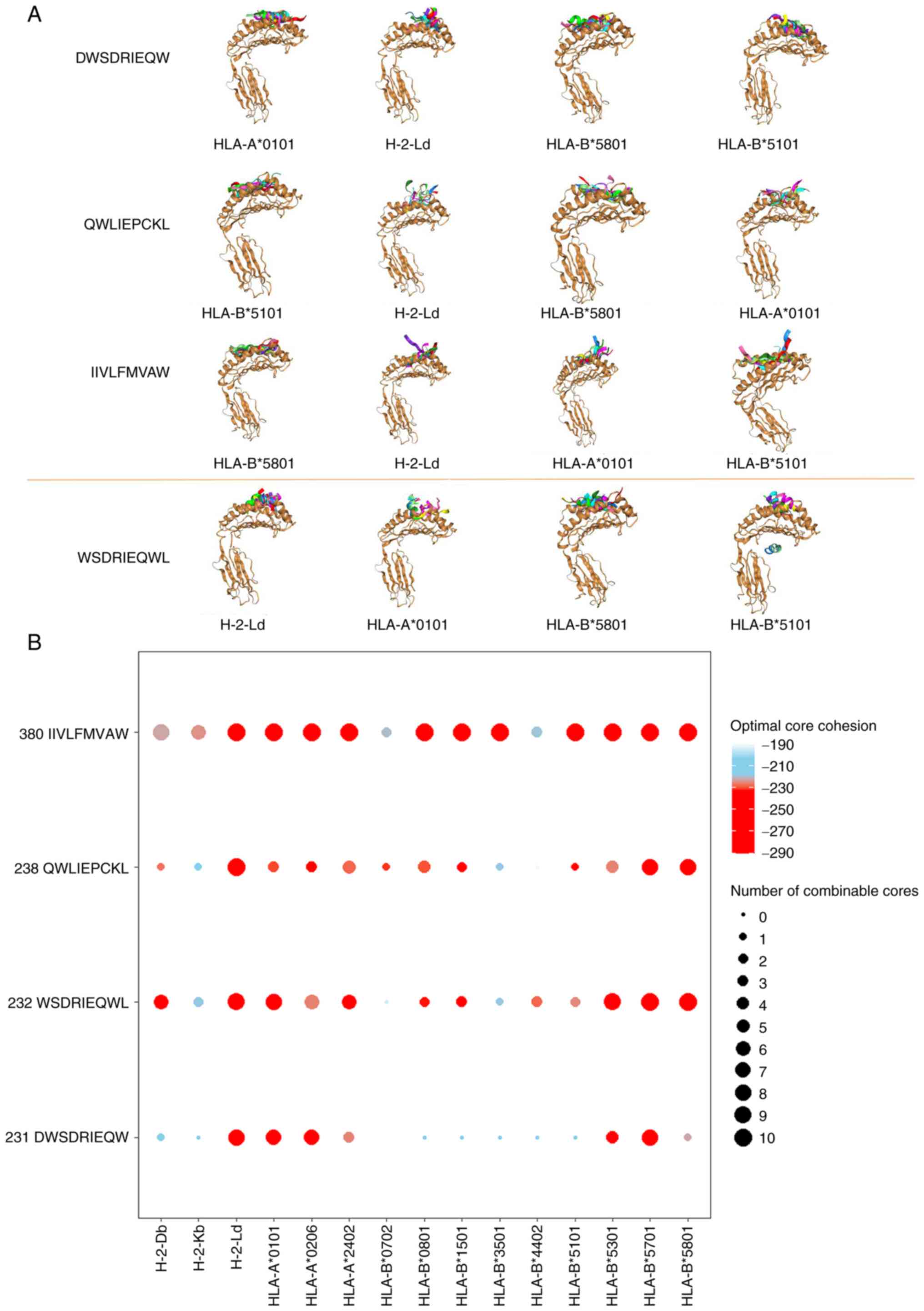
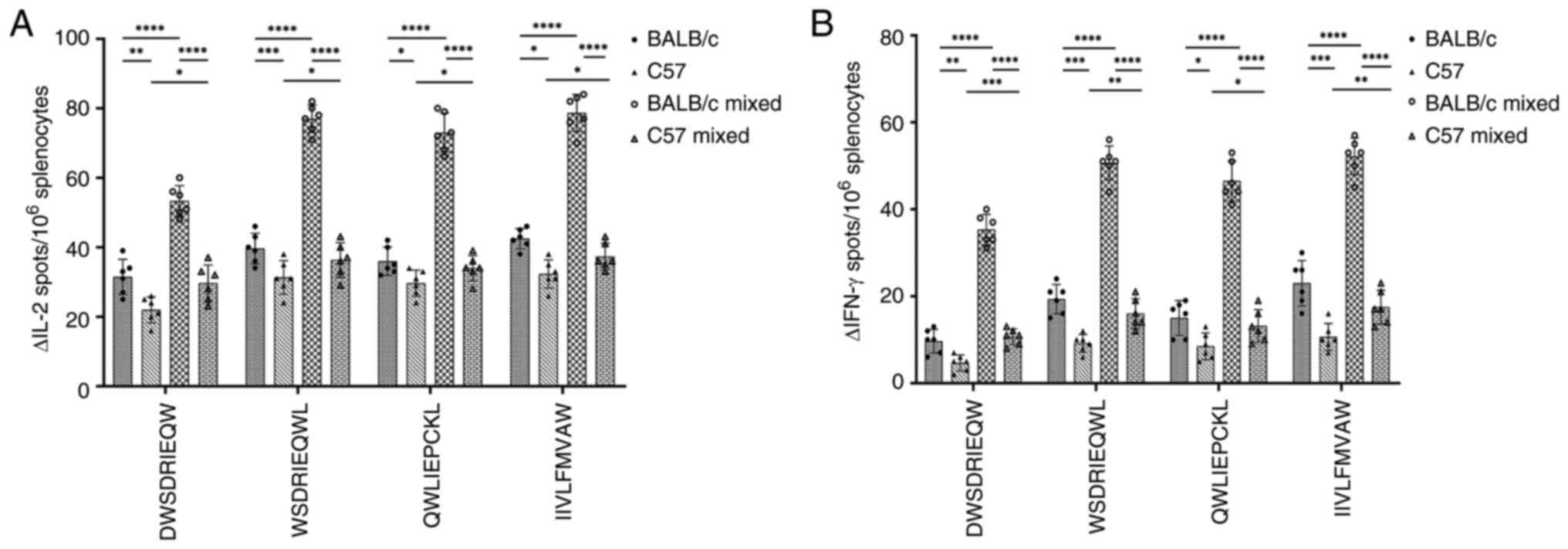
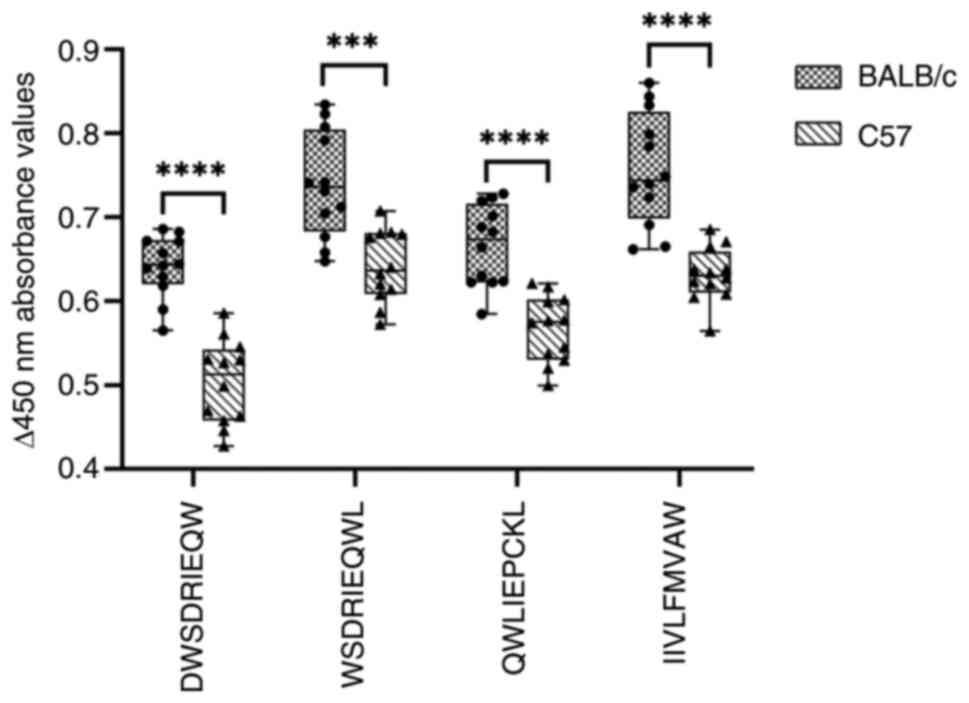
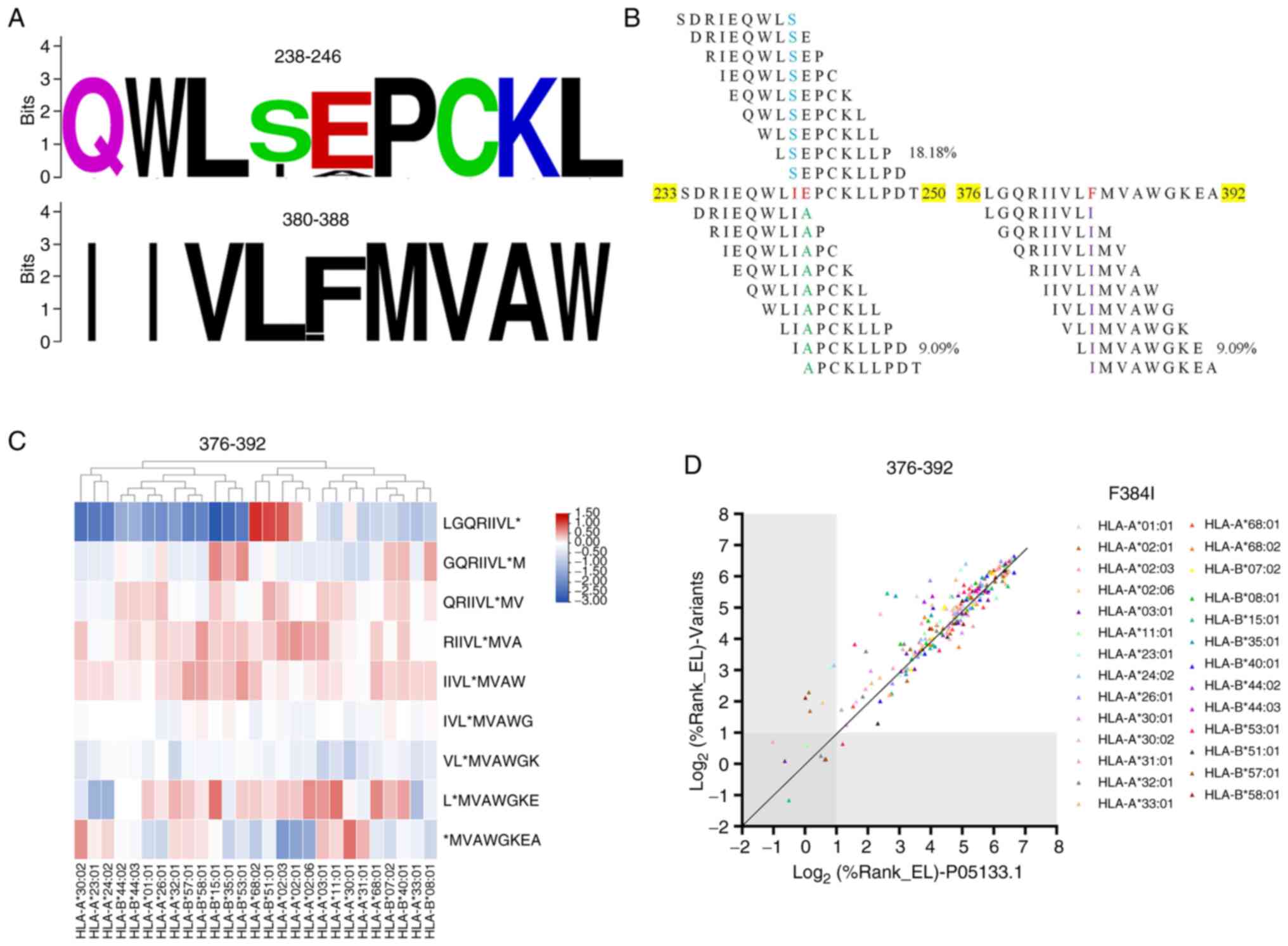
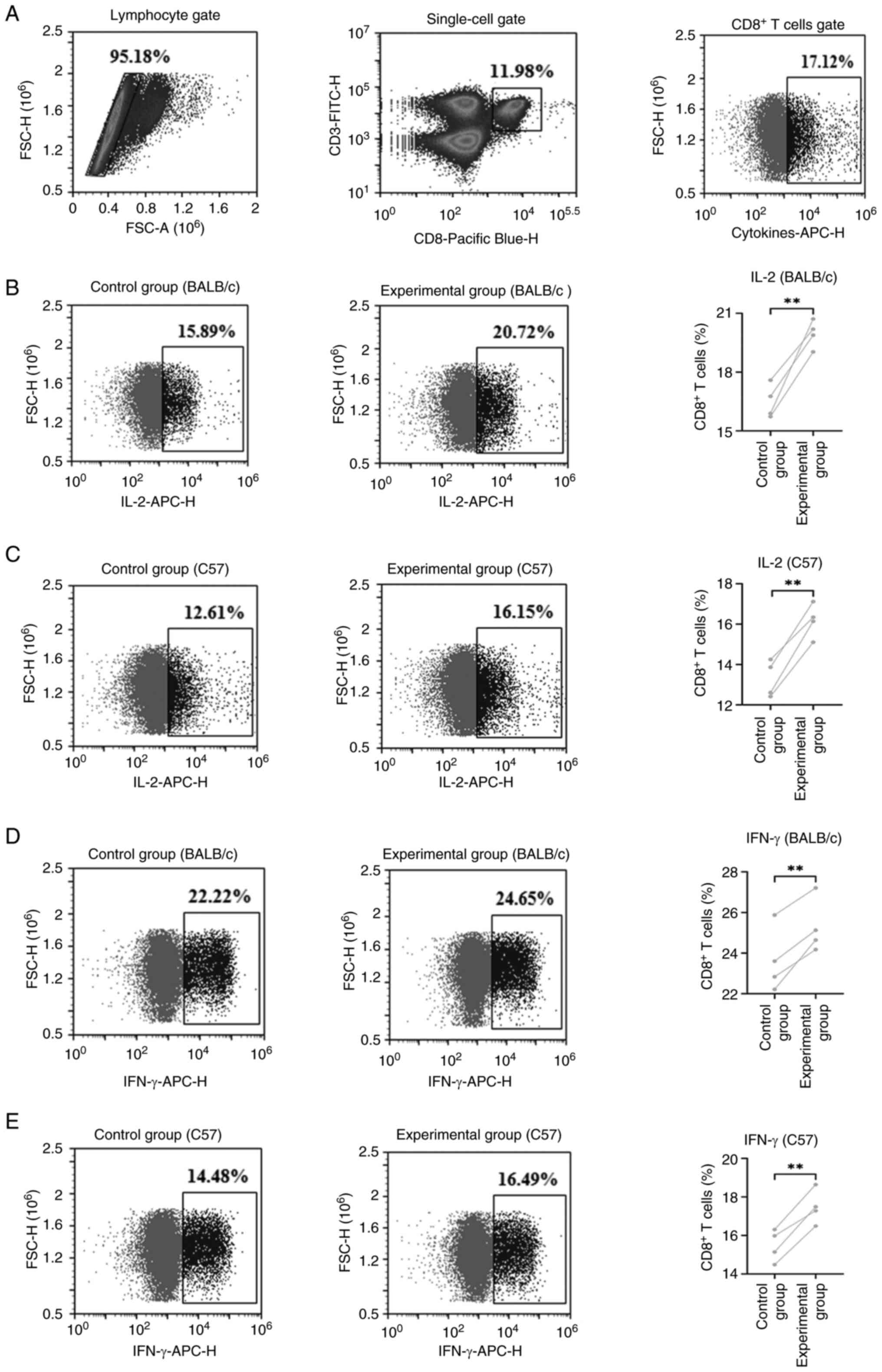
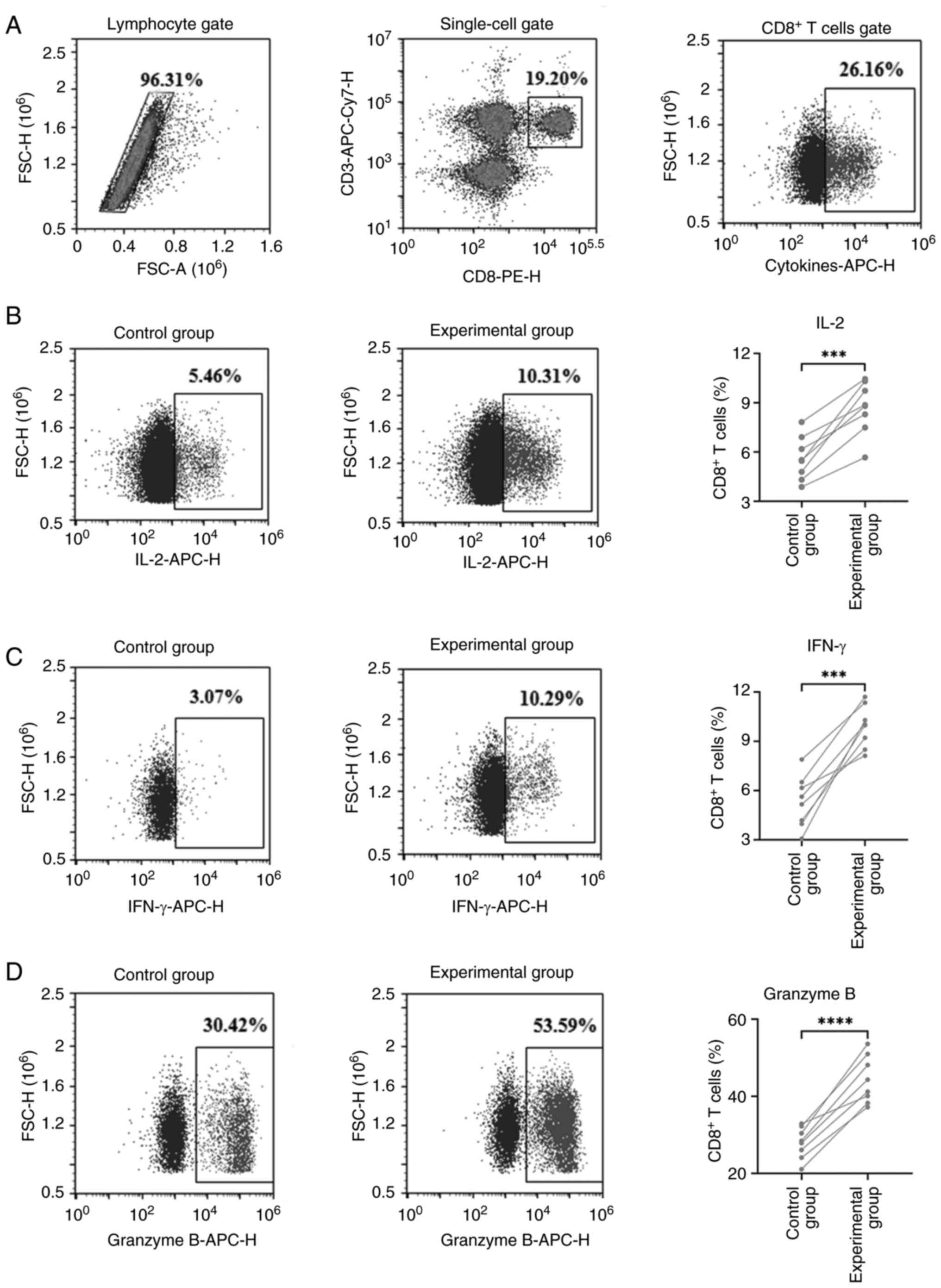
|
1
|
Brocato RL, Wu H, Kwilas SA, Principe LM,
Josleyn M, Shamblin J, Chivukula P, Bausch C, Luke T, Sullivan EJ
and Hooper JW: Preclinical evaluation of a fully human,
quadrivalent-hantavirus polyclonal antibody derived from a
non-human source. mBio. 15:e1600242024. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He J, Wang Y, Wei X, Sun H, Xu Y, Yin W,
Wang Y and Zhang W: Spatial-temporal dynamics and time series
prediction of HFRS in mainland China: A long-term retrospective
