Gastrointestinal cancers (GICs) account for >25%
of all new cancer cases worldwide and contribute to more than
one-third of cancer-related deaths (1). These cancers primarily include
gastric cancer (GC), colorectal cancer (CRC), esophageal squamous
cell carcinoma (ESCC), hepatocellular carcinoma (HCC),
cholangiocarcinoma (CCA), gallbladder cancer (GBC) and pancreatic
cancer. Traditional treatments for GICs, including surgery
intervention, radiation therapy, chemotherapy, targeted therapies
and immunotherapies, have long been the primary approaches for
management (2,3). However, early diagnosis of GICs is
challenging and a proportion of patients are diagnosed in the first
instance with advanced-stage cancer. For patients diagnosed with
advanced-stage cancer, traditional therapies, although essential,
often fall short due to the inherent chemoresistance of GIC cells,
frequently leading to tumor recurrence and distant metastasis
(4), resulting in poor clinical
outcomes for patients with advanced GICs (5).
Ferroptosis, a type of regulated cell death (RCD),
characterized by excessive lipid peroxidation and iron
accumulation, has gained significant attention in recent years
given its critical role in cancer biology (6,7).
Unlike apoptosis or necrosis, ferroptosis occurs due to the buildup
of toxic lipid peroxides, resulting from impaired cellular
antioxidant systems, particularly disruptions in glutathione (GSH)
metabolism (6). Research has
increasingly linked impaired ferroptosis with cancer progression,
suggesting that triggering ferroptosis could be a potential
approach for combating tumors, particularly in overcoming
therapeutic resistance (8-16).
Epigenetic processes, including DNA methylation,
histone modifications and regulatory non-coding (nc)RNAs,
profoundly influence gene expression and can affect cell death
pathways, including ferroptosis (17-19). In GICs, these modifications may
either promote or inhibit ferroptosis, thereby affecting tumor
behavior and treatment responsiveness. For instance, epigenetic
modifications can regulate genes associated with iron metabolism,
antioxidant systems and lipid peroxidation, thereby affecting the
susceptibility of GIC cells to ferroptosis.
Understanding the intricate relationship between
epigenetic regulation and ferroptosis in GICs could reveal novel
therapeutic strategies and targeting epigenetic modifications to
induce ferroptosis may serve as a novel approach to suppress tumor
progression and overcome resistance to conventional treatments. The
present review explores the current understanding of how epigenetic
regulation influences ferroptosis in GICs, highlighting its
implications for cancer progression, treatment and potential future
therapies.
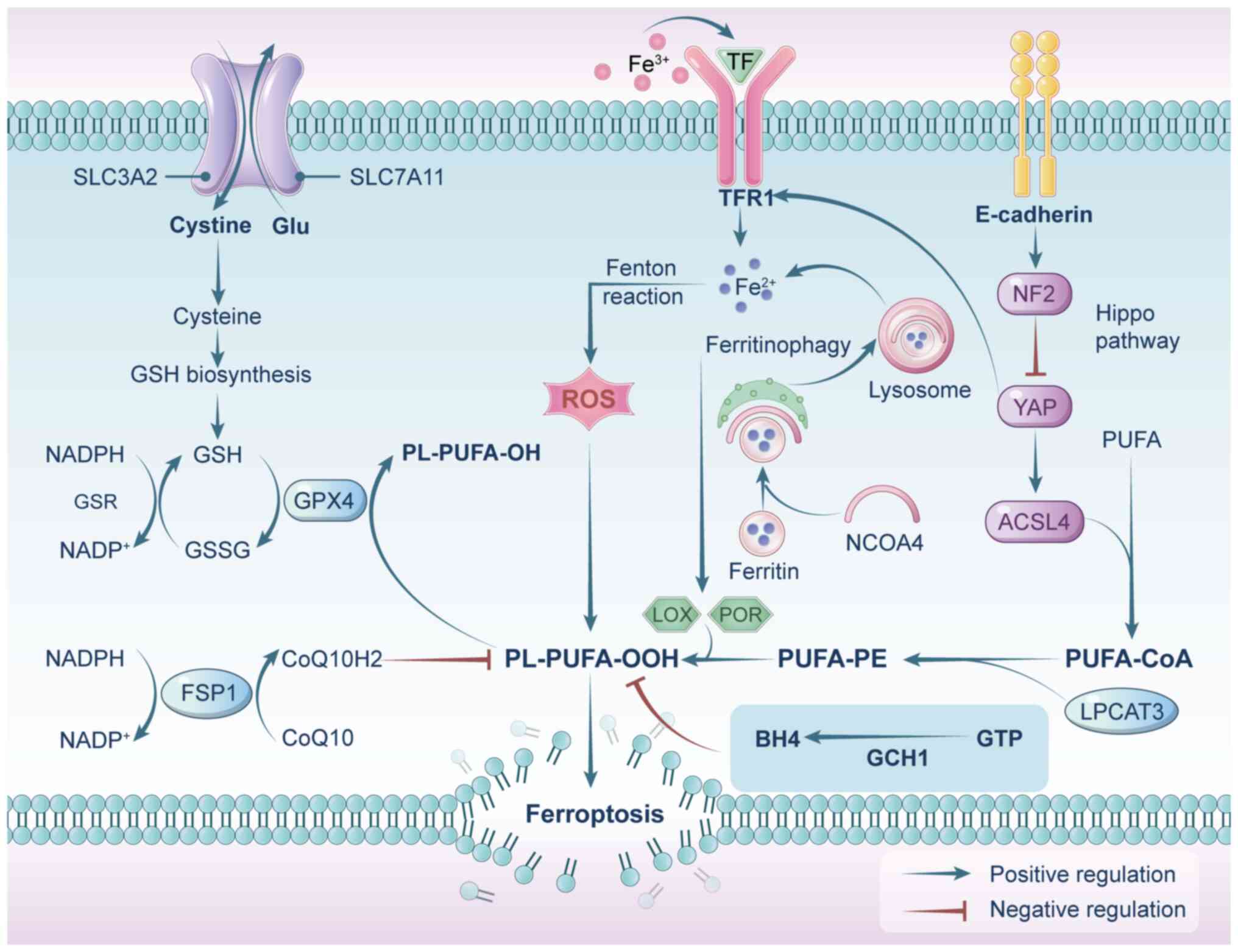
Ferritin, a protein complex responsible for storing
iron, consists of two components: Ferritin heavy chain 1 (FTH1) and
ferritin light chain (FTL). A specialized form of autophagy, known
as ferritinophagy, enhances the susceptibility of a cell to
ferroptosis by increasing the intracellular bioavailability of
Fe2+ (37-40). More specifically, ferritin is
identified by nuclear receptor coactivator 4 (NCOA4), which directs
FTH1 to autophagosomes for lysosomal breakdown, resulting in the
liberation of free iron (Fig. 1)
(39). Reducing lysosomal
activity (41) or downregulating
NCOA4 expression has been demonstrated to block ferroptosis
effectively (37,38). For instance, a relatively
recently discovered ferroptosis inhibitor called Compound 9a can
directly bind to recombinant NCOA4 protein. This binding disrupts
the NCOA4-FTH1 interaction, reducing the intracellular
bioavailability of Fe2+ and ultimately blocking
ferroptosis (42). As a key
regulator of ferroptosis, NCOA4 represents a potential target for
therapeutic intervention. Of note, ferritin and FPN can be
simultaneously upregulated by ATM, thereby rescuing ferroptosis in
multiple cancer cells triggered by cystine deprivation or erastin
(43).
Polyunsaturated fatty acids (PUFAs), such as adrenic
acid and arachidonic acid, are highly susceptible to peroxidation.
Enzymes like lysophosphatidylcholine acyltransferase 3 and Acyl-CoA
synthetase long-chain family member 4 (ACSL4) play essential roles
in incorporating these PUFAs into membrane phospholipids, thereby
promoting lipid peroxidation and ferroptotic cell death (44-47) (Fig. 1).
Lipid peroxidation can proceed through two pathways:
Non-enzymatic, driven by free radicals like ROS, and enzymatic
pathways, catalyzed by arachidonate lipoxygenases (ALOXs) and other
enzymes (48-51).
Non-enzymatic lipid peroxidation is a process driven
by free radicals, where ROS initiate the oxidation of PUFAs
(23). The most potent ROS is
the hydroxyl radical. This radical is primarily formed through
Fenton or Fenton-like reactions, which result from the interaction
of hydrogen peroxide (H2O2) with free
Fe2+ (52,53).
ALOXs hold a crucial role in catalyzing enzymatic
lipid peroxidation. This family of iron-containing enzymes
encompasses six distinct isoforms (ALOXE3, ALOX5, ALOX12, ALOX12B,
ALOX15 and ALOX15B), which facilitate the dioxygenation of PUFAs
and PUFA-containing lipids within biological membranes to produce a
variety of lipid hydroperoxides in a highly specific manner
(54). This suggests the
potential involvement of ALOXs in initiating ferroptosis. Previous
studies have indicated that certain pharmacological ALOX inhibitors
can effectively hinder ferroptosis (55,56). However, it is noteworthy that the
deletion of ALOX15 did not successfully rescue ferroptosis
triggered by the knockout of glutathione peroxidase 4 (GPX4)
(57). These results imply the
presence of compensatory mechanisms that could offset the loss of
ALOX15 activity (58). Although
ALOXs are crucial for enzymatic lipid peroxidation, their
contribution to ferroptosis appears context-dependent. ALOX12 is
crucial for p53-induced ferroptosis, but it is not involved in
ferroptosis triggered by erastin or GPX4 inhibitors (59), suggesting differential regulation
of ferroptosis by ALOX isoforms.
Consequently, ALOXs may specifically participate in
certain scenarios within the domain of ferroptosis. To gain a more
precise understanding, further research is needed to clarify the
precise function of ALOXs in ferroptosis. Of note, ALOXs may not be
the only factors governing lipid peroxidation during ferroptosis.
In fact, it has been indicated that cytochrome P450 oxidoreductase
actively contributes to the initiation of lipid peroxidation
(60).
Under physiological conditions, the process of
iron-driven lipid oxidation is carefully regulated by cellular
antioxidant defense systems. A central player in this defense
mechanism is GPX4, a pivotal antioxidant enzyme that plays a
crucial role in reducing membrane lipid hydroperoxides to non-toxic
lipid alcohols (9,61). GPX4 accomplishes this task by
relying on reduced GSH as an essential cofactor (62). Thus, the proper functioning of
GPX4 is intricately linked to the availability of GSH (9,63). GSH is produced through the
combination of three amino acids: Cysteine, glycine and glutamic
acid. Notably, the availability of cysteine stands out as the
primary limiting factor in this biosynthesis process (64,65). Within mammalian cells, the
cystine/glutamate antiporter system xc−,
composed of solute carrier family 7 member 11 (SLC7A11) and SLC3A2,
is responsible for transporting extracellular cystine (the oxidized
form of cysteine) into the cell while releasing intracellular
glutamate in a 1:1 exchange (66,67). The inhibition of system
xc− or GPX4, either pharmacologically or
genetically, triggers lipid peroxidation and ferroptosis (48,68,69). Furthermore, SLC7A11 expression
and function are enhanced by nuclear factor E2-related factor 2
(70) but suppressed by tumor
suppressors, including TP53 (71), BRCA1-associated protein 1 (BAP1)
(72) and beclin 1 (73). Therefore, SLC7A11 serves as a
critical regulatory point in the initiation of ferroptosis, making
it an important factor in the control of this cellular process.
Ferroptosis suppressor protein 1 (FSP1) functions as
a GSH-independent coenzyme Q (CoQ) oxidoreductase and serves as an
essential antioxidant defense system alongside GPX4, making it a
key regulator of ferroptosis (50,51,74). FSP1 utilizes NADH/NADPH to
convert CoQ10 into its reduced, active form, CoQ10H2. In this
reduced form, CoQ10H2 functions as a lipid-soluble antioxidant,
neutralizing free radicals and inhibiting lipid peroxidation,
thereby protecting cells from ferroptosis (50,75). In addition to GPX4/GSH and FSP1,
guanosine triphosphate cyclohydrolase 1 (GCH1) provides an
alternative ferroptosis defense by producing tetrahydrobiopterin
(BH4), which operates independently of the GPX4 system (76).
Epigenetics refers to regulatory mechanisms that
modify gene expression through DNA methylation, histone
modifications and ncRNAs without changing the underlying DNA
sequence (77,78).
In eukaryotic cells, DNA methylation predominantly
occurs at the 5′ position of the cytosine ring within CpG
dinucleotides, exerting regulatory control over gene transcription
by influencing promoters and enhancers (79). This modification involves the
enzymatic actions of DNA methyltransferases (DNMTs) and DNA
demethyltransferases (also named TET enzymes) (80-83). CpG dinucleotides are concentrated
in specific regions known as CpG islands (CGIs), and 50-60% of gene
promoters are situated within these CGIs. In healthy cells, CGIs,
particularly those associated with promoters, tend to remain
unmethylated. However, disruptions in DNA methylation processes are
intricately involved in cancer development (84). Promoter hypermethylation-induced
transcriptional silencing of tumor suppressor genes is a common
observation in the majority of cancers (85-87).
