Introduction
Lung cancer is the leading cause of cancer deaths
world-wide (1) and the majority of
cases (70–85%) are non-small cell lung cancers (NSCLC) (2). Most patients with NSCLC are diagnosed
after their cancer has metastasised and this is associated with a
poor prognosis (3,4). As metastatic disease is currently
incurable, new therapeutic approaches to lung cancer are urgently
required (4). The molecular
mechanisms involved in NSCLC tumourigenesis and metastatic
progression are largely unknown, however, recent studies have
demonstrated that non-coding RNAs are key regulators of these
processes (5–9). Less than 2% of the human genome is
transcribed into protein-coding mRNAs, while approximately 90% is
transcribed into non-protein coding RNAs (ncRNAs) and many are of
unknown function (9,10). Non-coding RNAs, therefore, provide
promising targets for the development of novel therapeutics for
cancer (11).
Here, we report the identification, genomic
organisation and initial characterisation of the candidate
antisense long ncRNA gene, GHSROS, which is located on the
opposite strand of the gene for the ghrelin receptor, the growth
hormone secretagogue receptor (GHSR) gene. We demonstrate
that GHSROS is expressed at a higher level in human lung
tumours compared to other tissue types and, when over-expressed in
lung cancer cell lines, promotes cell migration. The ability of
cancer cells to migrate and ultimately to metastasise to distant
sites in the body is a key hallmark of cancer (12,13).
We hypothesise that, like other recently described lncRNAs,
GHSROS contributes to lung cancer progression by stimulating
cancer cell migration.
Materials and methods
Sequence analysis and database
searches
Multiple sequence alignments were generated using
the Evolutionary Conserved Regions (ECR) Browser (14) and Clustal W2.0 (15). To examine putative coding sequences
we used the ExPASy Translate Tool (http://www.expasy.ch/tools/dna.html) and the Coding
Potential Calculator (16). The
presence of transposable elements was examined using CENSOR v4.2.8
(17).
Human tissues and RNA
Normal lung and lung tumour specimens were obtained
from the Ontario Tumour Bank (Toronto, Canada; Table I). Commercial total RNA was
obtained from human stomach, ovary (FirstChoice, Invitrogen,
Carlsbad, CA), cerebellum, thymus, whole brain, lung, testis,
foetal brain, lung adenocarcinoma and pancreas (Clontech, Mountain
View, CA).
 | Table I.Specimen characteristics of a range
of paired normal and non-small cell lung carcinoma tumour biopsies
from patients diagnosed with NSCLC. |
Table I.
Specimen characteristics of a range
of paired normal and non-small cell lung carcinoma tumour biopsies
from patients diagnosed with NSCLC.
| Sample ID | Age/gender | Source of
specimen | Histological tumour
type | Tumour size
(cm) |
Grade/differentiation | Tumour | Node | Metastasis |
|---|
| N1 | 77/M | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T1 | 77/M | Primary tumour | Squamous
carcinoma | 4.5 | II | T2 | N0 | MX |
| N2 | 73/M | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T2 | 73/M | Primary tumour | Adenocarcinoma | 2.5 | II | T2 | N0 | MX |
| N3 | 73/M | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T3 | 73/M | Primary tumour | Squamous
carcinoma | 3 | III | T2 | N0 | M0 |
| N4 | 70/M | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T4 | 70/M | Primary tumour | Squamous
carcinoma | 11 | II | T2 | N0 | M0 |
| N5 | 52/F | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T5 | 52/F | Primary tumour | Adenocarcinoma | 4 | III | T2 | N0 | MX |
| N6 | N/A | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T6 | N/A | N/A | Adenocarcinoma | N/A | N/A | N/A | N/A | N/A |
| N7 | N/A | Adjacent normal
tissue | N/A | N/A | N/A | N/A | N/A | N/A |
| T8 | 84/M | Primary tumour | Adenocarcinoma | 6 | III | T2 | N0 | M0 |
Cell lines
The A549 cell line (American Type Culture
Collection, ATCC 10801, Rockville, MD) was cultured in DMEM/F12
(Invitrogen), while the NCI-H1299 (ATCC CRL-5803) and Beas-2B (ATCC
CRL-9609) cell lines were cultured in RPMI-1640 medium
(Invitrogen). The complete medium contained 10% cosmic calf serum
(HyClone, ThermoFisher Scientific, Waltham, MA), 50 U/ml penicillin
and 50 μg/ml streptomycin (Invitrogen). Cells were incubated
at 37°C in air and 5% CO2 and free of Mycoplasma
contamination.