study. J Med Virol. 95:e282692023. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mittler E, Wec AZ, Tynell J,
Guardado-Calvo P, Wigren-Byström J, Polanco LC, O'Brien CM, Slough
MM, Abelson DM, Serris A, et al: Human antibody recognizing a
quaternary epitope in the Puumala virus glycoprotein provides broad
protection against orthohantaviruses. Sci Transl Med.
14:eabl53992022. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim J, Park K, Kim K, Noh J, Kim S, Yang
E, Cho H, Lee S, No JS, Lee G, et al: High-resolution
phylogeographical surveillance of Hantaan orthohantavirus using
rapid amplicon-based Flongle sequencing, Republic of Korea. J Med
Virol. 96:e293462024. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chai S, Wang L, Du H and Jiang H:
Achievement and challenges in orthohantavirus vaccines. Vaccines
(Basel). 13:1982025. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sabsay KR and Te Velthuis AJW: Using
structure prediction of negative sense RNA virus nucleoproteins to
assess evolutionary relationships. Virus Evol. 10:veae0582024.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Noor F, Ashfaq UA, Asif M, Adeel MM,
Alshammari A and Alharbi M: Comprehensive computational analysis
reveals YXXΦ[I/L/M/F/V] motif and YXXΦ-like tetrapeptides across
HFRS causing hantaviruses and their association with viral
pathogenesis and host immune regulation. Front Immunol.
