Introduction
Bone is a highly dynamic tissue that undergoes
continuous remodeling to maintain skeletal integrity and support
various physiological functions (1,2).
This remodeling process, known as bone turnover, involves a
delicate balance between bone formation and resorption (3). Through this ongoing process, bone
homeostasis is achieved, which is critical for maintaining overall
skeletal health. Specifically, bone homeostasis is maintained
through the coordinated activities of osteoblasts, osteoclasts,
osteocytes and chondrocytes, which work together to ensure that
bone density and structure remain stable (4). Disruptions in this balance can lead
to various bone-related diseases. Bone-related diseases encompass a
variety of conditions such as osteoporosis, osteopetrosis,
osteonecrosis, osteoarthritis (OA), osteosarcoma and
chondrosarcoma, characterized by abnormalities in bone density,
structure and integrity (5).
These conditions can lead to grave corresponding clinical symptoms.
For instance, osteoporosis is characterized by increased bone loss
and low peak bone mass, which give rise to low bone density (bone
brittleness); with a propensity to fall and poor bone quality,
fractures are inexorable in this disease (6). Additionally, osteopetrosis
manifests with higher bone mass resulting in phenotypic features
such as macrocephaly and altered craniofacial morphology,
especially influencing other organs and tissues, notably the bone
marrow and nervous system (7).
While OA is a complex condition involving mechanical cartilage
degradation and matrix protease activities, resulting in pain and
loss of joint function, osteonecrosis occurs when there is a
lasting inflammatory environment due to distinguished detrimental
factors such as glucocorticoids or alcohol, triggering bone tissue
death (8,9). As for bone tumors, they are rare
and heterogeneous groups of neoplasms that occur in the bones
(10). Epigenetic research has
greatly enhanced our understanding of these diseases by focusing on
gene expression regulation rather than changes in the DNA sequence
itself (11). Key epigenetic
mechanisms involve DNA methylation, histone modifications,
non-coding RNAs (ncRNAs) and chromatin remodeling complexes
(12). For instance, ncRNAs,
including microRNAs (miRNAs) and long ncRNAs (lncRNAs), play
crucial roles in regulating bone formation, resorption processes
and tumorigenesis in bone cancer (13). These epigenetic factors
collectively impact the differentiation, proliferation and
apoptosis of bone cells, contributing to the development and
progression of bone-related diseases (14-19).
Chromatin is a densely packed and highly regulated
structure that packages DNA, facilitating numerous cellular
processes in eukaryotes, particularly in the context of gene
expression regulation, DNA replication and repair. The fundamental
unit of chromatin is the nucleosome, a highly stable structure that
wraps 145-147 bp of DNA around an octamer of histone proteins
(20). The 2-meter-long DNA is
compressed into chromatin within the nucleus of each cell, creating
a notable barrier to cellular processes such as transcription
(21). Chromatin remodeling
complexes are sophisticated multiprotein assemblies that serve as
pivotal regulators of chromatin architecture, mediating dynamic
changes that profoundly influence gene expression, DNA repair,
replication and a wide range of other critical cellular processes
(22). To date, four families of
chromatin remodeling complexes have been identified:
Switch/sucrose-non-fermenting (SWI/SNF), imitation switch (ISWI),
chromodomain-helicase-DNA binding (CHD) and inositol requiring
80/SWi2/snf2-related 1 (INO80/SWR) (23). Each of these families perform
unique, non-redundant roles within the cell. Chromatin remodeling
complexes are typically composed of three parts: Nucleosome, module
[ATPase module, actin-related protein (ARP) module or body module]
and proteins. However, different chromatin remodeling complexes
have notably different components, including diversities in the
nucleosomes and configurations. For example, the SWI/SWF family is
composed of the nucleosome, ATPase module, ARP module, body module
and diverse proteins (including snf2, RT102, Arp7 and Arp9)
(24). The ISWI family does not
entail an ARP module and contains dissimilar proteins, such as
Isw1, Ioc2, Ioc3 and Ioc4 (25).
For the CHD family, the ATPase module exclusively contains distinct
proteins. SWR1 contains three main modules and differing proteins
(26). Therefore, with such a
framework, chromatin remodeling complexes comprise crucial enzymes
or cooperate with other chromatin-related factors to play a key
role in dynamically regulating various cellular processes,
including transcription and DNA repair, by controlling access to
genomic DNA (26). Additionally,
these gene-mediated chromatin remodeling complexes are crucial
contributors to the molecular pathology of bone-related diseases by
engaging in bone epigenetic mechanisms such as differentiation,
proliferation and senescence. For instance, disturbances in
bromodomain-containing protein (BRD)9 within the SWI/SWF family can
lead to debilitating bone-related diseases such as osteoporosis,
osteopetrosis and osteonecrosis (14).
Therefore, chromatin remodeling complexes play a
pivotal role in bone-related diseases. The present review aims to
provide an overview of the mechanisms and functions of
gene-mediated chromatin remodeling complexes in bone-related
diseases. Through literature mining and investigation (Data S1), the present review will
examine the relationship between chromatin remodeling complexes and
bone-related diseases, and the potential clinical relevance of this
relationship, such as pathological mechanisms and disease targets,
ultimately aiming to generate further effective clinical
therapeutic applications, such as BRM inhibitors for osteoporosis
treatment and BRD9 inhibitors for advanced synovial sarcoma
treatment. The primary strength of the present review lies in its
thorough examination of the role of chromatin remodeling complexes
specifically in bone-related diseases. Additionally, the present
review emphasizes the potential therapeutic applications of
chromatin remodeling complexes, which has been a relatively
underexplored area in bone research. Unlike earlier reviews
(27-30), which tend to focus on a narrow
subset of chromatin remodeling complexes or bone diseases, the
present review covers a broader range of chromatin remodeling
complexes and bone-related diseases, including bone tumors, OA,
bone-fat disorder, adolescent idiopathic scoliosis (AIS) and
osteoporosis. A detailed review of the role and mechanisms of
chromatin remodeling complexes in bone metabolic processes, such as
stemness maintenance, chondrogenesis, osteogenesis,
osteoclastogenesis and angiogenesis, are also provided.
Additionally, the impact of chromatin remodeling complexes on key
bone biological functions, including proliferation, senescence and
programmed cell death are explored in the present review. Another
strength of this review is its focus on therapeutic implications,
such as how chromatin remodeling complexes could be targeted for
intervention in bone-related diseases.
Classification, mechanisms and regulations
of chromatin remodeling complexes
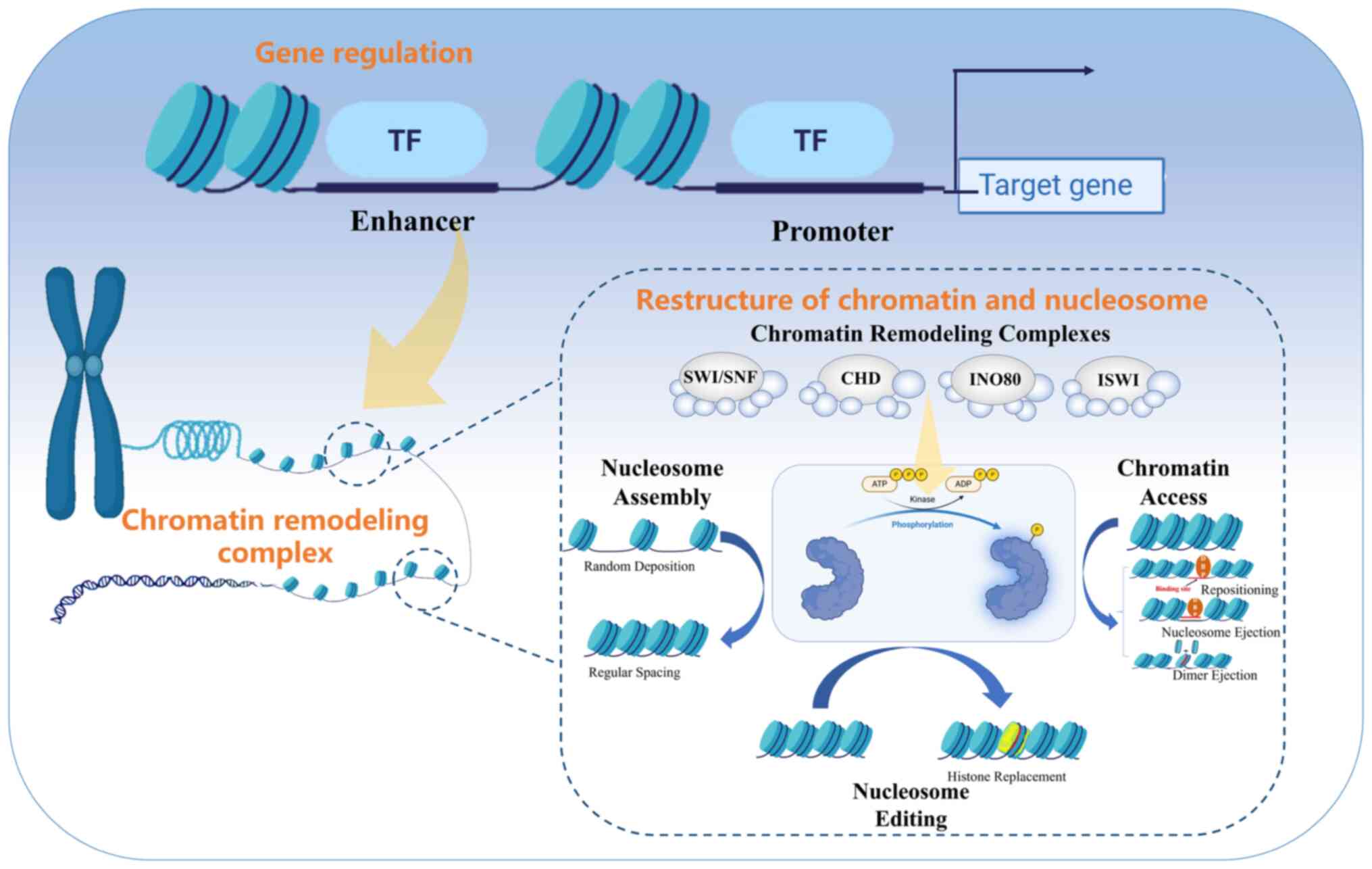
Chromatin remodeling complexes primarily operate
through three fundamental mechanisms (Fig. 1). First, these complexes can edit
assembled nucleosomes by replacing, moving or removing specific
nucleosomes, thus altering chromatin structure and function.
Second, chromatin remodeling complexes are involved in the assembly
and reorganization of nucleosomes from random deposition to regular
space. Third, chromatin remodeling complexes alter the structure of
chromatin to make specific regions of DNA more accessible to
transcription factors and other regulatory proteins (22,31,32). This is achieved through various
mechanisms, including repositioning nucleosomes, nucleosome
ejection and dimer ejection, which all contribute to the dynamic
remodeling of chromatin (22,31,32). Collectively, these mechanisms
serve as the driving force behind chromatin activation and
represent a key component of the hereditary code governing diverse
epigenetic regulations.
Based on the sequence homology of the catalytic
ATPase and the accessory subunits, chromatin remodeling complexes
are classified into four distinct families (Fig. 2): SWI/SNF, ISWI, CHD and
INO80/SWR (33,34). The SWI/SNF complex in human cells
is characterized by a notable diversity of assemblies. There are
several variants of this complex, including canonical BAF (cBAF),
polybromo BAF (PBAF) and non-canonical BAF (ncBAF/gBAF), each
distinguished by the presence of unique subunits (26). However, the ISWI and CHD family
have only one assembly (22).
Although INO80 and SWR belong to the same family of chromatin
remodelers, they are classified into different subfamilies with
distinct components (22,35,36).
To this point, each family of chromatin remodeling complexes
performs specialized functions within the cell (26). The SWI/SNF family, for instance,
includes the BRG1 ATPase subunit, which can independently remodel
nucleosome positioning on nucleosomal templates. A minimal
four-subunit complex composed of BRG1, BAF155, BAF170 and BAF47
efficiently achieves chromatin remodeling in vitro (37). ISWI complexes facilitate the
maturation of initial histone-DNA complexes (pre-nucleosomes) into
canonical octameric nucleosomes and help space nucleosomes at
relatively fixed distances (38,39). CHD family members contain a
conserved ATPase domain, comprising SNF2_N and Helicase_C PFAM
domains, which act as a motor to drive dynamic interactions with
chromatin and nucleosome substrates (40,41). However, INO80, features multiple
ATPases that provide catalytic activity and enable ATP-dependent
nucleosome sliding within the cell (42,43). Additionally, Ino80p serves as a
docking site for several core subunits, including actin, ARPs and
the Rvb AAA+ ATPases (44).
While these chromatin remodeling complex families implement diverse
functions in vivo, they share a common mission: Facilitating
genome utilization, which includes processes such as transcription
factor binding, DNA replication and repair (45,46). It is important to note that
ATP-dependent chromatin remodeling complexes can be distinguished
not only by the composition of their subunits but also by the
domain organization of their catalytic subunits (32). Overall, chromatin remodeling
complexes play a crucial role in epigenetic regulation.
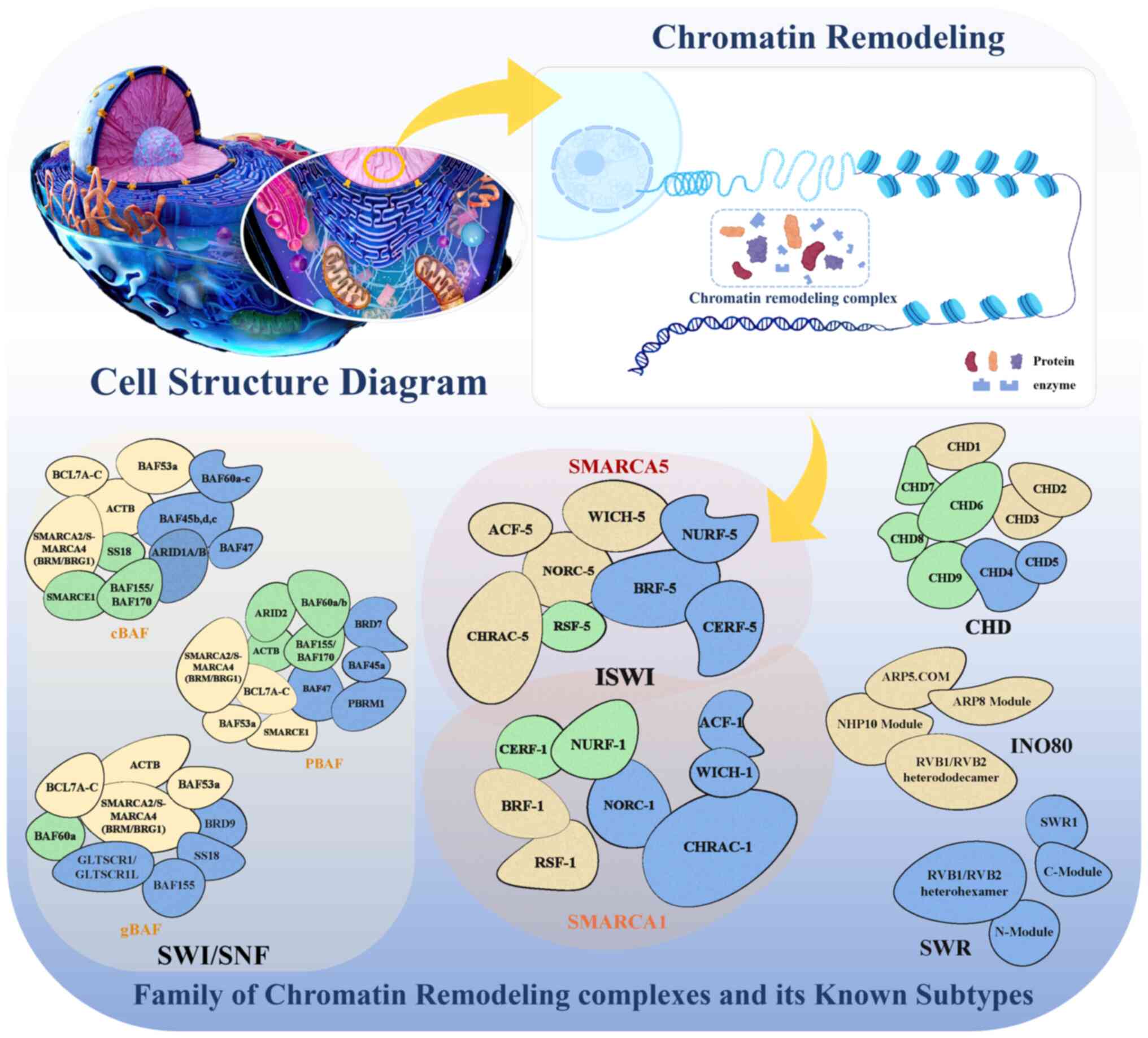 | Figure 2Chromatin remodeling complexes have
diverse subtypes to implement their indispensable functions.
Chromatin remodeling complexes are categorized into four distinct
families: SWI/SNF, ISWI, CHD and INO80/SWR. The SWI/SNF family
complexes in human cells are characterized by a marked diversity of
assemblies. There are several variants of this complex, including
the cBAF, PBAF and ncBAF/gBAF subtypes, which are large assemblies
with core subunits such as BRG1/BRM (ATPase), BAF155/BAF170
(scaffold proteins), BAF47 (tumor suppressor) and BAF60 (linking
proteins to transcription factors). The ISWI family complexes,
including NURF, CHRAC and ACF, feature key subunits such as the
ATPase ISWI (SMARCA1/SMARCA5), the large chromatin-binding NURF301
(BPTF), the accessory proteins CHRAC15/CHRAC17 and the scaffold
protein ACF1 (BAZ1A). The CHD family complexes, ranging from CHD1
to CHD9, include subunits such as CHD1/CHD2 (chromodomains for
histone binding) and CHD3/CHD4 (NuRD complex) with ATPase activity.
