Introduction
DNA methylation is a common feature often seen in
tumor suppressor and DNA repair genes (1). Methylation of CpG sites in the
promoters of these genes frequently causes loss of expression,
affecting cell cycle regulation, cell adhesion or DNA reparation.
Several genes have been reported to be involved, through DNA
methylation, in sporadic colorectal cancer (CRC) (2,3).
Here, we focus on five of these, O6-MGMT,
p14ARF, p16INK4a,
RASSF1A and APC1A(4–9).
Studies on the methylation status of the promoter regions of these
genes in CRC development reported frequent promoter
hypermethylation in tumor tissue DNA, whereas normal tissue DNA
remained unmethylated (10,11).
Methylation specific PCR, a common technique to
study DNA methylation, assays the methylation status of a few CpG
sites (i.e., those interfering with the PCR primer binding) and
only gives a qualitative indication (methylated or not). Like all
‘allele specific’ PCR methods, it depends largely on a combination
of the number of PCR optimization experiments and individual
judgment of presence or absence of bands. We do not know which
particular CpG sites will silence these suppressor genes when
methylated because the standard methods assay only a few of the
sometimes 100 or so CpG sites (12). These methodological concerns stall
our understanding of the clinical role of DNA methylation.
We have therefore developed
Pyrosequencing® assays (13) to get a DNA sequence-specific as
well as quantitative measure of DNA methylation of the promoter CpG
sites as well as the adjacent CpG sites, of the DNA repair gene
O6-MGMT and the 4 tumor suppressor genes
p14ARF, p16INK4a,
RASSF1A and APC1A previously studied by MS-PCR
(10,11). We report our data on tumor biopsies
from subjects diagnosed with colorectal cancer or adenomas and
adjacent normal mucosa, and furthermore we assessed if these assays
had any long-term prognostic value.
Materials and methods
Subjects
The study included 111 randomly selected patients
with primary colorectal adenocarcinoma who underwent surgical
resection at Linköping Hospital, Linköping and Vrinnevi Hospital,
Norköping, Sweden. In 46 of the patients, normal mucosa specimens
taken from the margin of the resected tumor were also available.
The study also included 10 patients with colorectal adenomas, from
7 of whom matched normal mucosa specimens were also available. The
patients’ gender, age, tumor location and stage were obtained from
surgical and/or pathological records at Linköping and Vrinnevi
Hospitals. Tumor differentiation was graded into well moderately or
poorly differentiated. The study was approved by the Regional
Ethics Review Board in Linköping and an informed consent document
was signed by participants.
DNA isolation and bisulfite treatment of
tissue DNA
Genomic DNA was isolated from 20 mg of colorectal
tumor tissue (n=111) by means of the Wizard® SV Genomic
DNA Purification System according to the manufacturer’s
instructions (Promega, Madison, WI, USA). For some of these
patients (n=46) distant normal colorectal mucosa tissue was also
available and DNA was isolated in the same way. Genomic DNA was
also isolated from 20 mg of colorectal adenoma tissue (n=10
subjects), and for several of these subjects (n=7) also from 20 mg
of distant normal colorectal mucosa, by means of the QIAamp DNA
Mini Kit according to the manufacturer’s instructions (Qiagen Inc.,
Valencia, CA, USA).
Approximately 1,000 ng of isolated and precipitated
DNA was used for the bisulfite treatment. The bisulfite treatment
was performed with EZ DNA Methylation kit according to the
instructions by the manufacturer (Zymo Research, Orange, CA, USA)
except that the incubation time was shortened to 10 h. In short,
DNA was diluted with M-Dilution buffer and was incubated for 15 min
at 37°C. After the incubation CT conversion reagent was added and
the samples were incubated at 50°C for 10 h. The samples were then
incubated on ice for 10 min and then M-Binding buffer was added.
The samples were centrifuged and then washed using centrifugation
and M-Wash buffer. The bisulfite treated DNA was eluted in 10
μl M-Elution buffer and then diluted 4 times with TE buffer
(10 mmol/l Tris-HCl, 0.05 mmol/l EDTA, pH 7.5).
PCR and Pyrosequencing
PCR and Pyrosequencing of the promoter regions of
the O6-MGMT, p14ARF,
p16INK4a, RASSF1A and APC1A genes
was performed as previously described (13). Pyrosequencing technology was used
to sequence-specifically quantitate each of the CpG sites, which
are analyzed in practice as C/T-polymorphisms, where a 100%
C-reading denotes a fully methylated C (MeC=100%) in the
original gDNA sample whereas a 100% T-reading denotes that this
locus was unmethylated (MeC=0%) in the original gDNA.
Intermediate MeC percentages denote partial methylation
at the level of the sample. Partial methylation, when present, is
presumed to be partly owing to admixture of unmethylated
non-neoplastic cell types present in the tissues to a varying
extent.
Statistical analysis
The significance of the difference of promoter
region hypermethylation on the O6-MGMT,
p14ARF, p16INK4a,
RASSF1A and APC1A genes in between normal mucosa
samples and primary tumors was tested by χ2 or McNemar’s
method. The relationships between the promoter region
hypermethylation and other factors were examined by the
χ2 test. The relationship between the expression and
survival was tested using Cox’s Proportional Hazard Model. Survival
curves were calculated using the Kaplan-Meier method. Two-sided
p-values of <0.05 were considered as statistically
significant.