13:10316082022. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ying Q, Zhang X, Wang S, Gu T, Zhang J,
Feng W, Li D, Dong Y, Wu X and Wang F: A novel HTNV budding
inhibitor interferes the interaction between viral glycoprotein and
host ESCRT accessory protein ALIX. J Med Virol. 97:e701822025.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen QZ, Wang X, Luo F, Li N, Zhu N, Lu S,
Zan YX, Zhong CJ, Wang MR, Hu HT, et al: HTNV sensitizes host
toward TRAIL-Mediated Apoptosis-A pivotal anti-hantaviral role of
TRAIL. Front Immunol. 11:10722020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brunnberg J, Barends M, Frühschulz S,
Winter C, Battin C, de Wet B, Cole DK, Steinberger P and Tampé R:
Dual role of the peptide-loading complex as proofreader and limiter
of MHC-I presentation. Proc Natl Acad Sci USA. 121:e23216001212024.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang X, Zhang H, Wang Y, Bramasole L, Guo
K, Mourtada F, Meul T, Hu Q, Viteri V, Kammerl I, et al: DNA
sensing via the cGAS/STING pathway activates the immunoproteasome
and adaptive T-cell immunity. EMBO J. 42:e1105972023. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Barbet G, Nair-Gupta P, Schotsaert M,
Yeung ST, Moretti J, Seyffer F, Metreveli G, Gardner T, Choi A,
Tortorella D, et al: TAP dysfunction in dendritic cells enables
noncanonical cross-presentation for T cell priming. Nat Immunol.
