The skin, as the largest organ of the human body,
provides a protective barrier against mechanical, microbial,
chemical and allergenic insults (1). The gastrointestinal tract, a
crucial mucosal immune organ, maintains immune homeostasis through
dynamic interactions between the microbiome and the intestinal
immune system (2). The
intestinal mucosal immune function is mediated by the mucus layer,
epithelial barrier and resident immune cells, all of which engage
with the gut microbiota (3). The
collective genome of gut microbes is termed the 'gut microbiome,'
which is intimately linked to long-term health (4). As interfaces with the external
environment, both the gut and skin host diverse microbial
communities and are richly innervated and vascularized. As early as
1930, John H. Stokes and Donald M. Pillsbury proposed an intrinsic
relationship between gut microbiota and skin inflammation,
conceptualizing the 'gut-skin axis' (5). Recent advances in microbiology and
immunology have begun to clarify the mechanisms through which gut
microbiota influences skin health. Lee and Sung (6) identified immune pathways by which
alterations in gut and skin microbiota contribute to
dermatopathology, while Szanto et al (7) suggested that gut microbiota
modulation may hold therapeutic potential for specific skin
disorders. These findings not only validate the gut-skin axis
theory but also open new avenues for clinical dermatology. In the
context of increasing antibiotic resistance, therapeutic strategies
are shifting toward alternatives such as probiotics, prebiotics and
dietary interventions, which can restore microbial balance and
modulate immune responses to improve skin health (8,9).
This review examines novel strategies for treating skin diseases
from the perspective of the gut-skin axis and explores future
translational research directions.
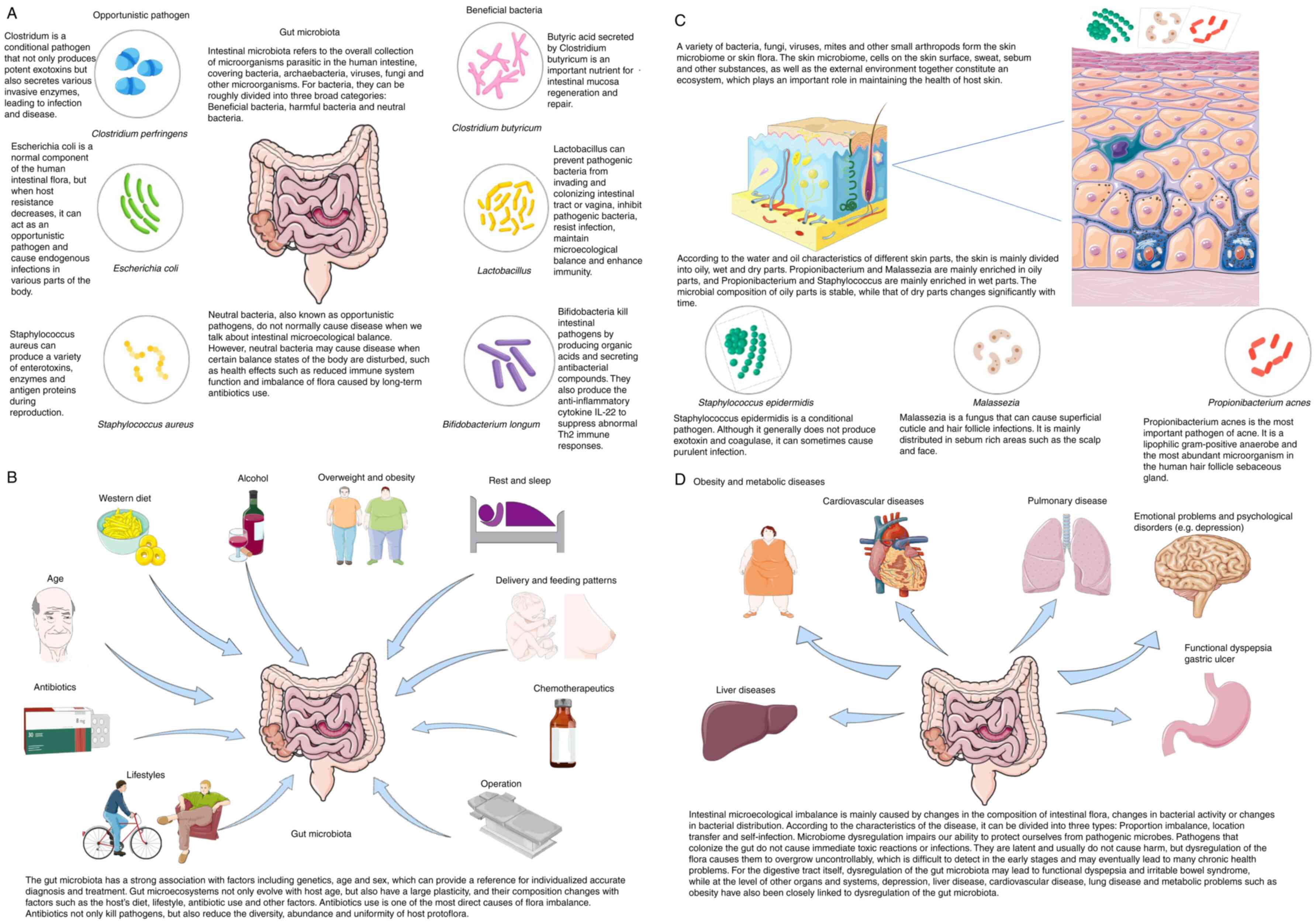
The gut microbiota comprises diverse and dynamic
microbial communities residing in the human gastrointestinal tract
(10) (Fig. 1A). Anatomically and functionally,
the gut is divided into the small and large intestines, each with
distinct physiological conditions and microbial populations
(11). Numerous factors
influence the gut microbiota (Fig.
1B), with diet being a primary determinant of its structure and
function (12). The gut
microbiota evolves from infancy and changes in composition and
diversity over time within an individual (13). There is considerable
interindividual variation in gut microbial composition. Through
host-microbe coevolution, the gut microbiome plays a critical role
in regulating host physiology, including metabolism, immune
development and behavioral responses (14,15).
As the body's outermost barrier, the skin is
continuously exposed to environmental factors and serves as the
first line of immune defense (16). The skin microbiota consists of
microbial communities adapted to the cutaneous environment through
long-term colonization, persisting in the chemical milieu of the
stratum corneum, sweat and sebaceous secretions (17). Environmental factors such as
ultraviolet radiation, temperature, humidity, sebum levels, oxygen
availability and pH create distinct ecological niches across skin
regions, leading to spatial variation in microbial composition
(18). Based on physiological
characteristics, Mahmud et al (19) classified skin into sebaceous
(e.g., between eyebrows), moist (e.g., forearm flexure) and dry
(e.g., palmar forearm) types. Lipophilic bacteria dominate
sebum-rich areas, whereas dry regions may support a more diverse
microbiota (Fig. 1C).
The 'gut-skin axis' theory is part of the broader
'gut-organ axis' framework, emphasizing bidirectional communication
between the gut and other organs via neurological, endocrine and
immune pathways (23). This
theory integrates multiple organs, the gut and the immune system
with the gut microbiota (24).
The gut and skin share structural and functional similarities,
including embryonic origin, symbiotic microbial communities,
innervation patterns and immune functions (19). As internal and external surfaces
in contact with the environment, they utilize similar signaling and
innervation pathways. T-cell-mediated immune responses often
manifest in both intestinal and cutaneous tissues (25).
The gut microbial community is central to
maintaining gut-skin homeostasis and underpins the gut-skin axis
theory (26). The gut microbiota
includes bacteria, fungi, parasites, protozoa and viruses, with
bacteria predominating (27).
Over 90% of gut bacteria belong to the Bacteroidetes and Firmicutes
phyla (28). The
Bacteroidota-to-Firmicutes ratio is commonly used to assess gut
microbiota characteristics and diversity (29). Gut bacteria can be categorized as
beneficial (e.g., Bifidobacteria, Lactobacillus) or opportunistic
pathogens (e.g., Staphylococci, Clostridia), which may cause
infection under certain conditions (30).
Alterations in gut microbiome composition,
metabolism and immunity can impact skin health. Manos (31) identified interspecies
communication within microbial communities as a key factor in
maintaining cutaneous homeostasis and responding to environmental
stressors, with dysregulation contributing to skin disease.
External factors such as genetics, diet, antimicrobials, and
lifestyle influence microbial diversity (32) (Fig. 2A).