RNA extraction
Total RNA was harvested from tissues and cultured
cells using an RNeasy Plus mini kit (QIAGEN, Germantown, MD)
according to the manufacturer’s instructions.
Quantitative real-time RT-PCR
cDNA was synthesised using a GHSROS
strand-specific primer (GHSROS-RT), and quantitative RT-PCR
of GHSR antisense mRNA was performed as previously described
(18,19) using the Prism 7000 Sequence
Detection System (Applied Biosystems, Foster City, CA). Data were
analysed using the comparative 2−ΔΔCt method with
normalisation to the 18S housekeeping gene (20). The primers used are listed in
Table II. All RT-PCR products were
purified using PureLink (Invitrogen) or MinElute (QIAGEN) PCR
Purification Kits, cloned into pCR-XL-TOPO (Invitrogen), or
pGEM-T Easy (Promega, Fitchburg, WI), transformed into One
Shot MAX Efficiency DH5α-T1R chemically competent cells
(Invitrogen) and sequenced at the Australian Genome Research
Facility (AGRF, Brisbane, Australia) using BigDye III (Applied
Biosystems).
 | Table II.Designations and sequences of
oligonucleotides. |
Table II.
Designations and sequences of
oligonucleotides.
| Name | Sequence
(5′-3′) | Ta | PCR cycles |
|---|
| GHSROS-RT |
CGACTGGAGCACGAGGACACTGACAACAGAATTCACTACTTCCCCAAA | | |
| GHSROS-F |
ACATTCAGCAAATCCAGTTATGACA | 60 | 40 |
| LK |
CGACTGGAGCACGAGGACACTGA | | |
| 18S-F |
TTCGGAACTGAGGCCATGAT | 60 | 40 |
| 18S-R |
CGAACCTCCGACTTTCGTTCT | | |
|
5′-RACE-adapter |
GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGGCUUUGAUGAAA | | |
| 5′-RACEout-F |
ATGGCGATGAATGAACACTG | 58 | 35 |
| 5′-RACEout-R |
GGGATCACTAAAGTGTTACAACGAC | | |
| 5′-RACE-in-F |
AATGAACACTGCGTTTGCTG | 57 | 35 |
| 5′-RACE-in-R |
ATTTTTCCCTGATTTCTGAATTT | | |
|
3′-RACE-adapter |
GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN | | |
| 3′-RACE-F |
GTTTCCACAAAGTCTCCTTTCC | 59 | 35 |
| 3′-RACE-R |
GCACAGAATTAATACGACTCAC | | |
| 3′-RACE-in-F |
ACTTCACTGATTTGTCACTG | 58 | 35 |
| Northern-F |
CGTTGTAACACTTTAGTGATCCC | 58 | 35 |
| Northern-T7-R | CTAATACGACTCACTATAGGGAGATTTACAGTTCCTGGATCCTC | | |
|
GHSROS-pTargeT-F |
CAGAGGATTGAATATACAGTTAGG | 58 | 35 |
|
GHSROS-pTargeT-R |
ATAATTGCATCAATTCTGTTTACC | | |
GHSR antisense transcript mapping
5′-RLM-RACE was performed using the FirstChoice
RLM-RACE kit (Invitrogen) according to the manufacturer’s
instructions, except that heat-labile shrimp alkaline phosphatase
(SAP; Fermentas, Burlington, ON, Canada) was used to
dephosphorylate degraded RNA. For 3′-RACE, 2 μg total RNA
was reverse transcribed using Transcriptor reverse transcriptase
(Roche, Penzberg, Germany) and a 3′-RACE adapter primer (Table II).
