Introduction
Gastric cancer is one of the most common types of
human cancer (1) and the second
most common cause of cancer mortality in the world (2). Gastric cancer has a wide variation in
geographical distribution and the incidence is particularly high in
Asia, South America and Eastern Europe (3–5).
Gastric cancer patients are frequently diagnosed in advanced
stages. Approximately 50% of all patients present with
unresectable, locally advanced or metastatic diseases (6,7) and
recurrence after curative gastrectomy remains high (8,9). The
prognosis for patients with advanced gastric cancer remains dismal,
and little improvement in the survival rates has been achieved in
recent years (6,8,9).
Gastric tumor metastases, during which the cancer cells spread to
distal sites such as the liver, omentum, and lymph nodes, are the
leading cause of cancer mortality. Gastric tumor of diffuse type is
poorly differentiated and comprises dissemination-prone gastric
cancer cells lacking the abilities of tissue cohesion and gastric
biological functions. This particular type of gastric cancer cells
demonstrates an increased propensity for metastatic spread via the
intra- and trans-peritoneal wall and, therefore, have been
associated with poor prognosis (7). The somatic inactivation of E-cadherin
in diffuse type gastric cancer also increases
epithelial-mesenchymal transition (EMT) ability and leads to an
enhanced metastasis potential of this gastric cancer type (10). There is currently no effective
chemotherapeutics, nor is a molecular-targeted drug available, for
treating the advanced gastric cancerous diseases. Further research
into molecular targeting drugs against gastric tumor metastasis is
warranted.
In the search for target molecules for the
development of anticancer drugs, research efforts to identify
biological molecules involved in the gastric cancer cell specific
proliferation, invasion and metastasis have been extensive.
Elevated plasma osteopontin was associated with gastric cancer
development and patient survival (11). Overexpression of c-met (12), LOXL2 (13), EGFR (14), HER2 (15), TNS4 (16) and phosphorylated mTOR (17) in gastric tumors has been well
correlated with the prognostic factors of tumor staging, the depth
of tumor invasion and/or lymph node metastasis. On the other hand,
reduced expressions of nm23 (18),
galectin-3 (19), or TIP30
(20), loss of RUNX3 (21) or E-cadherin (14) expressions, and abnormal expression
of E-cadherin (22) in gastric
tumors has been associated with gastric tumor metastasis and poor
prognosis. More systemically investigated, array-based technologies
were utilized to analyze and identify genes associated with gastric
cancer metastasis. Several studies compared human primary to
metastatic gastric cancer cell lines (23,24),
human gastric cancer cell lines to a normal gastric cell line
(25), unpaired tumors to normal
tissues (26), paired patient
gastric tumors to normal gastric tissues (25,27–31),
and patient gastric tumors with different clinical stages (32). However, differences in the genomic
backgrounds of patients and the mix of the tumor and its
surrounding cells may increase the complexities and difficulties
for the identification of specific genes and for database analyses.
Results of these array-based studies individually suggested that
putative metastasis-associated genes are not entirely the same, nor
are they similar, to a great extent, to each other. Further studies
are, therefore, needed to identify biologically relevant molecular
targets against which effective drugs for human gastric cancer may
be discovered.
In the present study, we generated, from a single
parental cell, a set of cell sublines exhibiting different
metastatic abilities and used them to identify the genes whose
expression levels are correlated to their metastatic abilities. We
applied a genome-wide scale cDNA microarray analysis using a
biological functional approach different from those previously
reported. A human gastric cancer cell MKN45 originally derived from
a poorly differentiated gastric adenocarcinoma of the stomach of a
62-year-old woman (33,34) was used as a model cell for its
diffuse phenotype potentially prone to metastasis. A series of the
human gastric cancer MKN45 cell subclones with varying invasion
capabilities were established. A number of putative tumor
metastasis-associated genes were identified using cDNA microarray
analysis and verified with correlated mRNA levels and protein
expressions.
Materials and methods
Cell line and transfection
The human diffuse-type gastric cancer cell line
MKN45 (JCRB0254) was purchased from the Japanese Collection of
Research Bioresources/Human Science Research Resources Bank (Osaka,
Japan). The cells were grown in RPMI-1640 medium (Gibco, product
no. 31800105) with 10% fetal bovine serum (FBS) (Invitrogen,
Carlsbad, CA, USA) and 2 mM L-glutamine at 37°C, 5% CO2.
One day before the transfection, 8×103 MKN45 cells were
seeded into 24-well culture plate and grown for 16–24 h to 70–90%
confluence for transfection. A mixture of enhanced green
fluorescent protein (GFP) expression plasmid (pEGFP-C1, cat. no.
6084-1) encoding under the control of cytomegalovirus promoter,
OPTI-MEM and Lipofectamine™ 2000 from Invitrogen at a ratio of 2
μg:2 μl:100 μl was added to the MKN45 cells.
The cells were harvested following incubation for 48 h at 37°C
followed by selection of the GFP-expressing clones by incubating
with 1 mg/ml G418 (Invitrogen) and isolated with limiting dilution
method to obtain stably GFP-expressing MKN45-GFP sublines.
Selection of highly invasive MKN45-GFP
sublines
The MKN45-GFP cells of the isolated clone were
selected for differential invasive characteristics using
Transwell® plates from Corning (Acton, MA, USA) with a
method modified from one previously described (35) Briefly, Transwell of the 24-well
inserts with semi-permeable polycarbonate membrane (8 μm in
pore size, Corning, cat. no. CLS3422, Corning, NY, USA) were coated
with 50 μl per well of reconstituted basement-membrane
matrix Matrigel™ (1:1 diluted with growth medium) from BD
Biosciences (San Jose, CA, USA). MKN45-GFP cells in RPMI-1640
containing 10% FBS were seeded at 1×105 cells/200
μl/well onto the upper surface of the Matrigel-coated
membrane. Following incubation for 72 h at 37°C, the inserts were
removed. The cells that migrated and invaded through the membrane
and attached to the surface of bottom well were harvested
aseptically and subsequently amplified for further selection
processes. The harvested invasive MKN45-GFP cell sublines after
each selection round were named with a number correspondingly.
In vitro invasion assay
The basement membrane matrix coated Transwell system
was used to measure the invasion ability of tumor cells as
previously reported (36).
Transwell of 24-well plates with a porous polycarbonate membrane of
8-μm pore size from Corning (Corning, NY) were coated with
50 μl Matrigel diluted (1:20 = v:v) with RPMI-1640 culture
medium. Cells (5×105) suspended in 200 μl
RPMI-1640 with 1% FBS were seeded into upper chamber. The lower
chamber was placed with 600 μl RPMI-1640 with 10% FBS.
Following incubation for 72 h at 37°C, the cells that invaded
through the Matrigel-coated membrane to the lower surface of the
membrane and the bottom surface of the culture well were fixed with
3.7% formaldehyde in phosphate-buffered saline (PBS) for 1 h. The
cells that remained on the upper surface of the membrane were
scraped with a cotton bud completely and invaded cells attached on
the opposite surface of the membrane in the lower chamber were
stained with hematoxylin for 1 h. The invaded cancer cells were
then visualized and counted from 5 different viewing areas under
100-fold magnification using a DM IRB inverted microscope from
Leica Microsystems (Wetzlar, Germany).
Cell proliferation activity
Cancer cell proliferation activity was measured
using a colorimetric
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
salt (MTS)/phenazine methosulfate (PMS) assay system as previously
reported (37). Cancer cells were
seeded at 2,000, 4,000, and 6,000 cells/well in 96-well culture
plates (Corning, Acton, MA) in triplicates. The cells were
harvested and the cell numbers were measured every 24 h or 6
consecutive days by a colorimetric reaction using MTS/PMS mixture,
2 mg/ml MTS (Promega, Madison, WI, USA) and 0.38 mg/ml PMS (Sigma,
St. Louis, MO, USA), in phenol red-free RPMI-1640 medium (Gibco,
product no. 11835-030) followed by optical density measurements at
490 nm after a 90-min incubation at 37°C. The cell doubling time
was obtained by nonlinear regression with the equation:
N=Ao*2^(T/Td) using SigmaPlot®
from SPSS Inc. (Chicago, IL, USA), in which N is the total cell
number at time T; Ao is the initial cell number; and
Td is the cell doubling time.
Orthotopic tumor growth and metastasis in
nude mice
Adult male athymic Nu-Fox1nu nude mice
purchased from BioLasco (Ilan, Taiwan) were housed in sterilized
cages under 12-h light/dark cycles with water and food ad
libitum. As previously described (38), nude mice were intraperitoneally
inoculated with 5×106 MKN45-GFP or MKN45-GFP-12
cells/mouse, suspended in 0.2 ml phenol-red free RPMI-1640 medium
using 24G syringe needles at 35 nude mice per cancer cell type.
After cancer cell inoculation, the occurrence of ascites was
monitored weekly. The progression of the tumor growth was monitored
by a fluorescence stereomicroscope MZ FLIII from Leica and animal
body weights were recorded twice a week. GFP-expressing tumors were
visualized with a D470/40× bandpass filter and a 495 DCLP dichroic
filter. On the second week after the inoculation, 7 nude mice per
cell type were euthanized weekly and the disseminated and
metastasized tumors growing in the peritoneal cavities were
visualized after laparotomy and the numbers of tumor nodules were
counted using a fluorescent imaging system LT-9500 Illumatool TLS
from Lightools Research (Encinitas, CA, USA). Metastasized tumors
that had invaded into the livers of the nude mice were surgically
removed and sectioned for microscopic examination following
hematoxylin and eosin staining. The use of the animals was approved
by The Institutional Animal Care and Use Committee of The National
Health Research Institutes.
cDNA microarray assay
Total RNAs were isolated from the cancer cells using
TRIzol® reagent (product no. 15596026, Invitrogen)
according to the manufacturer’s protocol. The purified RNA was
quantified in optical density at 260 nm by an ND-1000
spectrophotometer from Nanodrop Technology (Wilmington, DE, USA)
and qualified by Agilent 2100 Bioanalyzer. Total RNA of 0.5
μg was amplified by a low RNA input fluorescent linear
amplification kit (cat. no. 5184–3523) from Agilent Technologies
(Santa Clara, CA, USA) using Cyanine 3 (Cy3)-labeled CTP (cat. no.
NEL580001EA) or Cyanine 5 (Cy5)-labeled CTP (cat. no. NEL581001EA)
from Perkin Elmer (Waltham, MA, USA) during the in vitro
transcription. The cancer cell RNAs were labeled with Cy5 and
Universal Human Reference RNAs (cat. no. 636538) from Clontech
(Mountain View, CA, USA) were labeled with Cy3. The Cyanine-labeled
RNAs (cRNAs) of 2 μg were fragmented to an average size of
50–100 nucleotides by incubation with fragmentation buffer at 60°C
for 30 min. The fragmented labeled cRNAs were then pooled and
hybridized to Human 1A Oligo Microarray (V2) from Agilent
Technologies at 60°C for 17 h. After they were washed, the arrays
were dried using a nitrogen-filled air gun and scanned with Agilent
dual DNA microarray scanner at 535 nm for Cy3 and 625 nm for Cy5,
respectively. The scanned images were extracted by Feature
Extraction 8.1 from Agilent Technologies and analyzed to quantify
signal and background intensity and to substantially normalize the
data by rank-consistency-filtering LOWESS method. The microarray
data, including 4 analyzed samples (MKN45-GFP, MKN45-GFP-4,
MKN45-GFP-10, and MKN45-GFP-12), have been deposited to the Gene
Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/; accession number
GSE33570).
RT-PCR analysis
Samples of the total RNA (1–5 μg) from the
cancer cells were reverse-transcribed in a total volume of 20
μl by the SuperScript III First-Strand Synthesis System
(cat. no. 18080–400) from Invitrogen. The reverse transcription
products (1 μg) were used directly for PCR amplification.
PCR amplification was performed with the PCR Reagent System (cat.
no. 10198-018) from Invitrogen according to the manufacturer’s
instructions in a PC818 Program Temp. Control System from ASTEC
(Fukuoka, Japan). Oligonucleotide primers used for the
amplification of cDNAs for oxytocin receptor (OTR) were
5′-CCTTCATCGTGTGCTGGACG-3′ (forward) and 5′-CTAGGAGCAGAGCACTTATG-3′
(reverse); leucine-rich repeat-containing G protein-coupled
receptor 4/G protein-coupled receptor 48 (LGR4/GPR48) were
5′-GGGAAGCT GGATGATTCGTCTTACT-3′ (forward) and 5′-GAAAAGGG
GAAAACAGCCTGCT-3′ (reverse); trefoil factor 3 (TFF3) were
5′-AGAGCCTTCCCCAAGCAAACA-3′ (forward) and
5′-GCAGGGGCTTGAAACACCAA-3′ (reverse); brain expressed X-linked 2
(BEX2) were 5′-CCTTGGCCCTACCT TTGAATGT-3′ (forward) and
5′-TGCTGACTGCCCGCA AACTA-3′ (reverse); sarcoglycan-ε (SGCE)
were 5′-TTCTCC AAGGTACACTCCGATCG-3′ (forward) and 5′-GGCCGAT
GTGATGTTTATGGC-3′ (reverse); insulin-like growth factor binding
protein 3 (IGFBP3) were 5′-ACGAGTCTCAGAGC ACAGATACCC-3′
(forward) and 5′-TATCCACACACCAG CAGAAGCC-3′ (reverse); and internal
control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were
5′-TCCACCACC CTGTTGCTGTA-3′ (forward) and 5′-ACCACAGTCCATGC
CATCAC-3′ (reverse), respectively. The reaction mixtures were
subjected to PCR cycles of denaturation at 94°C for 1 min,
annealing at 54–59°C for 45 sec, and extension at 72°C for 1.5 min.