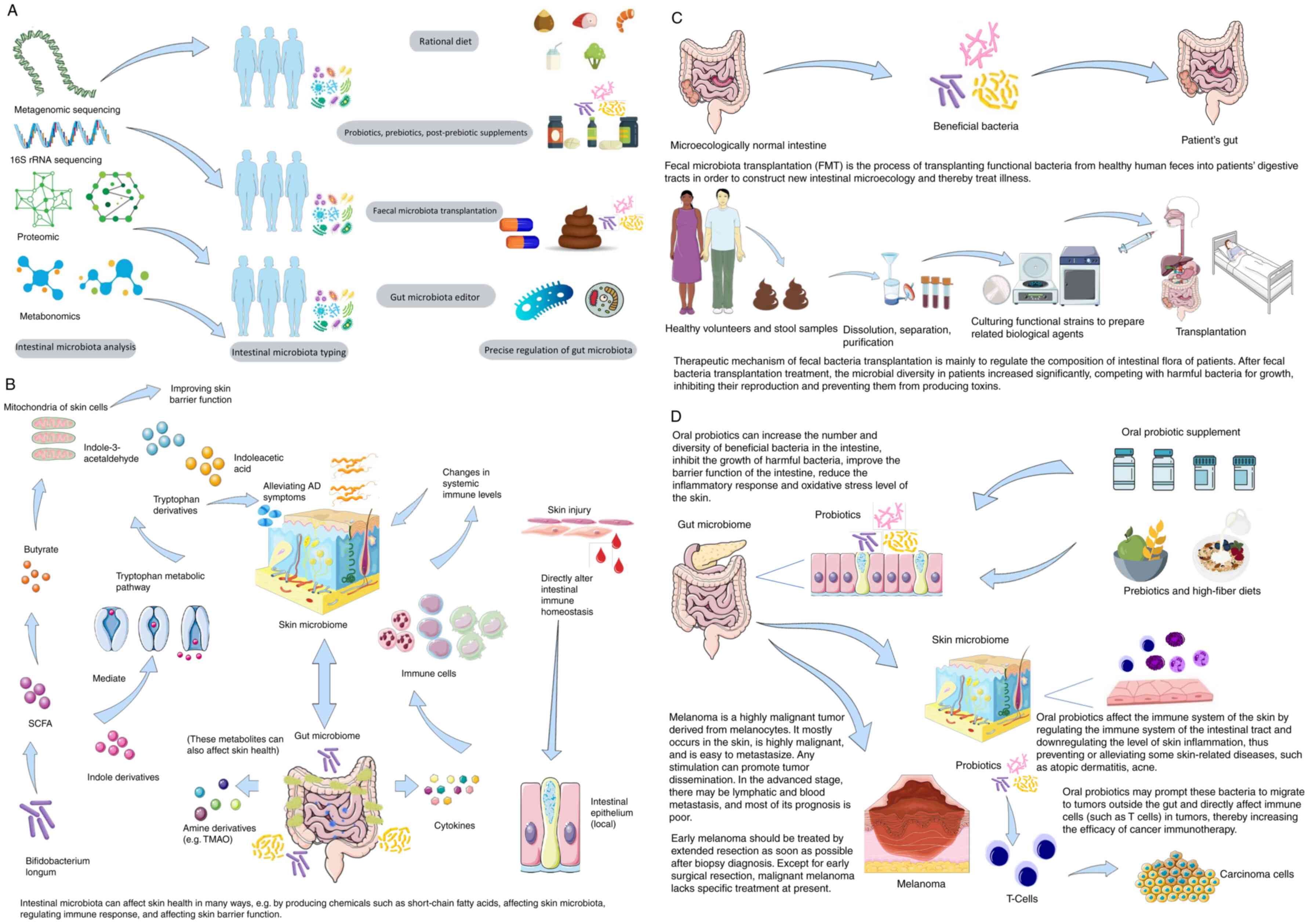
Gut microbiota-derived metabolites mediate skin
interactions through short-chain fatty acids (SCFAs), tryptophan
metabolites and amine derivatives (e.g., trimethylamine N-oxide),
which exert systemic effects via specific receptors (36). SCFAs play important roles in
immune regulation; Trompette et al (37) demonstrated that gut-derived
butyrate enhances skin barrier function by modifying mitochondrial
metabolism in keratinocytes. Fang et al (38) reported that Bifidobacterium
longum produces indole derivatives that alleviate AD via the
tryptophan pathway. Tryptophan metabolites (e.g., indoleacetic
acid) are critical for maintaining intestinal and systemic immune
homeostasis (39) (Fig. 2B).
Bidirectional interaction between the gut microbiome
and skin health is central to the gut-skin axis theory. While most
evidence highlights gut microbes influencing skin health, Dokoshi
et al (43) demonstrated
that skin injury directly remodels the gut microbiome (Fig. 2B), providing experimental support
for bidirectional signaling and indicating that skin damage may
impair gut immune homeostasis. Long et al (44), in a two-sample Mendelian
randomization study, established a causal relationship between gut
microbiota and four common inflammatory skin diseases: Eczema,
acne, psoriasis and rosacea.
Understanding gut microbiota-skin interactions
offers novel mechanistic insights for managing dermatological
conditions. Clinical evidence reveals frequent comorbidity of skin
and intestinal disorders, such as enteropathic acrodermatitis (zinc
malabsorption causing dermatitis, alopecia and diarrhea) (45), and celiac disease (associated
with eczema, psoriasis and urticaria) (46).
Restoring gut microbiota balance enhances intestinal
barrier integrity and regulates immune responses (47). This involves upregulating
immunomodulatory cytokines (e.g., IL-10) (48) and suppressing pro-inflammatory
mediators (e.g., TNF-α) (49).
Clinical interventions include probiotics, prebiotics, synbiotics
and fecal microbiota transplantation (FMT) (50).
FMT involves transferring processed stool from
healthy donors to patients to rebuild gut microbiota. Initially
used for digestive diseases (51-53), FMT's efficacy depends on donor
and recipient factors (immune status, genetic diversity, gut
microbial composition) and treatment protocols (stool amount,
number of infusions, delivery route and adjuvant therapy) (54) (Fig. 2C).
While most FMT research focuses on gastrointestinal
disorders, recent trials suggest it may modulate systemic immune
responses, including in skin cancers like melanoma. Kim et
al (55) showed that FMT
improved AD symptoms in mice by restoring gut microbiota. FMT
enhances immunotherapy efficacy in cancer patients by modulating
gut microbiota (56). Melanoma,
the most common and prognostically worst skin cancer, is increasing
globally (57). Baruch et
al (58) observed that FMT
induced favorable changes in immune cell infiltration and gene
expression in intestinal and tumor microenvironments. Liu et
al (59) demonstrated that
FMT is effective for moderate to severe AD in adults, altering gut
microbiota composition and function. However, FMT carries risks;
Eshel et al (60) found
that while FMT capsules rarely transmit bloodstream pathogens
directly, they may indirectly promote bacterial translocation via
gut inflammation, potentially compromising intestinal barrier
integrity and increasing infection risk. Further in-depth studies
are required to ascertain the efficacy and safety of FMT in
treating other skin diseases.
Prebiotics are indigestible food components that
selectively stimulate beneficial bacteria (e.g.,
Bifidobacterium, Lactobacillus) while inhibiting
pathogenic overgrowth, thereby improving intestinal microecology
(61). Probiotics are live
microorganisms that, when administered in adequate amounts, confer
health benefits through intestinal colonization (62). These interventions show promise
beyond gastrointestinal and cardiovascular diseases (63,64) with emerging applications in
dermatology.
In oncology, gut microbiome modulation may enhance
immunotherapy efficacy. Specific probiotics influence
T-cell-mediated therapies (e.g., anti-programmed cell death 1,
chimeric antigen receptor-T) (68), though strain-specific effects
require further validation. Bender et al (69) demonstrated that Lactobacillus
reuteri translocates to melanoma sites in mice, secreting
indole-3-aldehyde to activate CD8+ T-cell receptors and potentiate
immunotherapy. Such mechanisms hold potential for future
immunomodulatory strategies pending resolution of delivery
challenges and dose optimization.
Psoriasis is an immune-mediated skin disease
characterized by abnormal keratinocyte proliferation and
differentiation, posing significant physical and psychological
burdens (70). Previous studies
suggested gut microbiota dysbiosis in patients with psoriasis, but
the relationship remained vague (71). Zang et al (72) used bidirectional Mendelian
randomization to identify Pasteurellaceae, Brucella
and Methanobrevibacter smithii as potential pathogenic
contributors, suggesting microbial targets for therapy (Table I). Zhao et al (73) demonstrated through metabolomics
that gut microbiota transplantation from severely affected mice
exacerbated skin inflammation in mild cases, while a
phosphodiesterase-4 inhibitor alleviated symptoms and restored the
gut microbiota, providing a theoretical basis for gut
microbiota-skin axis-based psoriasis treatment (Table I).
