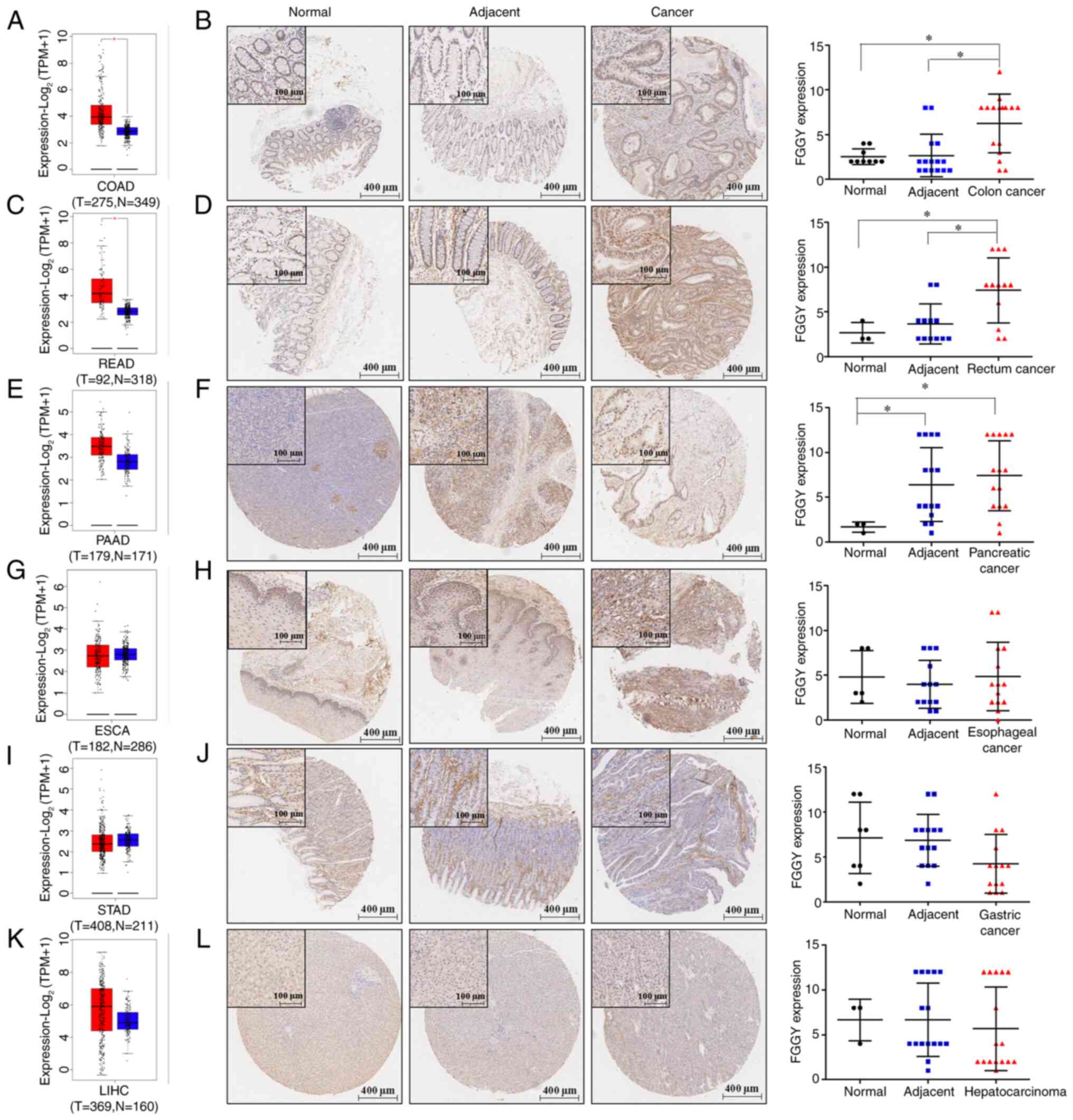
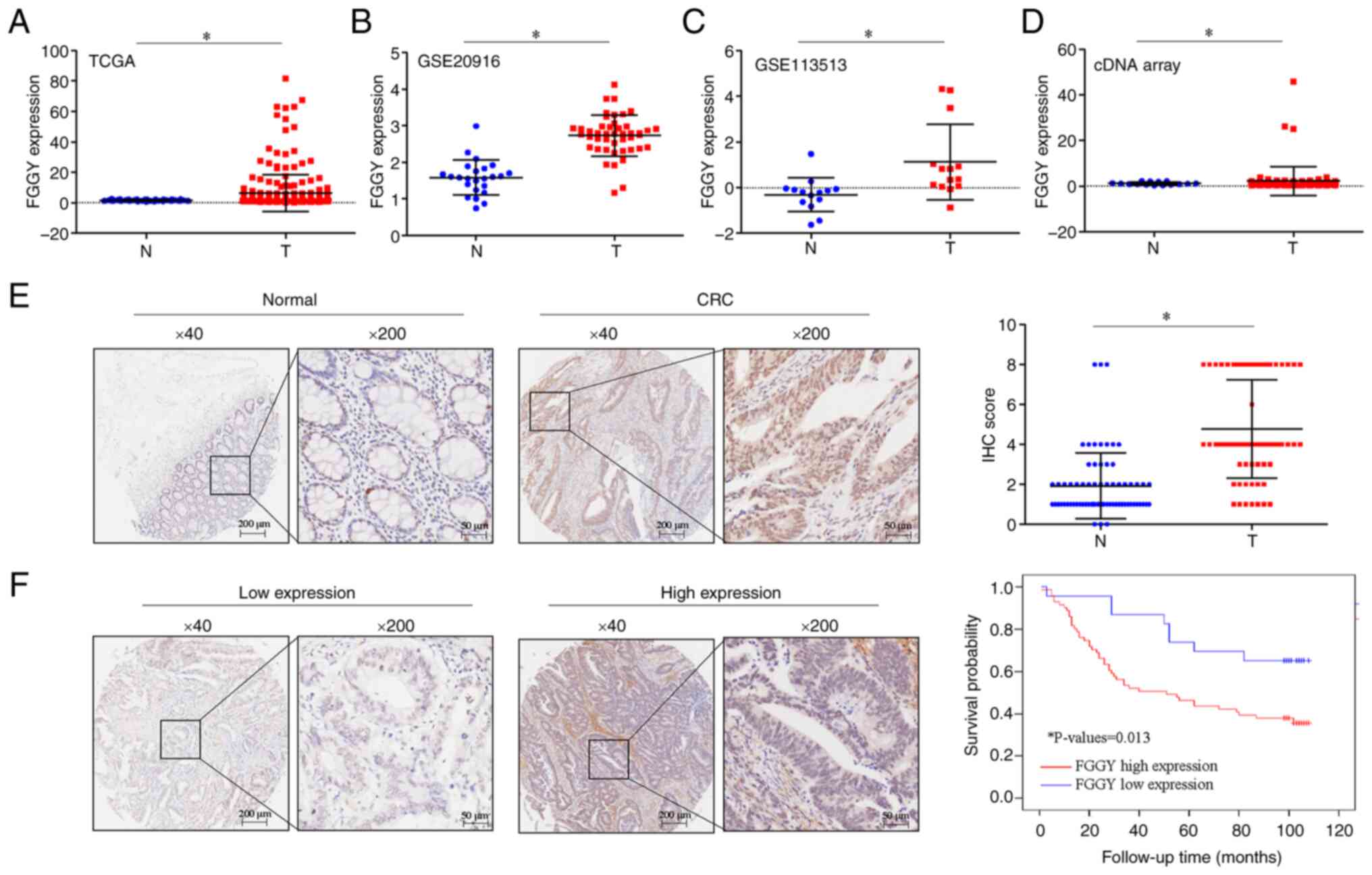
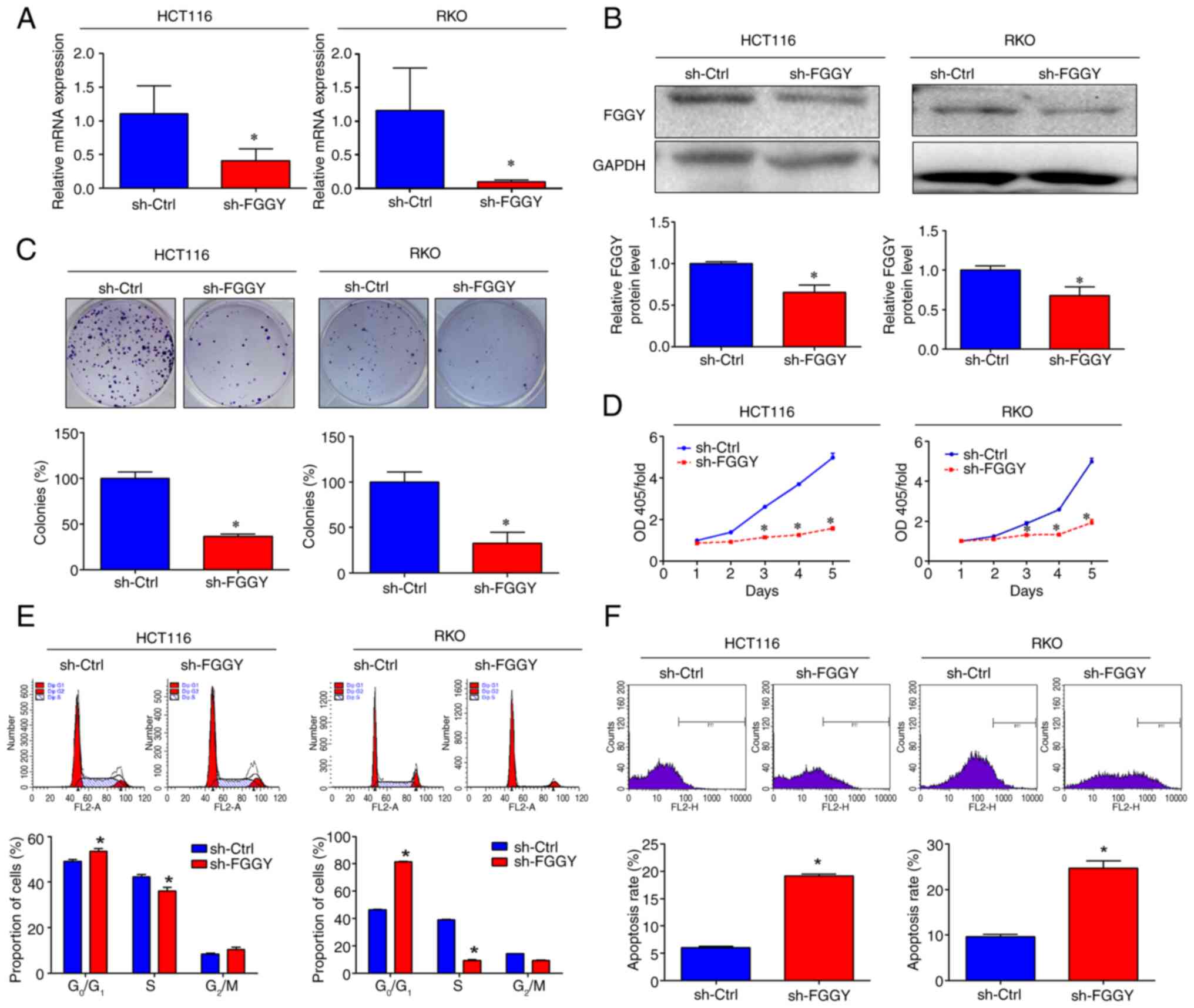
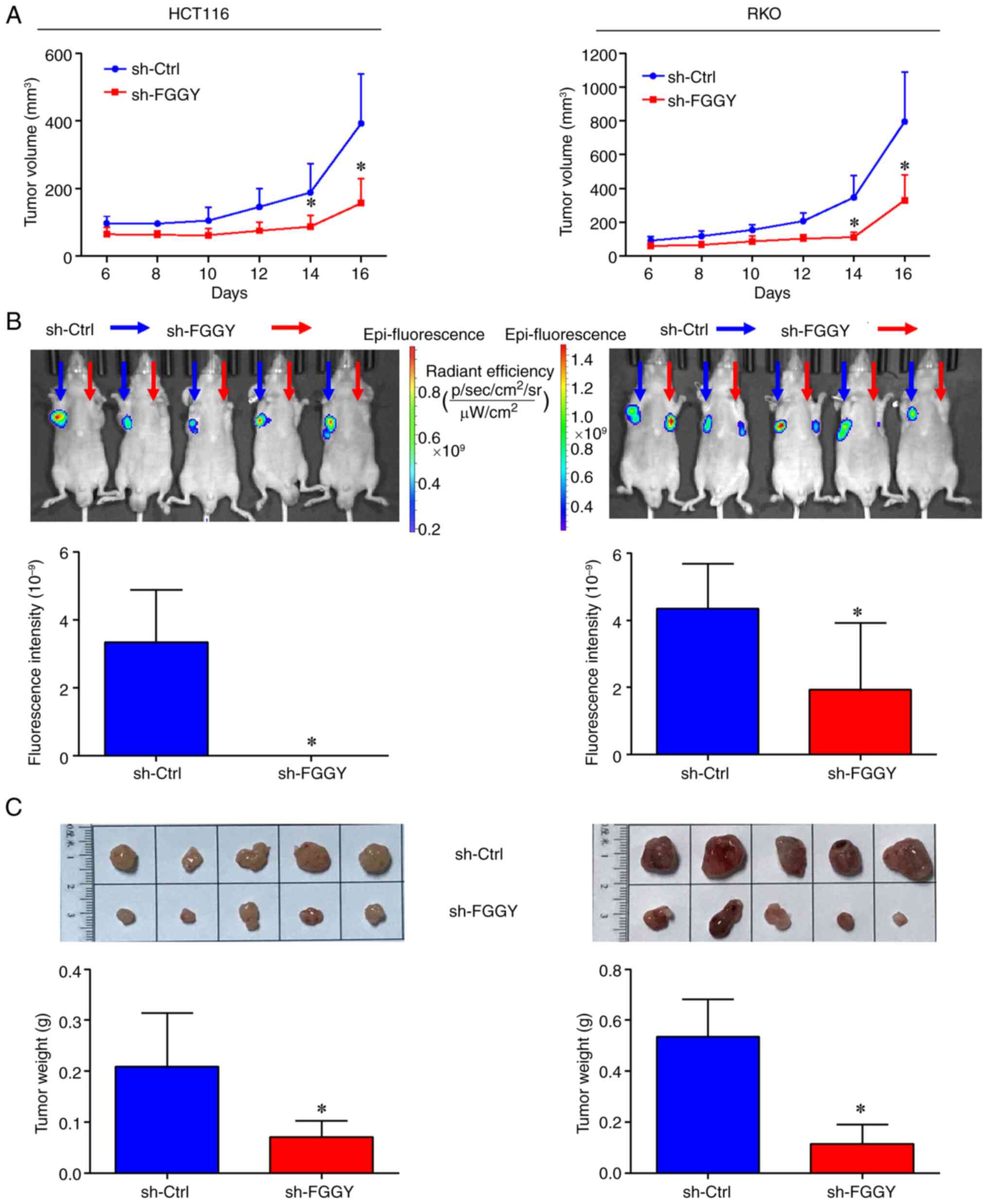
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA cancer J Clin. 73:17–48. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ladabaum U, Dominitz JA, Kahi C and Schoen
RE: Strategies for colorectal cancer screening. Gastroenterology.
158:418–432. 2020. View Article : Google Scholar
|
|
4
|
Kanth P and Inadomi JM: Screening and
prevention of colorectal cancer. BMJ. 374:n18552021. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen A, Chen Y, Liu L, Huang Y, Chen H, Qi
F, Lin J, Shen Z, Wu X, Wu M, et al: EBF1-mediated upregulation of
ribosome assembly factor PNO1 contributes to cancer progression by
negatively regulating the p53 signaling pathway. Cancer Res.
79:2257–2270. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Zagnitko O, Rodionova I, Osterman
A and Godzik A: The FGGY carbohydrate kinase family: Insights into
the evolution of functional specificities. PLoS Comput Biol.
7:e10023182011. View Article : Google Scholar
|
|
7
|
Omelchenko MV, Makarova KS, Wolf YI,
Rogozin IB and Koonin EV: Non-homologous isofunctional enzymes: A
systematic analysis of alternative solutions in enzyme evolution.
