Introduction
Tumor suppressor proteins are negative regulators of
cell growth and can inhibit neoplastic transformation of cells.
Unfortunately, most of the genes encoding various tumor suppressor
proteins are inactivated during the course of carcinogenesis
(1). Transducer of ErbB-2.1
(Tob1), is a tumor suppressor protein encoded by the Tob
gene located on chromosome 17q21 (2). Tob1 is a member of the Tob/B cell
translocation gene (BTG) family proteins, which upon overexpression
arrests cell cycle in G1 phase (3–5).
Tob1 binds to ErbB2 receptor and functions as a negative regulator
of cell growth by blocking ErbB2-mediated cell signaling pathways
(2). Yoshida et al(6) reported that mice lacking Tob
gene developed spontaneous tumors primarily in the lung, liver and
lymph nodes. Subsequent studies demonstrated that the expression of
Tob1 is lost in various cancers including those of the breast
(7), pancreas (8), thyroid (9) and stomach (10). While the loss of Tob1 has been
implicated in the progression of human papillary thyroid carcinomas
(9), ectopic expression of Tob1
inhibited cell cycle progression and reduced the growth of human
breast cancer cell xenografts in nude mice (7). The anti-proliferative effect of Tob1
has been ascribed to its ability to inhibit the promoter activity
of cyclin D1 (11).
The tumor suppressor function of Tob1 has largely
been corroborated to its anti-proliferative effect on various
cancer cells. However, the role of Tob1 in the migration and
invasion of gastric cancer cells and its underlying mechanisms are
yet to be investigated. Since Tob1 expression has been shown to be
lost in human gastric carcinomas (10), we examined whether the restoration
of Tob1 could prevent gastric cancer progression, and to elucidate
the molecular mechanisms associated with its tumor suppressor
function. It has been reported that Tob1 acts as a transcriptional
co-activator by binding with Smad4 (11) and as a transcriptional co-repressor
by interacting with β-catenin (12). Since functional inactivation of
Smad4 (13–14) and aberrant expression of β-catenin
(15) play pivotal roles in
gastric carcinogenesis, we examined whether Tob1 could modulate the
expression of Smad4 and β-catenin, thereby preventing gastric
cancer progression. In the present study, we report that
overexpression of Tob1 induces apoptosis and inhibits the
proliferation, migration and invasion of gastric cancer cells, at
least in part, by activating the expression of Smad4 and its target
gene p15 as well as by downregulating the expression of β-catenin
and its target genes, such as cyclin D1, uPAR and PPARδ.
Materials and methods
Cell culture and transfection
Human gastric cancer cell lines (AGS, MKN1 and
MKN28) were purchased from the Korean Cell Line Bank (Seoul, Korea)
and were maintained in RPMI-1640 medium supplemented with 10% fetal
bovine serum at 37°C in 5% CO2. Cells were routinely
checked for mycoplasma contamination. MKN28 and AGS cells were
transiently transfected with myc-Tob1 plasmid (2 or 4
μg), which was a generous gift from Professor Jun-Mo Yang,
Sungkyunkwan University, Seoul, Korea, using Genefectine™ reagent
(Genetrone Biotech, Seoul, Korea) following the manufacturer’s
instructions. Silencing of Tob1 in MKN1 and AGS cells was performed
by transiently transfecting cells with 25 or 50 nM Tob1 si-RNA
(Santa Cruz Biotechnology, Santa Cruz, CA, USA) by using DharmaFECT
(Dharmacon, Lafayette, CO, USA) according to the manufacturer’s
instructions.
Western blot analysis
Cells were harvested and lysed with RIPA buffer [150
mM NaCl, 10 mM Tris (pH 7.2), 0.1% sodium dodecyl sulphate (SDS),
1% Triton X-100, 1% deoxycholate and 5 mM
ethylenediaminetetraacetic acid (EDTA)] enriched with a complete
protease inhibitor cocktail tablet (Roche Diagnostics, Mannheim,
Germany), and then incubated on ice for 30 min with regular
vortexing before centrifuging at 14,000 rpm at 4°C for 15 min.
Protein concentration was determined by using bichinconinic acid
(BCA) protein assay kit (Pierce Biotechnology, Rockford, IL, USA).
The protein samples were boiled in 1X SDS sample buffer for 5 min
for complete denaturation and were resolved on a 10 to 15%
SDS-polyacrylamide gel according to the protocol described earlier
(16). After electrophoresis,
proteins were transferred onto polyvinyl difluoride (PVDF)
membrane, which was blocked with 5% nonfat dry milk in 1X TBST
(Tris-buffered saline with 0.1% Tween-20) and incubated with
primary antibody at the appropriate final concentration followed by
hybridization with horseradish peroxidase-conjugated anti-rabbit or
anti-mouse secondary antibodies (1:5,000). Finally, western blot
images were developed on photographic film using enhanced
chemiluminescence (ECL) reagents. For each step, the membrane was
washed with 1X TBST three times for 10 min. The primary antibodies
used were: β-actin, β-catenin, Smad4, p27, CDK4, uPAR, Bax and
Bcl-2 (Santa Cruz Biotechnology), Tob1 (Clone-4B1, Sigma Aldrich,
St. Louis, MO, USA), p15, PPARδ (Abcam, Cambridge, UK), and Akt,
pAkt, GSK3β, p-GSK3β (Ser-9), cleaved-polyadiporibosyl polymerase
(PARP) and cyclin D1 (Cell Signaling Inc., Beverly, MA, USA).
Total-RNA preparation and qRT-PCR
Total-RNA was extracted using TRIzol (Invitrogen,
Carlsbad, CA, USA) and reversely transcribed to cDNA using the
Superscript™ II First-Strand Synthesis System (Invitrogen).
Following cDNA synthesis, qRT-PCR was performed as described
(17) in a dual system LightCycler
(Roche Applied Science, Mannheim, Germany) using the primers for
p15, Tob1, cyclin D1, uPAR and
PPARδ. HPRT was used for normalizing gene expression. All
PCR primers and probe sequences (Universal Probe Library, Roche
Applied Science) including the HPRT TaqMan probe (TIB
Molbiol, Berlin, Germany) are listed in Table I.
 | Table IPrimer sequences for quantitative RT
PCR. |
Table I
Primer sequences for quantitative RT
PCR.