In INO80/SWR family complexes, INO80 and SWR are presented
separately since, although they belong to the same family of
chromatin remodelers, they are classified into different
subfamilies with distinct components. Crucial for histone exchange,
these complexes include subunits such as the INO80 ATPase,
RVB1/RVB2 for structural integrity and, within the SWR1 complex,
the SWR1 ATPase, Swc2 for binding H2A.Z and Swc5/Swc6 for histone
variant incorporation. cBAF, canonical BAF; ncBAF, non-canonical
BAF; BCL7A-C, B-cell lymphoma 7A-C; BAF, brahma-associated factor;
ACTB, β actin; SS18, synovial sarcoma translocation protein 18;
ARID1A/B, AT-rich interactive domain 1A/B; BRD,
bromodomain-containing protein; PBRM1, polybromo-1; PBAF,
polybromo-associated BAF; GLTSCR1/GLTSCR1L, glioma tumor suppressor
candidate region gene 1/glioma tumor suppressor candidate region
gene 1-like; gBAF, germinal center B-cell-specific BAF; SWI/SNF,
switching defective/sucrose non-fermenting; ACF-5, ATP-dependent
chromatin assembly and remodeling factor-5; WICH, WSTF-ISWI
chromatin-remodeling complex; NURF, nucleosome remodeling factor;
NORC, nucleolar remodeling complex; RSF, remodeling and spacing
factor; CERF-5, chromatin-associated Ets-related factor-5; ISWI,
imitation SWI; ACE, activating chromatin element; CERF-1,
chromatin-associated Ets-related factor-1; CHRAC-1, chromatin
accessibility complex-1; CHD, Chromodomain-helicase-DNA-binding;
ARP5. COM, actin-related protein 5 complex; ARP8 module,
actin-related protein 8 module; NHP10 module, non-histone protein
10 module; INO80, inositol-requiring 80; SWR1, Swi2/Snf2-related 1;
C-Module, C-terminal module; N-Module, N-terminal module. This
figure was created with BioRender.com. |
Chromatin remodeling complexes are regulated by
several factors that govern their own ultimate epigenetic functions
(Fig. 3). First, ncRNAs, miRNAs
and lncRNAs exert a pivotal influence on chromatin remodeling
complexes. miRNAs can target the mRNA of chromatin remodeling
complex subunits pre-transcriptionally, leading to their
degradation, and can also inhibit the translation of these
subunits. lncRNAs can interact with chromatin remodeling subunits
post-transcriptionally in several ways including binding to these
subunits to inhibit the assembly of the complex, acting as guiding
lncRNAs to recruit chromatin remodeling complexes and as decoy
lncRNAs, limiting the chromatin localization of the complexes
(47-51). Second, actin and ARPs are
integral components of chromatin remodeling complexes and regulate
ATPase activity, facilitate complex assembly and enhance DNA
binding and nucleosome repositioning through direct interaction
with ATPase subunits. Additionally, ARPs interact with histones and
serve as molecular bridges between chromatin-modifying complexes,
coordinating higher-order chromatin regulation (22,52). Lastly, chromatin remodeling
complexes are under the strict regulation of posttranslational
modifications, namely, they are modulated by phosphorylation,
acetylation and PARylation (22,53,54). For example, during mitosis, human
SWI/SNF is phosphorylated and this modification inhibits its
remodeling activity, which may contribute to the widespread
inhibition of chromatin remodeling during cell division (54). What's more, the human nucleosome
acetyltransferase of the histone H4 (NuA4)/Tat-interactive protein
60 kDa coactivator complex, a homologous fusion of the yeast SWR1
and NuA4 complexes, facilitates the incorporation of the histone
variant H2A.Z into nucleosomes while acetylating histones H4, H2A
and H2A.Z. This dual function plays a crucial role in gene
regulation and genome stability maintenance (52). Additionally, Sala et al
(55) found that PARylated ISWI
displays a reduced nucleosome-binding affinity, as well as lower
DNA- and nucleosome-stimulated ATPase activity. Therefore,
chromatin remodeling complexes must first be regulated by upstream
modulators before exerting their functions. Under normal
conditions, chromatin remodeling complexes serve as facilitators
and protectors of cellular molecular activities, but aberrations in
their functions can lead to various diseases, such as cancer,
Coffin-Siris syndrome and fibromuscular dysplasia (56). Chromatin remodeling complexes
play a pivotal role in bone pathology (11,28). Numerous studies have identified a
variety of relevant chromatin remodeling complexes and their
subunits as well as elucidated their specific functions (26,37). For instance, the SWI/SWF complex
includes subunits such as BRG1, BRM, BAF45, BAF47, BAF155, BAF170,
BAF180, BAF200, BAF250a, BRD7, BRD9 and SS18. The ISWI complex is
characterized by the presence of the bromodomain adjacent to zinc
finger domain 1A (BAZ1A) subunit. The CHD family comprises subunits
such as CHD1, CHD4, CHD7 and CHD9, and the INO80/SWR complex is
composed of multiple subunits, such as INO80, SWR1, ARP4, ARP5,
RvB1 and RvB2 that participate in chromatin remodeling. Each
subunit is involved in distinct functions, with some overlapping
roles and others being unique (28). The relationship between chromatin
remodeling complexes and bone biology and bone-related diseases is
dissected further below.
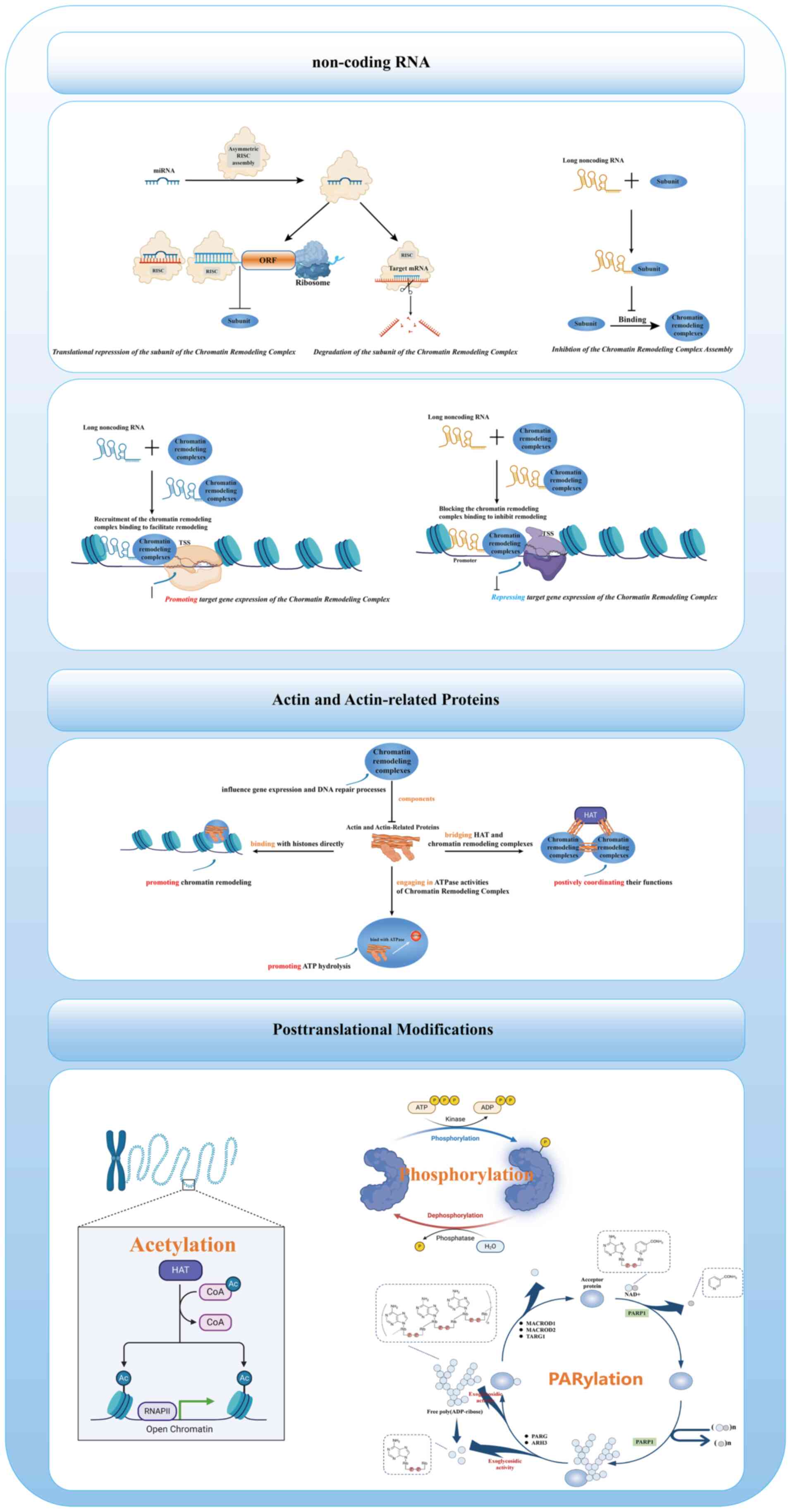 | Figure 3Chromatin remodeling complexes are
under three main regulation factors: Non-coding RNA, actin and
actin-related proteins, post-translational modifications.
Non-coding RNA: i) miRNAs mediate translational repression or
target mRNA degradation by assembling the RISC, thereby inhibiting
the expression of subunits of the chromatin remodeling complex; ii)
lncRNA inhibits the assembly of the chromatin remodeling complex by
binding to its subunits; iii) lncRNA facilitates the recruitment of
chromatin remodeling complexes to target gene promoter regions,
thereby promoting gene expression; and iv) lncRNA represses gene
expression by preventing the binding of chromatin remodeling
complexes, thereby inhibiting chromatin remodeling. Actin and
actin-related proteins: These proteins serve as components of
chromatin remodeling complexes, influencing gene expression and DNA
repair processes. They facilitate chromatin remodeling by directly
binding to histones and promoting ATP hydrolysis through
interaction with ATPase. Additionally, actin-related proteins
bridge HAT and chromatin remodeling complexes, thereby positively
coordinating their functions. Posttranslational modifications:
Chromatin remodeling complexes are regulated by the histone
post-translational modifications including phosphorylation,
acetylation and PARylation. RISC, RNA-induced silencing complex;
ORF, open reading frame; TSS, transcription start site; HAT,
histone acetyltransferase; Ac, acetyl; MACROD1,
macrodomain-containing protein 1; TARG1, tankyrase-associated
RING-containing protein 1; PARG, poly (ADP-ribose) glycohydrolase;
ARH3, ADP-ribosylhydrolase 3; PARP1, poly (ADP-ribose) polymerase
1. This figure was created with BioRender.com. |
Role of chromatin remodeling complexes in
bone metabolic processes
Chromatin remodeling complexes are intimately linked
to bone metabolic processes, performing various regulatory
mechanisms and functions in the modulation of gene expression
(Figs. 4 and 5). For instance, their expression at
specific stages of bone biology directs progenitor cells to
differentiate and proliferate into osteoblasts. Conversely, these
complexes can also contribute to bone-related diseases when their
functions become aberrant (28,29,57). Therefore, in this section, the
specific functional properties of these chromatin remodeling
complexes in normal bone metabolic processes as well as aberrant
settings are discussed (Table
I).
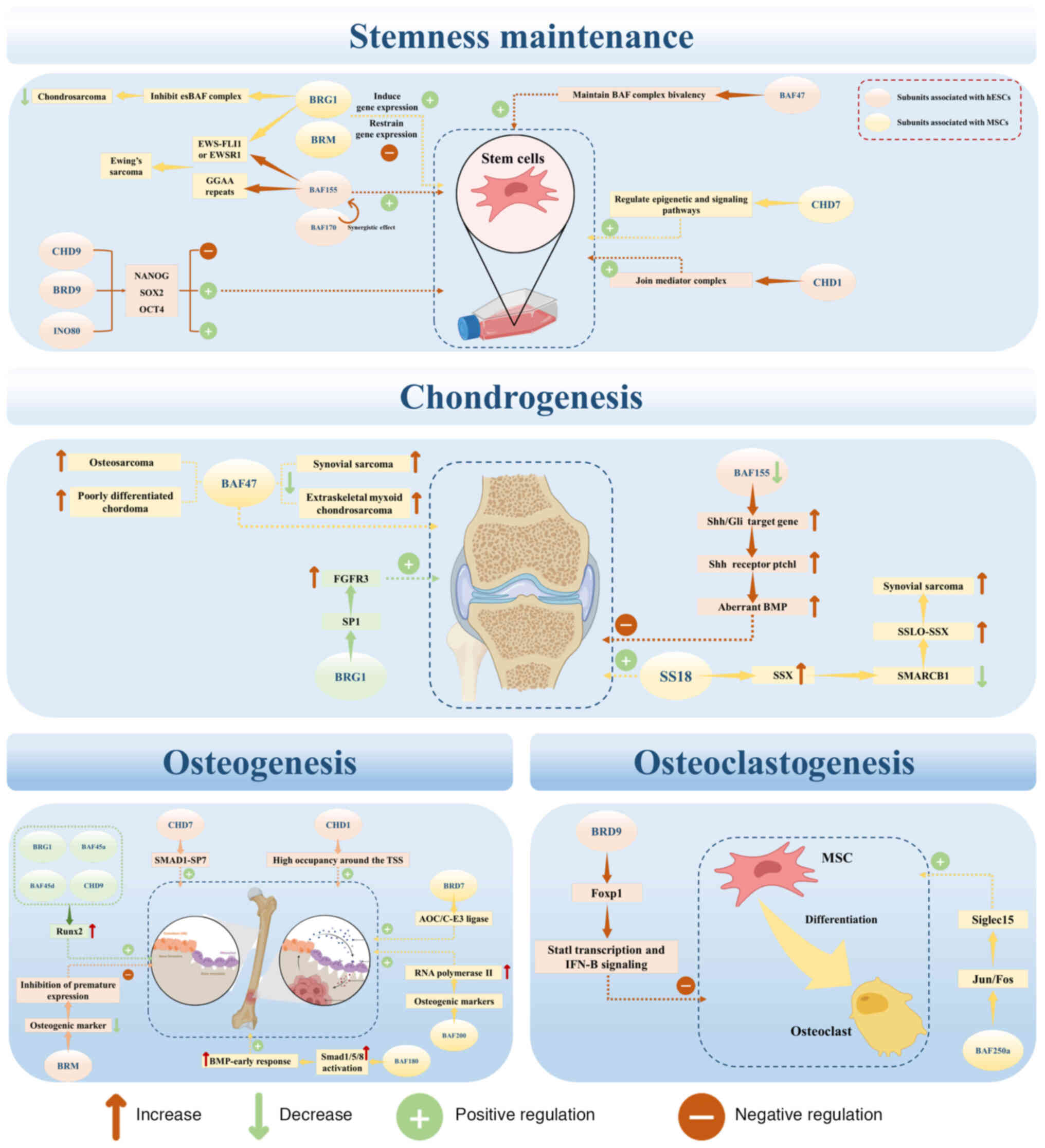 | Figure 4Epigenetic roles of chromatin
remodeling complexes in bone metabolic processes, including
stemness maintenance, chondrogenesis, osteogenesis and
osteoclastgenesis. This figure shows how each relevant subunit,
through its unique molecular mechanism, performs epigenetic changes
including in normal and pathological conditions. AOC/C-E3 ligase,
activator of c-Jun/C-E3 ligase; BAF, brahma-associated factor; BMP,
bone morphogenetic protein; BRD, bromodomain-containing protein;
BRG1, breast cancer susceptibility gene 1-associated protein; BRM,
bromodomain-containing protein BRM; CHD,
chromodomain-helicase-DNA-binding protein; EWS-FLI1, Ewing sarcoma
breakpoint region 1-Friend leukemia virus integration 1
transcription factor; EWSR1, Ewing sarcoma breakpoint region 1;
FGFR3, fibroblast growth factor receptor 3; Foxp1, forkhead Box
Protein P1; INO80, inositol-requiring 80; IFN-B, interferon-β; MSC,
mesenchymal stem cell; OCT4, octamer-binding transcription factor
4; Ptch1, Patched-1; Runx2, Runt-related transcription factor 2;
Shh/Gli, Sonic hedgehog/glioma-associated oncogene family
zinc-finger protein; Siglec15, sialic acid-binding
immunoglobulin-like lectin 15; SMAD1-SP7, SMAD family protein
1-specificity protein 7; SOX2, Sex-determining region Y-box 2; SP1,
specificity protein 1; SS18, synovial sarcoma translocation protein
18; SSLO-SSX, synovial sarcoma X-breakpoint gene; TSS,
transcription start site. This figure was created with BioRender.com. |
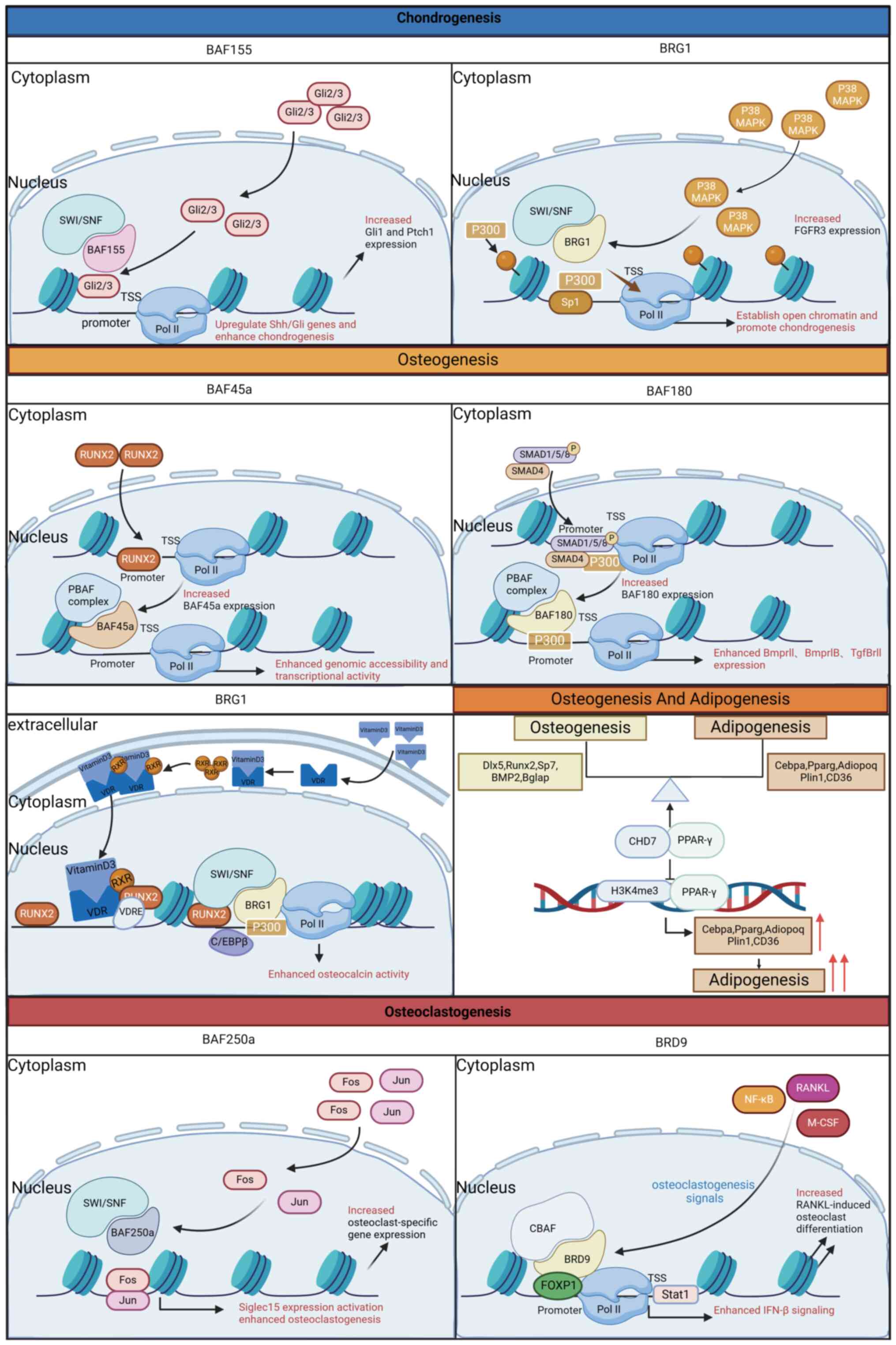 | Figure 5Chromatin remodeling complexes employ
various biological regulatory mechanisms that contribute to the
balance of bone development. This figure illustrates eight
well-established mechanisms involved in chondrogenesis,
osteogenesis, osteoclastogenesis and adipogenesis, each
characterized by its unique features. Gli2/3, glioma-associated
oncogene family zinc-finger protein 2/3; SWI/SNF, switch/sucrose
non-fermentable; BAF, brahma-associated factor; Ptch1, Patched-1;
Pol II, RNA polymerase II; RISC, RNA-induced silencing complex;
ORF, open reading frame; TSS, transcription start site; HAT,
histone acetyltransferase; Ac, acetyl group; MACROD,
macrodomain-containing protein; TARG1, tumor necrosis factor α
receptor-associated protein 1; PARG, Poly (ADP-ribose)
glycohydrolase; ARH3, ADP-ribosylhydrolase 3; PARP1, Poly
(ADP-ribose) polymerase 1; P38 MAPK, p38 mitogen-activated protein
kinase; BRG1, brahma-related gene 1; Sp1, specificity protein 1;
FGFR3, fibroblast growth factor receptor 3; RUNX2, Runt-related
transcription factor 2; PBAF, polybromo-associated BAF; RXR,
retinoid X receptor; VDR, vitamin D receptor; VDRE, vitamin D
response element; Dlx5, Distal-less homeobox 5; BMP2, bone
morphogenetic protein 2; Bglap, Bone γ-carboxyglutamate-containing
protein; CHD7, chromodomain-helicase-DNA-binding protein 7;
H3K4me3, Histone H3 lysine 4 trimethylation; PPAR-γ, peroxisome
proliferator-activated receptor γ; Cebpa, CCAAT-enhancer-binding
protein α; Adiopoq, adiponectin; Plin1, perilipin 1; CD36, NF-κB,
nuclear factor-κB; RANKL, receptor activator of NF-κB ligand;
M-CSF, macrophage colony-stimulating Factor; CBAF, chromatin
assembly factor; BRD9, bromodomain-containing protein 9; FOXP1,
forkhead box protein P1. This figure was created with BioRender.com. |
 | Table IDiverse role of chromatin remodeling
complexes in the bone metabolic process and the biological
functions of bone. |
Table I
Diverse role of chromatin remodeling
complexes in the bone metabolic process and the biological
functions of bone.