22:497–509. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Baljon JJ and Wilson JT: Bioinspired
vaccines to enhance MHC class-I antigen cross-presentation. Curr
Opin Immunol. 77:1022152022. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ma Y, Wang J, Yuan B, Wang M, Zhang Y, Xu
Z, Zhang C, Zhang Y, Liu B, Yi J, et al: HLA-A2 and B35 restricted
hantaan virus nucleoprotein CD8+ T-cell epitope-specific immune
response correlates with milder disease in hemorrhagic fever with
renal syndrome. PLoS Negl Trop Dis. 7:e20762013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma Y, Tang K, Zhang Y, Zhang C, Zhang Y,
Jin B and Ma Y: Design and synthesis of HLA-A*02-restricted Hantaan
virus multiple-antigenic peptide for CD8+ T cells. Virol
J. 17:152020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan Z, Kim K, Kim H, Ha B, Gambiez A,
Bennett J, de Almeida Mendes MF, Trevizani R, Mahita J, Richardson
E, et al: Next-generation IEDB tools: A platform for epitope
prediction and analysis. Nucleic Acids Res. 52((W1)): W526–W532.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Reynisson B, Alvarez B, Paul S, Peters B
and Nielsen M: NetMHCpan-4.1 and NetMHCIIpan-4.0: Improved
predictions of MHC antigen presentation by concurrent motif
deconvolution and integration of MS MHC eluted ligand data. Nucleic
Acids Res. 48((W1)): W449–W454. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Rammensee H, Bachmann J, Emmerich NP,
Bachor OA and Stevanovic S: SYFPEITHI: Database for MHC ligands and
peptide motifs. Immunogenetics. 50:213–219. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Reche PA, Glutting JP, Zhang H and
Reinherz EL: Enhancement to the RANKPEP resource for the prediction
of peptide binding to MHC molecules using profiles. Immunogenetics.
56:405–419. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim Y, Sidney J, Pinilla C, Sette A and
Peters B: Derivation of an amino acid similarity matrix for
peptide: MHC binding and its application as a Bayesian prior. BMC
Bioinformatics. 10:3942009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Polonskaya Z, Savage PB, Finn MG and
Teyton L: High-affinity anti-glycan antibodies: Challenges and
strategies. Curr Opin Immunol. 59:65–71. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bahrami AA, Payandeh Z, Khalili S, Zakeri
A and Bandehpour M: Immunoinformatics: In silico approaches and
computational design of a multi-epitope, immunogenic protein. Int
Rev Immunol. 38:307–322. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Saethang T, Hirose O, Kimkong I, Tran VA,
Dang XT, Nguyen LAT, Le TKT, Kubo M, Yamada Y and Satou K: PAAQD:
Predicting immunogenicity of MHC class I binding peptides using
amino acid pairwise contact potentials and quantum topological
molecular similarity descriptors. J Immunol Methods. 387:293–302.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou P, Jin B, Li H and Huang S: HPEPDOCK:
A web server for blind peptide-protein docking based on a
hierarchical algorithm. Nucleic Acids Res. 46((W1)): W443–W450.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Eberhardt J, Santos-Martins D, Tillack AF
and Forli S: AutoDock vina 1.2.0: New docking methods, expanded
force field, and python bindings. J Chem Inf Model. 61:3891–3898.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen C, Chen H, Zhang Y, Thomas HR, Frank
MH, He Y and Xia R: TBtools: An integrative toolkit developed for
interactive analyses of big biological data. Mol Plant.