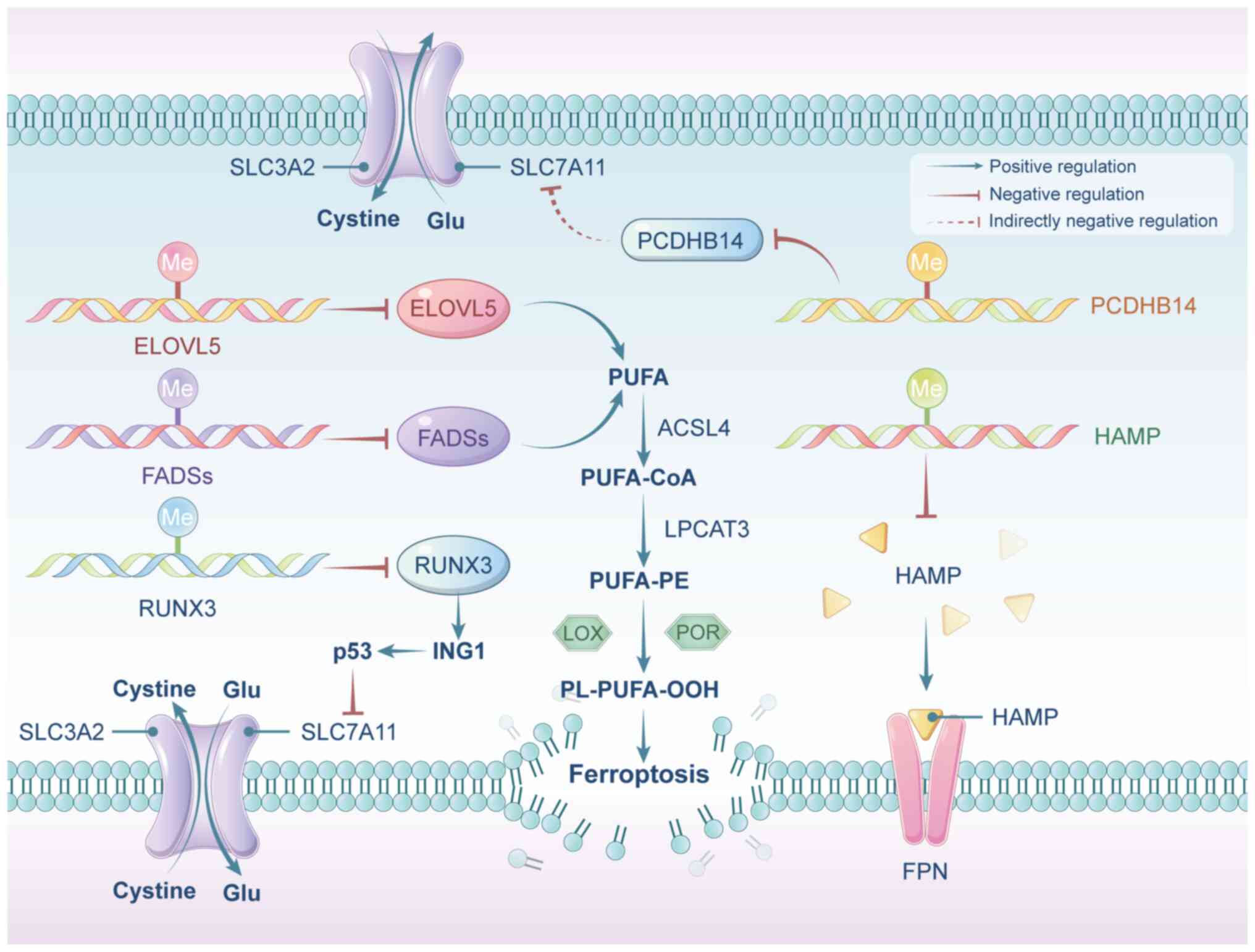
DNA methylation is a key regulatory mechanism that
influences ferroptosis by modulating the expression of critical
genes associated with this pathway (88). In intestinal GC cells, DNA
methylation silences fatty acid desaturase 1 and elongation of very
long-chain fatty acid protein 5, reducing PUFA synthesis and
inhibiting lipid peroxidation, thus suppressing ferroptosis
(89). Similarly, aberrant
hypermethylation of the protocadherin gene 14 promoter in HCC leads
to its inactivation, but p53 activation can reverse this, promoting
ferroptosis by reducing SLC7A11 expression (90). Iron homeostasis is largely
controlled by the HAMP-FPN axis, where HAMP plays a crucial role in
regulating iron absorption, distribution and overall balance
(91). In individuals with HCC,
HAMP expression is suppressed, coinciding with hypermethylation of
a highly conserved CpG island within its promoter region (92,93). In GBC, DNMT1-mediated methylation
downregulates runt-related transcription factor 3 (RUNX3),
impairing ferroptosis. However, activating RUNX3 induces
ferroptosis by enhancing inhibitor of growth 1 transcription, which
in turn downregulates SLC7A11 in a p53-dependent manner (94) (Fig. 2).
Histone modifications play a key role in shaping
chromatin structure and thus impact gene expression and cancer
development (95,96). This discussion will primarily
center on how histone acetylation, methylation and ubiquitination
contribute to the regulation of ferroptosis.
Histone acetylation modification refers to the
process of adding acetyl groups to histone proteins, which are
crucial for regulating chromatin structure and gene expression.
Acetylation of histone is typically associated with open chromatin
conformation, allowing for increased accessibility of DNA to
transcriptional machinery, and thus, increased gene transcription.
This process is facilitated by histone acetyltransferases, which
add acetyl groups, while histone deacetylases (HDACs) remove them,
leading to chromatin compaction and suppression of gene
expression.
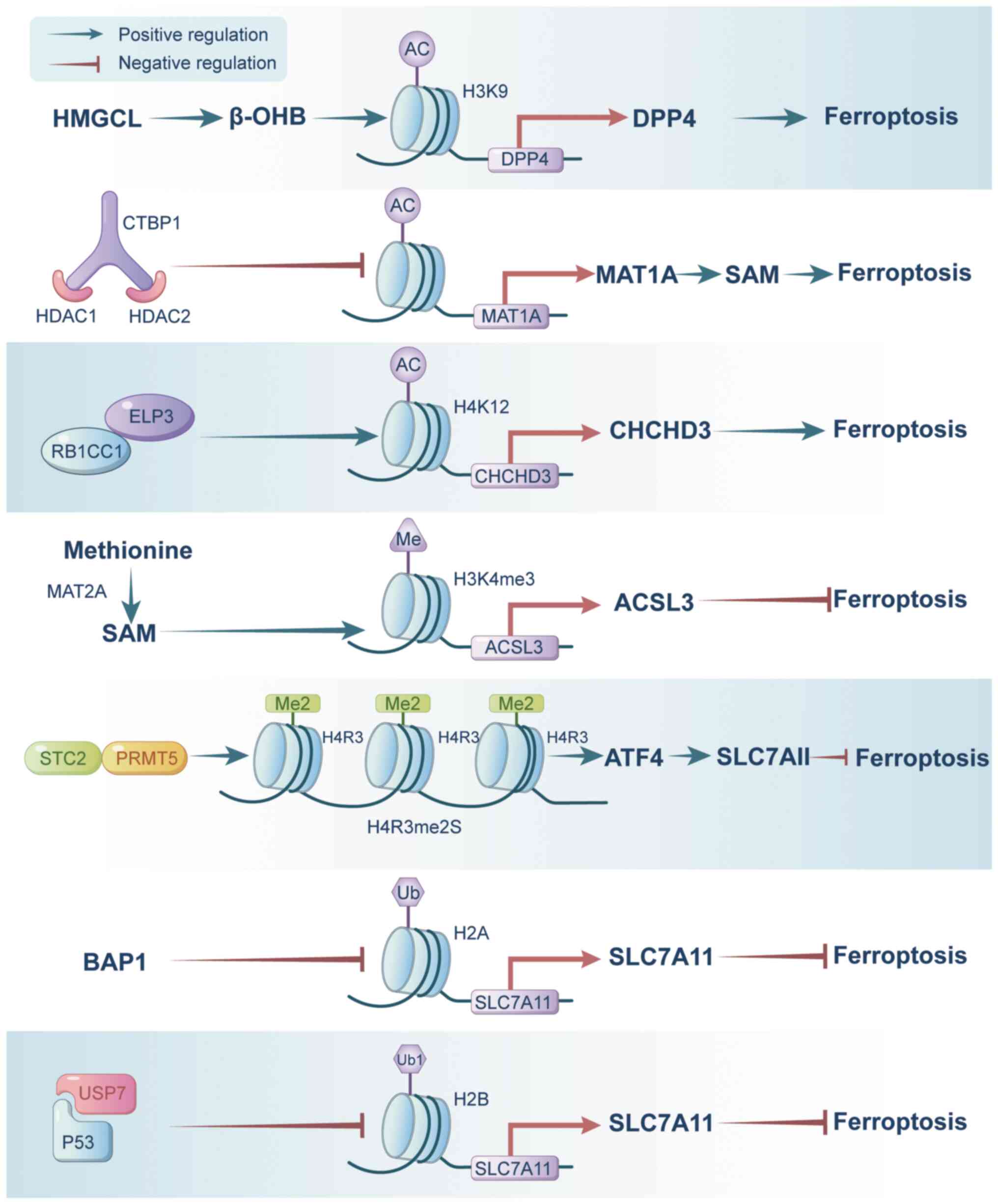
Hydroxymethylglutaryl-CoA lyase (HMGCL) catalyzes
the conversion of HMG-CoA into β-hydroxy-butyric acid (β-OHB) and
acetoacetate. Notably, β-OHB acts as a natural inhibitor of HDAC,
promoting elevated histone acetylation levels (97). Recent studies have demonstrated
that HMGCL, via β-OHB, enhances H3K9 acetylation, thereby
upregulating the expression of dipeptidyl peptidase 4 (DPP4). Of
note, DPP4, crucial for intracellular lipid peroxidation, renders
HCC cells susceptible to ferroptosis (98). In addition, C-terminal-binding
protein 1 binds with HDAC1 and HDAC2, creating a transcriptional
complex that downregulates the expression of methionine
adenosyltransferase 1A (MAT1A) through histone deacetylation
modification. Consequently, decreased MAT1A levels reduce
ferroptosis in HCC cells, potentially through direct mechanisms or
by impairing CD8+ T-cell and interferon-γ production, thereby
facilitating the progression of HCC (99). Furthermore, RB1-inducible
coiled-coil 1 facilitates the recruitment of elongator
acetyltransferase complex subunit 3 to enhance H4K12Ac at
ferroptosis-associated enhancers, promoting the transcription of
ferroptosis-associated genes, such as coiled-coil-helix-coile
d-coil-helix domain containing 3 (CHCHD3), and sensitizing cancer
cells to ferroptosis (100)
(Fig. 3).
Histone methylation modification involves the
addition of methyl groups to histone proteins, altering their
structure and affecting gene expression. This process can take
place on various amino acid residues within histone tails,
including lysine and arginine. Depending on the specific site and
extent of methylation, it can either activate or repress gene
transcription.
In the context of ferroptosis, histone methylation
modifications regulate the expression of genes involved in this
process. For instance, MAT2A promotes S-adenosylmethionine
production, leading to elevated H3K4me3 levels at the promoter
region of acyl-CoA synthetase long-chain family member 3, thereby
conferring resistance against ferroptosis in GC cells (101). Similarly, in ESCC,
stanniocalcin 2 induces the activation of protein methyltransferase
5 (PRMT5), which enhances the symmetric dimethylation of histone H4
at arginine 3 (H4R3me2s). This activation, in turn, triggers the
PRMT5-ATF4-SLC3A2/SLC7A11 axis, effectively inhibiting ferroptosis
in ESCC. Furthermore, this process also induces radioresistance by
facilitating DNA damage repair mechanisms (102) (Fig. 3). To date, to the best of our
knowledge, there has been no study on the regulation of ferroptosis
by H3K9me3 in GICs.
Histone ubiquitination modification is a key
epigenetic process that modulates gene expression and various
cellular functions. It involves attaching ubiquitin molecules to
histone proteins, which can alter chromatin structure and affect
transcriptional activity. Additionally, aberrant histone
ubiquitination has been linked to the development of several types
of cancer, including GICs.
SLC7A11, a pivotal gene involved in ferroptosis, is
regulated by histone ubiquitination modification. Tumor
suppressors, such as p53 and BAP, impact the expression of histone
deubiquitination enzymes, thus modulating the histone
ubiquitination modification in the regulatory region of SLC7A11
(Fig. 3). Specifically, BAP1
functions as a nuclear deubiquitinating enzyme that specifically
removes ubiquitin from histone H2A at the SLC7A11 promoter,
resulting in reduced SLC7A11 transcription. This decrease in
SLC7A11 levels leads to increased lipid peroxidation, ultimately
triggering ferroptosis in cancerous cells (72). Similarly, p53 facilitates the
nuclear import of the deubiquitinase USP7, which in turn reduces
H2Bub1 levels at the regulatory region of the SLC7A11 gene, thereby
repressing SLC7A11 expression and promoting ferroptosis in human
hepatoma cells (103).
ncRNAs encompass microRNAs (miRNAs/miRs), long
ncRNAs (lncRNAs) and circular RNAs (circRNAs) (104,105). ncRNAs have been widely
implicated in the regulation of ferroptosis.
miRNAs regulate gene expression at the
post-transcriptional level by binding to the 3′untranslated regions
of mRNA molecules, which either leads to mRNA degradation or blocks
its translation into a protein. They directly modulate the
expression of genes involved in ferroptosis. Specifically, certain
miRNAs may target genes related to lipid metabolism, iron
homeostasis or antioxidant defense systems.
GPX4 is essential for cell survival, as it reduces
harmful lipid peroxides into their non-toxic alcohol forms, thereby
preventing ferroptosis. In CRC and HCC, miR-15a-3p and miR-214-3p
directly target GPX4, suppressing its expression and facilitating
ferroptosis (111,112). Additionally, miRNAs exert an
indirect influence on GPX4. For instance, in CRC, miR-539 activates
the stress-activated protein kinase/c-Jun N-terminal kinase pathway
by targeting TNF-α-induced protein 8, which leads to reduced GPX4
levels and the induction of ferroptosis (113).