Results
Prevalence and extent of DNA
hypermethylation
Samples from 111 colorectal cancer tumors, 10
colorectal adenomas and 53 matched normal tissues from the same
patients (46 from CRC patients, 7 from adenoma patients) were
analyzed. Assay failure rate was 0% for all 5 genes studied. All
the 53 available paired normal tissues, both from tumor and adenoma
tissues were found to be 100% unmethylated on the promoter regions
of all the genes. The pyrograms of many of the tumor samples, on
the other hand, showed methylation peaks, amounting to a consistent
but individual-specific mean percentage of methylated fraction of
gene promoter CpG sites. In Table
I the number of patients with methylation-positive tumors or
adenomas, as well as their mean methylated fractions
(%MeC) of CpG sites, are shown. Promoter
hypermethylation in the CRC samples was commonest for the
O6-MGMT gene (34% of the patients) and least
frequent for RASSF1A (14%). Adenomas showed a similar
pattern.
 | Table I.Mean methylated fraction and standard
deviation (SD) for all colorectal cancer and adenoma specimens that
showed methylation. |
Table I.
Mean methylated fraction and standard
deviation (SD) for all colorectal cancer and adenoma specimens that
showed methylation.
| Gene | CRC
| Adenomas
|
|---|
| No. (%) | %MeC