| Gene | Orientation | Sequences | Tm (°C) |
|---|
| Tob1 | Forward |
5′-TCTGTATGGGCTTGGCTTG-3′ | 54.9 |
| Reverse |
5′-TGTTGCTGCTGTGGTGGT-3′ | 54.3 |
| CDKN2B | Forward |
5′-CAACGGAGTCAACCGTTTC-3′ | 54.9 |
| Reverse |
5′-GGTGAGAGTGGCAGGGTCT-3′ | 54.2 |
| Cyclin
D1 | Forward |
5′-GAAGATCGTCGCCACCTG-3′ | 56.6 |
| Reverse |
5′-GACCTCCTCCTCGCACTTCT-3′ | 59.5 |
| uPAR | Forward |
5′-ACACCACCAAATGCAACGA-3′ | 52.7 |
| Reverse |
5′-CCCCTTGCAGCTGTAACAC-3′ | 57.1 |
| PPARδ | Forward |
5′-GGGAAAAGTTTTGGCAGGAG-3′ | 55.4 |
| Reverse |
5′-TGCCCAAAACACTGTACAACA-3′ | 53.9 |
| HPRT | Forward |
5′-CTCAACTTTAACTGGAAAGAATGTC-3′ | 54.1 |
| Reverse |
5′-TCCTTTTCACCAGCAAGCT-3′ | 55.5 |
Cell proliferation assay
The effect of Tob1 overexpression on cell
proliferation was measured by the water soluble tetrazolium salts
(WST) method (EZ-Cytox kit; Daeil Lab Service, Seoul, Korea). MKN28
and AGS cells (1×105), transfected with either
myc-Tob1 (2 or 4 μg) or vector alone, were incubated
in triplicate in a 12-well plate for 24 h at 37°C. MKN1 and AGS
cells (1×105) were transfected with either control
si-RNA or Tob1 si-RNA (25 or 50 nM) in triplicate in a 12-well
plate for 48 h at 37°C. EZ-Cytox solution (200 μl) was added
to each well and incubated for 80 min. The number of viable cells
was measured in a 96-well plate at an optical density of 492 nm on
a Sunrise reader (Tecan Trading AG, Mannedorf, Switzerland). Cell
viability was described as the percentage of empty vector- or
control si-RNA-transfected cells.
Cell migration and invasion assay
Cells (5×104) transfected with empty
vector, myc-Tob1 plasmid (2 or 4 μg),
Tob1si-RNA (25 or 50 nM) or control si-RNA, were subjected
to Millipore’s (Billerica, MA, USA) 24-well Chemicon QCM™ cell
migration assay and QCM™ fluorimetric cell invasion assay systems.
After incubation at 37°C for 24 h, cell number was detected with a
GENios Pro microplate reader (Tecan Trading AG) using 485/535 nm
filter set. All migration and invasion assays were performed in
triplicate in at least three independent experiments. Values are
expressed as percentages of control (18).
Wound healing assay
In vitro wound healing assay was performed to
examine the migration of MKN28 and AGS cells transfected with
either a control vector or myc-Tob1. In another experiment,
AGS cells were transfected with either control si-RNA or Tob1
si-RNA. Transfected cells were grown on 6-well plates with their
respective culture media. After the growing cell layers had reached
confluence, wounds were prepared by a single scratch on the
monolayer using a yellow pipette tip and washed the wounded layers
with PBS to remove cell debris. We measured the closure or filling
of the wounds at 0, 12 or 24 h using an Olympus IX71 fluorescence
microscope with a TH4-200 camera. All experiments were performed in
triplicate.
Luciferase reporter gene assay
Cells were seeded into 12-well plates at a density
of 1×105 cells per well prior to transfection. Cells
were transfected with TOPflash or FOPflash plasmid (kindly provided
by Professor Sung-Hee Baek, College of Natural Sciences, Seoul
National University, Seoul, Korea) or p15 promoter (a kind gift
from Dr Joan Massague, Howard Hughes Medical Institute, Memorial
Sloan-Kettering Cancer Center, NY, USA) together with an empty
vector or myc-Tob1 using Genefectin transfection reagent.
pRL-TK (Promega, Madison, WI, USA) was used as a normalization
control. After a further 24-h culture, the luciferase activity was
measured using the Dual-Luciferase® Reporter Assay
System (Promega). For si-RNA experiments, cells were seeded into
12-well plates at a density of 1×105 cells per well
prior to transfection with either control-siRNA or Tob1-siRNA for
24 h followed by transfection with TOPflash or FOPflash plasmid
(AGS cells) or p15 promoter construct (MKN1 and AGS cells) together
with an normalization control pRL-TK for additional 24 h using
Genefectin transfection reagent. The luciferase activity was
measured by using the Dual-Luciferase Reporter Assay System
according to the manufacturer’s instructions (Promega).
The caspase-3 activity assay
The activity of caspase-3 in Tob1-transfected
MKN28 and AGS cells was detected using Caspase-3 Colorimetric
Activity Assay Kit (Millipore). The assay was performed in 96-well
plates by incubating cell lysates (50 μg) in 100 μl
reaction buffer containing caspase-3 substrate Ac-DEVD-pNA at 37°C
for 2 h 30 min. The amount of p-nitroanilide, released by caspase
activation, was quantified by measuring the optical density at 405
nm with a GENios Pro microplate reader (Tecan Trading AG). Lysis
buffer: 20 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, 0.5%
Nonidet P-40, 1 mM EDTA, 10 mM sodium fluoride, 10 mM
β-glycerophosphate, 0.5 mM sodium orthovanadate, and 0.1 mM
phenylmethylsulfonyl fluoride.
Flow cytometry
In Tob1-transfected cells apoptosis was
evaluated by flow cytometry. MKN28 or AGS (2×105) cells
were transfected with myc-Tob1 plasmid (2 or 4 μg)
and cultured for 24 h. After 24 h, cells were harvested and fixed
in cold 90% ethanol overnight at −20°C, and resuspended in staining
buffer consisting of 100 mg/ml of RNase A (Quiagen, Valencia, CA,
USA), 20 μg/ml of propidium iodide (BD-Biosciences, San
Jose, CA, USA), and 0.1% Nonidet P-40. The DNA content was analyzed
by flow cytometry (BD-Biosciences).
Annexin V staining
Annexin V staining was performed using FITC-Annexin
V staining kit (BD-Biosciences) following the manufacturer’s
instructions. Briefly, myc-Tob1-transfected cells were
washed with PBS and resuspended in binding buffer containing
Annexin V and propidium iodide. Flourescence intensity was measured
using flow cytometry (BD-Biosciences).
Statistical analysis
When necessary, data were expressed as mean ± SD of
at least three independent experiments, and statistical analysis
for single comparison was performed using the Student’s t-test. The
criterion for statistical significance was *p<0.05,
**p<0.01 and ***p<0.001, respectively,
compared to corresponding control vector or control si-RNA.
