Introduction
Colorectal cancer (CRC) is one of the highest
incidence cancers worldwide (1).
In the US, CRC is the fourth most common cancer in men and women
(1) and approximately 3,000 new
cases are diagnosed annually both in the Jewish male and female
population (2–4). It is uncommon in the non-Jewish
(Arab) population. This is attributed to different genetic
susceptibility and/or lifestyle and diet (2–4).
Gene mutation or genetic variations, whether it is at the single
nucleotide polymorphism (SNP) or structural level, could also
contribute to the development of CRC as shown in a number of Genome
Wide Association (GWAS) and related studies (5–9). The
glycosylphosphatidylinositol transamidase (GPIT) complex is a
family of proteins that assists in attaching a diverse group of
macromolecules to the plasma membrane of eukaryotes (10). The GPIT complex is composed of 5
subunits namely PIGU, PIGT, GPAA1, PIGS and PIGK (10). Among these 5 subunits, PIGK (GPI8)
plays a crucial role in protein-GPI anchoring (10,11).
It is well established that PIGK catalyzes the endoproteolysis and
amidation steps involved in transfer of GPI to proteins (10). However, the role of PIGK in
tumorigenesis remains largely unknown. Recently, we reported a low
expression of PIGK protein in some CRC, hepatocellular (HCC) and
urothelial cell carcinoma (UCC) patients (11). We hypothesize that the altered
expression of PIGK could be due to mutation in the coding regions
of this gene. As a pilot study, we examined all 10 exons of the
PIGK gene in 45 CRC patients by direct sequencing.
Immunohistochemistry was performed on the same 45 CRC patients to
examine corresponding protein expression. We did not detect any
mutation in the exons, but a SNP (C/C→C/G or G/G; rs1048575) was
found in the 3′UTR immediately after the stop codon of the PIGK
gene in 67% (30/45) of the samples. We also observed a
significantly low (P<0.002) expression of PIGK protein in all
patients with the altered alleles (C/G or G/G) compared to the
ancestral alleles (C/C). Similar observations were made on PIGK
expression in a several transformed human colon cell lines with the
ancestral (C/C) and altered (C/G) genotype. These results suggest
an association between SNP-1048575 and low PIGK expression in CRC
patients.
Materials and methods
Patient history and tissue samples
Matched adjacent normal and tumor tissues from 45
CRC patients and the HCC samples were collected at the Johns
Hopkins University (JHU) as per IRB approved protocol.
Formalin-fixed paraffin-embedded (FFPE) tumor tissue sections (5
μm) were procured from all 45 CRC patients for immunohistochemical
(IHC) analysis. All the subsequent analysis of tissue samples, IHC
staining was performed at JHU. Detailed patient information is
shown in Table I.
 | Table IColorectal cancer patients with the
PIGK genotype and corresponding PIGK protein expression. |
Table I
Colorectal cancer patients with the
PIGK genotype and corresponding PIGK protein expression.
| Patient ID | Genotype | Stage | PIGK
expressiona |
|---|
| C592 | CC | IV | +++ |
| C127 | CC | IV | +++ |
| C758 | CC | I | +++ |
| C358 | CC | IIA | +++ |
| C155 | CC | IIA | +++ |
| C277 | CC | I | +++ |
| C193 | CC | IIB | +++ |
| C410 | C/G | IIIB | + |
| C307 | C/G | IIIB | ++ |
| C183 | C/G | III | ++ |
| C301 | C/G | IIA | + |
| C345 | C/G | IIIC | + |
| C374 | C/G | IIIA | + |
| C330 | C/G | IIA | + |
| C217 | C/G | IIA | - |
| C206 | C/G | IIIB | + |
| C485 | C/G | IIIB | ++ |
| C80 | C/G | IV | + |
| C123 | C/G | IIA | + |
| C414 | G/G | III | + |
| C918 | CC | III | + |
| C850 | C/G | III | - |
| C870 | C/G | IV | ++ |
| C1015 | C/G | IV | + |
| C998 | C/G | IV | ++ |
| C1023 | CC | - | +++ |
| C1025 | C/G | III | + |
| C863 | CC | V | + |
| C837 | CC | - | +++ |
| C1014 | C/G | IV | + |
| C1021 | C/G | IV | + |
| C1003 | C/G | IV | ++ |
| C968 | C/G | - | ++ |
| C1032 | C/G | - | ++ |
| C989 | C/G | - | ++ |
| C980 | C/G | IV | + |
| C990 | C/G | II | +++ |
| C947 | C/G | IV | + |
| C935 | C/G | IV | + |
| C804 | CC | | +++ |
| C1019 | C/G | III | + |
| C1017 | CC | I | +++ |
| C1002 | C/G | III | + |
| C1016 | CC | II | +++ |
| C866 | CC | I | +++ |
Cell lines
We obtained 2 human non-tumorigenic colonic cell
lines; CCD841 and FHC and two hepatocellular carcinoma (HCC) lines
H2pG2 and Hep3B from ATCC. All cell lines were cultured in ATCC
recommended medium. The cell lines were periodically chacked for
mycoplasma contamination using Mycoplasma Detection kit from Sigma
(MP-0025). The tissue culture media and reagents were purchased
either from ATCC or Invitrogen.