Northern blot analysis
Northern blot analysis was carried out on mRNA
purified from A549 cells, using the Oligotex mRNA mini-kit (QIAGEN)
and the NorthernMax-Gly kit (Invitrogen) according to the
manufacturer’s instructions. Briefly, 250 ng mRNA and 50 ng RNA
Molecular Weight Marker II (Roche) were electrophoresed through a
1% glyoxal agarose gel, transferred onto a positively charged nylon
membrane (Roche) and probed with 10 ng/ml 331 nt
GHSROS-specific riboprobe. The riboprobe was generated from
A549 genomic DNA using PCR (primers Northern-F and Northern-T7-R,
Table II). The PCR product was
purified using a MinElute PCR Purification kit (QIAGEN), and the
cRNA probe was synthesised using a digoxigenin RNA labelling kit
(Roche). The membrane was hybridised overnight at 70°C with the
riboprobe using ULTRAhyb buffer (Invitrogen), followed by high
stringency washing conditions (70°C).
Stable transfection of GHSROS cDNA in
cell lines
Full-length GHSROS was generated by RT-PCR
from A549 cell line mRNA (using primers GHSROS-pTargeT-F and
GHSROS-pTargeT-R; Table II), and
cloned into the pTargeT mammalian expression vector
(Promega). Cells were transfected with DNA (linearised
GHSROS-pTargeT, or vector alone/mock) using Lipofectamine
LTX reagent (Invitrogen). After 48 h, stable polyclonal cell
populations were generated by culturing in complete medium
containing G418 (Invitrogen), at 1,000 μg/ml for the A549
and the Beas-2B cell lines and 600 μg/ml for the NIH-H1299
cell line. Cells were grown in the presence of G418 for at least
two weeks before functional analyses were performed. GHSROS
expression was verified twice weekly using quantitative real-time
RT-PCR, as described above.
Cell migration assays
Migration assays were performed using Transwell
inserts with polycarbonate membranes (8 μm pore size; BD
Biosciences, Franklin Lakes, NJ) in 12-well plates. Cells were
added to the upper chamber of the Transwell inserts in serum-free
medium and medium with 10% cosmic calf serum was used as a
chemo-attractant in the lower chamber. Control inserts containing
medium only were used to determine background staining. Cells were
cultured for 6–24 h and cells remaining on the upper surface of the
inserts were removed. The number of cells that migrated to the
underside of the inserts was quantified by fixing the cells (100%
methanol) and staining with 1% crystal violet. The stain was
extracted using 10% (v/v) acetic acid and absorbance measured at
595 nm. Cell migration in GHSROS overexpressing cells was
compared to cells expressing the vector alone. Each experiment
consisted of three replicates and was repeated independently at
least three times.
Cell proliferation assays
Cell proliferation was assessed by quantifying both
metabolic activity (WST-1 assay, Roche) and DNA synthesis (CyQUANT
NF Cell Proliferation Assay Kit; Invitrogen). Cells were cultured
in replicate 96-well plates (BD Biosciences) for 4, 24 and 48 h.
Absorbance was measured at 440 nm with a reference wavelength of
600 nm for the WST-1 assay and with excitation at 485 nm and
emission at 530 nm for the CyQUANT assay. All proliferation
experiments were performed independently at least three times, with
10 replicates each.
Statistical analyses
Statistical significance was determined using
Student’s t-test, with a p-value <0.05 considered to be
statistically significant.
