Introduction
Colon cancer is one of the most common cancers and
is the third leading cause of cancer-related mortality worldwide
(1). Up to now, the most efficient
treatment of colon cancer is the radical surgery combined with
chemotherapy, for local tumors, the 5-year survival rate is
approximately 93% (2), while
almost half of patients had developed metastasis on admission
(3), even the localized disease,
more than one-third of patients develop recurrence after surgery
(4). Despite advances in surgical
techniques and other treatments, even after the novel molecular
agents in recent years, colon cancer high mortality rate remains
because of the frequent recurrence and metastasis (5–7).
Currently, the mechanisms underlying the preocess of colon
tumorigenesis remain obscure, understanding the related mechanism
is important for us to develop new stratege and find new efficient
target for efficient treatment of colon cancer.
MicroRNAs (miRNAs) are a class of 19–24 nt short
non-coding RNAs, which regulate translation and degraduation of
target mRNAs by direct interactions with 3′-untranslated region
(3′UTR) of the target mRNAs. Approximately 1,500 miRNA sequences
have been found (8) and it was
reported that >60% of genes are conserved targets of microRNA in
the human genome (9). It has been
demonstrated that miRNAs can regulate the expression of a wide rage
of genes, and play important roles in cell progression such as
development (10), differentiation
(11), proliferation (12,13)
and apoptosis (14,15). Increasing evidence has suggested
that miRNAs play a crucial role in carcinogenesis and tumor
progression (16,17), and many miRNAs were found
upregulated or downregulated in various tumor types (18,19).
MicroRNA-31 (miR-31) plays important roles in
malignant tumors, dysregulation of it has been demonstrated to be
involved in many tumors, for example, it was upregulated in head
and neck cancer, hepato cellular carcinoma, lung cancer and colon
cancer (20–24). Many reports have revealed that
miR-31 was correlated to the ability of cancer metastasis.
Inhibition of miR-31 contributed to induction of metastasis in
breast cancer (25).
Interestingly, Laurila et al found both inhibition and
enhanced expression of miR-31 lead to reduced migration and
invasion of pancreatic cancer cells (26). In colon cancer, miR-31 was reported
to be significantly upregulated, and the expression level of which
was correlated with the stage of colon cancer (20,21)
and miR-31 was also found to be associated with colon cancer
invasion and metastasis, supression of miR-31 affected migration
and invasion of colon cancer cells (27). It was also found that for colon
cancer at early stage, supression of miR-31 could increase
sensitivity to 5-FU (28).
RhoBTB proteins belong to a subfamily of the Rho
family of small GTPases. The RhoBTB subfamily comprises three
members, RhoTBT1, RhoTBT2 and RhoTBT3. RhoTBT2 was first proposed
as a tumor supressor, it has been reported that in breast cancer,
lack of RHOBTB2 transcripts results in growth inhibition (29). Studies also found high rates of
loss of heterozygosity at the RHOTBT2 locus in gastric tumors and
bladder tumors (30,31). Similarly to RhoTBT2, recently
RhoTBT1 was also proposed as a tumor supressor in a study on head
and neck cancer (32).
Athough miR-31 was demonstrated to be envolved in
colon cancer, its mechanisms are still not clear. In this study, we
found miR-31 was upregulated in colon cancer, and demonstrated for
the first time that RhoBTB1 was a target of miR-31. We also
explored the role of miR-31 in colon and revealed that miR-31 could
promote colon tumorigenesis and invasiveness, at least partly
through targeting RhoBTB1.
Meterials and methods
Tumor samples
Colon cacer tissues and mathed adjacent normal
tissues were collected from 17 colon cancer patients. All carcinoma
samples were obtained at the time of operation and obtained with
the informed consent of the patients. All human materials were used
in accordance with the policies of the institutional review board
at China-Japan Union Hospital of Jilin University, China.
Cell culture and transfection
The HT29 and SW480 cell lines were from American
Type Culture Collection (ATCC) and cultured under conditions
provided by the manufacturer. Twenty-four hours before transfection
cells were seeded, then miR-31 antisense locked nucleic acid (LNA
anti-miR31) and control oligonucleotides (LAN control) or RhoBTB1
siRNA and control siRNA were transfected with Lipofectamine 2000
(Invitrogen, Carlsbad, CA) according to the manufacturer’s
instructions. Twenty-four or forty-eight hours later, cells were
collected and subjected to further analysis. The human RhoBTB1
siRNA and control siRNA were purchased from Origene.
RNA extraction and real-time RT-PCR
Total RNA was extracted from cells and tumor tissues
by using TRIzol (Invitrogen). For miR-31 detection, stem-loop
RT-PCR assay was performed. U6 snRNA was used as an internal
control. The following stem-loop primers were used for miRNA-31:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAGCTAT-3′ and U6
snRNA:
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATATGGAAC-3′. For
RhoBTB1 mRNA detection, M-MLV Reverse Transcriptase system
(Promega) was used to reverse-transcribe RNA into cDNA. The qRT-PCR
was carried out using the SYBR Green qPCR Master Mix (Tiangen) on
ABI 7300 (Applied Biosystems, Foster, CA). Glyceraldehyde
3-phosphate dehydrogenase was used as an internal control. The
primers used for detection of miR-31 and U6 were, forward:
5′-GGAGAGGCAAGATGCTGGCA-3′; U6-forward:
5′-CGCAAGGATGACACGCAAATTC-3′, and a universal downstream reverse
primer, 5′-GTGCAGGGTCCGAGGT-3′. The primers used for detection of
RhoBTB1, forward: 5′-GGAGTGAAGGAGCCTGTGAG-3′; reverse:
5′-TGCCAATGAACCCCTTACTC-3′. Internal control GAPDH primers for
qRT-PCR, forward: 5′-ACGGATTTGGTCGTATTGGG-3′, reverse:
5′-TGATTTTGGAGGGATCTCGC-3′.
