Introduction
Malignant mesothelioma (MM) is a rare and aggressive
tumour, accounting for less than 1% of all cancer deaths in the
world (1). Although malignant
mesothelioma is rare, it is a particularly aggressive cancer and
despite the exploration of a variety of new therapeutic approaches,
treatment remains palliative at best (2).
The platinum based compound cisplatin is a widely
used chemotherapeutic agent which acts primarily via DNA damage and
subsequent induction of apoptotic programs (3). Cisplatin based chemotherapy, either
as a single agent or in combination, is commonly employed in
therapy of malignant mesothelioma, although its efficacy is limited
(2). The antineoplastic activity
of cisplatin is rather complex and in light of the published
reports it appears that cisplatin may activate several parallel
pathways leading to cell cycle arrest and apoptosis depending on
the treatment conditions, cell type or concentration (4). Due to this complexity and lack of
cytotoxic efficiency in mesothelioma, our knowledge about
particular molecular targets of cisplatin in this type of
malignancy remains limited despite this compound being widely used
in its treatment. Recent evidence from clinical samples has
suggested that as for other malignancies inhibitor of apoptosis
proteins (IAPs) are upregulated in mesothelioma and may play a role
the resistance of these cells to therapy (5–7).
Previous research conducted in our laboratory has
characterised the mechanisms of cisplatin-induced apoptosis in
malignant mesothelioma cell lines derived from mice (8). The initial aim of this study was to
characterise the in vitro culture models of human MM, for
mechanisms involved in the action of cisplatin (9). Thus in our study we wanted to explore
in detail the possible proapoptotic mechanisms of cisplatin in
highly aggressive mesothelioma cell lines while also examining the
role of IAPs in resistance of this tumour. Various therapies are
under development targeting IAPs and in particular XIAP and
survivin in a number of malignancies (10) and we wanted to investigate in
detail the potential of such therapies in mesothelioma using a
panel of cell lines. We found that in these cell lines IAPs were
not a factor in cisplatin resistance and that cisplatin instead
targets caspase-independent pathways.
Materials and methods
Cell culture and reagents
The malignant mesothelioma cell lines JU77, LO68 and
ONE58 were used in this study. These cell lines were originally
derived from pleural effusions of different patients presenting
with malignant pleural mesothelioma (9). All cells were cultured and maintained
in medium R5, which is RPMI-1640 plus 5% heat-inactivated fetal
bovine serum (FBS), 300 mM L-glutamine, 120 μg/ml penicillin
and 100 μg/ml gentamicin (all supplied by Invitrogen,
Victoria, Australia). All cell cultures were grown at 37°C in a 5%
CO2 humidified atmosphere. Mesothelial cell RNA and
protein was kindly provided by Dr Steve Mutsaers (Lung Institute of
Western Australia).
Cell viability
An MTT assay was used to quantitate cell
death/viability following 24 h exposure to cisplatin. Cells were
seeded into 96-well plates at a density of 20,000 cells/well.
Following 24 h incubation, cisplatin was added at concentrations
from 0–100 μg/ml and the cells incubated for a further 24 h.
The assay was terminated by aspirating the media, 100 μl of
1 mg/ml MTT (Sigma-Aldrich, NSW, Australia) in RPMI was added and
the plates incubated for 60 min at 37°C. Following removal of the
supernatant 100 μl of DMSO was added to solubilise the dye
and absorbance measured at 595 nm with a microplate reader (Model
3550, Bio-Rad Laboratories, CA, USA). IC50 was defined
as the concentration causing a 50% reduction in absorbance relative
to the negative control. IC50 was determined by
non-linear regression analysis using Graphpad Prism v4 (Graphpad
Software, CA, USA).
Annexin-V and propidium iodide
staining
Annexin-V and Propidium Iodide (PI) staining was
performed using an Annexin-V-Fluos kit (Roche Diagnostics, NSW,
Australia). Cells were seeded into 6-well plates at a density of
5×105 cells/well. Following 24 h incubation cisplatin
was added at concentration 0–100 μg/ml and the cells
incubated for a further 24 h. Cells were harvested by
trypsinization, collected by centrifugation, washed once with 1 ml
of PBS and stained according to the manufacturers’ protocol and
analysed using a FACS Calibur Flow Cytometer (BD Biosciences, NSW,
Australia).
Determination of the mitochondrial
transmembrane potential
ΔΨm was analysed using the cationic dye
JC-1 (5, 5′, 6, 6′-tetrachloro-1, 1′, 3,
3′-tetraethyl-benzimidazolcarbocyanine iodide) which exhibits
potential-dependent accumulation in mitochondria by fluorescence
emission shift from green (∼525 nm) to red (∼590 nm). Consequently,
mitochondrial depolarization is indicated by a decrease in the
red/green fluorescence intensity ratio. Cells were seeded into
black 96-well plates (Greiner, Germany) at a density of 10,000
cells/well in 100 μl R5. Following 24 h incubation,
cisplatin was added at concentrations from 0–100 μg/ml and
the cells were incubated for a further 24 h. The media was then
aspirated and 50 μl of staining solution added [33 μM
JC-1 (Invitrogen) in serum free R-5], and the plates incubated for
1 h at 37°C, 5% CO2. The staining solution was then
removed and 200 μl of PBS with 5% BSA was added. After a
further 5-min incubation, PBS/BSA was removed and 100 μl PBS
was added. The plates were read on a Fluostar Optima plate reader
(excitation filter 485 nm and emission filters 520 nm /595 nm) (BMG
Laboratories, Victoria, Australia). Data are presented as the ratio
of red to green. FCCP [carbonyl cyanide
4-(trifluoromethoxy)phenylhydrazone] (Sigma-Aldrich) was used as a
positive control for maximal mitochondrial depolarisation.
Caspase activity assay
Activation of effector caspases during apoptosis was
determined using the Caspase-3 Assay kit#2 (Invitrogen) with a
protocol modified for a direct homogeneous assay. Cells were seeded
into black 96-well plates (Greiner) at a density of 20,000
cells/well in 100 μl medium. Following 24 h incubation,
cisplatin was added at concentrations from 0–100 μg/ml and
the cells incubated for a further 24 h. The assay was terminated by
the addition of 100 μl of reagent buffer (5 μl 20X
cell lysis buffer, 20 μl 5X reaction buffer, 0.5 μl
substrate z-DEVD-R110, 0.5 μl of 1M DTT, 74 μl
dH2O) and the fluorescent signal measured after 1 h
(Fluostar Optima, BMG Laboratories). Appropriate blank and control
samples were included in each run as recommended by the assay
manufacturer. Caspase inhibitors were the pan-caspase inhibitor
z-VAD-fmk and the caspase 3/7 inhibitor z-DEVD-cmk (Calbiochem,
Victoria, Australia).
