Introduction
Lung cancer was the most commonly diagnosed cancer
as well as the leading cause of cancer death in males in 2008
globally (1). Male lung cancer
death rates are decreasing in most Western countries, including
many European countries, North America and Australia (2). In contrast, lung cancer rates are
increasing in countries such as China and several other countries
in Asia and Africa (3,4). Although chemotherapy and radiation
therapy have yielded modest improvements in patient outcomes,
overall survival of lung cancer patients remains poor (5,6).
Therefore, new therapeutic targets are urgently needed.
KISS1 was originally identified in melanoma by Lee
et al (7) in experiments
designed to identify the molecules responsible for the
anti-metastatic effect of human chromosome 6. The KISS1 gene
is located on chromosome 1 near q32.1 with regulatory elements
localized in chromosome 6 at 6q16.3-q23 (8). The KISS1 product is a 145 amino acid
peptide, known as kisspeptin, which is cleaved to smaller peptides,
including a 54 amino acid known as metastatin (8). Associations between loss of KISS1
expression and increased tumor progression and poor prognosis were
found in various solid tumors, such as pancreatic, breast, bladder,
brain, epithelial ovarian and gastric cancer (9–14).
KISS1 receptor (KISS1R, also named GPR54) coupled to kisspeptins,
has been revealed to play a pivotal role for the onset of puberty
and to suppress cancer metastasis (15–17).
Kisspeptins regulate cell proliferation, migration, and invasion in
different cell lines via KISS1R/GPR54 (18–20).
The role of GPR54 in cancer has been difficult to discern. A recent
study has shown that the expression of KISS1 and GPR54 correlates
with breast tumor progression and poor patient prognosis (10). Ikeguchi et al (21) reported that overexpression of KISS1
and GPR54 was correlated with the progression of HCC. Zhang et
al (22) and Hata et al
(13) surveyed RNA expression of
the KISS1 and GPR54 in ovarian cancer and observed a trend towards
favorable prognosis where KISS1/GPR54 RNA expression is
elevated.
To our knowledge, little information is known about
KISS1 and KISSR1 expression in the Chinese Han people with
non-small cell lung cancer (NSCLC). In this study, we investigated
the expression of KISS1 and KISSR1 in 56 cases of NSCLC to
determine the relationship between their expression levels and
survival of lung cancer patients.
Materials and methods
Subjects
A total of 56 patients with NSCLC were obtained from
Department of Thoracic Surgery, the First Affiliated Hospital of
China Medical University (January 2006 to December 2010).
Twenty-eight patients had stage IIIB NSCLC (locally advanced) and
other 28 patients had stage IV NSCLC (metastatic) according to the
International Association for the Study of Lung Cancer (IASLC)
staging committee (23). All
patients underwent standard laboratory tests (cytology and
histology), confirmed computerized tomography of the thorax. None
of the patients underwent radiotherapy or chemotherapy before
operation. The study was approved by our University Ethics
Committee and was conducted in accordance with the Helsinki
Declaration. All patients gave their written informed consent to
participate in the study.
RNA isolation and Reverse
transcriptase-polymerase chain reaction (RT-PCR)
Total RNA was isolated using an RNeasy mini kit
(Biomed; Beijing, China). First strand cDNA was reverse transcribed
with 1 μg of total RNA, using Takara Reverse Transcription
kit (Takara; Dalian, China) and oligo (dT) 15 primers (Takara). The
KISS1 primers were: 5′-TGAACTC ACTGGTTTCTTGGC-3′ (forward
primer) and 5′-CAGCCT GGCAGTAGCAGCT-3′ (reverse primer). The
KISS1R primers were: 5′-ATCTACGTCATCTGCCGCCAC-3′ (forward
primer) and 5′-TCACGTACCAGCGGTCCACAC-3′ (reverse primer). The
house-keeping gene, GAPDH was used as an internal control
for normalization of the results. The GAPDH primers were:
5′-AGAAGGCTGGGGCTCATTTG-3′ (forward primer) and 5′-AGG
GGCCATCCACAGTCTTC-3′ (reverse primer). PCR amplification of cDNA
was performed in 20 μl mixtures. Finally, amplicons were
electrophoresed in 2% agarose gel with ethidium bromide and
visualized under UV illumination.
Methylation-specific PCR (MSP)
Genomic DNA was extracted from lung cancer specimens
using a TissueGen DNA kit (CWbiotech; Beijing, China). Genomic DNA
(2 μg) was denatured with 0.2 M NaOH. Then, 10 mM
hydroquinone (Sigma-Aldrich, St. Louis, MO, USA) and 3 M
sodium-bisulfite (Sigma-Aldrich) were added. The solution was
incubated at 55°C for 16 h. DNA samples were then purified using a
WizardDNA purification resin (Promega; Madison, WI, USA). In this
procedure unmethylated (but not methylated) cytosines can convert
to uracil, which is then converted to thymidine during subsequent
PCR to give sequence differences between methylated and
unmethylated DNA. The modified DNA was used as a template both for
MSP and USP. The primer sequences for the methylated KISS1
gene were: 5′-CGGGTTGGAAGT TTTAGC-3′ (forward primer) and
5′-GCTTCGACAAACGA AAAAC-3′ (reverse primer), and for the
unmethylated allele were: 5′-TTTTGGGTTGGAAGTTTTAG-3′ (forward
primer) and 5′-ACTTCAACAAACAAAAAACAAC-3′ (reverse primer). The PCR
products were separated in 2% agarose gel with ethidium bromide and
visualized under UV illumination.
Western blot analysis
Tissues were lysed in lysis buffer (20 mM Tris-HCl,
150 mM NaCl, 2 mM EDTA, 1% Triton X-100) containing a protease
inhibitor cocktail (Sigma-Aldrich). Extract protein amounts were
quantified using the BCA protein assay kit (CWbiotech). Equivalent
amounts of protein (40 μg) were separated using 10% SDS-PAGE
and transferred to a PVDF membrane (Millipore Corporation;
Billerica, MA, USA). Western blot analysis was performed using
primary antibodies: KISS1 (sc-15400), KISS1R (sc-134499) and
β-actin (sc-130657, Santa Cruz Biotechnology; Santa Cruz, CA, USA).
Each specific antibody binding was detected with horseradish
peroxidase (HRP)-conjugated respective secondary antibodies
(Amersham Biosciences; Amersham, UK) and ECL solutions (Amersham
Biosciences).
Immunohistochemical staining for KISS1
and KISS1R
Formalin-fixed, paraffin-embedded tissue sections
were cut into 4 μm-thick sequential sections. After
deparaffinization and rehydration, sections were boiled in citrate
buffer (0.01 M, pH 6.0) for antigen retrieval. Sections were then
incubated with 3% H2O2 and 5% serum to block
endogenous peroxidase activity and non-specific binding. For KISS1
protein, sections were incubated with rabbit anti-human KISS1
polyclonal antibody (sc-101246). For the KISS1R protein, sections
were incubated with mouse anti-human KISS1R monoclonal antibody
(H-048-61, Phoenix Pharmaceuticals; Burlingame, CA, USA). The
sections were then incubated with biotinylated secondary antibodies
and visualized by DAB. Counterstaining was carried out with
hematoxylin. The sections were dehydrated in alcohol and
coverslipped. For the negative controls, PBS replaced the primary
antibody.