Luciferase reporter assays
Twenty-four hours before transfection HT29 cells
were seeded into a 24-well plate (2.5–3×104/well), then
the cells were cotransfected with Renilla luciferase and luciferase
reporter plasmids containing miR-31 or vector control and wild-type
or mutated target gene 3′UTR with Lipofectamine 2000 (Invitrogen).
Forty-eight hours after transfection, luciferase activities were
measured using dual-luciferase reporter assay system (Promega).
Firefly luciferase activities were normalized to Renilla
luciferase.
Western blotting
After transfection as indicated, cells were
harvested and lysed in RIPA lysis buffer [150 mM NaCl, 10 mM Tris,
pH 7.5, 1% NP40, 1% deoxycholate, 0.1% SDS, protease inhibitor
cocktail (Sigma)] and total protein was measured using the Bradford
protein assay (Bio-Rad, Hercules, CA). Equal amounts of protein
sample was subjected to 10% SDS-PAGE and transferred to PVDF
membrane (Millipore, Billerica, MA, USA), after blocking in 5%
skimmed milk for 30 min at room temperature, the membrane was
incubated in anti-RhoBTB1 (1:2000, Abcam) for 2 h at room
temperature, followed by incubating with horseradish
peroxidase-linked secondary antibody for 1 h at room temerature and
visualized with ECL (Millipore). β-actin (1:5000, Abcam, Inc.,
Cambridge, MA) was used as loading control.
Immunohistochemistry (ICH) and in situ
hybridization
Tumor or matched normal tissues were fixed with 4%
paraformaldehyde, then the samples were embeded in paraffin and
sectioned. Followed incubated with RhoBTB1 antibody (1:500), the
bound antibodies were detected with the
biotin-streptavidinperoxidase system (Vector Labs, Burlingame, CA,
USA) using diaminobenzidine (Sigma-Aldrich) as chromogen.
For in situ hybridization assay, DIG-labeled
locked nucleic acid (LNA)-based probe specific for miR-31 (Exiqon,
Vedbaek, Denmark) was introduced. The in situ hybridization
signal was detected by incubation with horseradish peroxidase
(HRP)-conjugated anti-digoxigenin (1:200, Abcam Inc., Cambridge,
MA).
MTT
Cell proliferation rates were measured using
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
reduction assay according to the manufacturer’s instructios. In
brief, HT29 cells were seeded into 96-well plates 24 h before
transfection and treated as described previously. Then MTT solution
was added in each well, and the cells were cultured in incubator
for another 4 h. The medium was discarded and 100 μl of DMSO
was added to each cell, after shaking for 15 min the absorbance at
595 nm was measured with microplate reader (Bio-Rad).
Soft agar colony formation assay
Pre-warmed 1 ml of 2X RPMI-1640 medium with 20%
fetal calf serum (FBS) and 1 ml of 1.2% Bacto-agar (Difco) solution
were mixed and transferred onto a 3-cm dish, and then incubated at
4°C for 30 min to allow the bottom agar layer to solidify. Next,
mixed 0.5 ml melted 0.6% agar at 50°C with equal volume of 2X
medium and stored in 40°C water bath, then 1×103 cells
were added and mixed quickly and poured onto the bottom agar. Cells
were left to grown for 2 weeks in the incubator, then cell colonies
were observed under a microscope, and colonies of >50 cells were
counted.
Statistical analysis
Data are presented as mean ± SEM. Student’s t-test
(two-tailed) was used to compare two groups (P<0.05 was
considered significant) unless otherwise indicated (χ2
test).
Results
miR-31 expression is upregulated in human
colon cancer tissues and cell lines
It has been reported by many studies that miR-31 was
dysregulated in various cancers, so we examined miR-31 expression
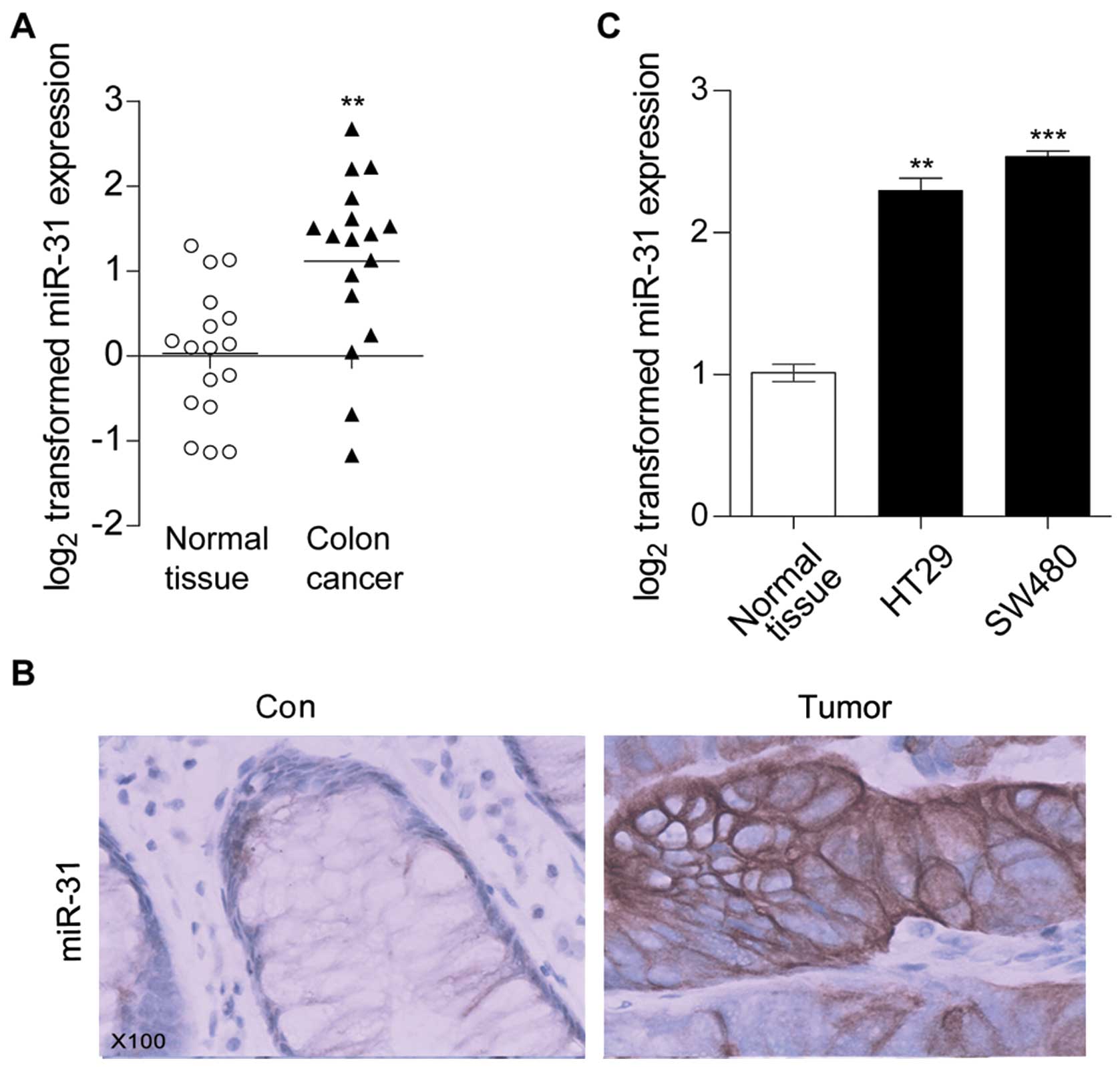
level in human colon cancer tissues by using qRT-PCR. As showed in
Fig. 1A, compared with the normal
colon tissues miR-31 was significantly increased in tumor tissues.