Nuclear factor erythroid 2-related factor 2 (NRF2),
an essential transcription factor in the antioxidant response, is
inhibited by the ubiquitin ligase complex formed by Kelch-like
ECH-associated protein 1 (KEAP1) and Cullin 3. NRF2 drives the
expression of antioxidant enzymes, including heme oxygenase 1,
NAD(P)H:quinone oxidoreductase 1 and superoxide dismutase, which
are essential for counteracting ROS, preventing lipid peroxidation
and ultimately inhibiting ferroptosis (114). Several miRNAs, including
miR-124-3p, miR-507, miR-634, miR-450a, miR-129-5p and miR-142-5p,
directly target the 3′-UTR of NRF2 mRNA, leading to decreased NRF2
expression and increased ROS in GC and ESCC (115-117). Furthermore, miRNAs indirectly
regulate NRF2 by targeting KEAP1. In ESCC, HCC and GC cells,
miR-432-3p, miR-200a, miR-141 and miR-328-3p target KEAP1,
resulting in NRF2 upregulation (118-122).
Under conditions of stress, ATF4 expression is
upregulated, leading to the induction of SLC7A11 and subsequent
inhibition of ferroptosis (123,124). Conversely, in HCC, miR-214-3p
directly targets ATF4, thereby promoting ferroptosis (125).
ACSL4 facilitates the incorporation of PUFAs into
phospholipids, thereby increasing the susceptibility of cells to
ferroptosis. By modulating ACSL4, miRNAs play a significant role in
controlling ferroptosis in GICs. For instance, miR-552-5p and
miR-23a-3p specifically target the 3′-UTR of ACSL4 mRNA, leading to
the inhibition of ferroptosis in HCC cells and contributing to
resistance against sorafenib (126,127). In addition, exosomes derived
from cancer-associated fibroblasts (CAFs) transport miR-3173-5p to
gemcitabine-treated pancreatic ductal adenocarcinoma (PDAC) cells,
where miR-3173-5p targets ACSL4, thereby inhibiting ferroptosis and
mediating resistance to gemcitabine (128). Similarly, exosomes released by
GC cells deliver miR-214-3p to vascular endothelial cells, where
miR-214-3p targets zinc finger protein A20, resulting in the
negative regulation of ACSL4 and subsequent inhibition of
ferroptosis in these cells (129). Apart from ASCL4, miRNAs have
the potential to modulate additional molecules or pathways linked
to lipid metabolism, influencing the regulation of ferroptosis. The
mevalonate (MVA) pathway, responsible for cholesterol and CoQ10
synthesis, is among the pathways subject to miRNA regulation.
Considering the ability of hydroxyacyl-CoA dehydrogenase α subunit
(HADHA) to enhance the expression of key MVA pathway enzymes,
miR-612 may exert a suppressive effect on this pathway by directly
targeting HADHA. This suppression could lead to reduced CoQ10
levels and elevated cellular PUFA levels, which can trigger
ferroptosis in HCC cells (130). The family of ALOXs is regarded
as the key mediator of lipid peroxidation production, which
eventually leads to ferroptosis (56,131). Previous studies have
demonstrated that by targeting ALOX15, exosomal miR-522, secreted
by CAFs, inhibits ferroptosis in GC cells (132).
miRNAs can regulate ferroptosis in GIC cells by
influencing intracellular iron metabolism. Specifically, miR-545
targets TF (133), while
several other miRNAs, such as miR-31, miR-141, miR-145, miR-182,
miR-194, miR-758, miR-22, miR-200a, miR-320 and miR-152, target TFR
in CRC and HCC, thereby reducing TF uptake (134-136). Notably, miR-194 not only
inhibits TFR expression but also inhibits FPN expression in CRC
(134). FPN serves as the
primary means of ferrous iron export (137), and is targeted by miR-150 and
miR-485-3p in HCC (138). In
CRC, miR-133a targets FTL, one of the components of ferritin
responsible for iron storage (134). In addition, miR-19a targets
iron-responsive element-binding protein 2, thus suppressing
ferroptosis in CRC (139).
These intricate regulatory interactions highlight the multifaceted
role of miRNAs in modulating ferroptosis through the regulation of
intracellular iron dynamics in GICs. Table I gives a comprehensive and
detailed overview of how miRNAs regulate ferroptosis in GICs.
lncRNAs are long RNA molecules typically >200
nucleotides in length, which have been recognized as key regulators
of ferroptosis in tumor cells (140). These molecules exhibit a dual
capability in modulating ferroptosis in GICs: They can directly
target key genes or molecules associated with ferroptosis or
indirectly modulate them by acting as miRNA sponges (141). For instance, lncRNAs have been
shown to directly or indirectly modulate the expression of SLC7A11,
impacting ferroptosis in HCC and GC (142-145), as well as modulate the
expression of GPX4, influencing ferroptosis in ESCC, CCA and liver
cancer cells (112,146,147). Additionally, lncRNAs can
directly bind to molecules such as FSP1 or Keap1, thereby
inhibiting ferroptosis in GICs (148,149).
Furthermore, specific lncRNAs, such as
URB1-antisense RNA 1 (AS1), have been implicated in driving
ferritin phase separation, which helps lower cellular free iron
levels and suppressed sorafenib-induced ferroptosis in HCC
(150). Additionally, lncRNA
NEAT1 promotes the expression of Myo-inositol oxygenase, leading to
increased ROS production and a reduction in intracellular NADPH and
GSH levels, thereby intensifying ferroptosis in HCC (151,152). Another crucial enzyme involved
is Stearoyl-CoA desaturase 1 (SCD1), which converts saturated fatty
acids into monounsaturated fatty acids (MUFAs), affecting cellular
membrane composition and susceptibility to lipid
peroxidation-induced ferroptosis (153-156). In colon cancer, lncRNA
LINC01606 has been reported to enhance SCD1 expression by competing
for miR-423-5p, thereby protecting cancer cells from ferroptosis
(157).
Furthermore, in hypoxic tumor microenvironments
(TMEs), lncRNAs are crucial regulators of ferroptosis. For
instance, hypoxia-inducible factor 1α (HIF-1α) activation leads to
the transcriptional upregulation of lncRNA PMAN, which enhances the
output of ELAV-like RNA binding protein 1 (ELAVL1) in the
cytoplasm. ELAVL1 then stabilizes SLC7A11 mRNA, increasing its
expression and preventing ferroptosis in GC peritoneal metastasis
(145). Similarly, HIF-1α
induces lncRNA-CBSLR, which eventually protects GC cells from
ferroptosis and confers chemoresistance to GC cells under hypoxic
conditions by modulating the stability of cystathionine β-synthase
(CBS) mRNA and ACSL4 protein (158).
Furthermore, lncRNAs can utilize exosomes as
carriers to regulate ferroptosis in target cells. Exosomes secreted
by CAFs carry lncRNA DACT3-AS1 into GC cells, where it targets the
miR-181a-5p/sirtuin 1 (SIRT1) axis, inducing ferroptosis and
sensitizing GC cells to oxaliplatin (159). These examples illustrate the
diverse mechanisms through which lncRNAs modulate ferroptosis in
GICs. Table II provides a more
comprehensive overview.
circRNAs, which are single-stranded, covalently
closed ncRNA molecules, exhibit distinct characteristics from other
ncRNAs (160).
In GICs, numerous circRNAs have been identified as
crucial regulators of ferroptosis. circRNAs typically function as
miRNA sponges, indirectly influencing the expression of their
target mRNAs (161-164), including ferroptosis-associated
genes like GPX4 (165) and
SLC7A11. These genes are well-established critical regulators of
ferroptosis and studies have shown that several circRNAs modulate
their expression by sequestering miRNAs, thereby playing a
significant role in controlling ferroptosis in GICs (108,166-171). Notably, circRHOT1 facilitates
the recruitment of KAT5 (acetyltransferase Tip60), increasing
H3K27-mediated acetylation of the GPX4 gene, which promotes its
transcription and inhibits ferroptosis in GC (172).
In addition to acting as miRNA sponges, circRNAs can
directly bind to RNA-binding proteins involved in ferroptosis
signaling pathways, thereby modulating ferroptosis in GICs. For
instance, ELAVL1, an RNA-binding protein, enhances the translation
efficiency of target mRNAs by interacting with AU-rich elements in
their 3′UTRs (173). circRHBDD1
upregulates SCD expression by promoting ELAVL1 binding to SCD mRNA,
leading to the suppression of ferroptosis in CRC (174).
A recent study also elucidated the mechanism by
which circRNA may contribute to the regulation of ferroptosis.
circPDE3B was shown to function as both an miR-516b-5p sponge,
resulting in increased CBS expression, and as a direct binder with
RNA-binding protein heterogeneous nuclear ribonucleoprotein K,
resulting in the stabilization of SLC7A11. Consequently, this dual
action of circPDE3B ultimately resulted in the suppression of
ferroptosis in ESCC (175). A
comprehensive and detailed overview of how circRNAs regulate
ferroptosis in GICs is provided in Table III.
It is worth noting that certain key genes or
molecules related to ferroptosis are not only regulated by a single
epigenetic modification but may be influenced by multiple
epigenetic modifications simultaneously. For instance, in cancer
cells, the upstream region of the GPX4 gene exhibits low DNA
methylation, high levels of H3K4 trimethylation and H3K27
acetylation (176), indicating
that GPX4 is regulated not only by DNA methylation but also by
histone modifications. These modifications work together to create
a more complex regulatory framework for ferroptosis.
Aurora kinase A (AURKA), a serine/threonine kinase
responsible for regulating mitotic spindle function (177,178), is another key player.
miR-4715-3p can directly target the 3′-UTR of AURKA, suppressing
its expression, which in turn inhibits GPX4 and promotes
ferroptosis (179). However, in
upper gastrointestinal cancers, miR-4715-3p itself is downregulated
due to hypermethylation of its gene promoter (179). This epigenetic interplay
further complicates the regulation of ferroptosis in these types of
cancer.
ROS are critical in driving ferroptosis and have
been implicated in regulating epigenetic processes, including DNA
methylation and histone acetylation (180,181). In cancer cells, elevated ROS
levels enhance DNA hypermethylation, resulting in the repression of
tumor suppressor and antioxidant genes, which promotes cancer cell
proliferation under oxidative stress (180,181). A notable example is the
epigenetic silencing of RUNX3, a well-established tumor suppressor,
which is frequently downregulated in cancer cells due to
ROS-induced DNA hypermethylation of its promoter. This methylation
process is mediated by DNMT1 and HDAC1, whose expression and
activity are upregulated in response to ROS. DNMT1 and HDAC1 bind
to the RUNX3 promoter, driving its hypermethylation and silencing
(182). Importantly, treatment
with the ROS scavenger N-acetylcysteine or the DNMT1 inhibitor
5-aza-2-deoxycytidine has been shown to reverse this epigenetic
modification, highlighting the potential for therapeutic
intervention. Similarly, ROS-induced promoter hypermethylation has
been observed in other tumor suppressor genes. In CRC, ROS enhances
the methylation of the caudal type homeobox-1 (CDX1) promoter,
leading to its suppression and facilitating cancer progression
(183). In HCC, ROS elevates
Snail expression, which promotes hypermethylation of the E-cadherin
promoter, thereby contributing to tumor growth (184). Overall, ROS-induced epigenetic
modifications play a significant role in cancer progression.
Targeting ROS represents a promising therapeutic strategy-not only
to reverse these epigenetic changes but also to enhance ferroptosis
sensitivity in GICs. Future research should further explore the
mechanisms of ROS-mediated epigenetic silencing and develop
ROS-modulating therapies to improve treatment outcomes in these
malignancies.
The TME is a complex ecosystem consisting of immune
cells, stromal cells, tumor cells and various other elements. Among
immune cells, CD8+ T-cells play a crucial role by releasing
interferon-γ, which suppresses SLC7A11 expression while enhancing
ACSL4 levels, thereby facilitating ferroptosis in cancer cells
(185,186).
CAFs, another critical component of the TME,
significantly influence both innate and adaptive immunity (187). CAFs suppress ferroptosis in GC
cells by secreting miR-522 (132). In GICs, anoctamin 1 inhibits
ferroptosis by activating the PI3K-Akt signaling pathway, promoting
tumor progression and facilitating CAF recruitment via TGF-β
release (188). This disrupts
CD8+ T cell-driven anti-tumor responses, contributing to
immunotherapy resistance (188). These findings highlight the
intricate interactions between cancer cells, immune cells and CAFs
in regulating ferroptosis.
One unique feature of the TME is its metabolic
profile, marked by high lactate accumulation and restricted glucose
supply (189,190). Cancer cells exploit lactate to
counteract ferroptosis by either increasing MUFA-phospholipid
synthesis or enhancing the acidity of the TME (191,192).
The reversible nature of epigenetic modifications
in cancer makes them a valuable target for therapeutic strategies
(193). Since ferroptosis plays
a key role in limiting cancer progression, leveraging epigenetic
mechanisms to regulate ferroptosis has gained interest as a
potential treatment strategy, particularly for GICs. Epigenetic
drugs have shown promising therapeutic potential in targeting
ferroptosis-related cancers, including GICs. A series of
preclinical and clinical studies have explored the efficacy of
various epigenetic modulators, such as HDAC inhibitors (HDACi),
enhancer of zeste homolog 2 (EZH2) inhibitors and IDH inhibitors,
in regulating ferroptosis and improving cancer therapy outcomes
(Table IV).
Preclinical research has demonstrated that the
HDACi BEBT-908 can induce ferroptosis in colon adenocarcinoma by
hyperacetylating p53, a critical tumor suppressor protein involved
in ferroptosis regulation (194). CRC is notoriously resistant to
ferroptosis, but preclinical studies have shown that vorinostat, an
HDACi, significantly sensitizes CRC cells to ferroptosis. When
combined with ferroptosis inducers, vorinostat synergistically
suppresses CRC growth (195).
Additionally, entinostat, another HDACi, has been shown to promote
ferroptosis in GC cells (196).
These studies suggest that HDACis may increase ferroptosis
susceptibility in GICs, representing a potential strategy for
overcoming resistance to therapy.