Blood samples and ELISA for plasma
metastin
Plasma levels of metastin were measured by ELISA, by
the methods of Katagiri et al (24). Baseline metastin serum levels were
measured in all patients. All blood samples were collected between
08.00 and 10.00 in the morning. Blood samples for metastin were
collected before surgery, placed in a chilled tube containing
aprotinin (500 KIU/ml) and EDTA (1.2 mg/ml), and immediately
centrifuged at 3,000 rpm for 20 min. Circulating serum metastin was
determined using a sandwich enzyme immunoassay (CSB-EL012373HU,
Cusabio Life Science, Wuhan, China) with a sensitivity of 0.078
ng/ml, while the variations of intra-assay precision (precision
within an assay) and inter-assay precision (precision between
assays) were less than 8 and 10%, respectively, according to the
manufacturer.
Cell cycle and apoptosis analysis
Cancer samples were subjected to chemical digestion
by incubating with 0.5% pepsin at 37°C in water bath for 30 min
with intermittent stirring. Disaggregated tissues were filtered
through a 50 μM nylon mesh. Cells were collected in PBS and
fixed on ice with 1% paraformaldehyde, followed by 70% cold ethanol
containing 10 μg/ml RNase. Then the cells were stained with
50 μg/ml propidium iodide (PI, KeyGen; Nanjing, China) for
15 min at room temperature for cell cycle analysis. The apoptotic
cells were detected with Annexin V-FITC/PI double staining.
Following the manufacturer’s instructions for the Apoptosis Assay
kit (KeyGen), the stained cells were analyzed by flow cytometry.
Data analysis was performed with CellQuest software (BD
Biosciences; Rockville, MD, USA).
Statistics and survival analysis
Association between KISS1 or KISS1R expression and
clinical covariates was assessed univariately. Overall survival
rates were determined using the Kaplan-Meier estimator.
Kaplan-Meier survival plots were generated and comparisons were
made with log-rank statistics. Cox proportional hazard model was
used to identify significant factors correlated with prognosis in
multivariate analysis. For all analyses, only P<0.05 were
considered significant. All the statistical analyses and graphics
were performed with GraphPad Prism 5.
Results
KISS1 protein expression relative to the
clinical and pathological variables