Biol Direct. 5:312010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Daoud H, Valdmanis PN, Dion PA and Rouleau
GA: Analysis of DPP6 and FGGY as candidate genes for amyotrophic
lateral sclerosis. Amyotroph Lateral Scler. 11:389–391. 2010.
View Article : Google Scholar
|
|
9
|
Van Es MA, Van Vught PW, Veldink JH,
Andersen PM, Birve A, Lemmens R, Cronin S, Van Der Kooi AJ, De
Visser M, Schelhaas HJ, et al: Analysis of FGGY as a risk factor
for sporadic amyotrophic lateral sclerosis. Amyotroph Lateral
Scler. 10:441–447. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taylor JA, Shioda K, Mitsunaga S, Yawata
S, Angle BM, Nagel SC, Vom Saal FS and Shioda T: Prenatal exposure
to bisphenol a disrupts naturally occurring bimodal dna methylation
at proximal promoter of fggy, an obesity-relevant gene encoding a
carbohydrate kinase, in gonadal white adipose tissues of cd-1 mice.
Endocrinology. 159:779–794. 2018. View Article : Google Scholar :
|
|
11
|
Zhang R, Zhang F, Sun Z, Liu P, Zhang X,
Ye Y, Cai B, Walsh MJ, Ren X, Hao X, et al: LINE-1
retrotransposition promotes the development and progression of lung
squamous cell carcinoma by disrupting the tumor-suppressor gene
fggy. Cancer Res. 79:4453–4465. 2019.PubMed/NCBI
|
|
12
|
Sun Z, Zhang R, Zhang X, Sun Y, Liu P,
Francoeur N, Han L, Lam WY, Yi Z, Sebra R, et al: LINE-1 promotes
tumorigenicity and exacerbates tumor progression via stimulating
metabolism reprogramming in non-small cell lung cancer. Mol Cancer.
21:1472022.PubMed/NCBI
|
|
13
|
Skrzypczak M, Goryca K, Rubel T, Paziewska
A, Mikula M, Jarosz D, Pachlewski J, Oledzki J and Ostrowski J:
Modeling oncogenic signaling in colon tumors by multidirectional
analyses of microarray data directed for maximization of analytical
reliability. PLoS One. 5:e130912010.PubMed/NCBI
|
|
14
|
Tomczak K, Czerwińska P and Wiznerowicz M:
The cancer genome atlas (TCGA): An immeasurable source of
knowledge. Contemp Oncol (Pozn). 19:A68–A77. 2015.PubMed/NCBI
|
|
15
|
Marisa L, de Reyniès A, Duval A, Selves J,
Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D,
Ayadi M, et al: Gene expression classification of colon cancer into
molecular subtypes: Characterization, validation, and prognostic
value. PLoS Med. 10:e10014532013.PubMed/NCBI
|
|
16
|
Li C, Tang Z, Zhang W, Ye Z and Liu F:
GEPIA2021: integrating multiple deconvolution-based analysis into
GEPIA. Nucleic Acids Res. 49:W242–W246. 2021.PubMed/NCBI
|
|
17
|
Rao X, Huang X, Zhou Z and Lin X: An
improvement of the 2ˆ(-delta delta CT) method for quantitative
real-time polymerase chain reaction data analysis. Biostat
Bioinforma Biomath. 3:71–85. 2013.PubMed/NCBI
|
|
18
|
Shen A, Liu L, Chen H, Qi F, Huang Y, Lin
J, Sferra TJ, Sankararaman S, Wei L, Chu J, et al: Cell division
cycle associated 5 promotes colorectal cancer progression by
activating the ERK signaling pathway. Oncogenesis.