RT-PCR-amplified cDNAs were visualized on 1.2% agarose gels stained
with ethidium bromide and the images were captured using UVP
GDS-8000 BioImaging System (Ultra Violet Products, Cambridge, UK)
for further analysis using Image Pro Plus 6.0 software. The
integrated optical density (IOD) = area × average density of each
gene expression band was measured and normalized to that of the
corresponding GAPDH expression. The RNA expression levels in the
cancer cells relative to those in MKN45-GFP cells were
calculated.
Western blot analysis
The cancer cells were lysed by mixing with
Radio-Immunoprecipitation Assay (cat. no. R0278) buffer containing
Proteinase Inhibitor Cocktail (cat. no. P8340) from Sigma. The
protein amounts in the cell lysates were quantified by
bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA)
and mixed with electrophoresis loading buffer (50 mM Tris-HCl, 2%
sodium dodecyl sulfate (SDS), 0.1% bromophenol blue, 10% glycerol,
and 1 mM dithiothreitol). The prepared samples were electrophoresed
on 12% SDS-polyacrylamide gel under reducing conditions. After
electrophoresis, the proteins were transferred electrophoretically
to the PVDF membrane (Immobilon™-P) from Millipore. The membrane
was blocked with 5% skim milk for 30 min at 37°C. After blocking,
the membrane was incubated overnight at 4°C with 1:200 diluted
rabbit polyclonal antibody H-300 (cat. no. sc-28215) against human
fanconi anemia complementation group A (FANCA) and C-19 (cat. no.
sc-540) against metallopanstimulin 1/TTK protein kinase (MPS1/TTK)
from Santa Cruz (Santa Cruz, CA, USA); and 1:500 diluted mouse
monoclonal antibodies clone 294216 (cat. no. MAB2050) against human
melanoma-inhibitory activity (MIA) and clone 84728 (cat. no.
MAB305) against IGFBP3 from R&D Systems (Minneapolis, MN, USA)
in PBS-T buffer. The membrane was washed 3 times with PBS with 0.5%
Triton X-100 buffer (PBS-T) for 5 min each, and then incubated with
1:5,000 diluted horse-radish peroxidase-conjugated bovine
anti-rabbit IgGs (cat. no. sc-2370) from Santa Cruz or
peroxidase-conjugated AffiniPure donkey anti-mouse IgGs (cat. no.
715035150) from Jackson ImmunoResearch (West Grove, PA, USA) in
PBS-T buffer for 1 h at room temperature. After washing with PBS-T,
the membrane was incubated with the Western Lightning™ ECL
detecting reagent from Perkin Elmer (Waltham, MA, USA) and exposed
to X-ray film (cat. no. 50880) from Fuji Medical (Tokyo, Japan).
The images were captured by scanning exposed X-ray film using an HP
Scanjet 4850 scanner (Hewlett-Packard, Palo Alto, CA, USA) and
further analyzed by ImagePro Plus 6.0 software. The IOD = area ×
averaged density of each expressed protein band was measured and
normalized to that of the corresponding GAPDH levels. The expressed
protein levels in the cancer cells relative to those in MKN45-GFP
cells were calculated.
Statistical analysis
Data were analyzed for significant differences using
ANOVA followed by multiple comparisons with Student-Newman-Keuls
test. A significant difference in the incidences of ascites and
liver metastasis among the groups of nude mice was examined by
using Fisher’s exact test. A difference in the averaged number of
tumor nodules per mouse between the two mouse groups of cancer
cells inoculated was detected using Wilcoxon signed rank test. All
statistical analyses were conducted using SPSS®
Statistics software from SPSS Inc. (Chicago, IL, USA). A
significant difference between groups was set at p<0.05.
Results
Stable GFP-expressing MKN45 cells
Following transfection with GFP expression plasmids
using lipofectamine 2000, human gastric cancer MKN45 cell clones
constitutively expressing GFP were collected. Stable GFP-expressing
MKN45 cell clones were selected by G418 in culture medium and a
stable GFP-expressing cell clone was isolated by the limiting
dilution process. A stable MKN45 cell clone with constitutive
expression of the enhanced GFP, MKN45-GFP, was established
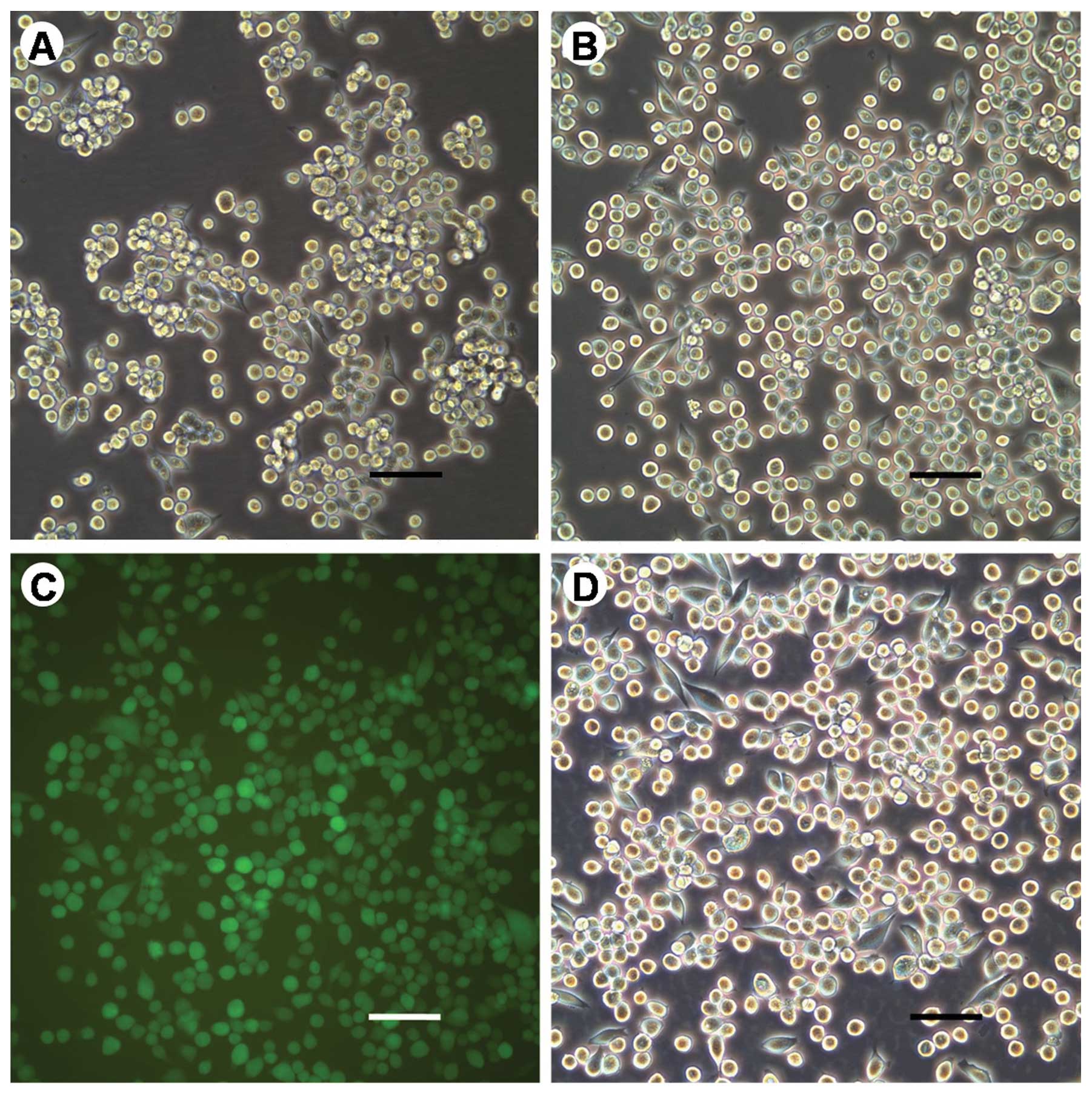
(Fig. 1). There was no difference
in morphology between the parental MKN45 and MKN45-GFP cells
(Fig. 1A and B).
Establishment of MKN45-GFP cell
sublines
The MKN45-GFP cells collected after the 4th, 10th,
and 12th round of 72 h-selection periods using Transwell system
with Matrigel-coated porous membrane were designated as
MKN45-GFP-4, MKN45-GFP-10, and MKN45-GFP-12, respectively. The cell
morphologies among MKN45-GFP and MKN45-GFP cell sublines were not
different from each other as shown in Fig. 1B (MKN45-GFP) and 1D (MKN45-GFP-12).
Furthermore, the cell proliferation doubling times of the parental
MKN45 and the established MKN45-GFP cell sublines ranged from 1.4
to 1.7 days, as shown in Table I,
and were not significantly different from each other (p=0.70).
 | Table IDoubling time of MKN45 and MKN45-GFP
sublines. |
Table I
Doubling time of MKN45 and MKN45-GFP
sublines.
| Cell line | Doubling time
(day) |
|---|
| MKN45 | 1.4±0.3* |
| MKN45-GFP | 1.7±0.1 |
| MKN45-GFP-4 | 1.5±0.5 |
| MKN45-GFP-10 | 1.7±0.4 |
| MKN45-GFP-12 | 1.5±0.4 |
MKN45-GFP cell sublines with different in
vitro invasion abilities
The three established MKN45-GFP sublines exhibited
different invasive abilities in an increasing tendency,
sequentially as MKN45-GFP, MKN45-GFP-4, MKN45-GFP-10 and
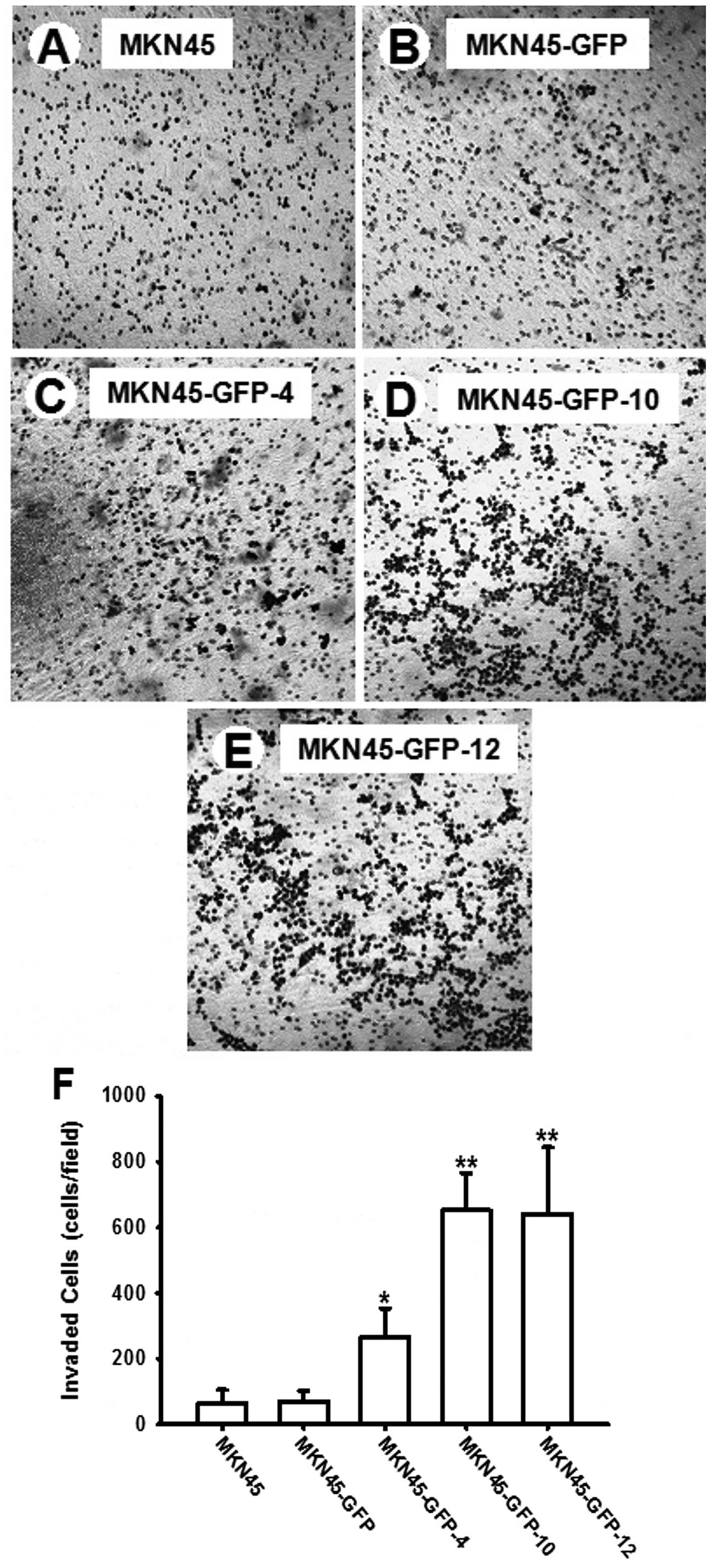
MKN45-GFP-12. Fig. 2A–E shows
representative images of hematoxylin-stained invaded cells on the
surface of the porous membrane of the lower chamber of Transwell
cultures. There was no difference in invasion ability between the
parental MKN45 and MKN45-GFP cells, whereas significantly more
MKN45-GFP-10 and MKN45-GFP-12 cells invaded through the membrane
onto the other side of the membrane compared to MKN45 and MKN45-GFP
cells. In comparison to that of MKN45-GFP, the invasion ability was
significantly increased by 2-, 6-, and 6-fold for MKN45-GFP-4,
MKN45-GFP-10 and MKN45-GFP-12 cells, respectively, as shown in
Fig. 2F.