The enhanced effectiveness of HDACis when combined
with other anticancer treatments has led to the development of
combination therapies in clinical trials. A phase I/II clinical
trial (NCT00943449) assessed the efficacy of combining the HDACi
resminostat with sorafenib in patients with HCC who exhibited
radiologically confirmed progression after first-line sorafenib
therapy. The combination treatment achieved a 62.5%
progression-free survival rate following six cycles of treatment
(12 weeks), whereas resminostat monotherapy resulted in a 12.5%
progression-free survival rate (197). Additionally, a notable phase II
clinical trial (NCT03250273) demonstrated that entinostat combined
with nivolumab (a programmed cell death-1 inhibitor) yielded
sustained responses in a limited group of patients with PDAC
(198). Furthermore, the
combination of entinostat with immunotherapeutic agents is being
tested in ongoing clinical trials for melanoma, non-small cell lung
cancer and CRC (ENCORE-601; NCT02437136). These studies highlight
the growing clinical interest in HDACis as key modulators of
ferroptosis and immune response, highlighting their potential to
enhance treatment efficacy across multiple cancer types.
Currently, most approved HDACis are pan-HDACis,
which can lead to significant side effects such as hematologic
toxicity (199),
gastrointestinal toxicity, cardiovascular toxicity (200), metabolic disturbances (201), fatigue and weakness (202), and immune-related side effects
(203). Therefore, there has
been a growing focus on developing selective HDACis to reduce the
toxic side effects of pan-HDACis. Another innovative strategy
involves proteolysis-targeting chimeras (PROTACs), small molecules
that promote the selective degradation of target proteins within
cells (204). HDAC-targeting
PROTACs are an emerging therapeutic strategy, offering several
advantages over traditional HDACi, including improved safety and
the potential to overcome drug resistance (205). Furthermore, it is noteworthy
that an increasing body of research demonstrates the effcacy of
combining low doses of HDACi with other medications to address the
issue of adverse effects.
EZH2 is a histone methyltransferase that represses
gene expression through H3K27 trimethylation (H3K27me3).
Preclinical studies suggested that EZH2 contributes to the
inhibition of ferroptosis in HCC by downregulating TFR2 expression
via H3K27me3 modifications. Of note, combining tazemetostat, an
EZH2 inhibitor, with sorafenib in sorafenib-resistant HCC cells
promotes ferroptosis and significantly enhances the therapeutic
effects of sorafenib (206).
This combination therapy approach demonstrates the potential of
EZH2 inhibition in overcoming resistance to standard treatments and
improving ferroptosis induction in GICs.
Alterations in IDH, particularly IDH1 and IDH2, are
common in several types of cancer, including CCA (207-209). These mutations result in the
accumulation of 2-hydroxyglutarate (210), a byproduct that suppresses
α-ketoglutarate-dependent dioxygenases, such as histone and DNA
demethylases (211-213). This disrupts cellular
differentiation and contributes to cancer development. Preclinical
studies suggest that mutant IDH1 can promote cancer cell survival
by supporting mitochondrial function and antioxidant defenses, thus
reducing the sensitivity to ferroptosis. Pharmacologic inhibition
of mutant IDH1 with ivosidenib has been shown to synergize with
conventional chemotherapeutics, enhancing ferroptosis sensitivity
in PDAC cells (214).
A phase III randomized clinical trial (NCT02989857)
demonstrated that Ivosidenib has significant overall survival
benefits in patients with advanced CCA harboring IDH1 mutations
(215). A current clinical
trial (NCT05209074) is testing the combination of ivosidenib with
chemotherapy in PDAC, demonstrating the promising potential of IDH
inhibitors in improving ferroptosis sensitivity and cancer
treatment outcomes.
In addition to the use of specific inhibitors,
epigenetic modifications such as histone modifications have been
identified as crucial regulators of ferroptosis in GICs. For
instance, anisomycin, an activator of the p38-MAPK pathway,
enhances histone H3 phosphorylation at serine 10 on the NCOA4
promoter. NCOA4 is crucial for ferroptosis as it facilitates
ferritinophagy, a mechanism that liberates iron from ferritin,
triggering lipid peroxidation. In HCC cells, anisomycin-induced
NCOA4 expression sensitizes the cells to ferroptosis, suggesting
that targeting epigenetic modifications like histone
phosphorylation could serve as a potential approach to overcoming
ferroptosis resistance in certain cancer types (216).
ncRNAs, particularly those contained in exosomes,
are essential regulators of ferroptosis and significantly impact
the survival and progression of GIC cells. These exosomal ncRNAs
can deliver miRNAs, lncRNAs and circRNAs directly to cancer cells,
offering significant therapeutic potential for GIC. Preclinical
studies have shown that exosomal lncRNAs, such as DACT3-AS1 derived
from CAFs, can be transferred to GC cells. There, DACT3-AS1 induces
ferroptosis through its regulation of the miR-181a-5p/SIRT1 pathway
(159). Additionally, heat
shock protein family B (small) member 1 (HSPB1) inhibits
ferroptosis by reducing iron uptake through TFRC (217). An interesting study utilized
small extracellular vesicles (sEV) to deliver miR-654-5p, resulting
in engineered vesicles (m654-sEV). The findings revealed that
m654-sEV efficiently transfers miR-654-5p to HCC cells, targeting
HSPB1 and amplifying sorafenib-induced ferroptosis (218). This combined approach using
m654-sEV and sorafenib holds the potential for overcoming sorafenib
resistance in HCC, thereby improving therapeutic efficacy. These
findings suggest that ncRNAs, which promote ferroptosis in GICs,
could be packaged into exosomes or sEVs to target and treat GICs
more effectively. Investigating the role of ncRNAs in modulating
ferroptosis in GICs may offer valuable therapeutic insights and
contribute to the development of more effective clinical strategies
for treating these cancers.
Numerous preclinical investigations and clinical
trials have explored the possibility of combining epigenetic drugs
with ferroptosis inducers in GICs. Sorafenib, a multi-target kinase
inhibitor that acts on Raf kinases, has been approved by the US
Food and Drug Administration as a first-line therapy for advanced
HCC (219). Research has shown
that sorafenib can induce ferroptosis in HCC cells by suppressing
the activity of system Xc−; therefore, sorafenib acts as
a ferroptosis inducer like erastin (220). However, the clinical
effectiveness of sorafenib in HCC is often compromised by both
primary and acquired resistance. A phase I/II clinical trial
involving patients with HCC who had experienced disease progression
with first-line sorafenib treatment investigated the combination of
sorafenib with the HDACi resminostat. The study reported a 62.5%
progression-free survival rate after six treatment cycles (12
weeks, primary endpoint) with the combination therapy, whereas
resminostat alone resulted in a 12.5% progression-free survival
rate (197). These results
indicate that epigenetic drugs combined with sorafenib can overcome
the resistance of HCC cells to sorafenib. Preclinical studies have
shown that lncRNAs play a crucial role in sorafenib resistance in
HCC, and lncRNA metastasis-associated lung adenocarcinoma
transcript 1 (MALAT1) is dysregulated in sorafenib-resistant HCC
cells. MALAT1 contributes to sorafenib resistance in HCC cells by
stabilizing SLC7A11 mRNA through direct interaction with ELAVL1,
promoting its translocation to the cytoplasm and subsequently
inhibiting sorafenib-induced ferroptosis (221). The combination of a MALAT1
inhibitor with sorafenib markedly improved the therapeutic
effectiveness of sorafenib in both in vitro and in
vivo models of HCC (221).
Ferroptosis has emerged as a critical mechanism in
the progression and treatment of GICs. The epigenetic regulation of
ferroptosis, involving DNA methylation, histone modifications and
ncRNAs, represents a complex but promising avenue for therapeutic
intervention. These modifications collectively influence key genes
involved in ferroptosis, including GPX4 and AURKA, which
underscores the potential of targeting epigenetic regulators to
modulate ferroptosis in GIC therapy. Current findings underscore
the importance of epigenetic drugs in modulating ferroptosis and
their potential to enhance therapeutic efficacy in GICs. However,
there remain significant challenges in translating these insights
into widely effective clinical therapies.
There is a need for more extensive research to map
the full range of epigenetic regulators involved in ferroptosis
across different types of GICs. Advanced technologies such as
single-cell RNA sequencing (seq) and multi-omics approaches,
including single-cell assay for transposase-accessible chromatin
seq (222) and chromatin
immunoprecipitation-seq (223),
may provide a high-resolution view of the epigenetic landscape
governing ferroptosis. These techniques can help elucidate
context-specific regulatory mechanisms and identify novel
therapeutic targets.
As the knowledge of the epigenetic mechanisms
governing ferroptosis expands, there is potential for the
development of more selective and effective epigenetic modulators.
The application of CRISPR/Cas9-based epigenome editing could enable
precise modifications of ferroptosis-related genes, allowing
researchers to dissect their functional roles and develop targeted
epigenetic therapies (224).
Furthermore, artificial intelligence (AI)-driven drug discovery and
high-throughput screening platforms could accelerate the
identification of small-molecule inhibitors or activators that
modulate ferroptosis through epigenetic mechanisms (225).
Identifying epigenetic markers that predict a
tumor's sensitivity to ferroptosis-inducing therapies could be
crucial for personalizing treatment. Techniques such as DNA
methylation arrays and bisulfite sequencing may be employed to
assess the methylation status of ferroptosis-related genes
(226,227). In addition, liquid biopsy
approaches using circulating tumor DNA and epigenetic profiling of
cell-free DNA may provide minimally invasive strategies for
monitoring ferroptosis sensitivity and treatment response in
real-time (228).
Resistance to ferroptosis-based therapies remains a
significant barrier. Investigating how epigenetic alterations
contribute to ferroptosis resistance could reveal new targets for
overcoming this challenge, particularly in refractory GICs. The
integration of CRISPR screens and epigenetic drug libraries could
help identify key regulatory elements driving resistance (229). Furthermore, single-cell
transcriptomics and spatial transcriptomics could provide insights
into intra-tumoral heterogeneity and epigenetic adaptations that
enable cancer cells to evade ferroptosis (230).
In conclusion, epigenetic regulation plays a
crucial role in modulating ferroptosis sensitivity in GICs,
offering a promising avenue for therapy. While epigenetic drugs
enhance ferroptosis and inhibit tumor growth, challenges remain in
clinical translation. Advancing technologies like single-cell
sequencing, CRISPR/Cas9 and AI-driven drug discovery may help
overcome resistance and enable personalized ferroptosis-targeted
therapies.
Not applicable.
LG, LW, SL, HY and PJ conceived the study. LG and
LW interpreted the relevant literature and wrote the original
manuscript. SZ, SX and XC prepared figures and tables. YZ and FL
revised the manuscript. SL, HY and PJ reviewed and edited the
manuscript. All authors have read and approved the final
manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This research was funded by the National Natural Science
Foundation of China (grant nos. 81602846 and 82272253), the Natural
Science Foundation of Shandong Province (grant no. ZR2021MH145),
the Taishan Scholar Project of Shandong Province (grant no.
tsqn201812159), the Science and Technology Program of Traditional
Chinese Medicine of Shandong Province (grant no. M-2022066), the
China International Medical Foundation (grant no. Z-2018-35-2002)
and the Key R&D Program of Jining (grant no. 2023YXNS003).
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Donohoe CL and Reynolds JV: Neoadjuvant
treatment of locally advanced esophageal and junctional cancer: The
evidence-base, current key questions and clinical trials. J Thorac
Dis. 9(Suppl 8): S697–S704. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schmidt B, Lee HJ, Ryeom S and Yoon SS:
Combining bevacizumab with radiation or chemoradiation for solid
tumors: A review of the scientific rationale, and clinical trials.
Curr Angiogenes. 1:169–179. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Madden EC, Gorman AM, Logue SE and Samali
A: Tumour cell secretome in chemoresistance and tumour recurrence.
Trends Cancer. 6:489–505. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smyth EC, Nilsson M, Grabsch HI, van
Grieken NC and Lordick F: Gastric cancer. Lancet. 396:635–648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dixon SJ, Lemberg KM, Lamprecht MR, Skouta
R, Zaitsev EM, Gleason CE, Patel DN, Bauer AJ, Cantley AM, Yang WS,
et al: Ferroptosis: An iron-dependent form of nonapoptotic cell
death. Cell. 149:1060–1072. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jia M, Qin D, Zhao C, Chai L, Yu Z, Wang
W, Tong L, Lv L, Wang Y, Rehwinkel J, et al: Redox homeostasis
maintained by GPX4 facilitates STING activation. Nat Immunol.
21:727–735. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou B, Liu J, Kang R, Klionsky DJ,
Kroemer G and Tang D: Ferroptosis is a type of autophagy-dependent
cell death. Semin Cancer Biol. 66:89–100. 2020. View Article : Google Scholar
|
|
9
|
Yang WS, SriRamaratnam R, Welsch ME,
Shimada K, Skouta R, Viswanathan VS, Cheah JH, Clemons PA, Shamji
AF, Clish CB, et al: Regulation of ferroptotic cancer cell death by
GPX4. Cell. 156:317–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu Y, Xie Y, Cao L, Yang L, Yang M, Lotze
MT, Zeh HJ, Kang R and Tang D: The ferroptosis inducer erastin
enhances sensitivity of acute myeloid leukemia cells to
chemotherapeutic agents. Mol Cell Oncol. 2:e10545492015. View Article : Google Scholar
|
|
11
|
Louandre C, Ezzoukhry Z, Godin C, Barbare
JC, Mazière JC, Chauffert B and Galmiche A: Iron-dependent cell
death of hepatocellular carcinoma cells exposed to sorafenib. Int J
Cancer. 133:1732–1742. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Louandre C, Marcq I, Bouhlal H, Lachaier
E, Godin C, Saidak Z, François C, Chatelain D, Debuysscher V,
Barbare JC, et al: The retinoblastoma (Rb) protein regulates
ferroptosis induced by sorafenib in human hepatocellular carcinoma
cells. Cancer Lett. 356:971–977. 2015. View Article : Google Scholar
|
|
13
|
Sun X, Niu X, Chen R, He W, Chen D, Kang R
and Tang D: Metallothionein-1G facilitates sorafenib resistance
through inhibition of ferroptosis. Hepatology. 64:488–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun X, Ou Z, Chen R, Niu X, Chen D, Kang R
and Tang D: Activation of the p62-Keap1-NRF2 pathway protects
against ferroptosis in hepatocellular carcinoma cells. Hepatology.