Results
Identification of a GHSR antisense
transcript
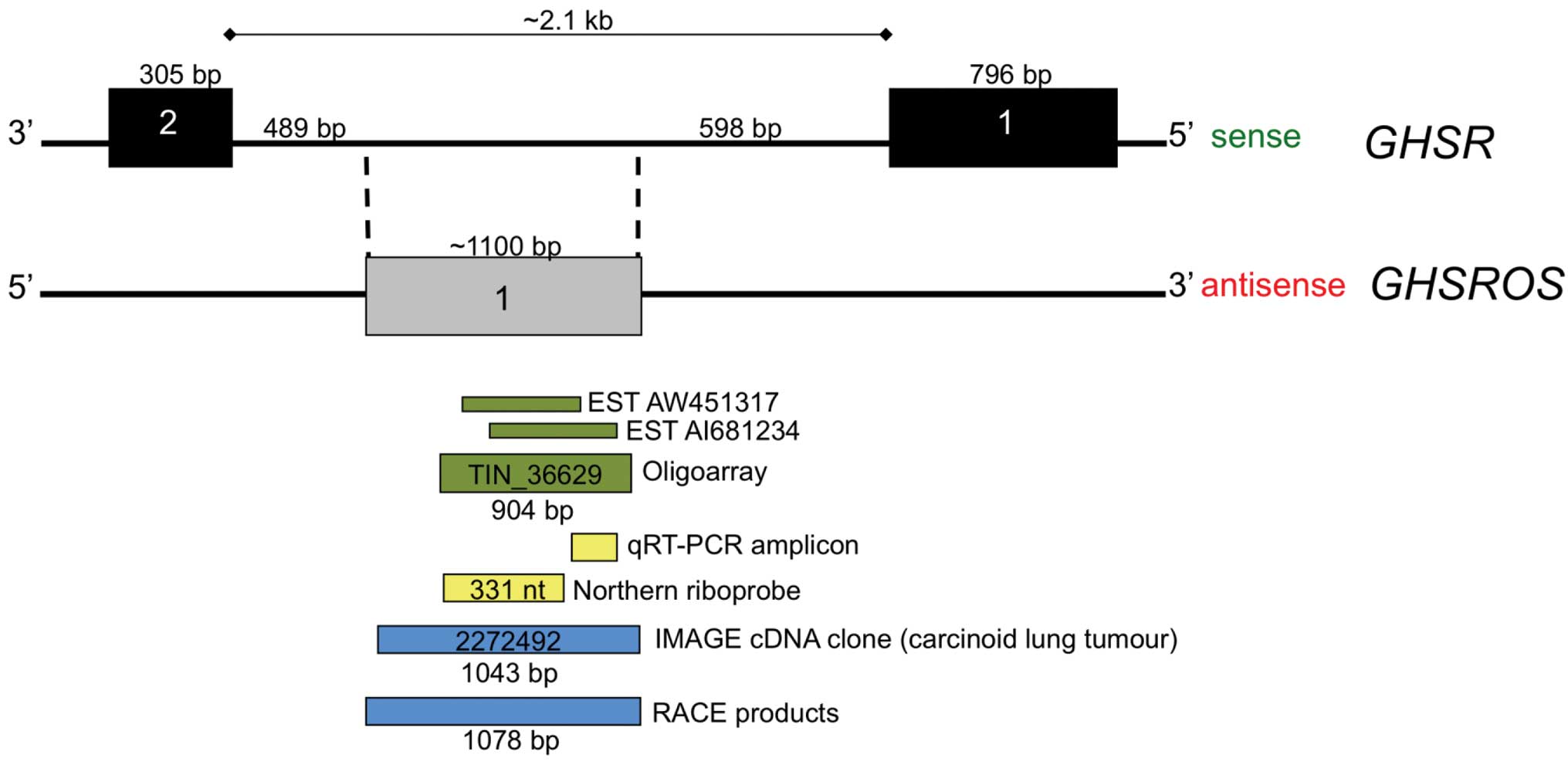
Inspection of the UCSC Genome Browser for Functional
RNA (21) revealed two overlapping
expressed sequence tags (ESTs) (GenBank entries AW451317 and
AI681234) which were antisense to the growth hormone secretagogue
receptor gene (GHSR) (Fig.
1). We named the putative antisense transcript growth hormone
secretagogue receptor opposite strand (GHSROS). These ESTs
were sequenced as a part of the Cancer Genome Anatomy Project
(http://cgap.nci.nih.gov) and were derived from
lung carcinoid tumour tissue, which is a rare, neuroendocrine lung
tumour type (22). The two
overlapping EST entries together span approximately 900 bp within
the 2.1 kb intron 1 of GHSR. Moreover, a 904 nucleotide
transcript (TIN_36629), derived from oligoarray analysis for
intronic non-coding RNAs of the liver, kidney and prostate
(23), also maps to the region
covered by the two ESTs (Fig. 1).
This 904 nucleotide GHSROS transcript was one of 55,000
transcripts denoted intronic non-coding RNA in a large-scale study
by Nakaya et al (23).