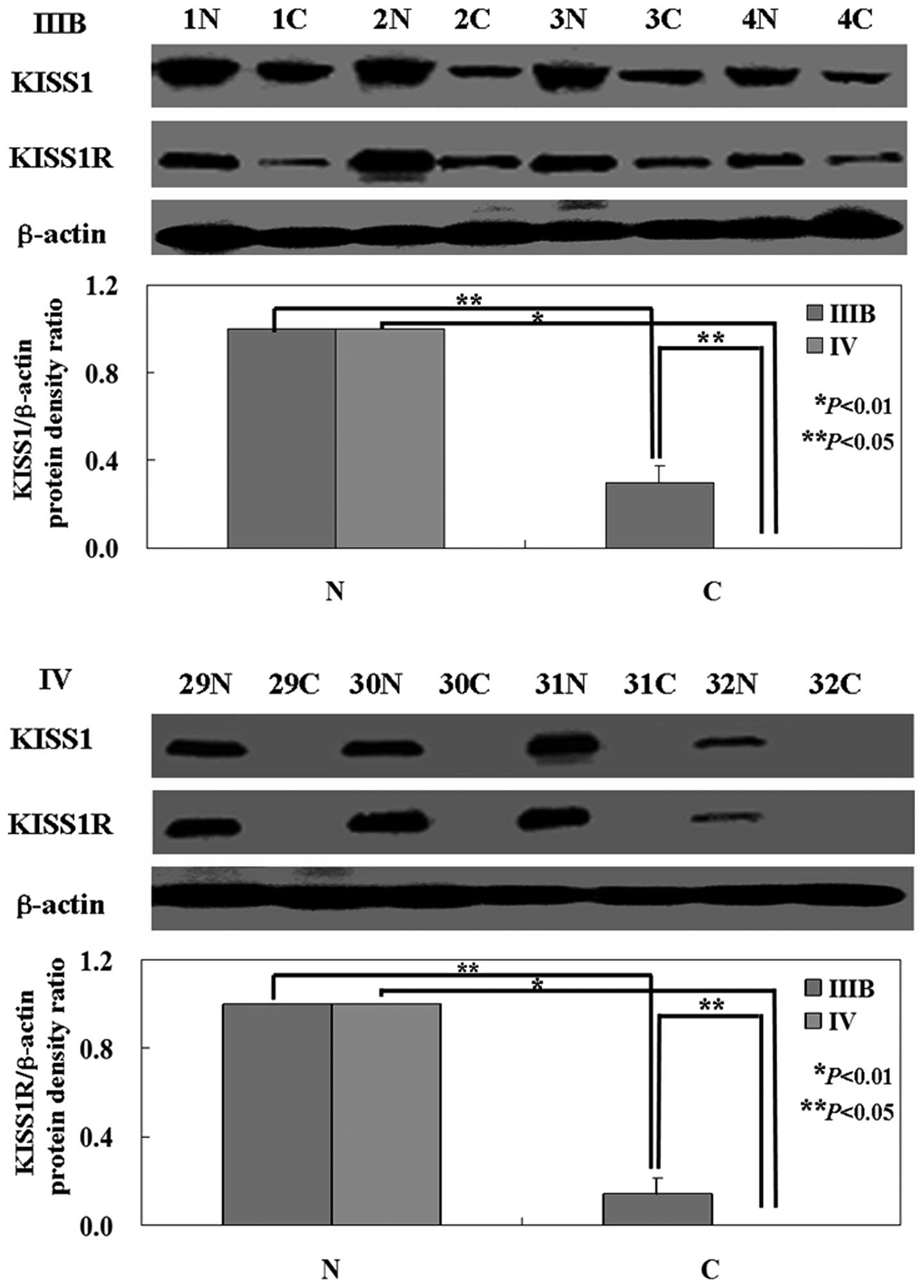
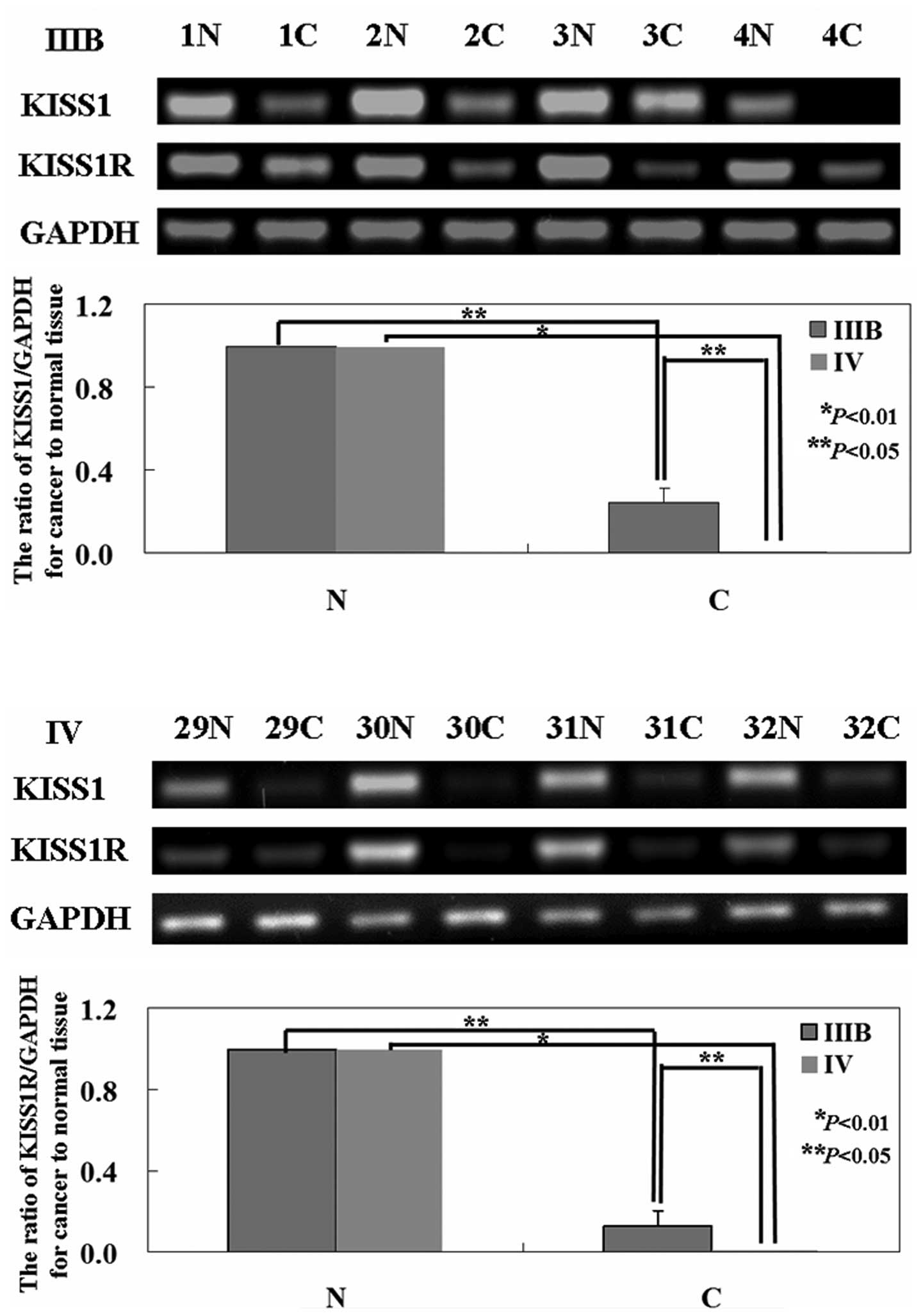
The levels of KISS1 mRNA and protein in tumor tissue
were both lower than that in normal tissue (P<0.05; Figs. 1 and 2) by using western blot analysis and
RT-PCR. Interesting, KISS1 expression was higher in the low stage
of NSCLC (IIIB) compared to advanced stage (IV) (P<0.05;
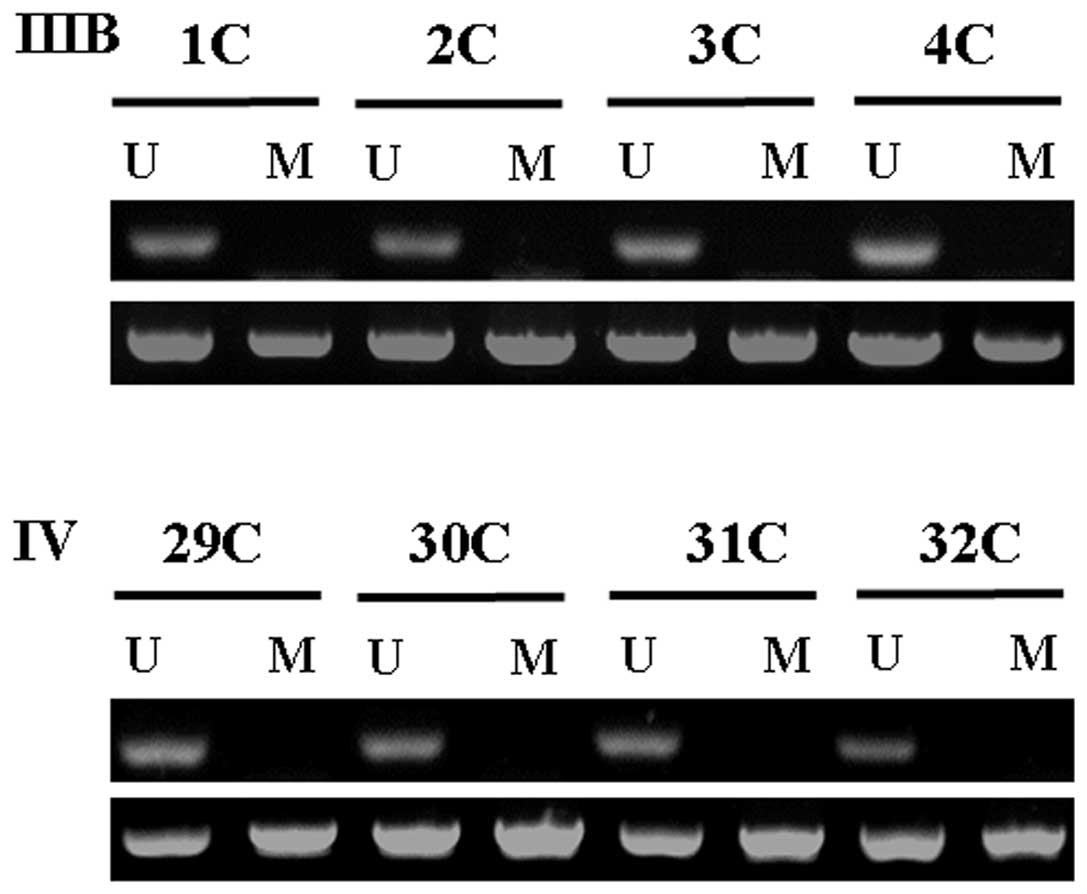
Figs. 1 and 2). Fifty-six tumor samples were examined
for KISS1 methylation by MSP. Representative examples are
illustrated in Fig. 3. We found a
correlation between CpG island KISS1 promoter methylation and
downregulated KISS1 mRNA levels in tumor samples. The
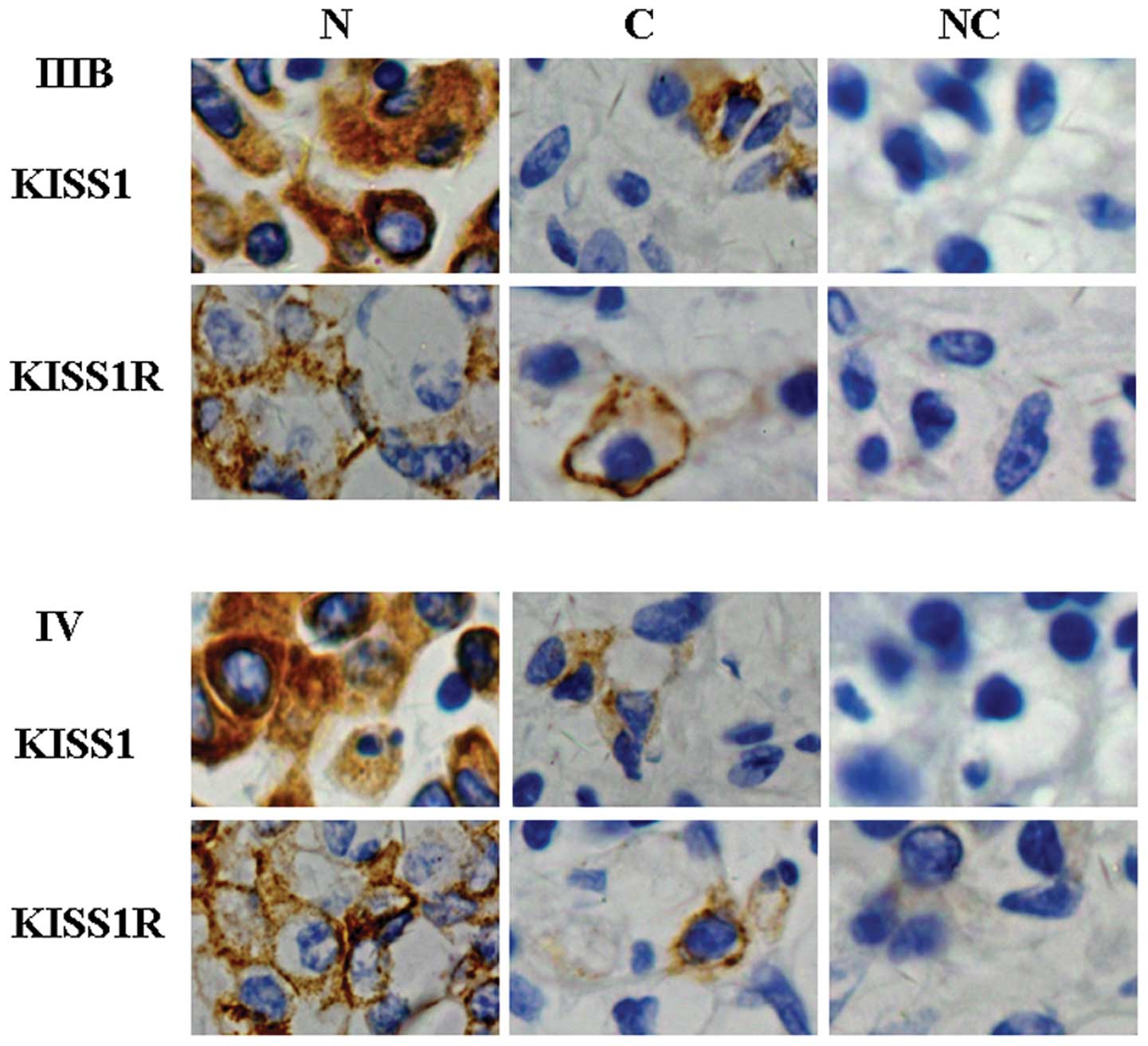
immunostaining results showed that KISS1 expression was distributed
in the cytoplasm of normal lung cells and lung cancer cells
(Fig. 4). Consistent with the
results of western blot analysis, the results of immunostaining
also showed that KISS1 protein was weakly expressed in lung cancer
specimens, but highly in normal parts of specimens. There was a
significant difference in KISS1 expression between the low stage of
NSCLC (IIIB) compared to advanced stage (IV). We then analyzed the
potential relationship between the expression of KISS1 and the
clinicopathological characteristics of these patients. The levels
of KISS1 protein had no relation to patient age or sex, tumor size,
lymphatic invasion, pN category, venous invasion or histological
type (P>0.05; Table I).
However, in addition, KISS1 expression was associated with the
differentiation of the patients with stage IV NSCLC (P<0.05;
Table I).
 | Table I.Relationship between KISS1 expression
and clinicopathological parameters of patients with stage IIIB and
IV NSCLC. |
Table I.
Relationship between KISS1 expression
and clinicopathological parameters of patients with stage IIIB and
IV NSCLC.
| Clinicopathological
features | KISS1 expression in
stage IIIB | KISS1 expression in
stage IV |
|---|
|
|
|---|
| n | − | + | PR (%) | χ2 | P-value | n | − | + | PR (%) | χ2 | P-value |
|---|
| Sex | | | | | 1.04 | 0.058 | | | | | 1.56 | 0.325 |
| Female | 8 | 5 | 3 | 37.5 | | | 10 | 7 | 3 | 30.0 | | |
| Male | 20 | 17 | 3 | 15.0 | | | 18 | 16 | 2 | 11.1 | | |
| Age (years) | | | | | 0.11 | 0.251 | | | | | 0.22 | 0.264 |