DNA extraction and PIGK exon
sequencing
Genomic DNA was extracted from the microdissected
tumor and normal tissue samples and cell lines as described
(12). The list of primers is
given in Table II. The PCR was
carried out in a 50 μl reaction volume with the conditions as
follows: 95°C for 5 min, 1 cycle; 94°C for 30 sec, 55°C for 30 sec,
72°C for 7 min, 40 cycles; and final extension at 72°C for 10 min.
All amplified products were analyzed on 1% agarose gels, cut,
purified and sequenced using the ABI Big Dye Cycle Sequencing kit
(Applied Biosystems). All experiments were carried out at JHU.
 | Table IIThe primers used to amplify the 10
exons of the human PIGK gene. |
Table II
The primers used to amplify the 10
exons of the human PIGK gene.
| Exon | Forward | Reverse |
|---|
| 1 |
GGATCAAGCAGAACAATTCTTTA |
CATATCTAATGTGAACAGCAAGGT |
| 2 |
TCAGGTGTGTACATCCCGATT |
TTACCTGTCAGGAATACCTAGCCTCT |
| 3 |
TTTTTCCCTAGTCACATTGTCC |
TACCTCGTAACTTCTATAATCCACTTC |
| 4 |
CCCCCTTAGGTAACTGTGGA |
TTCCATTTGCCTTTCTTTTCA |
| 5 |
ACAAGGGCATGGTGGAAAT |
ACCGTCTTTTCTGCCACATT |
| 6 |
TCATTTTTCTTTCAGCTACAATGAG |
ACAAATGGGGAAAATGCACA |
| 7 |
TTTTTCCCCCTTTCCATAGC |
CAAATCAAGTGAATACTTACAAGGTCA |
| 8 |
GCATTTATTATTATATATTT |
GAGACTAGTATTCTATTC |
| 9 |
TTTTCCAGCTATAAGGAAGACCA |
AAATGAAATGCAAACCTGGTG |
| 10-P1a |
GCAGAAACCGAAGCTGAAAG |
CCAAGTTTGCAGTCCTCCAT |
| P2 |
TTGCTGCCTGCCCTATTTAC |
TTTCACAGCACATTTCCAAAA |
| P3 |
TTTTGACTCCTGTGTTTCTGGA |
ACACTGAAGCAATAAGGCCAAT |
| P4 |
TAATGATGCCATCCTGCAAA |
ATGCTTCTTGTGGGAATCATC |
| P5 |
GCCTTTCTCACAGTGTTGTGG |
TGGGGGAGGAGAAAAAGAAC |
| P6 |
GCGGAGCTATTTAGGGTGGT |
AGGAACAACTGCCCTTTGAA |
| P7 |
GCTTTTTGCCAGCTCTGAAT |
GGGCAACATGGTCAATCTTT |
Genotyping assay for SNP rs1048575
TaqMan™ (Fluorogenic 5′ nuclease) assay was used for
typing our candidate SNP (rs1048575). The forward and reverse
primers and respective allele-specific probes were obtained from
Applied Biosystems (Applied Biosystems). PCR was conducted in ABI
9700 thermocycler, and the end-point results were auto-scored using
the ABI 7900HT Sequence Detection System (SDS; version 2.3). To
ensure reliability of genotyping data, each of the samples were
amplified and genotyped in triplicate along with positive and
negative control templates. Additionally, all the samples were
sequenced unidirectionally, and there was 100% concordance between
sequencing and genotyping data obtained through the TaqMan
genotyping assay. All experiments were carried out at JHU.