Reverse transcription PCR (RT-PCR) and
real-time RT-PCR
Total RNA was isolated from cultures of the cell
lines using Ultraspec RNA reagent (Biotec, TX, USA) according to
the manufacturer’s instructions. The RNA pellet was resuspended in
1 mM sodium citrate buffer and samples were stored at −80°C. Prior
to RT-PCR, contaminating DNA was removed from the RNA using RQ1
DNAse (Promega, NSW, Australia). First strand cDNA synthesis was
performed using SuperScript III First-Strand Synthesis System with
oligo dT primers (Invitrogen). Gene specific PCR primers were
designed using the Primer 3 software (11) and the sequences are shown in
Table I. Real-time PCR was
performed using a RotorGene real-time amplification instrument
(Corbett Research, NSW, Australia) using a SensiMix SYBR kit
(Bioline, NSW, Australia) according to the manufacturers’
instructions. A melt curve analysis was performed at end of each
experiment to confirm specificity.
 | Table I.The sequences used. |
Table I.
The sequences used.
| Gene | Primer sequence
5′-3′ | Genbank accn. |
|---|
| Survivin |
CTTGAAAGTGGCACCAGAGG | NM_001168 |
|
GGACCACCGCATCTCTACAT | |
| Bruce |
TCAACAGGTGAGTGCTCCAG | NM_016252 |
|
TCCAAAAGCAGCCAAAGAAT | |
| Livin |
CTGGTCAGAGCCAGTGTTCC | NM_139317 |
|
TCATAGAAGGAGGCCAGACG | |
| XIAP |
GGGGTTCAGTTTCAAGGA | NM_001167 |
|
CGCCTTAGCTGCTCTTCAGT | |
| IAP1 |
CCTGGATAGTCTACTAACTGCCT | NM_001166 |
|
GCTTCTTGCAGAGAGTTTCTGAA | |
| IAP2 |
CAGAATTGGCAAGAGCTGGT | NM_001165 |
|
ATTCGAGCTGCATGTGTCT | |
Gene expression data were normalised to levels of
expression of reference (housekeeping) genes GAPDH and HPRT1. PCR
primers were as follows: survivin (NM_001168) forward
(5′-cttgaaagtggcaccagagg-3′), reverse (5′-ggaccaccgcatct
ctacat-3′); XIAP (NM_001167) forward (5′-ggggttcagtttcaa gga-3′),
reverse (5′-cgccttagctgctcttcagt-3′); IAP1 (NM_001165) forward
(5′-cctggatagtctactaactgcct-3′), reverse (5′-gcttct
tcagagagtttctgaa-3′); IAP2 (NM_001160) forward (5′-cagaat
tggcaagagctggt-3′), reverse (5′-attcgagctgcatgtgtct-3′); HPRT1
(NM_000194) forward (5′-tgacactggcaaaacaatgca-3′), reverse
(5′-ggtccttttcaccagcaagct-3′) and GAPDH (NM_002046) forward
(5′-accacagtccatgccatcac-3′), reverse (5′-tccaccacc
ctgttgctgta-3′). Real-time assays were performed on the relevant
samples and from the reference gene data generated for each sample
using geNorm software (v3.3) essentially as described by
Vandesompele et al(12).
Relative expression of the target gene was normalised using this
factor and expressed as mean ± standard deviation relative to a
control or calibrator sample.
Immunoblot analysis
Cell monolayers were washed once with ice-cold
saline and lysed by addition of RiPa buffer [50 mM Tris-HCl (pH
8.0), 150 mM NaCl, 1% Triton X-100, 0.5% Na deoxycholate, 0.1% SDS)
with protease inhibitors (Roche Biochemicals)]. Incubated for 1 h
at 4°C with gentle agitation then harvested with a cell scraper and
transferred to a 1.5 ml tube. The lysate was then centrifuged at
12,000 g for 30 min. at 4°C and the supernatants recovered and
stored at −80°C. Protein concentrations were determined by DC
protein assay (Bio-Rad). Proteins were resolved on 4–12% NuPAGE
polyacrylamide gels (Invitrogen) and transferred to nitrocellulose
membrane (Pall, Victoria, Australia) and probed with the following
antibodies: anti-XIAP (Cell Signaling Technology, MA, USA),
anti-survivin (BioLegend, CA, USA) and anti-actin (Sigma-Aldrich).
After incubation with alkaline phosphatase conjugated secondary
antibody, colour was developed using 5-bromo-4-chloro-3-indolyl
phosphate/nitroblue tetrazolium reagents (Sigma-Aldrich).
Cell-based ELISA
A cell-based ELISA assay was used to monitor changes
in protein expression in response to RNAi duplex transfection.
Preliminary experiments were performed to optimise the conditions
for each cell line with respect to cell density and antibody
concentration. Following treatment in 96-well plates, the
experiment was terminated by removing the media and fixing the
cells with ice-cold methanol for 10 min. The methanol was aspirated
and the cells were rinsed 3 times for 5 min each with 200 μl
per well of 0.1% Triton X-100 in PBS. The cells were incubated for
5 min with 100 μl of 3% H2O2 (to block
endogenous peroxidase activity), washed twice with 0.5% Tween-20 in
PBS (wash buffer) and then blocked with 5% FBS in PBS for 30 min.
Cells were washed 3 times, incubated overnight with primary
antibody (as for immunoblot) and following three washes incubated
at room temperature for 1 h with HRP conjugated secondary antibody.
Detection was performed for 1 h with ABTS
(2,2′-azinodiethyl-benzthiazoline sulfonate) substrate
(Sigma-Aldrich), stopped with 1% SDS and absorbance measured at 405
nm. Appropriate background and antibody controls were included in
each assay.
RNAi-mediated gene silencing
The siRNA duplexes were Stealth Select 3 RNAi Sets
for human XIAP and survivin (Invitrogen) and the optimal duplex as
determined empirically for each cell line was used for the
experiments presented here. The negative control duplexes were the
appropriate (based upon GC content) Stealth siRNA negative controls
(Invitrogen). Transfection of RNAi duplexes was performed using
Lipofectamine (Invitrogen, Australia) essentially according to the
manufacturers recommendation with concentration determined
empirically.