| Chromatin
remodeling complex | Subunits | Key features | Functions in the
bone metabolic processes | Roles in the bone
biological functions |
|---|
| SWI/SNF
complex | BRG1 | ATPase activity,
essential for remodeling. | Regulates gene
expression during chondrogenesis and osteogenesis, while also
playing a role in the maintenance of stemness. | Sustains hESC
proliferation, activates autophagy, induces cell cycle arrest and
apoptosis in MSCs and prevents cellular senescence. |
| SWI/SNF
complex | BRM | A core
SWI2/SNF2-type ATPase, functions as a gene suppressor. | BRM-deficient
preosteoblasts show early expression of osteoblast markers,
indicating enhanced osteogenesis. | Regulates cell
proliferation. |
| SWI/SNF
complex | BAF250a/
BAF250b |
Bromodomain-containing, transcriptional
regulator, involved in targeting transcription factors and
recruitment. | Promotes osteoclast
differentiation by facilitating the cooperation between chromatin
and thetranscription factor Jun/Fos. | Loss of BAF250b
impairs MSC quiescence and proliferation through the pERK
pathway. |
| SWI/SNF
complex | BAF47 | Enhancer activation
and rescues BAF complex-mediated resolution of bivalency. | Regulates gene
expression during chondrogenesis while also playing a role in
stemness maintenance. | - |
| SWI/SNF
complex | BAF155 | Increase DNA
accessibility by remodeling nucleosomes. | Regulates target
genes during chondrogenesis and a crucial factor in stemness
maintenance. | Depletion of BAF155
in ESCs decreases proliferation and increases cell death. |
| SWI/SNF
complex | BAF170 | A subunit of the
BAF chromatin remodeling complex, working with BAF155 and BRG1 in
gene regulation and development. | Maintains
pluripotency and self-renewal in hESCs. | - |
| SWI/SNF
complex | BRD9 | A
bromodomain-containing protein, essential for SWI/SNF complex
stability. | Loss of BRD9
reduces NANOG and SOX2 levels, while its high expression suppresses
osteoclastogenesis by interacting with FOXP1 to enhance STAT1
transcription and activate IFN-β signaling. | Promotes
pyroptosis. |
| SWI/SNF
complex | SS18 | Involved in
SS18-SSX fusion from t(X;18) chromosomal translocation, inactivates
SMARCB1. | Regulates gene
expression during chondrogenesis and has a notable role in synovial
sarcoma. | - |
| SWI/SNF
complex | BAF45a and
BAF45d | Regulate chromatin
accessibility and gene expression through interactions with various
transcription factors, including PBAF-RUNX2 crosstalk. | Regulates gene
expression during osteogenesis | - |
| SWI/SNF
complex | BAF180 | A subunit of the
PBAF complex, crucial for chromatin remodeling and gene
regulation. | Facilitates the
progression of osteogenesis. | - |
| SWI/SNF
complex | BRD7 | A
bromodomain-containing protein, involved in chromatin
remodeling. | Regulates gene
expression in osteogenesis and osteosarcoma. | Upregulation of
BRD7 reduces apoptosis via the PI3K and YAP1 pathways. |
| CHD complex | CHD1 |
Chromodomain-containing, ATP-dependent,
engages in the transcriptional start site. | Facilitates
chromatin accessibility in hESCs, pre-initiates gene transcription
and induces pluripotent stem cells and osteogenesis | - |
| CHD complex | CHD7 | Interacts with
important osteogenesis and stem cell signaling pathways. | Regulates gene
expression in osteogenesis and stemness. | - |
| CHD complex | CHD9 | Binds to related
genomic domains, interacts with the transcriptional regulation of
Pol II and works synergistically with BRG1. | Regulates gene
expression in osteogenesis and the stemness of MSCs. | - |
| INO80/ SWR
complex | INO80 | Maintains an open
chromatin environment and facilitates the recruitment of
transcription factors and coactivators. | Maintains the
pluripotency of hESCs and stabilize the osteogenesis. | Regulates the
proliferation of ESCs. |
Chromatin remodeling complexes and
stemness maintenance
Stemness maintenance is crucial for bone development
and the prevention of bone-related diseases, as it ensures the
continuous supply of progenitor cells capable of differentiating
into osteoblasts and other bone-forming cells (58,59). In the following sections, the
specific mechanisms through which chromatin remodeling complexes
influence stemness maintenance, and their implications for bone
biology and disease, will be discussed.
BRG1 (also known as SMARCA4) and BRM
(also known as SMARCA2)
Well-established interactions between esBAF (BRG1 or
BRM) and polycomb complexes have been documented, highlighting that
both esBAF and polycomb group proteins play roles in maintaining
pluripotency through both antagonistic and synergistic acts in
polycomb group proteins (60,61). In osteoblast precursors,
BRG1-containing SWI/SNF typically induces the expression of
osteogenic genes, while BRM-specific complexes act in conjunction
with p130 and repressor E2Fs to primarily restrain differentiation.
This action helps sustain the committed precursor state until
pre-osteoblasts receive the appropriate differentiation signals
(60). Moreover, BRG1 and BRM
can function as either oncogenes or tumor suppressors. In
chondrosarcoma, both subunits in the esBAF complex exhibit
oncogenic properties when inhibited (62). Conversely, loss of BRG1 leads to
a loss of both self-renewal capacity and pluripotency (62,63), while the loss of BRM results in
enhanced osteoblast differentiation and impaired adipocyte
differentiation (64).
Additionally, a previous study found that BRG1 interacts with Ewing
sarcoma-friend leukemia integration 1 (EWS-FLI1) fusion protein or
Ewing sarcoma RNA binding protein 1 (EWSR1), with the EWS-FLI1
fusion protein being detected in >85% of Ewing sarcoma cases.
This interaction is a key regulator in the cell growth of Ewing
sarcoma (65).
BAF47 (also known as SMARCB1/INI1)
BAF47 plays a crucial role in ensuring the stability
and functionality of the BAF complex. Specifically, the presence of
BAF47 is essential for the SWI/SNF complex to effectively resolve
bivalency in the genome, highlighting its critical role in
regulating gene expression at bivalent promoters and enhancers
(66,67). Therefore, the loss of BAF47
decreases the chromatin affinity of the intact BAF complex, leading
to dysfunctional BAF complex activity, which is vital for every
step of bone development (67).
A study demonstrated that knockdown of BAF47 impedes
differentiation in human embryonic stem cells (hESCs), while
upregulation of BAF47 enhances differentiation (68,69). Therefore, BAF47 plays a notable
role in maintaining the balance of stemness in the human body.
BAF155 (also known as SMARCC1) and BAF170
(also known as SMARCC2)
Together with BRG1, BAF155 and BAF170 play pivotal
roles in maintaining pluripotency and self-renewal in hESCs. The
depletion of these three subunits leads to pluripotency dysfunction
in hESCs, significantly reducing the potential for future bone
development (62,70). Additionally, BAF155 interacts
with EWS-FLI1, which acts as an aberrant transcription factor that
drives the cellular transformation of Ewing sarcoma. Moreover, this
chimeric protein directly binds to GGAA repeats, thereby modifying
the transcriptional profile of Ewing sarcoma, eventually promoting
the progression of this disease (65).
BRD9
BRD9 is recruited by BRD4 to regions linked to naive
pluripotency genes in a bromodomain-dependent manner, playing a
crucial role in regulating and maintaining the naive pluripotent
state of ESCs (71). Inhibition
of BRD9 expression leads to decreased levels of NANOG and SOX2,
impairing self-renewal and altering differentiation, particularly
inhibiting mesoderm differentiation while promoting neural ectoderm
differentiation. Since the mesoderm is essential for the
development of various tissues, including cartilage and bone, its
disruption can affect skeletal formation (72). BRD9 also regulates pluripotency
gene expression and TGF-β/Nodal/Activin and Wnt signaling pathways
by forming a complex with BRD4, SMAD2/3, β-CATENIN and p300, and
modulating H3K27 acetylation to sustain hESC differentiation
(72). Furthermore, inhibition
of BRD9 enhances somatic cell reprogramming by downregulating
fibroblast genes, limiting chromatin accessibility at enhancers and
reducing reprogramming inhibitors such as MN1 and ZBTB38, thereby
facilitating reprogramming (73).
CHD1
CHD1 is an ATPase-dependent chromatin remodeling
protein that plays a crucial role in maintaining pluripotency and
stemness in ESCs and mesenchymal stem cells (MSCs) by facilitating
chromatin accessibility and regulating key gene expression. hESCs
possess chromatin accessibility that enables them to differentiate
into various daughter cell types, including hematopoietic, neural
stem, mesenchymal stem and other lineage cells (74-76). Notably, MSCs serve as progenitor
cells of osteoblasts, and osteoprogenitors originating from
mesenchymal stromal cells are also committed to their
differentiation into osteoblasts, the cells responsible for bone
formation and maintenance (77,78). In this context, CHD1 plays a
critical role in the mediator complex, facilitating chromatin
accessibility in hESCs, pre-initiating gene transcription and
inducing pluripotent stem cells by maintaining a
heterochromatin-poor pluripotent stem cell state (75). Specifically, CHD1 is crucial for
maintaining open chromatin and pluripotency in mouse ESCs; its
downregulation leads to heterochromatin accumulation, loss of
pluripotency and the impaired reprogramming of somatic cells to the
pluripotent state (74).
Moreover, CHD1 is essential for early mouse embryogenesis and
pluripotency, as its loss results in embryonic lethality after
implantation by significantly downregulating key regulators of cell
fate specification, such as Pou5f1 and Nanog, and impairing the
activation of Hmgpi during zygotic gene activation, which can be
rescued by Hmgpi mRNA microinjection (79). Furthermore, CHD1 is vital for
maintaining the integrity of the genome in ESCs by preventing the
accumulation of DNA double-stranded breaks, thereby further
supporting their pluripotency and overall stemness during
hypertranscription (76).
Additionally, a previous study demonstrated that CHD1 regulates the
stemness of MSCs, with higher levels promoting their tissue
regeneration and hematopoietic stem cell-supporting activity, while
lower levels are associated with a loss of clonogenic potential in
pathological conditions (2).
Ultimately, the loss of CHD1 leads to the transformation of
heterochromatin and a significant loss of pluripotency (28,80).
CHD7
CHD7 is a crucial gene for stemness maintenance as
it regulates epigenetic and signaling pathways and is highly
expressed in active gene expression signals within stem cells
(80). Research has shown that
CHD7 is also vital for maintaining open chromatin in ESCs,
regulated by the trithorax protein Ash2l, and the knockdown of
Ash2l reduces H3K4 methylation and promotes a silenced chromatin
state (81) Additionally, the
co-localization of BRG1 and CHD7 at distal regions in ESCs
influences the expression of key transcription factors such as
NANOG, SOX2 and OCT4, further affecting ESC pluripotency (82). Furthermore, it has been reported
that CHD7 interacts with lysine-specific histone demethylase 1
(LSD1) in mouse ESCs and is crucial for maintaining stemness and
proper differentiation. The increased co-occupancy of methylated
H3K4 and CHD7 on chromatin following LSD1 deletion underscores the
essential role of LSD1 in facilitating the binding of CHD7 to
chromatin and regulating differentiation (83). Finally, CHD7 knockout ESCs
exhibit defective expression of ectodermal markers that affect
pluripotency, which is vital for the stemness associated with
subsequent differentiation into bone, cartilage and skeletal muscle
(83). Notably, the depletion of
CHD7 in MSCs leads to the repression of osteogenic transcription
factors and impairs the osteogenesis ability of these cells
(84).
CHD9
CHD9 is dispensable for pluripotent marker
expression, including OCT4, SOX2, NANOG and SSEA-1 (85). The knockout of CHD9 promotes hESC
proliferation through binding to genomic domains that overlap with
DNA motifs recognized by cell cycle-related transcription factors
and interacting with the transcriptional regulation of Pol
I-controlled genes and nuclear receptors (85,86). Additionally, a study found that
CHD9 is transiently expressed during mesenchymal cell
differentiation both in vivo and in vitro, with
expression observed in osteoprogenitors and downregulation in
mature osteoblasts (78).
INO80
The INO80 is crucial for the self-renewal of hESCs,
as well as for reprogramming and embryonic development. INO80
specifically binds to the promoters of key pluripotency genes in
conjunction with transcription factors such as OCT4, NANOG, SOX2
and Krüppel-like factor 4, with its binding reliant on WD repeat
domain 5 (WDR5). INO80 helps maintain an open chromatin structure,
enabling the recruitment of the mediator complex and RNA polymerase
II, which enhances the expression of these pluripotency genes
(87). Furthermore, silencing
INO80 leads to the abnormal morphology of hESCs (87). While in mouse ESCs, INO80
deficiency causes a notable increase in the duration of the
G1-phase in ESCs cultured under primed conditions. Additionally,
INO80 directly associates with the transcription start site to
modulate the expression of genes involved in the cell cycle,
including Ccne1, Cdc25b and Cdkn1b (78).
Chromatin remodeling complexes and
chondrogenesis
Chondrogenesis is the process by which MSCs
differentiate into chondrocytes, ultimately forming cartilage
models (anlagen) that serve as precursors to future bones through
endochondral ossification while also secreting cartilage matrix to
form cartilage (88,89). In the following section, the
impact of chromatin remodeling complexes on chondrogenesis under
both normal and pathological conditions of cartilage will be
discussed.
BRG1
BRG1 is an indispensable chromatin regulator that
engages in numerous cellular activities and plays a core role in
DNA replication, repair, recombination and transcriptional
regulation (90,91). The expression of fibroblast
growth factor receptor 3 (FGFR3) is essential for cartilage
development and can be induced by bone morphogenic protein (BMP) 2.
This induction is mediated by the downstream factor Sp1, which is
influenced by BRG1. BRG1 promotes FGFR3 expression by remodeling
chromatin regions that have Sp1-binding sites at the FGFR3
transcriptional initiation sites. This alteration enhances Sp1's
binding to the proximal promoter, which facilitates p300
recruitment and leads to modifications in the 'histone code',
including phosphorylation and methylation changes (92). The natural small molecule
spermidine has been reported to protect against and restore
cartilage damage in OA (93-95), and BRG1 mediates its protective
effects by enhancing osteoarthritic cartilage through the
Nrf2/KEAP1 and STAT3 signaling pathways (96).
BAF47
Studies have shown that BAF47 serves as a genetic
hallmark in malignant tumors, and its loss is associated with
extraskeletal myxoid chondrosarcoma (EMC) featuring rhabdoid
cytological characteristics. However, in one case of EMC exhibiting
high-grade features such as increased cellularity and prominent
nucleoli, with no rhabdoid features present, the expression of
nuclear BAF47 remained intact (97,98). Additionally, BAF47 levels are
reduced in synovial sarcoma, and this reduction can be assessed
using immunohistochemical staining for diagnostic purposes
(29,99). In addition, compared with other
atypical teratoid tumors where BAF47 is absent or lost, in
mesenchymal chondrosarcoma, the level of BAF47 is retained
(100). Therefore, BAF47 is an
excellent marker to diagnose different tumor types but the
mechanisms remain unclarified (101).
SS18
Mutations in the BAF subunit across human cancer
types display a notable tissue-specific pattern, with BAF subunits
mutated in >20% of all human cancers (102). Notably, almost all cases of
synovial sarcoma are attributed to the SS18-SSX fusion due to the
t(X;18) translocation, whereas mutations in the SS18 BAF subunit
are infrequently observed in other types of cancer (103,104). The underlying mechanism
involves the SS18-SSX gene fusion, which indirectly inactivates the
function of SMARCB1 by binding to the chromatin remodeling complex,
thereby promoting the transcription of the oncoprotein, SS10-SSX,
and contributing to the development of synovial sarcoma (104,105).
BAF155
BAF155 is a component of SWI/SNF chromatin
remodeling complexes, which help make DNA more accessible by
altering the arrangement of nucleosomes during gene transcription
(106). The targeted deletion
of BAF155 leads to a failure in the transcriptional activation of
Sonic hedgehog (Shh)/Gli target genes within limb buds that are
undergoing development, such as the Ptch1 receptor for Shh and its
downstream effector Gli1 in the posterior region of the limb bud.
This disruption of the Hh pathway is associated with aberrant BMP
activity and the initiation of chondrogenesis (107).
Chromatin remodeling complexes and
osteogenesis
Osteogenesis is the biological process through which
new bone is formed, involving the differentiation of MSCs into
osteoblasts (108). This
complex process is regulated by various signaling pathways and
factors, including hormones, growth factors, transcription factors
and ncRNAs, ensuring proper bone development and homeostasis
(109,110). Regulating MSC osteogenic
differentiation is essential for advancing bone tissue engineering
(111). Several subunits of
chromatin remodeling complexes play a pivotal role in osteogenesis,
making them valuable for applications in bone tissue engineering
(84,112). For example, overexpression of
CHD7 can interact with downstream factors of BMP signaling, such as
SMAD1, thereby enhancing the osteogenic potential of MSCs (84). Additionally, BAF180 influences
MSC osteogenic differentiation by modulating BMP/Smad signaling,
improving chromatin accessibility, and increasing binding at key
osteogenic genes, including Alpl, BmprIb, TgfbrII and Runx2, in
BMP2-treated MSCs (112).
Overall, a deeper understanding of how chromatin remodeling
complexes regulate MSC osteogenic differentiation can optimize the
ability of MSCs to differentiate into osteoblasts, thereby
facilitating their application in bone tissue engineering and the
treatment of bone-related diseases (84).
BRG1
BRG1, a core component of the SWI/SNF chromatin
remodeling complex that functions as an ATPase, has been identified
in developing skeletal structures of the mouse embryo and in ex
vivo osteoblast cultures (113). A study has demonstrated that
BRG1 expression is consistently present throughout skeletal
components and is positively correlated with the key osteogenic
regulatory protein, runt related transcription factor 2 (RUNX2)
(57). These observations
suggest that SWI/SNF chromatin remodeling activity is essential for
supporting RUNX2-dependent skeletal gene expression. RUNX2 is
crucial for initiating osteogenesis upstream of SP7 in the
regulatory hierarchy of osteoblast development, ensuring
independent and autonomous regulation in cartilage and bone
development (57,114). In the context of BRG1
depletion, the induction of alkaline phosphatase, an osteogenic
marker, is impaired in pre-osteoblasts (115). Additionally, another study
found that the knockout of BRG1 severely damages the ability of
bone cells to mineralize and to express key markers of osteoblast
differentiation, such as osteocalcin and alkaline phosphatase
(60). Osteocalcin itself plays
an important role in determining bone size, shape and strength
(116,117). Furthermore, BRG1-containing
SWI/SNF complexes stimulate basal tissue and vitamin D3-enhanced
osteocalcin promoter activity via the transcription factor,
CCAAT/enhancer-binding protein β. This factor, together with RUNX2,
forms a stable complex that facilitates RNA polymerase II binding
and the activation of osteocalcin gene transcription in
osteoblastic cells (118).