63:173–184. 2016. View Article : Google Scholar
|
|
15
|
Eling N, Reuter L, Hazin J, Hamacher-Brady
A and Brady NR: Identification of artesunate as a specific
activator of ferroptosis in pancreatic cancer cells. Oncoscience.
2:517–532. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhu S, Zhang Q, Sun X, Zeh HJ III, Lotze
MT, Kang R and Tang D: HSPA5 regulates ferroptotic cell death in
cancer cells. Cancer Res. 77:2064–2077. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Berger SL, Kouzarides T, Shiekhattar R and
Shilatifard A: An operational definition of epigenetics. Genes Dev.
23:781–783. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Holliday R: The inheritance of epigenetic
defects. Science. 238:163–170. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou S, Liu J, Wan A, Zhang Y and Qi X:
Epigenetic regulation of diverse cell death modalities in cancer: a
focus on pyroptosis, ferroptosis, cuproptosis, and disulfidptosis.
J Hematol Oncol. 17:222024. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Stockwell BR, Friedmann Angeli JP, Bayir
H, Bush AI, Conrad M, Dixon SJ, Fulda S, Gascón S, Hatzios SK,
Kagan VE, et al: Ferroptosis: A regulated cell death nexus linking
metabolism, redox biology, and disease. Cell. 171:273–285. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kuang F, Liu J, Tang D and Kang R:
Oxidative damage and antioxidant defense in ferroptosis. Front Cell
Dev Biol. 8:5865782020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Torti SV and Torti FM: Iron and cancer:
More ore to be mined. Nat Rev Cancer. 13:342–355. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hassannia B, Vandenabeele P and Vanden
Berghe T: Targeting ferroptosis to iron out cancer. Cancer Cell.
35:830–849. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang WS, Kim KJ, Gaschler MM, Patel M,
Shchepinov MS and Stockwell BR: Peroxidation of polyunsaturated
fatty acids by lipoxygenases drives ferroptosis. Proc Natl Acad Sci
USA. 113:E4966–E4975. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Suttner DM and Dennery PA: Reversal of
HO-1 related cytoprotection with increased expression is due to
reactive iron. FASEB J. 13:1800–1809. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang WS and Stockwell BR: Synthetic lethal
screening identifies compounds activating Iron-dependent,
nonapoptotic cell death in oncogenic-RAS-harboring cancer cells.
Chem Biol. 15:234–245. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Liu Y, Liu J, Kang R and Tang D:
NEDD4L-mediated LTF protein degradation limits ferroptosis. Biochem
Biophys Res Commun. 531:581–587. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Drakesmith H, Nemeth E and Ganz T: Ironing
out Ferroportin. Cell Metab. 22:777–787. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Donovan A, Lima CA, Pinkus JL, Pinkus GS,
Zon LI, Robine S and Andrews NC: The iron exporter
ferroportin/Slc40a1 is essential for iron homeostasis. Cell Metab.
1:191–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Donovan A, Brownlie A, Zhou Y, Shepard J,
Pratt SJ, Moynihan J, Paw BH, Drejer A, Barut B, Zapata A, et al:
Positional cloning of zebrafish ferroportin1 identifies a conserved
vertebrate iron exporter. Nature. 403:776–781. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Nemeth E, Tuttle MS, Powelson J, Vaughn
MB, Donovan A, Ward DM, Ganz T and Kaplan J: Hepcidin regulates
cellular iron efflux by binding to ferroportin and inducing its
internalization. Science. 306:2090–2093. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
De Domenico I, Ward DM, Langelier C,
Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G and Kaplan J:
The molecular mechanism of hepcidin-mediated ferroportin
down-regulation. Mol Biol Cell. 18:2569–2578. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Qiao B, Sugianto P, Fung E,
Del-Castillo-Rueda A, Moran-Jimenez MJ, Ganz T and Nemeth E:
Hepcidin-induced endocytosis of ferroportin is dependent on
ferroportin ubiquitination. Cell Metab. 15:918–924. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ross SL, Tran L, Winters A, Lee KJ, Plewa
C, Foltz I, King C, Miranda LP, Allen J, Beckman H, et al:
Molecular mechanism of hepcidin-mediated ferroportin
internalization requires ferroportin lysines, not tyrosines or
JAK-STAT. Cell Metab. 15:905–917. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Geng N, Shi BJ, Li SL, Zhong ZY, Li YC,
Xua WL, Zhou H and Cai JH: Knockdown of ferroportin accelerates
erastin-induced ferroptosis in neuroblastoma cells. Eur Rev Med
Pharmacol Sci. 22:3826–3836. 2018.PubMed/NCBI
|
|
36
|
Tang Z, Jiang W, Mao M, Zhao J, Chen J and
Cheng N: Deubiquitinase USP35 modulates ferroptosis in lung cancer
via targeting ferroportin. Clin Transl Med. 11:e3902021. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gao M, Monian P, Pan Q, Zhang W, Xiang J
and Jiang X: Ferroptosis is an autophagic cell death process. Cell
Res. 26:1021–1032. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hou W, Xie Y, Song X, Sun X, Lotze MT, Zeh
HJ III, Kang R and Tang D: Autophagy promotes ferroptosis by
degradation of ferritin. Autophagy. 12:1425–1428. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mancias JD, Wang X, Gygi SP, Harper JW and
Kimmelman AC: Quantitative proteomics identifies NCOA4 as the cargo
receptor mediating ferritinophagy. Nature. 509:105–109. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang YQ, Chang SY, Wu Q, Gou YJ, Jia L,
Cui YM, Yu P, Shi ZH, Wu WS, Gao G and Chang YZ: The Protective
role of mitochondrial ferritin on Erastin-induced ferroptosis.
Front Aging Neurosci. 8:3082016. View Article : Google Scholar
|
|
41
|
Torii S, Shintoku R, Kubota C, Yaegashi M,
Torii R, Sasaki M, Suzuki T, Mori M, Yoshimoto Y, Takeuchi T and
Yamada K: An essential role for functional lysosomes in ferroptosis
of cancer cells. Biochem J. 473:769–777. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Fang Y, Chen X, Tan Q, Zhou H, Xu J and Gu
Q: Inhibiting ferroptosis through disrupting the NCOA4-FTH1
interaction: A new mechanism of action. ACS Cent Sci. 7:980–989.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen PH, Wu J, Ding CC, Lin CC, Pan S,
Bossa N, Xu Y, Yang WH, Mathey-Prevot B and Chi JT: Kinome screen
of ferroptosis reveals a novel role of ATM in regulating iron
metabolism. Cell Death Differ. 27:1008–1022. 2020. View Article : Google Scholar :
|
|
44
|
Kagan VE, Mao G, Qu F, Angeli JP, Doll S,
Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, et al: Oxidized
arachidonic and adrenic PEs navigate cells to ferroptosis. Nat Chem
Biol. 13:81–90. 2017. View Article : Google Scholar
|
|
45
|
Doll S, Proneth B, Tyurina YY, Panzilius
E, Kobayashi S, Ingold I, Irmler M, Beckers J, Aichler M, Walch A,
et al: ACSL4 dictates ferroptosis sensitivity by shaping cellular
lipid composition. Nat Chem Biol. 13:91–98. 2017. View Article : Google Scholar :
|
|
46
|
Yuan H, Li X, Zhang X, Kang R and Tang D:
Identification of ACSL4 as a biomarker and contributor of
ferroptosis. Biochem Biophys Res Commun. 478:1338–1343. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dixon SJ, Winter GE, Musavi LS, Lee ED,
Snijder B, Rebsamen M, Superti-Furga G and Stockwell BR: Human
haploid cell genetics reveals roles for lipid metabolism genes in
nonapoptotic cell death. ACS Chem Biol. 10:1604–1609. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Conrad M and Pratt DA: The chemical basis
of ferroptosis. Nat Chem Biol. 15:1137–1147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yin H, Xu L and Porter NA: Free radical
lipid peroxidation: Mechanisms and analysis. Chem Rev.
111:5944–5972. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Doll S, Freitas FP, Shah R, Aldrovandi M,
da Silva MC, Ingold I, Goya Grocin A, Xavier da Silva TN, Panzilius
E, Scheel CH, et al: FSP1 is a glutathione-independent ferroptosis
suppressor. Nature. 575:693–698. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bersuker K, Hendricks JM, Li Z, Magtanong
L, Ford B, Tang PH, Roberts MA, Tong B, Maimone TJ, Zoncu R, et al:
The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit
ferroptosis. Nature. 575:688–692. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ayala A, Muñoz MF and Argüelles S: Lipid
peroxidation: Production, metabolism, and signaling mechanisms of
malondialdehyde and 4-hydroxy-2-nonenal. Oxid Med Cell Longev.
2014:3604382014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Fenton HJH: LXXIII.-Oxidation of tartaric
acid in presence of iron. J Chemical Soc Transactions. 65:899–910.
1894. View Article : Google Scholar
|
|
54
|
Haeggström JZ and Funk CD: Lipoxygenase
and leukotriene pathways: Biochemistry, biology, and roles in
disease. Chem Rev. 111:5866–5898. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Li Y, Maher P and Schubert D: A role for
12-lipoxygenase in nerve cell death caused by glutathione
depletion. Neuron. 19:453–463. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Seiler A, Schneider M, Förster H, Roth S,
Wirth EK, Culmsee C, Plesnila N, Kremmer E, Rådmark O, Wurst W, et
al: Glutathione peroxidase 4 senses and translates oxidative stress
into 12/15-lipoxygenase dependent- and AIF-mediated cell death.
Cell Metab. 8:237–248. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Brütsch SH, Wang CC, Li L, Stender H,
Neziroglu N, Richter C, Kuhn H and Borchert A: Expression of
inactive glutathione peroxidase 4 leads to embryonic lethality, and
inactivation of the Alox15 gene does not rescue such knock-in mice.
Antioxid Redox Signal. 22:281–293. 2015. View Article : Google Scholar
|
|
58
|
Jiang X, Stockwell BR and Conrad M:
Ferroptosis: Mechanisms, biology and role in disease. Nat Rev Mol
Cell Biol. 22:266–282. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chu B, Kon N, Chen D, Li T, Liu T, Jiang
L, Song S, Tavana O and Gu W: ALOX12 is required for p53-mediated
tumour suppression through a distinct ferroptosis pathway. Nat Cell
Biol. 21:579–591. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Zou Y, Li H, Graham ET, Deik AA, Eaton JK,
Wang W, Sandoval-Gomez G, Clish CB, Doench JG and Schreiber SL:
Cytochrome P450 oxidoreductase contributes to phospholipid
peroxidation in ferroptosis. Nat Chem Biol. 16:302–309. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xie Y, Kang R, Klionsky DJ and Tang D:
GPX4 in cell death, autophagy, and disease. Autophagy.
19:2621–2638. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Yant LJ, Ran Q, Rao L, Van Remmen H,
Shibatani T, Belter JG, Motta L, Richardson A and Prolla TA: The
selenoprotein GPX4 is essential for mouse development and protects
from radiation and oxidative damage insults. Free Radic Biol Med.
34:496–502. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Brigelius-Flohé R and Maiorino M:
Glutathione peroxidases. Biochim Biophys Acta. 1830:3289–3303.
2013. View Article : Google Scholar
|
|
64
|
Meister A: Glutathione metabolism. Methods
Enzymol. 251:3–7. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Lu SC: Regulation of glutathione
synthesis. Mol Aspects Med. 30:42–59. 2009. View Article : Google Scholar :
|
|
66
|
Cao JY and Dixon SJ: Mechanisms of
ferroptosis. Cell Mol Life Sci. 73:2195–2209. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Sato H, Tamba M, Ishii T and Bannai S:
Cloning and expression of a plasma membrane cystine/glutamate
exchange transporter composed of two distinct proteins. J Biol
Chem. 274:11455–11458. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sato H, Shiiya A, Kimata M, Maebara K,
Tamba M, Sakakura Y, Makino N, Sugiyama F, Yagami K, Moriguchi T,
et al: Redox imbalance in cystine/glutamate transporter-deficient
mice. J Biol Chem. 280:37423–37429. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Friedmann Angeli JP, Schneider M, Proneth
B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch
A, Eggenhofer E, et al: Inactivation of the ferroptosis regulator
Gpx4 triggers acute renal failure in mice. Nat Cell Biol.
16:1180–1191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Chen D, Tavana O, Chu B, Erber L, Chen Y,
Baer R and Gu W: NRF2 is a major target of ARF in p53-independent
tumor suppression. Mol Cell. 68:224–232.e4. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Jiang L, Kon N, Li T, Wang SJ, Su T,
Hibshoosh H, Baer R and Gu W: Ferroptosis as a p53-mediated
activity during tumour suppression. Nature. 520:57–62. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zhang Y, Shi J, Liu X, Feng L, Gong Z,
Koppula P, Sirohi K, Li X, Wei Y, Lee H, et al: BAP1 links
metabolic regulation of ferroptosis to tumour suppression. Nat Cell
Biol. 20:1181–1192. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Song X, Zhu S, Chen P, Hou W, Wen Q, Liu
J, Xie Y, Liu J, Klionsky DJ, Kroemer G, et al: AMPK-mediated BECN1
phosphorylation promotes ferroptosis by directly blocking system
Xc-activity. Curr Biol. 28:2388–2399.e5. 2018. View Article : Google Scholar
|
|
74
|
Yang M, Tsui MG, Tsang JKW, Goit RK, Yao
KM, So KF, Lam WC and Lo ACY: Involvement of FSP1-CoQ(10)-NADH and
GSH-GPx-4 pathways in retinal pigment epithelium ferroptosis. Cell
Death Dis. 13:4682022. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zeng F, Chen X and Deng G: The
anti-ferroptotic role of FSP1: Current molecular mechanism and
therapeutic approach. Mol Biomed. 3:372022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kraft VAN, Bezjian CT, Pfeiffer S,
Ringelstetter L, Müller C, Zandkarimi F, Merl-Pham J, Bao X,
Anastasov N, Kössl J, et al: GTP Cyclohydrolase
1/Tetrahydrobiopterin counteract ferroptosis through lipid
remodeling. ACS Cent Sci. 6:41–53. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S,
Han J and Wei X: Targeting epigenetic regulators for cancer
therapy: Mechanisms and advances in clinical trials. Signal
Transduct Target Ther. 4:622019. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Logie E, Van Puyvelde B, Cuypers B,
Schepers A, Berghmans H, Verdonck J, Laukens K, Godderis L,
Dhaenens M, Deforce D, et al: Ferroptosis induction in multiple
myeloma cells triggers DNA methylation and histone modification
changes associated with cellular senescence. Int J Mol Sci.