(Mean ± SD) | %MeC
(Range) | No. (%) | %MeC
(Mean ± SD) | %MeC
(Range) |
|---|
|
O6-MGMT | 38 (34) | 28±11 | 13–56 | 3 (30) | 24±9 | 16–33 |
|
p14ARF | 32 (29) | 36±15 | 15–90 | 0 (0) | 0±0 | - |
|
p16INK4a | 31 (28) | 29±10 | 12–51 | 1 (10) | 22±0 | - |
| RASSF1A | 16 (14) | 31±10 | 16–61 | 0 (0) | 0±0 | - |
| APC1A | 30 (27) | 38±11 | 21–67 | 2 (20) | 44±6 | 41–48 |
The methylation pattern throughout the promoter
regions of all the genes was consistent within each individual
patient: if methylation was detected for a sample all the CpG sites
in the entire promoter region of that gene were methylated at
roughly the same proportion (%MeC, as obtained from the
Pyrograms). Representative patients showing non-methylated normal
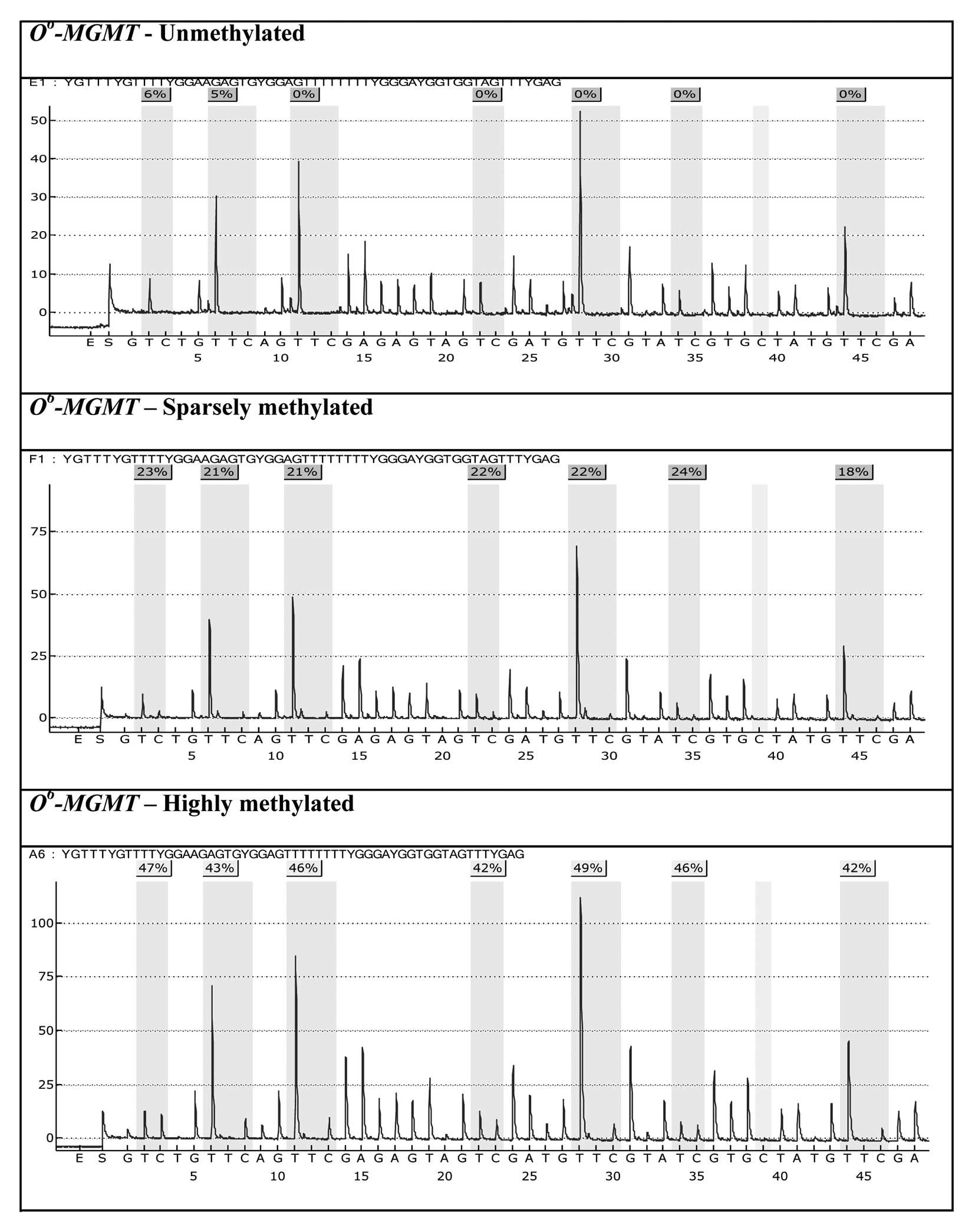
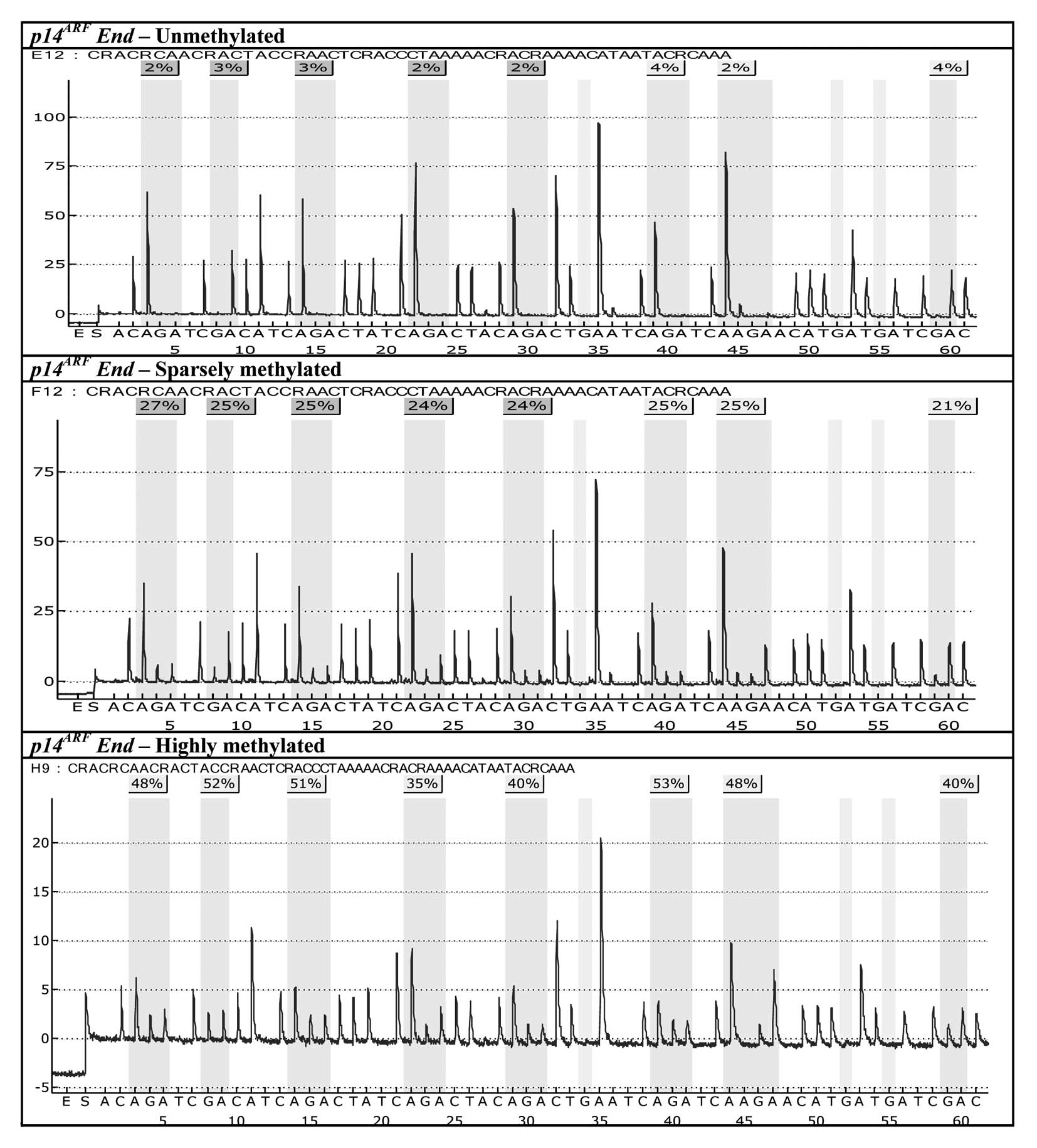
mucosa and tumor tissues methylated to varying degrees are
displayed for O6-MGMT and
p14ARF in Figs.
1 and 2. While the occurrence
of methylated promoter CpG sites varied (Table I), within each patient the
methylated fraction (%MeC) varied very little between
the different CpG sites of a particular gene. The mean methylated
fractions stated in Table I are
the inter-individual (total-sample) mean values of %MeC
of CpG sites though the entire promoter regions of all
methylation-positive tumor samples.