| <50 | 11 | 9 | 2 | 18.2 | | | 8 | 7 | 1 | 12.5 | | |
| ≥50 | 17 | 13 | 4 | 23.5 | | | 20 | 16 | 4 | 20.0 | | |
|
Differentiation | | | | | 0.84 | 0.162 | | | | | 5.38 | 0.027 |
| Well or
moderate | 9 | 8 | 1 | 11.1 | | | 6 | 3 | 3 | 50.0 | | |
| Poor | 19 | 14 | 5 | 26.3 | | | 22 | 20 | 2 | 9.1 | | |
| Lymphatic
invasion | | | | | 0.85 | 0.146 | | | | | 0.17 | 0.435 |
| − | 14 | 10 | 4 | 28.6 | | | 19 | 16 | 3 | 15.8 | | |
| + | 14 | 12 | 2 | 14.3 | | | 9 | 7 | 2 | 22.2 | | |
| Venous
invasion | | | | | 1.77 | 0.235 | | | | | 0.65 | 0.237 |
| − | 12 | 8 | 4 | 33.3 | | | 18 | 14 | 4 | 22.2 | | |
| + | 16 | 14 | 2 | 12.5 | | | 10 | 9 | 1 | 10.0 | | |
| Histological
type | | | | | 1.87 | 0.215 | | | | | 2.17 | 0.073 |
| Squamous
cell | 9 | 8 | 1 | 11.1 | | | 9 | 8 | 1 | 11.1 | | |
|
Adenocarcinoma | 8 | 5 | 3 | 37.5 | | | 9 | 6 | 3 | 33.3 | | |
| Small cell | 11 | 9 | 2 | 18.2 | | | 10 | 9 | 1 | 10.0 | | |
| Tumor size | | | | | 0.04 | 0.487 | | | | | 1.10 | 0.081 |
| <3 cm | 15 | 12 | 3 | 20.0 | | | 17 | 15 | 2 | 11.8 | | |
| ≥3 cm | 13 | 10 | 3 | 23.1 | | | 11 | 8 | 3 | 27.3 | | |
| pN category | | | | | 0.57 | 0.279 | | | | | 1.33 | 0.067 |
| pN0 | 6 | 5 | 1 | 16.7 | | | 6 | 5 | 1 | 16.7 | | |
| pN1 | 7 | 6 | 1 | 14.3 | | | 7 | 6 | 1 | 14.3 | | |
| pN2 | 8 | 6 | 2 | 25.0 | | | 6 | 4 | 2 | 33.3 | | |
| pN3 | 7 | 5 | 2 | 28.6 | | | 9 | 8 | 1 | 11.1 | | |
KISS1R protein expression relative to the
clinical and pathological variables
RT-PCR and western blot analysis were carried out to
investigate the levels of KISS1R mRNA and protein in NSCLC
specimens. As shown in the results, the levels of KISS1R mRNA and
protein were lower in tumor tissue than that in normal tissue
(P<0.05; Figs. 1 and 2). The levels of KISS1R mRNA and protein
were higher in cancer tissues from stage IIIB NSCLC than that from
stage IV NSCLC (P<0.05; Figs. 1
and 2). The immunostaining results
showed that KISS1R expression was distributed to the cytomembrane
(Fig. 4). However, KISS1R
expression showed no correlation with the clinical and pathological
variables of the patients (P>0.05; Table II).
 | Table II.Relationship between KISS1R
expression and clinicopathological parameters of patients with
stage IIIB and IV NSCLC. |
Table II.