ISH assay results further confirmed that miR-31 was induced in
colon cancer tissues (Fig. 1B).
Moreover, we detected miR-31 expression levels in colon cancer cell
lines HT29 and SW480. qRT-PCR results showed that compared with
normal colon tissues, miR-31 was also increased in HT29 and SW480
(Fig. 1C), this was in accordance
with the previous studies (19–21).
These results suggested that miR-31 might be involved in colon
tumor progression.
Repression of miR-31 affects human colon
cancer cell viability and anchorage-independent colony
formation
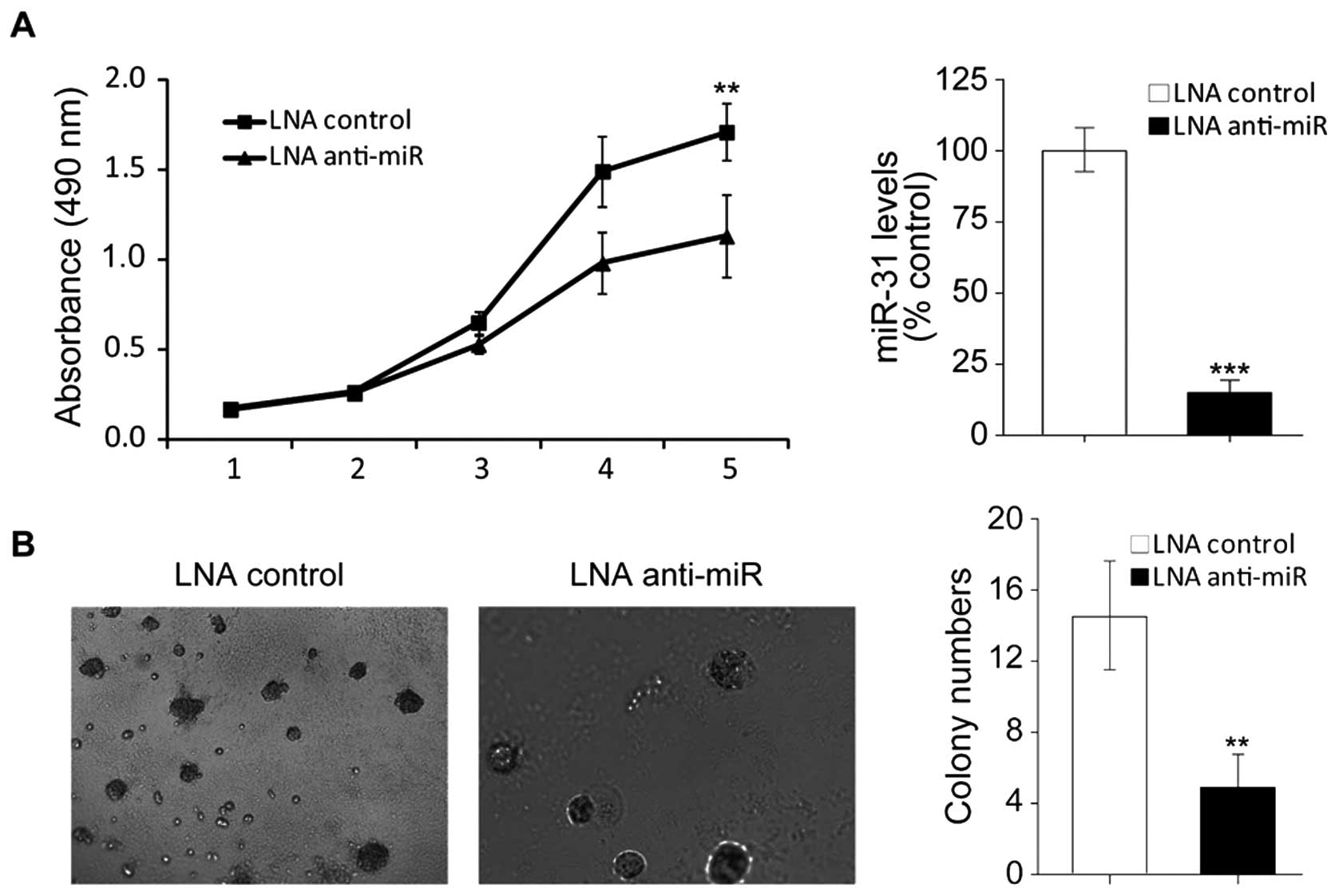
To examine the role of miR-31 in colon cancer
development, we inhibited miR-31 in HT29 cells by using locked
nucleic acid-modified anti-miR31 antisense (LNA anti-miR31). As
showed in Fig. 2A (right), after
tranfected with LNA anti-miR31, miR31 was effectively inhibited in
HT29 cells. MTT assays showed that inhibition of miR-31 reduced
viability of HT29 cells (Fig. 2A,
left), which indicates that upregulation of miR-31 might attribute
to colon tumorigenesis.