Immunohistochemistry
To study the expression of PIGK protein, we used
FFPE tissue samples collected from 45 samples described in Table I. Several serial sections (5 μm)
from different regions of the tumors were stained using rabbit
anti-human PIGK antibody (Abcam #38445) for determining protein
expression. Staining and evaluation was performed as described
earlier (13,14). For comparison, all sections were
processed in parallel. All experiments were carried out at JHU.
PIGK constructs and transfection
The wild-type (wt, C/C) human PIGK gene was
subcloned into SalI and NotI sites of the
phosphorylated cytomegalovirus (pCMV)/myc/ER plasmid (Invitrogen)
to express the exogenous protein in the endoplasmic reticulum (ER)
as PIGK is localized and functions in the ER (10). The resultant plasmids were
resequenced using the ABI BigDye cycle sequencing kit (Applied
Biosystems) for verification of the insert sequences. An empty
pCMV/myc/ER and a Green Fluorescence Protein (GFP) expressing
pCMV/myc/ER-GFP plasmids were also used as controls. In
transfections, HepG2 and Hep3B cells were transiently transfected
with wt-PIGK plasmid in the presence of the FuGene 6 transfection
reagent (Roche Applied Science). An empty pCMV/myc/ER vector was
used for mock transfection. We also used GFP expressing pCMV/myc/ER
plasmid vector as a control. Cells were analyzed 24 h post
transfection. Positive cell clones were confirmed to express
wt-PIGK by immunofluorescence analysis using the fusion protein tag
anti-myc-FITC antibody (Invitrogen).
Reverse-transcription PCR
Total RNA was extracted from wt-PIGK transfected and
non-transfected Hep3B cells after 24 h using Qiazol reagent
following the manufacturer’s instruction (Qiagen). Conventional