Incidences of ascites caused by MKN45-
GFP and MKN45-GFP-12 cells in nude mice
The incidences of ascites in the MKN45-GFP and
MKN45-GFP-12 cells inoculated nude mice were monitored as an index
of tumorigenic aggressiveness and tumor burden caused by the cells.
As the data summarized in Table II
show, MKN45-GFP-12 cells caused a significant on-set of ascites on
week 4 after the inoculation, which is one week earlier than that
caused by MKN45-GFP cells. The higher incidence of ascites at week
4 caused by MKN45-GFP-12 indicated a higher tumorigenic
aggressiveness of MKN45-GFP-12 than MKN45-GFP cells.
 | Table IIIncidence of ascites in the nude mice
intraperitoneally inoculated with the cancer cells. |
Table II
Incidence of ascites in the nude mice
intraperitoneally inoculated with the cancer cells.
| Week 2* | Week 3 | Week 4 | Week 5 |
|---|
|
|
|---|
| MKN45-GFP | 0/7 | 1/7 | 1/7 | 6/7 |
| MKN45-GFP-12 | 0/7 | 1/7 | 6/7** | 6/7 |
Orthotopic tumorigenesis and liver
metastasis of MKN45-GFP and MKN45-GFP-12 cells in nude mice
Adult male nude mice intraperitoneally inoculated
with MNK45-GFP or MKN45-GFP-12 cells were observed for
intraperitoneal tumorigenesis and the numbers of tumor nodules were
counted by visualizing the GFP-expressing tumors using a
fluorescence imaging system. The orthotopic intraperitoneal
inoculation of gastric cancer cells of diffuse type in mice is to
mimic the dissemination and metastasis conditions of the cancer
cells in the peritoneum of gastric cancer patients. After the
inoculation, MKN45-GFP-12 tumor cells grew inside the peritoneal
cavity and metastasized to and grew in the mouse liver.
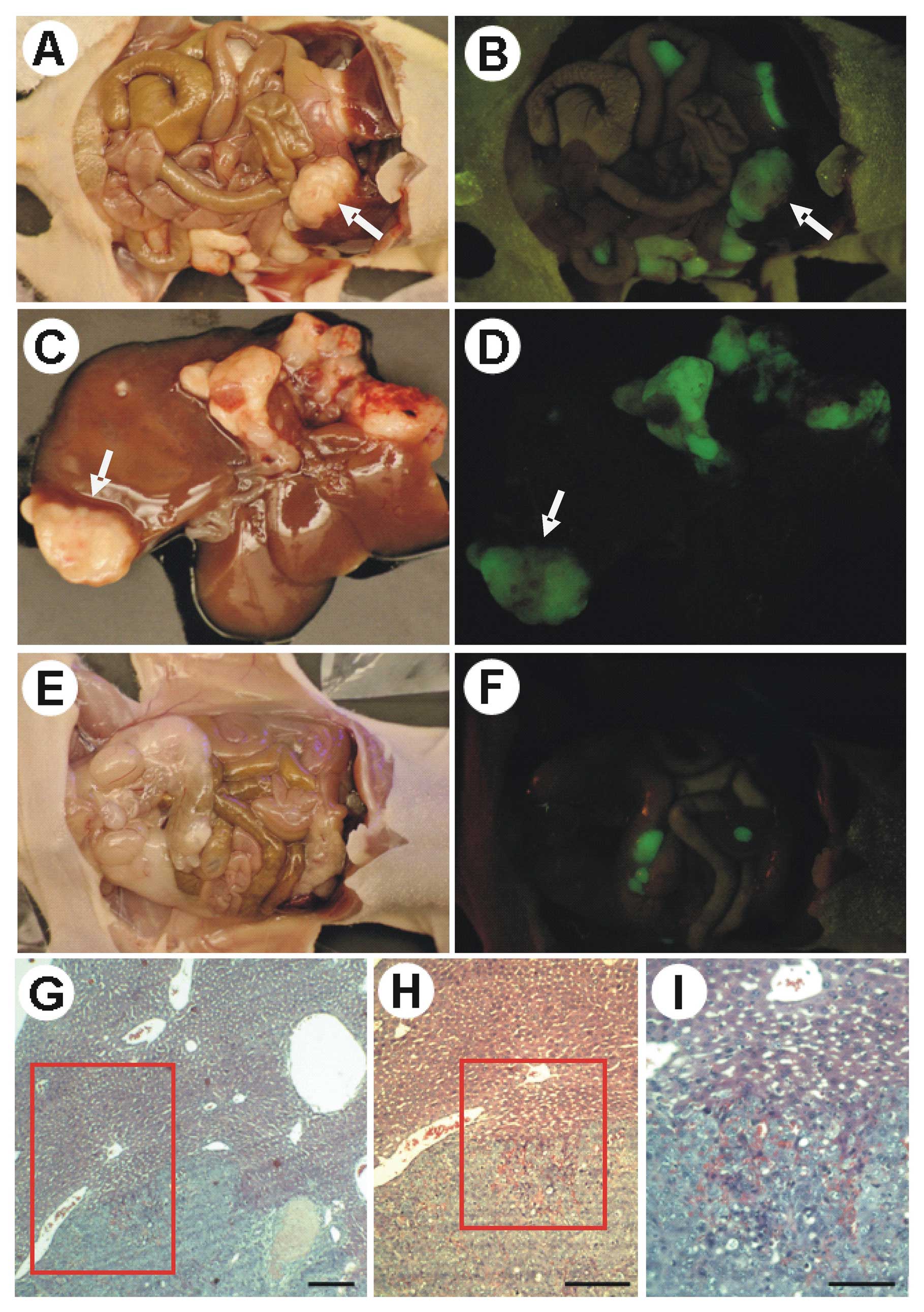
Representative images of the intraperitoneally disseminated growing
MKN45-GFP-12 tumors in a nude mouse after laparotomy in bright and
fluorescent fields are shown in Fig.
3A and B, respectively. Growing MKN45-GFP-12 tumors that
invaded into the mouse liver are shown in Fig. 3C (bright field) and D (fluorescent
field). MKN45-GFP cells also grew into tumors in the mouse
peritoneum as shown in Fig. 3E
(bright field) and F (fluorescent field). On Day 14 after the
cancer cell inoculation, we observed that MKN45-GFP cells caused
14–25 (median 17; mean 18; n=7) intraperitoneally orthotopic
growing tumors, whereas MKN45-GFP-12 cells caused 10–40 (median 25;
mean 24; n=7) intraperitoneally orthotopic growing tumors in the
laparotomized nude mice. The number of intraperitoneal tumor
nodules caused by the orthotopic inoculation was compared and
MKN45-GFP-12 cells were found to cause more, though not
statistically significant (p=0.128), intraperitoneally disseminated
growing tumors in the nude mice. Furthermore, both
intraperitoneally inoculated MKN45-GFP and MKN45-GFP-12 cells
caused metastatic tumor growth in the nude mouse livers during the
observation period (Fig. 3G–I).
MKN45-GFP-12 cells metastasized to the mouse liver and established
metastatic growth in more nude mouse livers (p=0.042) than
MKN45-GFP cells as detailed in the liver metastasis incidence
summary in Table III. The results
showed that MKN45-GFP-12 cells exhibited a higher metastasizing
potential in vivo than MKN45-GFP cells.
 | Table IIIIncidence of liver metastasis in the
nude mice intraperitoneally inoculated with the cancer cells. |
Table III
Incidence of liver metastasis in the
nude mice intraperitoneally inoculated with the cancer cells.
| No metastasis | Metastasis |
|---|
|
|
|---|
| MKN45-GFP | 33/35 (94%) | 2/35 (6%) |
| MKN45-GFP-12 | 27/35 (77%) | 8/35 (23%)* |
cDNA microarray analyses of genes in
MKN45-GFP cell sublines
Total RNAs from the established MKN45-GFP and the
selected MKN45-GFP-4, MKN45-GFP-10, and MKN45-GFP-12 cells were
cDNA microarray analyzed. The gene expression levels and patterns
of MKN45-GFP-4, MKN45-GFP-10, and MKN45-GFP-12 cells were compared
in relation to those of the MKN45-GFP cells (GEO accession number
GSE33570). We used the PvalueLogRatio method to sieve out
significant spots of whole microarray analysis. PvalueLogRatio
gives the statistical significance on the log ratio for each spot
between experimental and control signals. In order to focus on the
desired data, we setup the criteria for removal steps and filtering
procedure to identify positive or negative expression-correlated
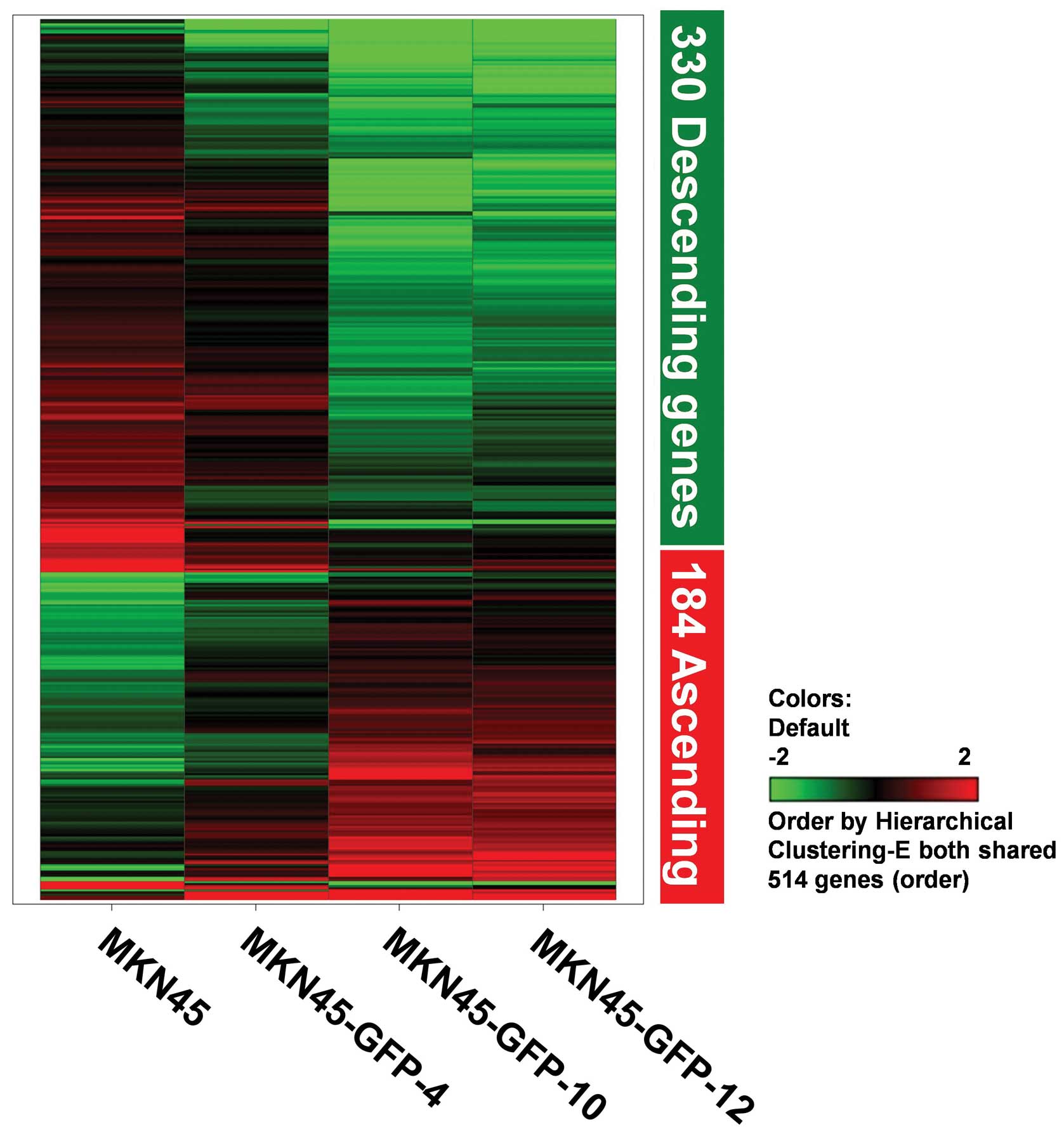
514 genes (184 ascending and 330 descending genes) of the examined
genes by hierarchical clustering calculation. We found that only
<3% of all genes may be involved in human gastric cancer
invasion and metastasis. Fig. 4
displays a hierarchical clustering analysis image with 514 genes
(accounting for 2.75% of the total number of 18,716 genes examined)
from significant expression levels by using the selection criteria
described above, which share a similar tendency containing 184
ascending (positively correlated with invasiveness) and 330
descending genes (negatively correlated with invasiveness). The
expression levels were pseudo-color encoded showing the descending
and ascending expression levels of genes in the MKN45-GFP cell
sublines. Based on the hierarchical clustering analyses, the
expression patterns of these 514 genes were subjected to regulating
tendency selection processes.