AD is a common chronic inflammatory skin disease
characterized by persistent itching and eczematous rash (74). It increases the risk of
comorbidities like food allergies, asthma, allergic rhinitis and
mental health disorders (75).
The pathogenesis involves hereditary immune dysregulation, skin
barrier dysfunction and environmental factors (76). The gut microbiota modulates
immune system development, implying a key role in AD (77).
First-line AD treatment includes topical
corticosteroids and calcineurin inhibitors (e.g., pimecrolimus,
tacrolimus). For moderate to severe cases, ultraviolet phototherapy
is used adjunctively (78).
Probiotic supplementation is increasingly used, particularly in
children, to modulate gut microbiota and regulate immune responses
(79,80). Meta-analyses indicate certain
probiotics reduce Scoring Atopic Dermatitis indices in adults
(81). Fang et al
(82) showed that
Bifidobacterium CCFM16 and Lactobacillus plantar
CCFM8610 specifically improve AD by altering gut microbiota
composition and function (Table
I).
AV is a chronic inflammatory skin disease affecting
sebaceous units, characterized by comedones, papules, pustules,
nodules and scars, typically on the face, upper trunk and
extremities (83). The global
prevalence is estimated at 8.96% in men and 9.81% in women
(84). The pathogenesis involves
multifactorial mechanisms, with pubertal sebum hypersecretion being
a key factor. AV often persists into adulthood with distinct
features (85). Androgens and
testosterone regulate sebum production, explaining why males often
experience more severe symptoms (86).
Gut dysbiosis may exacerbate AV through upregulated
insulin-like growth factor 1 signaling, insulin resistance and
systemic pro-inflammatory cytokines (87). First-line treatments include
topical retinoids, azelaic acid and benzoyl peroxide (88). The role of gut microbiota
modulation via probiotics in improving AV is underexplored. Jung
et al (89) found that
probiotic-antibiotic combination therapy synergistically reduced
inflammation and antibiotic side effects. Fabbrocini et al
(90) showed that
Lactobacillus rhamnosus SP1 (LSP1) probiotic normalized skin
insulin signaling gene expression. Eguren et al (91) conducted a 12-week trial with
Lactobacillus rhamnosus and Arthrospira in patients
with AV aged 12-30 years, finding the probiotic adjunct safe and
effective (Table I).
Rosacea is a chronic recurrent inflammatory skin
condition affecting the central face (forehead, nose, cheeks, chin)
(92). It is classified into
erythematotelangiectatic, papulopustular, phymatous and ocular
subtypes (93).
According to 2021 guidelines, urticaria is
classified as acute (lasting <6 weeks) or chronic urticaria
(CU). CU is a common inflammatory skin disorder involving mast
cell-mediated allergy and autoimmunity, characterized by recurrent
wheals, angioedema and itching lasting ≥6 weeks (99). Pathophysiology involves immune
dysregulation, inflammatory cascade imbalance and
coagulation-fibrinolytic activation (100). CU subtypes include chronic
spontaneous urticaria (CSU) and chronic inducible urticarial
(101).
Vitiligo is a depigmentation disorder affecting
0.5-2% globally, with a genetic component (106). It presents as white, scaleless
patches (107).
Pathophysiologically, vitiligo is an autoimmune condition where
melanocytes are sensitive to oxidative stress, triggering
inflammatory cytokine release and innate immune activation.
CD8+ T cells destroy melanocytes, driven by IFN-γ.
Oxidative stress may induce gut dysbiosis, promoting autoimmunity.
IFN-γ blockade therapies temporarily reverse depigmentation but
relapse occurs upon cessation (108). The chronic course of vitiligo
affects patients' appearance and psychological well-being;
Kussainova et al (109)
reported a 35.8% anxiety prevalence, higher in women.
Current evidence on the gut-vitiligo link is
limited. Emerging studies suggest gut microbiome involvement. Hadi
et al (110) reported
vitiligo-inflammatory bowel disease comorbidity. Ni et al
(111) documented a
Firmicutes/Bacteroidetes ratio reduction in patients
with vitiligo (Table I), while
Luan et al (112)
observed decreased microbial alpha diversity with altered
cysteine/galactose metabolism (Table
I). These findings support the gut-microbiota-skin hypothesis
but require further investigation for therapeutic applications.
Skin cancer is the fifth most common cancer
globally, with its incidence rising (113). It arises from genetic defects
or DNA mutations in skin cells, primarily due to ultraviolet
exposure (114). Skin cancers
are classified as malignant melanoma (MM) or non-melanoma skin
cancer (NMSC). MM originates from melanocytes, while NMSC (e.g.,
squamous cell carcinoma, basal cell carcinoma) arises from
epidermal cells (115).
Treatments include cryotherapy, radiotherapy and photodynamic
therapy, but novel approaches are needed (116).
MM is highly metastatic, has a poor prognosis and is
the leading cause of death among patients with skin cancer
(117). Mrazek et al
(118) found compositional
differences between melanoma and healthy skin microbiomes, with
elevated Fusobacterium and Eubacterium in tumors
(Table I). Mekadim et al
(119) demonstrated distinct
gut and skin microbiome profiles in melanoma models, addressing
knowledge gaps (Table I).
Several studies confirmed that the gut microbiota
can modulate the response to cancer immune checkpoint blockade
therapy (120-122). Spencer et al (123) reported improved
progression-free survival in ICB-treated patients with higher fiber
intake, particularly without probiotics; fiber/prebiotics enhanced
antitumor T-cell responses via microbiome modulation (Table I). Routy et al (124)'s phase I trial confirmed FMT
safety combined with first-line melanoma therapy, though efficacy
requires further validation (Fig.
2D) (Table I). Future
investigations of the gut-skin axis may yield innovative
therapeutic strategies for skin cancer.
Rare dermatoses like AIBD and LP receive limited
research due to funding and recruitment challenges (125). AIBD involve chronic
immune-mediated blistering, with fragile cutaneous/mucosal vesicles
rupturing into erosions. These include pemphigus vulgaris (PV),
bullous pemphigoid (BP) and mucous membrane pemphigoid (126). PV subtypes include vulgaris,
foliaceus, IgA pemphigus and paraneoplastic pemphigus. BP, the most
common pemphigoid, features tense, rupture-resistant subepidermal
blisters, often involving the oropharyngeal and ocular mucosa. It
primarily affects adults aged >50 years but can also occur in
younger individuals (127).
Pemphigus has a long course and poor prognosis,
severely affecting patients' quality of life, with most deaths due
to uncontrollable secondary infections (128). The pathogenesis involves
Th1/Th2 and Th17/Treg imbalances, leading to IgG autoantibodies
that target epidermal/mucosal antigens, causing loss of cell
adhesion and blister formation (129).
LP is a chronic inflammatory disease presenting as
violaceous pruritic papules on skin, mucosa or nails. Subtypes
include cutaneous LP (CLP) and oral LP (OLP), with possible
esophageal, genital or nail involvement (132). The prevalence of LP ranges from
0.22 to 1%, with OLP about five times more common than CLP
(133). Genital subtypes have
been reported (134), and
concurrent subtypes complicate diagnosis (135). The pathogenesis of CLP involves
cell-mediated immune responses against basal keratinocytes
(136). Georgescu et al
(137) reported an
oxidant-antioxidant imbalance in patients with LP, suggesting
oxidative stress plays a role.
Few studies have focused on the role of the 'gut
microbe-skin axis' in rare skin diseases such as LP and pemphigus
(22). Li et al (138) observed a distinct gut
microbiota in patients with active pemphigus, with
Prevotella spp. and Coriobacteriaceae abundance
correlating with autoantibodies. Roy et al (139) reviewed intestinal
microecological dysregulation in several rare diseases, including
LP, and highlighted the complexity of host-microbiota interactions,
emphasizing knowledge gaps, the need for improved study designs,
and the promise of microbiome-based therapeutics. Kamal et
al (140)'s trial on 60
patients with OLP showed a greater reduction in pain and
Thongprasom scores with combined clobetasol/probiotic therapy vs.
clobetasol alone. Despite research challenges, such as scarce
pathogenic data and small cohorts, elucidating microbiota-skin
interactions could yield novel diagnostics and therapies for rare
dermatoses.
Despite progress, translating gut microbiome
research into clinical applications remains challenging.