Anchorage-independence of cell growth abitity is a
property of cancer cells. To verify the involvement of miR-31 in
colon tumorigenesis we next performed soft agarose assays. The
results showed, after knockdown of miR-31 colony formation in soft
agarose was obviously supressed (Fig.
2B). Collectively the results demostrated that repression of
miR-31 inhibited human colon cancer cell viability and
anchorage-independent colony formation, which further confirmed
that miR-31 plays an important role in colon cancer
development.
miR-31 targets RhoBTB1
Since miRNAs regulate translation and degraduation
of target mRNAs by direct interactions with 3′UTR of the target
mRNAs, we investigated the targets of miR-31 to further uncover the
mechanism underlying its tumor promoting effect on colon
tumorigenesis. By using online miRNA target prediction databases
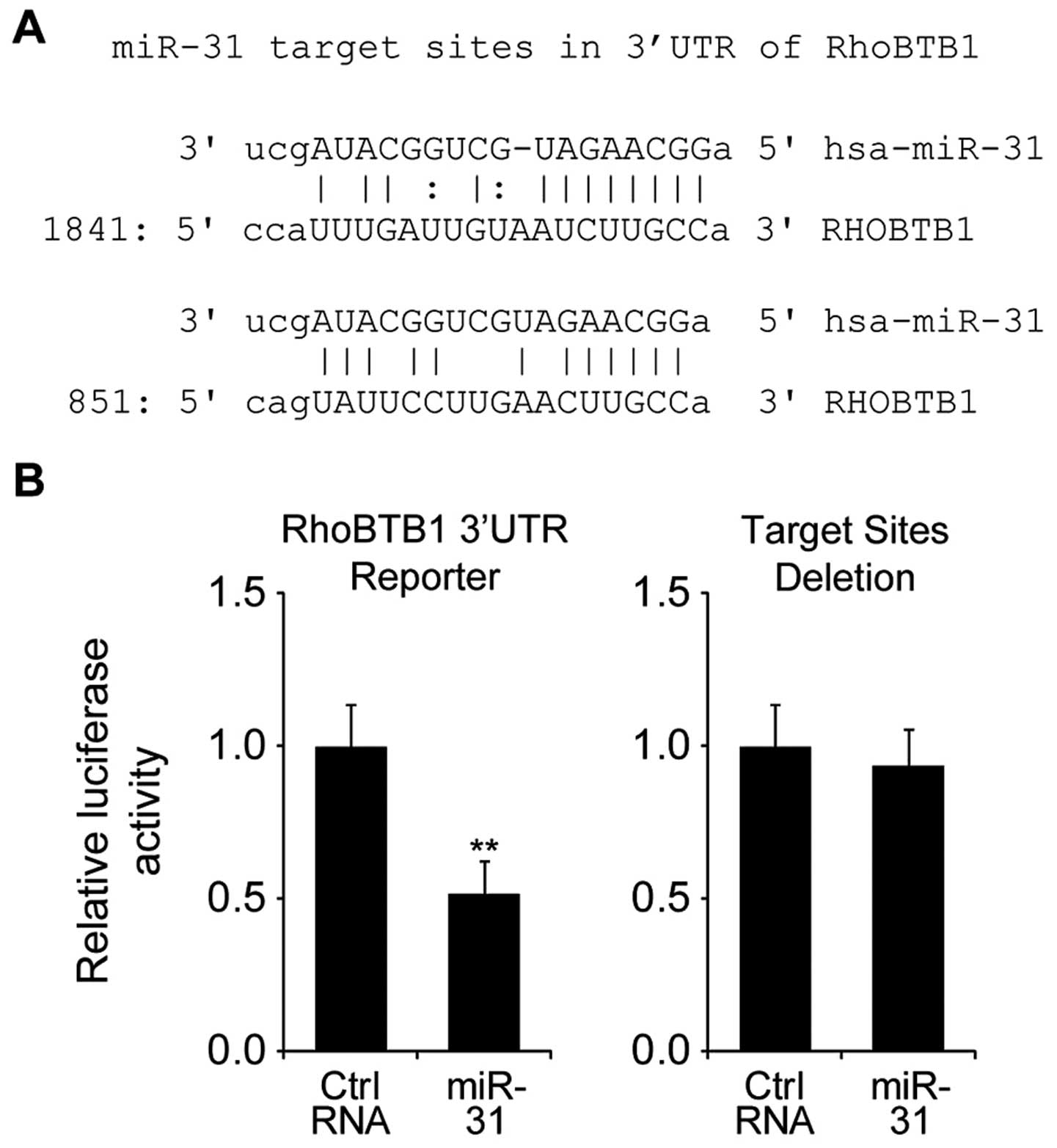
(http://www.targetscan.org), we
hypothesized that RhoBTB1 is a target of miR-31 (Fig. 3A). To investigate whether RhoBTB1
was directly targeted by miR-31, we performed luciferase reporter
gene assay in HT29 cells.