Concurrent methylation of two or more genes in the
colorectal cancer tumors are summarized in Table II. We found 42 patients (38%) to be
methylated on one gene, 18 (16%) were methylated on two genes, 15
(14%) on three genes, 5 (4.5%) were found to be methylated on four
genes and 1 (0.9%) was methylated on all the genes studied. Out of
the 111 tumors, 30 (27%) were thus found to be unmethylated on all
genes.
 | Table II.Number of colorectal cancer specimens
simultaneously methylated on more than one gene. |
Table II.
Number of colorectal cancer specimens
simultaneously methylated on more than one gene.
|
O6-MGMT |
p14ARF |
p16INK4a | RASSF1A | APC1A |
|---|
|
O6-MGMT | - | 15 | 10 | 10 | 12 |
|
p14ARF | | - | 16 | 11 | 7 |
|
p16INK4a | | | - | 7 | 9 |
| RASSF1A | | | | - | 5 |
| APC1A | | | | | - |
DNA hypermethylation and long-term
outcome
Survival plots for patients with or without promoter
methylation for each of the five genes were analysed. In univariate
analysis two genes reached statistical significance.
Hypermethylation of p14ARF was related to worse
survival (p=0.036) but its significance was attenuated in
multivariate analysis when adjusting for tumor stage and
differentiation (p=0.065). Hypermethylation of
O6-MGMT was associated with better survival
through the first 60 months of follow-up, the risk ratio was 0.36
(95% CI 0.15–0.87, p= 0.049) and still remained significant after
adjusting for tumor stage and differentiation (p=0.023), see
Table III.
 | Table III.Multivariate analysis of combined
promoter methylation of 06-MGMT, tumor stage and
differentiation in relation to patient survival through 60 months
post-surgery. |
Table III.
Multivariate analysis of combined
promoter methylation of 06-MGMT, tumor stage and
differentiation in relation to patient survival through 60 months
post-surgery.
| Variables | No. | Cancer death Rate
ratio | 95% CI | P-value |
|---|
| Methylation | | | | 0.023 |
| No | 71 | 1.00 | - | |
| Yes | 35 | 0.36 | 0.15–0.87 | |
| Stage | | | | <0.0001 |
| I | 17 | 1.00 | - | |
| II | 46 | 4.35 | 0.55–34.28 | |
| III | 24 | 8.65 | 1.08–69.08 | |
| IV | 19 | 40.05 | 4.98–321.7 | |
|
Differentiation | | | | 0.151 |
| Well | 26 | 1.00 | - | |
| Moderately | 62 | 0.68 | 0.29–1.58 | |
| Poorly | 18 | 0.85 | 0.30–2.40 | |
Since promoter hypermethylation of
p14ARF, RASSF1A and APC1A all were
associated with a similar tendency towards worse prognosis, and
many patients were methylated on more than one gene (Table II), we examined whether methylation
of any of the three genes (i.e., one or more), would improve
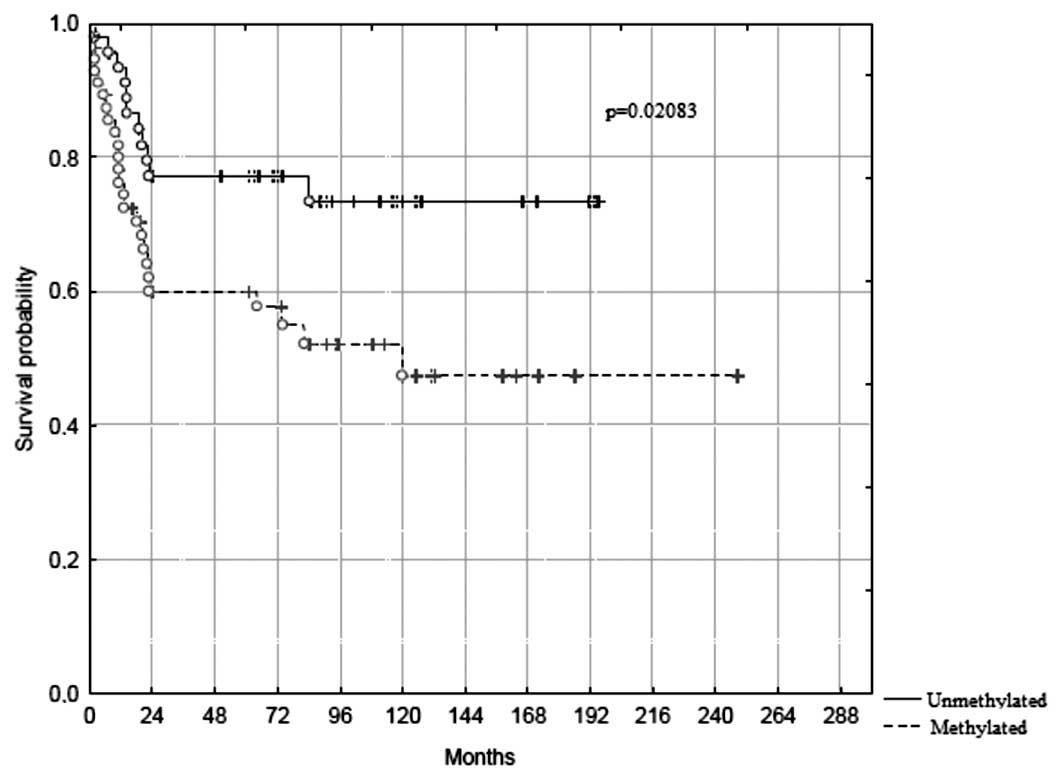
prediction of survival. Indeed, hypermethylation of one or more of
these three genes defined a set of patient with a significantly
(p=0.021) worse long-term survival (Fig. 3), where only ∼45% were still alive
in the methylated group by 20 years of follow-up, compared to ∼75%
in the unmethylated group. Adjusting for tumor stage and
differentiation did not attenuate this association (risk ratio
2.20; 95% CI, 1.05–4.62, p=0.037; Table IV). No association could be found
between promoter hypermethylation of these three genes and the
other clinicopathological factors including gender, age, tumor
location, or tumor stage and differentiation (p>0.05 for all
variables).