Methodological limitations in data collection, representativeness
and analysis are key barriers. Resources like GMrepo (https://gmrepo.humangut.info/home) and gutMGene
(https://bio-computing.hrbmu.edu.cn/gutmgene/#/home)
provide valuable data, but heterogeneity due to geographic,
demographic and lifestyle factors complicates sampling and
standardization (Fig. 2A). The
lack of a consensus definition for 'healthy gut microbiota' impedes
benchmark establishment (141,142).
Patient and provider acceptance barriers persist,
especially in culturally conservative settings. Probiotics, though
generally safer than FMT, carry risks such as bacteremia in
immunocompromised hosts and horizontal gene transfer (152,153). Cases like Lactobacillus
rhamnosus infections in immunosuppressed individuals underscore
the need for further safety research (154-156).
Preclinical gut microbiome research will continue
diversifying. International data-sharing collaborations may
overcome ethnographic limitations (157). Integrating medicine, biology
and informatics could expand the research scope, as seen in machine
learning applications for microbial community analysis (158). Current algorithms predict
health status and identify disease-microbiome associations through
differential abundance analysis (159).
In clinical translation, precision medicine
requires refinement. Longitudinal studies with larger cohorts may
better evaluate probiotic efficacy across populations, informing
targeted microbiome-based interventions using adaptive trial
designs (e.g., cluster randomized controlled trials). These
frameworks would strengthen disease prevention and treatment
strategies.
Emerging technologies like high-intensity
ultrasound show potential for enhancing functional food development
by improving probiotic stability and bioactivity (160), Future validation studies should
assess gut health optimization and personalized approaches
(161,162). Beyond dairy probiotics,
fermented foods and beverages are demonstrating health benefits
(163,164).
The following conclusions can be drawn from the
present review: i) The gut-skin axis framework highlights the
influence of gut microbiota on skin homeostasis, with dysbiosis
implicated in psoriasis, AD, acne and AIBD; ii) the gut microbiota
modulates skin health through immune regulation, microbial
metabolite production (e.g., SCFAs) and systemic inflammation
control. FMT, probiotics and prebiotics show promise in restoring
microbial balance and reducing inflammation, though larger trials
are needed; iii) current evidence primarily establishes
correlations, but causal relationships require validation via
multi-omics approaches integrating genomics, metabolomics and
immune profiling; iv) clinical translation faces challenges
including methodological limitations, ethical concerns with FMT and
insufficient long-term safety data for probiotics; v) mechanistic
insights into rare skin diseases via the gut-skin axis are scarce,
warranting targeted investigations; vi) future research should
prioritize patient-specific microbiome interventions,
machine-learning diagnostics and cross-disciplinary collaboration
(e.g., dermatology-gastroenterology-bioinformatics) to advance
precision dermatology; and vii) the gut-skin axis redefines
skincare paradigms, emphasizing gut health as integral to
dermatological wellness, with breakthroughs in microbiome
engineering and Artificial Intelligence-driven interventions
offering transformative potential.
Not applicable.
YZ performed the analyses and wrote the first draft
of the manuscript. CY, JZ, QY, XZ and XZ performed the literature
search and discussed and edited the manuscript. XZ and XZ
supervised the preparation of the manuscript. Data authentication
is not applicable. All authors have read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This study was partially supported by the National Natural
Science Foundation of China (grant nos. 32170915 and 82172931).
|
1
|
Karimzadeh F, Soltani Fard E, Nadi A,
Malekzadeh R, Elahian F and Mirzaei SA: Advances in skin gene
therapy: Utilizing innovative dressing scaffolds for wound healing,
a comprehensive review. J Mater Chem B. 12:6033–6062. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shi N, Li N, Duan X and Niu H: Interaction
between the gut microbiome and mucosal immune system. Mil Med Res.
4:142017.PubMed/NCBI
|
|
3
|
Wang J, He M, Yang M and Ai X: Gut
microbiota as a key regulator of intestinal mucosal immunity. Life
Sci. 345:1226122024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ezenabor EH, Adeyemi AA and Adeyemi OS:
Gut microbiota and metabolic syndrome: Relationships and
opportunities for new therapeutic strategies. Scientifica (Cairo).
2024:42220832024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Saarialho-Kere U: The gut-skin axis. J
Pediatr Gastroenterol Nutr. 39(Suppl 3): S734–S735. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HR and Sung JH: Multiorgan-on-a-chip
for the realization of gut-skin axis. Biotechnol Bioeng.
119:2590–2601. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Szanto M, Dozsa A, Antal D, Szabo K,
Kemeny L and Bai P: Targeting the gut-skin axis-Probiotics as new
tools for skin disorder management? Exp Dermatol. 28:1210–1218.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suaini NHA, Siah KTH and Tham EH: Role of
the gut-skin axis in IgE-mediated food allergy and atopic diseases.
Curr Opin Gastroenterol. 37:557–564. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alesa DI, Alshamrani HM, Alzahrani YA,
Alamssi DN, Alzahrani NS and Almohammadi ME: The role of gut
microbiome in the pathogenesis of psoriasis and the therapeutic
effects of probiotics. J Family Med Prim Care. 8:3496–3503. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gomaa EZ: Human gut microbiota/microbiome
in health and diseases: A review. Antonie Van Leeuwenhoek.
113:2019–2040. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Adak A and Khan MR: An insight into gut
microbiota and its functionalities. Cell Mol Life Sci. 76:473–493.
2019. View Article : Google Scholar
|
|
12
|
Zmora N, Suez J and Elinav E: You are what
you eat: Diet, health, and the gut microbiota. Nat Rev
Gastroenterol Hepatol. 16:35–56. 2019. View Article : Google Scholar
|
|
13
|
Milani C, Duranti S, Bottacini F, Casey E,
Turroni F, Mahony J, Belzer C, Delgado Palacio S, Arboleya Montes
S, Mancabelli L, et al: The first microbial colonizers of the human
gut: Composition, activities, and health implications of the infant
gut microbiota. Microbiol Mol Biol Rev. 81:e00036–17. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Aggarwal N, Kitano S, Puah GRY, Kittelmann
S, Hwang IY and Chang MW: Microbiome and human health: Current
understanding, engineering, and enabling technologies. Chem Rev.
123:31–72. 2023. View Article : Google Scholar :
|
|
15
|
Banaszak M, Gorna I, Wozniak D,
Przyslawski J and Drzymala-Czyz S: Association between gut
dysbiosis and the occurrence of SIBO, LIBO, SIFO and IMO.
Microorganisms. 11:5732023. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee HJ and Kim M: Skin barrier function
and the microbiome. Int J Mol Sci. 23:130712022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen YE, Fischbach MA and Belkaid Y: Skin
microbiota-host interactions. Nature. 553:427–436. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smythe P and Wilkinson HN: The skin
microbiome: Current landscape and future opportunities. Int J Mol
Sci. 24:39502023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mahmud MR, Akter S, Tamanna SK, Mazumder
L, Esti IZ, Banerjee S, Akter S, Hasan MR, Acharjee M, Hossain MS
and Pirttilä AM: Impact of gut microbiome on skin health: Gut-skin
axis observed through the lenses of therapeutics and skin diseases.
Gut Microbes. 14:20969952022. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Petersen C and Round JL: Defining
dysbiosis and its influence on host immunity and disease. Cell
Microbiol. 16:1024–1033. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Rygula I, Pikiewicz W, Grabarek BO, Wojcik
M and Kaminiow K: The role of the gut microbiome and microbial
dysbiosis in common skin diseases. Int J Mol Sci. 25:19842024.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Karimova M, Moyes D, Ide M and Setterfield
JF: The human microbiome in immunobullous disorders and lichen
planus. Clin Exp Dermatol. 47:522–528. 2022. View Article : Google Scholar
|
|
23
|
Ahlawat S, Asha and Sharma KK: Gut-organ
axis: A microbial outreach and networking. Lett Appl Microbiol.
72:636–668. 2021. View Article : Google Scholar
|
|
24
|
Guo Y, Chen X, Gong P, Li G, Yao W and
Yang W: The gut-organ-axis concept: Advances the application of
gut-on-chip technology. Int J Mol Sci. 24:40892023. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yano JM, Yu K, Donaldson GP, Shastri GG,
Ann P, Ma L, Nagler CR, Ismagilov RF, Mazmanian SK and Hsiao EY:
Indigenous bacteria from the gut microbiota regulate host serotonin
biosynthesis. Cell. 161:264–276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Salem I, Ramser A, Isham N and Ghannoum
MA: The gut microbiome as a major regulator of the gut-skin axis.