8:192019.PubMed/NCBI
|
|
19
|
Hubrecht RC and Carter E: The 3Rs and
humane experimental technique: Implementing change. Animals
(Basel). 9:7542019.PubMed/NCBI
|
|
20
|
Calderón-González KG, Valero Rustarazo ML,
Labra-Barrios ML, Bazán-Méndez CI, Tavera-Tapia A, Herrera-Aguirre
ME, Sánchez del Pino MM, Gallegos-Pérez JL, González-Márquez H,
Hernández-Hernández JM, et al: Determination of the protein
expression profiles of breast cancer cell lines by quantitative
proteomics using iTRAQ labelling and tandem mass spectrometry. J
Proteomics. 124:50–78. 2015.PubMed/NCBI
|
|
21
|
Ma YT, Xiao Q, Xu X, Shao and Wang H:
iTRAQ-based quantitative analysis of cancer-derived secretory
proteome reveals TPM2 as a potential diagnostic biomarker of
colorectal cancer. Front Med. 10:278–285. 2016.PubMed/NCBI
|
|
22
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000.
|
|
23
|
Milacic M, Beavers D, Conley P, Gong C,
Gillespie M, Griss J, Haw R, Jassal B, Matthews L, May B, et al:
The reactome pathway knowledgebase 2024. Nucleic Acids Res.
52:D672–D678. 2024.
|
|
24
|
Mi H and Thomas P: PANTHER pathway: An
ontology-based pathway database coupled with data analysis tools.
Methods Mol Biol. 563:123–140. 2009.PubMed/NCBI
|
|
25
|
Li F, Zhao D, Yang S, Wang J, Li Q, Jin X
and Wang W: ITRAQ-based proteomics analysis of triptolide on human
A549 lung adenocarcinoma cells. Cell Physiol Biochem. 45:917–934.
2018.PubMed/NCBI
|
|
26
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
|
|
27
|
Kulak NA, Pichler G, Paron I, Nagaraj N
and Mann M: Minimal, encapsulated proteomic-sample processing
applied to copy-number estimation in eukaryotic cells. Nat Methods.
11:319–324. 2014.PubMed/NCBI
|
|
28
|
Humphrey SJ, Azimifar SB and Mann M:
High-throughput phosphoproteomics reveals in vivo insulin signaling
dynamics. Nat Biotechnol. 33:990–995. 2015.PubMed/NCBI
|
|
29
|
Collado M, Blasco MA and Serrano M:
Cellular senescence in cancer and aging. Cell. 130:223–233.
2007.PubMed/NCBI
|
|
30
|
Kurz DJ, Decary S, Hong Y and Erusalimsky
JD: Senescence-associated (beta)-galactosidase reflects an increase
in lysosomal mass during replicative ageing of human endothelial
cells. J Cell Sci. 113:3613–3622. 2000.PubMed/NCBI
|
|
31
|
Bannister AJ, Zegerman P, Partridge JF,
Miska EA, Thomas JO, Allshire RC and Kouzarides T: Selective
recognition of methylated lysine 9 on histone H3 by the HP1 chromo
domain. Nature. 410:120–124. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kramer HB, Lai CF, Patel H, Periyasamy M,
Lin ML, Feller SM, Fuller-Pace FV, Meek DW, Ali S and Buluwela L:
LRH-1 drives colon cancer cell growth by repressing the expression
of the CDKN1A gene in a p53-dependent manner. Nucleic Acids Res.
44:582–594. 2016. View Article : Google Scholar :
|
|
35
|
Wang G, Fu Y, Hu F, Lan J, Xu F, Yang X,
Luo X, Wang J and Hu J: Loss of BRG1 induces CRC cell senescence by
regulating p53/p21 pathway. Cell Death Dis. 8:e26072017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shen A, Wu M, Liu L, Chen Y, Chen X,
Zhuang M, Xie Q, Cheng Y, Li J, Shen Z, et al: Targeting NUFIP1
suppresses growth and induces senescence of colorectal cancer
cells. Front Oncol. 11:6814252021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ohtani N, Mann DJ and Hara E: Cellular
senescence: Its role in tumor suppression and aging. Cancer Sci.
100:792–797. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Shen Y and White V: p53-dependent
apoptosis pathways. Adv Cancer Res. 82:55–84. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Stein GH, Drullinger LF, Soulard A and
Dulić V: Differential roles for cyclin-dependent kinase inhibitors
p21 and p16 in the mechanisms of senescence and differentiation in
human fibroblasts. Mol Cell Biol. 19:2109–2117. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Li XL, Zhou J, Chen ZR and Chng WJ: P53
mutations in colorectal cancer-molecular pathogenesis and
pharmacological reactivation. World J Gastroenterol. 21:84–93.
2015. View Article : Google Scholar : PubMed/NCBI
|