Moreover, p107, alongside retinoblastoma protein (pRB), plays a
distinct role in the induction of the osteogenic gene, Alpl, during
osteoblast differentiation, with p107 being crucial for the
efficient recruitment of the BRG1-SWI/SNF chromatin-remodeling
complex, necessary for Alpl activation (119).
BRM
Notably, unlike other subunits of the chromatin
remodeling complex, BRM-deficient preosteoblasts exhibit premature
expression of osteoblast differentiation markers suggesting an
enhancement of osteogenesis and subsequent bone formation (64,120).
BAF45a and BAF45d
In the BAF45 family, there are four homologs,
including BAF45a, BAF45b, BAF45c and BAF45d (68,121). Among these homologs, BAF45a and
BAF45d are more accessible during the induction of bone MSC
differentiation into osteoblasts compared with BAF45b and BAF45c,
as BAF45a and BAF45d are preferentially expressed in osteoblasts.
Notably, BAF45a serves as a vital subunit of the chromatin
regulatory complex and, through the bone tissue-specific
RUNX2-BAF45A-EZH2 molecular epigenetic axis, determines chromatin
accessibility during the early stage of commitment and
differentiation of mineralized cells (122). The deletion of BAF45a results
in decreased promoter accessibility for vital transcription factors
required for the induction and maturation of osteoblasts (15,122).
BAF180 (also known as PBRM1)
Mechanistically, BMP and osteogenic signaling
pathways induce the expression of BAF180 and PBAF in MSCs,
promoting ossification in vivo by affecting the activation
of SMAD1/5/8 through locus-specific epigenomic remodeling. This
involves PBRM1 bromodomains and the transcriptional regulation of
BMPR/TGFβRII, which together lead to the expression of
BMP-early-responsive genes and facilitate the progression of
osteogenesis (57,112).
BAF200 (also known as ARID2)
A study has suggested that BAF200 is crucial for
promoting osteoblast commitment and differentiation (123). BAF200 targets key osteogenic
markers, such as the Alpl promoter and Bglap, and in some cases,
significantly contributes to the recruitment of RNA polymerase II,
ultimately facilitating the commitment and differentiation of
osteoblasts. Conversely, the depletion of BAF200 expression during
the osteoblast differentiation process results in a failure to
mineralize into a calcified bone matrix (124,125).
BRD7
In addition to functioning alongside BAF180 and
BAF200 as components of the PBAF complex to promote osteogenesis
(112), BRD7 has also been
identified as a novel substrate of the APC/C-E3 ligase during the
cell cycle. This role may provide a therapeutic target for treating
osteosarcoma and serves as a crucial regulator for stabilizing and
suppressing tumor progression in osteosarcoma (126).
CHD1
As a member of the CHD family, CHD1 is required for
the induction of osteoblast-specific gene expression, extracellular
matrix (ECM) mineralization and ectopic bone formation in
vivo. CHD1 is closely associated with transcription and
nucleosome turnover downstream of the transcriptional start site
(TSS); it exhibits high occupancy around the TSS of
differentiation-activated genes, thereby supporting the
differentiation of MSCs into osteoblasts (2,75).
CHD7
SMAD1 and SP7 are core components of the canonical
BMP signaling pathway. Previous studies have elucidated that CHD7
is required for the osteogenic differentiation of human bone MSCs
by interacting with SMAD1 and binding to the enhancer region of SP7
to promote differentiation. By contrast, the absence of CHD7
impairs bone formation in vivo (80,84). Additionally, other studies have
demonstrated that depletion of CHD7 in MSCs and preosteoblasts
results in a phenotype characterized by low bone mass and
significantly increased marrow adiposity. This occurs through the
enhancement of the peroxisome proliferator-activated receptor
(PPAR) signaling associated with methylated H3K4 patterns, which
subsequently activates the transcription of downstream lipogenic
genes. This disruption of the balance between osteogenesis and
lipogenesis ultimately results in bone and fat disorder (127-129).
CHD9
CHD9, a master regulator of ribosomal gene
transcription in MSCs and osteogenesis, works synergistically with
BRG1 to upregulate the expression of RUNX2 (86,130). Additionally, CHD9 binds to
skeletal tissue-specific promoters associated with key genes
involved in osteoblast differentiation, including glycan,
core-binding factor subunit α1, collagen II, osteocalcin and myosin
(85).
INO80
The only subunit in the INO80/SWR family that
contributes to osteogenesis is INO80. INO80 interacts with WDR5 in
MSCs, regulates canonical Wnt activity and stabilizes the
expression of key osteogenic markers, including RUNX2, SP7,
collagen type I α1 chain and osteopontin, all of which promote
osteogenesis (131).
Chromatin remodeling complexes and
osteoclastogenesis
Osteoclastogenesis plays a crucial role in bone
development and bone diseases by regulating the resorption of bone
tissue, thereby maintaining the balance between bone formation and
degradation (132). On the
other side of the bone remodeling balance, osteoclastogenesis
requires chromatin remodeling complexes to exert their influence
and contribute to the overall process.
BAF250a (also known as ARID1a/OSA1)
The expression of BAF250a facilitates the
cooperation between chromatin and the transcription factor,
Jun/Fos, leading to the upregulation of sialic acid-binding Ig-like
lectin 15 in osteoclast precursors, thereby triggering the
differentiation of osteoclasts (16). Moreover, BAF250a plays a critical
role in safeguarding osteoclast fate canalization during the
proliferation-differentiation switch by facilitating the formation
of transcriptional condensates with coactivator BRD4 and
lineage-specifying transcription factor PU.1 at the Nfatc1
super-enhancer (133).
Additionally, the antagonistic interactions between the ARID1A-cBAF
and BRD9-ncBAF complexes, along with the dependency on coactivator
BRD4, highlight the intricate balance required for proper cell fate
canalization during osteoclastogenesis (133). Furthermore, a recent study has
shown that the expression of BAF250a is downregulated in
osteosarcoma compared with non-tumor tissues, suggesting its
potential as a novel prognostic and therapeutic marker (134,135).
BRD9
BRD9 is upregulated during receptor activator of
NF-κB ligand-induced osteoclast differentiation and suppresses
osteoclastogenesis by interacting with the transcription factor,
FOXP1. This interaction enhances STAT1 transcription by increasing
chromatin accessibility at the STAT1 promoter and enhancer regions,
subsequently activating IFN-β signaling, which serves as a negative
feedback mechanism for osteoclastogenesis. Conversely, the absence
of BRD9 expression can lead to debilitating bone diseases such as
osteoporosis, osteopetrosis, osteonecrosis and Paget's disease
(14).
Chromatin remodeling complexes and
angiogenesis
Angiogenesis is crucial for bone formation and
resorption, as blood vessels play a vital role in bone regeneration
and are influenced by aging and pathological conditions (136,137). Vascular endothelial growth
factor (VEGF), which is expressed by osteoprogenitors, hypertrophic
chondrocytes and osteoblast precursors, regulates angiogenesis
within the skeletal system (138). In addition, the
hypoxia-inducible factor (HIF)-1α pathway is essential for
maintaining bone homeostasis and angiogenesis, significantly
impacting the development of bone metabolic diseases (138). Sena et al (139) demonstrated that BRM and BRG1
enhance the hypoxic induction of HIF1α and HIF2α genes, along with
their target genes, by recruiting BRG1 complexes to gene promoters.
Additionally, the SWI/SNF subunits, BRM and BRG1, enhance
HIF-1-mediated target gene activation in an ATPase-dependent manner
and are recruited to the VEGF gene promoter under hypoxia (140). CHD4 also acts as a crucial
coactivator of HIF1 and HIF2, amplifying HIF-driven transcriptional
programs (141). Although there
is currently no direct research exploring the role of chromatin
remodeling complexes in angiogenesis during bone development and
related diseases, their influence on angiogenesis-related genes
suggests that they may also play a significant role in these
processes.
During the embryonic stage, angiogenesis serves a
crucial role in bone development by providing essential signaling
molecules, such as VEGF, and supporting osteoblasts and bone marrow
stromal cells. This process not only promotes bone formation but
also contributes to bone function, making it a key regulatory
factor in bone development (142). It has been reported that
BRG1-deficient mutants die by embryonic day 11.5 and show abnormal
angiogenesis in the yolk sac (143). BRG1 deletion also reduces
COUP-TFII expression in venous endothelial cells and causes
improper arterial marker expression in veins during embryonic
vascular development (144).
CHD4 also plays a key role in embryonic vasculature development by
suppressing plasmin activation, with its loss leading to increased
urokinase-type plasminogen activator receptor expression and
decreased thrombospondin-1, both critical for plasmin activation
(145). While chromatin
remodeling complexes play a crucial role in embryonic vascular
development, their influence on angiogenesis during bone formation
and its overall impact on bone development remain largely
unexplored. Further research is needed to deepen the understanding
of these mechanisms and their relevance to bone-related
diseases.
Role of chromatin remodeling complexes in
bone biological functions
Normal bone development and aging are regulated by
a complex interplay of biological and physiological processes that
work in concert to sustain skeletal health throughout the lifespan
(146). This section mainly
focuses on chromatin remodeling complexes in three notable aspects:
Proliferation, senescence and programmed cell death (Table I and Fig. 6), as they play crucial roles in
regulating bone cell growth, influencing bone density and quality,
affecting bone formation and repair processes as well as
maintaining the balance of bone tissue (147).
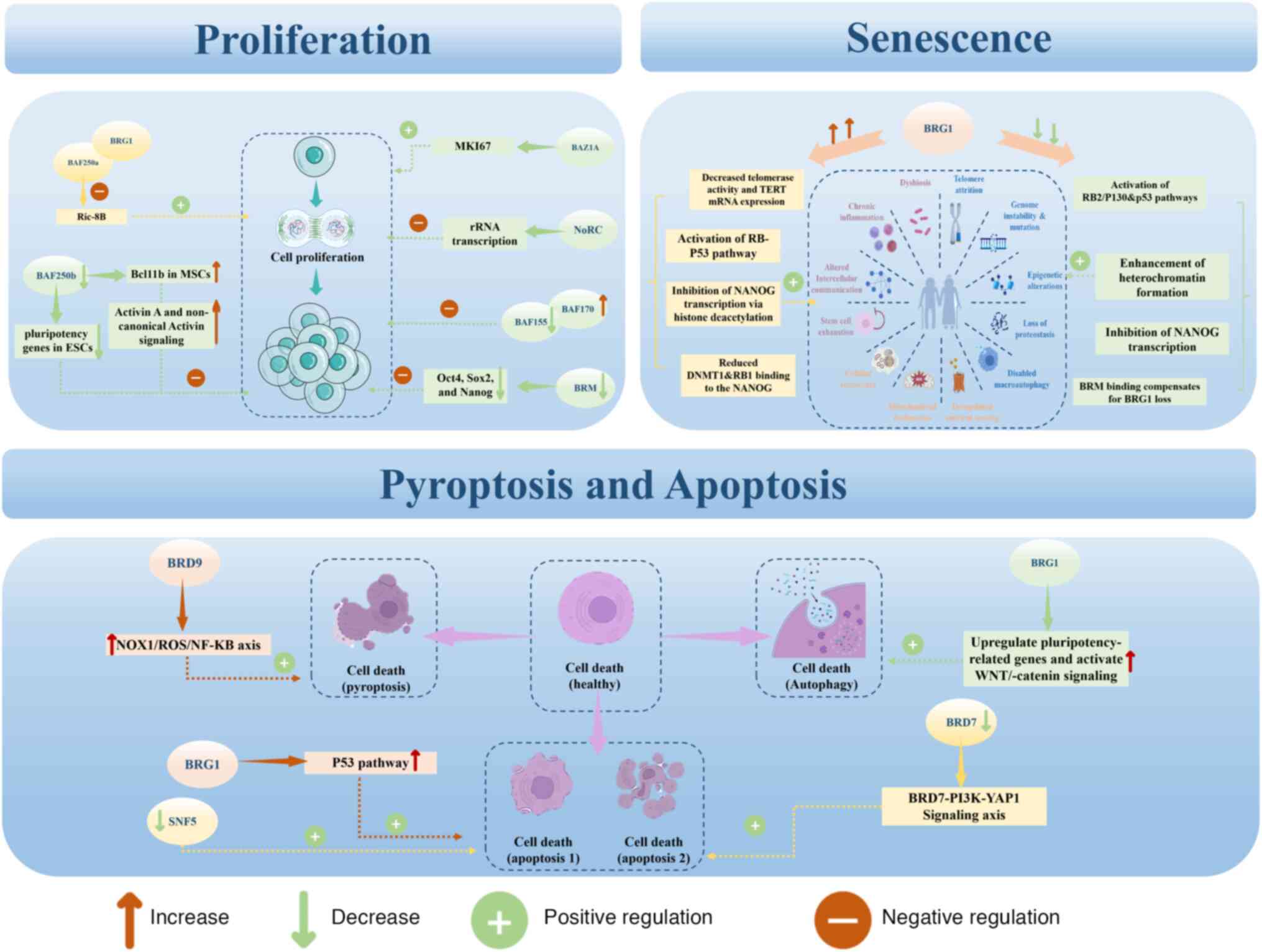 | Figure 6Epigenetic roles of chromatin
remodeling complexes in bone biological functions. There are three
other bone-related processes in which chromatin remodeling
complexes and their essential subunits are involved. This figure
demonstrates how each relevant subunit utilizes its distinct
molecular mechanism to achieve epigenetic modifications, both under
normal and pathological conditions. BAF, BRG1-associated factor;
Ric-8B, regulator of G-protein signaling 8B; BRG1, brahma-related
gene 1; Bcl11b, B-cell lymphoma/leukemia 11B; MKI67, marker of
proliferation Ki-67; BAZ1A, bromodomain-adjacent to Zinc finger
domain 1A; NoRC, nucleolar remodeling complex; Oct4,
octamer-binding transcription factor 4; Sox2, sex-determining
region Y-box 2; TERT, telomerase reverse transcriptase; DNMT1, DNA
methyltransferase 1; RB1, Retinoblastoma 1; BRD,
bromodomain-containing protein; NOX1, nicotinamide adenine
dinucleotide phosphate oxidase 1; ROS, reactive oxygen species;
NF-κB, nuclear factor-κB; YAP1, Yes-associated protein 1. |
Chromatin remodeling complexes and
proliferation
Bone formation and remodeling during development
are regulated by a network of transcription factors that facilitate
the transition from proliferation to differentiation in
osteoblastogenesis. Grandy et al (148) reported that the Ric-8B gene, a
pro-proliferative factor, is negatively regulated during osteoblast
differentiation by the transcription factor C/EBPb, which, along
with BRG1 and BRM, inhibits Ric-8B promoter activity. This
repression of Ric-8B expression is crucial as it influences
osteoblast proliferation and differentiation, ultimately impacting
bone development (148).
During bone development and remodeling, the proper
proliferation of MSCs and ESCs is essential for maintaining stem
cell pluripotency and regulating differentiation. However,
uncontrolled proliferation can lead to the loss of stem cell
function or abnormal differentiation. In this context, chromatin
remodeling complexes play a critical role in maintaining the
delicate balance between proliferation and differentiation. For
instance, the loss of BAF250b in MSCs results in the upregulation
of Bcl11b, which induces activin A and activates non-canonical
activin signaling through the pERK pathway, impairing MSC
quiescence and proliferation, thereby disrupting tissue homeostasis
(149). Similarly, the loss of
BRG1 initially leads to reduced proliferation, self-renewal and
altered colony morphology in ESCs. Long-term depletion results in
the complete loss of key ESC determinants, including OCT4, SOX2 and
NANOG (150). Additionally,
depletion of BAF155 in ESCs decreases proliferation and increases
cell death, while forced expression of BAF170 reduces the
competitive self-renewal ability of ESCs in immunocompromised mice
and impairs teratoma formation (150,151). Furthermore, biallelic
inactivation of BAF250b in ESCs leads to reduced proliferation,
downregulation of pluripotency genes, increased expression of
lineage-specific genes, including mesodermal differentiation
markers, and cell cycle abnormalities, along with downregulation of
the cell cycle regulator Cdc20 (152). Lastly, the nucleolar remodeling
complex (NoRC), assembled by the histone variant H2A.X at ribosomal
DNA (rDNA) promoters, recruits NoRC to rRNA genes, leading to the
formation of a heterochromatin-like structure that downregulates
rRNA transcription and ultimately reduces the proliferation rate of
ESCs (153). Therefore, these
findings highlight the intricate interplay between chromatin
remodeling complexes and transcription factors in regulating
proliferation, which is vital for proper bone development and
tissue homeostasis.
Chromatin remodeling complexes and
senescence
Cellular senescence, a pivotal aspect of biological
aging, significantly influences bone development and related
diseases (154). BRG1, serving
as the ATPase subunit of the SWI/SNF chromatin remodeling complex,
plays a crucial role in regulating cellular senescence in MSCs.
Specifically, prior research has indicated that BRG1 is upregulated
during the replicative senescence of MSCs, with forced BRG1
expression resulting in decreased telomerase activity and
telomerase reverse transcriptase mRNA expression, coupled with the
activation of RB- and p53-related pathways (155). Conversely, Alessio et al
(156) reported that
downregulation of BRG1 enhances senescence and heterochromatin
formation via the activation of the RB2/P130 and p53 pathways,
leading to reduced expression of stemness-related genes and
compromised cellular physiology. Despite these seemingly
inconsistent findings, the subsequent investigation by Squillaro
et al (157) provided
crucial insights, confirming that both forced upregulation and
silencing of BRG1 in MSCs trigger cellular senescence.
Specifically, in cells with silenced BRG1 expression, NANOG
transcription is inhibited by RB1 and/or RB2-mediated DNA
methyltransferase 1 (DNMT1) recruitment, causing methylation of the
NANOG promoter, with BRM binding serving as a compensatory
mechanism for the loss of BRG1. By contrast, in cells
overexpressing BRG1, NANOG transcription is mainly suppressed
through histone deacetylation, accompanied by reduced binding of
DNMT1 and RB1 to the promoter, while histone deacetylase 1 and RB2
remain bound at E2F sites (157). Given these intricate interplays
between BRG1 and cellular senescence in MSCs, it is evident that
the role of chromatin remodeling complexes, particularly the
SWI/SNF complex, in the senescence of osteoblasts, chondrocytes and
osteoclasts remains an area ripe for further exploration.
Chromatin remodeling complexes and
programmed cell death
Programmed cell death, including apoptosis,
pyroptosis, autophagy and ferroptosis, plays a crucial role in the
development, maintenance and repair of bone tissue (147). Apoptosis, a key form of
programmed cell death, is essential for normal cell turnover,
immune system function, embryonic development and the removal of
damaged cells, ensuring the proper regulation of cellular processes
(158). In bone development,
chromatin remodeling complexes have been shown to regulate
apoptosis, highlighting their critical role in maintaining cellular
homeostasis. For example, BRG1 induces cell cycle arrest and
apoptosis in MSCs, with p53 playing a central role in triggering
programmed cell death (155).
Similarly, SNF5 is vital for the survival of early embryonic cells,
and its loss leads to apoptotic cell death and extensive DNA
fragmentation (159). In the
context of bone degeneration, BRD7 expression is decreased in
severely degenerated NP tissues, and its knockdown promotes matrix
degradation and apoptosis. Upregulation of BRD7 reverses these
effects, enhancing matrix synthesis and reducing apoptosis through
the PI3K and YAP1 pathways (160). These findings suggest that
targeting the BRD7-PI3K-YAP1 axis, a chromatin remodeling pathway,
could offer a potential therapeutic approach for treating
intervertebral disc degeneration (IDD) by regulating apoptosis
pathways.