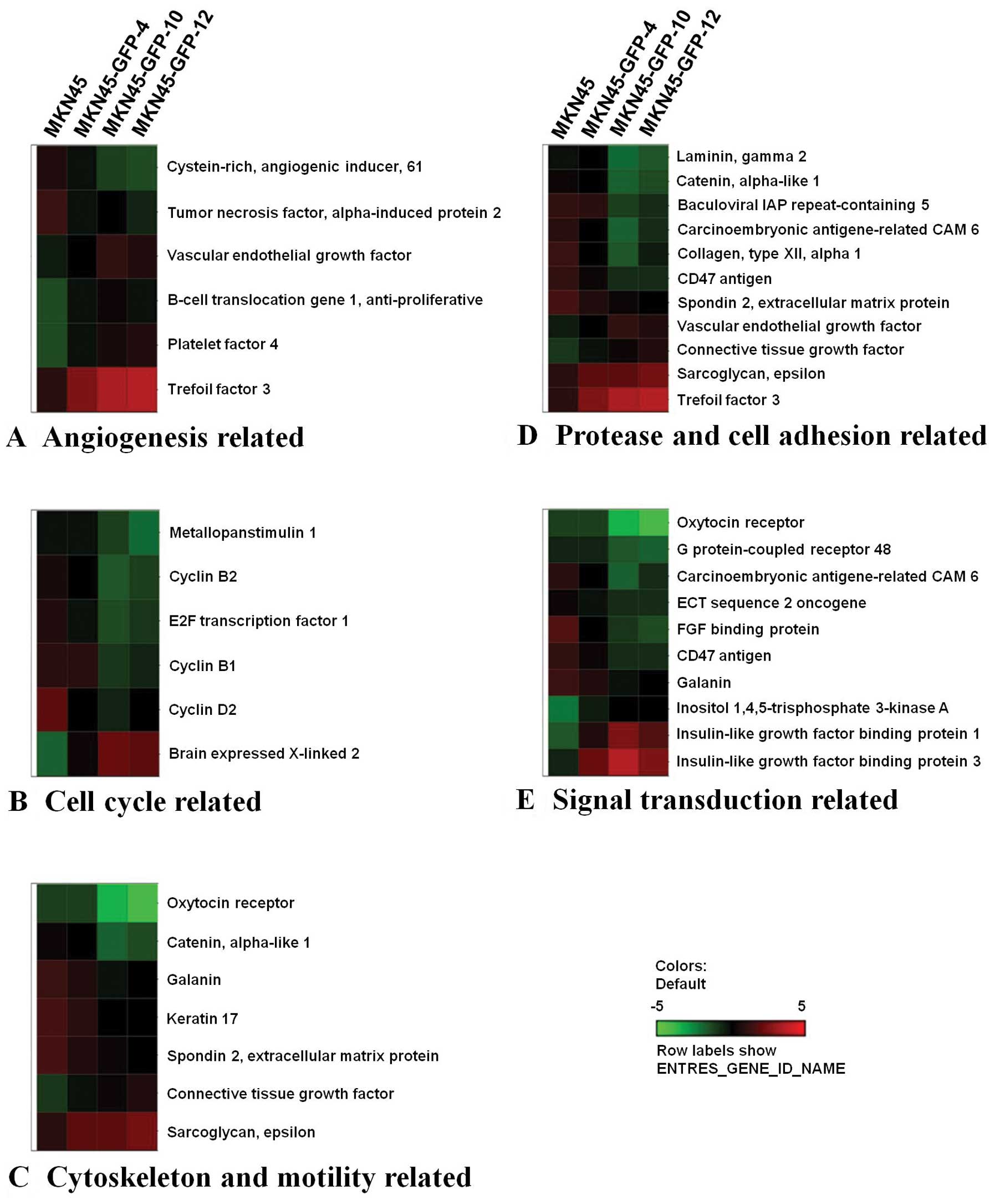
The 514 genes identified in the hierarchical
clustering analysis were further categorized into groups such as
angiogenesis-related genes, cell cycle regulators, cytoskeleton and
motility molecules, protease and adhesion proteins, and signal
transduction molecules, based on known molecular and biological
functions associated with tumor progression. A total of 133 genes
were found to be associated with these biological functions and 30
of them are shown in Fig. 5 and
Table IV. The 5 selective groups
were (A) angiogenesis-related genes such as angiogenic inducer
Cyr61 and vascular endothelial growth factor; (B) cell cycle
regulators such as MPS1/TTK and cyclin B2; (C) cytoskeleton
and motility molecules such as OTR and catenin α-like 1; (D)
protease and adhesion molecules such as laminin γ 2 and collagen
type XII α 1, and (E) signal transduction molecules such as
LGR4/GPR48 and heparin-binding growth factor binding
protein. Genes associated with multiple biological functions were
included in the corresponding categorized functional groups
accordingly. A number of genes that have not been previously
reported to correlate with cancer progression or tumor metastasis
were also identified and are listed in Table V. The levels of these genes
differed more than two-fold between highly metastatic and parental
cells, indicating that these genes may play significant roles in
gastric cancer metastasis.
 | Table IVGenes of known functions
differentially expressed between parental and highly metastatic
MKN45 cell lines. |
Table IV
Genes of known functions
differentially expressed between parental and highly metastatic
MKN45 cell lines.
| Probe name | Accession no. | Sequence homology
(GenBank/EMBL) | Gene symbol | Chromosome
location |
Log2[E12/Ep] |
|---|
| Angiogenesis
related genes |
|
| A_23_P257296 | NM_003226 | Trefoil factor
3 | TFF3 | 21q22.3 | 2.74 |
| A_23_P132987 | NM_002619 | Platelet factor
4 | PF4 | 4q12-q21 | 2.06 |
| A_23_P387668 | NM_003376 | Vascular
endothelial growth factor | VEGFA | 6p12 | 1.40 |
| A_23_P87564 | NM_001731 | B-cell
translocation gene 1, anti-proliferative | BTG1 | 12q22 | 1.24 |
| A_23_P421423 | NM_006291 | Tumor necrosis
factor, α-induced protein 2 | TNFAIP2 | 14q32 | −1.76 |
| A_23_P46429 | NM_001554 | Cystein-rich,
angiogenic inducer, 61 | CYR61 | 1p31-p22 | −2.12 |
|
| Cell cycle
regulation related genes |
|
| A_23_P22735 | NM_032621 | Brain expressed
X-linked 2 | BEX2 | Xq22 | 3.99 |
| A_23_P122197 | NM_031966 | Cyclin B1 | CCNB1 | 5q12 | −1.56 |
| A_23_P80032 | NM_005225 | E2F transcription
factor | E2F1 | 20q11.2 | −1.66 |
| A_23_P65757 | NM_004701 | Cyclin B2 | CCNB2 | 15q22.2 | −1.78 |
| A_23_P259586 | NM_003318 | Metallopanstimulin
1 | TTK | 6q13-q21 | −1.85 |
| A_23_P139881 | NM_001759 | Cyclin D2 | CCND2 | 12p13 | −2.33 |
|
| Cytoskeleton and
motility related genes |
|
| A_23_P19663 | NM_001901 | Connective tissue
growth factor | CTGF | 6q23.1 | 1.56 |
| A_23_P254626 | NM_003919 | Sarcoglycan,
epsilon | SGCE | 7q21-q22 | 1.46 |
| A_23_P374844 | NM_015973 | Galanin | GAL | 11q13.3 | −1.52 |
| A_23_P121533 | NM_012445 | Spondin 2,
extracellular matrix protein | SPON2 | 4p16.3 | −1.57 |
| A_23_P96149 | NM_000422 | Keratin 17 | KRT17 | 17q12-q21 | −1.59 |
| A_23_P157795 | NM_003798 | Catenin, α-like
1 | CTNNAL1 | 9q31.2 | −2.14 |
| A_23_P132619 | NM_000916 | Oxytocin
receptor | OXTR | 3p25 | −2.90 |
|
| Protease and cell
adhesion related genes |
|
| A_23_P257296 | NM_003226 | Trefoil factor
3 | TFF3 | 21q22.3 | 2.74 |
| A_23_P19663 | NM_001901 | Connective tissue
growth factor | CTGF | 6q23.1 | 1.56 |
| A_23_P254626 | NM_003919 | Sarcoglycan,
epsilon | SGCE | 7q21-q22 | 1.46 |
| A_23_P387668 | NM_003376 | Vascular
endothelial growth factor | VEGFA | 6p12 | 1.40 |
| A_23_P121533 | NM_012445 | Spondin 2,
extracellular matrix protein | SPON2 | 4p16.3 | −1.57 |
| A_23_P6935 | NM_001777 | CD47 antigen | CD47 | 3q13.1-q13.2 | −1.89 |
| A_23_P201636 | NM_005562 | Laminin, gamma
2 | LAMC2 | 1q25-q31 | −2.00 |
| A_23_P118815 | NM_001168 | Baculoviral IAP
repeat-containing 5 | BIRC5 | 17q25 | −2.12 |
| A_23_P157795 | NM_003798 | Catenin, α-like
1 | CTNNAL1 | 9q31.2 | −2.14 |
| A_23_P218441 | NM_002483 | Carcinoembryonic
antigene-related CAM 6 | CEACAM6 | 19q13.2 | −2.61 |
| A_23_P214168 | NM_004370 | Collagen, type XII,
α 1 | COL12A1 | 6q12-q13 | −2.74 |
|
| Signal transduction
related genes |
|
| A_23_P42869 | NM_000596 | Insulin-like growth
factor binding protein 1 | IGFBP1 | 7p13-p12 | 4.19 |
| A_23_P215634 | NM_000598 | Insulin-like growth
factor binding protein 3 | IGFBP3 | 7p13-p12 | 4.10 |
| A_23_P4714 | NM_006533 | Melanoma inhibitory
activity | MIA |
19q13.32-q13.33 | 2.73 |
| A_23_P65918 | NM_002220 | Inositol
1,4,5-trisphosphate 3-kinase A | ITPKA | 15q14-q21 | 2.44 |
| A_23_P9571 | NM_018098 | ECT sequence 2
oncogene | ECT2 | 3q26.1-q26.2 | −1.13 |
| A_23_P203767 | NM_018490 | G protein-coupled
receptor 48 | LGR4 | 11p14-p13 | −1.35 |
| A_23_P206441 | NM_000135 | Fanconi anemia,
complementation group A | FANCA | 16q24.3 | −1.38 |
| A_23_P374844 | NM_015973 | Galanin | GAL | 11q13.3 | −1.52 |
| A_23_P6935 | NM_001777 | CD47 antigen | CD47 | 3q13.1-q13.2 | −1.89 |
| A_23_P218441 | NM_002483 | Carcinoembryonic
antigene-related CAM 6 | CEACAM6 | 19q13.2 | −2.61 |
| A_23_P132619 | NM_000916 | Oxytocin
receptor | OXTR | 3p25 | −2.90 |
| A_23_P30126 | NM_005130 | FGF binding
protein | FGFBP1 | 4p16-p15 | −3.20 |
 | Table VNovel genes of unknown functions
differentially expressed between parental and highly metastatic
MKN45 cell lines. |
Table V
Novel genes of unknown functions
differentially expressed between parental and highly metastatic
MKN45 cell lines.