Relationship between KISS1R
expression and clinicopathological parameters of patients with
stage IIIB and IV NSCLC.
| Clinicopathological
features | KISS1R expression
in stage IIIB | KISS1R expression
in stage IV |
|---|
|
|
|---|
| n | − | + | PR (%) | χ2 | P-value | n | − | + | PR (%) | χ2 | P-value |
|---|
| Sex | | | | | 0.39 | 0.657 | | | | | 0.41 | 0.425 |
| Female | 8 | 6 | 2 | 25.0 | | | 10 | 8 | 2 | 20.0 | | |
| Male | 20 | 17 | 3 | 15.0 | | | 18 | 16 | 2 | 11.1 | | |
| Age (years) | | | | | 0.95 | 0.386 | | | | | 0.03 | 0.827 |
| <50 | 11 | 10 | 1 | 9.1 | | | 8 | 7 | 1 | 12.5 | | |
| ≥50 | 17 | 13 | 4 | 23.5 | | | 20 | 17 | 3 | 15.0 | | |
|
Differentiation | | | | | 0.41 | 0.648 | | | | | 0.04 | 0.732 |
| Well or
moderate | 9 | 8 | 1 | 11.1 | | | 6 | 5 | 1 | 16.7 | | |
| Poor | 19 | 15 | 4 | 21.1 | | | 22 | 19 | 3 | 13.6 | | |
| Lymphatic
invasion | | | | | 0.24 | 0.835 | | | | | 0.11 | 0.242 |
| − | 14 | 11 | 3 | 21.4 | | | 19 | 16 | 3 | 15.8 | | |
| + | 14 | 12 | 2 | 14.3 | | | 9 | 8 | 1 | 11.1 | | |
| Venous
invasion | | | | | 3.43 | 0.175 | | | | | 0.23 | 0.185 |
| − | 12 | 8 | 4 | 33.3 | | | 18 | 15 | 3 | 16.7 | | |
| + | 16 | 15 | 1 | 6.25 | | | 10 | 9 | 1 | 10.0 | | |
| Histological
type | | | | | 0.56 | 0.464 | | | | | 0.69 | 0.095 |
| Squamous
cell | 9 | 8 | 1 | 11.1 | | | 9 | 8 | 1 | 11.1 | | |
|
Adenocarcinoma | 8 | 6 | 2 | 25.0 | | | 9 | 7 | 2 | 22.2 | | |
| Small cell | 11 | 9 | 2 | 18.2 | | | 10 | 9 | 1 | 10.0 | | |
| Tumor size | | | | | 0.45 | 0.341 | | | | | 0.22 | 0.641 |
| <3 cm | 15 | 13 | 2 | 13.3 | | | 17 | 15 | 2 | 11.8 | | |
| ≥3 cm | 13 | 10 | 3 | 23.1 | | | 11 | 9 | 2 | 18.2 | | |
| pN category | | | | | 0.41 | 0.379 | | | | | 0.13 | 0.879 |
| pN0 | 6 | 5 | 1 | 16.7 | | | 6 | 5 | 1 | 16.7 | | |
| pN1 | 7 | 6 | 1 | 14.3 | | | 7 | 6 | 1 | 14.3 | | |
| pN2 | 8 | 6 | 2 | 25.0 | | | 6 | 5 | 1 | 16.7 | | |
| pN3 | 7 | 6 | 1 | 14.3 | | | 9 | 8 | 1 | 11.1 | | |
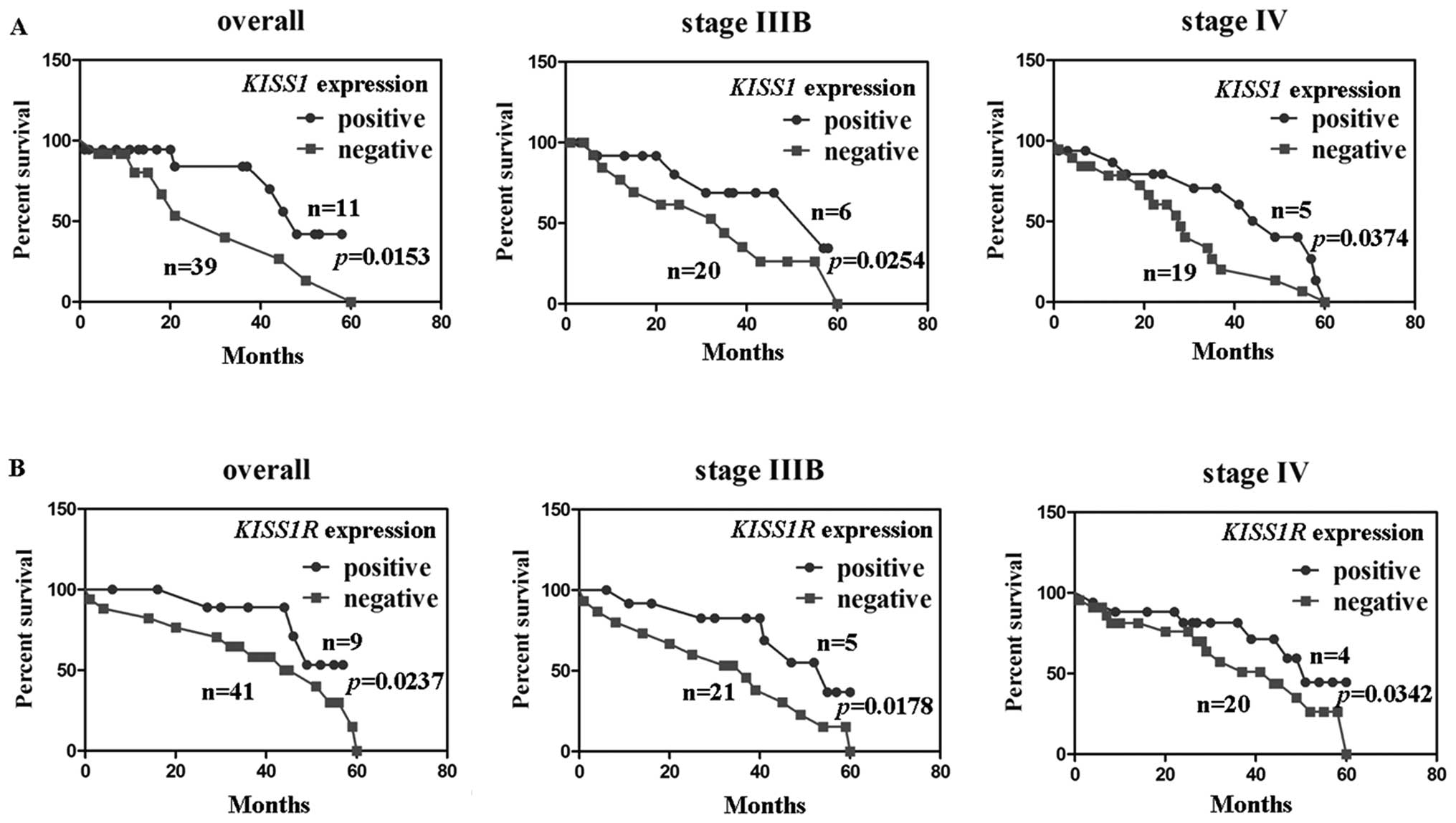
Kaplan-Meier survival analysis and Cox
proportional hazard analysis
To investigate the association of KISS1 expression
or KISS1R expression with patient survival, the survival data from
50 patients with NSCLC (6 missing follow-up) were assessed.
Comparison between KISS1 expression and 5-year survival rates
showed significant differences. In patients with stage IIIB and IV
NSCLC, comparison by the Kaplan-Meier method for low versus high
KISS1 expression showed a significant difference in the 5-year
survival rate (P<0.05; Fig.
5A). Comparisons between KISS1R expression and the 5-year
survival rate also showed significant differences. Kaplan-Meier
analysis showed that KISS1R expression was closely correlated with
the favorable prognosis of patients with NSCLC (P<0.05; Fig. 5B). Cox proportional hazard analysis
indicated that KISS1 and KISS1R were independent prognostic factors
for stage IIIB and IV NSCLC (P<0.05; Tables III and IV).
 | Table III.Multivariate analysis of clinical
variables for stage IIIB and IV NSCLC. |
Table III.