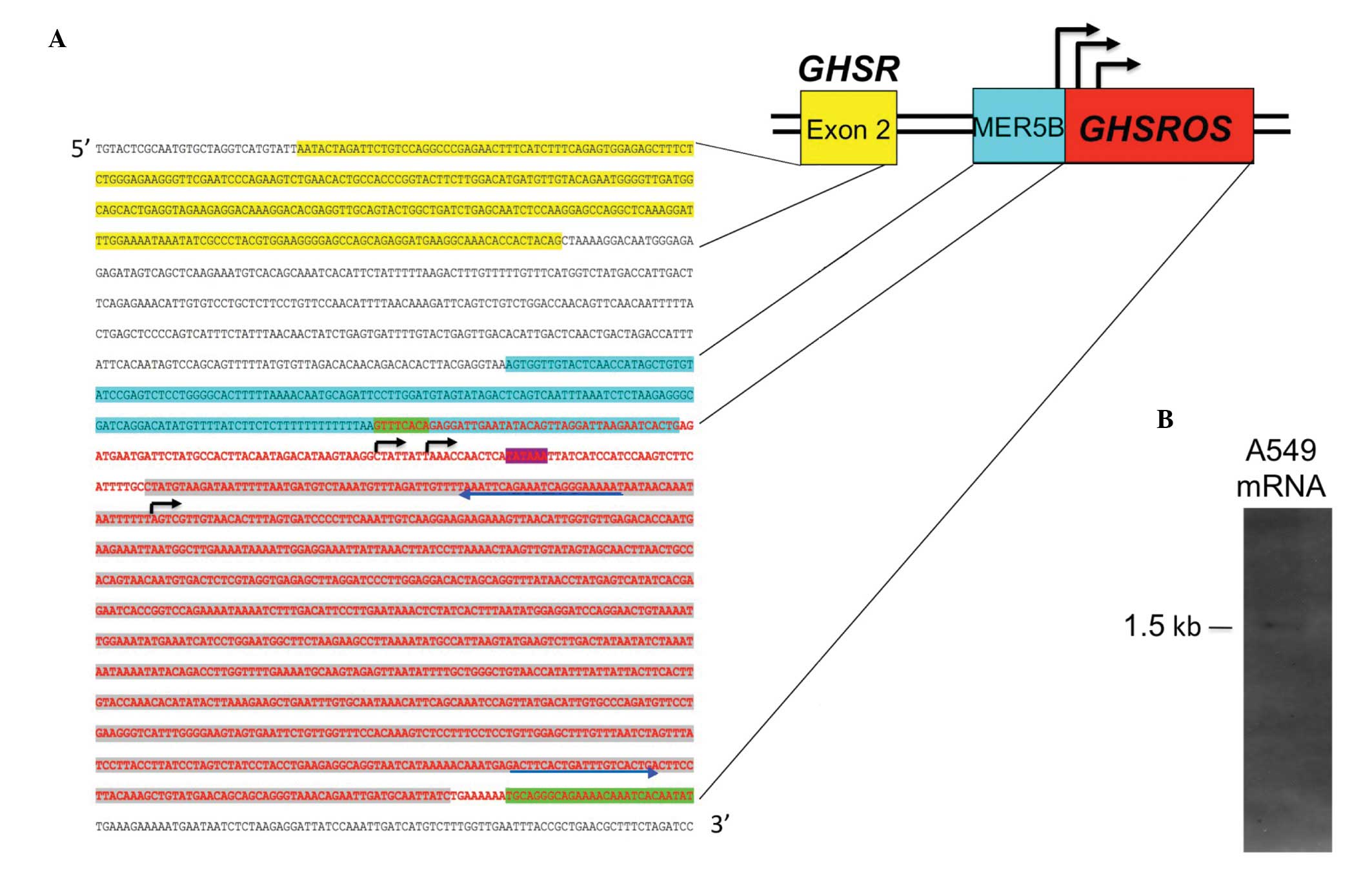
Structure of GHSROS
To map the full-length GHSROS transcript, a
multi-pronged approach was employed. As noted, the public domain
oligoarray-deduced sequence (TIN_36629, Fig. 1) spans 904 bp of genomic DNA. A
sequence conforming to the consensus TATA box motif (TATAAA)
(24) is present just upstream of
this sequence (Fig. 2A),
suggesting that an antisense promoter is present in the intron of
GHSR. To confirm the oligoarray data, 5′- and 3′-RACE PCR
products and a full-length cDNA clone from a lung carcinoid tumour
(Image cDNA clone 2272492) were sequenced. The sequenced
full-length transcript is 1078 nucleotides in size, consists of a
single exon and maps within the GHSR intron (Fig. 2A) (GenBank entries FJ355932,
FJ355933 and GU289929). Northern blot analysis of mRNA from the
A549 NSCLC cell line showed that the polyadenylated, full-length
GHSROS is approximately 1,500 bp in size (Fig. 2B), and this corresponds closely to
the predicted size of GHSROS mRNA. As shown in Fig. 2A, GHSROS has three
transcription start sites: one just downstream of the consensus
TATA-box and two immediately upstream of a poly-T-repeat within an
ancient MER5B (medium reiteration frequency 5B) DNA transposable
element (25). This thymidine-rich
repeat is absent in non-primates (data not shown), suggesting that
a primate-specific antisense promoter in the GHSR intron may
have emerged through accumulated mutations (ab initio
generation) (26). Interestingly,
it is well known that poly-T-repeats in promoters can result in
highly efficient transcription by depleting repressive nucleosomes
from promoters (27). Together,
these observations suggest that an antisense promoter in the