Results
Tob1 inhibits proliferation of gastric
cancer cells
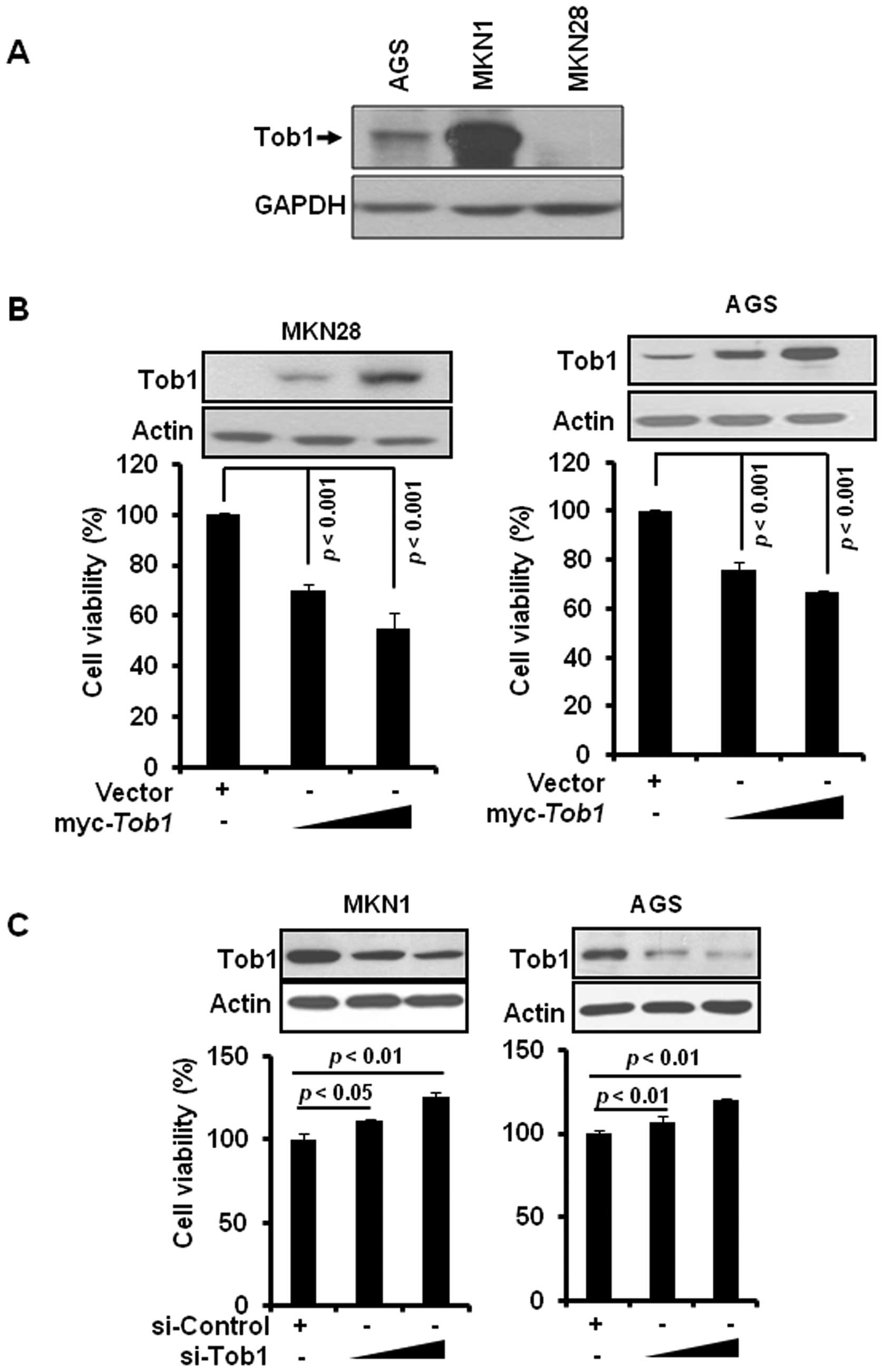
We first examined the expression of Tob1 in a
variety of gastric cancer cells including MKN28, AGS, and MKN1.
Tob1 was highly expressed in MKN1 cells and moderately expressed in
AGS cells, while it was not detectable in MKN28 cells (Fig. 1A). To examine the role of Tob1 in
gastric cancer, MKN28 and AGS cells were transiently transfected
with myc-Tob1. Tob1 significantly decreased cell viability
after 24 h in both MKN28 and AGS cells compared to vector alone
(Fig. 1B). Conversely, the
downregulation of Tob1 using specific si-RNA significantly
increased the viability of MKN1 and AGS cells (Fig. 1C).
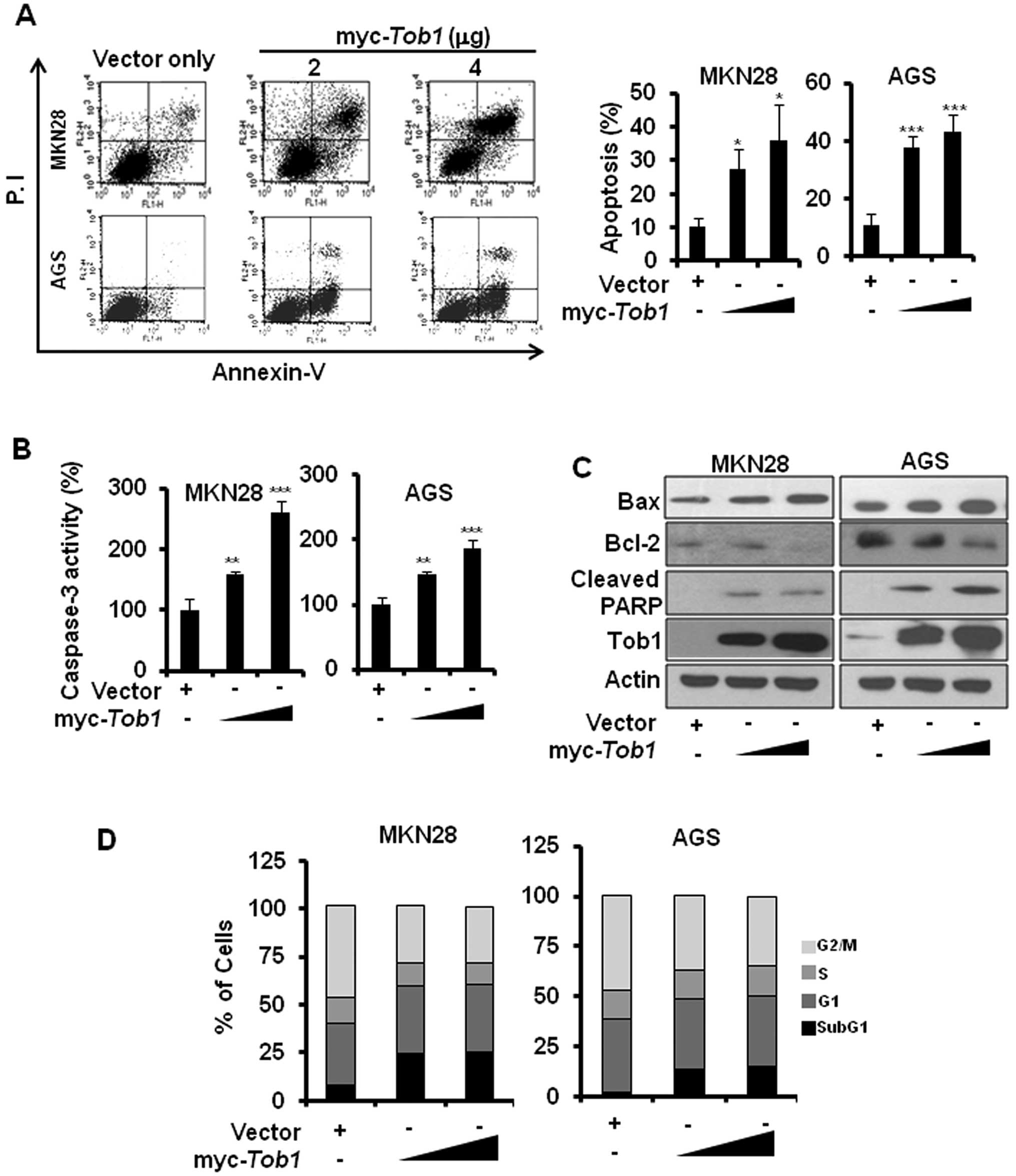
Restoration of Tob1 induces apoptosis in
gastric cancer cells
The inhibition of cell viability by ectopic
expression of Tob1 prompted us to examine if Tob1 overexpression
can induce apoptosis in gastric cancer cells. Annexin V staining
revealed that Tob1 significantly induced apoptosis in both MKN28
and AGS cells (Fig. 2A). The
caspase-3 activity assay using colorimetric substrate confirmed
that Tob1 significantly induced apoptosis (Fig. 2B). In addition, Tob1 showed
increased expression of Bax, reduced expression of Bcl-2 and
induction of PARP cleavage (Fig.