22:122342021. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Bird A: DNA methylation patterns and
epigenetic memory. Genes Dev. 16:6–21. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Parry A, Rulands S and Reik W: Active
turnover of DNA methylation during cell fate decisions. Nat Rev
Genet. 22:59–66. 2021. View Article : Google Scholar
|
|
81
|
Tahiliani M, Koh KP, Shen Y, Pastor WA,
Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et
al: Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in
mammalian DNA by MLL partner TET1. Science. 324:930–935. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Ito S, D'Alessio AC, Taranova OV, Hong K,
Sowers LC and Zhang Y: Role of Tet proteins in 5mC to 5hmC
conversion, ES-cell self-renewal and inner cell mass specification.
Nature. 466:1129–1133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Ito S, Shen L, Dai Q, Wu SC, Collins LB,
Swenberg JA, He C and Zhang Y: Tet proteins can convert
5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine.
Science. 333:1300–1303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Robertson KD: DNA methylation and human
disease. Nat Rev Genet. 6:597–610. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Esteller M: Epigenetic gene silencing in
cancer: The DNA hypermethylome. Hum Mol Genet. 16(Spec No 1):
R50–R59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Veigl ML, Kasturi L, Olechnowicz J, Ma AH,
Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL,
Sedwick WD, et al: Biallelic inactivation of hMLH1 by epigenetic
gene silencing, a novel mechanism causing human MSI cancers. Proc
Natl Acad Sci USA. 95:8698–8702. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Klutstein M, Nejman D, Greenfield R and
Cedar H: DNA methylation in cancer and aging. Cancer Res.
76:3446–3450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu Z, Zhao Q, Zuo ZX, Yuan SQ, Yu K,
Zhang Q, Zhang X, Sheng H, Ju HQ, Cheng H, et al: Systematic
analysis of the aberrances and functional implications of
ferroptosis in cancer. iScience. 23:1013022020. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Lee JY, Nam M, Son HY, Hyun K, Jang SY,
Kim JW, Kim MW, Jung Y, Jang E, Yoon SJ, et al: Polyunsaturated
fatty acid biosynthesis pathway determines ferroptosis sensitivity
in gastric cancer. Proc Natl Acad Sci USA. 117:32433–32442. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Liu Y, Ouyang L, Mao C, Chen Y, Li T, Liu
N, Wang Z, Lai W, Zhou Y, Cao Y, et al: PCDHB14 promotes
ferroptosis and is a novel tumor suppressor in hepatocellular
carcinoma. Oncogene. 41:3570–3583. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Bolotta A, Abruzzo PM, Baldassarro VA,
Ghezzo A, Scotlandi K, Marini M and Zucchini C: New insights into
the Hepcidin-ferroportin axis and iron homeostasis in iPSC-derived
cardiomyocytes from Friedreich's ataxia patient. Oxid Med Cell
Longev. 2019:76230232019. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Udali S, Castagna A, Corbella M,
Ruzzenente A, Moruzzi S, Mazzi F, Campagnaro T, De Santis D,
Franceschi A, Pattini P, et al: Hepcidin and DNA promoter
methylation in hepatocellular carcinoma. Eur J Clin Invest.
48:e128702018. View Article : Google Scholar
|
|
93
|
Udali S, Guarini P, Ruzzenente A,
Ferrarini A, Guglielmi A, Lotto V, Tononi P, Pattini P, Moruzzi S,
Campagnaro T, et al: DNA methylation and gene expression profiles
show novel regulatory pathways in hepatocellular carcinoma. Clin
Epigenetics. 7:432015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Cai C, Zhu Y, Mu J, Liu S, Yang Z, Wu Z,
Zhao C, Song X, Ye Y, Gu J, et al: DNA methylation of RUNX3
promotes the progression of gallbladder cancer through repressing
SLC7A11-mediated ferroptosis. Cell Signal. 108:1107102023.
View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Cedar H and Bergman Y: Linking DNA
methylation and histone modification: Patterns and paradigms. Nat
Rev Genet. 10:295–304. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Du Q, Luu PL, Stirzaker C and Clark SJ:
Methyl-CpG-binding domain proteins: Readers of the epigenome.
Epigenomics. 7:1051–1073. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Shimazu T, Hirschey MD, Newman J, He W,
Shirakawa K, Le Moan N, Grueter CA, Lim H, Saunders LR, Stevens RD,
et al: Suppression of oxidative stress by β-hydroxybutyrate, an
endogenous histone deacetylase inhibitor. Science. 339:211–214.
2013. View Article : Google Scholar
|
|
98
|
Cui X, Yun X, Sun M, Li R, Lyu X, Lao Y,
Qin X and Yu W: HMGCL-induced β-hydroxybutyrate production
attenuates hepatocellular carcinoma via DPP4-mediated ferroptosis
susceptibility. Hepatol Int. 17:377–392. 2023. View Article : Google Scholar
|
|
99
|
Li Y, Hu G, Huang F, Chen M, Chen Y, Xu Y
and Tong G: MAT1A suppression by the CTBP1/HDAC1/HDAC2
transcriptional complex induces immune escape and reduces
ferroptosis in hepatocellular carcinoma. Lab Invest.
103:1001802023. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xue X, Ma L, Zhang X, Xu X, Guo S, Wang Y,
Qiu S, Cui J, Guo W, Yu Y, et al: Tumour cells are sensitised to
ferroptosis via RB1CC1-mediated transcriptional reprogramming. Clin
Transl Med. 12:e7472022. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Ma M, Kong P, Huang Y, Wang J, Liu X, Hu
Y, Chen X, Du C and Yang H: Activation of MAT2A-ACSL3 pathway
protects cells from ferroptosis in gastric cancer. Free Radic Biol
Med. 181:288–299. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Jiang K, Yin X, Zhang Q, Yin J, Tang Q, Xu
M, Wu L, Shen Y, Zhou Z, Yu H, et al: STC2 activates PRMT5 to
induce radioresistance through DNA damage repair and ferroptosis
pathways in esophageal squamous cell carcinoma. Redox Biol.
60:1026262023. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Wang Y, Yang L, Zhang X, Cui W, Liu Y, Sun
QR, He Q, Zhao S, Zhang GA, Wang Y, et al: Epigenetic regulation of
ferroptosis by H2B monoubiquitination and p53. EMBO Rep.
20:e475632019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016. View Article : Google Scholar
|
|
105
|
Guttman M and Rinn JL: Modular regulatory
principles of large Non-coding RNAs. Nature. 482:339–346. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Ni H, Qin H, Sun C, Liu Y, Ruan G, Guo Q,
Xi T, Xing Y and Zheng L: MiR-375 reduces the stemness of gastric
cancer cells through triggering ferroptosis. Stem Cell Res Ther.
12:3252021. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Mao SH, Zhu CH, Nie Y, Yu J and Wang L:
Levobupivacaine Induces Ferroptosis by miR-489-3p/SLC7A11 Signaling
in Gastric Cancer. Front Pharmacol. 12:6813382021. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Dong X, Chen X, Zhao Y, Wu Q and Ren Y:
CircTMEM87A promotes the tumorigenesis of gastric cancer by
regulating the miR-1276/SLC7A11 axis. J Gastroenterol Hepatol.
39:121–132. 2024. View Article : Google Scholar
|
|
109
|
Martino E, Balestrieri A, Aragona F,
Bifulco G, Mele L, Campanile G, Balestrieri ML and D'Onofrio N:
MiR-148a-3p promotes colorectal cancer cell ferroptosis by
targeting SLC7A11. Cancers (Basel). 15:43422023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Elrebehy MA, Abdelghany TM, Elshafey MM,
Gomaa MH and Doghish AS: miR-509-5p promotes colorectal cancer cell
ferroptosis by targeting SLC7A11. Pathol Res Pract. 247:1545572023.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Liu L, Yao H, Zhou X, Chen J, Chen G, Shi
X, Wu G, Zhou G and He S: MiR-15a-3p regulates ferroptosis via
targeting glutathione peroxidase GPX4 in colorectal cancer. Mol
Carcinog. 61:301–310. 2022. View Article : Google Scholar
|
|
112
|
He GN, Bao NR, Wang S, Xi M, Zhang TH and
Chen FS: Ketamine induces ferroptosis of liver cancer cells by
Targeting lncRNA PVT1/miR-214-3p/GPX4. Drug Des Devel Ther.
15:3965–3978. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Yang Y, Lin Z, Han Z, Wu Z, Hua J, Zhong
R, Zhao R, Ran H, Qu K, Huang H, et al: miR-539 activates the
SAPK/JNK signaling pathway to promote ferropotosis in colorectal
cancer by directly targeting TIPE. Cell Death Discov. 7:2722021.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Dodson M, Castro-Portuguez R and Zhang DD:
NRF2 plays a critical role in mitigating lipid peroxidation and
ferroptosis. Redox Biol. 23:1011072019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Zheng F, Bi JC, Wei YY, Wang Y, Zhang Q,
Liang CL, Wu J and Dai Z: MiR-124-3p mediates gastric cancer cell
ferroptosis induced by an anti-cancer drug polyphyllin I. Front
Pharmacol. 14:12857992023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yamamoto S, Inoue J, Kawano T, Kozaki K,
Omura K and Inazawa J: The impact of miRNA-based molecular
diagnostics and treatment of NRF2-stabilized tumors. Mol Cancer
Res. 12:58–68. 2014. View Article : Google Scholar
|
|
117
|
Xiao S, Liu N, Yang X, Ji G and Li M:
Polygalacin D suppresses esophageal squamous cell carcinoma growth
and metastasis through regulating miR-142-5p/Nrf2 axis. Free Radic
Biol Med. 164:58–75. 2021. View Article : Google Scholar
|
|
118
|
Akdemir B, Nakajima Y, Inazawa J and Inoue
J: miR-432 Induces NRF2 stabilization by directly targeting KEAP1.
Mol Cancer Res. 15:1570–1578. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Liu M, Hu C, Xu Q, Chen L, Ma K, Xu N and
Zhu H: Methylseleninic acid activates Keap1/Nrf2 pathway via
up-regulating miR-200a in human oesophageal squamous cell carcinoma
cells. Biosci Rep. 35:e002562015. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Li P, Liu X, Xing W, Qiu H, Li R, Liu S
and Sun H: Exosome-derived miR-200a promotes esophageal cancer cell
proliferation and migration via the mediating Keap1 expression. Mol
Cell Biochem. 477:1295–1308. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Shi L, Wu L, Chen Z, Yang J, Chen X, Yu F,
Zheng F and Lin X: MiR-141 activates Nrf2-dependent antioxidant
pathway via Down-regulating the expression of keap1 conferring the
resistance of hepatocellular carcinoma cells to 5-Fluorouracil.
Cell Physiol Biochem. 35:2333–2348. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Xiao Z, Zheng YB, Dao WX, Luo JF, Deng WH,
Yan RC and Liu JS: MicroRNA-328-3p facilitates the progression of
gastric cancer via KEAP1/NRF2 axis. Free Radic Res. 55:720–730.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Lewerenz J and Maher P: Basal levels of
eIF2alpha phosphorylation determine cellular antioxidant status by
regulating ATF4 and xCT expression. J Biol Chem. 284:1106–1115.
2009. View Article : Google Scholar :
|
|
124
|
He F, Zhang P, Liu J, Wang R, Kaufman RJ,
Yaden BC and Karin M: ATF4 suppresses hepatocarcinogenesis by
inducing SLC7A11 (xCT) to block stress-related ferroptosis. J
Hepatol. 79:362–377. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Bai T, Liang R, Zhu R, Wang W, Zhou L and
Sun Y: MicroRNA-214-3p enhances Erastin-induced ferroptosis by
targeting ATF4 in hepatoma cells. J Cell Physiol. 235:5637–5648.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yang H, Sun W, Bi T, Sun J, Lu Z, Li J and
Wei H: ZNF8-miR-552-5p axis modulates ACSL4-Mediated ferroptosis in
hepatocellular carcinoma. DNA Cell Biol. 42:336–347. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lu Y, Chan YT, Tan HY, Zhang C, Guo W, Xu
Y, Sharma R, Chen ZS, Zheng YC, Wang N, et al: Epigenetic
regulation of ferroptosis via ETS1/miR-23a-3p/ACSL4 axis mediates
sorafenib resistance in human hepatocellular carcinoma. J Exp Clin
Cancer Res. 41:32022. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Qi R, Bai Y, Li K, Liu N, Xu Y, Dal E,
Wang Y, Lin R, Wang H, Liu Z, et al: Cancer-associated fibroblasts
suppress ferroptosis and induce gemcitabine resistance in
pancreatic cancer cells by secreting exosome-derived
ACSL4-targeting miRNAs. Drug Resist Updat. 68:1009602023.
View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang W, Wang T, Zhang Y, Deng T, Zhang H
and Ba YI: Gastric cancer secreted miR-214-3p inhibits the
anti-angiogenesis effect of apatinib by suppressing ferroptosis in
vascular endothelial cells. Oncol Res. 32:489–502. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Xing K, Bian X, Shi D, Dong S, Zhou H,
Xiao S, Bai J and Wu W: miR-612 Enhances RSL3-induced ferroptosis
of hepatocellular carcinoma cells via mevalonate pathway. J
Hepatocell Carcinoma. 10:2173–2185. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Yang WS and Stockwell BR: Ferroptosis:
Death by lipid peroxidation. Trends Cell Biol. 26:165–176. 2016.