Firstly, we constructed luciferase reporter plasmids
containing RhoBTB1 3′UTR sequence (wt-RhoBTB1) or with deleted
putative miR-31 target sites (mut-RhoBTB1). As shown in Fig. 3B, after co-tranfected with
wt-RhoBTB1 plasmid, the luciferase activity of miR-31 transfected
cells was significantly reduced compared to control miR tranfected
cells, while co-tranfected with mut-RhoBTB1 plasmid did not change
the luciferase activity. Taken together, these results indicated
that miR-31 directly regulated RhoBTB1 by targeting its 3′UTR.
miR-31 downregulates RhoBTB1 expression
in HT29 cells
RhoBTB family belongs to the Rho family of small
GTPases, which comprises three members, of which RhoBTB1 and
RhoBTB2 have been proposed as tumor suppressors. Here we found that
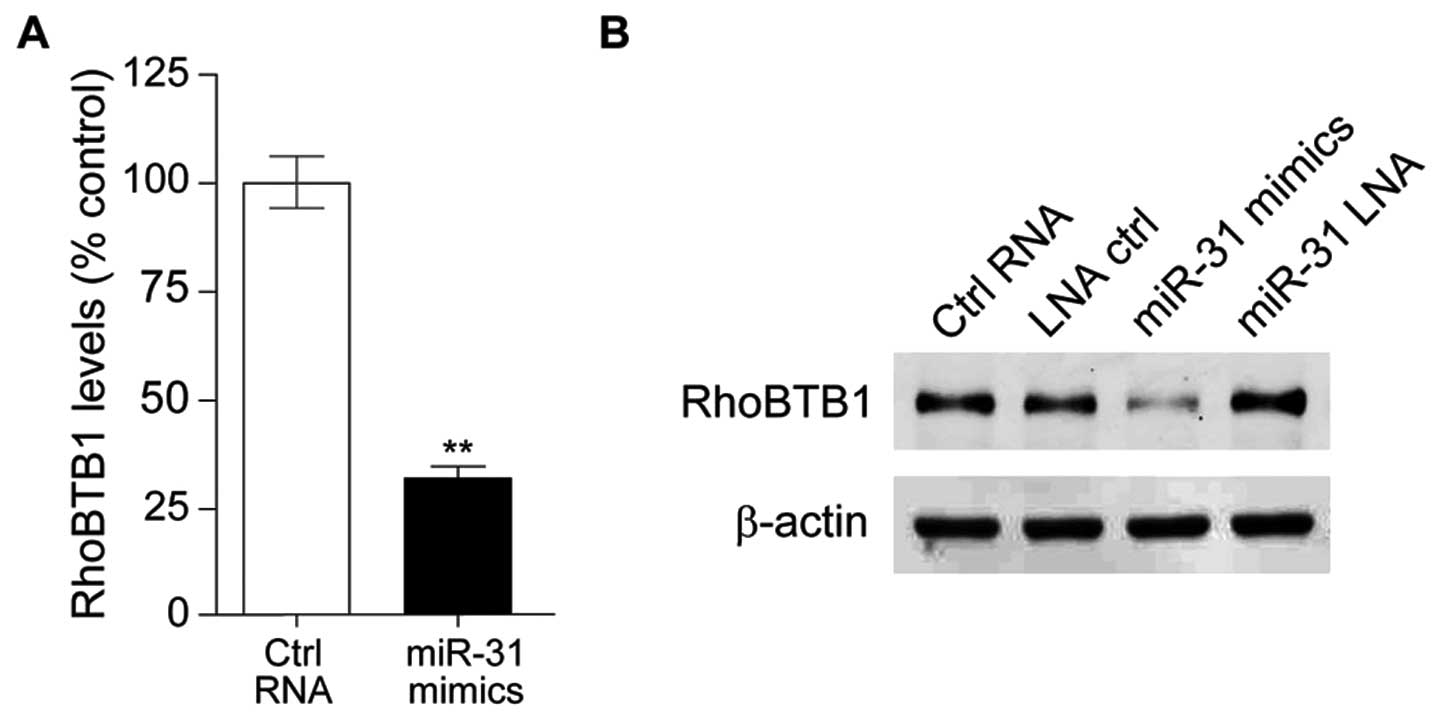
RhoBTB1 was a target of miR-31 in the colon cancer cell line HT29,
then we investigated the effect of miR-31 on RhoBTB1. Compared with
HT29 transfected with control RNA, RhoBTB1 mRNA level was
significantly downregulated in miR-31 mimic-tansfected HT29 cells