Multivariate analysis of clinical
variables for stage IIIB and IV NSCLC.
| Clinicopathological
parameters | KISS1 expression in
stage IIIB | KISS1 expression in
stage IV |
|---|
|
|
|---|
| Relative risk (95%
CI) | P-value | Relative risk (95%
CI) | P-value |
|---|
| Sex (male) | 0.683
(0.45–1.05) | 0.224 | 0.498
(0.33–0.77) | 0.407 |
| Age (>50
years) | 0.727
(0.48–1.12) | 0.231 | 0.712
(0.47–1.10) | 0.236 |
|
Differentiation | 0.654
(0.43–1.01) | 0.342 | 0.622
(0.41–0.96) | 0.325 |
| Lymphatic
invasion | 0.715
(0.47–1.10) | 0.267 | 0.643
(0.42–0.99) | 0.281 |
| Venous
invasion | 0.592
(0.39–0.91) | 0.453 | 0.518
(0.34–0.80) | 0.463 |
| Lymph node
metastasis | 0.788
(0.52–1.21) | 0.231 | 0.678
(0.44–1.05) | 0.354 |
| Tumor size (≥3
cm) | 0.573
(0.37–0.88) | 0.463 | 0.624
(0.41–0.96) | 0.442 |
| KISS1 expression (+
to +++) | 0.894
(0.58–0.38) | 0.045 | 1.122
(0.73–1.73) | 0.038 |
| KISS1R expression
(+ to +++) | 0.921
(0.60–1.42) | 0.037 | 1.352
(0.88–2.09) | 0.023 |
 | Table IV.Comparison of the NSCLC patients with
positive for metastin, and those negative, by using ELISA. |
Table IV.
Comparison of the NSCLC patients with
positive for metastin, and those negative, by using ELISA.
| Clinicopathological
features | Positive for
metastin (n=11) | Negative for
metastin (n=45) | P-value |
|---|
| Age (years) | 60.3±6.7 | 58.2±6.2 | 0.467 |
| Sex | | | 0.432 |
| Female | 4 | 14 | |
| Male | 7 | 31 | |
|
Differentiation | | | 0.162 |
| Well or
moderate | 5 | 10 | |
| Poor | 6 | 35 | |
| Lymphatic
invasion | | | 0.321 |
| − | 6 | 27 | |
| + | 5 | 18 | |
| Venous
invasion | | | 0.154 |
| − | 2 | 28 | |
| + | 9 | 17 | |
| Histological
type | | | 0.752 |
| Squamous
cell | 3 | 15 | |
|
Adenocarcinoma | 4 | 13 | |
| Small cell | 4 | 17 | |
| Tumor size | | | 0.534 |
| <3 cm | 4 | 28 | |
| ≥3 cm | 7 | 17 | |
| pN category | | | 0.576 |
| pN0 | 2 | 10 | |
| pN1 | 1 | 13 | |
| pN2 | 4 | 10 | |
| pN3 | 4 | 12 | |
| Stage | | | 0.034 |
| IIIB | 9 | 19 | |
| IV | 2 | 26 | |
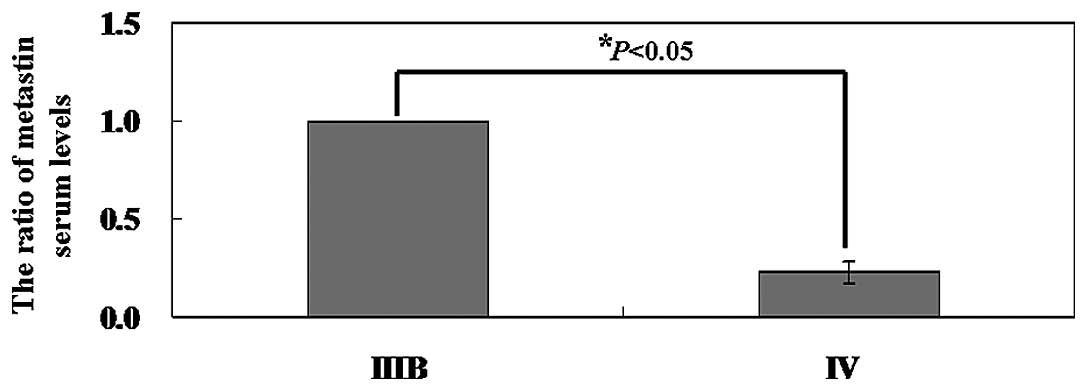
Correlations of metastin serum levels at
diagnosis
There was significant correlation between serum
metastin levels in patients and the stages of NSCLC. When we
examined separately the patients with NSCLC stage IIIB and the
patients with stage IV NSCLC, a statistically significant
difference was observed between circulating metastin levels
(P<0.05; Fig. 6). The plasma
level of metastin in patients with stage IIIB NSCLC ranged from
0.53 to 2.1 ng/ml (mean, 1.07±0.08 ng/ml) and the mean plasma level
of metastin in patients with stage IV NSCLC was 0.24±0.03
ng/ml.
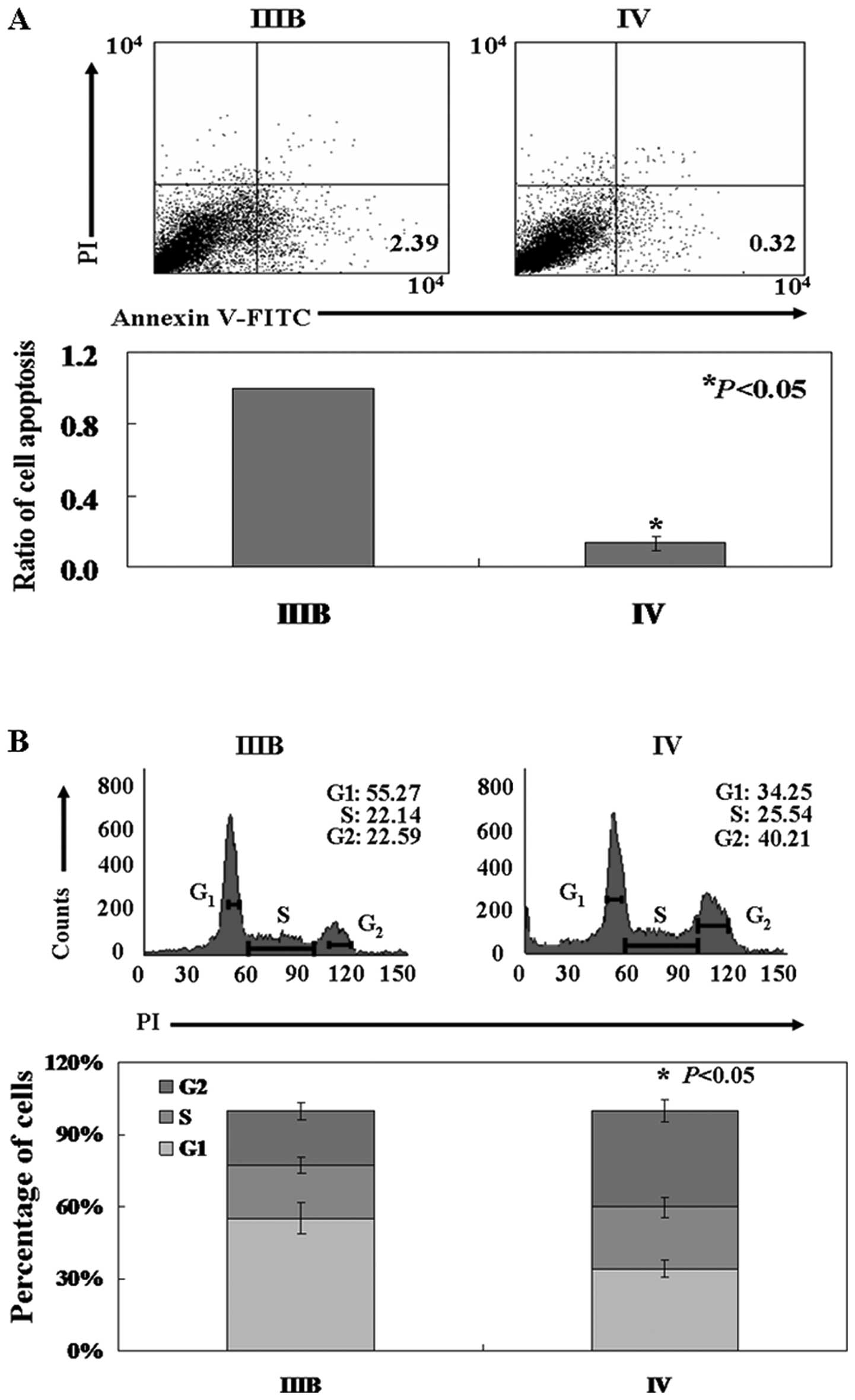
The assessment of tumors by flow
cytometry
We compared the apoptotic percentage of cells
collected from stage IIIB and stage IV NSCLC tissues by using
Annexin V-FITC and propidium iodide (PI) double staining. As shown
in Fig. 7A, the percentage of
apoptotic cells in stage IIIB was 2.39±0.42%, whereas 0.32±0.03% of
cells in stage IV were undergoing apoptosis (P<0.05). Cells from
stage IIIB cancer tissue had a higher ratio in the G1
phase than the ones from stage IV cancer tissue. The mean value of
G1 phase fraction of stage IIIB and stage IV cells was
55.3±5.7 and 34.3±5.8%, respectively (P<0.05, Fig. 7B).