View Article : Google Scholar :
|
|
132
|
Zhang H, Deng T, Liu R, Ning T, Yang H,
Liu D, Zhang Q, Lin D, Ge S, Bai M, et al: CAF secreted miR-522
suppresses ferroptosis and promotes acquired chemo-resistance in
gastric cancer. Mol Cancer. 19:432020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zheng S, Hu L, Song Q, Shan Y, Yin G, Zhu
H, Kong W and Zhou C: miR-545 promotes colorectal cancer by
inhibiting transferring in the non-normal ferroptosis signaling.
Aging (Albany NY). 13:26137–26147. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Hamara K, Bielecka-Kowalska A,
Przybylowska-Sygut K, Sygut A, Dziki A and Szemraj J: Alterations
in expression profile of iron-related genes in colorectal cancer.
Mol Biol Rep. 40:5573–5585. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Greene CM, Varley RB and Lawless MW:
MicroRNAs and liver cancer associated with iron overload:
Therapeutic targets unravelled. World J Gastroenterol.
19:5212–5226. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Kindrat I, Tryndyak V, de Conti A,
Shpyleva S, Mudalige TK, Kobets T, Erstenyuk AM, Beland FA and
Pogribny IP: MicroRNA-152-mediated dysregulation of hepatic
transferrin receptor 1 in liver carcinogenesis. Oncotarget.
7:1276–1287. 2016. View Article : Google Scholar :
|
|
137
|
Galy B, Conrad M and Muckenthaler M:
Mechanisms controlling cellular and systemic iron homeostasis. Nat
Rev Mol Cell Biol. 25:133–155. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Sangokoya C, Doss JF and Chi JT:
Iron-responsive miR-485-3p regulates cellular iron homeostasis by
targeting ferroportin. PLoS Genet. 9:e10034082013. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Fan H, Ai R, Mu S, Niu X, Guo Z and Liu L:
MiR-19a suppresses ferroptosis of colorectal cancer cells by
targeting IREB2. Bioengineered. 13:12021–12029. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Balihodzic A, Prinz F, Dengler MA, Calin
GA, Jost PJ and Pichler M: Non-coding RNAs and ferroptosis:
Potential implications for cancer therapy. Cell Death Differ.
29:1094–1106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Wu C, Hou X, Li S and Luo S: Long
noncoding RNA ZEB1-AS1 attenuates ferroptosis of gastric cancer
cells through modulating miR-429/BGN axis. J Biochem Mol Toxicol.
37:e233812023. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Shi Z, Li Z, Jin B, Ye W, Wang L, Zhang S,
Zheng J, Lin Z, Chen B, Liu F, et al: Loss of LncRNA DUXAP8
synergistically enhanced sorafenib induced ferroptosis in
hepatocellular carcinoma via SLC7A11 de-palmitoylation. Clin Transl
Med. 13:e13002023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Zhang B, Bao W, Zhang S, Chen B, Zhou X,
Zhao J, Shi Z, Zhang T, Chen Z, Wang L, et al: LncRNA HEPFAL
accelerates ferroptosis in hepatocellular carcinoma by regulating
SLC7A11 ubiquitination. Cell Death Dis. 13:7342022. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Wang J, Jia Q, Jiang S, Lu W and Ning H:
POU6F1 promotes ferroptosis by increasing lncRNA-CASC2
transcription to regulate SOCS2/SLC7A11 signaling in gastric
cancer. Cell Biol Toxicol. 40:32024. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Lin Z, Song J, Gao Y, Huang S, Dou R,
Zhong P, Huang G, Han L, Zheng J, Zhang X, et al: Hypoxia-induced
HIF-1α/lncRNA-PMAN inhibits ferroptosis by promoting the
cytoplasmic translocation of ELAVL1 in peritoneal dissemination
from gastric cancer. Redox Biol. 52:1023122022. View Article : Google Scholar
|
|
146
|
Yang R, Wan J, Ma L, Zhou F, Yang Z, Li Z,
Zhang M and Ming L: TMEM44-AS1 promotes esophageal squamous cell
carcinoma progression by regulating the IGF2BP2-GPX4 axis in
modulating ferroptosis. Cell Death Discov. 9:4312023. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Lei S, Cao W, Zeng Z, Zhang Z, Jin B, Tian
Q, Wu Y, Zhang T, Li D, Hu C, et al: JUND/linc00976 promotes
cholangiocarcinoma progression and metastasis, inhibits ferroptosis
by regulating the miR-3202/GPX4 axis. Cell Death Dis. 13:9672022.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Yuan J, Lv T, Yang J, Wu Z, Yan L, Yang J
and Shi Y: HDLBP-stabilized lncFAL inhibits ferroptosis
vulnerability by diminishing Trim69-dependent FSP1 degradation in
hepatocellular carcinoma. Redox Biol. 58:1025462022. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Han Y, Gao X, Wu N, Jin Y, Zhou H, Wang W,
Liu H, Chu Y, Cao J, Jiang M, et al: Long noncoding RNA LINC00239
inhibits ferroptosis in colorectal cancer by binding to Keap1 to
stabilize Nrf2. Cell Death Dis. 13:7422022. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Gao Y, Tong M, Wong TL, Ng KY, Xie YN,
Wang Z, Yu H, Loh JJ, Li M and Ma S: Long noncoding RNA
URB1-antisense RNA 1 (AS1) suppresses sorafenib-induced ferroptosis
in hepatocellular carcinoma by driving ferritin phase separation.
ACS Nano. 17:22240–22258. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Zhang Y, Luo M, Cui X, O'Connell D and
Yang Y: Long noncoding RNA NEAT1 promotes ferroptosis by modulating
the miR-362-3p/MIOX axis as a ceRNA. Cell Death Differ.
29:1850–1863. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Guan L, Wang F, Wang M, Han S, Cui Z, Xi
S, Xu H and Li S: Downregulation of HULC induces ferroptosis in
hepatocellular carcinoma via targeting of the miR-3200-5p/ATF4
axis. Oxid Med Cell Longev. 2022:96130952022. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Wang H, Klein MG, Zou H, Lane W, Snell G,
Levin I, Li K and Sang BC: Crystal structure of human
stearoyl-coenzyme A desaturase in complex with substrate. Nat
Struct Mol Biol. 22:581–585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li J, Condello S, Thomes-Pepin J, Ma X,
Xia Y, Hurley TD, Matei D and Cheng JX: Lipid desaturation is a
metabolic marker and therapeutic target of ovarian cancer stem
cells. Cell Stem Cell. 20:303–314.e5. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Sen U, Coleman C and Sen T: Stearoyl
coenzyme A desaturase-1: Multitasker in cancer, metabolism, and
ferroptosis. Trends Cancer. 9:480–489. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Tesfay L, Paul BT, Konstorum A, Deng Z,
Cox AO, Lee J, Furdui CM, Hegde P, Torti FM and Torti SV:
Stearoyl-CoA desaturase 1 protects ovarian cancer cells from
ferroptotic cell death. Cancer Res. 79:5355–5366. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Luo Y, Huang S, Wei J, Zhou H, Wang W,
Yang J, Deng Q, Wang H and Fu Z: Long noncoding RNA LINC01606
protects colon cancer cells from ferroptotic cell death and
promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop
signalling. Clin Transl Med. 12:e7522022. View Article : Google Scholar
|
|
158
|
Yang H, Hu Y, Weng M, Liu X, Wan P, Hu Y,
Ma M, Zhang Y, Xia H and Lv K: Hypoxia inducible lncRNA-CBSLR
modulates ferroptosis through m6A-YTHDF2-dependent modulation of
CBS in gastric cancer. J Adv Res. 37:91–106. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Qu X, Liu B, Wang L, Liu L, Zhao W, Liu C,
Ding J, Zhao S, Xu B, Yu H, et al: Loss of cancer-associated
fibroblast-derived exosomal DACT3-AS1 promotes malignant
transformation and Ferroptosis-mediated oxaliplatin resistance in
gastric cancer. Drug Resist Updat. 68:1009362023. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar :
|
|
161
|
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen
B, Luo Y, Lyu D, Li Y, Shi G, et al: Circular RNA profiling reveals
an abundant circHIPK3 that regulates cell growth by sponging
multiple miRNAs. Nat Commun. 7:112152016. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Wang Y, Chen H and Wei X: Circ_0007142
downregulates miR-874-3p-mediated GDPD5 on colorectal cancer cells.
Eur J Clin Invest. 51:e135412021. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Gao X and Wang XL: Dexmedetomidine
promotes ferroptotic cell death in gastric cancer via
hsa_circ_0008035/miR-302a/E2F7 axis. Kaohsiung J Med Sci.
39:390–403. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Yao W, Wang J, Meng F, Zhu Z, Jia X, Xu L,
Zhang Q and Wei L: Circular RNA CircPVT1 Inhibits 5-Fluorouracil
chemosensitivity by regulating ferroptosis through MiR-30a-5p/FZD3
axis in esophageal cancer cells. Front Oncol. 11:7809382021.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Zhai H, Zhong S, Wu R, Mo Z, Zheng S, Xue
J, Meng H, Liu M, Chen X, Zhang G, et al: Suppressing
circIDE/miR-19b-3p/RBMS1 axis exhibits promoting-tumour activity
through upregulating GPX4 to diminish ferroptosis in hepatocellular
carcinoma. Epigenetics. 18:21924382023. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Xu Q, Zhou L, Yang G, Meng F, Wan Y, Wang
L and Zhang L: CircIL4R facilitates the tumorigenesis and inhibits
ferroptosis in hepatocellular carcinoma by regulating the
miR-541-3p/GPX4 axis. Cell Biol Int. 44:2344–2356. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Liu Y and Li J: Circular RNA 0016142
knockdown induces ferroptosis in hepatocellular carcinoma cells via
modulation of the MicroRNA-188-3p/Glutathione peroxidase 4 axis.
Biochem Genet. 62:333–351. 2024. View Article : Google Scholar
|
|
168
|
Tan YR, Jiang BH, Feng WJ, He ZL, Jiang
YL, Xun Y, Wu XP, Li YH and Zhu HB: Circ0060467 sponges miR-6805 to
promote hepatocellular carcinoma progression through regulating
AIFM2 and GPX4 expression. Aging (Albany NY). 16:1796–1807. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Lyu N, Zeng Y, Kong Y, Chen Q, Deng H, Ou
S, Bai Y, Tang H, Wang X and Zhao M: Ferroptosis is involved in the
progression of hepatocellular carcinoma through the
circ0097009/miR-1261/SLC7A11 axis. Ann Transl Med. 9:6752021.
View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Li Q, Li K, Guo Q and Yang T: CircRNA
circSTIL inhibits ferroptosis in colorectal cancer via
miR-431/SLC7A11 axis. Environ Toxicol. 38:981–989. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Liu J, Yang H, Deng J, Jiang R, Meng E and
Wu H: CircRPPH1 promotes the stemness of gastric cancer cells by
targeting miR-375/SLC7A11 axis. Environ Toxicol. 38:115–125. 2023.
View Article : Google Scholar
|
|
172
|
Wang H, Breadner DA, Deng K and Niu J:
CircRHOT1 restricts gastric cancer cell ferroptosis by
epigenetically regulating GPX4. J Gastrointest Oncol. 14:1715–1725.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Majumder M, Chakraborty P, Mohan S,
Mehrotra S and Palanisamy V: HuR as a molecular target for cancer
therapeutics and immune-related disorders. Adv Drug Deliv Rev.
188:1144422022. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Long F, Zhong C, Long Q, Zhu K, Wang J, Yu
Y, Xie C and Hu G: Circular RNA RHBDD1 regulates tumorigenicity and
ferroptosis in colorectal cancer by mediating the ELAVL1/SCD mRNA
interaction. Cancer Gene Ther. 31:237–249. 2024. View Article : Google Scholar
|
|
175
|
Zhou P, Wu Z, Zhang Q, Wang L, Zhang W and
Han X: A novel link between circPDE3B and ferroptosis in esophageal
squamous cell carcinoma progression. Genomics. 116:1107612024.
View Article : Google Scholar
|
|
176
|
Zhang X, Sui S, Wang L, Li H, Zhang L, Xu
S and Zheng X: Inhibition of tumor propellant glutathione
peroxidase 4 induces ferroptosis in cancer cells and enhances
anticancer effect of cisplatin. J Cell Physiol. 235:3425–3437.
2020. View Article : Google Scholar
|
|
177
|
Katsha A, Belkhiri A, Goff L and El-Rifai
W: Aurora kinase A in gastrointestinal cancers: Time to target. Mol
Cancer. 14:1062015. View Article : Google Scholar : PubMed/NCBI
|
|
178
|
Du R, Huang C, Liu K, Li X and Dong Z:
Targeting AURKA in cancer: Molecular mechanisms and opportunities
for cancer therapy. Mol Cancer. 20:152021. View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Gomaa A, Peng D, Chen Z, Soutto M,
Abouelezz K, Corvalan A and El-Rifai W: Epigenetic regulation of
AURKA by miR-4715-3p in upper gastrointestinal cancers. Sci Rep.
9:169702019. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Prasad S, Gupta SC and Tyagi AK: Reactive
oxygen species (ROS) and cancer: Role of antioxidative
nutraceuticals. Cancer Lett. 387:95–105. 2017. View Article : Google Scholar
|
|
181
|
Zhang X, Ma L and Wang J: Cross-regulation
between redox and epigenetic systems in tumorigenesis: Molecular
mechanisms and clinical applications. Antioxid Redox Signal.
39:445–471. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Kang KA, Zhang R, Kim GY, Bae SC and Hyun
JW: Epigenetic changes induced by oxidative stress in colorectal
cancer cells: methylation of tumor suppressor RUNX3. Tumour Biol.