RT-PCR was carried out in 50 μl reaction volume as described
earlier (14). The primers used
for PIGK were forward 5′-CCACTTTGCCTTTCTCTCCA-3′; reverse
5′-GCCCCATTCAGATCCTCTTC-3′.
Statistical analysis
χ2 or Fisher’s exact tests were used as
appropriate. Student’s t-test or non-parametric Wilcoxon rank sum
test was also used as appropriate. All p-values are two-sided and
all confidence intervals are at the 95% level. Computations for all
the analysis were performed using the Statistical Analysis System
(SAS).
Results
Distribution of SNP 1048575 among the CRC
patients
In an earlier study, we observed a low expression of
PIGK protein in tumors compared to paired normal tissues of some
CRC patients (11). We
hypothesized that such low expression could be due to mutation in
the coding regions of the PIGK gene. Therefore in this pilot study,
we examined all 10 exons of the PIGK gene by direct sequencing in
45 primary CRC tumors. No mutations were detected in the coding
regions, however, we discovered a SNP (rs1048575; C/C→C/G or G/G)
in the 3′UTR of the PIGK gene in 67% (30/45) of the CRC patients
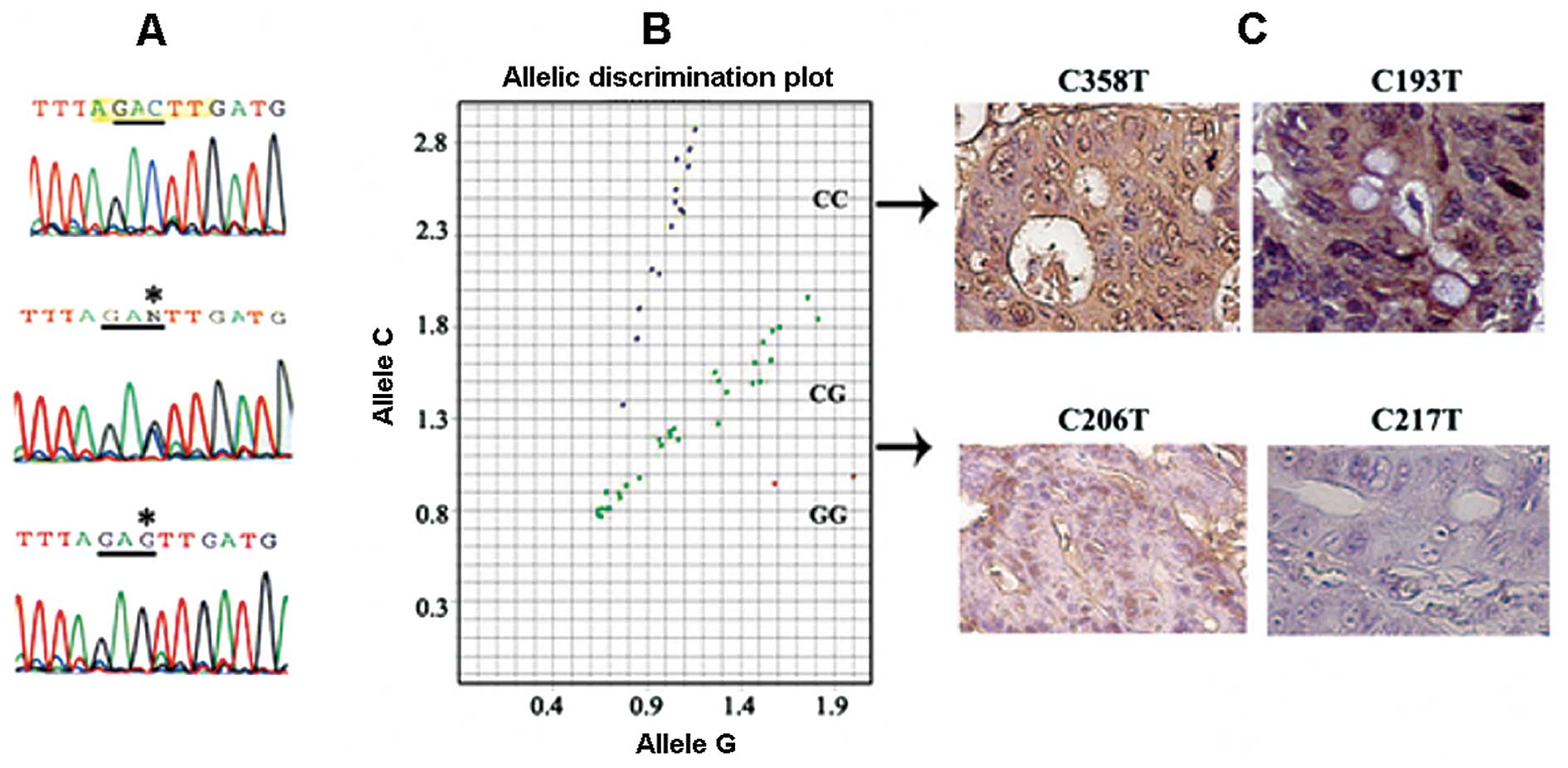
(Table I and Fig. 1). Schematic representation of the
location of the ancestral and altered allele in PIGK is shown in
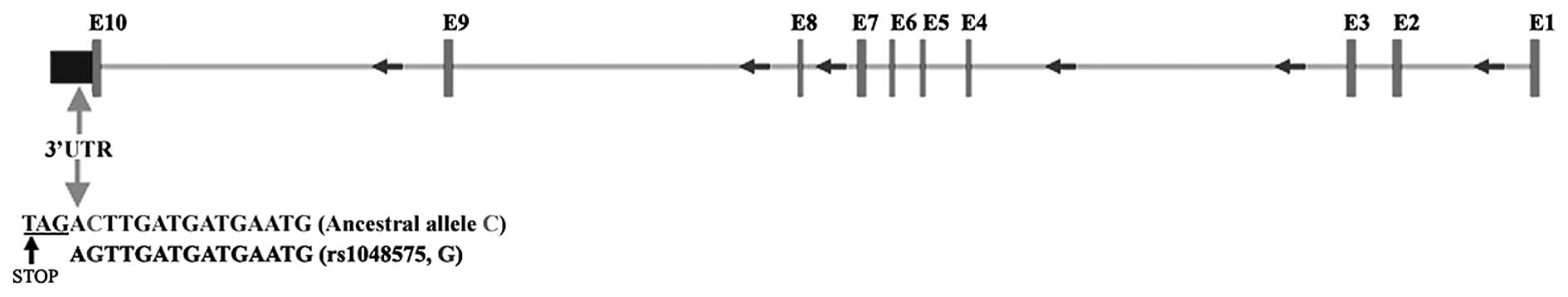
Fig. 2. This observation was
further confirmed by genotyping the SNP-1048575 in all the above
samples (Table I, Fig. 1B). Of the 45 CRC patients, 26 were
of Jewish and 17 were of Arabian origin. The ethnicity of the
remainder of the 3 patients was unknown. Eighty-five percent
(22/26) of the Jewish patients showed the altered alleles (C/G or
G/G) whereas 47% (8/17) of the Arabian patient populations was
detected with the altered alleles (Table I, Fig.