(Fig. 4A), and western blotting
results showed RhoBTB1 protein expression level was also
downregulated with miR-31 overexpression (Fig. 4B). These observations suggested
that miR-31 upregulation in colon tumor might depress RhoBTB1.
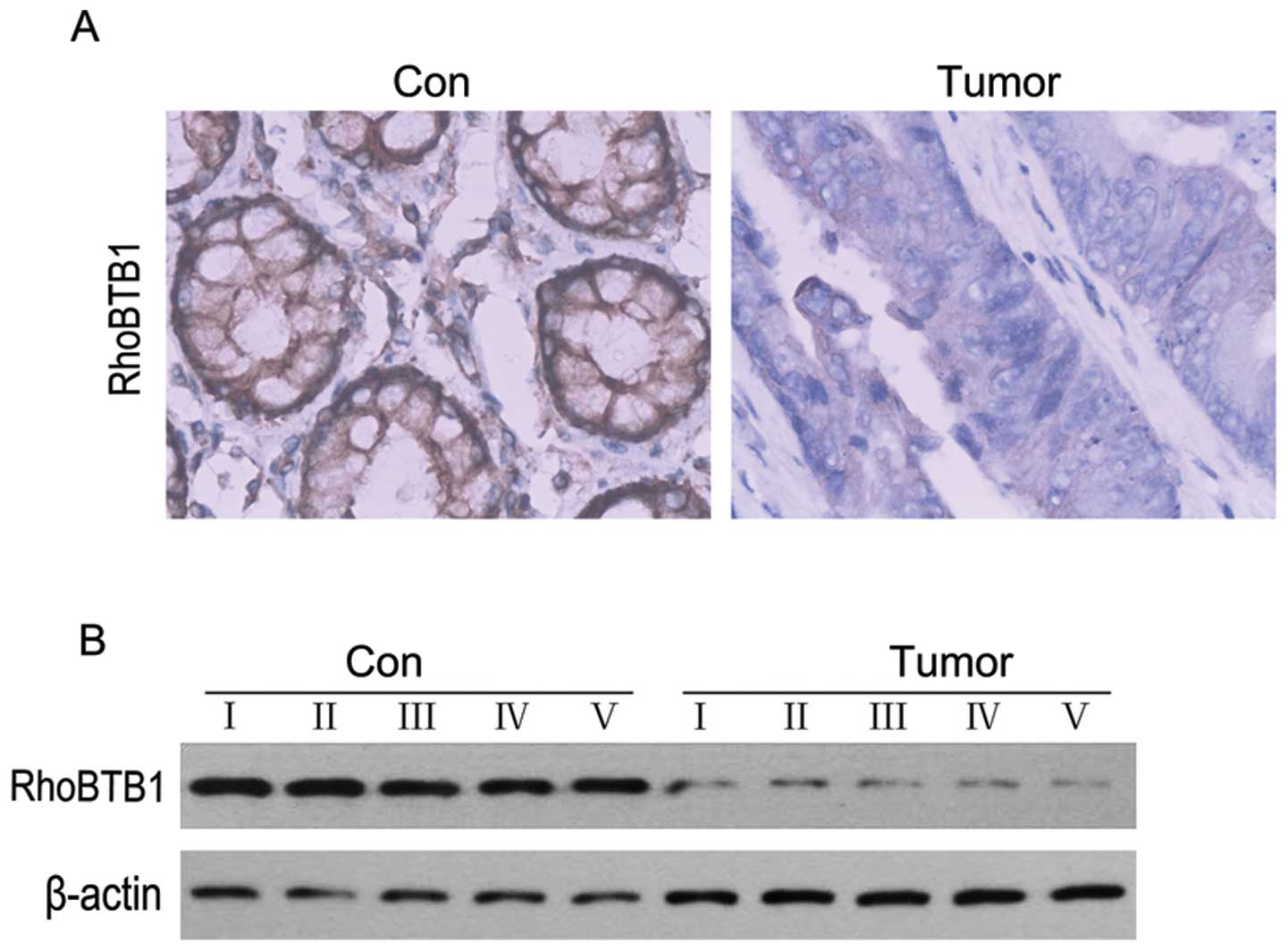
RhoBTB1 expression is downregulated in
human colon cancer tissues
As mentioned above, RhoTBT1 was proposed as a tumor
supressor (32), and we also found
that upregulation of miR-31 in colon cancer cell line HT29 could
depressed RhoBTB1, then we performed ICH and western blotting
experiments to examine the expression level of RhoBTB1 in colon
cancer tissues. As showed in Fig.
5, RhoBTB1 was clearly decreased compared to normal colon
tissues. These results suggested that the reduction of RhoBTB1
might attribute to colon tumorigenesis.
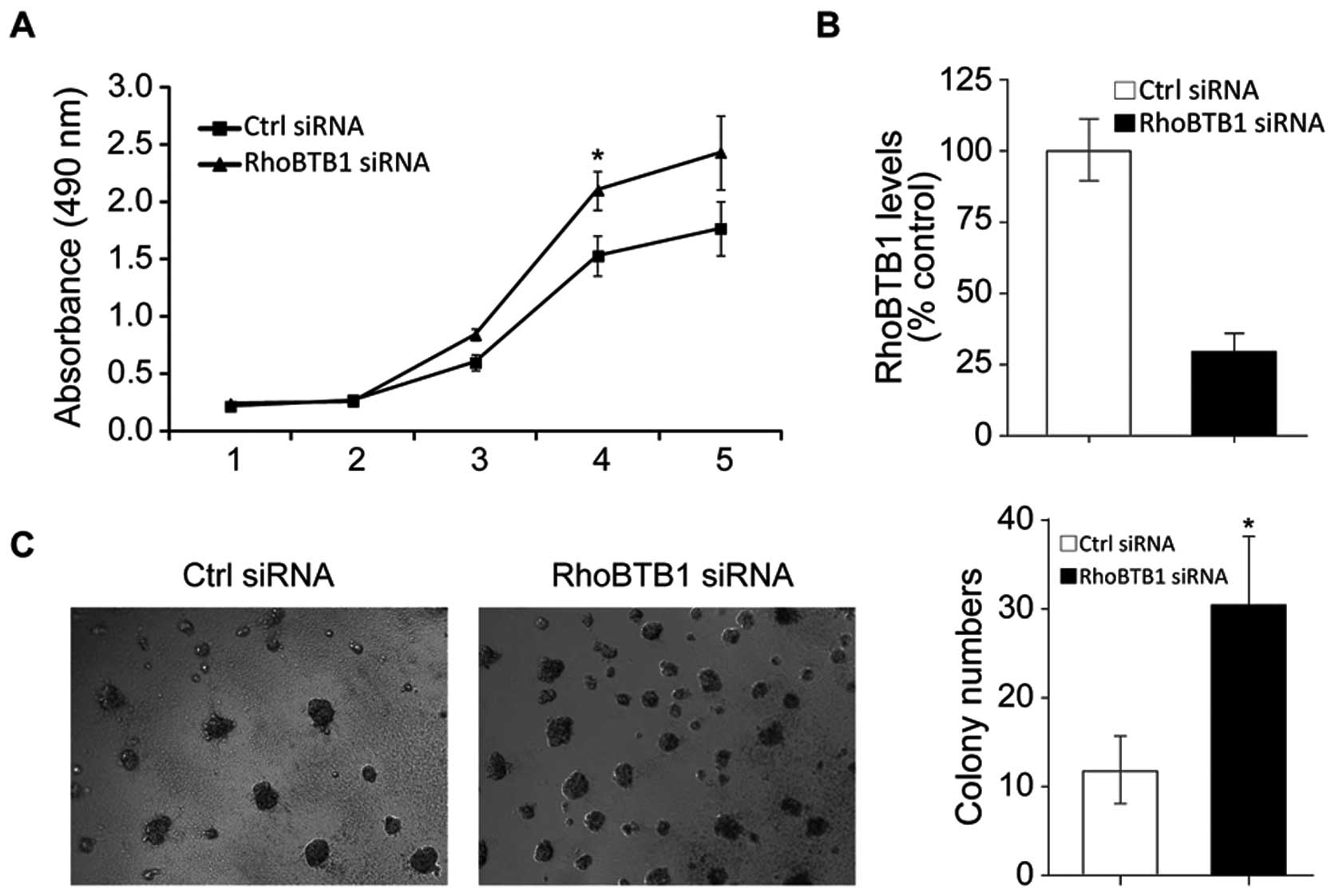
Inhibition of RhoBTB1 is responsible for
the tumor promoting effects of miR-31
To further confirm that miR-31-induced colon tumor
progression is mediated by RhoBTB1, we knockdown RhoBTB1 in HT29
cells by RNAi. As shown in Fig.