Front Microbiol. 9:14592018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Komine M: Recent advances in psoriasis
research; The clue to mysterious relation to gut microbiome. Int J
Mol Sci. 21:25822020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Buhas MC, Gavrilas LI, Candrea R, Catinean
A, Mocan A, Miere D and Tătaru A: Gut microbiota in psoriasis.
Nutrients. 14:29702022. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Frioux C, Ansorge R, Ozkurt E, Ghassemi
Nedjad C, Fritscher J, Quince C, Waszak SM and Hildebrand F:
Enterosignatures define common bacterial guilds in the human gut
microbiome. Cell Host Microbe. 31:1111–1125 e6. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Quaglio AEV, Grillo TG, De Oliveira ECS,
Di Stasi LC and Sassaki LY: Gut microbiota, inflammatory bowel
disease, and colorectal cancer. World J Gastroenterol.
28:4053–4060. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Manos J: The human microbiome in disease
and pathology. APMIS. 130:690–705. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wu J, Wang K, Wang X, Pang Y and Jiang C:
The role of the gut microbiome and its metabolites in metabolic
diseases. Protein Cell. 12:360–373. 2021. View Article : Google Scholar :
|
|
33
|
Chen Y, Zhou J and Wang L: Role and
mechanism of gut microbiota in human disease. Front Cell Infect
Microbiol. 11:6259132021. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Olejniczak-Staruch I, Ciazynska M,
Sobolewska-Sztychny D, Narbutt J, Skibinska M and Lesiak A:
Alterations of the skin and gut microbiome in psoriasis and
psoriatic arthritis. Int J Mol Sci. 22:39982021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Park DH, Kim JW, Park HJ and Hahm DH:
Comparative analysis of the microbiome across the gut-skin axis in
atopic dermatitis. Int J Mol Sci. 22:42282021. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Stec A, Sikora M, Maciejewska M,
Paralusz-Stec K, Michalska M, Sikorska E and Rudnicka L: Bacterial
metabolites: A link between gut microbiota and dermatological
diseases. Int J Mol Sci. 24:34942023. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Trompette A, Pernot J, Perdijk O,
Alqahtani RAA, Domingo JS, Camacho-Munoz D, Wong NC, Kendall AC,
Wiederkehr A, Nicod LP, et al: Gut-derived short-chain fatty acids
modulate skin barrier integrity by promoting keratinocyte
metabolism and differentiation. Mucosal Immunol. 15:908–926. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fang Z, Pan T, Li L, Wang H, Zhu J, Zhang
H, Zhao J, Chen W and Lu W: Bifidobacterium longum mediated
tryptophan metabolism to improve atopic dermatitis via the gut-skin
axis. Gut Microbes. 14:20447232022. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su X, Gao Y and Yang R: Gut
microbiota-derived tryptophan metabolites maintain gut and systemic
homeostasis. Cells. 11:22962022. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kinashi Y and Hase K: Partners in leaky
gut syndrome: intestinal dysbiosis and autoimmunity. Front Immunol.
12:6737082021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Marrs T, Jo JH, Perkin MR, Rivett DW,
Witney AA, Bruce KD, Logan K, Craven J, Radulovic S, Versteeg SA,
et al: Gut microbiota development during infancy: Impact of
introducing allergenic foods. J Allergy Clin Immunol. 147:613–621
e9. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Jiao Y, Wu L, Huntington ND and Zhang X:
Crosstalk between gut microbiota and innate immunity and its
implication in autoimmune diseases. Front Immunol. 11:2822020.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dokoshi T, Chen Y, Cavagnero KJ, Rahman G,
Hakim D, Brinton S, Schwarz H, Brown EA, O'Neill A, Nakamura Y, et
al: Dermal injury drives a skin-to-gut axis that disrupts the
intestinal microbiome and intestinal immune homeostasis in mice.
Nat Commun. 15:30092024. View Article : Google Scholar
|
|
44
|
Long J, Gu J, Yang J, Chen P, Dai Y, Lin
Y, Wu M and Wu Y: Exploring the association between gut microbiota
and inflammatory skin diseases: A two-sample mendelian
randomization analysis. Microorganisms. 11:25862023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Glutsch V, Hamm H and Goebeler M: Zinc and
skin: An update. J Dtsch Dermatol Ges. 17:589–596. 2019.PubMed/NCBI
|
|
46
|
Therrien A, Kelly CP and Silvester JA:
Celiac disease: Extraintestinal manifestations and associated
conditions. J Clin Gastroenterol. 54:8–21. 2020. View Article : Google Scholar
|
|
47
|
Ni Q, Zhang P, Li Q and Han Z: Oxidative
stress and gut microbiome in inflammatory skin diseases. Front Cell
Dev Biol. 10:8499852022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee SY, Jhun J, Woo JS, Lee KH, Hwang SH,
Moon J, Park G, Choi SS, Kim SJ, Jung YJ, et al: Gut
microbiome-derived butyrate inhibits the immunosuppressive factors
PD-L1 and IL-10 in tumor-associated macrophages in gastric cancer.
Gut Microbes. 16:23008462024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xiao P, Hu Z, Lang J, Pan T, Mertens RT,
Zhang H, Guo K, Shen M, Cheng H, Zhang X, et al: Mannose metabolism
normalizes gut homeostasis by blocking the TNF-α-mediated
proinflammatory circuit. Cell Mol Immunol. 20:119–130. 2023.
View Article : Google Scholar
|
|
50
|
Yadegar A, Bar-Yoseph H, Monaghan TM,
Pakpour S, Severino A, Kuijper EJ, Smits WK, Terveer EM, Neupane S,
Nabavi-Rad A, et al: Fecal microbiota transplantation: current
challenges and future landscapes. Clin Microbiol Rev.
37:e00060222024. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Joachim A, Schwerd T, Holz H, Sokollik C,
Konrad LA, Jordan A, Lanzersdorfer R, Schmidt-Choudhury A, Hünseler
C and Adam R: Fecal Microbiota Transfer (FMT) in children and
adolescents-review and statement by the GPGE microbiome working
group. Z Gastroenterol. 60:963–969. 2022.PubMed/NCBI
|
|
52
|
Porcari S, Severino A, Rondinella D, Bibbo
S, Quaranta G, Masucci L, Maida M, Scaldaferri F, Sanguinetti M,
Gasbarrini A, et al: Fecal microbiota transplantation for recurrent
Clostridioides difficile infection in patients with concurrent
ulcerative colitis. J Autoimmun. 141:1030332023. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Porcari S, Baunwall SMD, Occhionero AS,
Ingrosso MR, Ford AC, Hvas CL, Gasbarrini A, Cammarota G and Ianiro
G: Fecal microbiota transplantation for recurrent C. difficile
infection in patients with inflammatory bowel disease: A systematic
review and meta-analysis. J Autoimmun. 141:1030362023. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Porcari S, Benech N, Valles-Colomer M,
Segata N, Gasbarrini A, Cammarota G, Sokol H and Ianiro G: Key
determinants of success in fecal microbiota transplantation: From
microbiome to clinic. Cell Host Microbe. 31:712–733. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kim JH, Kim K and Kim W: Gut microbiota
restoration through fecal microbiota transplantation: A new atopic
dermatitis therapy. Exp Mol Med. 53:907–916. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu YH, Chen J, Chen X and Liu H: Factors
of faecal microbiota transplantation applied to cancer management.
J Drug Target. 32:101–114. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Arnold M, Singh D, Laversanne M, Vignat J,
Vaccarella S, Meheus F, Cust AE, de Vries E, Whiteman DC and Bray
F: Global burden of cutaneous melanoma in 2020 and projections to
2040. JAMA Dermatol. 158:495–503. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Baruch EN, Youngster I, Ben-Betzalel G,
Ortenberg R, Lahat A, Katz L, Adler K, Dick-Necula D, Raskin S,
Bloch N, et al: Fecal microbiota transplant promotes response in
immunotherapy-refractory melanoma patients. Science. 371:602–609.
2021. View Article : Google Scholar
|
|
59
|
Liu X, Luo Y, Chen X, Wu M, Xu X, Tian J,
Gao Y, Zhu J, Wang Z, Zhou Y, et al: Fecal microbiota
transplantation against moderate-to-severe atopic dermatitis: A
randomized, double-blind controlled exploratory trial. Allergy.
80:1377–1388. 2025. View Article : Google Scholar
|
|
60
|
Eshel A, Sharon I, Nagler A, Bomze D,
Danylesko I, Fein JA, Geva M, Henig I, Shimoni A, Zuckerman T, et
al: Origins of bloodstream infections following fecal microbiota
transplantation: A strain-level analysis. Blood Adv. 6:568–573.