Results
Mechanisms of cisplatin induced cell
death in mesothelioma cells
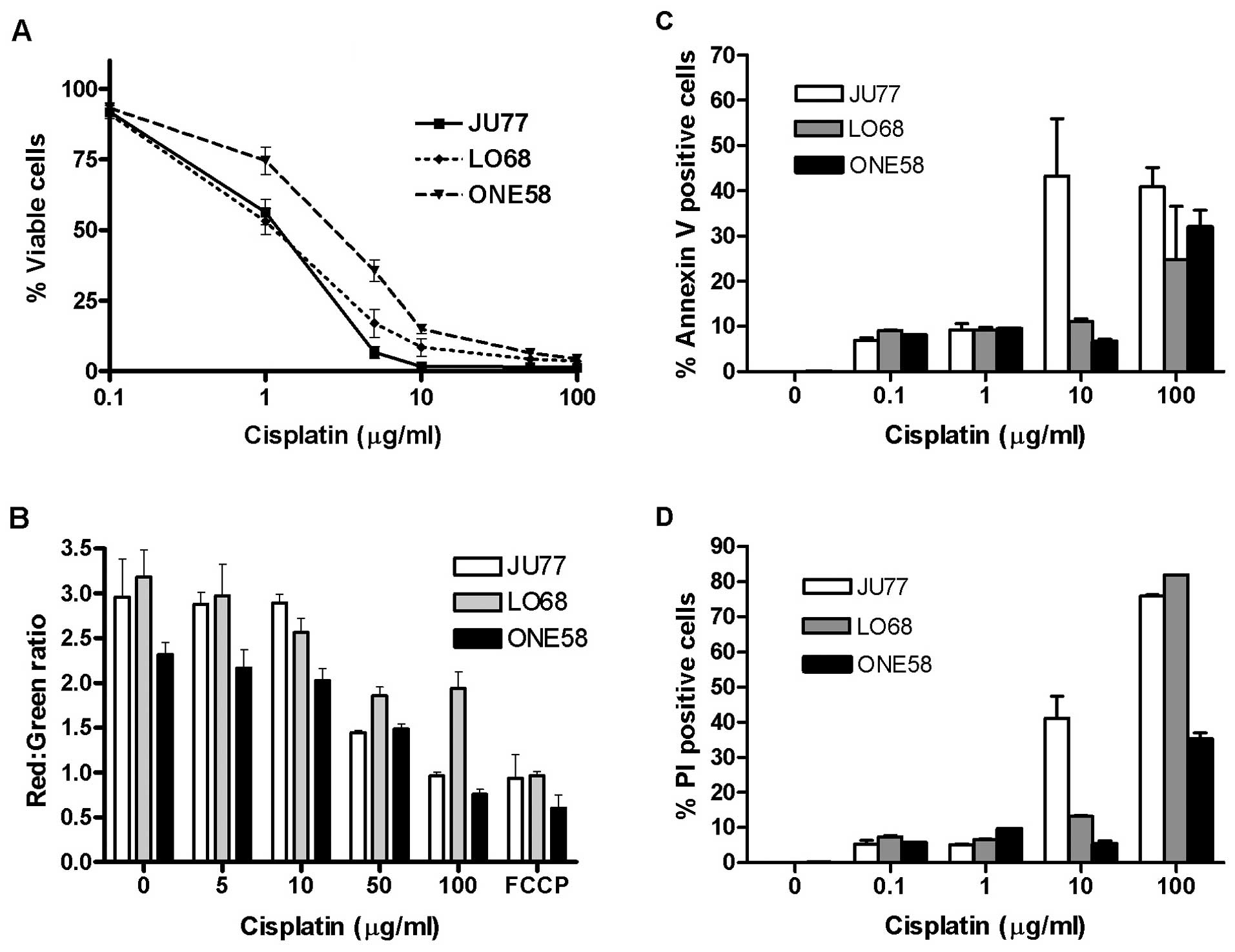
In order to examine the mechanisms of cisplatin
effect upon mesothelioma we initially evaluated the in vitro
sensitivity of three mesothelioma cell lines, LO68, ONE58 and JU77
to 24 h exposure to cisplatin (0.1–100 μg/ml) by MTT assay.
Results were determined by constructing dose-response curves
(Fig. 1A) and the IC50
calculated by further non-linear regression analysis of the data.
The results showed that JU77 (IC50 of 1.14±0.21
μg/ml) and LO68 (IC50 of 1.2±0.45 μg/ml)
were the most cisplatin sensitive cell lines when compared to ONE58
(IC50 of 3.08±0.83 μg/ml).
Given the importance of mitochondria in cell death
regulation and apoptotic signalling we examined mitochondrial
functional integrity in cisplatin treated mesothelioma cells.
Mitochondrial disruption and the loss of mitochondrial membrane
potential (MMP) was assayed using the fluorescent mitochondrial
potential sensor (JC-1). We found that all three mesothelioma cell
lines exhibited dose-dependent loss of MMP following cisplatin
treatment (Fig. 1B).
Interestingly, at 100 μg/ml in both JU77 and ONE58 cisplatin
inhibited the function of mitochondria to the same extent as the
positive control FCCP (a protonophore that dissipates the H+
gradient across the inner membrane of mitochondria) whereas a
lesser effect was seen in LO68 cells.
In order to further explore the specific mechanisms
associated with cisplatin-induced cell death in this tumour cell
type, the biochemical changes in the cell membrane associated with
apoptosis were measured (Fig. 1C and
D). All three cell lines demonstrated increased Annexin-V
binding indicative of phosphatidylserine translocation in response
to cisplatin treatment, which was most pronounced in JU77 cells
(Fig. 1C). When PI staining
showing loss of membrane integrity was determined, JU77 cells also
showed response at a lower dose than LO68 or ONE58 cells (Fig. 1D). These results were consistent
with cell viability data showing JU77 as the most sensitive
(Fig. 1A).