2C). Furthermore, cell cycle analysis revealed the increased
accumulation of Tob1-transfected MKN28 and AGS cells at the
sub-G1 phase (Fig. 2D).
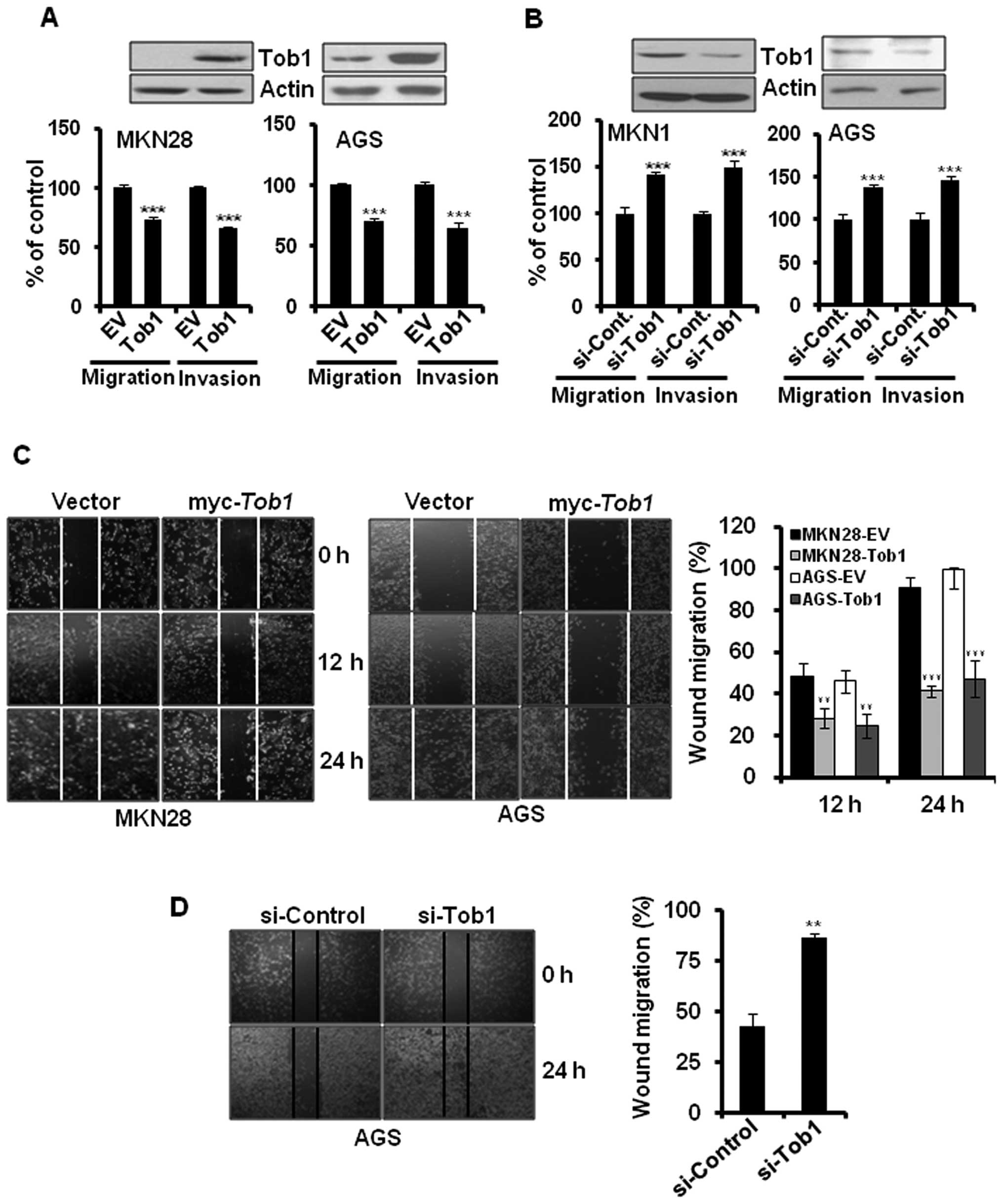
Tob1 suppresses the migration and
invasion of gastric cancer cells
To evaluate the role of Tob1 in gastric cancer
progression, myc-Tob1-transfected MKN28 and AGS cells were
subjected to migration and invasion assay. Overexpression of Tob1
significantly inhibited the migration and invasion of MKN28 and AGS
cells (Fig. 3A). In addition,
knock-down of Tob1 using specific si-RNA increased the migration
and invasion of MKN1 and AGS cells by about 40% (Fig. 3B). These findings were further
confirmed by the wound healing assay. The overexpression of Tob1
significantly inhibited the migration of MKN28 and AGS cells at 24
h after transfection (Fig. 3C). In
addition, we examined the knock-down effect of Tob1 in AGS cells
alone because MKN28 cells did not express endogenous Tob1. The
downregulation of Tob1 using specific si-RNA increased cell
migration compared to control si-RNA (Fig. 3D).
Overexpression of Tob1 increases the
expression of Smad4 and its target gene the p15
It has been reported that Tob1 interacts with Smad4
(11), a tumor suppressor protein
that is inactivated during gastric cancer progression (14). Therefore, we investigated whether
Tob1 could regulate the expression of Smad4. Ectopic expression of
Tob1 significantly elevated the expression of Smad4 in a
dose-dependent manner in both MKN28 and AGS cells (Fig. 4A), but did not alter Smad4 mRNA
expression (data not shown). In addition, we examined the effect of
Tob1 on the expression of Smad4 target genes. As shown in Fig. 4A, transient overexpression of Tob1
increased the expression of p15, but not p27. qRT-PCR analysis
showed that the expression of p15 was also increased at
transcriptional level in both MKN28 and AGS cells (Fig. 4B). The luciferase activity assay
using p15 promoter further confirmed that Tob1 significantly
increased the p15 promoter activity (Fig. 4C). The role of Tob1 in the
regulation of the expression of Smad4 and its target gene p15 was
further confirmed by transfecting MKN1 and AGS cells with Tob1
siRNA. The knock-down of Tob1 significantly reduced the expression
of Smad4 and p15 in both MKN1 and AGS cells as compared to cells
harboring control si-RNA (Fig.
4D). In addition, in comparison to control si-RNA, the relative
expression of p15 mRNA (Fig. 4E)
and the p15 promoter activity (Fig.
4F) were significantly decreased in both MKN1 and AGS cells
transfected with Tob1 siRNA.
