–5750. 2023. View Article : Google Scholar
|