1A and B). The number of Jewish patients with the altered
alleles was significantly higher (Student’s t-test; p=0.03)
compared to the Arabian patient populations.
Association between SNP-1048575 and PIGK
protein expression in CRC patients
To examine whether there was any correlation between
the altered alleles (C/G, G/G) and PIGK protein expression in the
CRC patients, we performed IHC on FFPE tissue sections from all 45
CRC patients. The expression of PIGK protein was significantly
lower (p<0.002) in all of the patients with altered genotype
(C/G or G/G; 30/45, 67%) compared to the ancestral genotype (C/C;
15/45, 33%) (Table I and Fig. 1C).
Association between SNP-1048575 and PIGK
protein expression in transformed colon cell lines
We also determined the SNP-1048575 allele
distribution and corresponding PIGK expression in a couple of
transformed colon cell lines. One colon line (CCD841) exhibited the
ancestral alleles (C/C), whereas the other line (FHC) exhibited the
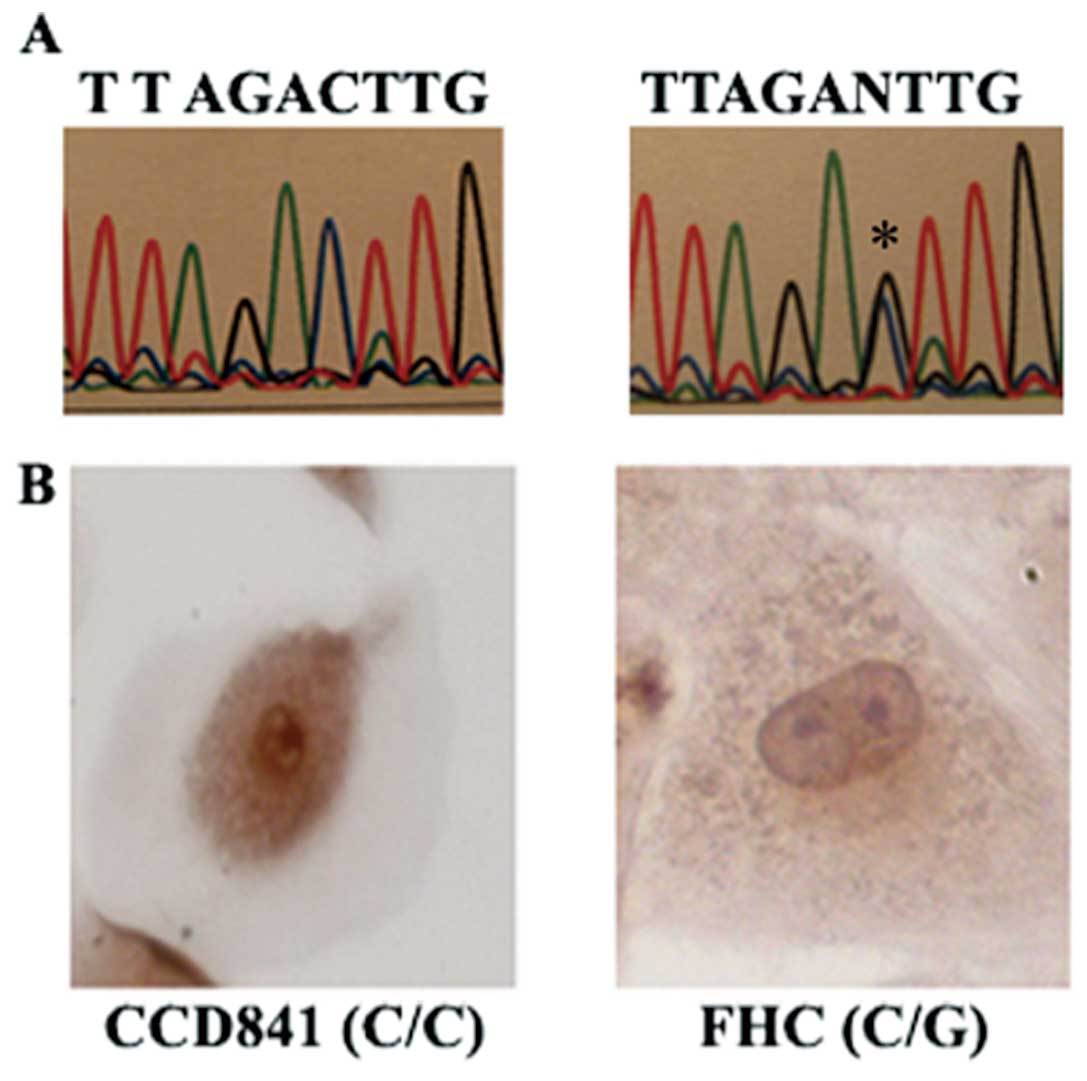
altered alleles (C/G) (Fig. 3A).