 | Table IV.Multivariate analysis of combined
promoter methylation of p14ARF, RASSF1A
and APC1A, tumor stage and differentiation in relation to
patient survival. |
Table IV.
Multivariate analysis of combined
promoter methylation of p14ARF, RASSF1A
and APC1A, tumor stage and differentiation in relation to
patient survival.
| Variables | No. | Cancer death Rate
ratio | 95% CI | P-value |
|---|
| Methylation | | | | 0.037 |
| No | 48 | 1.00 | - | |
| Yes | 58 | 2.20 | 1.05–4.62 | |
| Stage | | | | <0.0001 |
| I | 18 | 1.00 | - | |
| II | 46 | 3.82 | 0.48–30.2 | |
| III | 24 | 7.72 | 0.96–61.8 | |
| IV | 18 | 31.50 | 3.99–248.6 | |
|
Differentiation | | | | 0.330 |
| Well | 28 | 1.00 | - | |
| Moderately | 62 | 0.78 | 0.34–1.78 | |
| Poorly | 16 | 1.14 | 0.42–3.10 | |
Discussion
Aging and environmental factors may lead to
neoplasia and cancer. This process usually involves changed
expression pattern of genes involved in adhesion, proliferation,
differentiation, cell growth, migration and apoptosis, and can be
due to mutations, genetic rearrangements, chromosomal instability
or promoter hypermethylation (14). In this study we have focused on
five such genes, previously suggested to be involved in the
development of CRC; O6-MGMT,
p14ARF, p16INK4a,
RASSF1A and APC1A(3–12).
Methylation of the promoter regions of some of these genes in
tissues or serum from patients with CRC has been reported (15–25),
but data on the long-term prognostic implications of the whole set
is limited.
By using Pyrosequencing, a technique that offers a
unique opportunity to quantitate, site-specifically, the methylated
fraction in partially methylated CpG sites, we demonstrated that
the promoter regions of one or more of the genes analyzed are
methylated in tumor tissue from a majority (73%) of patients
diagnosed with CRC, range 14–34% for the different genes. None of
these genes was methylated in the 46 paired normal mucosa samples.
Interestingly, adenoma tissue also appeared to be methylated in
about a third of the patients and the paired normal mucosal tissue
unmethylated, although caution is prudent here since we have
analysed rather few adenomas.
Prognosis and methylation of
p14ARF, p16INK4a, APC1A and RASSF1A
When correlating the methylation of the promoter
regions of these genes with survival of the CRC patients, we found
that methylation of p14ARF was significantly
associated with shorter survival compared to patients that has this
gene unmethylated. When adjusting for tumor stage and tumor
differentiation this significance was attenuated somewhat. We also
saw clear trends towards poorer prognosis when methylation was
found in the promoter regions of RASSF1A and
APC1A.
In some of the subjects several of the genes had
their promoter regions hypermethylated concurrently, implicating
that signalling pathways such as Wnt where APC1A are
involved, p16INK4a-Rb and
p14ARF-p53, and normal cell mechanisms such as
alkylation and mitotic progression could be altered all at once.
Since it has been suggested that CRC evolves from alterations of
several of these pathways (26),
we thought it relevant to investigate whether this concurrent
methylation could affect the outcome for the patient. Grouping
together all individuals showing methylation of one or more of the
p14ARF, RASSF1A and APC1A genes, we
obtained an association between promoter hypermethylation and
shorter survival which remained statistically significant even
after adjusting for tumor stage and differentiation (Fig. 3, Table
IV). Thus, promoter hypermethylation of one or more of the
genes p14ARF, RASSF1A and APC1A,
when defined by Pyrosequencing assays, might serve as a marker of
poor prognosis which we suggest as a novel, relevant stratification
factor in future prospective and interventional studies on CRC.
In other recent studies, poor survival has been
reported for patients methylated on the p14ARF
gene (25,27) in agreement with our findings and
for APC1A one report based on measurements of methylation in
plasma DNA reported worse prognosis (28) but another study claimed a better
survival in patients with methylated tumor tissue
APC1A(17). Several reports
have claimed predictive value of p16INK4a
methylation (19,21–23,27,29,30)
which we could not replicate in our cohort. Data on the prognostic
implications of RASSF1A methylation are sparse, one study
reported a higher prevalence of methylation in liver metastases
than in the primary tumor (31).
Prognosis and methylation of
O6-MGMT
Promoter methylation in the
O6-MGMT gene has in some studies on glioblastoma
been associated with longer survival especially in therapeutic
trials using alkylating agents (32–36).