13:1194–1202. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Crooks GE, Hon G, Chandonia J and Brenner
SE: WebLogo: A sequence logo generator. Genome Res. 14:1188–1190.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Rathore AS, Choudhury S, Arora A, Tijare P
and Raghava GPS: ToxinPred 3.0: An improved method for predicting
the toxicity of peptides. Comput Biol Med. 179:1089262024.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sharma N, Patiyal S, Dhall A, Pande A,
Arora C and Raghava GPS: AlgPred 2.0: An improved method for
predicting allergenic proteins and mapping of IgE epitopes. Brief
Bioinform. 22:bbaa2942021. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang D, Ma Z, Zhang J, Sun Y, Bai T, Liu
R, Wang Y, Guan L, Fu S, Sun Y, et al: Immunoreactivity analysis of
MHC-I epitopes derived from the nucleocapsid protein of SARS-CoV-2
via computation and vaccination. Vaccines (Basel). 12:12142024.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li Z, Sun Y, Sun B, Zhang J, Wang J, Fang
Z, Li Y, Ding W, Zhou B, Cai S, et al: Comparative immunobiology
and cross-species validations of pan-MHC-II epitopes on Hantaan
virus nucleocapsid protein. Int Immunopharmacol. 158:1148652025.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Woo GJ, Chun EY, Kim KH and Kim W:
Analysis of immune responses against nucleocapsid protein of the
Hantaan virus elicited by virus infection or DNA vaccination. J
Microbiol. 43:537–545. 2005.PubMed/NCBI
|
|
33
|
Park JM, Cho SY, Hwang YK, Um SH, Kim WJ,
Cheong HS and Byun SM: Identification of H-2K(b)-restricted T-cell
epitopes within the nucleocapsid protein of Hantaan virus and
establishment of cytotoxic T-cell clones. J Med Virol. 60:189–199.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lü X, Zhang F, Li Y, Xue X, Yin W and Xu
Z: Antigenic characterization of expressed complete and different
truncated recombinant nucleocapsid proteins of hantaan virus by
monoclonal antibodies. Hybridoma (Larchmt). 30:445–450. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sun B, Zhang J, Wang J, Liu Y, Sun H, Lu
Z, Chen L, Ding X, Pan J, Hu C, et al: Comparative immunoreactivity
analyses of hantaan virus glycoprotein-derived MHC-I epitopes in
vaccination. Vaccines (Basel). 10:5642022. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang H, Liu H, Wei J, Dang Y, Wang Y,
Yang Q, Zhang L, Ye C, Wang B, Jin X, et al: Single dose
recombinant VSV based vaccine elicits robust and durable
neutralizing antibody against Hantaan virus. NPJ Vaccines.
9:282024. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Rissmann M, Noack D, Spliethof TM, Vaes
VP, Stam R, van Run P, Clark JJ, Verjans GMGM, Haagmans BL, Krammer
F, et al: A pan-orthohantavirus human lung xenograft mouse model
and its utility for preclinical studies. PLoS Pathog.
21:e10128752025. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rakib A, Sami SA, Mimi NJ, Chowdhury MM,
Eva TA, Nainu F, Paul A, Shahriar A, Tareq AM, Emon NU, et al:
Immunoinformatics-guided design of an epitope-based vaccine against
severe acute respiratory syndrome coronavirus 2 spike glycoprotein.
Comput Biol Med. 124:1039672020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sun Y, Pumroy RA, Mallik L, Chaudhuri A,
Wang C, Hwang D, Danon JN, Dasteh Goli K, Moiseenkova-Bell VY and
Sgourakis NG: CryoEM structure of an MHC-I/TAPBPR peptide-bound
intermediate reveals the mechanism of antigen proofreading. Proc
Natl Acad Sci USA. 122:e24169921222025. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Goddery EN, Fain CE, Lipovsky CG, Ayasoufi
K, Yokanovich LT, Malo CS, Khadka RH, Tritz ZP, Jin F, Hansen MJ
and Johnson AJ: Microglia and perivascular macrophages act as
antigen presenting cells to promote CD8 T cell infiltration of the
brain. Front Immunol. 12:7264212021. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mcauliffe J, Panetti S, Steffke E, Wicki
A, Pereira-Almeida V, Noblecourt L, Hu Y, Guo SYW, Lesenfants J,
Ramirez-Valdez RA, et al: Novel H-2Db-restricted CD8
epitope derived from mouse MAGE-type antigen P1A mediates antitumor
immunity in C57BL/6 mice. J Immunother Cancer. 12:e0089982024.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim JA, Kim WK, No JS, Lee SH, Lee SY, Kim
JH, Kho JH, Lee D, Song DH, Gu SH, et al: Genetic diversity and
reassortment of hantaan virus tripartite RNA genomes in nature, the
Republic of Korea. PLoS Negl Trop Dis. 10:e00046502016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun H, Lu Z, Xuan G, Liu N, Wang T, Liu Y,
Lan M, Xu J, Feng Y, Xu S, et al: Integrative analysis of HTNV
glycoprotein derived MHC II epitopes by in silico prediction and
experimental validation. Front Cell Infect Microbiol.
11:6716942021. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang J, Sun B, Shen W, Wang Z, Liu Y, Sun
Y, Zhang J, Liu R, Wang Y, Bai T, et al: In silico analyses,
experimental verification and application in DNA vaccines of
ebolavirus GP-derived pan-MHC-II-restricted epitopes. Vaccines
(Basel). 11:16202023. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Jiang D, Zhang J, Shen W, Sun Y, Wang Z,
Wang J, Zhang J, Zhang G, Zhang G, Wang Y, et al: DNA vaccines
encoding HTNV GP-derived Th epitopes benefited from a
LAMP-targeting strategy and established cellular immunoprotection.