33:403–412. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Zhang R, Kang KA, Kim KC, Na SY, Chang WY,
Kim GY, Kim HS and Hyun JW: Oxidative stress causes epigenetic
alteration of CDX1 expression in colorectal cancer cells. Gene.
524:214–219. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Lim SO, Gu JM, Kim MS, Kim HS, Park YN,
Park CK, Cho JW, Park YM and Jung G: Epigenetic changes induced by
reactive oxygen species in hepatocellular carcinoma: Methylation of
the E-cadherin promoter. Gastroenterology. 135:2128–2140. e1–e8.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Liao P, Wang W, Wang W, Kryczek I, Li X,
Bian Y, Sell A, Wei S, Grove S, Johnson JK, et al: CD8+
T cells and fatty acids orchestrate tumor ferroptosis and immunity
via ACSL4. Cancer Cell. 40:365–378.e366. 2022. View Article : Google Scholar
|
|
186
|
Wang W, Green M, Choi JE, Gijón M, Kennedy
PD, Johnson JK, Liao P, Lang X, Kryczek I, Sell A, et al:
CD8+ T cells regulate tumour ferroptosis during cancer
immunotherapy. Nature. 569:270–274. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Sahai E, Astsaturov I, Cukierman E,
DeNardo DG, Egeblad M, Evans RM, Fearon D, Greten FR, Hingorani SR,
Hunter T, et al: A framework for advancing our understanding of
cancerassociated fibroblasts. Nat Rev Cancer. 20:174–186. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Jiang F, Jia K, Chen Y, Ji C, Chong X, Li
Z, Zhao F, Bai Y, Ge S, Gao J, et al: ANO1-mediated inhibition of
cancer ferroptosis confers immunotherapeutic resistance through
recruiting cancer-associated fibroblasts. Adv Sci (Weinh).
10:e23008812023. View Article : Google Scholar : PubMed/NCBI
|
|
189
|
Leone RD and Powell JD: Metabolism of
immune cells in cancer. Nat Rev Cancer. 20:516–531. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Angelin A, Gil-de-Gómez L, Dahiya S, Jiao
J, Guo L, Levine MH, Wang Z, Quinn WJ III, Kopinski PK, Wang L, et
al: Foxp3 reprograms T cell metabolism to function in low-glucose,
high-lactate environments. Cell Metab. 25:1282–1293.e7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Zhao Y, Li M, Yao X, Fei Y, Lin Z, Li Z,
Cai K, Zhao Y and Luo Z: HCAR1/MCT1 regulates tumor ferroptosis
through the lactate-mediated AMPK-SCD1 activity and its therapeutic
implications. Cell Rep. 33:1084872020. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Yang Z, Su W, Wei X, Qu S, Zhao D, Zhou J,
Wang Y, Guan Q, Qin C, Xiang J, et al: HIF-1α drives resistance to
ferroptosis in solid tumors by promoting lactate production and
activating SLC1A1. Cell Rep. 42:1129452023. View Article : Google Scholar
|
|
193
|
Babar Q, Saeed A, Tabish TA, Pricl S,
Townley H and Thorat N: Novel epigenetic therapeutic strategies and
targets in cancer. Biochim Biophys Acta Mol Basis Dis.
1868:1665522022. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Fan F, Liu P, Bao R, Chen J, Zhou M, Mo Z,
Ma Y, Liu H, Zhou Y, Cai X, et al: A dual PI3K/HDAC inhibitor
induces immunogenic ferroptosis to potentiate cancer immune
checkpoint therapy. Cancer Res. 81:6233–6245. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Yang Z, Su W, Zhang Q, Niu L, Feng B,
Zhang Y, Huang F, He J, Zhou Q, Zhou X, et al: Lactylation of HDAC1
confers resistance to ferroptosis in colorectal cancer. Adv Sci
(Weinh). 12:e24088452025. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Jenke R, Oliinyk D, Zenz T, Körfer J,
Schäker-Hübner L, Hansen FK, Lordick F, Meier-Rosar F, Aigner A and
Büch T: HDAC inhibitors activate lipid peroxidation and ferroptosis
in gastric cancer. Biochem Pharmacol. 225:1162572024. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Bitzer M, Horger M, Giannini EG, Ganten
TM, Wörns MA, Siveke JT, Dollinger MM, Gerken G, Scheulen ME, Wege
H, et al: Resminostat plus sorafenib as second-line therapy of
advanced hepatocellular carcinoma-The SHELTER study. J Hepatol.
65:280–288. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Baretti M, Danilova L, Durham JN, Betts
CB, Cope L, Sidiropoulos DN, Tandurella JA, Charmsaz S, Gross N,
Hernandez A, et al: Entinostat in combination with nivolumab in
metastatic pancreatic ductal adenocarcinoma: A phase 2 clinical
trial. Nat Commun. 15:98012024. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Olsen EA, Kim YH, Kuzel TM, Pacheco TR,
Foss FM, Parker S, Frankel SR, Chen C, Ricker JL, Arduino JM and
Duvic M: Phase IIb multicenter trial of vorinostat in patients with
persistent, progressive, or treatment refractory cutaneous T-cell
lymphoma. J Clin Oncol. 25:3109–3115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Piekarz RL, Frye AR, Wright JJ, Steinberg
SM, Liewehr DJ, Rosing DR, Sachdev V, Fojo T and Bates SE: Cardiac
studies in patients treated with depsipeptide, FK228, in a phase II
trial for T-cell lymphoma. Clin Cancer Res. 12:3762–3773. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Duvic M, Talpur R, Ni X, Zhang C, Hazarika
P, Kelly C, Chiao JH, Reilly JF, Ricker JL, Richon VM, et al: Phase
2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA)
for refractory cutaneous T-cell lymphoma (CTCL). Blood. 109:31–39.
2007. View Article : Google Scholar
|
|
202
|
Whittaker SJ, Demierre MF, Kim EJ, Rook
AH, Lerner A, Duvic M, Scarisbrick J, Reddy S, Robak T, Becker JC,
et al: Final results from a multicenter, international, pivotal
study of romidepsin in refractory cutaneous T-cell lymphoma. J Clin
Oncol. 28:4485–4491. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
203
|
O'Connor OA, Heaney ML, Schwartz L,
Richardson S, Willim R, MacGregor-Cortelli B, Curly T, Moskowitz C,
Portlock C, Horwitz S, et al: Clinical experience with intravenous
and oral formulations of the novel histone deacetylase inhibitor
suberoylanilide hydroxamic acid in patients with advanced
hematologic malignancies. J Clin Oncol. 24:166–173. 2006.
View Article : Google Scholar
|
|
204
|
Chen S, Zheng Y, Liang B, Yin Y, Yao J,
Wang Q, Liu Y and Neamati N: The application of PROTAC in HDAC. Eur
J Med Chem. 260:1157462023. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Cheng B, Pan W, Xiao Y, Ding Z, Zhou Y,
Fei X, Liu J, Su Z, Peng X and Chen J: HDAC-targeting epigenetic
modulators for cancer immunotherapy. Eur J Med Chem.
265:1161292024. View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Lai Y, Han X, Xie B, Xu Y, Yang Z, Wang D,
Li W, Xie Y, Song W, Zhang X, et al: EZH2 suppresses ferroptosis in
hepatocellular carcinoma and reduces sorafenib sensitivity through
epigenetic regulation of TFR2. Cancer Sci. 115:2220–2234. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Yan H, Parsons DW, Jin G, McLendon R,
Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ,
et al: IDH1 and IDH2 mutations in gliomas. N Engl J Med.
360:765–773. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath
SD, et al: Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Amary MF, Bacsi K, Maggiani F, Damato S,
Halai D, Berisha F, Pollock R, O'Donnell P, Grigoriadis A, Diss T,
et al: IDH1 and IDH2 mutations are frequent events in central
chondrosarcoma and central and periosteal chondromas but not in
other mesenchymal tumours. J Pathol. 224:334–343. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Dang L, White DW, Gross S, Bennett BD,
Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, et
al: Cancer-associated IDH1 mutations produce 2-hydroxyglutarate.
Nature. 462:739–744. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim
SH, Ito S, Yang C, Wang P, Xiao MT, et al: Oncometabolite
2-hydroxyglutarate is a competitive inhibitor of
α-ketoglutarate-dependent dioxygenases. Cancer Cell. 19:17–30.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
212
|
Turcan S, Rohle D, Goenka A, Walsh LA,
Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al: IDH1
mutation is sufficient to establish the glioma hypermethylator
phenotype. Nature. 483:479–483. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS,
Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, et
al: Leukemic IDH1 and IDH2 mutations result in a hypermethylation
phenotype, disrupt TET2 function, and impair hematopoietic
differentiation. Cancer Cell. 18:553–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Zarei M, Hajihassani O, Hue JJ, Loftus AW,
Graor HJ, Nakazzi F, Naji P, Boutros CS, Uppin V, Vaziri-Gohar A,
et al: IDH1 inhibition potentiates chemotherapy efficacy in
pancreatic cancer. Cancer Res. 84:3072–3085. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
215
|
Zhu AX, Macarulla T, Javle MM, Kelley RK,
Lubner SJ, Adeva J, Cleary JM, Catenacci DVT, Borad MJ, Bridgewater
JA, et al: Final overall survival efficacy results of ivosidenib
for patients with advanced cholangiocarcinoma with IDH1 mutation:
The phase 3 randomized clinical ClarIDHy trial. JAMA Oncol.
7:1669–1677. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
216
|
Chen W, Yang W, Zhang C, Liu T, Zhu J,
Wang H, Li T, Jin A, Ding L, Xian J, et al: Modulation of the p38
MAPK pathway by anisomycin promotes ferroptosis of hepatocellular
carcinoma through phosphorylation of H3S10. Oxid Med Cell Longev.
2022:69864452022. View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Sun X, Ou Z, Xie M, Kang R, Fan Y, Niu X,
Wang H, Cao L and Tang D: HSPB1 as a novel regulator of ferroptotic
cancer cell death. Oncogene. 34:5617–5625. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
218
|
Sun J, Liu Q, Jiang Y, Cai Z, Liu H and
Zuo H: Engineered small extracellular vesicles loaded with
miR-654-5p promote ferroptosis by targeting HSPB1 to alleviate
sorafenib resistance in hepatocellular carcinoma. Cell Death
Discov. 9:3622023. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Cheng AL, Kang YK, Chen Z, Tsao CJ, Qin S,
Kim JS, Luo R, Feng J, Ye S, Yang TS, et al: Efficacy and safety of
sorafenib in patients in the Asia-Pacific region with advanced
hepatocellular carcinoma: A phase III randomised, double-blind,
placebo-controlled trial. Lancet Oncol. 10:25–34. 2009. View Article : Google Scholar
|
|
220
|
Dixon SJ, Patel DN, Welsch M, Skouta R,
Lee ED, Hayano M, Thomas AG, Gleason CE, Tatonetti NP, Slusher BS,
et al: Pharmacological inhibition of cystine-glutamate exchange
induces endoplasmic reticulum stress and ferroptosis. Elife.
3:e025232014. View Article : Google Scholar : PubMed/NCBI
|
|
221
|
Shi CJ, Pang FX, Lei YH, Deng LQ, Pan FZ,
Liang ZQ, Xie T, Wu XL, Wang YY, Xian YF, et al: 5-methylcytosine
methylation of MALAT1 promotes resistance to sorafenib in
hepatocellular carcinoma through ELAVL1/SLC7A11-mediated
ferroptosis. Drug Resist Updat. 78:1011812025. View Article : Google Scholar
|
|
222
|
Terekhanova NV, Karpova A, Liang WW,
Strzalkowski A, Chen S, Li Y, Southard-Smith AN, Iglesia MD, Wendl
MC, Jayasinghe RG, et al: Epigenetic regulation during cancer
transitions across 11 tumour types. Nature. 623:432–441. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
223
|
Rotem A, Ram O, Shoresh N, Sperling RA,
Goren A, Weitz DA and Bernstein BE: Single-cell ChIP-seq reveals
cell subpopulations defined by chromatin state. Nat Biotechnol.
33:1165–1172. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Wang SW, Gao C, Zheng YM, Yi L, Lu JC,
Huang XY, Cai JB, Zhang PF, Cui YH and Ke AW: Current applications
and future perspective of CRISPR/Cas9 gene editing in cancer. Mol
Cancer. 21:572022. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Pun FW, Ozerov IV and Zhavoronkov A:
AI-powered therapeutic target discovery. Trends Pharmacol Sci.
44:561–572. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
226
|
Pratt D, Sahm F and Aldape K: DNA
methylation profiling as a model for discovery and precision
diagnostics in neuro-oncology. Neuro Oncol. 23(23 Suppl 5):
S16–S29. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Dai Q, Ye C, Irkliyenko I, Wang Y, Sun HL,
Gao Y, Liu Y, Beadell A, Perea J, Goel A, et al: Ultrafast
bisulfite sequencing detection of 5-methylcytosine in DNA and RNA.
Nat Biotechnol. 42:1559–1570. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
228
|
Lone SN, Nisar S, Masoodi T, Singh M,
Rizwan A, Hashem S, El-Rifai W, Bedognetti D, Batra SK, Haris M, et
al: Liquid biopsy: A step closer to transform diagnosis, prognosis
and future of cancer treatments. Mol Cancer. 21:792022. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Zhao J, Cui X, Zhan Q, Zhang K, Su D, Yang
S, Hong B, Wang Q, Ju J, Cheng C, et al: CRISPR-Cas9 library
screening combined with an exosome-targeted delivery system
addresses tumorigenesis/TMZ resistance in the mesenchymal subtype
of glioblastoma. Theranostics. 14:2835–2855. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Gulati GS, D'Silva JP, Liu Y, Wang L and
Newman AM: Profiling cell identity and tissue architecture with
single-cell and spatial transcriptomics. Nat Rev Mol Cell Biol.
26:11–31. 2025. View Article : Google Scholar
|