2022. View Article : Google Scholar :
|
|
61
|
Gibson GR and Roberfroid MB: Dietary
modulation of the human colonic microbiota: Introducing the concept
of prebiotics. J Nutr. 125:1401–1412. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Hill C, Guarner F, Reid G, Gibson GR,
Merenstein DJ, Pot B, Morelli L, Canani RB, Flint HJ, Salminen S,
et al: Expert consensus document. The International Scientific
Association for Probiotics and Prebiotics consensus statement on
the scope and appropriate use of the term probiotic. Nat Rev
Gastroenterol Hepatol. 11:506–514. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Manzoor S, Wani SM, Ahmad Mir S and Rizwan
D: Role of probiotics and prebiotics in mitigation of different
diseases. Nutrition. 96:1116022022. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Oniszczuk A, Oniszczuk T, Gancarz M and
Szymanska J: Role of gut microbiota, probiotics and prebiotics in
the cardiovascular diseases. Molecules. 26:11722021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rahmayani T, Putra IB and Jusuf NK: The
effect of oral probiotics on the interleukin-10 serum levels of
acne vulgaris. Open Access Maced J Med Sci. 7:3249–5322. 2019.
View Article : Google Scholar
|
|
66
|
Ouyang W and O'Garra A: IL-10 family
cytokines IL-10 and IL-22: From basic science to clinical
translation. Immunity. 50:871–891. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Buhas MC, Candrea R, Gavrilas LI, Miere D,
Tataru A, Boca A and Cătinean A: Transforming psoriasis care:
Probiotics and prebiotics as novel therapeutic approaches. Int J
Mol Sci. 24:112252023. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Stein-Thoeringer CK, Saini NY, Zamir E,
Blumenberg V, Schubert ML, Mor U, Fante MA, Schmidt S, Hayase E,
Hayase T, et al: A non-antibiotic-disrupted gut microbiome is
associated with clinical responses to CD19-CAR-T cell cancer
immunotherapy. Nat Med. 29:906–916. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bender MJ, McPherson AC, Phelps CM, Pandey
SP, Laughlin CR, Shapira JH, Medina Sanchez L, Rana M, Richie TG,
Mims TS, et al: Dietary tryptophan metabolite released by
intratumoral Lactobacillus reuteri facilitates immune checkpoint
inhibitor treatment. Cell. 186:1846–1862 e26. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Raharja A, Mahil SK and Barker JN:
Psoriasis: A brief overview. Clin Med (Lond). 21:170–173. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Hidalgo-Cantabrana C, Gomez J, Delgado S,
Requena-Lopez S, Queiro-Silva R, Margolles A, Coto E, Sánchez B and
Coto-Segura P: Gut microbiota dysbiosis in a cohort of patients
with psoriasis. Br J Dermatol. 181:1287–1295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Zang C, Liu J, Mao M, Zhu W, Chen W and
Wei B: Causal associations between gut microbiota and psoriasis: A
mendelian randomization study. Dermatol Ther (Heidelb).
13:2331–2343. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Zhao Q, Yu J, Zhou H, Wang X, Zhang C, Hu
J, Hu Y, Zheng H, Zeng F, Yue C, et al: Intestinal dysbiosis
exacerbates the pathogenesis of psoriasis-like phenotype through
changes in fatty acid metabolism. Signal Transduct Target Ther.
8:402023. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Sroka-Tomaszewska J and Trzeciak M:
Molecular mechanisms of atopic dermatitis pathogenesis. Int J Mol
Sci. 22:41302021. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Langan SM, Irvine AD and Weidinger S:
Atopic dermatitis. Lancet. 396:345–360. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Schuler CF IV, Tsoi LC, Billi AC, Harms
PW, Weidinger S and Gudjonsson JE: Genetic and immunological
pathogenesis of atopic dermatitis. J Invest Dermatol. 144:954–968.
2024. View Article : Google Scholar :
|
|
77
|
Lee SY, Lee E, Park YM and Hong SJ:
Microbiome in the gut-skin axis in atopic dermatitis. Allergy
Asthma Immunol Res. 10:354–362. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Frazier W and Bhardwaj N: Atopic
dermatitis: Diagnosis and treatment. Am Fam Physician. 101:590–598.
2020.PubMed/NCBI
|
|
79
|
Jiang W, Ni B, Liu Z, Liu X, Xie W, Wu IXY
and Li X: The role of probiotics in the prevention and treatment of
atopic dermatitis in children: An updated systematic review and
meta-analysis of randomized controlled trials. Paediatr Drugs.
22:535–549. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
D'Elios S, Trambusti I, Verduci E,
Ferrante G, Rosati S, Marseglia GL, Drago L and Peroni DG:
Probiotics in the prevention and treatment of atopic dermatitis.
Pediatr Allergy Immunol. 31(Suppl 26): S43–S45. 2020. View Article : Google Scholar
|
|
81
|
Umborowati MA, Damayanti D, Anggraeni S,
Endaryanto A, Surono IS, Effendy I and Prakoeswa CRS: The role of
probiotics in the treatment of adult atopic dermatitis: A
meta-analysis of randomized controlled trials. J Health Popul Nutr.
41:372022. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Fang Z, Lu W, Zhao J, Zhang H, Qian L,
Wang Q and Chen W: Probiotics modulate the gut microbiota
composition and immune responses in patients with atopic
dermatitis: A pilot study. Eur J Nutr. 59:2119–2130. 2020.
View Article : Google Scholar
|
|
83
|
Sanchez-Pellicer P, Navarro-Moratalla L,
Nunez-Delegido E, Ruzafa-Costas B, Aguera-Santos J and
Navarro-Lopez V: Acne, microbiome, and probiotics: The gut-skin
axis. Microorganisms. 10:13032022. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Vos T, Flaxman AD, Naghavi M, Lozano R,
Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V,
et al: Years lived with disability (YLDs) for 1160 sequelae of 289
diseases and injuries, 1990-2010: A systematic analysis for the
Global Burden of Disease Study 2010. Lancet. 380:2163–2196. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Kutlu O, Karadag AS and Wollina U: Adult
acne versus adolescent acne: A narrative review with a focus on
epidemiology to treatment. An Bras Dermatol. 98:75–83. 2023.
View Article : Google Scholar :
|
|
86
|
Chilicka K, Rogowska AM, Szygula R,
Dziendziora-Urbinska I and Taradaj J: A comparison of the
effectiveness of azelaic and pyruvic acid peels in the treatment of
female adult acne: A randomized controlled trial. Sci Rep.
10:126122020. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Bowe W, Patel NB and Logan AC: Acne
vulgaris, probiotics, and the gut-brain-skin axis: From anecdote to
translational medicine. Beneficial Microbes. 5:185–199. 2014.
View Article : Google Scholar
|
|
88
|
Mohsin N, Hernandez LE, Martin MR, Does AV
and Nouri K: Acne treatment review and future perspectives.
Dermatol Ther. 35:e157192022. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Jung GW, Tse JE, Guiha I and Rao J:
Prospective, randomized, open-label trial comparing the safety,
efficacy, and tolerability of an acne treatment regimen with and
without a probiotic supplement and minocycline in subjects with
mild to moderate acne. J Cutan Med Surg. 17:114–122. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Fabbrocini G, Bertona M, Picazo O,
Pareja-Galeano H, Monfrecola G and Emanuele E: Supplementation with
Lactobacillus rhamnosus SP1 normalises skin expression of genes
implicated in insulin signalling and improves adult acne. Benef
Microbes. 7:625–630. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Eguren C, Navarro-Blasco A, Corral-Forteza
M, Reolid-Perez A, Seto-Torrent N, Garcia-Navarro A, Prieto-Merino
D, Núñez-Delegido E, Sánchez-Pellicer P and Navarro-López V: A
randomized clinical trial to evaluate the efficacy of an oral
probiotic in acne vulgaris. Acta Derm Venereol. 104:adv332062024.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
van Zuuren EJ, Arents BWM, van der Linden
MMD, Vermeulen S, Fedorowicz Z and Tan J: Rosacea: New concepts in
classification and treatment. Am J Clin Dermatol. 22:457–465. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Ivanic MG, Oulee A, Norden A, Javadi SS,
Gold MH and Wu JJ: Neurogenic rosacea treatment: A literature
review. J Drugs Dermatol. 22:566–575. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Haber R and El Gemayel M: Comorbidities in
rosacea: A systematic review and update. J Am Acad Dermatol.