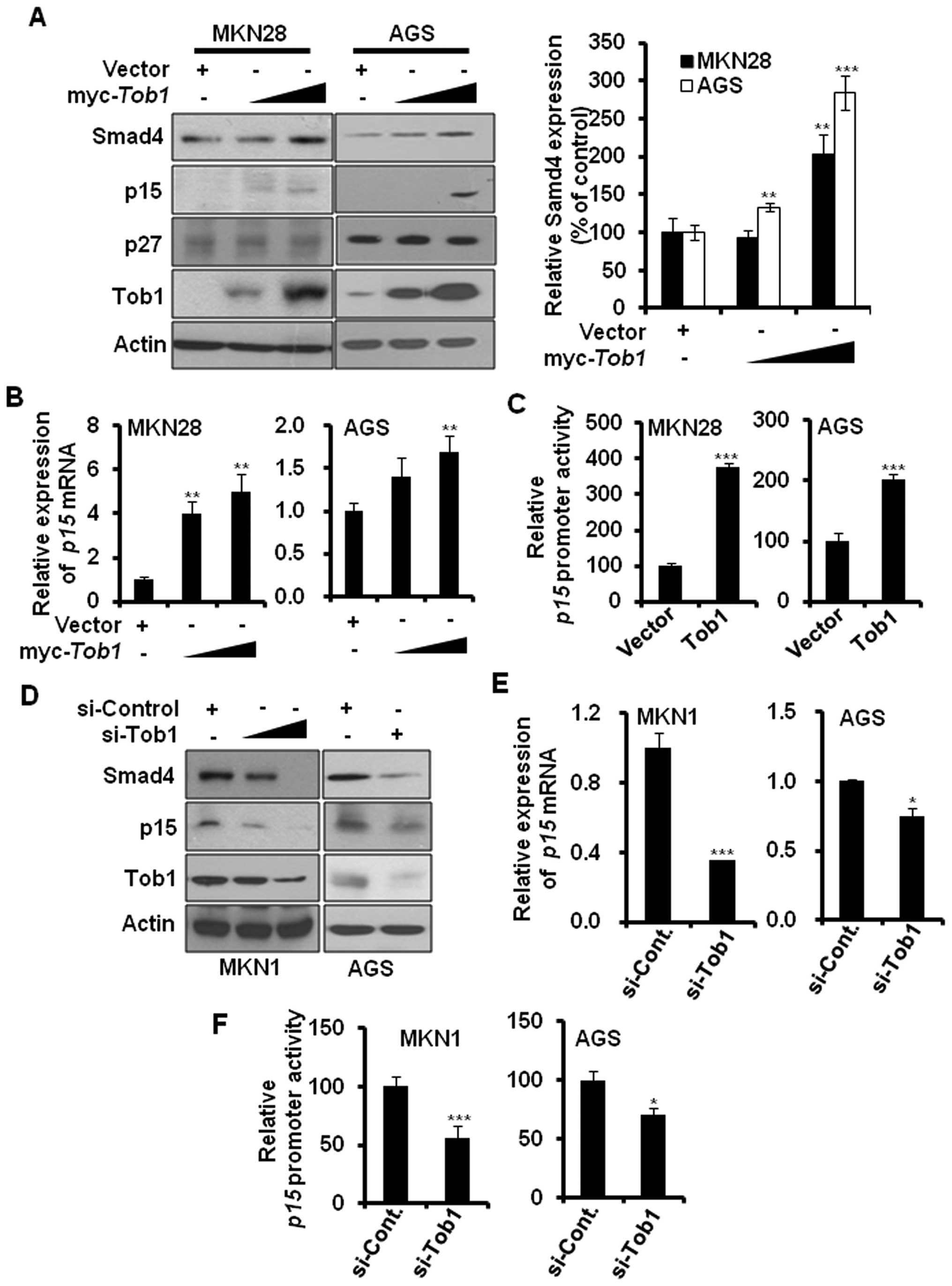 | Figure 4Tob1 enhances the expression of Smad4
and p15 in gastric cancer cells. (A) Protein lysates (30 μg)
from MKN28 or AGS cells transfected with either control vector or
myc-Tob1 (2 or 4 μg) were separated by 10–15%
SDS-PAGE, and the expression of Smad4 and its target gene products,
p15 and p27, was detected by immunoblot analysis. Data are
representative of three independent experiments.
**p<0.01 and ***p<0.001 compared to
respective control vector. (B) Total-RNA was isolated from MKN28 or
AGS cells transfected with either vector alone or myc-Tob1
(2 or 4 μg) and qRT-PCR was performed to assess the
expression of p15 mRNA. HPRT was used for normalizing
gene expression. **p<0.01, compared to control
vector. (C) The luciferase activity assay using p15 promoter was
performed in the presence or absence of myc-Tob1 (0.4
μg). ***p<0.001, compared to control vector.
(D) MKN1 and AGS cells were transiently transfected with either
control si-RNA (50 nM) or Tob1 si-RNA (25 and 50 nM for MKN1, 50 nM
for AGS) for 48 h, and cell lysates were separated by SDS-PAGE and
immunoblotted using their specific antibodies. Data are
representative of three independent experiments. (E) Total-RNA was
isolated from MKN1 or AGS cells transfected with either control
si-RNA (50 nM) or Tob1 si-RNA, and qRT-PCR was performed to assess
the expression of p15 mRNA. HPRT was used as internal
control. *p<0.05 and ***p<0.001, as
compared to respective control si-RNA. (F) The luciferase activity
assay using p15 promoter was performed after transfection of
control si-RNA (50 nM) or Tob1 si-RNA (50 nM) for 48 h. The p15
promoter activity assay was performed in triplicate.
*p<0.05 and ***p<0.001, compared to
control si-RNA. |
Tob1 inhibits β-catenin-mediated
signaling in gastric cancer cells
Aberrant activation of β-catenin-mediated signaling
has been implicated in gastric cancer progression (15). Overexpression of Smad4 has been
reported to diminish β-catenin signaling, and, hence, prevent tumor
progression (19). Since
restoration of Tob1 elevated the expression of Smad4, we
investigated the role of Tob1 in the regulation of
β-catenin-mediated signaling. Overexpression of Tob1 decreased the
expression of β-catenin in MKN28 and AGS cells (Fig. 5A). β-catenin is degraded by GSK3β,
an enzyme inactivated via phosphorylation of its serine-9 residue
by upstream kinase Akt (20). We
examined whether the decreased expression of β-catenin in
Tob1-overexpressing gastric cancer cells could result from
the reduced phosphorylation of Akt and GSK3β. As expected, Tob1
decreased the phosphorylation of Akt and GSK3β (Fig. 5A). Ectopic expression of Tob1 not
only reduced the expression of β-catenin but also attenuated
β-catenin-mediated transcriptional activity. The luciferase
activity assay using TopFlash or FopFlash along with control vector
or myc-Tob1 showed that Tob1 significantly reduced the
transcriptional activity of β-catenin/T cell factor (TCF) complex
(Fig. 5B), which resulted in
decreased expression of β-catenin target genes, such as cyclin
D1, uPAR and PPARδ in both qRT-PCR and western
blot analyses (Fig. 5C and D). The
knock-down of Tob1 using specific si-RNA increased the
phosphorylation of Akt and GSK3β (serine-9), resulting in the
increased expression of β-catenin and its target gene in AGS cells
(Fig. 5E). In addition, the
luciferase activity assay using TopFlash vector after knock-down of
Tob1 using specific si-RNA in AGS cells significantly increased the
transcriptional activity of β-catenin (Fig. 5F), verifying again that Tob1
negatively regulates β-catenin signaling pathway.