Caspase activation and IAP expression in
MM cells
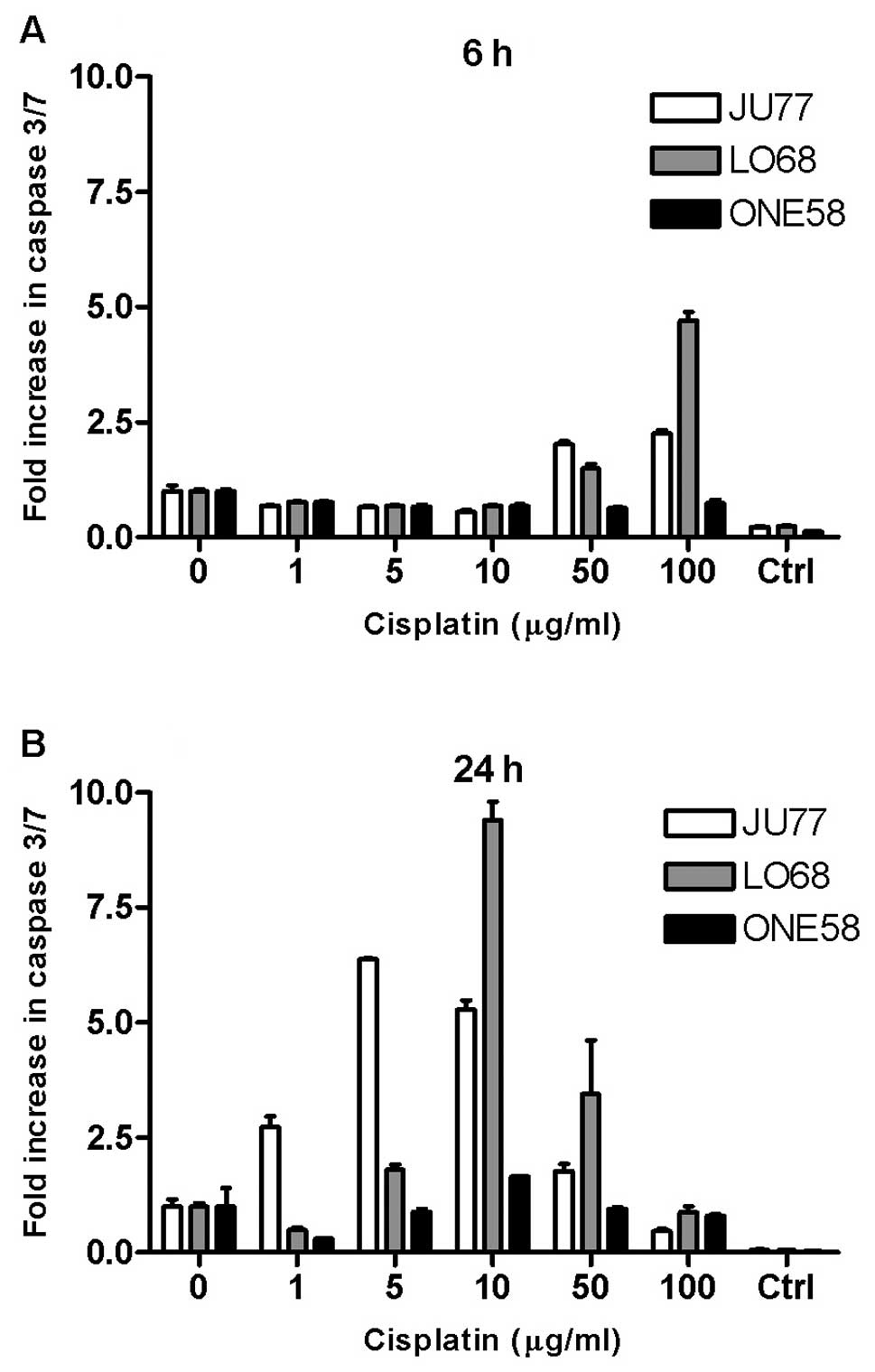
We char-acterised time- and dose-dependent patterns
of cisplatin induced caspase-3/7 activation in these cell lines.
Cultures of JU77, LO68 and ONE58 were treated with cisplatin and
dose-related changes in caspase 3/7 activity were assayed at 0, 3,
6 and 24 h. All MM cell lines demonstrated an increase in caspase
3/7 activation in response to cisplatin (Fig. 2), although no effect was seen at 3
h (not shown). Caspase 3/7 activation occurs at a later time with
lower doses of cisplatin (Fig.
2B), whereas with higher doses of cisplatin, caspase 3/7
activation occurs earlier (Fig.
2A). In JU77 increases in caspase 3/7 activity was demonstrated
at low concentrations of cisplatin (1–10 μg/ml) after 24 h
of exposure (Fig. 2A).
Interestingly, the most pronounced upregulation in caspase activity
was seen in LO68 cells. In contrast, overall caspase 3/7 activation
in ONE58 cells was only seen at 10 μg/ml at 24 h and in no
other sample.