78:786–792 e8. 2018. View Article : Google Scholar
|
|
95
|
Jun YK, Yu DA, Han YM, Lee SR, Koh SJ and
Park H: The relationship between rosacea and inflammatory bowel
disease: A systematic review and meta-analysis. Dermatol Ther
(Heidelb). 13:1465–1475. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Li M, He SX, He YX, Hu XH and Zhou Z:
Detecting potential causal relationship between inflammatory bowel
disease and rosacea using bi-directional Mendelian randomization.
Sci Rep. 13:149102023. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Manzhalii E, Hornuss D and Stremmel W:
Intestinal-borne dermatoses significantly improved by oral
application of Escherichia coli Nissle 1917. World J Gastroenterol.
22:5415–5421. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Fortuna MC, Garelli V, Pranteda G,
Romaniello F, Cardone M, Carlesimo M and Rossi A: A case of scalp
rosacea treated with low-dose doxycycline and probiotic therapy and
literature review on therapeutic options. Dermatol Ther.
29:249–251. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Zuberbier T, Abdul Latiff AH, Abuzakouk M,
Aquilina S, Asero R, Baker D, Ballmer-Weber B, Bangert C,
Ben-Shoshan M, Bernstein JA, et al: The international
EAACI/GA(2)LEN/EuroGuiDerm/APAAACI guideline for the definition,
classification, diagnosis, and management of urticaria. Allergy.
77:734–766. 2022. View Article : Google Scholar
|
|
100
|
Kaplan A, Lebwohl M, Gimenez-Arnau AM,
Hide M, Armstrong AW and Maurer M: Chronic spontaneous urticaria:
Focus on pathophysiology to unlock treatment advances. Allergy.
78:389–401. 2023. View Article : Google Scholar
|
|
101
|
Kolkhir P, Gimenez-Arnau AM, Kulthanan K,
Peter J, Metz M and Maurer M: Urticaria. Nat Rev Dis Primers.
8:612022. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Wang D, Guo S, He H, Gong L and Cui H: Gut
microbiome and serum metabolome analyses identify unsaturated fatty
acids and butanoate metabolism induced by gut microbiota in
patients with chronic spontaneous urticaria. Front Cell Infect
Microbiol. 10:242020. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Luo Z, Jin Z, Tao X, Wang T, Wei P, Zhu C
and Wang Z: Combined microbiome and metabolome analysis of gut
microbiota and metabolite interactions in chronic spontaneous
urticaria. Front Cell Infect Microbiol. 12:10947372023. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Fu HY, Yu HD, Bai YP, Yue LF, Wang HM and
Li LL: Effect and safety of probiotics for treating urticaria: A
systematic review and meta-analysis. J Cosmet Dermatol.
22:2663–2670. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Bi XD, Lu BZ, Pan XX, Liu S and Wang JY:
Adjunct therapy with probiotics for chronic urticaria in children:
Randomised placebo-controlled trial. Allergy Asthma Clin Immunol.
17:392021. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Spritz RA and Santorico SA: The genetic
basis of vitiligo. J Invest Dermatol. 141:265–273. 2021. View Article : Google Scholar
|
|
107
|
Bergqvist C and Ezzedine K: Vitiligo: A
review. Dermatology. 236:571–592. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Frisoli ML, Essien K and Harris JE:
Vitiligo: Mechanisms of pathogenesis and treatment. Annu Rev
Immunol. 38:621–648. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Kussainova A, Kassym L, Akhmetova A,
Glushkova N, Sabirov U, Adilgozhina S, Tuleutayeva R and Semenova
Y: Vitiligo and anxiety: A systematic review and meta-analysis.
PLoS One. 15:e02414452020. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Hadi A, Wang JF, Uppal P, Penn LA and
Elbuluk N: Comorbid diseases of vitiligo: A 10-year cross-sectional
retrospective study of an urban US population. J Am Acad Dermatol.
82:628–633. 2020. View Article : Google Scholar
|
|
111
|
Ni Q, Ye Z, Wang Y, Chen J, Zhang W, Ma C,
Li K, Liu Y, Liu L, Han Z, et al: Gut microbial dysbiosis and
plasma metabolic profile in individuals with vitiligo. Front
Microbiol. 11:5922482020. View Article : Google Scholar :
|
|
112
|
Luan M, Niu M, Yang P, Han D, Zhang Y, Li
W, He Q, Zhao Y, Mao B, Chen J, et al: Metagenomic sequencing
reveals altered gut microbial compositions and gene functions in
patients with non-segmental vitiligo. BMC Microbiol. 23:2652023.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Siegel RL, Miller KD, Wagle NS and Jemal
A: Cancer statistics, 2023. CA Cancer J Clin. 73:17–48.
2023.PubMed/NCBI
|
|
114
|
Arivazhagan N, Mukunthan MA,
Sundaranarayana D, Shankar A, Vinoth Kumar S, Kesavan R,
Chandrasekaran S, Shyamala Devi M, Maithili K, Barakkath Nisha U
and Abebe TG: Analysis of skin cancer and patient healthcare using
data mining techniques. Comput Intell Neurosci. 2022:22502752022.
View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Hasan N, Nadaf A, Imran M, Jiba U, Sheikh
A, Almalki WH, Almujri SS, Mohammed YH, Kesharwani P and Ahmad FJ:
Skin cancer: Understanding the journey of transformation from
conventional to advanced treatment approaches. Mol Cancer.
22:1682023. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Jindal M, Kaur M, Nagpal M, Singh M,
Aggarwal G and Dhingra GA: Skin cancer management: Current scenario
and future perspectives. Curr Drug Saf. 18:143–158. 2023.
View Article : Google Scholar
|
|
117
|
Abbas O, Miller DD and Bhawan J: Cutaneous
malignant melanoma: Update on diagnostic and prognostic biomarkers.
Am J Dermatopathol. 36:363–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Mrazek J, Mekadim C, Kucerova P, Svejstil
R, Salmonova H, Vlasakova J, Tarasová R, Čížková J and Červinková
M: Melanoma-related changes in skin microbiome. Folia Microbiol
(Praha). 64:435–442. 2019. View Article : Google Scholar
|
|
119
|
Mekadim C, Skalnikova HK, Cizkova J,
Cizkova V, Palanova A, Horak V and Mrazek J: Dysbiosis of skin
microbiome and gut microbiome in melanoma progression. BMC
Microbiol. 22:632022. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Jia D, Wang Q, Qi Y, Jiang Y, He J, Lin Y,
Sun Y, Xu J, Chen W, Fan L, et al: Microbial metabolite enhances
immunotherapy efficacy by modulating T cell stemness in pan-cancer.
Cell. 187:1651–1665 e21. 2024. View Article : Google Scholar
|
|
121
|
Zhou CB, Zhou YL and Fang JY: Gut
microbiota in cancer immune response and immunotherapy. Trends
Cancer. 7:647–660. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Andrews MC, Duong CPM, Gopalakrishnan V,
Iebba V, Chen WS, Derosa L, Khan MAW, Cogdill AP, White MG, Wong
MC, et al: Gut microbiota signatures are associated with toxicity
to combined CTLA-4 and PD-1 blockade. Nat Med. 27:1432–1441. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Spencer CN, McQuade JL, Gopalakrishnan V,
McCulloch JA, Vetizou M, Cogdill AP, Khan MAW, Zhang X, White MG,
Peterson CB, et al: Dietary fiber and probiotics influence the gut
microbiome and melanoma immunotherapy response. Science.
374:1632–1640. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Routy B, Lenehan JG, Miller WH Jr, Jamal
R, Messaoudene M, Daisley BA, Hes C, Al KF, Martinez-Gili L,
Punčochář M, et al: Author correction: Fecal microbiota
transplantation plus anti-PD-1 immunotherapy in advanced melanoma:
A phase I trial. Nat Med. 30:6042024. View Article : Google Scholar
|
|
125
|
Ingram J: Editor's choice: Rare skin
diseases themed issue. Br J Dermatol. 182:ix2020. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Buonavoglia A, Leone P, Dammacco R, Di
Lernia G, Petruzzi M, Bonamonte D, Vacca A, Racanelli V and
Dammacco F: Pemphigus and mucous membrane pemphigoid: An update
from diagnosis to therapy. Autoimmun Rev. 18:349–358. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Malik AM, Tupchong S, Huang S, Are A, Hsu
S and Otaparthi K: An updated review of pemphigus diseases.
Medicina (Kaunas). 57:10802021. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Kridin K: Pemphigus group: overview,
epidemiology, mortality, and comorbidities. Immunol Res.