| Probe name | Accession no. | Sequence homology
(GenBank/EMBL) | Gene symbol | Chromosome
location |
Log2[E12/Ep] |
|---|
| A_23_P209360 | NM_052920 | Kelch-like 29 | KLHL29 | 2p24.1 | 3.26 |
| A_23_P20876 | NM_152422 | Protein tyrosine
phosphatase domain containing 1 | PTPDC1 | 9q22.32 | 2.51 |
| A_23_P83339 | NM_145051 | Ring finger protein
183 | RNF183 | 9q32 | 2.14 |
| A_23_P108823 | NM_145739 | Oxysterol binding
protein-like 6 | OSBPL6 | 2q32.1 | 1.92 |
| A_23_P154740 | NM_018474 | Polo-like kinase 1
substrate 1 | PLK1S1 | 20p11.23 | 1.80 |
| A_23_P79302 | NM_177964 | LY6/PLAUR domain
containing 6B | LYPD6B | 2q23.2 | 1.44 |
| A_23_P52727 | NM_182964 | Neuron navigator
2 | NAV2 | 11p15.1 | −1.49 |
| A_23_P51966 | NM_003035 | SCL/TAL1
interrupting locus | STIL | 1q32; 1p32 | −1.55 |
| A_23_P379778 | NM_017594 | DIRAS family,
GTP-binding RAS-like 2 | DIRAS2 | 9q22.2 | −1.56 |
| A_23_P69179 | NM_018192 | Leprecan-like
1 | LEPREL1 | 3q28 | −1.57 |
| A_23_P340909 | NM_145061 | Spindle and
kinetochore associated complex subunit 3 | SKA3 | 13q12.11 | −1.66 |
| A_23_P118582 | NM_013290 | PSMC3 interacting
protein | PSMC3IP | 17q21.2 | −1.70 |
| A_23_P48835 | NM_138555 | Kinesin family
member 23 | KIF23 | 15q23 | −1.80 |
| A_23_P166526 | NM_015653 | RIB43A domain with
coiled-coils 2 | RIBC2 | 22q13.31 | −1.90 |
| A_23_P80718 | NM_144642 | Synaptoporin | SYNPR | 3p14.2 | −1.92 |
| A_23_P49878 | NM_019013 | Family with
sequence similarity 64, member A | FAM64A | 17p13.2 | −1.93 |
| A_23_P323751 | NM_030919 | Family with
sequence similarity 83, member D | FAM83D | 20q11.22-q12 | −1.95 |
| A_23_P42811 | NM_176813 | Anterior gradient
homolog 3 | AGR3 | 7p21.1 | −2.06 |
| A_23_P388812 | NM_152515 | Cytoskeleton
associated protein 2-like | CKAP2L | 2q13 | −2.07 |
| A_23_P25964 | NM_000153 |
Galactosylceramidase | GALC | 14q31 | −2.16 |
| A_23_P355525 | NM_173497 | HECT domain
containing 2 | HECTD2 | 10q23.32 | −2.39 |
| A_23_P250853 | N/A | SM3A_HUMAN (Q14563)
Semaphorin 3A precursor (Semaphorin III) (Sema III), partial
(8%) | THC2052903 | N/A | −2.41 |
| A_23_P35995 | NM_024769 | Adipocyte-specific
adhesion molecule | ASAM | 11q24.1 | −2.49 |
| A_23_P3302 | NM_018365 | Meiosis-specific
nuclear structural 1 | MNS1 | 15q21.3 | −2.50 |
| A_23_P217785 | NM_016500 | Chromosome X open
reading frame 26 | CXorf26 | Xq13.3 | −2.54 |
| A_23_P130027 | NM_017957 | Epsin 3 | EPN3 | 17q21.33 | −2.60 |
| A_23_P301360 | NM_152412 | Zinc finger protein
572 | ZNF572 | 8q24.13 | −2.62 |
| A_23_P254842 | NM_012080 | Haloacid
dehalogenase-like hydrolase domain containing 1 | HDHD1 | Xp22.32 | −2.68 |
| A_23_P329254 | NM_153032 | Archaelysin family
metallopeptidase 2 pseudogene 1 | AMZ2P1 | 17q24.1 | −2.69 |
| A_23_P35871 | NM_024680 | E2F transcription
factor 8 | E2F8 | 11p15.1 | −2.75 |
| A_23_P154279 | NM_032309 |
Coiled-coil-helix-coiled-coil-helix domain
containing 5 | CHCHD5 | 2q13 | −3.01 |
| A_23_P252432 | NM_004617 | Transmembrane 4 L
six family member 4 | TM4SF4 | 3q25 | −3.06 |
| A_23_P75915 | NM_024557 | Resistance to
inhibitors of cholinesterase 3 homolog (C. elegans) | RIC3 | 11p15.4 | −3.06 |
| A_23_P252388 | NM_024867 | Sperm flagellar
2 | SPEF2 | 5p13.2 | −3.20 |
| A_23_P396765 | NM_173582 | Phosphoglucomutase
2-like 1 | PGM2L1 | 11q13.4 | −3.41 |
| A_23_P139604 | NM_003805 | CASP2 and RIPK1
domain containing adaptor with death domain | CRADD | 12q21.33-q23.1 | −4.71 |
mRNA transcript levels in MKN45-GFP cell
sublines
Several putative tumor metastasis-associated genes
were further examined by using RT-PCR analysis. Four genes,
TFF3, BEX2, SGCE and IGFBP3, positively
correlated with the invasion ability, and two genes, OTR and
LGR4/GPR48, negatively correlated with the invasion ability
of the gastric cancer MKN45-GFP cell sublines, were RT-PCR
analyzed. The housekeeping gene GAPDH was selected as the internal
control. Results showed that all four examined genes TFF3,
BEX2, SGCE, and IGFBP3 exhibited an increasing
order of the mRNA transcript levels from MKN45-GFP, MKN45-GFP-4,
MKN45-GFP-10, to MKN45-GFP-12 cells. On the other hand, mRNA
transcript levels of the two genes OTR and LGR4/GPR48
were in a decreasing order in these MKN45-GFP cell sublines in
relation to their invasion ability (Fig. 6). Results of the RT-PCR analyses on
these selective genes were consistent with those from the cDNA
microarray analyses.
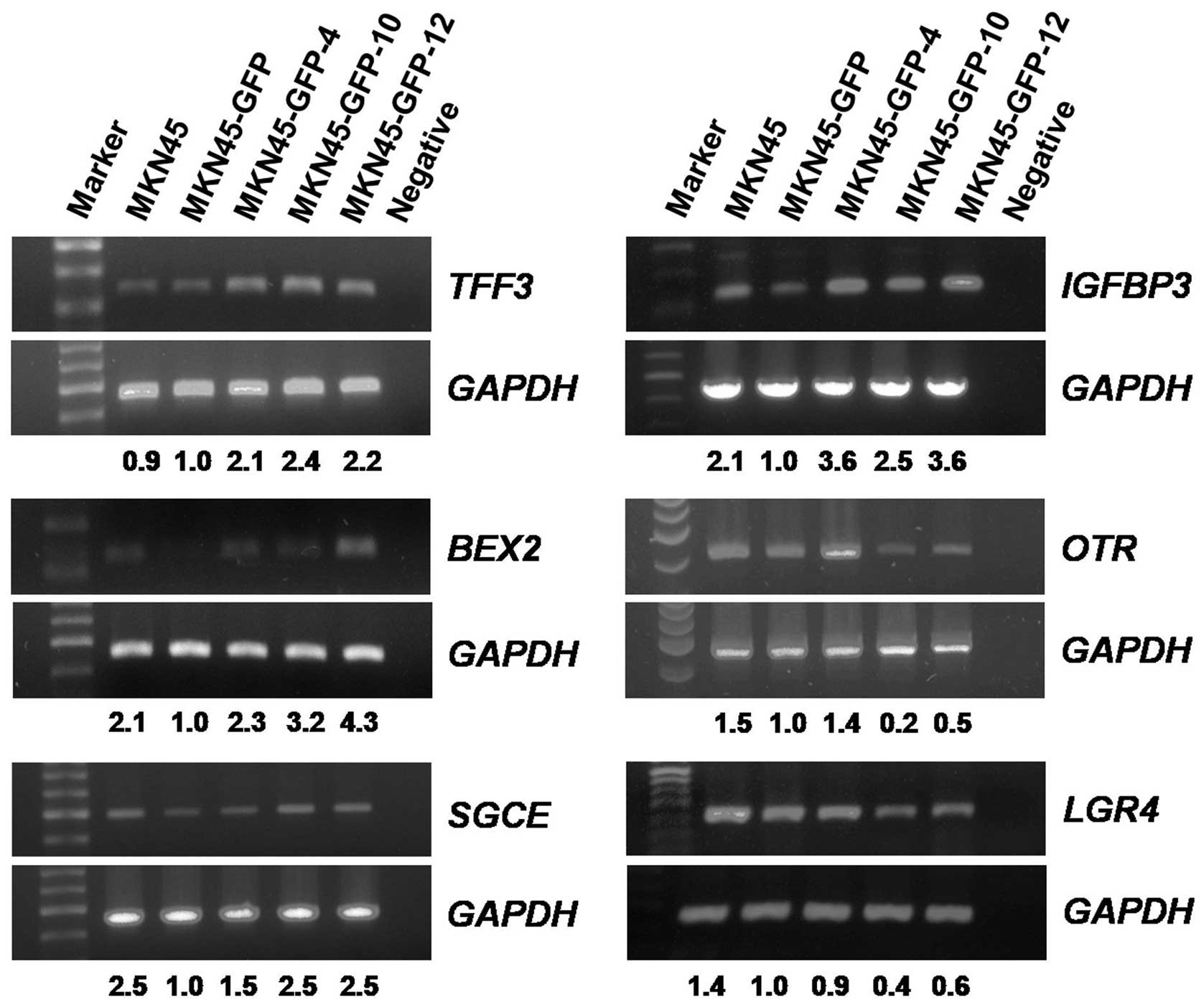 | Figure 6Reverse transcriptase PCR analysis of
mRNA levels in MKN45-GFP cell sublines. Four genes (TFF3, BEX2,
SGCE, IGFBP3) with ascending mRNA levels and two genes (OTR,
LGR4/GPR48) with descending mRNA levels were analyzed. The
level of GAPDH mRNA was used as the internal control for
normalization. The relative mRNA levels of the genes compared to
that of MKN45-GFP were indicated accordingly. (TFF3, trefoil
factor 3; BEX2, brain expressed X-linked 2; SGCE,
sarcoglycan, epsilon; IGFBP3, insulin-like growth factor
binding protein 3; OTR, oxytocin receptor;
LGR4/GPR48, leucine-rich repeat-containing G protein-coupled
receptor 4/G protein-coupled receptor 48). |
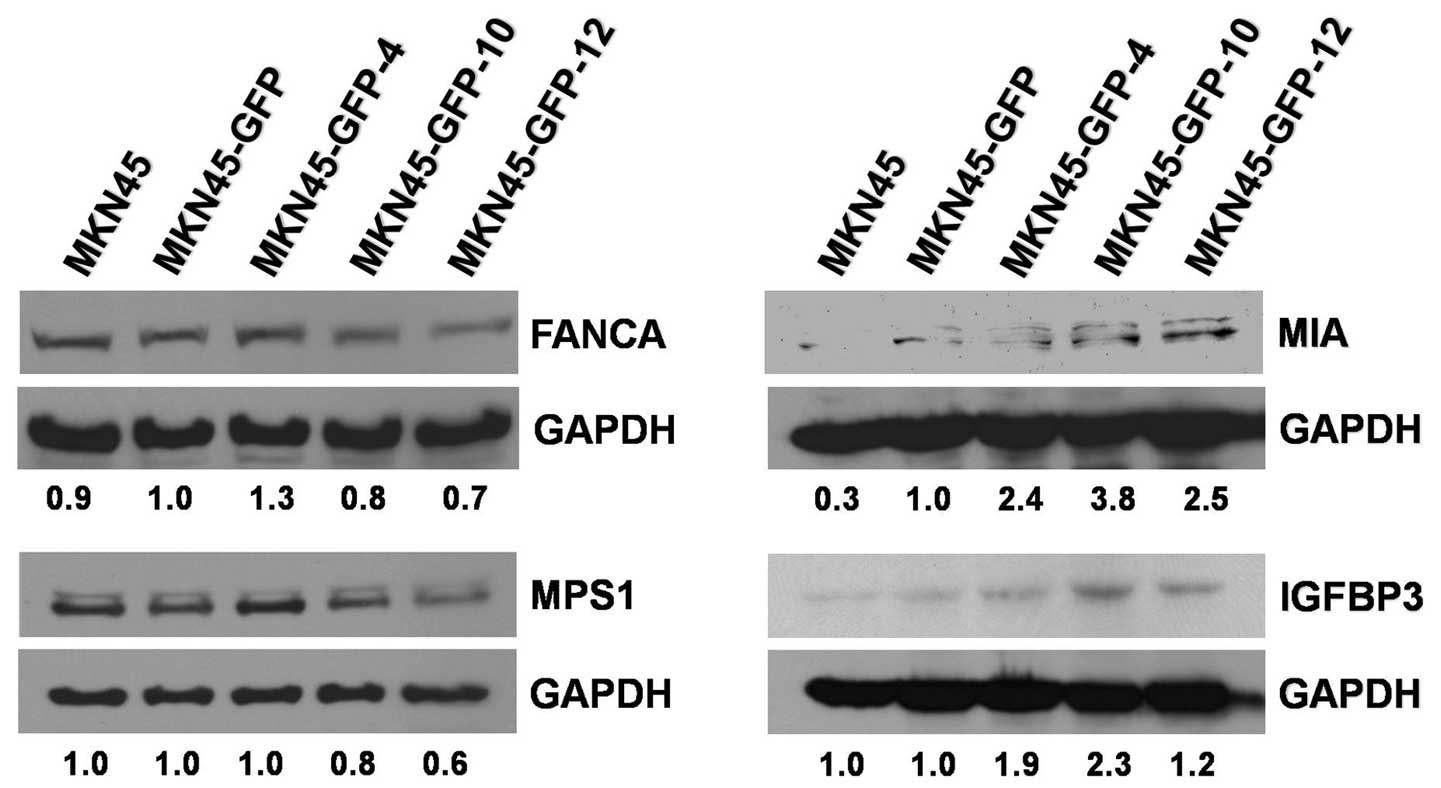
Protein expression levels in MKN45-GFP
cell sublines
The expression levels of several proteins of the
microarray-identified genes were also examined using western blot
analysis. FANCA, MPS1/TTK, MIA, and IGFBP3 protein levels in the
MKN45-GFP cell sublines were analyzed in triplicates. FANCA and
MPS1/TTK protein levels were downregulated with the increase of the
invasive potentials of the MKN45-GFP cell sublines. By contrast,
the protein levels of (precursor and/or mature) MIA and IGFBP3 were
upregulated with the increase of the invasive potentials of the
MKN45-GFP cell sublines (Fig. 7).