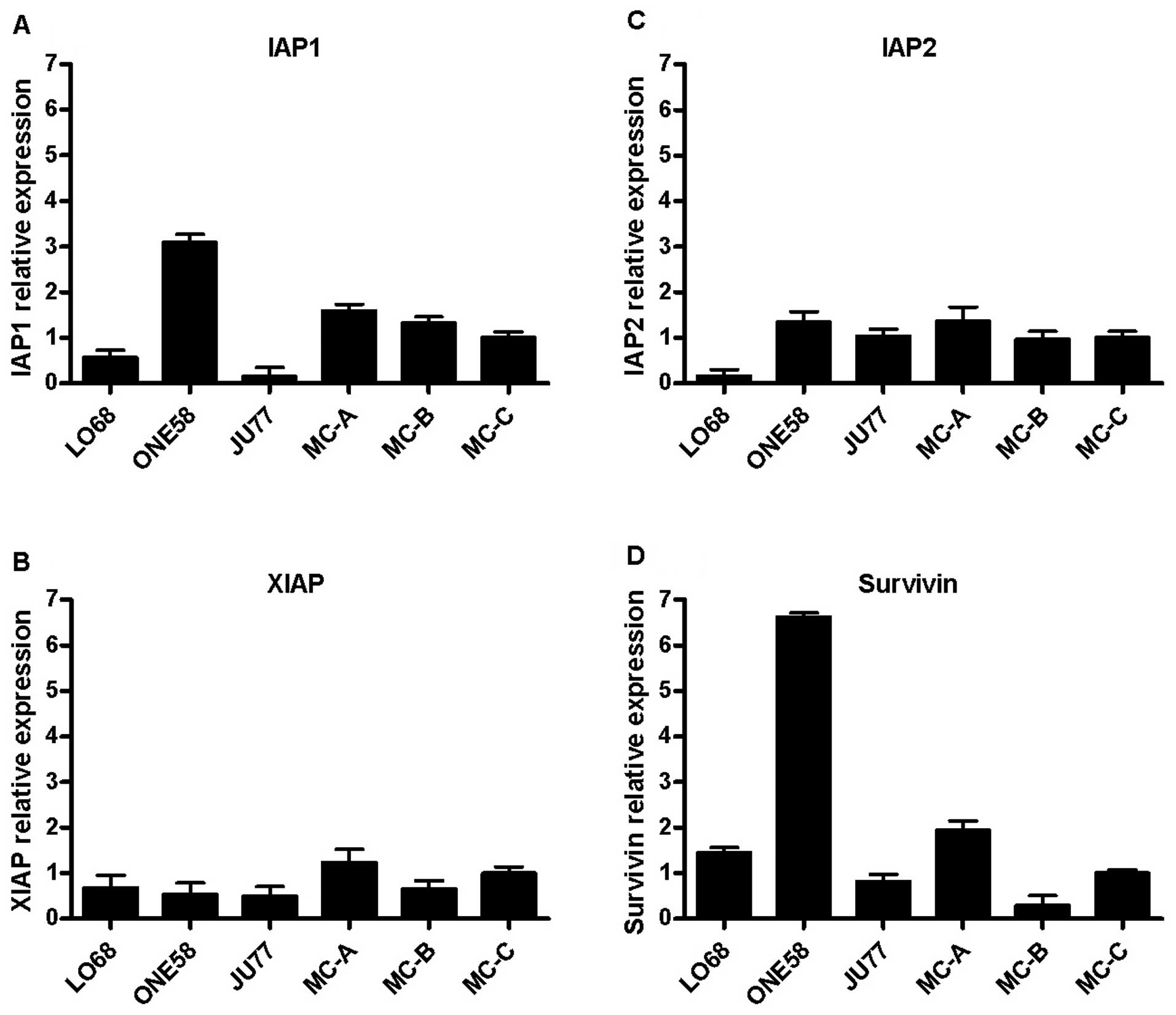
We next investigated the expression of IAPs in these
cells since reports have indicated overexpression of some of these
molecules in MM (5,6) with implications for apoptosis
resistance. Using conventional PCR we found that IAP-1, IAP-2,
XIAP, and survivin mRNAs were expressed in all MM cell lines (data
not shown) and we went on to examine differential expression of
these genes in MM cells and primary mesothelial cell cultures
(Fig. 3). We found that gene
expression of both IAP1 and survivin were upregulated in ONE58
cells (Fig. 3A and D) which may
account for the lack of caspase activation in these cells but that
otherwise there was no clear pattern of expression difference
between the MM cell lines and mesothelial cultures. Since some
authors (13,14) have reported that cisplatin can
downregulate IAP expression in tumour cells we also examined the
mRNA levels of IAPs by real-time PCR following 3, 6 and 24 h of
treatment with 10 μg/ml cisplatin but we did not see any
significant changes in IAP expression in MM cells (data not shown).
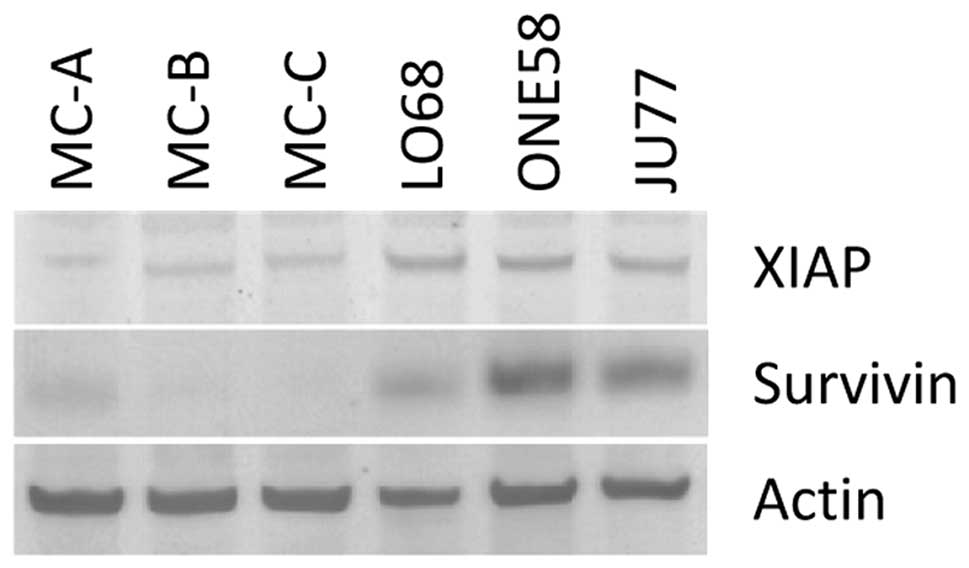
To further investigate IAP expression in these cells we performed
immunoblotting studies of XIAP and survivin protein expression
(Fig. 4) which demonstrated higher
levels of both these proteins in the MM tumour cell lines than in
primary cells. Given the results of the mRNA analysis of these
genes (Fig. 3B and D) this
suggests a level of translational regulation of these
molecules.
Silencing of XIAP and survivin in
mesothelioma cells
Since immunoblotting studies indicated differential
expression of both XIAP and survivin protein in mesothelioma cell
lines (Fig. 4) we further
investigated the role of these proteins in resistance to cisplatin
using RNAi mediated knockdown. Previously, in our laboratory
(unpublished data) we observed that mRNA knockdown by RNAi may not
correlate with protein. Therefore, we employed cell based ELISA
assays for XIAP and survivin to optimise for RNAi mediated
knockdown at the protein level.