Discussion
Previous reports showed that the KISS1/KISS1R system
plays an important role in tumor progression in a wide variety of
tumor types (8). However, to date,
the presence and potential role of the KISS1/KISS1R system in NSCLC
has not been reported yet. In this work, we analyzed the
differential expression of KISS1 and KISS1R in 28 patients with
stage IIIB NSCLC and 28 patients with stage IV NSCLC and found that
KISS1 and KISS1R expression was higher in stage III disease
compared to stage IV disease. The results indicated an inverse
correlation between KISS1 and KISS1R expression and NSCLC
progression. KISS1 was originally identified as a metastasis
suppressor by microcell-mediated transfer in melanoma lines,
responsible for tumor cell invasive and migratory properties
without affecting their tumourigenicity (25). Loss of KISS1 expression was found
to be a significant predictor and a potential biomarker of lymph
node metastasis in esophageal squamous cell carcinoma (26). Dhar et al (27) found that gastric cancers with low
KISS1 had a frequent venous invasion, distant metastasis and tumor
recurrence. Schmid et al (28) studied the expression of
KISS1 gene in HCC and its role in invasion, metastasis and
prognosis of human HCC by immunohistochemistry. Another study
showed that KISS1 expression in NSCLC was significantly higher in
the primary tumors compared to the secondary metastatic site
(29). However, in our study, we
did not find the relationship between KISS1 and metastasis of
NSCLC. Consistent with our study, Karapanagiotou et al
(30) found that KISS1 is not
involved in metastatic potential of non-small cell lung cancer.
Data from our study suggest that KISS1 is less likely to serve as a
diagnostic marker for NSCLC or metastatic disease. Possible
explanations for our findings could be the role of KISS1 seems to
be different in different types of cancer. Another explanation
could be either the lack of KISS1 expression in lung tissue or the
lack of KISS1R expression. In an attempt to uncover the mechanisms
by which KISS1 is lost in NSCLC, we tested the promoter
hypermethylation of KISS1. Consistent with our results,
Cebrian et al (31) found
that KISS1 hypermethylation was frequent in bladder cancer
cells analyzed by methylation-specific PCR and bisulfite sequencing
and was associated with low gene expression. Another study showed
that KISS1 hypermethylation correlated with transcript and
protein expression loss, being increased in vitro by
azacytidine (32).
KISS1R is a G protein-coupled receptor with a common
structure of seven transmembrane α-helices (33). The binding of KISS1 results in
conformation of KISS1R, which leads to signalling via G-proteins
and downstream effectors to prevent invasion and metastasis
(17,34). Martin et al (10) found that GPR54 expression increased
in the invasive ductal tumor. Higher levels of GPR54 mRNA were
observed in the moderately differentiated tumors compared to the
poorly differentiated high-grade cancers (35).Consistent with previous studies, we
confirmed that the KISS1R was lower in low stage of NSCLC (IIIB)
compared to advanced stage (IV). Another study conducted by Zajac
et al (36) suggested that
KISS1/GPR54 signaling is pro-migratory and invasive in breast
cancer cells. However, in our study, we did not find the
relationship between KISS1R expression and metastasis of NSCLC.
Most studies have shown that the KISS1/GPR54 system is negatively
correlated with tumor progression. The prognostic relevance of
KISS1 and GPR54 has been investigated in some solid tumors
(9–17). We also confirmed the KISS1 or
KISS1R expression was closely correlated with the favorable
prognosis of patients with NSCLC.
The bioactive sequence of the KISS-1 gene product
metastin is the C-terminal 10 amino acids (37). Metastin was initially purified from
human placenta (17). An enormous
increase in circulating metastin levels has been detected during
pregnancy (38). No statistically
significant difference in circulating metastin levels was observed
between healthy volunteers and patients with resectable pancreatic
cancer (39). Data from
Karapanagiotou et al study suggested that metastin is less
likely to serve as a diagnostic marker for NSCLC or metastatic
disease (30). However, in our
study, we confirmed the plasma metastin levels of the patients with
stage IIIB NSCLC were significantly higher than that of patients
with stage IV NSCLC. The possible explanation for the difference
between Karapanagiotou et al results (30) and ours could be that the subjects
derived from different ethnic groups. In our future studies, we
will further investigate this problem.
We confirmed cells collected from stage IIIB tumor
had higher apoptotic ratio than the ones from stage IV tumor
specimens. In addition, we also found the KISS1/GPR54 system could
induce G1 arrest in lung cancer cells. These results
indicated that the KISS1/GPR54 system may not only inhibit invasion
and migration of cancer cells but also induce apoptosis and cell
cycle arrest in the cells.
In conclusion, the expression of KISS1 or KISS1R was
associated with better survival of patients with NSCLC. KISS1
hypermethylation was identified in cancer tissues, providing a
potential mechanistic explanation for the observed loss of KISS1.
Furthermore, the serum metastin level could become an independent
prognostic tool for the Chinese Han people with NSCLC.
Acknowledgements
We are indebted to Yu Yan for his
technical supports and constructive suggestions in the preparation
of this manuscript.
References
|
1.
|
Jemal A, Bray F, Center MM, et al: Global
cancer statistics. CA Cancer J Clin. 61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Bray FI and Weiderpass E: Lung cancer
mortality trends in 36 European countries: secular trends and birth
cohort patterns by sex and region 1970–2007. Int J Cancer.