Both the mRNA transcript and protein expression levels of the
IGFBP3 gene were positively correlated with the invasive
abilities of the MKN45-GFP cell sublines. The results demonstrated
that the expression levels of these proteins examined were
consistent with those from the cDNA microarray analyses.
Discussion
It is well known that differential expression of
multiple genes and dynamic interactions of a number of cellular
proteins are involved in the multiple progressive steps of cancer
metastasis including gastric cancer. cDNA microarray has been
applied to explore the expression profiles of a massive number of
genes between testing cancer samples. In the present study, we used
cDNA microarray to identify gastric cancer
invasion/metastasis-associated genes on a genome-wide scale in
human gastric cancer sublines that have demonstrated different
meta-static abilities. As the poorly differentiated diffuse type
gastric cancer remains an unmet medical need, a gastric cancer cell
line of diffuse type MKN45, originally isolated from a poorly
differentiated adenocarcinoma of a patient’s stomach (33,34),
was used as the model cell. By taking advantage of the
fluorescence-emitting ability of GFP, we established an MKN45 cell
line constitutively expressing GFP which can be visualized, and
applied it to select the MKN45-GFP sublines with differential
invasion abilities by using the Transwell system. Genes in relation
to the invasiveness of these sublines were identified using cDNA
microarray analysis and verified by RT-PCR and western blot
analyses. Furthermore, the invasiveness of the highly invasive
subline MKN45-GFP-12 was in vivo demonstrated by the
incidence of ascites and organ invasive abilities in nude mice.
Meanwhile, we observed that a number of the identified genes are
also related to angiogenesis, cell cycle, cytoskeleton and
motility, protease and cell adhesion, and signal transduction,
which are cellular events known to be associated with invasion and
tumor metastasis (39).
Accordingly, a number of novel genes without previously known
cancer-related biological activities were also identified in
relation to gastric tumor metastasis in the present study.
Systemically analyzed using genomic microarray
technologies and biological activity assays, genes putatively
associated with tumor cell invasion and metastasis have been
identified in various human cancer cells, including gastric cancer
cells. Many studies utilized microarray technologies to discover
genes specifically associated with tumor invasion and metastasis.
Hippo et al observed a number of differentially expressed
genes by using a set of human gastric scirrhous cancer cells prone
to peritoneal and lymph node metastasis in nude mice (23). Although the array-based
oligonucleotide hybridization method is a powerful tool for
identifying potential genes associated with tumor metastasis, the
genes reported in these above-mentioned studies are different from
each other. The intrinsic heterogeneity of the cancer cell
populations in patient tumor tissues likely mixed with normal
stromal and/or vascular cells may contribute to these observed
discrepancies in the gene expression profiles (40). Herein we used a human gastric
cancer cell line of diffuse type in vitro to normalize and
minimize the potentially existing heterogeneity in order to
identify human gastric tumor metastasis-associated genes.
Chronic infection of Helicobacter P. has been
proven to be an important etiologic factor of gastric cancer
(41) and accounts for 18% of all
cancers caused by infections (42). Helicobacter infection has been
found to be associated with β-catenin, MMP-1, ERK (p38), EGFR,
AP-1, Shh, CDX2, E-cadherin (CDH1), p16, APC, MLH1, COX2, IL-1,
IL-6, IL-10, CTLR4, BCL-2, FOXP1 and p53 (43), and IL-8, ras family and Ras p21 are
involved in the TNFα/NF-κB/IκB-mediated gastric tumorigenesis
(44). Helicobacter colonization
is usually asymptomatic and gastric tumorigenesis occurs only in a
subset of individuals depending on the host response and genetic
variation of the bacteria. The infection alone could not attribute
entirely to the gastric tumorigenesis as the clinically observed
human gastric cancers are genetically and histologically
diversifying (43,45). Although several studies reported
that Helicobacter infection increases tumor invasiveness and
metastasis via multiple molecular pathways (46–50),
in the present study we did not find significant changes in the
expressions of these mentioned genes and/or the genes in the
TNFα/NF-κB/IκB-mediated signaling pathways (43,44).
Therefore, the results suggest that the genes identified in the
present study are not Helicobacter infection-related and are thus
directly correlated to the metastatic abilities of human gastric
cancer tumors.
GFP has been utilized as a reporter molecule and has
been widely applied in vitro and in vivo to visualize
the developmental processes of tumors, including primary and
metastatic tumor growths, tumor cell metastases, tumor
angiogenesis, and interactions between tumor and the host
microenvironment (38,51–54).
Chu et al established a set of human lung cancer cells with
differential invasive abilities (35), Chen et al identified a
number of genes associated with human lung cancer metastasis
(55), and Chen et al
demonstrated a significant clinical association for some of these
identified genes (56).
Accordingly, we took advantage of the fluorescence-emitting GFP and
established the human GFP-expressing MKN45 sublines with
differential invasive abilities in the present study. The
morphology and proliferation rates were not different from each
other among these established MKN45-GFP sublines including the
parental MKN45 cells. One of the important advantages is that we
are able to monitor the tumor growth and peritoneal dissemination
in individual nude mice without laparotomy. The appropriate timing
for animal euthanization can then easily be determined and
therefore the number of animals used in a study can be reduced. The
identification of the genes previously known with biological
activities related to tumor progression and metastasis suggests the
validity of the approach utilized in this study. The observations
indicate that the genes differentially expressed among these
sublines are likely to be associated with their invasive and, thus,
metastatic abilities.
In the present study, 184 and 330 genes were found
positively and negatively expressed, respectively, with regard to
the increasing order of the invasive abilities of these model cell
sublines. The identified genes are likely to be associated with
cancer progression and/or tumor metastasis. They are found to be
associated with the biological activities pertaining to tumor
metastasis such as angiogenesis-, cell cycle regulation-,
cytoskeleton and motility-, protease and cell adhesion-, and signal
transduction-related genes as shown in Fig. 5 and Table IV. Angiogenesis is one of the
essential steps of tumor growth and metastasis (39). Proteins of the trefoil factor
family genes may induce tumor vascularity and be associated with
tumor metastatic phenotype and recurrence in human gastric cancer
(57). Trefoil factor 3 (TFF3) is
connected with multiple oncogenic pathways such as COX2 and EGFR
and participates in the blood vessel formation during tumor
progression (58). IGFBP3 is a
protein of multi-functions. IGFBP3 induces IGF-independent
anti-angiogenic effects and exerts tumor suppressive effects
against prostate cancer (59) and
non-small cell lung cancer (60),
whereas its overexpression is associated with the increasing
metastatic ability of well-differentiated pancreatic endocrine
tumors (61). We report here that
IGFBP3 expression is positively correlated with the invasive
ability of human gastric cancer cells suggesting that IGFBP3 is
involved in gastric cancer metastasis. In agreement with a previous
study (62), we found that
vascular endothelial growth factor (VEGF) genes were positively
regulated in accordance with the development of tumor invasiveness.
Overexpression of B-cell translocation gene 1 (BTG1) increases tube
formation and migration of the endothelial cells (63). Accordingly, we observed that BTG1
is upregulated during the development of human gastric cancer cell
invasiveness. Further studies are required to explore the
pathological roles of these angiogenesis-related genes during
gastric tumor progression.
During cell cycle progression, MPS1/TTK is a protein
kinase and is involved in the regulation of centrosomal control of
cytokinesis in the mitotic checkpoint process (64). MPS1/TTK kinase affects the
chromosomal stabilities in the M phase of the cell cycles in zebra
fish and mammalian cells (65,66).
Chromosomal instability may induce changes in the cellular
functions and thus increase the invasive and metastatic abilities
of tumor cells (67). In
accordance with the implications of these observations, we found
that MPS1/TTK expression was downregulated in the invasive cell
sublines. BEX2 participates in G1 phase regulation during cell
cycle progression and its over-expression protects breast cancer
cells against mitochondrial apoptosis (68). We herein observed that upregulation
of BEX2 was associated with the development of the invasion and
metastasic abilities of gastric tumor cells.
Cellular cytoskeleton and motility also play
important roles in tumor cell metastasis. Connective tissue growth
factor (CTGF), whose transcript levels were moderately increased,
as shown in the present study, modulates the motility of human
breast cancer cells through activation of the ERK1/2 signaling
pathway (69). Catenin-α is
anti-metastatic in human squamous carcinoma and breast cancer by
forming a complex with E-cadherin and thus enhancing the cell-cell
adhesion (70,71). In agreement with these
observations, catenin-α was down-regulated in the invasive cell
sublines in this study. Oxytocin receptor was found in the smooth
muscles of the gastric/intestinal system and mediates gastric
motility (72). A previous study
showed that oxytocin inhibited ovarian carcinoma cell metastasis
in vitro and in vivo (73). Furthermore, we observed that
oxytocin receptors are downregulated in the highly metastatic
gastric cancer cell sublines. The results suggest that the
oxytocin/oxytocin receptor system may play a significant role in
gastric cancer metastasis.
Proteases and adhesion proteins play key roles in
gastric tumor metastasis. The ability of tumor cells to detach from
a primary tumor tissue and degradation of the extracellular matrix
and basement membrane structures are associated with the progress
of tumor cell invasion and metastasis. TFF3 modulates
E-cadherin/catenin-mediated cell-cell contact and increases cell
motility in cancer progression (74). We found that TFF3 was upregulated
in the gastric cancer cells with high invasive ability. Involvement
of SGCE in adhesion activity has been reported to inhibit the
invasion of colorectal tumors (75). Contrary to that observed in
colorectal tumors, SGCE expression was positively correlated with
the gastric cancer cell invasion ability shown in the present
study. Laminin family proteins involved in the cell-laminin
interactions are related to tumor angiogenesis, invasion and
metastasis (76). We found that
laminin-γ was downregulated in the gastric cancer cells of high
invasiveness. Collagen XII is an adhesion protein reported to have
critical roles in the interactions between cancer cells and bone
marrow, in cell adhesion, and in cell motility and is thus involved
in the bone metastasis of prostate cancer (77). In gastric cancer cells, we observed
that collagen XII expression is inversely correlated with cancer
cell invasion ability.