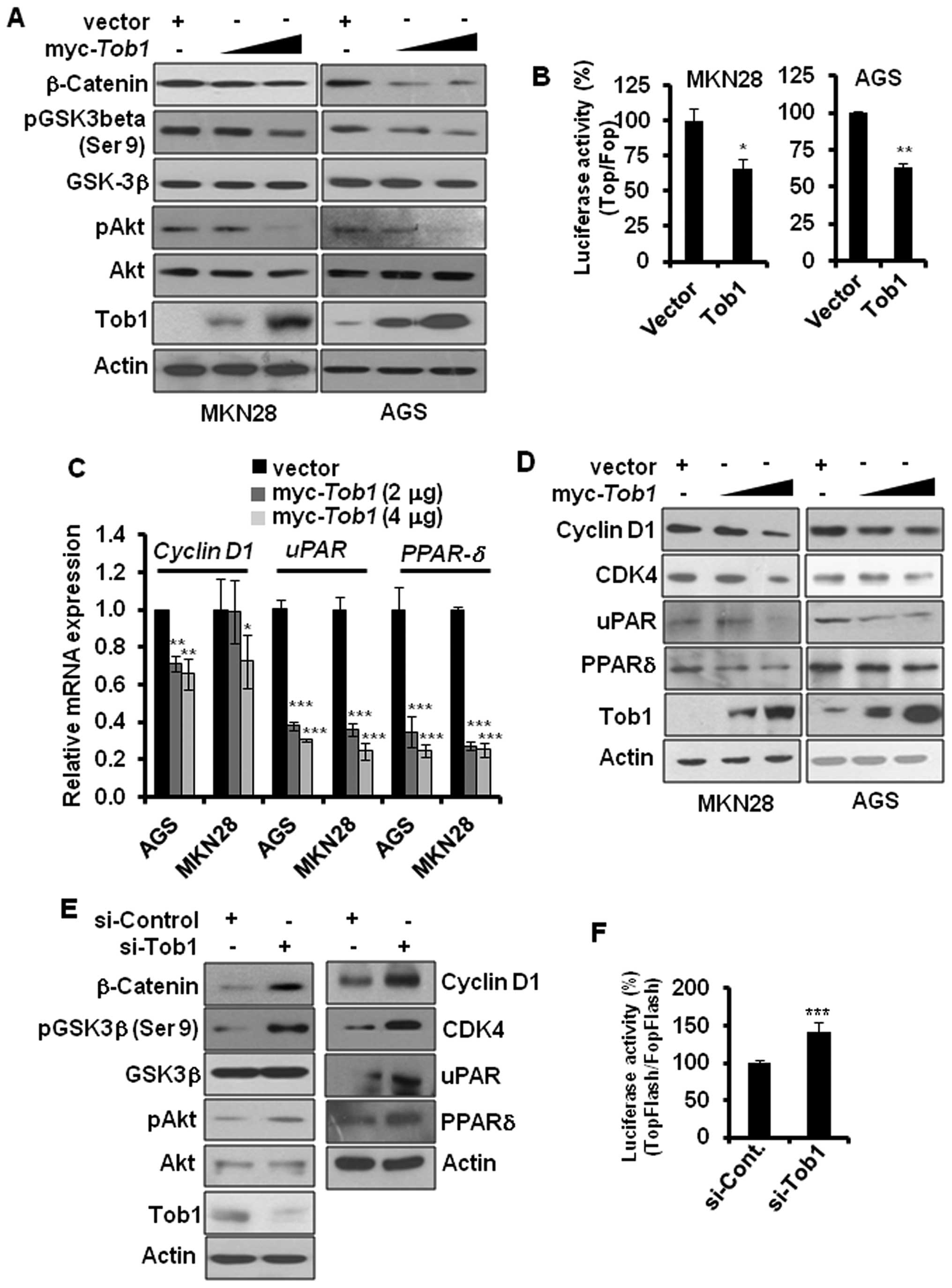 | Figure 5Tob1 inhibits β-catenin signaling in
gastric cancer cells. (A) Protein lysates (15 μg) from MKN28
or AGS cells transfected with myc-Tob1 (2 or 4 μg)
were separated by 10% SDS-PAGE, and the expression of β-catenin,
pAkt and pGSK3β-(serine-9) was detected using their specific
antibodies. Data are representative of three different experiments.
(B) Cells (MKN28 and AGS) were transfected with luciferase
constructs harboring TopFlash or FopFlash along with control vector
or myc-Tob1 (0.4 μg). The luciferase activity assay
was performed as described in the Materials and methods.
*p<0.05 and **p<0.01 compared to
respective control vector. (C) Total-RNA was isolated from MKN28 or
AGS cells transfected with either vector alone or myc-Tob1
(2 or 4 μg) and qRT-PCR was performed to assess the mRNA
expression of cyclin D1, uPAR and PPARδ. Gene
expression was normalized to the level of HPRT.
*p<0.05, **p<0.01 and
***p<0.001 compared to control vector. (D) Protein
lysates (30 μg) from MKN28 and AGS cells transfected with
either control vector or myc-Tob1 (2 or 4 μg) were
subjected to western blot analysis to detect the expression of
cyclin D1, CDK4, uPAR and PPARδ. Data are representative of three
different experiments. (E) AGS cells were transiently transfected
with either control si-RNA or Tob1 si-RNA for 48 h, and cell
lysates were separated by 10% SDS-PAGE to determine the expression
of β-catenin, pAkt, pGSK-3β-(serine-9), cyclin D1, CDK4, uPAR and
PPARδ. Data are representative of three independent experiments.
(F) AGS cells were transfected with luciferase constructs harboring
TopFlash or FopFlash with control si-RNA (25 nM) or Tob1 si-RNA (25
nM). The luciferase activity assay was performed as described in
the Materials and methods. *p<0.001 (vs. control
si-RNA). |
Discussion
Gastric cancer is the second leading cause of
cancer-related deaths in the world (21). Inactivation of tumor-suppressor
genes is one of the critical events in the development and
progression of gastric cancer (14). Thus, restoration of tumor
suppressor gene function is considered as a rational approach for
therapeutic intervention of carcinogenesis. For instance, the
pharmacological activation of a tumor suppressor protein, p53, is
being widely studied for the development of new cancer
chemotherapeutics (22). Like many
other tumor suppressor proteins, the expression of Tob1 is
frequently lost in various cancers. A recent study by Yu et
al(10) demonstrated that Tob1
is either absent or expressed at reduced level in 75% of primary
gastric cancer cases. However, the role of Tob1 in gastric cancer
progression and its underlying mechanisms have not been fully
clarified. Thus, we attempted to investigate whether ectopic
expression of Tob1 could inhibit gastric cancer progression and to
elucidate its underlying mechanisms. Here, we report for the first
time that ectopic expression of Tob1 induces apoptosis and inhibits
the proliferation of gastric cancer cells in vitro. Our
results showed that Tob1 induced Bax expression and inhibited Bcl-2
expression in gastric cancer cells. Similar effects of Tob1 on Bax
and Bcl-2 expression in human breast cancer cells have been
reported earlier (23). In
addition, we found a novel mechanism showing that Tob1 induced PARP
cleavage by promoting caspase-3 activity, leading to the apoptosis
of gastric cancer cells. The increased migration and invasion of
cancer cells has been known to be directly associated with poor
prognosis. Thus, the restoration of Tob1 in gastric cancer patients
could delay the disease progression and improve the prognosis by
inhibiting migration and invasion of cancer cells.