6B, RhoBTB1 mRNA was effectively inhibited after RhoBTB1 siRNA
transfected, and MTT assay results showed HT29 proliferation was
induced with suppression of RhoBTB1. Soft agarose colony forming
assay results showed inhibition of RhoBTB1 promoted colon cancer
cell clonal growth. These data proved that the miR-31 promoted
tumor development at least partly through suppressing tumor
supressor RhoBTB1.
Discussion
miRNAs are endogenously expressed small non-coding
RNAs, which control gene expression by targeting messenger RNAs. It
has been estimated that in human, genes code for miRNAs account for
approximately 3%, which may regulate 30% of the protein-coding
genes (33,34). Convincing evidence has shown that
miRNAs are often dysregulated in human cancers and play pivotal
roles in tumorigenesis and progression (35,36).
miR-31 was found to be aberrant in many cancers,
including colon cancer (20–26).
In this study, we demostrated that miR-31 was significantly
upregulated in human colon cancer. As shown in Fig. 1, compared to normal colon tissues,
miR-31 was increased in colon cancer tissues and colon cancer cell
line HT29 and SW480. ISH assays further confirmed that miR-31 was
overexpressed in colon cancer tissues. The results were in
consistent with the previous studies. miR-31 has been reported to
be associated with metastasis in many tumor types. For example,
miR-31 was markedly upregulated in oral squamous cell carcinoma
(OSCC) and mediated oral oncogenesis (37), and it has been reported that miR-31
promotes lung tumorigenesis by targeting tumor supressors LATS2 and
PPP2R2A (23). To examine effect
of miR-31 depression on colon cancer growth and proliferation, we
used LNA anti-miR31 to knockdown miR-31 in HT29 cells. MTT assay
results showed HT29 cells proliferation was increased after LAN
miR-31 transfection (Fig. 2A).
With knockdown of miR-31, HT29 clonal formation ability was
inhibited (Fig. 2B). The results
above suggested that miR-31 acts primarily as an oncogene in colon
cancer. Since miRNAs affect cellular activities by targeting their
downstream targets, we next looked for the targets of miR-31.
In order to identify miR-31 target genes, we used
TargetScan software, the results showed that tumor supressor
RhoBTB1 might be a target of miR-31. RhoTBT1 belongs to RhoBTB
subfamily. RhoBTB proteins are atypical members of Rho family of
small GTPases. Rho GTPases act as molecular switches in regulating
actin filament system, and are considered as major regulators of
cytoskeletal remodeling, while increasing evidence points to
participation also in the regulation of cell cycle progression and
apoptosis, and Rho GTPases were also reported to be involved in
tumor development (38–40).
The RhoBTB subfamily was identified during the study
of the genes encoding Rho-related proteins in the lower eukaryote
Dictyostelium discoideum in 2001 (41), thus it was the most recent addition
to the Rho family. Unlike other typical RhoGTPases, RhoBTB proteins
do not act as a main regulator in actin filament system. RhoBTB2
and RhoBTB1, have been proposed as tumor supressors. RhoBTB2 was
the first to be proposed as a candidate tumor suppressor being
identified as the gene homozygously deleted at 8p21 in breast
cancer and resulted in cell growth inhibition. RhoBTB1, which is
located at the 10q21 region, was suggested to have a role in head
an neck squamous cell carcinomas (HNSCC) development. Levent and
coworkers found that in 52 HNSCC, 42% showed LOH at 10q23.1, and in
5 tumors decreased expression of RhoBTB1 was accompanied by
LOH.
Although it has been demostrated that RhoBTB1 was
involved in tumorigenesis, scarce knowledge exists on its role in
colon cancer, so we examined the level of RhoBTB1 protein
expression in colon cancer tissues. ICH results showed that,
compared to normal colon tissues, RhoBTB1 was dramatically
decreased in colon cancer (Fig.
5). To further examine whether the depressed RhoBTB1 was
responsible for the tumor promoting effects of miR-31, RhoBTB1 was
silenced by RNAi, as indicated in Fig.
6A, supression of RhoTBT1 in HT29 inhibited cell proliferation,
which mimics the function of miR-31. The knockdown of RhoTBT1 also
promoted HT29 cells clonal growth.
In conclusion, our study suggests high levels of
miR-31 are involved in colon cancer development, and we discovered
tumor supressor RhoBTB1 as a new target of miR-31. Over-expression
of miR-31 promotes tumor proliferation through depressing the tumor
supressor RhoBTB1. These observations shed new light on mechanisms
underlying development of colon cancer and supply potential novel
therapeutic targets in inhibiting colon tumorigenesis.
Acknowledgements
This study was supported by grants
from Jilin Provincial Sciecce and Technology Department (no.
20070727-01) and the International Partnership.
References
|
1.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2.
|
Colorectal Cancer Survival by Stage:
National Cancer Intelligence Network. 2009
|
|
3.
|
Figer A, Perez-Staub N, Carola E, et al:
FOLFOX in patients aged between 76 and 80 years with metastatic
colorectal cancer: an exploratory cohort of the OPTIMOX1 study.
Cancer. 110:2666–2671. 2007.PubMed/NCBI
|
|
4.
|
Becker H: Surgery of colorectal carcinoma.