It is now proposed that tests for O6-MGMT
methylation status should be included in all future clinical trials
in malignant glioma if treatment includes alkylating agents, since
it is anticipated that those tests may guide choice of future
therapy (37). To asses
O6-MGMT promoter methylation in the mentioned
glioma studies, several different methods have been employed, as
was recently pointed out by van den Bent et al(12), who stressed the point that the CpG
island in that part of the gene actually contains almost 100
individual CpG sites and that the used methods only give
information on the methylation status of a few of these sites. It
has not been clear how many CpG loci, or which ones of them, have
to be methylated to achieve O6-MGMT gene
silencing. Our method utilizes a sequence-specific and quantitative
assay, Pyrosequencing, to classify the methylation status (13) and thus provides a tool to answer
this question. Our assays cover 25 different CpG sites, 18 of which
have never been assayed before, and we show here that in the
individual patient, the %MeC of each of these sites is a
characteristic feature of each individual tumor sample, and varies
very little from CpG site to CpG site within the same tumor tissue
sample (Fig. 1; the same
observation holds for p14ARF, cf. Fig. 2). On the other hand, a considerable
inter-individual difference was seen which could be owing to
differing degrees of admixture of unmethylated non-neoplastic
cells, to true differences in the biology of the tumor, or to a
combination of these factors.
There are a few reports on CRC prognosis in relation
to methylation of O6-MGMT, claiming basically no
relation (17,29,38).
We show here for the first time that like in glioma, promoter
methylation of O6-MGMT in CRC patients tended to
confer better long-term survival (Table III), in a context where alkylating
agents are rarely an option. This suggests that
O6-MGMT testing might be of a more general
interest and warrants to be included not only in planning of glioma
treatment but in studies on other malignant neoplasias as well.
General observations and limitations
A number of methodological differences between the
various studies need to be pointed out. Most of the data come from
methylation-specific PCR-based methods, not from sequencing-based
techniques. Many studies are made on DNA isolated from FFPE
samples, which may have decayed due to harsh conditions. We used
DNA isolated from fresh tumor tissue, and analysis was performed by
bisulphite pyrosequencing (13).
The fraction of patients showing methylation of our set of genes
ranged from 14% to 34% for the different genes, figures which are
both higher (29) and lower
(17) than those reported earlier.
For instance, one study claiming no prognostic value of
O6-MGMT in CRC classified 60% as
methylation-positive by a MS-PCR method (17), as against our figure of 34% using
Pyrosequencing which agrees better with the figure of Ogino and
coworkers of 38% (38). In future
studies, more attention should be payed to the source of DNA and to
methods used to classify promoter methylation status, and we
contend that Pyrosequencing has earned a place among the methods of
choice.
The concept of CIMP-positivity (CpG Island
Methylator Phenotype) needs to be relativised based on our
findings. The number of subjects simultaneously methylated on 2 or
more of the selected set of genes was rather small (Table II) and any predictions based on
such a subset of the patient population will therefore have a very
limited utility. In contrast, we found a statistically significant
negative relation with survival, adjusted for tumor stage and
differentiation based on a combined methylation variable defined as
having any one or more of the genes p14ARF,
RASSF1A, APC1A methylated (Table IV, Fig. 3). Employing the same Pyrosequencing
assays (13) as in the present
paper, we recently showed that APC1A promoter methylation
was associated with poor prognosis also in cervical cancer
(39).
Moreover, methylation of one of the genes in our
set, O6-MGMT, showed a significant association
with better survival through the first 60 months following primary
surgery. For all these reasons, the concept of a CIMP as a unified
set of methylated genes accompanying poor prognosis appears to have
limited prospects as a valid prognostic tool in most CRC patients.
Perhaps it may ultimately be replaced by a more precise molecular
signature associated with poor prognosis (40,41),
possibly even tailor-made on the individual basis which might
better reflect the random stochastic nature of the processes that
characterise sporadic CRC. Such signatures would likely include
both chromosomal instability, microsatellite instability, and
methylation of a larger set of cancer-related genes than has been
currently studied (28,29,30,42,43)
as well as less explored features such as histone modifications,
nucleosomal occupancy and remodeling, chromatin looping, and
non-coding RNAs (44). Hopefully,
DNA methylation data may in the future also be useful in guiding
adjuvant treatment in CRC as suggested in a recent article
(45).
In conclusion, this is the first study to report a
significant correlation between CRC patient survival and promoter
methylation of the genes p14ARF, RASSF1A
and APC1A, as defined by Pyrosequencing assay (13), as well as a protective role of
O6-MGMT methylation. Such biomarkers of prognosis
in CRC could be utilized as a relevant stratification factor in
future prospective and interventional studies on CRC, and might
serve as a tool when tailoring treatment for the individual
patient. Finally, we maintain that choice of assay methodology may
have a determining effect on proper classification of methylation
status in the individual patient.
Acknowledgements
Financial support by Örebro County
Council, Nyckelfonden, Örebro, and Lions Cancerfond, Uppsala is
gratefully acknowledged.
References
|
1.
|
Das PM and Singal R: DNA methylation and
cancer. J Clin Oncol. 22:4632–4642. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Esteller M, Corn PG, Baylin SB and Herman
JG: A gene hyper-methylation profile of human cancer. Cancer Res.