Pyroptosis, an inflammatory form of programmed cell
death typically triggered by inflammasomes and executed by
gasdermin proteins in response to danger signals and pathogen
infections, plays several crucial roles development, maintenance
and repair, such as in osteogenic processes (161). Chai et al (162) discovered that tumor necrosis
factor-α (TNF-α) induces pyroptosis in preosteoblastic MC3T3-E1
cells, negatively affecting osteogenic differentiation.
Additionally, inhibiting Caspase-1-mediated pyroptosis promotes
osteogenic differentiation, underscoring the complex relationship
between these cell death pathways and bone development (163). A recent study has also shown
that BRD9 contributes to bone degeneration through the regulation
of inflammatory pathways via regulating pyroptosis (164). BRD9 expression increases as IDD
progresses, and its inhibition reduces TNF-α-induced matrix
breakdown, reactive oxygen species (ROS) production and pyroptosis
in rat NP cells. BRD9 promotes IDD through the NADPH oxidase
1/ROS/NF-κB pathway, and in vivo inhibition of BRD9 helps
mitigate IDD progression in a rat model (164). These findings suggest that
targeting BRD9 and its chromatin remodeling functions may offer
potential treatments for IDD, linking chromatin remodeling
complexes and pyroptosis in bone development and degeneration.
Autophagy is a cellular process that facilitates
the removal and recycling of damaged or unnecessary components to
maintain bone homeostasis and function (165). A recent study has shown that
BRG1 plays a crucial role in regulating bone development by
upregulating pluripotency-related genes and activating pathways
such as WNT/β-catenin signaling and autophagy, with inhibition of
BRG1 leading to the opposite effect, suggesting an interconnected
role of chromatin remodeling and autophagy in bone development and
maintenance (166).
Role of chromatin remodeling complexes in
the pathogenesis of bone-related diseases
The role of chromatin remodeling complexes in the
pathogenesis of bone-related diseases is a critical area of study.
Dysregulation of these complexes can impact bone cell
differentiation, proliferation and apoptosis, resulting in a range
of bone-related diseases, such as bone tumors, osteoporosis and OA.
For example, low levels of BRD7 are associated with a poor
prognosis in patients with osteosarcoma, while upregulation of
BAF155 triggers OA (Table II).
Understanding the mechanisms by which chromatin remodeling
complexes contribute to these pathologies could reveal new
therapeutic targets for treating bone-related diseases.
 | Table IIKey chromatin remodeling complexes in
the pathogenesis of bone-related diseases. |
Table II
Key chromatin remodeling complexes in
the pathogenesis of bone-related diseases.
| Disease | Related
subunits | Alias | Family | Expression | Targets | Biological
function | (Refs.) |
|---|
| Ewing sarcoma | BRG1 | SMARCA4 | mSWI/SWF
complex | Up | EWS-FLI1 or
EWSR1 | Contributes to
Ewing sarcoma progression. | (65) |
| Ewing sarcoma | BAF155 | SMARCC1 | mSWI/SWF
complex | Up | EWS-FLI1 | Contributes to
Ewing sarcoma progression. | (65) |
| Ewing sarcoma | BAF250a | ARID1a/ OSA1 | mSWI/SWF
complex | Up | EWS-FLI1 | serves as an
interface between EWS-FLI1 and the BAF complex. | (177) |
| Ewing sarcoma | CHD4 | Mi-2β | CHD complex | Up | EWS-FLI1 | Promotes Ewing
sarcoma cell survival and its oncogenic gene expression
program. | (176) |
| Synovial
sarcoma | BAF47 | SMARCB1/ INI1 | mSWI/SWF
complex | Down | - | Synovial sarcoma
progression. | (29,99,104,105) |
| Synovial
sarcoma | SS18 | SSXT/SYT | mSWI/SWF
complex | Up | SS18-SSX
(t(X;18) | Synovial sarcoma
progression. | (103-105) |
| Chondrosarcoma | BRG1 | SMARCA4 | mSWI/SWF
complex | Up | esBAF complex | Chondrosarcoma
progression. | (62) |
| Chondrosarcoma | BRM | SMARCA2 | mSWI/SWF
complex | Up | esBAF complex | Chondrosarcoma
progression. | (62) |
| Extraskeletal
myxoid chondrosarcoma | BAF47 | SMARCB1/ INI1 | mSWI/SWF
complex | Down | - | - | (98) |
| Osteosarcoma | BAF47 | SMARCB1/ INI1 | mSWI/SWF
complex | Down | - | - | (69,184) |
| Osteosarcoma | BRD7 | - | mSWI/SWF
complex | Down | APC/C-E3
ligase | Stabilizes and
suppresses osteosarcoma progression. | (126) |
| Osteosarcoma | BAZ1A | ACF1 | ISWI complex | Up | - | Induces senescence
of osteosarcoma. | (198) |
| Osteosarcoma | BAF250a | ARID1a | mSWI/SWF
complex | Down | - | - | (134,135) |
| Osteosarcoma | BRG1 | SMARCA4 | mSWI/SWF
complex | Down | - | - | (134) |
| Poorly
differentiated chordoma | BAF47 | SMARCB1/ INI1 | mSWI/SWF
complex | Down | - | Binds with mSWI/
SNF complexes at different sites in the genome. | (183-185,190) |
| Skull base
chordomas | BAF180 | PBRM1 | mSWI/SWF
complex | Down | - | - | (191-193) |
| Osteoarthritis | BAF155 | SMARCC1 | mSWI/SWF
complex | Up | IL-1β in
chondrocytes | Silencing BAF155
impairs the pro-inflammatory response induced by IL-1β in
chondrocytes. | (200) |
| Osteoarthritis | BRD7 | - | mSWI/SWF
complex | Up | - | Causes ferroptosis
in osteoarthritis. | (201) |
| Bone-fat
disorder | CHD7 | - | CHD complex | Down | PPARγ signaling
with H3K4me patterns | Disrupts the
balance between osteogenesis and lipogenesis. | (127-129) |
| Adolescent
idiopathic scoliosis | CHD7 | - | CHD complex | Down | - | - | (205) |
| Osteoporosis | BRM | - | mSWI/SWF
complex | Up | Osteoblast
progenitors | BRM depletion
results in an increased proportion of cells that express markers
for osteoblast precursors and provides substantial resistance to
osteoporosis. | (64) |
| Osteoporosis | BRD9 | - | mSWI/SWF
complex | Down | Osteoblast
progenitors | BRD9 deficiency
promotes the specification of the osteoclast lineage and increases
bone resorption. | (14) |
| Postmenopausal
osteoporosis | CHD1 | - | CHD complex | Up | - | - | (210) |
Chromatin remodeling complexes and bone
tumors
Bone tumors refer to abnormal cellular
proliferation occurring in the bone or surrounding tissues (such as
cartilage and bone marrow) and can be classified as either benign
or malignant (10,167). Although malignant bone tumors
are rare [the annual incidence rate of osteosarcoma is 4-5
individuals per million (168)], the early symptoms are often
subtle, leading to misdiagnosis as 'growing pains' or arthritis,
which delays treatment (169-171). Common types of bone tumors
include benign tumors, such as chondroma and giant cell tumor of
the bone, the latter of which may undergo malignant transformation
(172). Malignant bone tumors
can be primary, including osteosarcoma (predominantly affecting
adolescents), chondrosarcoma (more common in middle-aged and
elderly individuals) and Ewing sarcoma (frequently seen in children
and adolescents) (173,174). Alternatively, malignant bone
tumors can be metastatic, originating from cancer types such as
lung or breast cancer and subsequently spreading to the bone
(175).
Chromatin remodeling complexes play a critical role
in regulating tumor cell proliferation, survival and metastasis
across various bone tumors. In Ewing sarcoma, for example, CHD4
depletion leads to increased DNA accessibility, which in turn
increases DNA damage (176).
Moreover, Ewing sarcoma is driven by the EWS-FLI1 fusion protein,
which alters gene expression through the prion-like domains of EWS
and ARID1A, facilitating the formation of biomolecular condensates
crucial for tumor progression. ARID1A condensates localize to
EWS/FLI1 target enhancers, driving long-range chromatin remodeling
and the formation of functional hubs at oncogenic genes (177). Selvanathan et al
(178) describe a novel
feed-forward cycle in Ewing sarcoma, where EWS-FLI1 promotes the
splicing of the ARID1A isoform protein variant ARID1A-L, driving
tumor growth, while ARID1A-L, in turn, stabilizes EWS-FLI1,
revealing potential targets for cancer-specific therapies.
Additionally, in the presence of EWS-FLI1, multimerization at GGAA
repeats and the prion-like domains of EWSR1 facilitate the
recruitment of BAF complexes to tumor-specific enhancers, driving
oncogenic gene expression (179). Autocrine neural EGFL like 2
(NELL2) signaling in Ewing sarcoma cells regulates BAF chromatin
remodeling complexes and promotes cell proliferation by inhibiting
Cdc42 and upregulating EWS-FLI1, with NELL2, CD133 and EWS-FLI1
positively regulating each other to enhance proliferative capacity
(180). Similarly, in synovial
sarcoma, the SS18-SSX fusion protein disrupts normal chromatin
remodeling, leading to tumorigenesis (181). Additionally, ncBAF complexes
uniquely localize to CTCF sites and promoters, acting as synthetic
lethal targets in synovial sarcoma, with BRD9 subunit depletion
effectively reducing cell proliferation (182). In chondrosarcoma, proline-rich
polypeptide-1 inhibits chondrosarcoma cancer stem cell
proliferation by targeting the SWI/SNF chromatin-remodeling
complex, reducing the expression of key oncogenic players such as
BRG, BAF170 and BRM (62). EMC
is characterized by alterations in BAF47, although the precise
mechanisms remain unclear (98,99). Osteosarcoma, the most prevalent
primary bone sarcoma, is associated with reduced levels of BAF47
and BAF250a, suggesting their potential involvement in tumor
progression (69,173,183-185). BRD7 also functions as a tumor
suppressor in osteosarcoma and is degraded by the APC/C complex
during the cell cycle. A BRD7 mutant resistant to APC/C degradation
more effectively suppresses tumor growth, suggesting that targeting
the APC/C-BRD7 pathway, along with APC/C inhibitors such as
proTAME, could provide a novel therapeutic approach for
osteosarcoma (126).
Additionally, in response to doxorubicin treatment in U2OS cells,
the INO80 complex regulates the removal of H2A.Z at the p21
promoter, facilitating p21 activation and contributing to the DNA
damage response in osteosarcoma (186). Another study uncovered that pRB
directly activates the osteoblast differentiation marker, Alpl, by
recruiting the BRG1-containing SWI/SNF chromatin-remodeling
complex, which displaces BRM-containing complexes, marking the
onset of differentiation and highlighting the critical role of pRB
in osteosarcoma susceptibility (187). Moreover, cohesin and PBAF play
a crucial role in suppressing gene transcription near DNA
double-strand breaks during interphase, and their absence results
in extensive genome alterations, potentially driving genomic
instability in osteosarcoma (188). The PHD domain of the KAP1
corepressor acts as an intramolecular E3 SUMO ligase, facilitating
sumoylation of the adjacent bromodomain, which is crucial for
KAP1-mediated gene silencing in osteosarcoma by recruiting SETDB1
and the CHD3/Mi2 NuRD complex, thereby enhancing histone
methyltransferase activity (189). Similarly, poorly differentiated
chordoma also exhibits BAF47 loss, implicating its role in tumor
biology (69,183,185,190). In skull base chordomas, BAF180
mutations are linked to poor prognosis, underscoring the importance
of chromatin remodeling complexes in determining tumor
aggressiveness (191-193).
Given their central role in tumor progression,
chromatin remodeling complexes present viable targets for
therapeutic intervention. Trabectedin, for instance, has been shown
to evict the SWI/SNF complex from chromatin in Ewing sarcoma,
redistributing EWS-FLI1 and altering histone modifications to
suppress tumor growth (194).
The loss of CHD4 in Ewing sarcoma not only delays tumor growth and
enhances overall survival but also, when combined with PARP
inhibition using olaparib, significantly boosts the antitumor
effects of genotoxic agents, positioning CHD4 as a promising
therapeutic target (171). In
synovial sarcoma, BRD9 inhibitors, such as FHD-609 (NCT04965753)
(195) and CFT8634
(NCT05355753), have been developed to degrade BRD9, effectively
impairing tumor proliferation (196,197). In osteosarcoma, BAZ1A and BRD7
have emerged as potential therapeutic targets, with BAZ1A
influencing senescence and BRD7 stabilizing tumor suppression
pathways (126,198). These findings suggest that
targeting specific chromatin remodeling subunits could provide
novel treatment strategies for sarcomas driven by epigenetic
dysregulation.
Several chromatin remodeling components are
frequently mutated or lost in bone and soft tissue sarcomas,
highlighting their diagnostic and prognostic significance. In
osteosarcoma, BRG1 mutations and the downregulation of BAF47 and
BAF250a suggest a disrupted chromatin landscape (69,173,183-185). Similarly, synovial sarcoma is
characterized by the SS18-SSX fusion, which affects SMARCB1
function (104,105). BAF47 loss is a hallmark of
poorly differentiated chordoma and EMC, whereas BAF180 mutations in
skull base chordomas are associated with poor prognosis (98,99,191-193). These genetic alterations could
serve as valuable biomarkers for the early detection, risk
stratification and treatment response monitoring, reinforcing the
clinical relevance of chromatin remodeling complexes in sarcoma
pathology.
Chromatin remodeling complexes and
OA
OA, the most common joint disorder primarily
affecting the aging population, is a leading musculoskeletal cause
of impaired mobility, with the majority of individuals >65 years
experiencing this condition (199). Wang et al (200) reported that interferon
regulatory factor 1 (IRF1) regulates BAF155 expression, playing a
crucial role in OA development, as knockdown of either IRF1 or
BAF155 alleviates OA-like symptoms and inflammatory responses.
Epigenetic modifications near the BAF155 promoter, mediated by GCN5
and SETD2, are key drivers of its expression, with upregulation of
these enzymes worsening OA symptoms (200). Additionally, a study found that
reduced levels of BRD7 activates the main immunodeficiency pathway.
By contrast, increased levels of BRD7 may activate the ECM receptor
interaction pathway and inhibit the primary immunodeficiency
pathway, ultimately leading to ferroptosis in OA (201).
Chromatin remodeling complexes and
bone-fat disorder
Bone marrow is the only tissue where bone and fat
are adjacent and collaborate in various processes such as metabolic
homeostasis, hemopoiesis and osteogenesis (202). Successful osteogenesis and
adipogenesis are tightly regulated processes that must maintain a
balance for proper skeletal and metabolic function. It has been
reported that PPARγ serves as a differentiation switch between
osteoblastogenesis and adipogenesis in the bone marrow, inhibiting
the process of osteoblastogenesis (128). Depletion of CHD7 in MSCs and
preosteoblasts disrupts this balance, leading to low bone mass and
increased marrow adiposity through enhanced PPARγ signaling and
trimethylated H3K4-mediated activation of lipogenic genes (127-129).
Chromatin remodeling complexes and
AIS
AIS is defined by a lateral curvature of the spine
(Cobb angle) of at least 10 degrees, occurring in the absence of
any underlying congenital or neuromuscular abnormalities (203). The Cobb angle is calculated by
drawing lines along the most tilted vertebrae at the curve's top
and bottom and measuring the intersection (204). Studies have found that genetic
variants of CHD7 play a vital role in AIS. According to a
comparison of genotype and allele frequency between patients with
AIS and healthy controls, rs121434341 of CHD7 is significantly
associated with AIS susceptibility (205). Meanwhile, the rs1017861
polymorphism in the CHD7 gene is associated with susceptibility
idiopathic scoliosis in Caucasian female patients, averaging
16.8±4.2 years old (range, 12.3-50.1). Additionally, the rs1017861
and rs4738813 polymorphisms are associated with higher curvature
severity and progression rate (206). Lower expression of CHD7 has
also been observed in patients with AIS compared with controls,
which may be associated with the etiology of aberrant bone mass in
AIS, but the pathogenetic mutation mechanism remains to be
determined (205,207).
Chromatin remodeling complexes and
osteoporosis
During senescence, the bone marrow has a reduced
number of MSCs and an impaired balance between osteogenesis and
osteoclastogenesis, leading to age-related osteoporosis (131,208,209). Therefore, the chromatin
remodeling complex that contributes to osteogenesis and
osteoclastogenesis may play a significant role in osteoporosis,
such as BRM, BRD9 and CHD1 (14,64,210). BRM plays a crucial role in
maintaining the balance of MSC lineage commitment between
adipocytes and osteoblasts. BRM-null mice show a marked resistance
to osteoporosis as the population of osteoblast progenitors is
enhanced at the expense of increased adipocyte differentiation
(64). Glucocorticoids, while
effective for treating inflammatory and autoimmune disorders, cause
bone loss by repressing osteogenic genes through the glucocorticoid
receptor (GR), particularly involving the BRM. A study has revealed
that BRM, unlike the essential ATPase BRG1, cooperates with the GR
to repress genes such as Bglap, Per3 and FasL, disrupting
osteoblast differentiation and promoting bone degradation (211). Moreover, BRD9 deficiency
enhances osteoclast commitment and accelerates the development of
osteoporosis (14).
Postmenopausal bone loss, related to estrogen deficiency, is the
predominant contributor to osteoporosis. Serum levels of CHD1 are
elevated during postmenopausal osteoporosis (PMOP) and may play a
role in pathogenesis. This elevation could potentially serve as an
effective marker for diagnosing PMOP (210).
Conclusions and perspectives
Chromatin remodeling complexes are crucial for
maintaining normal bone development and are implicated in various
bone-related diseases. The present review provides a detailed
examination of the diverse roles of these complexes, revealing
their functions as regulators, promoters, inhibitors and
stabilizers in bone biology. The present review emphasizes that
understanding the specific mechanisms by which chromatin remodeling
complexes influence bone homeostasis is essential for developing
targeted therapies. For instance, certain subunits of these
complexes may play critical roles in modulating gene expression
during osteoblast differentiation, and disruptions in their
function could lead to conditions such as osteoporosis or other
metabolic bone diseases. Identifying these specific subunits and
their interactions will be vital for designing interventions that
can restore normal bone function.
However, the significant gap in research regarding
the impact of aging on the functionality of chromatin remodeling
complexes in bone health should be noted. The few studies available
suggest that age-related changes in these complexes may contribute
to impaired bone regeneration and increased fracture risk. This
highlights the need for longitudinal studies that examine how the
activity and composition of chromatin remodeling complexes evolve
with age, and how these changes correlate with clinical outcomes in
bone health. Understanding these dynamics will be crucial for
developing strategies to mitigate the effects of aging on bone
integrity.
Furthermore, it is important to consider the
potential therapeutic implications of targeting chromatin
remodeling complexes. Given their central role in regulating gene
expression, pharmacological modulation of these complexes could
provide a novel approach to treating bone-related diseases. For
example, small molecules that enhance the activity of specific
chromatin remodeling complexes may promote osteoblast
differentiation and function, offering a new avenue for
osteoporosis treatment. Conversely, small molecule inhibitors that
target dysfunctional subunits of these complexes could help restore
normal gene expression patterns, thereby addressing the underlying
causes of metabolic bone diseases, such as using dBRD9 to treat
synovial sarcoma and trabectedin for Ewing sarcoma (194,196). Controlled lineage selection in
MSCs offers a promising strategy for cell engineering and tissue
regeneration. For instance, CHD7-silenced MSCs show comprised
osteogenic ability when cultured with a scaffold in vivo
(84). Since BRM remains
unchanged during normal osteogenesis, targeting it may enhance bone
formation, and developing BRM inhibitors holds promise as a
potential osteoporosis treatment (64). Targeting BRM, a key mediator of
GR-induced repression in osteogenesis, may also offer a therapeutic
strategy to counteract glucocorticoid-induced bone loss and
potentially aid in reversing age-related osteoporosis by relieving
repression on osteogenic genes (211). Furthermore, a study has shown
that transfection of MSCs with small interfering RNA targeting
INO80 results in impaired mineral deposition under osteogenic
induction conditions, and mice implanted with INO80-silenced MSCs
also exhibit reduced bone formation. This highlights the critical
role of the INO80 complex in MSC osteogenic differentiation and its
potential applications in clinical tissue engineering and
osteoporosis treatment (131).