66:255–270. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Yamagami J: B-cell targeted therapy of
pemphigus. J Dermatol. 50:124–131. 2023. View Article : Google Scholar
|
|
130
|
Huang S, Mao J, Zhou L, Xiong X and Deng
Y: The imbalance of gut microbiota and its correlation with plasma
inflammatory cytokines in pemphigus vulgaris patients. Scand J
Immunol. 90:e127992019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Han Z, Fan Y, Wu Q, Guo F, Li S, Hu X and
Zuo YG: Comparison of gut microbiota dysbiosis between pemphigus
vulgaris and bullous pemphigoid. Int Immunopharmacol.
128:1114702024. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Ioannides D, Vakirlis E, Kemeny L,
Marinovic B, Massone C, Murphy R, Nast A, Ronnevig J, Ruzicka T,
Cooper SM, et al: European S1 guidelines on the management of
lichen planus: A cooperation of the European Dermatology Forum with
the European academy of dermatology and venereology. J Eur Acad
Dermatol Venereol. 34:1403–1414. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Chen K, Qin Y, Yan L, Dong Y, Lv S, Xu J,
Kang N, Luo Z, Liu Y, Pu J, et al: Variations in salivary
microbiota and metabolic phenotype related to oral lichen planus
with psychiatric symptoms. BMC Oral Health. 25:9932025. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Jacques L, Kornik R, Bennett DD and
Eschenbach DA: Diagnosis and management of vulvovaginal lichen
planus. Obstet Gynecol Surv. 75:624–635. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
van Hees CLM and van der Meij EH: Lichen
planus. Ned Tijdschr Tandheelkd. 130:221–226. 2023.In Dutch.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Husein-ElAhmed H, Gieler U and Steinhoff
M: Lichen planus: A comprehensive evidence-based analysis of
medical treatment. J Eur Acad Dermatol Venereol. 33:1847–1862.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Georgescu SR, Mitran CI, Mitran MI,
Nicolae I, Matei C, Ene CD, Popa GL and Tampa M: Oxidative stress
in cutaneous lichen planus narrative review. J Clin Med.
10:26922021. View Article : Google Scholar
|
|
138
|
Li SZ, Wu QY, Fan Y, Guo F, Hu XM and Zuo
YG: Gut microbiome dysbiosis in patients with pemphigus and
correlation with pathogenic autoantibodies. Biomolecules.
14:8802024. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Roy S, Nag S, Saini A and Choudhury L:
Association of human gut microbiota with rare diseases: A close
peek through. Intractable Rare Dis Res. 11:52–62. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Kamal Y, Abdelwhab A, Salem ST and Fakhr
M: Evaluation of the efficacy of supplementary probiotic capsules
with topical clobetasol propionate 0.05% versus topical clobetasol
propionate 0.05% in the treatment of oral lichen planus (a
randomized clinical trial). BMC Oral Health. 25:3442025. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Shanahan F, Ghosh TS and O'Toole PW: The
healthy microbiome-what is the definition of a healthy gut
microbiome? Gastroenterology. 160:483–494. 2021. View Article : Google Scholar
|
|
142
|
Shalon D, Culver RN, Grembi JA, Folz J,
Treit PV, Shi H, Rosenberger FA, Dethlefsen L, Meng X, Yaffe E, et
al: Profiling the human intestinal environment under physiological
conditions. Nature. 617:581–591. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Schmidt TSB, Raes J and Bork P: The human
gut microbiome: From association to modulation. Cell.
172:1198–1215. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
McCallum G and Tropini C: The gut
microbiota and its biogeography. Nat Rev Microbiol. 22:105–118.
2024. View Article : Google Scholar
|
|
145
|
Wang J and Jia H: Metagenome-wide
association studies: fine-mining the microbiome. Nat Rev Microbiol.
14:508–522. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Uitterlinden AG: An introduction to
genome-wide association studies: GWAS for dummies. Semin Reprod
Med. 34:196–204. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Li L, Yang S, Li R, Su J, Zhou X, Zhu X
and Gao R: Unraveling shared and unique genetic causal relationship
between gut microbiota and four types of uterine-related diseases:
Bidirectional mendelian inheritance approaches to dissect the
'gut-uterus axis'. Ann Epidemiol. 100:16–26. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Asnicar F, Thomas AM, Passerini A, Waldron
L and Segata N: Machine learning for microbiologists. Nat Rev
Microbiol. 22:191–205. 2024. View Article : Google Scholar
|
|
149
|
Ma J, Fang Y, Li S, Zeng L, Chen S, Li Z,
Ji G, Yang X and Wu W: Interpretable machine learning algorithms
reveal gut microbiome features associated with atopic dermatitis.
Front Immunol. 16:15280462025. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Al-Bakri AG, Akour AA and Al-Delaimy WK:
Knowledge, attitudes, ethical and social perspectives towards fecal
microbiota transplantation (FMT) among Jordanian healthcare
providers. BMC Med Ethics. 22:192021. View Article : Google Scholar
|
|
151
|
Benech N, Barbut F, Fitzpatrick F, Krutova
M, Davies K, Druart C, Cordaillat-Simmons M, Heritage J, Guery B
and Kuijper E; ESGCD and ESGHAMI: Update on microbiota-derived
therapies for recurrent Clostridioides difficile infections. Clin
Microbiol Infect. 30:462–468. 2024. View Article : Google Scholar
|
|
152
|
Sada RM, Matsuo H, Motooka D, Kutsuna S,
Hamaguchi S, Yamamoto G and Ueda A: Clostridium butyricum
Bacteremia associated with probiotic use, Japan. Emerg Infect Dis.
30:665–671. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Zawistowska-Rojek A and Tyski S: Are
probiotics safe for humans? Pol J Microbiol. 67:251–258. 2018.
View Article : Google Scholar
|
|
154
|
Gouriet F, Million M, Henri M, Fournier PE
and Raoult D: Lactobacillus rhamnosus bacteremia: An emerging
clinical entity. Eur J Clin Microbiol Infect Dis. 31:2469–2480.
2021. View Article : Google Scholar
|
|
155
|
Harty DW, Oakey HJ, Patrikakis M, Hume EB
and Knox KW: Pathogenic potential of lactobacilli. Int J Food
Microbiol. 24:179–189. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Bartalesi F, Veloci S, Baragli F,
Mantengoli E, Guidi S, Bartolesi AM, Mannino R, Pecile P and
Bartoloni A: Successful tigecycline lock therapy in a Lactobacillus
rhamnosus catheter-related bloodstream infection. Infection.
40:331–334. 2012. View Article : Google Scholar
|
|
157
|
Jin DM, Morton JT and Bonneau R:
Meta-analysis of the human gut microbiome uncovers shared and
distinct microbial signatures between diseases. mSystems.
9:e00295242024. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Camacho DM, Collins KM, Powers RK,
Costello JC and Collins JJ: Next-generation machine learning for
biological networks. Cell. 173:1581–1592. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Su Q, Liu Q, Lau RI, Zhang J, Xu Z, Yeoh
YK, Leung TWH, Tang W, Zhang L, Liang JQY, et al: Faecal
microbiome-based machine learning for multi-class disease
diagnosis. Nat Commun. 13:68182022. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Chuang YF, Fan KC, Su YY, Wu MF, Chiu YL,
Liu YC and Lin CC: Precision probiotics supplement strategy in an
aging population based on gut microbiome composition. Brief
Bioinform. 25:bbae3512024. View Article : Google Scholar
|
|
161
|
Guimaraes JT, Balthazar CF, Scudino H,
Pimentel TC, Esmerino EA, Ashokkumar M, Freitas MQ and Cruz AG:
High-intensity ultrasound: A novel technology for the development
of probiotic and prebiotic dairy products. Ultrason Sonochem.
57:12–21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Balthazar CF, Guimaraes JF, Coutinho NM,
Pimentel TC, Ranadheera CS, Santillo A, Albenzio M, Cruz AG and
Sant'Ana AS: The future of functional food: Emerging technologies
application on prebiotics, probiotics, and postbiotics. Compr Rev
Food Sci Food Saf. 21:2560–2586. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Kucukgoz K and Trzaskowska M: Nondairy
probiotic products: Functional foods that require more attention.
Nutrients. 14:7532022. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Shumye Gebre T, Admassu Emire S, Okomo
Aloo S, Chelliah R, Vijayalakshmi S and Hwan Oh D: Unveiling the
potential of African fermented cereal-based beverages: Probiotics,
functional drinks, health benefits and bioactive components. Food
Res Int. 191:1146562024. View Article : Google Scholar : PubMed/NCBI
|