Immunohistochemical analysis also revealed a significantly lower
(Student’s t-test, p<0.002) expression of the PIGK protein in
the FHC-colon line carrying the altered alleles (C/G) compared to
the CCD-841 cell line with the ancestral alleles (C/C) (Fig. 3B).
Association between SNP-1048575 and PIGK
protein expression in HCC patients and cell lines
Similar to the CRC patients, our earlier study also
demonstrated a low expression of PIGK in some primary HCC tumors
(11). Therefore, we also examined
5 HCC patients and two HCC cell lines (Hep3B and HepG2) for PIGK
genotype (SNP-1048575) and corresponding protein expression. We
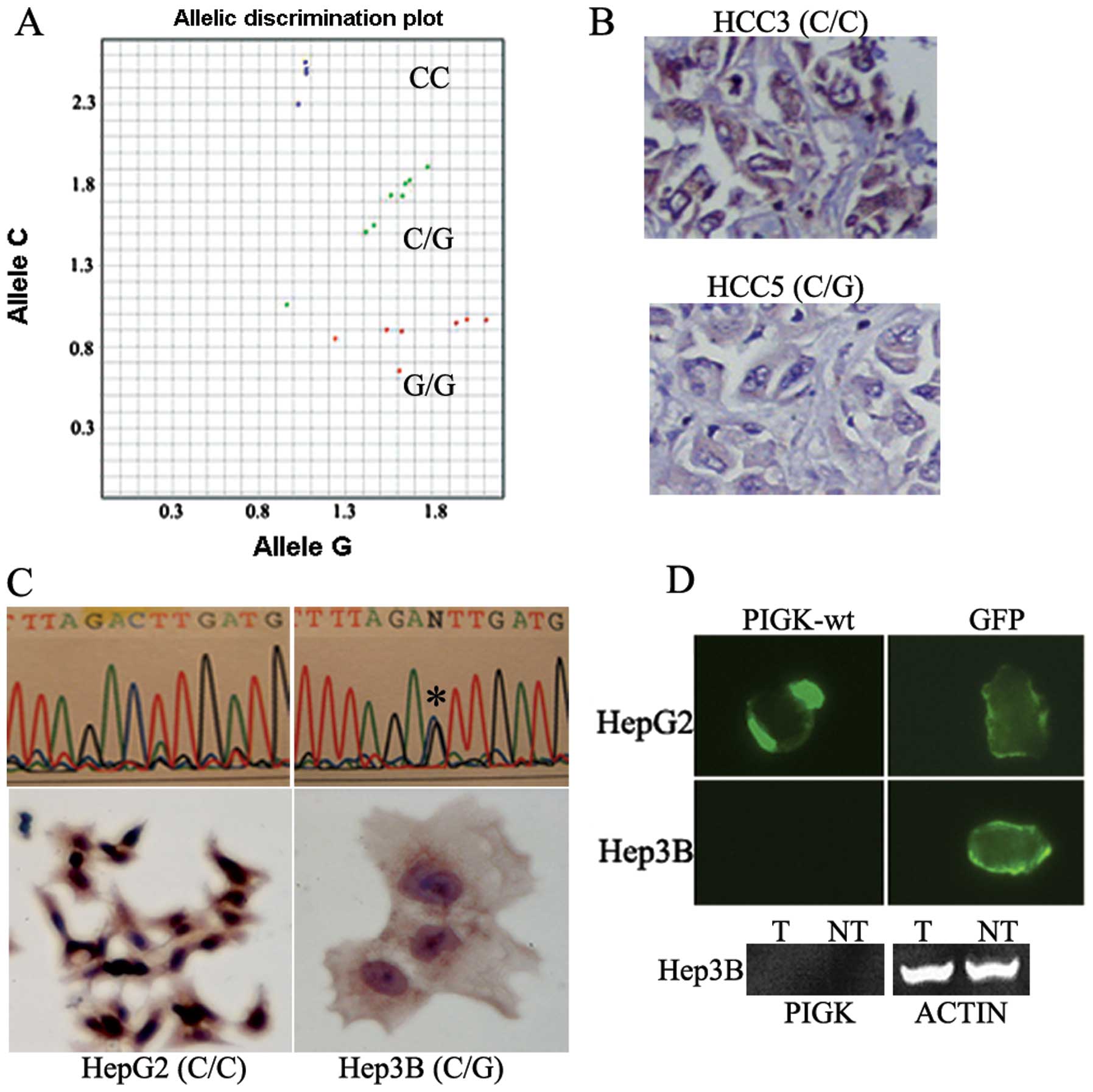
observed altered alleles (C/G or G/G) (Fig. 4A) and corresponding low PIGK
protein expression (Fig. 4B) in 4
out of 5 (80%) primary HCC tumors. Among the HCC cell lines, HepG2
line exhibited ancestral C/C alleles, whereas Hep3B showed altered
C/G alleles (Fig. 4C, upper
panel). Similar to the HCC patients, Hep3B line with the
altered alleles (C/G) exhibited significantly low (Student’s
t-test, P<0.002) PIGK protein expression compared to the Hep3B
line carrying the ancestral (C/C) alleles (Fig. 4C, lower panel).