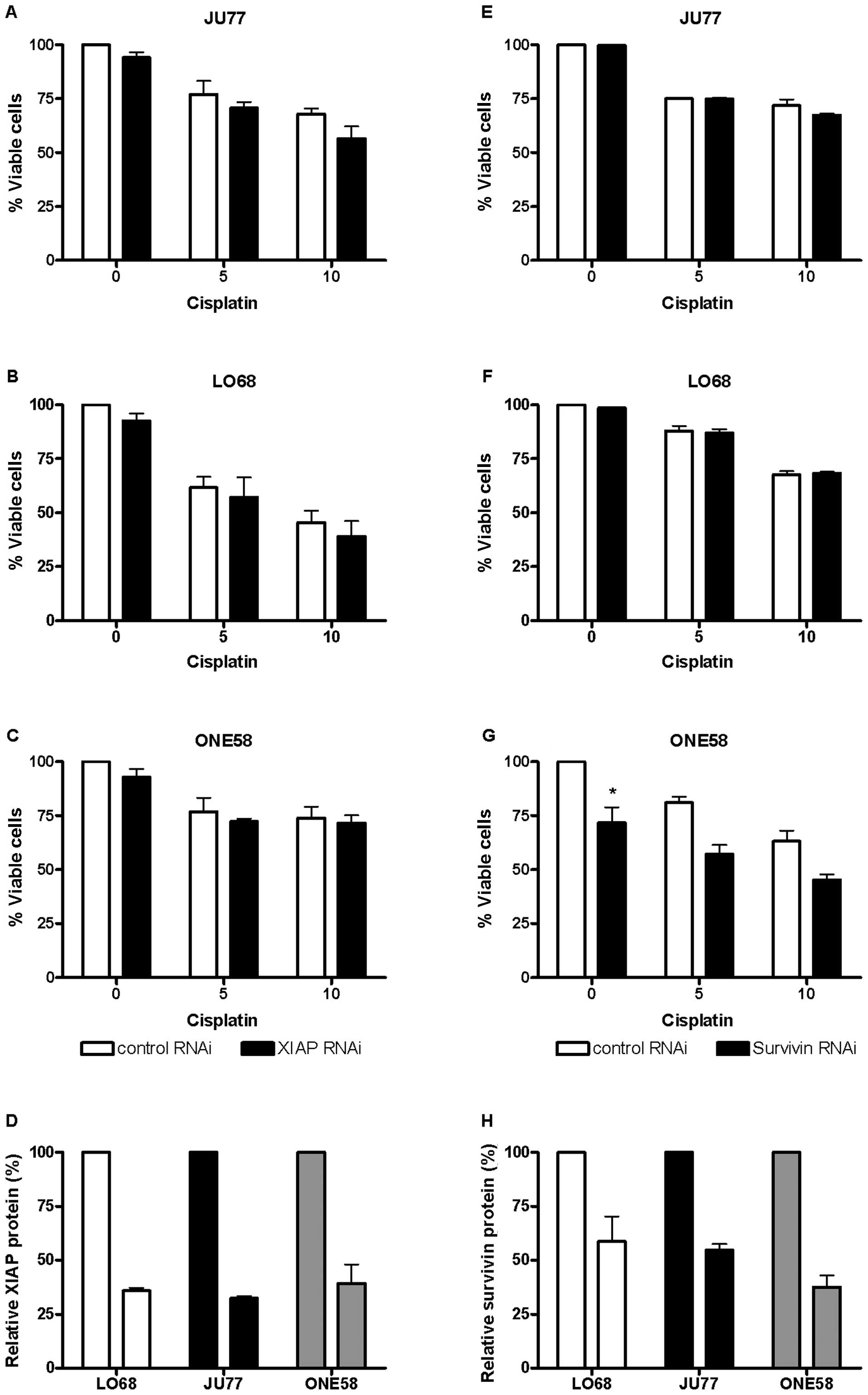
In all three cell lines we were able to establish
conditions which achieved optimal knockdown to at least 40% of XIAP
(Fig. 5D) 48–72 h after
transfection. Experiments in JU77, LO68 and ONE58 showed that
knockdown of XIAP had little effect upon the sensitivity of these
cell lines to cisplatin (Fig.
5A–C). Subsequently a similar series of experiments was
conducted using RNAi mediated knockdown of survivin. It proved more
difficult in these cells to knock down survivin at the protein
level (Fig. 5H) than XIAP,
although the effect in ONE58 cells (the highest expressing) was
comparable. Similarly the sensitivity of JU77 and LO68 cells
(Fig. 5E and F) to cisplatin was
not affected by survivin knockdown. In ONE58 cells survivin protein
downregulation had a significant effect upon baseline cell
viability (Fig. 5G) although this
was independent of cisplatin treatment.
Caspase independence of cisplatin-induced
cell death in mesothelioma cells
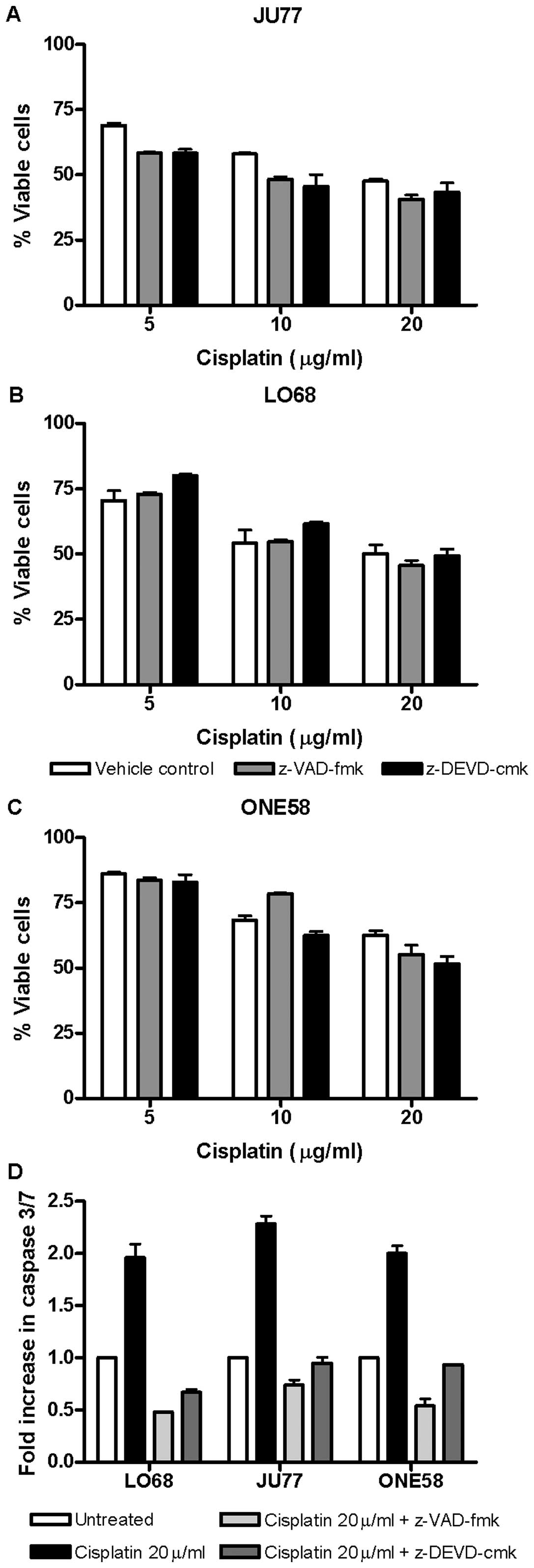
Given the lack of effect of knockdown of XIAP and
survivin upon drug cytotoxicity we hypothesised that caspase
activation may be unnecessary for the effect of cisplatin in these
cells. Therefore, we investigated the effect of both the
pan-caspase inhibitor z-VAD-fmk and the caspase 3/7 inhibitor
z-DEVD-cmk upon these cells. Both of these inhibitors effectively
blocked caspase 3/7 activation by cisplatin (Fig. 6D). Neither broad nor the more
narrow spectrum caspase inhibition significantly influenced
cisplatin induced cytotoxicity in mesothelioma cells (Fig. 6A–C). These results suggest that in
these cell lines caspase activation is not essential for cell death
due to cisplatin and most likely caspase-independent pathways
predominate.