GHSR intron gives rise to single-exon GHSROS
transcripts that are processed into mRNA (5′ capped and 3′
polyadenylated).
GHSROS is a candidate long non-coding
RNA
While it is difficult to predict and experimentally
prove that a transcript either codes for very small peptides or is
a non-coding RNA, a number of parameters can be assessed (28). Analysis using the coding potential
calculator (CPC) tool predicted that GHSROS is a non-coding
transcript. The CPC tool is a highly accurate algorithm that takes
into account multiple features, including putative peptide length,
amino acid composition, secondary structure, the conservation of
protein homologues and alignment information (28). GHSROS demonstrates a number
of features typical of a non-coding RNA. As the open reading frames
are very short, GHSROS would encode very small peptides
(with 13 open reading frames which are 6–46 amino acids in size).
GHSROS also has a high frequency of stop codons throughout
the 1.1 kb GHSROS sequence in all three reading frames
(Table III). Moreover,
GHSROS open reading frames have poor consensus to the
translation initiation sequence proposed by Kozak (29) (Table
III). Multiple sequence alignments show that the GHSROS
nucleotide sequence is highly conserved in the chimpanzee, while
there is low nucleotide and open reading frame conservation
compared to the mouse (data not shown). The current data,
therefore, suggests that GHSROS is a non-protein-coding RNA
gene.
 | Table III.Open reading frames (ORFs) in the
GHSROS transcript determined using the ExPASy translate tool
(http://www.expasy.ch/tools/dna.html). |
Table III.
Open reading frames (ORFs) in the
GHSROS transcript determined using the ExPASy translate tool
(http://www.expasy.ch/tools/dna.html).