61:3225–3229. 2001.PubMed/NCBI
|
|
3.
|
van Engeland M, Roemen GM, Brink M, et al:
K-ras mutations and RASSF1A promoter methylation in colorectal
cancer. Oncogene. 21:3792–3795. 2002.
|
|
4.
|
Preuss I, Eberhagen I, Haas S, Eibl RH,
Kaufmann M, von Minckwitz G and Kaina B:
O6-methylguanine-DNA methyltransferase activity in
breast and brain tumors. Int J Cancer. 61:321–326. 1995.
|
|
5.
|
Newcomb EW, Alonso M, Sung T and Miller
DC: Incidence of p14ARF gene deletion in high-grade
adult and pediatric astrocytomas. Hum Pathol. 31:115–119. 2000.
|
|
6.
|
Serrano M, Hannon GJ and Beach D: A new
regulatory motif in cell-cycle control causing specific inhibition
of cyclin D/CDK4. Nature. 366:704–707. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Song MS, Song SJ, Ayad NG, et al: The
tumor suppressor RASSF1A regulates mitosis by inhibiting the
APC-Cdc20 complex. Nat Cell Biol. 6:129–137. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Rubinfeld B, Souza B, Albert I, et al:
Association of the APC gene product with beta-catenin. Science.
262:1731–1734. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Su LK, Vogelstein B and Kinzler KW:
Association of the APC tumor suppressor protein with catenins.
Science. 262:1734–1737. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
van den Donk M, Pellis L, Crott JW, et al:
Folic acid and vitamin B-12 supplementation does not favorably
influence uracil incorporation and promoter methylation in rectal
mucosa DNA of subjects with previous colorectal adenomas. J Nutr.
137:2114–2120. 2007.
|
|
11.
|
van Engeland M, Weijenberg MP, Roemen GM,
et al: Effects of dietary folate and alcohol intake on promoter
methylation in sporadic colorectal cancer: the Netherlands cohort
study on diet and cancer. Cancer Res. 63:3133–3137. 2003.PubMed/NCBI
|
|
12.
|
van den Bent MJ, Dubbink HJ, Sanson M, et
al: MGMT promoter methylation is prognostic but not predictive for
outcome to adjuvant PCV chemotherapy in anaplastic oligodendroglial
tumors: a report from EORTC Brain Tumor Group Study 26951. J Clin
Oncol. 27:5881–5886. 2009.PubMed/NCBI
|
|
13.
|
Löf-Öhlin ZM and Nilsson TK:
Pyrosequencing assays to study promoter CpG site methylation of the
O6-MGMT, hMLH1, p14ARF,
p16INK4a, RASSF1A and APC1A genes. Oncol
Rep. 21:721–729. 2009.PubMed/NCBI
|
|
14.
|
Aoyagi H, Iida S, Uetake H, et al: Effect
of classification based on combination of mutation and methylation
in colorectal cancer prognosis. Oncol Rep. 25:789–794.
2011.PubMed/NCBI
|
|
15.
|
Esteller M, Tortola S, Toyota M, Capella
G, Peinado MA, Baylin SB and Herman JG: Hypermethylation-associated
inactivation of p14(ARF) is independent of p16(INK4a) methylation
and p53 mutational status. Cancer Res. 60:129–133. 2000.PubMed/NCBI
|
|
16.
|
Kamiyama H, Noda H, Takata O, Suzuki K,
Kawamura Y and Konishi F: Promoter hypermethylation of
tumor-related genes in peritoneal lavage and the prognosis of
patients with colorectal cancer. J Surg Oncol. 100:69–74. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Chen SP, Chiu SC, Wu CC, et al: The
association of methylation in the promoter of APC and MGMT and the
prognosis of Taiwanese CRC patients. Genet Test Mol Biomarkers.
13:67–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Noda H, Mashima R, Kamiyama H, Okada S,
Kawamura YJ and Konishi F: Promoter hypermethylation of
tumor-related genes in sporadic colorectal cancer in young
patients. J Exp Clin Cancer Res. 26:521–526. 2007.PubMed/NCBI
|
|
19.
|
Krtolica K, Krajnovic M, Usaj-Knezevic S,
Babic D, Jovanovic D and Dimitrijevic B: Comethylation of p16 and
MGMT genes in colorectal carcinoma: correlation with
clinicopathological features and prognostic value. World J
Gastroenterol. 13:1187–1194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Kohonen-Corish MR, Daniel JJ, Chan C, et
al: Low microsatellite instability is associated with poor
prognosis in stage C colon cancer. J Clin Oncol. 23:2318–2324.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nakayama G, Hibi K, Kodera Y, Koike M,
Fujiwara M and Nakao A: P16 methylation in serum as a potential
marker for the malignancy of colorectal carcinoma. Anticancer Res.
27:3367–3370. 2007.PubMed/NCBI
|
|
22.
|
Lee M, Han WS, Kim OK, Sung SH, Cho MS,
Lee SN and Koo H: Prognostic value of p16INK4a and
p14ARF gene hypermethylation in human colon cancer.