Discussion
In the present study, we have characterised the
cisplatin chemosensitivity of malignant mesothelioma cell lines
(JU77, LO68 and ONE58), and investigated in detail the mechanisms
of cell death induced by this chemotherapeutic which is commonly
employed in the clinical treatment of MM. Conventionally the best
understood pathway of cisplatin toxicity is activation of DNA
damage signalling pathways which trigger mitochondrial apoptosis
(15). Our initial experiments
established the cytotoxic effect and relative sensitivity of 3
mesothelioma cells lines to cisplatin. Examination of biochemical
mechanisms induced by cisplatin demonstrated changes generally
associated with apoptotic cell death including mitochondrial
depolarisation and characteristic membrane changes. The active
involvement of caspases is considered one of the hallmarks of
cytotoxic drug-induced apoptosis. We focussed upon the executioner
caspase 3/7 and found activation in both JU77 and LO68 but much
less in ONE58 (the most resistant cell line).
We next investigated basal expression of IAPs in
these cells given the purported role of these molecules in
resistance to drug induced cytotoxicity (10). Survivin mRNA was upregulated in
ONE58 consistent with the low caspase activation in these cells.
However, apart from this we did not see the difference in XIAP and
survivin expression that was expected from the literature (5–7).
While upregulation of basal IAPs have been reported in numerous
studies few have examined changes in IAP gene expression in
response to cisplatin treatment. We did not find regulation of IAP
gene expression in response to cisplatin treatment in the present
study. Results of other studies have provided conflicting evidence
for the regulation of IAPs in response to chemotherapeutic drugs
including cisplatin (13,14,16).
Our results are consistent with this being a cell type- and
drug-dependent mechanism which does not operate downstream of
cisplatin in mesothelioma cells.
Surprisingly, immunoblotting revealed differential
expression of both XIAP and survivin at the protein level which was
not seen in mRNA, with the exception of survivin in ONE58 cells.
While many studies have focussed upon differences in mRNA levels of
these proteins there is also evidence for significant regulation of
both at the protein level (10).
Both XIAP (17) and survivin
(18) have been shown as regulated
through mechanisms controlling the proteasome degradation pathway.
In addition there is evidence for other posttranslational
mechanisms functioning to regulate these proteins (reviewed in
refs. 19 and 20).
Our results for protein expression were consistent
with the few studies which have investigated IAP molecules in
clinical samples of mesothelioma (6,7).
However, a role for these proteins in resistance to cisplatin was
not borne out by our subsequent experiments. The XIAP protein
knockdown achieved by us was at a level consistent with similar
studies in other cell types which showed effects upon viability and
drug sensitization (21,22). Our data indicate that inhibition of
caspase activation by XIAP was not likely to be important in the
resistance of mesothelioma cell lines to cisplatin.
The frequent overexpression of survivin in cancer is
well recognised and often associated with resistance to therapy
poor prognosis and an aggressive progression (10), therapies targeting this molecule
are in various stages of development (23). We achieved a 40–60% knockdown of
this protein, however, as with XIAP, there was no effect upon
cisplatin sensitivity. Most studies have merely assessed cell
viability directly following survivin knockdown rather than in
conjunction with drug treatment and we did observe a significant
effect upon cell viability in ONE58 cells. Targeting survivin by
RNAi in other cell types combined with drugs has shown conflicting
results with sensitization (24)
or no effect (25). Several
previous studies have examined the functional effects of inhibition
of survivin in mesothelioma cell lines finding enhanced caspase
activation (7,26) and apoptotic morphology (7) in response to cisplatin. In the
present study we used a panel of three distinct mesothelioma cell
lines and assessed viability along with other apoptotic mechanisms,
caspase activation was observed (unpublished data) but this did not
affect cell viability.
In addition to more recently described mechanisms,
inhibition of caspase activation either directly by XIAP or
indirectly by survivin remains an important function of these
proteins (10). We speculated that
the absence of effect by RNAi in our experiments may relate to
caspase activation. Our finding that cisplatin induced cell death
in mesothelioma cells did not require caspase activation confirmed
this. To our knowledge this is the first time that
caspase-independent pathways have been identified as prominent in
mesothelioma cells response to chemotherapy.
Classical apoptosis pathways involving caspase
activation are the most widely described cisplatin induced
mechanisms (15) although there is
a growing body of evidence for caspase independent pathways
(27–30). These pathways are increasingly of
interest because they represent a level of redundancy in cell death
signalling which might be therapeutically exploited in tumours
which are inherently resistant to apoptosis (31). Notable in the present study was the
commonality of the findings regarding caspase independence in all
three cell lines under investigation. Current studies in our
laboratory are directed at elucidating the alternate pathways
operating in these cells.
Acknowledgements
I.L.C was recipient of a University of
Western Australia International Postgraduate Research Scholarship.
The authors would like to thank Erin Bolitho for technical
assistance. We are grateful to Steven Mutsaers for the provision of
mesothelial cell protein and RNA.
References
|
1.
|
Spugnini EP, Bosari S, Citro G, Lorenzon
I, Cognetti F and Baldi A: Human malignant mesothelioma: molecular
mechanisms of pathogenesis and progression. Int J Biochem Cell
Biol. 38:2000–2004. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Ray M and Kindler HL: Malignant pleural
mesothelioma: an update on biomarkers and treatment. Chest.