Forced overexpression of wild-type PIGK
in liver cancer cell lines
To examine the exogenous PIGK protein expression
status, we transiently transfected both HepG2 (C/C alleles) and
Hep3B (C/G alleles) cell lines with wt-PIGK constructs. As shown in
Fig. 4D (upper panel), we could
detect exogenously expressed PIGK protein in HepG2 (C/C) cells, but
no PIGK expression was detectable in Hep3B (C/G) cells. The control
GFP protein was detectable in both cell lines using the same
expression cassette. We also examined the mRNA expression level of
the exogenously expressed wt-PIGK in Hep3B (C/G) cells. As shown in
Fig. 4D (lower panel), no mRNA
expression was detected in Hep3B cells.
Discussion
The human PIGK gene is involved in the key step of
transferring GPI-anchor to the respective protein molecules in the
plasma membrane (10). However,
its precise role in tumorigenesis remains poorly defined. In an
earlier study, we observed a significantly low expression of the
PIGK protein in some human CRC, HCC and UCC patients (11). Thus, we hypothesized that somatic
mutation in the coding regions of PIGK might be associated with
such low expression as shown for other genes in human cancers
(15). Therefore, we examined the
coding regions of the PIGK gene and corresponding protein
expression in 45 CRC patients of both Jewish and Arabic ethnicity
where CRC incidences are high, particularly among the Jewish
population (2–4). No somatic mutations were detected in
the coding regions of PIGK, but we discovered an SNP (rs1048575)
located immediately after the stop codon at the 3′ UTR in a
majority of the CRC patients with a concomitant decrease in PIGK
protein expression. As per the HapMap database (16; http://www.ncbi.nlm.nih.gov), the altered SNP-1048575
was reported in different population with C allele (wild-type)
being more frequent. However, no association of the altered allele
G with any diseases has so far been reported. Our observation in
CRC patients correlates well with the results obtained in a several
transformed colon cell lines, further suggesting that altered PIGK
expression may be an early event in colon carcinogenesis. In a
recent study, 3′ UTR polymorphisms in the BRCA1 gene were shown to
be associated with reduced gene activity and this alteration was
proposed as a possible breast and ovarian cancer risk factor
(17). Similar to the CRC
patients, we observed altered alleles (C/G or G/G) and
corresponding low PIGK protein expression in some HCC patients and
cell lines. Moreover, by forced overexpression, wild-type PIGK
protein was detectable in the HCC cell line HepG2 with the
ancestral (C/C) alleles but not in Hep3B carrying the altered
alleles (C/G). This could be a consequence of transcriptional
deregulation of the PIGK gene as we could not detect the
exogenously expressed PIGK mRNA. Another possible mechanism could
be regulation of PIGK by potential micro-RNAs through perfect or
imperfect complementarity mechanism(s) at the sites located within
the 3′ UTR of this gene (18,19).
In summary, our results demonstrate for the first
time a link between deregulated expression of PIGK and SNP-1048575
in CRC patients and also suggest a possible association between
altered PIGK expression and CRC susceptibility.