Pathol Res Pract. 202:415–424. 2006.
|
|
23.
|
Sanz-Casla MT, Maestro ML, Vidaurreta M,
Maestro C, Arroyo M and Cerdan J: p16 Gene methylation in
colorectal tumors: correlation with clinicopathological features
and prognostic value. Dig Dis. 23:151–155. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Zou HZ, Yu BM, Wang ZW, et al: Detection
of aberrant p16 methylation in the serum of colorectal cancer
patients. Clin Cancer Res. 8:188–191. 2002.
|
|
25.
|
Dominguez G, Silva J, Garcia JM, et al:
Prevalence of aberrant methylation of p14ARF over
p16INK4a in some human primary tumors. Mutat Res.
530:9–17. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Olschwang S, Hamelin R, Laurent-Puig P, et
al: Alternative genetic pathways in colorectal carcinogenesis. Proc
Natl Acad Sci USA. 94:12122–12127. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Shen L, Catalano PJ, Benson AB III,
O’Dwyer P, Hamilton SR and Issa JP: Association between DNA
methylation and shortened survival in patients with advanced
colorectal cancer treated with 5-fluorouracil based chemotherapy.
Clin Cancer Res. 13:6093–6098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Hoffmann AC, Vallböhmer D, Prenzel K, et
al: Methylated DAPK and APC promoter DNA detection in peripheral
blood is a significantly associated with apparent residual tumor
and outcome. J Cancer Res Clin Oncol. 135:1231–1237. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Kim JC, Choi JS, Roh SA, Cho DH, Kim TW
and Kim YS: Promoter methylation of specific genes is associated
with the phenotype and progression of colorectal adenocarcinomas.
Ann Surg Oncol. 17:1767–1776. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Wettergren Y, Odin E, Nilsson S, Carlsson
G and Gustavsson B: p16INK4a gene promoter
hypermethylation in mucosa as a prognostic factor for patients with
colorectal cancer. Mol Med. 14:412–421. 2008. View Article : Google Scholar
|
|
31.
|
Tommasi S, Pinto R, Petriella D, et al:
Oncosuppressor methylation: a possible key role in colon metastatic
progression. J Cell Physiol. 226:1934–1939. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Weller M, Felsberg J, Hartmann C, et al:
Molecular predictors of progression-free and overall survival in
patients with newly diagnosed glioblastoma: a prospective
translational study of the German Glioma Network. J Clin Oncol.
27:5743–5750. 2009. View Article : Google Scholar
|
|
33.
|
Hegi ME, Diserens AC, Gorlia T, et al:
MGMT gene silencing and benefit from temozolomide in glioblastoma.
N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Esteller M, Garcia-Foncillas J, Andion E,
et al: Inactivation of the DNA-repair gene MGMT and the clinical
response of gliomas to alkylating agents. N Engl J Med.
343:1350–1354. 2000. View Article : Google Scholar
|
|
35.
|
Jaeckle KA, Eyre HJ, Townsend JJ, et al:
Correlation of tumor O6 methylguanine-DNA
methyltransferase levels with survival of malignant astrocytoma
patients treated with bis-chloroethylnitrosourea: a Southwest
Oncology Group study. J Clin Oncol. 16:3310–3315. 1998.
|
|
36.
|
Hegi ME, Diserens AC, Godard S, et al:
Clinical trial substantiates the predictive value of
O-6-methylguanine-DNA methyltransferase promoter methylation in
glioblastoma patients treated with temozolomide. Clin Cancer Res.
10:1871–1874. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Stupp R and Hegi ME: Methylguanine
methyltransferase testing in glioblastoma: when and how? J Clin
Oncol. 25:1459–1460. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Shima K, Morikawa T, Baba Y, et al: MGMT
promoter methylation, loss of expression and prognosis in 855
colorectal cancers. Cancer Causes Control. 22:301–309. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Löf-Öhlin ZM, Sorbe B, Wingren S and
Nilsson TK: Hypermethylation of promoter regions of the
APC1A and p16INK4a genes in relation to
prognosis and tumor characteristics in cervical cancer patients.
Int J Oncol. 39:683–688. 2011.
|
|
40.
|
Carmona FJ and Esteller M: Moving closer
to a prognostic DNA methylation signature in colon cancer. Clin
Cancer Res. 17:1215–1217. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Yi JM, Dhir M, Van Neste L, et al: Genomic
and epigenomic integration identifies a prognostic signature in
colon cancer. Clin Cancer Res. 17:1535–1545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Cui T, Chen Y, Yang L, Knösel T, Zöller K,
Huber O and Petersen I: DSC3 expression is regulated by p53, and
methylation of DSC3 DNA is a prognostic marker in human colorectal
cancer. Br J Cancer. 104:1013–1019. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Tong JD, Jiao NL, Wang YX, Zhang YW and
Han F: Downregulation of fibulin-3 gene by promoter methylation in
colorectal cancer predicts adverse prognosis. Neoplasma.
58:441–448. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
van Engeland M, Derks S, Smits KM, Meijer
GA and Herman JG: Colorectal cancer epigenetics: complex
simplicity. J Clin Oncol. 29:1382–1391. 2011.PubMed/NCBI
|
|
45.
|
Jover R, Nguyen TP, Pérez-Carbonell L, et
al: 5-Fluorouracil adjuvant chemotherapy does not increase survival
in patients with CpG island methylator phenotype colorectal cancer.
Gastroenterology. 140:1174–1181. 2011. View Article : Google Scholar : PubMed/NCBI
|