136:888–896. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Rabik CA and Dolan ME: Molecular
mechanisms of resistance and toxicity associated with platinating
agents. Cancer Treat Rev. 33:9–23. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Koberle B, Tomicic MT, Usanova S and Kaina
B: Cisplatin resistance: preclinical findings and clinical
implications. Biochim Biophys Acta. 1806:172–182. 2010.PubMed/NCBI
|
|
5.
|
Davidson B: Expression of
cancer-associated molecules in malignant mesothelioma. Biomark
Insights. 2:173–184. 2007.PubMed/NCBI
|
|
6.
|
Kleinberg L, Lie AK, Florenes VA, Nesland
JM and Davidson B: Expression of inhibitor-of-apoptosis protein
family members in malignant mesothelioma. Hum Pathol. 38:986–994.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Zaffaroni N, Costa A, Pennati M, et al:
Survivin is highly expressed and promotes cell survival in
malignant peritoneal mesothelioma. Cell Oncol. 29:453–466.
2007.PubMed/NCBI
|
|
8.
|
Fox SA, Kusmiaty, Loh SS, Dharmarajan AM
and Garlepp MJ: Cisplatin and TNF-alpha downregulate transcription
of Bcl-xL in murine malignant mesothelioma cells. Biochem Biophys
Res Commun. 337:983–991. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Manning LS, Whitaker D, Murch AR, et al:
Establishment and characterization of five human malignant
mesothelioma cell lines derived from pleural effusions. Int J
Cancer. 47:285–290. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Altieri DC: Survivin and IAP proteins in
cell-death mechanisms. Biochem J. 430:199–205. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Rozen S and Skaletsky H: Primer3 on the
WWW for general users and for biologist programmers. Methods Mol
Biol. 132:365–386. 2000.PubMed/NCBI
|
|
12.
|
Vandesompele J, De Preter K, Pattyn F, et
al: Accurate normalization of real-time quantitative RT-PCR data by
geometric averaging of multiple internal control genes. Genome
Biol. 3:Research 0034. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Matsumiya T, Imaizumi T, Yoshida H, Kimura
H and Satoh K: Cisplatin inhibits the expression of
X-chromosome-linked inhibitor of apoptosis protein in an oral
carcinoma cell line. Oral Oncol. 37:296–300. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Nomura T, Mimata H, Yamasaki M and Nomura
Y: Cisplatin inhibits the expression of X-linked inhibitor of
apoptosis protein in human LNCaP cells. Urol Oncol. 22:453–460.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Siddik ZH: Cisplatin: mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Andjilani M, Droz JP, Benahmed M and
Tabone E: Down-regulation of FAK and IAPs by laminin during
cisplatin-induced apoptosis in testicular germ cell tumors. Int J
Oncol. 28:535–542. 2006.PubMed/NCBI
|
|
17.
|
Golovine K, Makhov P, Uzzo RG, et al:
Cadmium down-regulates expression of XIAP at the
post-transcriptional level in prostate cancer cells through an
NF-kappaB-independent, proteasome-mediated mechanism. Mol Cancer.
9:1832010. View Article : Google Scholar
|
|
18.
|
Zhao J, Tenev T, Martins LM, Downward J
and Lemoine NR: The ubiquitin-proteasome pathway regulates survivin
degradation in a cell cycle-dependent manner. J Cell Sci. 113(Pt
23): 4363–4371. 2000.PubMed/NCBI
|
|
19.
|
Altieri DC: New wirings in the survivin
networks. Oncogene. 27:6276–6284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Holcik M: Translational upregulation of
the X-linked inhibitor of apoptosis. Ann N Y Acad Sci.
1010:249–258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Li Y, Jian Z, Xia K, et al: XIAP is
related to the chemoresistance and inhibited its expression by RNA
interference sensitize pancreatic carcinoma cells to
chemotherapeutics. Pancreas. 32:288–296. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Zhang Y, Wang Y, Gao W, et al: Transfer of
siRNA against XIAP induces apoptosis and reduces tumor cells growth
potential in human breast cancer in vitro and in vivo. Breast
Cancer Res Treat. 96:267–277. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Mita AC, Mita MM, Nawrocki ST and Giles
FJ: Survivin: key regulator of mitosis and apoptosis and novel
target for cancer therapeutics. Clin Cancer Res. 14:5000–5005.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Liu WS, Yan HJ, Qin RY, et al: siRNA
directed against survivin enhances pancreatic cancer cell
gemcitabine chemosensitivity. Dig Dis Sci. 54:89–96. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Yamaguchi Y, Shiraki K, Fuke H, et al:
Targeting of X-linked inhibitor of apoptosis protein or survivin by
short interfering RNAs sensitize hepatoma cells to TNF-related
apoptosis-inducing ligand- and chemotherapeutic agent-induced cell
death. Oncol Rep. 14:1311–1316. 2005.
|
|
26.
|
Hopkins-Donaldson S, Belyanskaya LL,
Simoes-Wust AP, et al: p53-induced apoptosis occurs in the absence
of p14(ARF) in malignant pleural mesothelioma. Neoplasia.
8:551–559. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Cummings BS, Kinsey GR, Bolchoz LJ and
Schnellmann RG: Identification of caspase-independent apoptosis in
epithelial and cancer cells. J Pharmacol Exp Ther. 310:126–134.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Kim R, Emi M and Tanabe K:
Caspase-dependent and -independent cell death pathways after DNA
damage (Review). Oncol Rep. 14:595–599. 2005.PubMed/NCBI
|
|
29.
|
Liu L, Xing D and Chen WR: Micro-calpain
regulates caspase-dependent and apoptosis inducing factor-mediated
caspase-independent apoptotic pathways in cisplatin-induced
apoptosis. Int J Cancer. 125:2757–2766. 2009. View Article : Google Scholar
|
|
30.
|
Lock EA, Reed CJ, Kinsey GR and
Schnellmann RG: Caspase-dependent and -independent induction of
phosphatidylserine externalization during apoptosis in human renal
carcinoma Cak(1)-1 and A-498 cells. Toxicology. 229:79–90. 2007.
View Article : Google Scholar
|
|
31.
|
Delavallee L, Cabon L, Galan-Malo P,
Lorenzo HK and Susin SA: AIF-mediated caspase-independent
necroptosis: a new chance for targeted therapeutics. IUBMB Life.
63:221–232. 2011. View
Article : Google Scholar : PubMed/NCBI
|