Acknowledgements
The study was supported by a grant
from Flight Attendant Medical Research Institute and Elsa U. Pardee
Foundation to B. Trink; US-Egypt Joint Science and Technology fund
(58-3148-169), A.D. Williams (646299) and Elsa U. Pardee Foundation
(548424) to S. Dasgupta. PBF holds the Thelma Newmeyer Corman Chair
in Cancer Research in the VCU Massey Cancer Center.
References
|
1
|
Markowitz SD and Bertagnoli MM: Molecular
basis of colorectal cancer. N Engl J Med. 361:2449–2460. 2010.
View Article : Google Scholar
|
|
2
|
Barchana M, Liphshitz I and Rozen P:
Trends in colorectal cancer incidence and mortality in the Israeli
Jewish ethnic populations. Fam Cancer. 3:207–214. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rozen P, Rosner G, Liphshitz I and
Barchana M: The changing incidence and sites of colorectal cancer
in the Israeli Arab population and their clinical implications. Int
J Cancer. 120:147–151. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rozen P, Liphshitz I and Barchana M:
Changing sites of colorectal cancer in the Israeli Jewish ethnic
populations and its clinical implications. Eur J Cancer Prev.
16:1–9. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Poynter JN, Figueiredo JC, Conti DV, et
al: Colon CFR: Variants on 9p24 and 8q24 are associated with risk
of colorectal cancer: results from the Colon Cancer Family
Registry. Cancer Res. 67:11128–11132. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pittman AM, Webb E, Carvajal-Carmona L, et
al: Refinement of the basis and impact of common 11q23.1 variation
to the risk of developing colorectal cancer. Hum Mol Genet.
17:3720–3727. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tomlinson IP, Webb E, Carvajal-Carmona L,
et al: A genome-wide association study identifies colorectal cancer
susceptibility loci on chromosomes 10p14 and 8q23.3. Nat Genet.
40:623–630. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tenesa A, Farrington SM, Prendergast JG,
et al: Genome-wide association scan identifies a colorectal cancer
susceptibility locus on 11q23 and replicates risk loci at 8q24 and
18q21. Nat Genet. 40:631–637. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cheah PY: Recent advances in colorectal
cancer genetics and diagnostics. Crit Rev Oncol Hematol. 69:45–55.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zacks MA and Garg N: Recent developments
in the molecular, biochemical and functional characterization of
GPI8 and the GPI-anchoring mechanism. Mol Membr Biol. 23:209–225.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nagpal JK, Dasgupta S, Jadallah S, Chae
YK, Ratovitski EA, Toubaji A, Netto GJ, Nissan A, Sidransky D and
Trink B: Profiling the expression pattern of GPI transamidase
complex subunits in human cancer. Mod Pathol. 8:979–991. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dasgupta S, Mukherjee N, Roy S, Roy A,
Sengupta A, Roychowdhury S and Panda CK: Mapping of the candidate
tumor suppressor genes’ loci on human chromosome 3 in head and neck
squamous cell carcinoma of an Indian patient population. Oral
Oncol. 38:6–15. 2002.
|
|
13
|
Dasgupta S, Bhattacharya-Chatterjee M,
O’Malley BW Jr and Chatterjee SK: Reversal of immune suppression in
an orthotopic murine model of head and neck squamous cell carcinoma
following vaccination with recombinant vaccinia virus expressing
IL-2. Cancer Ther. 2:375–388. 2004.
|
|
14
|
Dasgupta S, Bhattacharya-Chatterjee M,
O’Malley BW Jr and Chatterjee SK: Inhibition of NK cell activity
through TGF-1 by down regulation of NKG2D in a murine model of head
and neck cancer. J Immunol. 175:5541–5550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ali MA and Sjoblom T: Molecular pathways
in tumor progression: from discovery to functional understanding.
Mol Biosyst. 5:902–908. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
http://www.ncbi.nlm.nih.gov.
|
|
17
|
Pongsavee M, Yamkamon V, Dakeng S,
O-charoenrat P, Smith DR, Saunders GF and Patmasiriwat P: The BRCA1
3′-UTR: 5711+421T/T_5711+1286T/T genotype is a possible breast and
ovarian cancer risk factor. Genet Test Mol Biomarkers. 13:307–317.
2009.
|
|
18
|
Olena AF and Patton JG: Genomic
organization of microRNAs. J Cell Physiol. 222:540–545.
2010.PubMed/NCBI
|
|
19
|
Iorio MV and Croce CM: MicroRNAs in
cancer: small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|