The expression of Smad4 has been known to be reduced
in gastric cancer (14), and
re-expression of Smad4 has been reported to inhibit tumor
progression (24–25). In this study, we found that Tob1
induced the expression of Smad4, which increased the expression of
p15, a CDK inhibitor, at transcriptional level. Since Tob1 has been
known to increase the Smad4-mediated transcriptional activity by
interacting with Smad4 (11), the
induction of p15 expression by Tob1 may be due to the enhancement
of transcriptional activity of Tob1-Smad4 complex. Thus, the
induction of Smad4 expression by Tob1 in MKN28 and AGS cells could
inhibit gastric cancer progression through activation of
Smad4-mediated signaling. In addition, Smad4 has been shown to
inhibit the expression of β-catenin (19), which is implicated in gastric
cancer progression (15). The
downregulation of β-catenin expression by Tob1 suggests that Tob1
can interfere with β-catenin-mediated signal transduction pathways.
Our study revealed that Tob1 decreased the expression of
β-catenin-target genes including cyclin D1, CDK4, uPAR, and PPARδ.
While cyclin D1 and CDK4 are well known biomarker of cell
proliferation, uPAR (26) and
PPARδ have been reported to play role in the migration and invasion
of gastric cancer cells (27).
Thus, the anti-proliferative effect of Tob1 in gastric cancer might
be due to the decreased expression of cyclin D1 and CDK4, and the
increased expression of p15, while the anti-migratory and
anti-invasive potential of Tob1 may be mediated through the
inhibition of uPAR and PPARδ in gastric cancer cells. Besides uPAR,
which degrades extracellular matrix and facilitates gastric cancer
progression (28,29), matrix metalloproteinases (MMPs)
have also been implicated in the migration and invasion of gastric
cancer (30). However, the
expression and the activity of MMP-2/-9 in gastric cancer cells
remained unaffected by Tob1 overexpression (data not shown).
A recent study demonstrated that overexpression of
Tob1 in lung cancer cells increased the expression of another tumor
suppressor protein phosphatase and tensin homolog (PTEN) (31), which is a negative regulator of
Akt. In addition, re-expression of PTEN in prostate cancer (PC3)
cells downregulated β-catenin expression (32). Thus, it would be worthwhile to
investigate whether the decreased phosphorylation of Akt and the
reduced expression of β-catenin upon overexpression of Tob1 in
gastric cancer cells also results from the induction of PTEN.
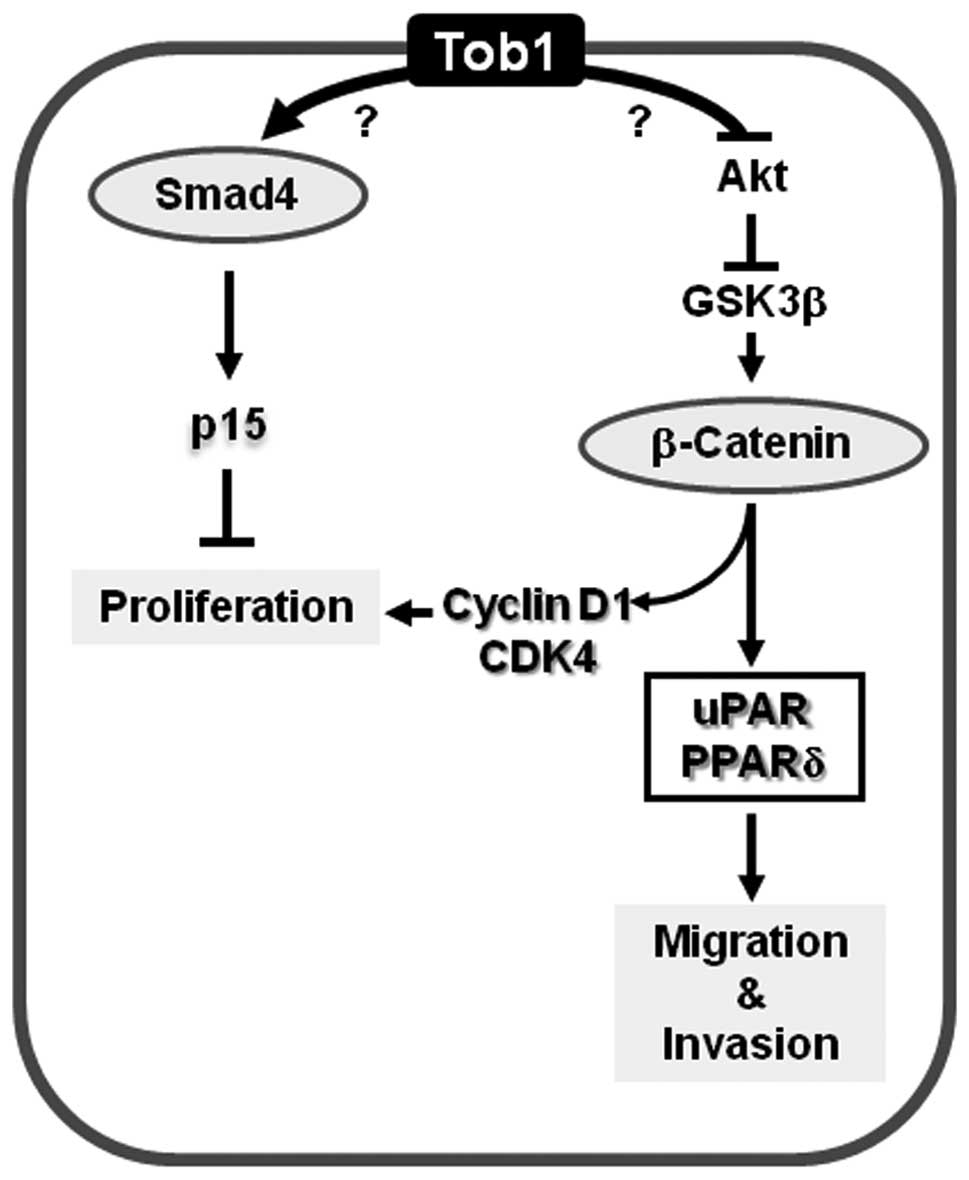
In conclusion, the present study demonstrates that
Tob1 functions as a tumor suppressor protein in gastric cancer
cells, at least in part, by inducing apoptosis and inhibiting
proliferation, migration and invasion via the activation of Smad4-
and suppression of β-catenin-mediated signaling pathways (Fig. 6).