BAF250a, another subunit, acts as a key link between EWs:Fli1 and
the BAF complex; its capacity to form condensates is crucial for
the dysregulated gene expression that fuels growth. As a result,
the absence of condensate-competent BAF250a significantly obstructs
tumor advancement, highlighting it as a possible therapeutic
target. Nonetheless, the transient nature of these interactions
complicates efforts to effectively target condensate formation,
posing challenges in creating suitable inhibitors (177). In clinical trials, some new
drugs are still under development and in the early phase such as
FHD-609, a heterobifunctional degrader of BRD9 that treats advanced
synovial sarcoma or SMARCB1-deficient tumors, which is in phase I
study (195). Moreover,
tazemetostat has demonstrated some efficacy in treating certain
SMARCB1-deficient tumors in a subset of patients. However, it still
faces limitations such as inconsistent efficacy, unclear
indications and ambiguous dosing guidelines (212). Therefore, the potential
therapeutic role of chromatin remodeling complexes in bone-related
diseases needs further studies and research.
Additionally, through the continuous exploration of
the assembly and 3D structures of chromatin remodeling complex
subunits, there is potential for the discovery and development of
specific chemical modulators that can optimize therapeutic
outcomes. Understanding the structural nuances of these complexes
will facilitate the design of targeted interventions that can more
effectively influence their activity in bone biology. By leveraging
structural biology techniques, such as X-ray crystallography and
cryo-electron microscopy, researchers can gain insights into the
conformational changes and interactions that govern the function of
these complexes. For example, a potent and selective BRD9
bromodomain inhibitor series was developed based on a novel
pyridinone-like scaffold, with crystallographic data guiding the
optimization of its potency for BRD9 (213). Moreover, the integration of
artificial intelligence in structural analysis presents a
transformative opportunity for advancing our understanding of
chromatin remodeling complexes. This approach can significantly
accelerate the discovery of small molecules that specifically
target the active or allosteric sites of chromatin remodeling
complexes, enhancing their therapeutic efficacy.
In conclusion, while there has been progress in
understanding the role of chromatin remodeling complexes in
bone-related diseases, our knowledge remains limited and
fragmented. We consider that utilizing advanced techniques, such as
CRISPR-based gene editing and high-throughput sequencing, will
allow for a more comprehensive exploration of these complexes and
their interactions within the bone microenvironment. Additionally,
integrating insights from systems biology could help elucidate the
complex regulatory networks involving chromatin remodeling in bone
biology. This deeper understanding could ultimately lead to
innovative therapeutic strategies aimed at enhancing bone health
and effectively treating degenerative bone conditions, thereby
improving patient outcomes and quality of life.
Supplementary Data
Availability of data and materials
Not applicable.
Authors' contributions
WW prepared the first draft of the manuscript; YW,
YN and CZ performed the literature search; WW and YC wrote and
edited the manuscript; WW and QY drew the figures; WS and QY
supervised and polished the manuscript. All authors read and
approved the final version of the manuscript. Data authentication
is not applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
lncRNAs
|
long non-coding RNAs
|
|
SWI/SNF
|
switch/sucrose-non-fermenting
|
|
ISWI
|
imitation switch
|
|
CHD
|
chromodomain-helicase-DNA binding
|
|
INO80/SWR
|
inositol requiring
80/SWi2/snf2-related 1
|
|
EWS-FLI1
|
Ewing sarcoma-friend leukemia
integration 1
|
|
EWSR1
|
Ewing sarcoma RNA binding protein
1
|
|
hESCs
|
human embryonic stem cells
|
|
FGFR3
|
fibroblast growth factor receptor
3
|
|
BMP
|
bone morphogenic protein
|
|
MSCs
|
mesenchymal stem cells
|
|
PBAF
|
polybromo BAF
|
|
TSS
|
transcriptional start site
|
|
PPAR
|
peroxisome proliferator-activated
receptor
|
|
IDD
|
intervertebral disc degeneration
|
|
NP
|
nucleus pulposus
|
|
OA
|
osteoarthritis
|
|
AIS
|
adolescent idiopathic scoliosis
|
|
PMOP
|
postmenopausal osteoporosis
|
Acknowledgments
Not applicable.
Funding
This research was supported by Sichuan Science and Technology
Program (grant nos. 2022YFS0609 and 2022NSFSC1368), Scientific
Research Foundation of Southwest Medical University (grant no.
2021ZKMS009) and Shenzhen Science and Technology Projects (grant
nos. JCYJ20210324103604013, JSGG20220831110400001 and
KCXFZ20230731093059012).
References
|
1
|
Florencio-Silva R, Sasso GR, Sasso-Cerri
E, Simões MJ and Cerri PS: Biology of bone tissue: Structure,
function, and factors that influence bone cells. Biomed Res Int.
2015:4217462015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee HR, Yang SJ, Choi HK, Kim JA and Oh
IH: The chromatin remodeling complex CHD1 regulates the primitive
state of mesenchymal stromal cells to control their stem cell
supporting activity. Stem Cells Dev. 30:363–373. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schini M, Vilaca T, Gossiel F, Salam S and
Eastell R: Bone turnover markers: Basic biology to clinical
applications. Endocr Rev. 44:417–473. 2023. View Article : Google Scholar :
|
|
4
|
Hadjidakis DJ and Androulakis II: Bone
remodeling. Ann N Y Acad Sci. 1092:385–396. 2006. View Article : Google Scholar
|
|
5
|
Liang J, Yi Q, Liu Y, Li J, Yang Z and Sun
W and Sun W: Recent advances of m6A methylation in skeletal system
disease. J Transl Med. 22:1532024. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lane NE: Epidemiology, etiology, and
diagnosis of osteoporosis. Am J Obstet Gynecol. 194(2 Suppl):
S3–S11. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stark Z and Savarirayan R: Osteopetrosis.
Orphanet J Rare Dis. 4:52009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng J, Yao Z, Xue L, Wang D and Tan Z:
The role of immune cells in modulating chronic inflammation and
osteonecrosis. Front Immunol. 13:10642452022. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Glyn-Jones S, Palmer AJ, Agricola R, Price
AJ, Vincent TL, Weinans H and Carr AJ: Osteoarthritis. Lancet.
386:376–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Choi JH and Ro JY: The 2020 WHO
classification of tumors of bone: An updated review. Adv Anat
Pathol. 28:119–138. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park-Min KH: Epigenetic regulation of bone
cells. Connect Tissue Res. 58:76–89. 2017. View Article : Google Scholar :
|
|
12
|
Zhang Y, Wang Q, Xue H, Guo Y, Wei S, Li
F, Gong L, Pan W and Jiang P: Epigenetic regulation of autophagy in
bone metabolism. Function (Oxf). 5:zqae0042024. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sikora M, Marycz K and Smieszek A: Small
and Long Non-coding RNAs as functional regulators of bone
homeostasis, acting alone or cooperatively. Mol Ther Nucleic Acids.
21:792–803. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Du J, Liu Y, Wu X, Sun J, Shi J, Zhang H,
Zheng A, Zhou M and Jiang X: BRD9-mediated chromatin remodeling
suppresses osteoclastogenesis through negative feedback mechanism.
Nat Commun. 14:14132023. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Busby T, Chen Y, Godfrey TC, Rehan M,
Wildman BJ, Smith CM and Hassan Q: Baf45a mediated chromatin
remodeling promotes transcriptional activation for osteogenesis and
odontogenesis. Front Endocrinol (Lausanne). 12:7633922022.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhang Y, Sun H, Huang F, Chen Y, Ding X,
Zhou C, Wu Y, Zhang Q, Ma X, Wang J, et al: The chromatin
remodeling factor Arid1a cooperates with Jun/Fos to promote
osteoclastogenesis by epigenetically upregulating Siglec15
expression. J Bone Miner Res. 39:775–790. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wei W, Tang X, Jiang N, Ni C, He H, Sun S,
Yu M, Yu C, Qiu M, Yan D, et al: Chromatin remodeler Znhit1
controls bone morphogenetic protein signaling in embryonic lung
tissue branching. J Biol Chem. 298:1024902022. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xue Y, Morris JL, Yang K, Fu Z, Zhu X,
Johnson F, Meehan B, Witkowski L, Yasmeen A, Golenar T, et al:
SMARCA4/2 loss inhibits chemotherapy-induced apoptosis by
restricting IP3R3-mediated Ca(2+) flux to mitochondria. Nat Commun.
12:54042021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bosch PJ, Fuller LC and Weiner JA: A
critical role for the nuclear protein Akirin2 in the formation of
mammalian muscle in vivo. Genesis. 57:e232862019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gamarra N and Narlikar GJ; Collaboration
through chromatin: motors of transcription and chromatin structure.
J Mol Biol. 433:1668762021. View Article : Google Scholar
|
|
21
|
El Hadidy N and Uversky VN: Intrinsic
disorder of the BAF complex: Roles in chromatin remodeling and
disease development. Int J Mol Sci. 20:52602019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Clapier CR and Cairns BR: The biology of
chromatin remodeling complexes. Annu Rev Biochem. 78:273–304. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fröb F and Wegner M: The role of chromatin
remodeling complexes in Schwann cell development. Glia.
68:1596–1603. 2020. View Article : Google Scholar
|
|
24
|
Han Y, Reyes AA, Malik S and He Y: Cryo-EM
structure of SWI/SNF complex bound to a nucleosome. Nature.
579:452–455. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vary JC Jr, Gangaraju VK, Qin J, Landel
CC, Kooperberg C, Bartholomew B and Tsukiyama T: Yeast Isw1p forms
two separable complexes in vivo. Mol Cell Biol. 23:80–91. 2003.
View Article : Google Scholar
|
|
26
|
Reyes AA, Marcum RD and He Y: Structure
and function of chromatin remodelers. J Mol Biol. 433:1669292021.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nacev BA, Jones KB, Intlekofer AM, Yu JSE,
Allis CD, Tap WD, Ladanyi M and Nielsen TO: The epigenomics of
sarcoma. Nat Rev Cancer. 20:608–623. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du W, Guo D and Du W: ATP-Dependent
chromatin remodeling complex in the lineage specification of
mesenchymal stem cells. Stem Cells Int. 2020:88397032020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wojcik J and Cooper K: Epigenetic
alterations in bone and soft tissue tumors. Adv Anat Pathol.
24:362–371. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chakraborty S, Sinha S and Sengupta A:
Emerging trends in chromatin remodeler plasticity in mesenchymal
stromal cell function. FASEB J. 35:e212342021. View Article : Google Scholar
|
|
31
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Clapier CR, Iwasa J, Cairns BR and
Peterson CL: Mechanisms of action and regulation of ATP-dependent
chromatin-remodelling complexes. Nat Rev Mol Cell Biol. 18:407–422.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tyagi M, Imam N, Verma K and Patel AK:
Chromatin remodelers: We are the drivers! Nucleus. 7:388–404. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Gong H, Wang P, Zhu Y, Peng H, Cui
Y, Li H, Liu J and Wang Z: The emerging role of ISWI chromatin
remodeling complexes in cancer. J Exp Clin Cancer Res. 40:3462021.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Centore RC, Sandoval GJ, Soares LMM,
Kadoch C and Chan HM: Mammalian SWI/SNF chromatin remodeling
complexes: Emerging mechanisms and therapeutic strategies. Trends
Genet. 36:936–950. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Willhoft O and Wigley DB: INO80 and SWR1
complexes: The non-identical twins of chromatin remodelling. Curr
Opin Struct Biol. 61:50–58. 2020. View Article : Google Scholar :
|
|
37
|
Pulice JL and Kadoch C: Composition and
function of mammalian SWI/SNF chromatin remodeling complexes in
human disease. Cold Spring Harb Symp Quant Biol. 81:53–60. 2016.
View Article : Google Scholar
|
|
38
|
Judd J, Duarte FM and Lis JT: Pioneer-like
factor GAF cooperates with PBAP (SWI/SNF) and NURF (ISWI) to
regulate transcription. Genes Dev. 35:147–156. 2021. View Article : Google Scholar :
|
|
39
|
Poli J, Gasser SM and Papamichos-Chronakis
M: The INO80 remodeller in transcription, replication and repair.
Philos Trans R Soc Lond B Biol Sci. 372:201602902017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Trujillo JT, Long J, Aboelnour E, Ogas J
and Wisecaver JH: CHD Chromatin Remodeling Protein Diversification
Yields Novel Clades and Domains Absent in Classic Model Organisms.
Genome Biol Evol. 14:evac0662022. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Iyer J, Gentry LK, Bergwell M, Smith A,
Guagliardo S, Kropp PA, Sankaralingam P, Liu Y, Spooner E, Bowerman
B and O'Connell KF: The chromatin remodeling protein CHD-1 and the
EFL-1/DPL-1 transcription factor cooperatively down regulate CDK-2
to control SAS-6 levels and centriole number. PLoS Genet.
18:e10097992022. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ebbert R, Birkmann A and Schüller HJ: The
product of the SNF2/SWI2 paralogue INO80 of Saccharomyces
cerevisiae required for efficient expression of various yeast
structural genes is part of a high-molecular-weight protein
complex. Mol Microbiol. 32:741–751. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
van Attikum H, Fritsch O and Gasser SM:
Distinct roles for SWR1 and INO80 chromatin remodeling complexes at
chromosomal double-strand breaks. EMBO J. 26:4113–4125. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Conaway RC and Conaway JW: The INO80
chromatin remodeling complex in transcription, replication and
repair. Trends Biochem Sci. 34:71–77. 2009. View Article : Google Scholar
|
|
45
|
Saha A, Wittmeyer J and Cairns BR:
Mechanisms for nucleosome movement by ATP-dependent chromatin
remodeling complexes. Results Probl Cell Differ. 41:127–148. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Barisic D, Stadler MB, Iurlaro M and
Schübeler D: Mammalian ISWI and SWI/SNF selectively mediate binding
of distinct transcription factors. Nature. 569:136–140. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wang Y, He L, Du Y, Zhu P, Huang G, Luo J,
Yan X, Ye B, Li C, Xia P, et al: The long noncoding RNA lncTCF7
promotes self-renewal of human liver cancer stem cells through
activation of Wnt signaling. Cell Stem Cell. 16:413–425. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang Y, Wang J, Lian Y, Fan C, Zhang P, Wu
Y, Li X, Xiong F, Li X, Li G, et al: Linking long non-coding RNAs
and SWI/SNF complexes to chromatin remodeling in cancer. Mol
Cancer. 16:422017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Patty BJ and Hainer SJ: Non-Coding RNAs
and nucleosome remodeling complexes: An intricate regulatory
relationship. Biology (Basel). 9:2132020.PubMed/NCBI
|
|
50
|
Han P, Li W, Lin CH, Yang J, Shang C,
Nuernberg ST, Jin KK, Xu W, Lin CY, Lin CJ, et al: A long noncoding
RNA protects the heart from pathological hypertrophy. Nature.
514:102–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Atala A: Re: The long noncoding RNA
SChLAP1 promotes aggressive prostate cancer and antagonizes the
SWI/SNF complex. J Urol. 192:6132014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang Z, Mameri A, Cattoglio C, Lachance C,
Florez Ariza AJ, Luo J, Humbert J, Sudarshan D, Banerjea A, Galloy
M, et al: Structural insights into the human NuA4/TIP60
acetyltransferase and chromatin remodeling complex. Science.
385:eadl58162024. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tallant C, Valentini E, Fedorov O,
Overvoorde L, Ferguson FM, Filippakopoulos P, Svergun DI, Knapp S
and Ciulli A: Molecular basis of histone tail recognition by human
TIP5 PHD finger and bromodomain of the chromatin remodeling complex
NoRC. Structure. 23:80–92. 2015. View Article : Google Scholar :
|
|
54
|
Charles GM, Chen C, Shih SC, Collins SR,
Beltrao P, Zhang X, Sharma T, Tan S, Burlingame AL, Krogan NJ, et
al: Site-specific acetylation mark on an essential
chromatin-remodeling complex promotes resistance to replication
stress. Proc Natl Acad Sci USA. 108:10620–10625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Sala A, La Rocca G, Burgio G, Kotova E, Di
Gesù D, Collesano M, Ingrassia AM, Tulin AV and Corona DF: The
nucleosome-remodeling ATPase ISWI is regulated by
poly-ADP-ribosylation. PLoS Biol. 6:e2522008. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Bure IV and Nemtsova MV: Mutual regulation
of ncRNAs and chromatin remodeling complexes in normal and
pathological conditions. Int J Mol Sci. 24:78482023. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Young DW, Pratap J, Javed A, Weiner B,
Ohkawa Y, van Wijnen A, Montecino M, Stein GS, Stein JL, Imbalzano
AN and Lian JB: SWI/SNF chromatin remodeling complex is obligatory
for BMP2-induced, Runx2-dependent skeletal gene expression that
controls osteoblast differentiation. J Cell Biochem. 94:720–730.
2005. View Article : Google Scholar
|
|
58
|
Kuhn NZ and Tuan RS: Regulation of
stemness and stem cell niche of mesenchymal stem cells:
Implications in tumorigenesis and metastasis. J Cell Physiol.
222:268–277. 2010. View Article : Google Scholar
|
|
59
|
Im GI and Shin KJ: Epigenetic approaches
to regeneration of bone and cartilage from stem cells. Expert Opin
Biol Ther. 15:181–193. 2015. View Article : Google Scholar
|
|
60
|
Flowers S, Nagl NG Jr, Beck GR Jr and
Moran E: Antagonistic roles for BRM and BRG1 SWI/SNF complexes in
differentiation. J Biol Chem. 284:10067–10075. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Xu Y, Zhang J and Chen X: The activity of
p53 is differentially regulated by Brm- and Brg1-containing SWI/SNF
chromatin remodeling complexes. J Biol Chem. 282:37429–37435. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Moran A, Hoyt A, Sedani A, Granger C,
Saigh S, Blonska M, Zhao-Ju L, Conway SA, Pretell J, Brown J and
Galoian K: Proline-rich polypeptide-1 decreases cancer stem cell
population by targeting BAFF chromatin-remodeling complexes in
human chondrosarcoma JJ012 cells. Oncol Rep. 44:393–403. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kidder BL, Palmer S and Knott JG:
SWI/SNF-Brg1 regulates self-renewal and occupies core
pluripotency-related genes in embryonic stem cells. Stem Cells.
27:317–328. 2009. View Article : Google Scholar
|
|
64
|
Nguyen KH, Xu F, Flowers S, Williams EA,
Fritton JC and Moran E: SWI/SNF-mediated lineage determination in
mesenchymal stem cells confers resistance to osteoporosis. Stem
Cells. 33:3028–3038. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Kitagawa T, Kobayashi D, Baron B, Okita H,
Miyamoto T, Takai R, Paudel D, Ohta T, Asaoka Y, Tokunaga M, et al:
AT-hook DNA-binding motif-containing protein one knockdown
downregulates EWS-FLI1 transcriptional activity in Ewing's sarcoma
cells. PLoS One. 17:e02690772022. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Wang X, Lee RS, Alver BH, Haswell JR, Wang
S, Mieczkowski J, Drier Y, Gillespie SM, Archer TC, Wu JN, et al:
SMARCB1-mediated SWI/SNF complex function is essential for enhancer
regulation. Nat Genet. 49:289–295. 2017. View Article : Google Scholar :
|
|
67
|
Nakayama RT, Pulice JL, Valencia AM,
McBride MJ, McKenzie ZM, Gillespie MA, Ku WL, Teng M, Cui K,
Williams RT, et al: SMARCB1 is required for widespread BAF
complex-mediated activation of enhancers and bivalent promoters.