Cellular and subcellular signaling pathways regulate
multiple and complex cellular activities, including tumor cell
proliferation, invasion and metastasis. IGFBP3 plays significant
roles in tumor cell proliferation, invasion and metastasis in
different human cancer diseases. Compared to the primary melanoma
cells, IGFBP3 was overexpressed in the metastatic melanoma cells
and may serve as a biomarker in the early diagnosis of melanoma
(78). On the other hand, Torng
et al reported that IGFBP3 plays a major role as an
invasion-metastasis suppressor via an insulin growth
factor-independent pathway in human ovarian endometrioid carcinomas
(79). These previous observations
suggested possible dual biological roles of the IGFBP3 signal
pathways in various cancers (59–61,78,79).
Nonetheless, our results showed a positive correlation between
IGFBP3 protein expression and metastasis potential of the gastric
cancer cells. Contrary to that reported in colorectal cancer HCT116
cells (80), we found in gastric
cancer MKN45 cells that LGR4 expression was inversely correlated
with the cell invasion ability.
A number of the genes identified in our studies have
been verified for their transcriptional or translational activities
in the mRNA or protein levels by semi-quantified RT-PCR and western
blotting, respectively, such as TFF3, IGFBP3, BEX2, OTR, SGCE,
LGR4, FANCA, MIA, and MPS1/TTK. FANCA has been reported to
participate in the BRCA signaling pathway and is involved in the
chromosome stability of cancer cells (81). MIA proteins binding to integrins
promote detachment of melanoma cells from extracellular matrix
structures and increase the migration ability of the cells, which
could be inhibited by treatments with chemicals (82). MIA may play an important role in
gastric tumor cell invasion and metastasis. The mRNA or protein
expression profiles verified in the present study are in agreement
with those previously observed.
In addition, we have also identified a number of
novel genes, listed in Table V,
using the same evaluation processes. These novel genes have not
been previously demonstrated with any biological activity nor are
they known with any cancer progression-related or tumor
metastasis-associated activities. Similarly, these novel genes are
likely to be involved in important biological processes in the
development and progression of human gastric tumor invasion and
metastasis. Their possible cellular/subcellular mechanisms
involving tumor progression or metastasis are to be further
explored in the future. By using gene-silencing approaches such as
siRNA treatments to the MKN45-GFP-12 cells, one may further
investigate the functional roles of the pro-metastasis genes in the
tumor metastatic process and their underlying molecular mechanisms.
For the genes against metastasis, one can restore the downregulated
gene functions by gene transfection in MKN45-GFP-12 cells to
demonstrate a decrease in their metastatic abilities. Furthermore,
one can look into the gene expression levels in the clinical
patient specimens and search for clinical relevance in terms of
tumor progression as well as patient survival.
Furthermore, we established a highly aggressive
MKN45-GFP-12 cell line and adopted an orthotopic model in which
human gastric cancer cells of diffuse type were intraperitoneally
inoculated in nude mice to mimic the pathological condition of
peritoneal metastasis in patients. The malignancies of MKN45-GFP
and MKN45-GFP-12 cells were compared using the orthotopic model.
Compared to MKN45-GFP cells, MKN45-GFP-12 cells tend to invade
internal organs, such as the liver, and to cause ascites at a
shorter onset time, indicating a higher malignant property of the
MKN45-GFP-12 cells. More tumor nodules were growing in the
abdominal cavity of MKN45-GFP-12-inoculated nude mice at Day 14
(data not show). By using an in vivo selection process, we
previously established a human gastric cancer cell line
MKN45-GFP-ip4 (38). Whether the
two malignant cell lines MKN45-GFP12 and MKN45-GFP-ip4 established
via the in vitro and in vivo selections,
respectively, share a similar gene expression profile requires
further investigation.
In this study, we established several human diffuse
type gastric cancer cell sublines with different invasion abilities
and identified a number of putative gastric tumor
metastasis-associated genes, including novel genes without any
known biological activity (GEO accession number GSE33570). These
genes and protein products are to be further investigated for their
roles in the molecular and cellular actions related to tumor
metastasis. The molecular profiles of these identified genes, gene
transcripts, and/or proteins in the patient specimens are likely to
be useful for diagnostic, therapeutic, and/or prognostic purposes.
In particular, they may be used as molecular targets for the
development of drugs against tumor metastasis. These further
research efforts could ultimately lead to the identification of
tumor metastasis-associated molecules from which diagnostics and
therapeutics for the human gastric cancer may be discovered.
Acknowledgements
This study was supported by the
following grants, NHRIBP-091-CF03, NHRI-BP-093-CB06,
NHRI-BP-094-PP11, NHRI-BP-094-PP13, NHRI-095-PP07, and
NSC-92-2218-E-400-001, awarded to C.T. Chen from The National
Health Research Institutes, Miaoli and The National Science
Council, Taiwan, R.O.C.
References
|
1.
|
Archie V, Kauh J, Jones DV Jr, Cruz V,
Karpeh MS Jr and Thomas CR Jr: Gastric cancer: standards for the
21st century. Crit Rev Oncol Hematol. 57:123–131. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Methods Mol Biol. 472:467–477.
2009. View Article : Google Scholar
|
|
3.
|
Yeh KH and Cheng AL: Recent advances in
therapy for gastric cancer. J Formos Med Assoc. 103:171–185.
2004.
|
|
4.
|
Parkin DM: The global health burden of
infection-associated cancers in the year 2002. Int J Cancer.
118:3030–3044. 2006.PubMed/NCBI
|
|
5.
|
Vital and Health Statistics Division,
Statistics and Information Department, Ministry of Health, Labour
and Welfare: Vital statistics in Japan (1950–2007.
|
|
6.
|
Horner MJ, Ries LAG, Krapcho M, Neyman N,
Aminou R, Howlader N, Altekruse SF, Feuer EJ, Huang L, Mariotto A,
Miller BA, Lewis DR, Eisner MP, Stinchcomb DG and Edwards BK: SEER
Cancer statistics review, 1975–2006. National Cancer Institute;
Bethesda: 2008
|
|
7.
|
Lee KH, Lee JH, Cho JK, Kim TW, Kang YK,
Lee JS, Kim WK, Chung JG, Lee IC and Sun HS: A prospective
correlation of Laurén’s histological classification of stomach
cancer with clinicopathological findings including DNA flow
cytometry. Pathol Res Pract. 197:223–229. 2001.
|
|
8.
|
Whiting J, Sano T, Saka M, Fukagawa T,
Katai H and Sasako M: Follow-up of gastric cancer: a review.
Gastric Cancer. 9:74–81. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
The EUROCARE-4 database on cancer survival
in Europe. [http://www.eurocare.it/Results/tabid/79/Default.aspx#eu4dB].
|
|
10.
|
Guarino M: Epithelial-mesenchymal
transition and tumour invasion. Int J Biochem Cell Biol.
39:2153–2160. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Wu CY, Wu MS, Chiang EP, Wu CC, Chen YJ,
Chen CJ, Chi NH, Chen GH and Lin JT: Elevated plasma osteopontin
associated with gastric cancer development, invasion and survival.
Gut. 56:782–789. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Kuniyasu H, Yasui W, Yokozaki H, Kitadai Y
and Tahara E: Aberrant expression of c-met mRNA in human gastric
carcinomas. Int J Cancer. 55:72–75. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Ran Y, Peng L, Hu H, Yu L, Liu Q, Zhou Z,
Sun YM, Sun LC, Pan J, Sun LX, Zhao P and Yang ZH: Secreted LOXL2
is a novel therapeutic target that promotes gastric cancer
metastasis via the Src/FAK pathway. Carcinogenesis. 30:1660–1669.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kim JH, Kim MA, Lee HS and Kim WH:
Comparative analysis of protein expressions in primary and
metastatic gastric carcinomas. Hum Pathol. 40:314–322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Gravalos C and Jimeno A: HER2 in gastric
cancer: a new prognostic factor and a novel therapeutic target. Ann
Oncol. 19:1523–1529. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Sakashita K, Mimori K, Tanaka F, Kamohara
Y, Inoue H, Sawada T, Hirakawa K and Mori M: Prognostic relevance
of Tensin4 expression in human gastric cancer. Ann Surg Oncol.
15:2606–2613. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Yu G, Wang J, Chen Y, Wang X, Pan J, Li G,
Jia Z, Li Q, Yao JC and Xie K: Overexpression of phosphorylated
mammalian target of rapamycin predicts lymph node metastasis and
prognosis of Chinese patients with gastric cancer. Clin Cancer Res.
15:1821–1829. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Nakayama H, Yasui W, Yokozaki H and Tahara
E: Reduced expression of nm23 is associated with metastasis of
human gastric carcinomas. Jpn J Cancer Res. 84:184–190. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Okada K, Shimura T, Suehiro T, Mochiki E
and Kuwano H: Reduced galectin-3 expression is an indicator of
unfavorable prognosis in gastric cancer. Anticancer Res.
26:1369–1376. 2006.PubMed/NCBI
|
|
20.
|
Li X, Zhang Y, Cao S, Chen X, Lu Y, Jin H,
Sun S, Chen B, Liu J, Ding J, Wu K and Fan D: Reduction of TIP30
correlates with poor prognosis of gastric cancer patients and its
restoration drastically inhibits tumor growth and metastasis. Int J
Cancer. 124:713–721. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hsu PI, Hsieh HL, Lee J, Lin LF, Chen HC,
Lu PJ and Hsiao M: Loss of RUNX3 expression correlates with
differentiation, nodal metastasis, and poor prognosis of gastric
cancer. Ann Surg Oncol. 16:1686–1694. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kim DY, Joo JK, Park YK, Ryu SY, Kim HS,
Noh BK, Lee HK and Lee HJ: E-cadherin expression in early gastric
carcinoma and correlation with lymph node metastasis. J Surg Oncol.
96:429–435. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Hippo Y, Yashiro M, Ishii M, Taniguchi H,
Tsutsumi S, Hirakawa K, Kodama T and Aburatani H: Differential gene
expression profiles of scirrhous gastric cancer cells with high
metastatic potential to peritoneum or lymph nodes. Cancer Res.
61:889–895. 2001.PubMed/NCBI
|
|
24.
|
Wang J and Chen S: Screening and
identification of gastric adenocarcinoma metastasis-related genes
using cDNA micro-array coupled to FDD-PCR. J Cancer Res Clin Oncol.
128:547–553. 2002. View Article : Google Scholar
|
|
25.
|
Kim JM, Sohn HY, Yoon SY, Oh JH, Yang JO,
Kim JH, Song KS, Rho SM, Yoo HS, Kim YS, Kim JG and Kim NS:
Identification of gastric cancer-related genes using a cDNA
microarray containing novel expressed sequence tags expressed in
gastric cancer cells. Clin Cancer Res. 11:473–482. 2005.PubMed/NCBI
|
|
26.
|
Myllykangas S, Junnila S, Kokkola A, Autio
R, Scheinin I, Kiviluoto T, Karjalainen-Lindsberg ML, Hollmén J,
Knuutila S, Puolakkainen P and Monni O: Integrated gene copy number
and expression microarray analysis of gastric cancer highlights
potential target genes. Int J Cancer. 123:817–825. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Hippo Y, Taniguchi H, Tsutsumi S, Machida
N, Chong JM, Fukayama M, Kodama T and Aburatani H: Global gene
expression analysis of gastric cancer by oligonucleotide
microarrays. Cancer Res. 62:233–240. 2002.PubMed/NCBI
|
|
28.
|
Inoue H, Matsuyama A, Mimori K, Ueo H and
Mori M: Prognostic score of gastric cancer determined by cDNA
microarray. Clin Cancer Res. 8:3475–3479. 2002.PubMed/NCBI
|
|
29.
|
Hasegawa S, Furukawa Y, Li M, Satoh S,
Kato T, Watanabe T, Katagiri T, Tsunoda T, Yamaoka Y and Nakamura
Y: Genome-wide analysis of gene expression in intestinal-type
gastric cancers using a complementary DNA microarray representing
23,040 genes. Cancer Res. 62:7012–7017. 2002.PubMed/NCBI
|
|
30.
|
Yang S, Jeung HC, Jeong HJ, Choi YH, Kim
JE, Jung JJ, Rha SY, Yang WI and Chung HC: Identification of genes
with correlated patterns of variations in DNA copy number and gene
expression level in gastric cancer. Genomics. 89:451–459. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Chang W, Ma L, Lin L, Gu L, Liu X, Cai H,
Yu Y, Tan X, Zhai Y, Xu X, et al: Identification of novel hub genes
associated with liver metastasis of gastric cancer. Int J Cancer.
125:2844–2853. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Oue N, Aung PP, Mitani Y, Kuniyasu H,
Nakayama H and Yasui W: Genes involved in invasion and metastasis
of gastric cancer identified by array-based hybridization and
serial analysis of gene expression. Oncology. 69:17–22. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Motoyama T, Hojo H and Watanabe H:
Comparison of seven cell lines derived from human gastric
carcinomas. Acta Pathol Jpn. 36:65–83. 1986.PubMed/NCBI
|
|
34.
|
Yokozaki H: Molecular characteristics of
eight gastric cancer cell lines established in Japan. Pathol Int.