Praxis. 84:1371–1372. 1995.
|
|
5.
|
Jemal A, Siegel R, Ward E, Murray T, Xu J,
Smigal C and Thun MJ: Cancer statistics, 2006. CA Cancer J Clin.
56:106–130. 2006. View Article : Google Scholar
|
|
6.
|
Tsai HL, Yeh YS, Yu FJ, Lu CY, Chen CF,
Chen CW, Chang YT and Wang JY: Predicting factors of postoperative
relapse in T2-4N0M0 colorectal cancer patients via harvesting a
minimum of 12 lymph nodes. Int J Colorectal Dis. 24:177–183. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Longo WE and Johnson FE: The preoperative
assessment and postoperative surveillance of patients with colon
and rectal cancer. Surg Clin North Am. 82:1091–1108. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Baraniskin A, Birkenkamp-Demtroder K,
Maghnouj A, et al: MiR-30a-5p suppresses tumor growth in colon
carcinoma by targeting DTL. Carcinogenesis. 33:732–739. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Chen CZ, Li L, Lodish HF and Bartel DP:
MicroRNAs modulate hematopoietic lineage differentiation. Science.
303:83–86. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Lai EC: Micro RNAs are complementary to
3′UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002.
|
|
13.
|
Engels BM and Hutvagner G: Principles and
effects of microRNA-mediated post-transcriptional gene regulation.
Oncogene. 25:6163–6169. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Calin GA, Ferracin M, Cimmino A, et al: A
MicroRNA signature associated with prognosis and progression in
chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar
|
|
17.
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
18.
|
Slaby O, Svoboda M, Michalek J and Vyzula
R: MicroRNAs in colorectal cancer: translation of molecular biology
into clinical application. Mol Cancer. 8:1022009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Schetter AJ and Harris CC: Alterations of
microRNAs contribute to colon carcinogenesis. Semin Oncol.
38:734–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Bandres E, Cubedo E, Agirre X, et al:
Identification by real-time PCR of 13 mature microRNAs
differentially expressed in colorectal cancer and non-tumoral
tissues. Mol Cancer. 5:292006. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Slaby O, Svoboda M, Fabian P, Smerdova T,
Knoflickova D, Bednarikova M, Nenutil R and Vyzula R: Altered
expression of miR-21, miR-31, miR-143 and miR-145 is related to
clinicopathologic features of colorectal cancer. Oncology.
72:397–402. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Wong QW, Lung RW, Law PT, Lai PB, Chan KY,
To KF and Wong N: MicroRNA-223 is commonly repressed in
hepatocellular carcinoma and potentiates expression of Stathmin1.
Gastroenterology. 135:257–269. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Liu X, Sempere LF, Ouyang H, et al:
MicroRNA-31 functions as an oncogenic microRNA in mouse and human
lung cancer cells by repressing specific tumor suppressors. J Clin
Invest. 120:1298–1309. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Liu X, Chen Z, Yu J, Xia J and Zhou X:
MicroRNA profiling and head and neck cancer. Comp Funct Genomics.
8375142009.PubMed/NCBI
|
|
25.
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szasz AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Laurila EM, Sandström S, Rantanen LM,
Autio R and Kallioniemi A: Both inhibition and enhanced expression
of miR-31 lead to reduced migration and invasion of pancreatic
cancer cells. Genes Chromosomes Cancer. 51:557–568. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Cottonham CL, Kaneko S and Xu L: miR-21
and miR-31 converge on TIAM 1 to regulate migration and invasion of
colon carcinoma cells. J Biol Chem. 285:35293–35302. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Wang CJ, Stratmann J, Zhou ZG and Sun XF:
Suppression of microRNA-31 increases sensitivity to 5-FU at an
early stage, and affects cell migration and invasion in HCT-116
colon cancer cells. BMC Cancer. 10:6162010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Hamaguchi M, Meth JL, von Klitzing C, et
al: DBC2, a candidate for a tumor suppressor gene involved in
breast cancer. Proc Natl Acad Sci USA. 99:13647–13652. 2002.
View Article : Google Scholar
|
|
30.
|
Knowles MA, Aveyard JS, Taylor CF, Harnden
P and Bass S: Mutation analysis of the 8p cand idate tumour
suppressor genes DBC2 (RHOBT B2) and LZTS1 in bladder cancer.
Cancer Lett. 225:121–130. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Cho YG, Choi BJ, Kim CJ, Song JH, Zhang C,
Nam SW, Lee JY and Park WS: Genetic analysis of the DBC2 gene in
gastric cancer. Acta Oncol. 47:366–371. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Beder LB, Gunduz M, Ouchida M, et al:
Identification of a candidate tumor suppressor gene RHOBTB1 located
at a novel allelic loss region 10q21 in head and neck cancer. J
Cancer Res Clin Oncol. 132:19–27. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Hwang HW and Mendel JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Cho WC: MicroRNAs in cancer from research
to therapy. Biochim Biophys Acta. 1805:209–217. 2010.PubMed/NCBI
|
|
36.
|
Cho WC: MicroRNAs: potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Liu CJ, Tsai MM, Hung PS, et al: miR-31
ablates expression of the HIF regulatory factor FIH to activate the
HIF pathway in head and neck carcinoma. Cancer Res. 70:1635–1644.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Sahai E and Marshall CJ: RHO-GTPases and
cancer. Nat Rev Cancer. 2:133–142. 2002. View Article : Google Scholar
|
|
39.
|
Gómez del Pulgar T, Benitah SA, Valerón
PF, Espina C and Lacal JC: Rho GTPase expression in tumourigenesis:
evidence for a significant link. Bioessays. 27:602–613.
2005.PubMed/NCBI
|
|
40.
|
Ellenbroek SI and Collard JG: RhoGTPases:
functions and association with cancer. Clin Exp Metastasis.
24:657–672. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Rivero F, Dislich H, Glockner G and Noegel
AA: The Dictyostelium discoideum family of Rho-related proteins.
Nucleic Acids Res. 29:1068–1079. 2001. View Article : Google Scholar : PubMed/NCBI
|