Nat Genet. 49:1613–1623. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang H, Wang X, Li J, Shi R and Ye Y: BAF
Complex in embryonic stem cells and early embryonic development.
Stem Cells Int. 2021:66688662021. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Antonelli M, Raso A, Mascelli S, Gessi M,
Nozza P, Coli A, Gardiman MP, Arcella A, Massimino M, Buttarelli FR
and Giangaspero F: SMARCB1/INI1 involvement in pediatric chordoma:
A mutational and immunohistochemical analysis. Am J Surg Pathol.
41:56–61. 2017. View Article : Google Scholar
|
|
70
|
Zhang X, Li B, Li W, Ma L, Zheng D, Li L,
Yang W, Chu M, Chen W, Mailman RB, et al: Transcriptional
repression by the BRG1-SWI/SNF complex affects the pluripotency of
human embryonic stem cells. Stem Cell Reports. 3:460–474. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Gatchalian J, Malik S, Ho J, Lee DS, Kelso
TWR, Shokhirev MN, Dixon JR and Hargreaves DC: A non-canonical
BRD9-containing BAF chromatin remodeling complex regulates naive
pluripotency in mouse embryonic stem cells. Nat Commun. 9:51392018.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Wang X, Song C, Ye Y, Gu Y, Li X, Chen P,
Leng D, Xiao J, Wu H, Xie S, et al: BRD9-mediated control of the
TGF-β/Activin/Nodal pathway regulates self-renewal and
differentiation of human embryonic stem cells and progression of
cancer cells. Nucleic Acids Res. 51:11634–11651. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sevinç K, Sevinç GG, Cavga AD, Philpott M,
Kelekçi S, Can H, Cribbs AP, Yıldız AB, Yılmaz A, Ayar ES, et al:
BRD9-containing non-canonical BAF complex maintains somatic cell
transcriptome and acts as a barrier to human reprogramming. Stem
Cell Reports. 17:2629–2642. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Gaspar-Maia A, Alajem A, Polesso F,
Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT,
Plath K, Meshorer E and Ramalho-Santos M: Chd1 regulates open
chromatin and pluripotency of embryonic stem cells. Nature.
460:863–868. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Baumgart SJ, Najafova Z, Hossan T, Xie W,
Nagarajan S, Kari V, Ditzel N, Kassem M and Johnsen SA: CHD1
regulates cell fate determination by activation of
differentiation-induced genes. Nucleic Acids Res. 45:7722–7735.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Bulut-Karslioglu A, Jin H, Kim YK, Cho B,
Guzman-Ayala M, Williamson AJK, Hejna M, Stötzel M, Whetton AD,
Song JS and Ramalho-Santos M: Chd1 protects genome integrity at
promoters to sustain hypertranscription in embryonic stem cells.
Nat Commun. 12:48592021. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Caplan AI: Mesenchymal stem cells. J
Orthop Res. 9:641–650. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Benayahu D, Shacham N and Shur I: Insights
on the functional role of chromatin remodelers in osteogenic cells.
Crit Rev Eukaryot Gene Expr. 17:103–113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Suzuki S, Nozawa Y, Tsukamoto S, Kaneko T,
Manabe I, Imai H and Minami N: CHD1 acts via the Hmgpi pathway to
regulate mouse early embryogenesis. Development. 142:2375–2384.
2015.PubMed/NCBI
|
|
80
|
Liu C, Kang N, Guo Y and Gong P: Advances
in chromodomain helicase DNA-binding (CHD) proteins regulating stem
cell differentiation and human diseases. Front Cell Dev Biol.
9:7102032021. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Wan M, Liang J, Xiong Y, Shi F, Zhang Y,
Lu W, He Q, Yang D, Chen R, Liu D, et al: The trithorax group
protein Ash2l is essential for pluripotency and maintaining open
chromatin in embryonic stem cells. J Biol Chem. 288:5039–5048.
2013. View Article : Google Scholar :
|
|
82
|
Yang P, Oldfield A, Kim T, Yang A, Yang
JYH and Ho JWK: Integrative analysis identifies co-dependent gene
expression regulation of BRG1 and CHD7 at distal regulatory sites
in embryonic stem cells. Bioinformatics. 33:1916–1920. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Malla S, Martinez-Gamero C, Kumari K,
Achour C, Mermelekas G, Martinez-Delgado D, Coego A, Guallar D,
Roman AC and Aguilo F: Cooperative role of LSD1 and CHD7 in
regulating differentiation of mouse embryonic stem cells. Sci Rep.
14:284952024. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chen Y, Wang M, Chen D, Wang J and Kang N:
Chromatin remodeling enzyme CHD7 is necessary for osteogenesis of
human mesenchymal stem cells. Biochem Biophys Res Commun.
478:1588–1593. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Yoo H, La H, Lee EJ, Choi HJ, Oh J, Thang
NX and Hong K: ATP-dependent chromatin remodeler CHD9 controls the
proliferation of embryonic stem cells in a cell culture
condition-dependent manner. Biology (Basel). 9:4282020.PubMed/NCBI
|
|
86
|
Salomon-Kent R, Marom R, John S, Dundr M,
Schiltz LR, Gutierrez J, Workman J, Benayahu D and Hager GL: New
face for chromatin-related mesenchymal modulator: n-CHD9 localizes
to nucleoli and interacts with ribosomal genes. J Cell Physiol.
230:2270–2280. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Wang L, Du Y, Ward JM, Shimbo T, Lackford
B, Zheng X, Miao YL, Zhou B, Han L, Fargo DC, et al: INO80
facilitates pluripotency gene activation in embryonic stem cell
self-renewal, reprogramming, and blastocyst development. Cell Stem
Cell. 14:575–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Furumatsu T and Ozaki T: Epigenetic
regulation in chondrogenesis. Acta Med Okayama. 64:155–161.
2010.PubMed/NCBI
|
|
89
|
Berendsen AD and Olsen BR: Bone
development. Bone. 80:14–18. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Mashtalir N, D'Avino AR, Michel BC, Luo J,
Pan J, Otto JE, Zullow HJ, McKenzie ZM, Kubiak RL, St Pierre R, et
al: Modular organization and assembly of SWI/SNF family chromatin
remodeling complexes. Cell. 175:1272–1288.e20. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
You S, Zhang Y, Xu J, Qian H, Wu S, Wu B,
Lu S, Sun Y and Zhang N: The role of BRG1 in antioxidant and redox
signaling. Oxid Med Cell Longev. 2020:60956732020. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Sun F, Chen Q, Yang S, Pan Q, Ma J, Wan Y,
Chang CH and Hong A: Remodeling of chromatin structure within the
promoter is important for bmp-2-induced fgfr3 expression. Nucleic
Acids Res. 37:3897–3911. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sacitharan PK, Lwin S, Gharios GB and
Edwards JR: Spermidine restores dysregulated autophagy and
polyamine synthesis in aged and osteoarthritic chondrocytes via
EP300. Exp Mol Med. 50:1–10. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chen Z, Lin CX, Song B, Li CC, Qiu JX, Li
SX, Lin SP, Luo WQ, Fu Y, Fang GB, et al: Spermidine activates RIP1
deubiquitination to inhibit TNF-α-induced NF-κB/p65 signaling
pathway in osteoarthritis. Cell Death Dis. 11:5032020. View Article : Google Scholar
|
|
95
|
Guo X, Feng X, Yang Y, Zhang H and Bai L:
Spermidine attenuates chondrocyte inflammation and cellular
pyroptosis through the AhR/NF-κB axis and the NLRP3/caspase-1/GSDMD
pathway. Front Immunol. 15:14627772024. View Article : Google Scholar
|
|
96
|
Mao X, Yan B, Chen H, Lai P and Ma J: BRG1
mediates protective ability of spermidine to ameliorate
osteoarthritic cartilage by Nrf2/KEAP1 and STAT3 signaling pathway.
Int Immunopharmacol. 122:1105932023. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Wei S, Pei J, von Mehren M, Abraham JA,
Patchefsky AS and Cooper HS: SMARCA2-NR4A3 is a novel fusion gene
of extraskeletal myxoid chondrosarcoma identified by RNA
next-generation sequencing. Genes Chromosomes Cancer. 60:709–712.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Schaefer IM and Hornick JL: SWI/SNF
complex-deficient soft tissue neoplasms: An update. Semin Diagn
Pathol. 38:222–231. 2021. View Article : Google Scholar :
|
|
99
|
Rekhi B and Vogel U: Utility of
characteristic 'Weak to Absent' INI1/SMARCB1/BAF47 expression in
diagnosis of synovial sarcomas. Apmis. 123:618–628. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Fanburg-Smith JC, Auerbach A, Marwaha JS,
Wang Z, Santi M, Judkins AR and Rushing EJ: Immunoprofile of
mesenchymal chondrosarcoma: Aberrant desmin and EMA expression,
retention of INI1, and negative estrogen receptor in 22
female-predominant central nervous system and musculoskeletal
cases. Ann Diagn Pathol. 14:8–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Kohashi K, Oda Y, Yamamoto H, Tamiya S,
Matono H, Iwamoto Y, Taguchi T and Tsuneyoshi M: Reduced expression
of SMARCB1/INI1 protein in synovial sarcoma. Mod Pathol.
23:981–990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Shain AH and Pollack JR: The spectrum of
SWI/SNF mutations, ubiquitous in human cancers. PLoS One.
8:e551192013. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Kadoch C, Williams RT, Calarco JP, Miller
EL, Weber CM, Braun SM, Pulice JL, Chory EJ and Crabtree GR:
Dynamics of BAF-Polycomb complex opposition on heterochromatin in
normal and oncogenic states. Nat Genet. 49:213–222. 2017.
View Article : Google Scholar
|
|
104
|
Cheng Y, Shen Z, Gao Y, Chen F, Xu H, Mo
Q, Chu X, Peng CL, McKenzie TT, Palacios BE, et al: Phase
transition and remodeling complex assembly are important for
SS18-SSX oncogenic activity in synovial sarcomas. Nat Commun.
13:27242022. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
McBride MJ, Pulice JL, Beird HC, Ingram
DR, D'Avino AR, Shern JF, Charville GW, Hornick JL, Nakayama RT,
Garcia-Rivera EM, et al: The SS18-SSX fusion oncoprotein hijacks
BAF complex targeting and function to drive synovial sarcoma.
Cancer Cell. 33:1128–1141.e7. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Zhang N, Zhang Y, Chen Y, Qian H, Wu B, Lu
S, You S, Xu W, Zou Y, Huang X, et al: BAF155 promotes cardiac
hypertrophy and fibrosis through inhibition of WWP2-mediated PARP1
ubiquitination. Cell Discov. 9:462023. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Jeon S and Seong RH: Anteroposterior limb
skeletal patterning requires the bifunctional action of SWI/SNF
chromatin remodeling complex in hedgehog pathway. PLoS Genet.
12:e10059152016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Ju C, Liu R, Zhang YW, Zhang Y, Zhou R,
Sun J, Lv XB and Zhang Z: Mesenchymal stem cell-associated lncRNA
in osteogenic differentiation. Biomed Pharmacother. 115:1089122019.
View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Peng Y, Jiang H and Zuo HD: Factors
affecting osteogenesis and chondrogenic differentiation of
mesenchymal stem cells in osteoarthritis. World J Stem Cells.
15:548–560. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Toosi S and Behravan J: Osteogenesis and
bone remodeling: A focus on growth factors and bioactive peptides.
Biofactors. 46:326–340. 2020. View Article : Google Scholar
|
|
111
|
Gou Y, Huang Y, Luo W, Li Y, Zhao P, Zhong
J, Dong X, Guo M, Li A, Hao A, et al: Adipose-derived mesenchymal
stem cells (MSCs) are a superior cell source for bone tissue
engineering. Bioact Mater. 34:51–63. 2023.
|
|
112
|
Sinha S, Biswas M, Chatterjee SS, Kumar S
and Sengupta A: Pbrm1 steers mesenchymal stromal cell osteolineage
differentiation by integrating PBAF-dependent chromatin remodeling
and BMP/TGF-β signaling. Cell Rep. 31:1075702020. View Article : Google Scholar
|
|
113
|
Mardinian K, Adashek JJ, Botta GP, Kato S
and Kurzrock R: SMARCA4: Implications of an altered
chromatin-remodeling gene for cancer development and therapy. Mol
Cancer Ther. 20:2341–2351. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hojo H, Saito T, He X, Guo Q, Onodera S,
Azuma T, Koebis M, Nakao K, Aiba A, Seki M, et al: Runx2 regulates
chromatin accessibility to direct the osteoblast program at
neonatal stages. Cell Rep. 40:1113152022. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nagl NG Jr, Patsialou A, Haines DS, Dallas
PB, Beck GR Jr and Moran E: The p270 (ARID1A/SMARCF1) subunit of
mammalian SWI/SNF-related complexes is essential for normal cell
cycle arrest. Cancer Res. 65:9236–9244. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Bailey S, Karsenty G, Gundberg C and
Vashishth D: Osteocalcin and osteopontin influence bone morphology
and mechanical properties. Ann N Y Acad Sci. 1409:79–84. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Komori T: Functions of osteocalcin in
bone, pancreas, testis, and muscle. Int J Mol Sci. 21:75132020.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Villagra A, Cruzat F, Carvallo L, Paredes
R, Olate J, van Wijnen AJ, Stein GS, Lian JB, Stein JL, Imbalzano
AN and Montecino M: Chromatin remodeling and transcriptional
activity of the bone-specific osteocalcin gene require
CCAAT/enhancer-binding protein beta-dependent recruitment of
SWI/SNF activity. J Biol Chem. 281:22695–22706. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Flowers S, Patel PJ, Gleicher S, Amer K,
Himelman E, Goel S and Moran E: p107-Dependent recruitment of
SWI/SNF to the alkaline phosphatase promoter during osteoblast
differentiation. Bone. 69:47–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Reyes JC, Barra J, Muchardt C, Camus A,
Babinet C and Yaniv M: Altered control of cellular proliferation in
the absence of mammalian brahma (SNF2alpha). EMBO J. 17:6979–6991.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Middeljans E, Wan X, Jansen PW, Sharma V,
Stunnenberg HG and Logie C: SS18 together with animal-specific
factors defines human BAF-type SWI/SNF complexes. PLoS One.
7:e338342012. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Godfrey TC, Wildman BJ, Javed A, Lengner
CJ and Hassan MQ: Epigenetic remodeling and modification to
preserve skeletogenesis in vivo. Connect Tissue Res. 59(supp1):
S52–S54. 2018. View Article : Google Scholar
|
|
123
|
Hasenfratz M, Mellert K, Marienfeld R, von
Baer A, Schultheiss M, Roitman PD, Aponte-Tinao LA, Lehner B,
Möller P, Mechtersheimer G and Barth TFE: Profiling of three
H3F3A-mutated and denosumab-treated giant cell tumors of bone
points to diverging pathways during progression and malignant
transformation. Sci Rep. 11:57092021. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Xu F, Flowers S and Moran E: Essential
role of ARID2 protein-containing SWI/SNF complex in tissue-specific
gene expression. J Biol Chem. 287:5033–5041. 2012. View Article : Google Scholar :
|
|
125
|
Wilsker D, Patsialou A, Dallas PB and
Moran E: ARID proteins: A diverse family of DNA binding proteins
implicated in the control of cell growth, differentiation, and
development. Cell Growth Differ. 13:95–106. 2002.PubMed/NCBI
|
|
126
|
Hu K, Liao D, Wu W, Han AJ, Shi HJ, Wang
F, Wang X, Zhong L, Duan T, Wu Y, et al: Targeting the
anaphase-promoting complex/cyclosome (APC/C)-bromodomain containing
7 (BRD7) pathway for human osteosarcoma. Oncotarget. 5:3088–3100.
2014. View Article : Google Scholar :
|
|
127
|
Liu C, Xiong Q, Li Q, Lin W, Jiang S,
Zhang D, Wang Y, Duan X, Gong P and Kang N: CHD7 regulates bone-fat
balance by suppressing PPAR-γ signaling. Nat Commun. 13:19892022.
View Article : Google Scholar
|
|
128
|
Takada I, Yogiashi Y and Kato S: Signaling
crosstalk between PPARγ and BMP2 in mesenchymal stem cells. PPAR
Res. 2012:6071412012. View Article : Google Scholar
|
|
129
|
Schnetz MP, Bartels CF, Shastri K,
Balasubramanian D, Zentner GE, Balaji R, Zhang X, Song L, Wang Z,
Laframboise T, et al: Genomic distribution of CHD7 on chromatin
tracks H3K4 methylation patterns. Genome Res. 19:590–601. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Newton AH and Pask AJ: CHD9 upregulates
RUNX2 and has a potential role in skeletal evolution. BMC Mol Cell
Biol. 21:272020. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Zhou C, Zou J, Zou S and Li X: INO80 is
required for osteogenic differentiation of human mesenchymal stem
cells. Sci Rep. 6:359242016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Anwar A, Sapra L, Gupta N, Ojha RP, Verma
B and Srivastava RK: Fine-tuning osteoclastogenesis: An insight
into the cellular and molecular regulation of osteoclastogenesis. J
Cell Physiol. 238:1431–1464. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Du J, Liu Y, Sun J, Yao E, Xu J, Wu X, Xu
L, Zhou M, Yang G and Jiang X: ARID1A safeguards the canalization
of the cell fate decision during osteoclastogenesis. Nat Commun.
15:59942024. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Urban W, Krzystańska D, Piekarz M, Nazar J
and Jankowska A: Osteosarcoma's genetic landscape painted by genes'
mutations. Acta Biochim Pol. 70:671–678. 2023.PubMed/NCBI
|
|
135
|
Gaeta R, Morelli M, Lessi F, Mazzanti CM,
Menicagli M, Capanna R, Andreani L, Coccoli L, Aretini P and
Franchi A: Identification of new potential prognostic and
predictive markers in high-grade osteosarcoma using whole exome
sequencing. Int J Mol Sci. 24:100862023. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Tuckermann J and Adams RH: The
endothelium-bone axis in development, homeostasis and bone and
joint disease. Nat Rev Rheumatol. 17:608–620. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Bixel MG, Sivaraj KK, Timmen M,
Mohanakrishnan V, Aravamudhan A, Adams S, Koh BI, Jeong HW, Kruse
K, Stange R and Adams RH: Angiogenesis is uncoupled from
osteogenesis during calvarial bone regeneration. Nat Commun.
15:45752024. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Duan X, Murata Y, Liu Y, Nicolae C, Olsen
BR and Berendsen AD: Vegfa regulates perichondrial vascularity and
osteoblast differentiation in bone development. Development.
142:1984–1991. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Sena JA, Wang L and Hu CJ: BRG1 and BRM
chromatin-remodeling complexes regulate the hypoxia response by
acting as coactivators for a subset of hypoxia-inducible
transcription factor target genes. Mol Cell Biol. 33:3849–3863.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Wang F, Zhang R, Beischlag TV, Muchardt C,
Yaniv M and Hankinson O: Roles of Brahma and Brahma/SWI2-related
gene 1 in hypoxic induction of the erythropoietin gene. J Biol
Chem. 279:46733–46741. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Wang Y, Chen Y, Bao L, Zhang B, Wang JE,
Kumar A, Xing C, Wang Y and Luo W: CHD4 promotes breast cancer
progression as a coactivator of hypoxia-inducible factors. Cancer
Res. 80:3880–3891. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Collins JM, Lang A, Parisi C, Moharrer Y,
Nijsure MP, Thomas Kim JH, Ahmed S, Szeto GL, Qin L, Gottardi R, et
al: YAP and TAZ couple osteoblast precursor mobilization to
angiogenesis and mechanoregulation in murine bone development. Dev
Cell. 59:211–227.e5. 2024. View Article : Google Scholar :
|
|
143
|
Griffin CT, Brennan J and Magnuson T: The
chromatin-remodeling enzyme BRG1 plays an essential role in
primitive erythropoiesis and vascular development. Development.