Vaccines (Basel). 12:9282024. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Rissanen I, Krumm SA, Stass R, Whitaker A,
Voss JE, Bruce EA, Rothenberger S, Kunz S, Burton DR, Huiskonen JT,
et al: Structural basis for a neutralizing antibody response
elicited by a recombinant hantaan virus Gn immunogen. mBio.
12:e2531202021. View Article : Google Scholar
|
|
47
|
Rak A, Isakova-Sivak I and Rudenko L:
Nucleoprotein as a promising antigen for broadly protective
influenza vaccines. Vaccines (Basel). 11:17472023. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao W, Chen W, Li J, Chen M, Li Q, Lv M,
Zhou S, Bai S, Wang Y, Zhang L, et al: Status of humoral and
cellular immune responses within 12 months following CoronaVac
vaccination against COVID-19. mBio. 13:e00181222022. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ahmadi P, Hartjen P, Kohsar M, Kummer S,
Schmiedel S, Bockmann JH, Fathi A, Huber S, Haag F and Schulze Zur
Wiesch J: Defining the CD39/CD73 axis in SARS-CoV-2 infection: The
CD73− phenotype identifies polyfunctional cytotoxic
lymphocytes. Cells. 9:17502020. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shibata H, Xu N, Saito S, Zhou L, Ozgenc
I, Webb J, Fu C, Zolkind P, Egloff AM and Uppaluri R: Integrating
CD4+ T cell help for therapeutic cancer vaccination in a
preclinical head and neck cancer model. Oncoimmunology.
10:19585892021. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sameer Khan M, Gupta G, Alsayari A, Wahab
S, Sahebkar A and Kesharwani P: Advancements in liposomal
formulations: A comprehensive exploration of industrial production
techniques. Int J Pharm. 658:1242122024. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Kocabas BB, Almacioglu K, Bulut EA,
Gucluler G, Tincer G, Bayik D, Gursel M and Gursel I: Dual-adjuvant
effect of pH-sensitive liposomes loaded with STING and TLR9
agonists regress tumor development by enhancing Th1 immune
response. J Control Release. 328:587–595. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ismail S, Abbasi SW, Yousaf M, Ahmad S,
Muhammad K and Waheed Y: Design of a multi-epitopes vaccine against
hantaviruses: An immunoinformatics and molecular modelling
approach. Vaccines (Basel). 10:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Weng MT, Yang SF, Liu SY, Hsu YC, Wu MC,
Chou HC, Chiou LL, Liang JD, Wang LF, Lee HS and Sheu JC: In situ
vaccination followed by intramuscular poly-ICLC injections for the
treatment of hepatocellular carcinoma in mouse models. Pharmacol
Res. 188:1066462023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chen X, Xu Z, Li T, Thakur A, Wen Y, Zhang
K, Liu Y, Liang Q, Liu W, Qin J and Yan Y:
Nanomaterial-encapsulated STING agonists for immune modulation in
cancer therapy. Biomark Res. 12:22024. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Americo JL, Cotter CA, Earl PL, Liu R and
Moss B: Intranasal inoculation of an MVA-based vaccine induces IgA
and protects the respiratory tract of hACE2 mice from SARS-CoV-2
infection. Proc Natl Acad Sci USA. 119:e22020691192022. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chatsiricharoenkul S, Niyomnaitham S,
Posen HJ, Toh ZQ, Licciardi PV, Wongprompitak P, Duangchinda T,
Pakchotanon P, Chantima W and Chokephaibulkit K: Safety and
immunogenicity of intradermal administration of fractional dose
CoronaVac®, ChAdOx1 nCoV-19 and BNT162b2 as primary
series vaccination. Front Immunol. 13:10108352022. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Kopicki J, Saikia A, Niebling S, Günther
C, Anjanappa R, Garcia-Alai M, Springer S and Uetrecht C: Opening
opportunities for Kd determination and screening of MHC
peptide complexes. Commun Biol. 5:4882022. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Peng S, Xing D, Ferrall L, Tsai Y, Roden
RBS, Hung C and Wu T: Development of a spontaneous HPV16
E6/E7-expressing head and neck squamous cell carcinoma in HLA-A2
transgenic mice. mBio. 13:e3252212022. View Article : Google Scholar
|
|
60
|
Gupta S, Nerli S, Kutti Kandy S, Mersky GL
and Sgourakis NG: HLA3DB: Comprehensive annotation of peptide/HLA
complexes enables blind structure prediction of T cell epitopes.
Nat Commun. 14:63492023. View Article : Google Scholar : PubMed/NCBI
|