Acknowledgements
This study was supported by grants
from the National R&D Program for Cancer Control
(07200603-20982).
References
|
1
|
Sherr CJ: Principles of tumor suppression.
Cell. 116:235–246. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matsuda S, Kawamura-Tsuzuku J, Ohsugi M,
et al: Tob, a novel protein that interacts with p185erbB2, is
associated with anti-proliferative activity. Oncogene. 12:705–713.
1996.PubMed/NCBI
|
|
3
|
Guardavaccaro D, Corrente G, Covone F, et
al: Arrest of G(1)-S progression by the p53-inducible gene PC3 is
Rb dependent and relies on the inhibition of cyclin D1
transcription. Mol Cell Biol. 20:1797–1815. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ikematsu N, Yoshida Y, Kawamura-Tsuzuku J,
et al: Tob2, a novel anti-proliferative Tob/BTG1 family member,
associates with a component of the CCR4 transcriptional regulatory
complex capable of binding cyclin-dependent kinases. Oncogene.
18:7432–7441. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rouault JP, Rimokh R, Tessa C, et al:
BTG1, a member of a new family of antiproliferative genes. EMBO J.
11:1663–1670. 1992.PubMed/NCBI
|
|
6
|
Yoshida Y, Nakamura T, Komoda M, et al:
Mice lacking a transcriptional corepressor Tob are predisposed to
cancer. Genes Dev. 17:1201–1206. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O’Malley S, Su H, Zhang T, Ng C, Ge H and
Tang CK: TOB suppresses breast cancer tumorigenesis. Int J Cancer.
125:1805–1813. 2009.
|
|
8
|
Yanagie H, Tanabe T, Sumimoto H, et al:
Tumor growth suppression by adenovirus-mediated introduction of a
cell-growth-suppressing gene tob in a pancreatic cancer model.
Biomed Pharmacother. 63:275–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ito Y, Suzuki T, Yoshida H, et al:
Phosphorylation and inactivation of Tob contributes to the
progression of papillary carcinoma of the thyroid. Cancer Lett.
220:237–242. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu J, Liu P, Cui X, et al: Identification
of novel subregions of LOH in gastric cancer and analysis of the
HIC1 and TOB1 tumor suppressor genes in these subregions. Mol
Cells. 32:47–55. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tzachanis D, Freeman GJ, Hirano N, et al:
Tob is a negative regulator of activation that is expressed in
anergic and quiescent T cells. Nat Immunol. 2:1174–1182. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiong B, Rui Y, Zhang M, et al: Tob1
controls dorsal development of zebrafish embryos by antagonizing
maternal beta-catenin transcriptional activity. Dev Cell.
11:225–238. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Powell SM, Harper JC, Hamilton SR,
Robinson CR and Cummings OW: Inactivation of Smad4 in gastric
carcinomas. Cancer Res. 57:4221–4224. 1997.PubMed/NCBI
|
|
14
|
Wang LH, Kim SH, Lee JH, et al:
Inactivation of SMAD4 tumor suppressor gene during gastric
carcinoma progression. Clin Cancer Res. 13:102–110. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kolligs FT, Bommer G and Goke B:
Wnt/beta-catenin/tcf signaling: a critical pathway in
gastrointestinal tumorigenesis. Digestion. 66:131–144. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jung HS, Erkin OC, Kwon MJ, et al: The
synergistic therapeutic effect of cisplatin with human
papillomavirus E6/E7 short interfering RNA on cervical cancer cell
lines in vitro and in vivo. Int J Cancer. 130:1925–1936. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kwon MJ, Oh E, Lee S, et al:
Identification of novel reference genes using multiplatform
expression data and their validation for quantitative gene
expression analysis. PLoS One. 4:e61622009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gildea JJ, Harding MA, Gulding KM and
Theodorescu D: Transmembrane motility assay of transiently
transfected cells by fluorescent cell counting and luciferase
measurement. Biotechniques. 29:81–86. 2000.PubMed/NCBI
|
|
19
|
Tian X, Du H, Fu X, Li K, Li A and Zhang
Y: Smad4 restoration leads to a suppression of Wnt/beta-catenin
signaling activity and migration capacity in human colon carcinoma
cells. Biochem Biophys Res Commun. 380:478–483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Novak A and Dedhar S: Signaling through
beta-catenin and Lef/Tcf. Cell Mol Life Sci. 56:523–537. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Resende C, Thiel A, Machado JC and
Ristimaki A: Gastric cancer: basic aspects. Helicobacter. 16(Suppl
1): 38–44. 2011. View Article : Google Scholar
|
|
22
|
Athar M, Elmets CA and Kopelovich L:
Pharmacological activation of p53 in cancer cells. Curr Pharm Des.
17:631–639. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Jiao Y, Ge CM, Meng QH, Cao JP, Tong J and
Fan SJ: Adenovirus-mediated expression of Tob1 sensitizes breast
cancer cells to ionizing radiation. Acta Pharmacol Sin.
28:1628–1636. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwarte-Waldhoff I, Klein S,
Blass-Kampmann S, et al: DPC4/SMAD4 mediated tumor suppression of
colon carcinoma cells is associated with reduced urokinase
expression. Oncogene. 18:3152–3158. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shen W, Tao GQ, Li DC, Zhu XG, Bai X and
Cai B: Inhibition of pancreatic carcinoma cell growth in vitro by
DPC4 gene transfection. World J Gastroenterol. 14:6254–6260. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alpizar-Alpizar W, Nielsen BS, Sierra R,
et al: Urokinase plasminogen activator receptor is expressed in
invasive cells in gastric carcinomas from high- and low-risk
countries. Int J Cancer. 126:405–415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pollock CB, Rodriguez O, Martin PL, et al:
Induction of metastatic gastric cancer by peroxisome
proliferator-activated receptordelta activation. PPAR Res.
2010:5717832010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kawasaki K, Hayashi Y, Wang Y, et al:
Expression of urokinase-type plasminogen activator receptor and
plasminogen activator inhibitor-1 in gastric cancer. J
Gastroenterol Hepatol. 13:936–944. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Okusa Y, Ichikura T, Mochizuki H and
Shinomiya N: Urokinase type plasminogen activator and its receptor
regulate the invasive potential of gastric cancer cell lines. Int J
Oncol. 17:1001–1005. 2000.PubMed/NCBI
|
|
30
|
Choi BD, Jeong SJ, Wang G, et al:
Secretory leukocyte protease inhibitor is associated with MMP-2 and
MMP-9 to promote migration and invasion in SNU638 gastric cancer
cells. Int J Mol Med. 28:527–534. 2011.PubMed/NCBI
|
|
31
|
Jiao Y, Sun KK, Zhao L, Xu JY, Wang LL and
Fan SJ: Suppression of human lung cancer cell proliferation and
metastasis in vitro by the transducer of ErbB-2.1 (TOB1). Acta
Pharmacol Sin. 33:250–260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Persad S, Troussard AA, McPhee TR,
Mulholland DJ and Dedhar S: Tumor suppressor PTEN inhibits nuclear
accumulation of beta-catenin and T cell/lymphoid enhancer factor
1-mediated transcriptional activation. J Cell Biol. 153:1161–1174.
2001. View Article : Google Scholar : PubMed/NCBI
|