50:767–777. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Chu YW, Yang PC, Yang SC, Shyu YC, Hendrix
MJ, Wu R and Wu CW: Selection of invasive and metastatic
subpopulations from a human lung adenocarcinoma cell line. Am J
Respir Cell Mol Biol. 17:353–360. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Huang YC, Chen CT, Chen SC, Lai PH, Liang
HC, Chang Y, Yu LC and Sung HW: A natural compound (Ginsenoside Re)
isolated from Panax Ginseng as a novel angiogenic agent for tissue
regeneration. Pharm Res. 22:636–646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Li WT, Hwang DR, Chen CP, Shen CW, Huang
CL, Chen TW, Lin CH, Chang YL, Chang YY, Lo YK, et al: Synthesis
and biological evaluation of N-heterocyclic indolyl glyoxylamides
as orally active anticancer agents. J Med Chem. 46:1706–1715. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Tuan TF, Tsai ML, Yeh KC, Huang HC, Chung
CT, Huang CL, Han CH, Chen CP, Wang MH, Shen CC, et al: Intravenous
paclitaxel against metastasis of human gastric tumors of diffuse
type. Cancer Chemother Pharmacol. 66:773–783. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Geiger TR and Peeper DS: Metastasis
mechanisms. Biochim Biophys Acta. 1796:293–308. 2009.PubMed/NCBI
|
|
40.
|
Chen X, Leung SY, Yuen ST, Chu KM, Ji J,
Li R, Chan AS, Law S, Troyanskaya OG, Wong J, So S, Botstein D and
Brown PO: Variation in gene expression patterns in human gastric
cancers. Mol Biol Cell. 14:3208–3215. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Hansson LE, Engstrand L, Nyrén O, Evans DJ
Jr, Lindgren A, Bergström R, Andersson B, Athlin L, Bendtsen O and
Tracz P: Helicobacter pylori infection: independent risk indicator
of gastric adenocarcinoma. Gastroenterology. 105:1098–1103.
1993.PubMed/NCBI
|
|
42.
|
Piazuelo MB, Epplein M and Correa P:
Gastric cancer: an infectious disease. Infect Dis Clin North Am.
24:853–869. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Ferreira AC, Isomoto H, Moriyama M,
Fujioka T, Machado JC and Yamaoka Y: Helicobacter and gastric
malignancies. Helicobacter. 13(Suppl 1): 28–34. 2008. View Article : Google Scholar
|
|
44.
|
Suganuma M, Kuzuhara T, Yamaguchi K and
Fujiki H: Carcinogenic role of tumor necrosis factor-alpha inducing
protein of Helicobacter pylori in human stomach. J Biochem Mol
Biol. 39:1–8. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Smith MG, Hold GL, Tahara E and El-Omar
EM: Cellular and molecular aspects of gastric cancer. World J
Gastroenterol. 12:2979–2990. 2006.
|
|
46.
|
Chan AO, Lam SK, Wong BC, Wong WM, Yuen
MF, Yeung YH, Hui WM, Rashid A and Kwong YL: Promoter methylation
of E-cadherin gene in gastric mucosa associated with Helicobacter
pylori infection and in gastric cancer. Gut. 52:502–506. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Iwamoto J, Mizokami Y, Takahashi K,
Nakajima K, Ohtsubo T, Miura S, Narasaka T, Takeyama H, Omata T,
Shimokobe K, Ito M, Takehara H and Matsuoka T: Expressions of
urokinase-type plasminogen activator, its receptor and plasminogen
activator inhibitor-1 in gastric cancer cells and effects of
Helicobacter pylori. Scand J Gastroenterol. 40:783–793. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Schmausser B, Endrich S, Brändlein S,
Schär J, Beier D, Müller-Hermelink HK and Eck M: The chemokine
receptor CCR7 is expressed on epithelium of non-inflamed gastric
mucosa, Helicobacter pylori gastritis, gastric carcinoma and its
precursor lesions and up-regulated by H. pylori. Clin Exp Immunol.
139:323–327. 2005. View Article : Google Scholar
|
|
49.
|
Yasumoto K, Koizumi K, Kawashima A, Saitoh
Y, Arita Y, Shinohara K, Minami T, Nakayama T, Sakurai H, Takahashi
Y, Yoshie O and Saiki I: Role of the CXCL12/CXCR4 axis in
peritoneal carcinomatosis of gastric cancer. Cancer Res.
66:2181–2187. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Zhao C, Lu X, Bu X, Zhang N and Wang W:
Involvement of tumor necrosis factor-alpha in the upregulation of
CXCR4 expression in gastric cancer induced by Helicobacter pylori.
BMC Cancer. 10:4192010. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Amoh Y, Katsuoka K and Hoffman RM:
Color-coded fluorescent protein imaging of angiogenesis: the
AngioMouse models. Curr Pharm Des. 14:3810–3819. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Bouvet M, Tsuji K, Yang M, Jiang P, Moossa
AR and Hoffman RM: In vivo color-coded imaging of the interaction
of colon cancer cells and splenocytes in the formation of liver
metastases. Cancer Res. 66:11293–11297. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Hoffman RM: The multiple uses of
fluorescent proteins to visualize cancer in vivo. Nat Rev Cancer.
5:796–806. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Hoffman RM: Dual-color imaging of tumor
angiogenesis. Methods Mol Biol. 515:45–61. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Chen JJ, Peck K, Hong TM, Yang SC, Sher
YP, Shih JY, Wu R, Cheng JL, Roffler SR, Wu CW and Yang PC: Global
analysis of gene expression in invasion by a lung cancer model.
Cancer Res. 61:5223–5230. 2001.PubMed/NCBI
|
|
56.
|
Chen HY, Yu SL, Chen CH, Chang GC, Chen
CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al: A five-gene
signature and clinical outcome in non-small-cell lung cancer. N
Engl J Med. 356:11–20. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Dhar DK, Wang TC, Tabara H, Tonomoto Y,
Maruyama R, Tachibana M, Kubota H and Nagasue N: Expression of
trefoil factor family members correlates with patient prognosis and
neoangiogenesis. Clin Cancer Res. 11:6472–6478. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Rodrigues S, Van Aken E, Van Bocxlaer S,
Attoub S, Nguyen QD, Bruyneel E, Westley BR, May FE, Thim L, Mareel
M, Gespach C and Emami S: Trefoil peptides as proangiogenic factors
in vivo and in vitro: implication of cyclooxygenase-2 and EGF
receptor signaling. FASEB J. 17:7–16. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Liu B, Lee KW, Anzo M, Zhang B, Zi X, Tao
Y, Shiry L, Pollak M, Lin S and Cohen P: Insulin-like growth
factor-binding protein-3 inhibition of prostate cancer growth
involves suppression of angiogenesis. Oncogene. 26:1811–1819. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Akhtar S, Meeran SM, Katiyar N and Katiyar
SK: Grape seed proanthocyanidins inhibit the growth of human
non-small cell lung cancer xenografts by targeting insulin-like
growth factor binding protein-3, tumor cell proliferation, and
angiogenic factors. Clin Cancer Res. 15:821–831. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Hansel DE, Rahman A, House M, Ashfaq R,
Berg K, Yeo CJ and Maitra A: Met proto-oncogene and insulin-like
growth factor binding protein 3 overexpression correlates with
metastatic ability in well-differentiated pancreatic endocrine
neoplasms. Clin Cancer Res. 10:6152–6158. 2004. View Article : Google Scholar
|
|
62.
|
Saaristo A, Karpanen T and Alitalo K:
Mechanisms of angiogenesis and their use in the inhibition of tumor
growth and metastasis. Oncogene. 19:6122–6129. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Iwai K, Hirata K, Ishida T, Takeuchi S,
Hirase T, Rikitake Y, Kojima Y, Inoue N, Kawashima S and Yokoyama
M: An anti-proliferative gene BTG1 regulates angiogenesis in vitro.
Biochem Biophys Res Commun. 316:628–635. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Fisk HA, Mattison CP and Winey M: A field
guide to the Mps1 family of protein kinases. Cell Cycle. 3:439–442.
2004.PubMed/NCBI
|
|
65.
|
Dorer RK, Zhong S, Tallarico JA, Wong WH,
Mitchison TJ and Murray AW: A small-molecule inhibitor of Mps1
blocks the spindle-checkpoint response to a lack of tension on
mitotic chromosomes. Curr Biol. 15:1070–1076. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Poss KD, Nechiporuk A, Stringer KF, Lee C
and Keating MT: Germ cell aneuploidy in zebrafish with mutations in
the mitotic checkpoint gene mps1. Genes Dev. 18:1527–1532. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
67.
|
Takayama T, Miyanishi K, Hayashi T, Sato Y
and Niitsu Y: Colorectal cancer: genetics of development and
metastasis. J Gastroenterol. 41:185–192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Naderi A, Liu J and Bennett IC: BEX2
regulates mitochondrial apoptosis and G1 cell cycle in breast
cancer. Int J Cancer. 126:1596–1610. 2010.PubMed/NCBI
|
|
69.
|
Chen PS, Wang MY, Wu SN, Su JL, Hong CC,
Chuang SE, Chen MW, Hua KT, Wu YL, Cha ST, et al: CTGF enhances the
motility of breast cancer cells via an
integrin-alphavbeta3-ERK1/2-dependent S100A4-upregulated pathway. J
Cell Sci. 120:2053–2065. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Bracke ME, Van Roy FM and Mareel MM: The
E-cadherin/catenin complex in invasion and metastasis. Curr Top
Microbiol Immunol. 213:123–161. 1996.PubMed/NCBI
|
|
71.
|
Mauro L, Bartucci M, Morelli C, Ando S and
Surmacz E: IGF-I receptor-induced cell-cell adhesion of MCF-7
breast cancer cells requires the expression of junction protein
ZO-1. J Biol Chem. 276:39892–39897. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Qin J, Feng M, Wang C, Ye Y, Wang PS and
Liu C: Oxytocin receptor expressed on the smooth muscle mediates
the excitatory effect of oxytocin on gastric motility in rats.
Neurogastroenterol Motil. 21:430–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Morita T, Shibata K, Kikkawa F, Kajiyama
H, Ino K and Mizutani S: Oxytocin inhibits the progression of human
ovarian carcinoma cells in vitro and in vivo. Int J Cancer.
109:525–532. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Meyer zum Büschenfelde D, Hoschützky H,
Tauber R and Huber O: Molecular mechanisms involved in TFF3
peptide-mediated modulation of the E-cadherin/catenin cell adhesion
complex. Peptides. 25:873–883. 2004.PubMed/NCBI
|
|
75.
|
Ortega P, Moran A, Fernandez-Marcelo T, De
Juan C, Frias C, Lopez-Asenjo JA, Sanchez-Pernaute A, Torres A,
Diaz-Rubio E, Iniesta P and Benito M: MMP-7 and SGCE as distinctive
molecular factors in sporadic colorectal cancers from the mutator
phenotype pathway. Int J Oncol. 36:1209–1215. 2010.PubMed/NCBI
|
|
76.
|
Patarroyo M, Tryggvason K and Virtanen I:
Laminin isoforms in tumor invasion, angiogenesis and metastasis.
Semi Cancer Biol. 12:197–207. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Wang J, Levenson AS and Satcher RL Jr:
Identification of a unique set of genes altered during cell-cell
contact in an in vitro model of prostate cancer bone
metastasis. Int J Mol Med. 17:849–856. 2006.PubMed/NCBI
|
|
78.
|
Xi Y, Nakajima G, Hamil T, Fodstad O,
Riker A and Ju J: Association of insulin-like growth factor binding
protein-3 expression with melanoma progression. Mol Cancer Ther.
5:3078–3084. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
79.
|
Torng PL, Lee YC, Huang CY, Ye JH, Lin YS,
Chu YW, Huang SC, Cohen P, Wu CW and Lin CT: Insulin-like growth
factor binding protein-3 (IGFBP-3) acts as an invasion-metastasis
suppressor in ovarian endometrioid carcinoma. Oncogene.
27:2137–2147. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Gao Y, Kitagawa K, Hiramatsu Y, Kikuchi H,
Isobe T, Shimada M, Uchida C, Hattori T, Oda T, Nakayama K,
Nakayama KI, Tanaka T, Konno H and Kitagawa M: Up-regulation of
GPR48 induced by down-regulation of p27Kip1 enhances carcinoma cell
invasiveness and metastasis. Cancer Res. 66:11623–11631. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
81.
|
D’Andrea AD and Grompe M: The Fanconi
anaemia/BRCA pathway. Nat Rev Cancer. 3:23–34. 2003.
|
|
82.
|
Schmidt J and Bosserhoff AK: Processing of
MIA protein during melanoma cell migration. Int J Cancer.
125:1587–1594. 2009. View Article : Google Scholar : PubMed/NCBI
|