135:493–500. 2008. View Article : Google Scholar
|
|
144
|
Davis RB, Curtis CD and Griffin CT: BRG1
promotes COUP-TFII expression and venous specification during
embryonic vascular development. Development. 140:1272–1281. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Ingram KG, Curtis CD, Silasi-Mansat R,
Lupu F and Griffin CT: The NuRD chromatin-remodeling enzyme CHD4
promotes embryonic vascular integrity by transcriptionally
regulating extracellular matrix proteolysis. PLoS Genet.
9:e10040312013. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Forriol F and Shapiro F: Bone development:
interaction of molecular components and biophysical forces. Clin
Orthop Relat Res. 432:14–33. 2005. View Article : Google Scholar
|
|
147
|
Komori T: Cell death in chondrocytes,
osteoblasts, and osteocytes. Int J Mol Sci. 17:20452016. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Grandy R, Sepulveda H, Aguilar R, Pihan P,
Henriquez B, Olate J and Montecino M: The Ric-8B gene is highly
expressed in proliferating preosteoblastic cells and downregulated
during osteoblast differentiation in a SWI/SNF- and
C/EBPbeta-mediated manner. Mol Cell Biol. 31:2997–3008. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Zhang M, Guo T, Pei F, Feng J, Jing J, Xu
J, Yamada T, Ho TV, Du J, Sehgal P and Chai Y: ARID1B maintains
mesenchymal stem cell quiescence via inhibition of BCL11B-mediated
non-canonical activin signaling. Nat Commun. 15:46142024.
View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Ho L, Ronan JL, Wu J, Staahl BT, Chen L,
Kuo A, Lessard J, Nesvizhskii AI, Ranish J and Crabtree GR: An
embryonic stem cell chromatin remodeling complex, esBAF, is
essential for embryonic stem cell self-renewal and pluripotency.
Proc Natl Acad Sci USA. 106:5181–5186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Schaniel C, Ang YS, Ratnakumar K, Cormier
C, James T, Bernstein E, Lemischka IR and Paddison PJ:
Smarcc1/Baf155 couples self-renewal gene repression with changes in
chromatin structure in mouse embryonic stem cells. Stem Cells.
27:2979–2991. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Yan Z, Wang Z, Sharova L, Sharov AA, Ling
C, Piao Y, Aiba K, Matoba R, Wang W and Ko MS: BAF250B-associated
SWI/SNF chromatin-remodeling complex is required to maintain
undifferentiated mouse embryonic stem cells. Stem Cells.
26:1155–1165. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Eleuteri B, Aranda S and Ernfors P: NoRC
Recruitment by H2A.X deposition at rRNA gene promoter limits
embryonic stem cell proliferation. Cell Rep. 23:1853–1866. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Li B, Lyu P, Tang J, Li J, Ouchi T, Fan Y,
Zhao Z and Li L: The potential role and therapeutic relevance of
cellular senescence in skeletal pathophysiology. J Gerontol A Biol
Sci Med Sci. 79:glae0372024. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Napolitano MA, Cipollaro M, Cascino A,
Melone MA, Giordano A and Galderisi U: Brg1 chromatin remodeling
factor is involved in cell growth arrest, apoptosis and senescence
of rat mesenchymal stem cells. J Cell Sci. 120(Pt 16): 2904–2911.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Alessio N, Squillaro T, Cipollaro M,
Bagella L, Giordano A and Galderisi U: The BRG1 ATPase of chromatin
remodeling complexes is involved in modulation of mesenchymal stem
cell senescence through RB-P53 pathways. Oncogene. 29:5452–5463.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Squillaro T, Severino V, Alessio N, Farina
A, Di Bernardo G, Cipollaro M, Peluso G, Chambery A and Galderisi
U: De-regulated expression of the BRG1 chromatin remodeling factor
in bone marrow mesenchymal stromal cells induces senescence
associated with the silencing of NANOG and changes in the levels of
chromatin proteins. Cell Cycle. 14:1315–1326. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Yuan J and Ofengeim D: A guide to cell
death pathways. Nat Rev Mol Cell Biol. 25:379–395. 2024. View Article : Google Scholar
|
|
159
|
Klochendler-Yeivin A, Fiette L, Barra J,
Muchardt C, Babinet C and Yaniv M: The murine SNF5/INI1 chromatin
remodeling factor is essential for embryonic development and tumor
suppression. EMBO Rep. 1:500–506. 2000. View Article : Google Scholar
|
|
160
|
Li S, Huang Z, Zhu Y, Yan J, Li J, Chen J,
Zhou J, Zhang Y, Chen W, Xu K and Ye W: Bromodomain-containing
protein 7 regulates matrix metabolism and apoptosis in human
nucleus pulposus cells through the BRD7-PI3K-YAP1 signaling axis.
Exp Cell Res. 405:1126582021. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
Li Z, Cheng W, Gao K, Liang S, Ke L, Wang
M, Fan J, Li D, Zhang P, Xu Z and Li N: Pyroptosis: A spoiler of
peaceful coexistence between cells in degenerative bone and joint
diseases. J Adv Res. 71:227–262. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Chai S, Yang Y, Wei L, Cao Y, Ma J, Zheng
X, Teng J and Qin N: Luteolin rescues postmenopausal osteoporosis
elicited by OVX through alleviating osteoblast pyroptosis via
activating PI3K-AKT signaling. Phytomedicine. 128:1555162024.
View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Ruan H, Zhang H, Feng J, Luo H, Fu F, Yao
S, Zhou C, Zhang Z, Bian Y, Jin H, et al: Inhibition of
Caspase-1-mediated pyroptosis promotes osteogenic differentiation,
offering a therapeutic target for osteoporosis. Int
Immunopharmacol. 124(Pt B): 1109012023. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Deng Z, Zhang Y, Zhu Y, Zhu J, Li S, Huang
Z, Qin T, Wu J, Zhang C, Chen W, et al: BRD9 inhibition attenuates
matrix degradation and pyroptosis in nucleus pulposus by modulating
the NOX1/ROS/NF-κB axis. Inflammation. 46:1002–1021. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Wang J, Zhang Y, Cao J, Wang Y, Anwar N,
Zhang Z, Zhang D, Ma Y, Xiao Y, Xiao L and Wang X: The role of
autophagy in bone metabolism and clinical significance. Autophagy.
19:2409–2427. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Ren X, Xu J, Xue Q, Tong Y, Xu T, Wang J,
Yang T, Chen Y, Shi D and Li X: BRG1 enhances porcine iPSC
pluripotency through WNT/β-catenin and autophagy pathways.
Theriogenology. 215:10–23. 2024. View Article : Google Scholar
|
|
167
|
Yang C, Tian Y, Zhao F, Chen Z, Su P, Li Y
and Qian A: Bone microenvironment and osteosarcoma metastasis. Int
J Mol Sci. 21:69852020. View Article : Google Scholar : PubMed/NCBI
|
|
168
|
Urlić I, Jovičić MŠ, Ostojić K and Ivković
A: Cellular and genetic background of osteosarcoma. Curr Issues Mol
Biol. 45:4344–4358. 2023. View Article : Google Scholar
|
|
169
|
Hang JF and Chen PC: Parosteal
osteosarcoma. Arch Pathol Lab Med. 138:694–699. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Qiu YQ and Chen YL: Primary meningeal
osteoblastic osteosarcoma containing fibroblast osteosarcoma:
Clinicopathological analysis and literature review. Osteoporos Int.
32:1007–1012. 2021. View Article : Google Scholar
|
|
171
|
Guan Y, Zhang W, Mao Y and Li S:
Nanoparticles and bone microenvironment: A comprehensive review for
malignant bone tumor diagnosis and treatment. Mol Cancer.
23:2462024. View Article : Google Scholar : PubMed/NCBI
|
|
172
|
Basu Mallick A and Chawla SP: Giant cell
tumor of bone: An update. Curr Oncol Rep. 23:512021. View Article : Google Scholar : PubMed/NCBI
|
|
173
|
Corre I, Verrecchia F, Crenn V, Redini F
and Trichet V: The osteosarcoma microenvironment: A complex but
targetable ecosystem. Cells. 9:9762020. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Grünewald TGP, Cidre-Aranaz F, Surdez D,
Tomazou EM, de Álava E, Kovar H, Sorensen PH, Delattre O and
Dirksen U: Ewing sarcoma. Nat Rev Dis Primers. 4:52018. View Article : Google Scholar : PubMed/NCBI
|
|
175
|
Weber K, Damron TA, Frassica FJ and Sim
FH: Malignant bone tumors. Instr Course Lect. 57:673–688.
2008.PubMed/NCBI
|
|
176
|
Graca Marques J, Pavlovic B, Ngo QA, Pedot
G, Roemmele M, Volken L, Kisele S, Perbet R, Wachtel M and Schäfer
BW: The chromatin remodeler CHD4 sustains ewing sarcoma cell
survival by controlling global chromatin architecture. Cancer Res.
84:241–257. 2024. View Article : Google Scholar
|
|
177
|
Sohn EJ and Libich DS: Hijacking the BAF
complex: The mechanistic interplay of ARID1A and EWS::FLI1 in Ewing
sarcoma. Mol Oncol. 19:961–964. 2025. View Article : Google Scholar :
|
|
178
|
Selvanathan SP, Graham GT, Grego AR, Baker
TM, Hogg JR, Simpson M, Batish M, Crompton B, Stegmaier K, Tomazou
EM, et al: EWS-FLI1 modulated alternative splicing of ARID1A
reveals novel oncogenic function through the BAF complex. Nucleic
Acids Res. 47:9619–9636. 2019.PubMed/NCBI
|
|
179
|
Boulay G, Sandoval GJ, Riggi N, Iyer S,
Buisson R, Naigles B, Awad ME, Rengarajan S, Volorio A, McBride MJ,
et al: Cancer-specific retargeting of BAF complexes by a prion-like
domain. Cell. 171:163–178.e19. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Jayabal P, Zhou F, Lei X, Ma X, Blackman
B, Weintraub ST, Houghton PJ and Shiio Y: NELL2-cdc42 signaling
regulates BAF complexes and Ewing sarcoma cell growth. Cell Rep.
36:1092542021. View Article : Google Scholar : PubMed/NCBI
|
|
181
|
Cyra M, Schulte M, Berthold R, Heinst L,
Jansen EP, Grünewald I, Elges S, Larsson O, Schliemann C,
Steinestel K, et al: SS18-SSX drives CREB activation in synovial
sarcoma. Cell Oncol (Dordr). 45:399–413. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
182
|
Michel BC, D'Avino AR, Cassel SH,
Mashtalir N, McKenzie ZM, McBride MJ, Valencia AM, Zhou Q, Bocker
M, Soares LMM, et al: A non-canonical SWI/SNF complex is a
synthetic lethal target in cancers driven by BAF complex
perturbation. Nat Cell Biol. 20:1410–1420. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
183
|
Shih AR, Cote GM, Chebib I, Choy E,
DeLaney T, Deshpande V, Hornicek FJ, Miao R, Schwab JH, Nielsen GP
and Chen YL: Clinicopathologic characteristics of poorly
differentiated chordoma. Mod Pathol. 31:1237–1245. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
184
|
Sergi CM: Commentary on: SMARCB1 as a
novel diagnostic and prognostic biomarker for osteosarcoma. Biosci
Rep. 42:BSR202200402022. View Article : Google Scholar : PubMed/NCBI
|
|
185
|
Mobley BC, McKenney JK, Bangs CD, Callahan
K, Yeom KW, Schneppenheim R, Hayden MG, Cherry AM, Gokden M,
Edwards MS, et al: Loss of SMARCB1/INI1 expression in poorly
differentiated chordomas. Acta Neuropathol. 120:745–753. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Ding J, Yu C, Sui Y, Wang L, Yang Y, Wang
F, Yao H, Xing F, Liu H, Li Y, et al: The chromatin remodeling
protein INO80 contributes to the removal of H2A.Z at the
p53-binding site of the p21 gene in response to doxorubicin. FEBS
J. 285:3270–3285. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Flowers S, Beck GR Jr and Moran E:
Transcriptional activation by pRB and its coordination with SWI/SNF
recruitment. Cancer Res. 70:8282–8287. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Meisenberg C, Pinder SI, Hopkins SR,
Wooller SK, Benstead-Hume G, Pearl FMG, Jeggo PA and Downs JA:
Repression of transcription at DNA breaks requires cohesin
throughout interphase and prevents genome instability. Mol Cell.
73:212–223.e7. 2019. View Article : Google Scholar :
|
|
189
|
Ivanov AV, Peng H, Yurchenko V, Yap KL,
Negorev DG, Schultz DC, Psulkowski E, Fredericks WJ, White DE, Maul
GG, et al: PHD domain-mediated E3 ligase activity directs
intramolecular sumoylation of an adjacent bromodomain required for
gene silencing. Mol Cell. 28:823–837. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
190
|
Walhart TA, Vacca B, Hepperla AJ, Hamad
SH, Petrongelli J, Wang Y, McKean EL, Moksa M, Cao Q, Yip S, et al:
SMARCB1 loss in poorly differentiated chordomas drives tumor
progression. Am J Pathol. 193:456–473. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Passeri T, Gutman T, Hamza A,
Adle-Biassette H, Girard E, Beaurepere R, Tariq Z, Mariani O,
Dahmani A, Bourneix C, et al: The mutational landscape of skull
base and spinal chordomas and the identification of potential
prognostic and theranostic biomarkers. J Neurosurg. 139:1270–1280.
2023. View Article : Google Scholar : PubMed/NCBI
|
|
192
|
Wu X, Lin X, Chen Y, Kong W, Xu J and Yu
Z: Response of metastatic chordoma to the immune checkpoint
inhibitor pembrolizumab: A case report. Front Oncol. 10:5659452020.
View Article : Google Scholar
|
|
193
|
Wang L, Zehir A, Nafa K, Zhou N, Berger
MF, Casanova J, Sadowska J, Lu C, Allis CD, Gounder M, et al:
Genomic aberrations frequently alter chromatin regulatory genes in
chordoma. Genes Chromosomes Cancer. 55:591–600. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Harlow ML, Chasse MH, Boguslawski EA,
Sorensen KM, Gedminas JM, Kitchen-Goosen SM, Rothbart SB, Taslim C,
Lessnick SL, Peck AS, et al: Trabectedin inhibits EWS-FLI1 and
evicts SWI/SNF from chromatin in a schedule-dependent manner. Clin
Cancer Res. 25:3417–3429. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Livingston JA, Blay JY, Trent J, Valverde
C, Agulnik M, Gounder M, Le Cesne A, McKean M, Wagner MJ,
Stacchiotti S, et al: A phase I study of FHD-609, a
heterobifunctional degrader of bromodomain-containing protein 9, in
patients with advanced synovial sarcoma or SMARCB1-deficient
tumors. Clin Cancer Res. 31:628–638. 2025. View Article : Google Scholar
|
|
196
|
Dreier MR, Walia J and de la Serna IL:
Targeting SWI/SNF complexes in cancer: Pharmacological approaches
and implications. Epigenomes. 8:72024. View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Gatchalian J, Liao J, Maxwell MB and
Hargreaves DC: Control of stimulus-dependent responses in
macrophages by SWI/SNF chromatin remodeling complexes. Trends
Immunol. 41:126–140. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Li X, Ding D, Yao J, Zhou B, Shen T, Qi Y,
Ni T and Wei G: Chromatin remodeling factor BAZ1A regulates
cellular senescence in both cancer and normal cells. Life Sci.
229:225–232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
199
|
Martel-Pelletier J, Barr AJ, Cicuttini FM,
Conaghan PG, Cooper C, Goldring MB, Goldring SR, Jones G, Teichtahl
AJ, Pelletier JP, et al: Osteoarthritis. Nat Rev Dis Primers.
2:160722016. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Wang D, Zhang Y, Zhang L, He D, Zhao L,
Miao Z, Cheng W, Zhu C, Zhu L, Zhang W, et al: IRF1 governs the
expression of SMARCC1 via the GCN5-SETD2 axis and actively engages
in the advancement of osteoarthritis. J Orthop Translat.
45:211–225. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Qiu Y, Yao J, Li L, Xiao M, Meng J, Huang
X, Cai Y, Wen Z, Huang J, Zhu M, et al: Machine learning identifies
ferroptosis-related genes as potential diagnostic biomarkers for
osteoarthritis. Front Endocrinol (Lausanne). 14:11987632023.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Devlin MJ and Rosen CJ: The bone-fat
interface: Basic and clinical implications of marrow adiposity.
Lancet Diabetes Endocrinol. 3:141–147. 2015. View Article : Google Scholar
|
|
203
|
Kuznia AL, Hernandez AK and Lee LU:
Adolescent idiopathic scoliosis: Common questions and answers. Am
Fam Physician. 101:19–23. 2020.PubMed/NCBI
|
|
204
|
Cheng JC, Castelein RM, Chu WC, Danielsson
AJ, Dobbs MB, Grivas TB, Gurnett CA, Luk KD, Moreau A, Newton PO,
et al: Adolescent idiopathic scoliosis. Nat Rev Dis Primers.
1:150302015. View Article : Google Scholar : PubMed/NCBI
|
|
205
|
Wu Z, Dai Z, Yuwen W, Liu Z, Qiu Y, Cheng
JC, Zhu Z and Xu L: Genetic variants of CHD7 are associated with
adolescent idiopathic scoliosis. Spine (Phila Pa 1976).
46:E618–E624. 2021. View Article : Google Scholar
|
|
206
|
Borysiak K, Janusz P, Andrusiewicz M,
Chmielewska M, Kozinoga M, Kotwicki T and Kotwicka M: CHD7 gene
polymorphisms in female patients with idiopathic scoliosis. BMC
Musculoskelet Disord. 21:182020. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Kulkarni S, Nagarajan P, Wall J, Donovan
DJ, Donell RL, Ligon AH, Venkatachalam S and Quade BJ: Disruption
of chromodomain helicase DNA binding protein 2 (CHD2) causes
scoliosis. Am J Med Genet A. 146A:1117–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
208
|
Bonyadi M, Waldman SD, Liu D, Aubin JE,
Grynpas MD and Stanford WL: Mesenchymal progenitor self-renewal
deficiency leads to age-dependent osteoporosis in Sca-1/Ly-6A null
mice. Proc Natl Acad Sci USA. 100:5840–5845. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Yu W, Wang HL, Zhang J and Yin C: The
effects of epigenetic modifications on bone remodeling in
age-related osteoporosis. Connect Tissue Res. 64:105–116. 2023.
View Article : Google Scholar
|
|
210
|
Li C, Pan H, Liu W, Jin G, Liu W, Liang C
and Jiang X: Discovery of novel serum biomarkers for diagnosing and
predicting postmenopausal osteoporosis patients by 4D-label free
protein omics. J Orthop Res. 41:2713–2720. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
Pico MJ, Hashemi S, Xu F, Nguyen KH,
Donnelly R, Moran E and Flowers S: Glucocorticoid receptor-mediated
cis-repression of osteogenic genes requires BRM-SWI/SNF. Bone Rep.
5:222–227. 2016. View Article : Google Scholar
|
|
212
|
Chi SN, Yi JS, Williams PM, Roy-Chowdhuri
S, Patton DR, Coffey BD, Reid JM, Piao J, Saguilig L, Alonzo TA, et
al: Tazemetostat for tumors harboring SMARCB1/SMARCA4 or EZH2
alterations: Results from NCI-COG pediatric MATCH APEC1621C. J Natl
Cancer Inst. 115:1355–1363. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Martin LJ, Koegl M, Bader G, Cockcroft XL,
Fedorov O, Fiegen D, Gerstberger T, Hofmann MH, Hohmann AF, Kessler
D, et al: Structure-based design of an in vivo active selective
BRD9 inhibitor. J Med Chem. 59:4462–4475. 2016. View Article : Google Scholar : PubMed/NCBI
|