126:1454–1466. 2010.PubMed/NCBI
|
|
3.
|
Lam WK, White NW and Chan-Yeung MM: Lung
cancer epidemiology and risk factors in Asia and Africa. Int J
Tuberc Lung Dis. 8:1045–1057. 2004.PubMed/NCBI
|
|
4.
|
Youlden DR, Cramb SM and Baade PD: The
International Epidemiology of Lung Cancer: geographical
distribution and secular trends. J Thorac Oncol. 3:819–831. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Herbst RS, Heymach JV and Lippman SM: Lung
cancer. N Engl J Med. 359:1367–1380. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Soon YY, Stockler MR, Askie LM, et al:
Duration of chemotherapy for advanced non-small cell lung cancer: a
systematic review and meta-analysis of randomized trials. J Clin
Oncol. 27:3277–3283. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lee JH, Miele ME, Hicks DJ, et al: KiSS-1,
a novel human malignant melanoma metastasis-suppressor gene. J Natl
Cancer Inst. 88:1731–1737. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Makri A, Pissimissis N, Lembessis P, et
al: The kisspeptin (KISS-1)/GPR54 system in cancer biology. Cancer
Treat Rev. 34:682–692. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Masui T, Doi R, Mori T, et al: Metastin
and its variant forms suppress migration of pancreatic cancer
cells. Biochem Biophys Res Commun. 315:85–92. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Martin TA, Watkins G and Jiang WG: KiSS-1
expression in human breast cancer. Clin Exp Metastasis. 22:503–511.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Nicolle G, Comperat E, Nicolaïew N, et al:
Metastin (KISS-1) and metastin-coupled receptor (GPR54) expression
in transitional cell carcinoma of the bladder. Ann Oncol.
18:605–606. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Zohrabian VM, Nandu H, Gulati N, et al:
Gene expression profiling of metastatic brain cancer. Oncol Rep.
18:321–328. 2007.PubMed/NCBI
|
|
13.
|
Hata K, Dhar DK, Watanabe Y, et al:
Expression of metastin and a G-protein-coupled receptor (AXOR12) in
epithelial ovarian cancer. Eur J Cancer. 43:1452–1459. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Yamashita S, Tsujino Y, Moriguchi K, et
al: Chemical genomic screening for methylation-silenced genes in
gastric cancer cell lines using 5-aza-2′-deoxycytidine treatment
and oligonucleotide microarray. Cancer Sci. 97:64–71.
2006.PubMed/NCBI
|
|
15.
|
Seminara SB, Messager S, Chatzidaki EE, et
al: The GPR54 gene as a regulator of puberty. N Engl J Med.
349:1614–1627. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
De Roux N, Genin E, Carel JC, et al:
Hypogonadotropic hypogonadism due to loss of function of the
KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci U S A.
100:10972–10976. 2003.
|
|
17.
|
Ohtaki T, Shintani Y, Honda S, et al:
Metastasis suppressor gene KiSS-1 encodes peptide ligand of a
G-protein-coupled receptor. Nature. 411:613–617. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Stafford LJ, Xia C, Ma W, et al:
Identification and characterization of mouse metastasis-suppressor
KiSS1 and its G-protein coupled receptor. Cancer Res. 62:5399–5404.
2002.PubMed/NCBI
|
|
19.
|
Mitchell DC, Stafford LJ, Li D, et al:
Transcriptional regulation of KiSS-1 gene expression in metastatic
melanoma by specificity protein-1 and its coactivator DRIP-130.
Oncogene. 26:1739–1747. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Mead EJ, Maguire JJ, Kuc RE, et al:
Kisspeptins: a multifunctional peptide system with a role in
reproduction, cancer and the cardiovascular system. Br J Pharmacol.
151:1143–1153. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Ikeguchi M, Hirooka Y and Kaibara N:
Quantitative reverse transcriptase polymerase chain reaction
analysis for KiSS-1 and orphan G-protein-coupled receptor
(hOT7T175) gene expression in hepatocellular carcinoma. J Cancer
Res Clin Oncol. 129:531–535. 2003. View Article : Google Scholar
|
|
22.
|
Zhang SL, Yu Y, Jiang T, et al: Expression
and significance of KiSS-1 and its receptor GPR54 mRNA in
epithelial ovarian cancer. Zhonghua Fu Chan Ke Za Zhi. 40:689–692.
2005.(In Chinese).
|
|
23.
|
Tanoue LT and Detterbeck FC: New TNM
classification for non-small-cell lung cancer. Expert Rev
Anticancer Ther. 9:413–423. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Katagiri F, Tomita K, Oishi S, et al:
Establishment and clinical application of enzyme immunoassays for
determination of luteinizing hormone releasing hormone and
metastin. J Pept Sci. 13:422–429. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Lee JH and Welch DR: Identification of
highly expressed genes in metastasis-suppressed chromosome 6/human
malignant melanoma hybrid cells using subtractive hybridization and
differential display. Int J Cancer. 71:1035–1044. 1997. View Article : Google Scholar
|
|
26.
|
Ikeguchi M, Yamaguchi K and Kaibara N:
Clinical significance of the loss of KiSS-1 and orphan
G-protein-coupled receptor (hOT7T175) gene expression in esophageal
squamous cell carcinoma. Clin Cancer Res. 10:1379–1383. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Dhar DK, Naora H, Kubota H, et al:
Downregulation of KiSS-1 expression is responsible for tumor
invasion and worse prognosis in gastric carcinoma. Int J Cancer.
111:868–872. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Schmid K, Wang X, Haitel A, et al: KiSS-1
overexpression as an independent prognostic marker in
hepatocellular carcinoma: an immunohistochemical study. Virchows
Arch. 450:143–149. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Zheng S, Chang Y, Hodges KB, et al:
Expression of KISS1 and MMP-9 in non-small cell lung cancer and
their relations to metastasis and survival. Anticancer Res.
30:713–718. 2010.PubMed/NCBI
|
|
30.
|
Karapanagiotou EM, Dilana KD, Gkiozos I,
et al: Metastin is not involved in metastatic potential of
non-small cell lung cancer. Med Oncol. 28:559–564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Cebrian V, Fierro M, Orenes-Piñero E, et
al: KISS1 methylation and expression as tumor stratification
biomarkers and clinical outcome prognosticators for bladder cancer
patients. Am J Pathol. 179:540–546. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Moya P, Esteban S, Fernandez-Suarez A, et
al: KiSS-1 methylation and protein expression patterns contribute
to diagnostic and prognostic assessments in tissue specimens for
colorectal cancer. Tumour Biol. 34:471–479. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Muir AI, Chamberlain L, Elshourbagy NA, et
al: AXOR12, a novel human G protein-coupled receptor, activated by
the peptide KiSS-1. J Biol Chem. 276:28969–28975. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Kotani M, Detheux M, Vandenbogaerde A, et
al: The metastasis suppressor gene KiSS-1 encodes kisspeptins, the
natural ligands of the orphan G protein-coupled receptor GPR54. J
Biol Chem. 276:34631–34636. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Jarząbek K, Kozłowski L, Milewski R, et
al: KiSS1/GPR54 and estrogen-related gene expression profiles in
primary breast cancer. Oncol Lett. 3:930–934. 2012.PubMed/NCBI
|
|
36.
|
Zajac M, Law J, Cvetkovic DD, et al: GPR54
(KISS1R) transactivates EGFR to promote breast cancer cell
invasiveness. PLoS One. 6:e215992011. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Niida A, Wang Z, Tomita K, et al: Design
and synthesis of downsized metastin (45–54) analogs with
maintenance of high GPR54 agonistic activity. Bioorg Med Chem Lett.
16:134–137. 2006.PubMed/NCBI
|
|
38.
|
Horikoshi Y, Matsumoto H, Takatsu Y, et
al: Dramatic elevation of plasma metastin concentrations in human
pregnancy: metastin as a novel placentaderived hormone in humans. J
Clin Endocrinol Metab. 88:914–919. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Katagiri F, Nagai K, Kida A, et al:
Clinical significance of plasma metastin level in pancreatic cancer
patients. Oncol Rep. 21:815–819. 2009.PubMed/NCBI
|