| ORF no.a | Frame | Start | Stop | Length (bp) | Length (AA) | Initiator
sequenceb |
|---|
| 1 | 1 | 538 | 567 | 36 | 11 |
TttAatATGG |
| 2 | 1 | 586 | 606 | 21 | 6 |
CCtGgaATGG |
| 3 | 1 | 610 | 633 | 24 | 7 |
TAaAatATGc |
| 4 | 2 | 47 | 159 | 123 | 40 |
ACtGagATGa |
| 5 | 2 | 302 | 322 | 21 | 6 |
ACacCaATGa |
| 6 | 2 | 461 | 517 | 57 | 18 |
taacCtATGa |
| 7 | 2 | 797 | 937 | 141 | 46 |
CCaGttATGa |
| 8 | 3 | 51 | 74 | 24 | 7 |
AgatgaATGa |
| 9 | 3 | 159 | 245 | 87 | 28 |
tttttaATGa |
| 10 | 3 | 570 | 596 | 27 | 8 |
GgaAatATGa |
| 11 | 3 | 678 | 755 | 78 | 25 |
TtgAaaATGc |
| 12 | 3 | 813 | 842 | 30 | 9 |
GCCcagATGt |
| 13 | 3 | 1005 | 1046 | 42 | 13 |
AgCtgtATGa |
| | | | | | A |
| | | | Consensus Kozak’s
rule: | GCC
CCATGG |
| | | | | | G |
GHSROS is overexpressed in lung
cancer
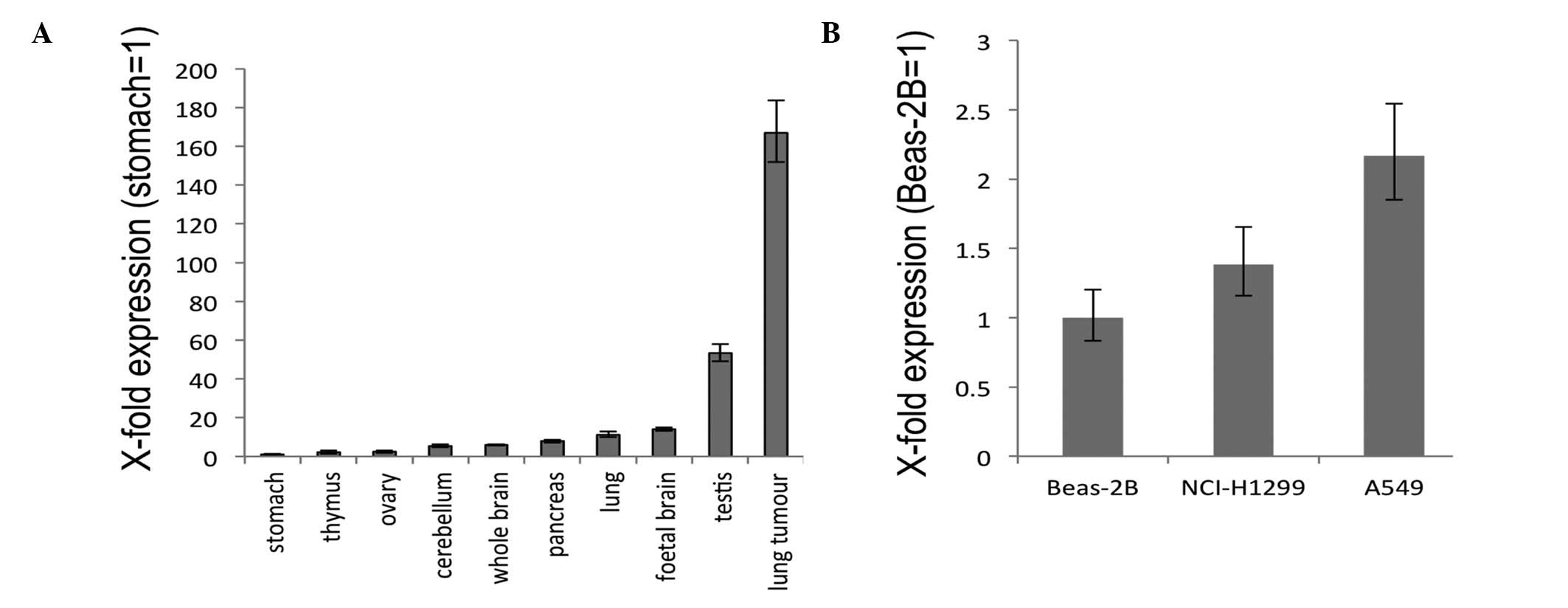
To examine the expression of GHSROS,
quantitative real-time RT-PCR was performed using commercially
available RNA from a range of normal human tissues. Stomach,
cerebellum, ovary, thymus, whole brain, lung, pancreas and foetal
brain displayed relatively low levels of GHSROS expression
with relatively moderate expression in the testis (Fig. 3A). In contrast, GHSROS was
highly expressed in a lung tumour sample (Fig. 3A). In the lung-derived cell lines
examined, the lowest level of GHSROS expression was seen in
the normal tissue-derived, Beas-2B bronchoepithelial cell line,
while higher expression levels were seen in the NCI-H1299 and A549
NSCLC cell lines (Fig. 3B).
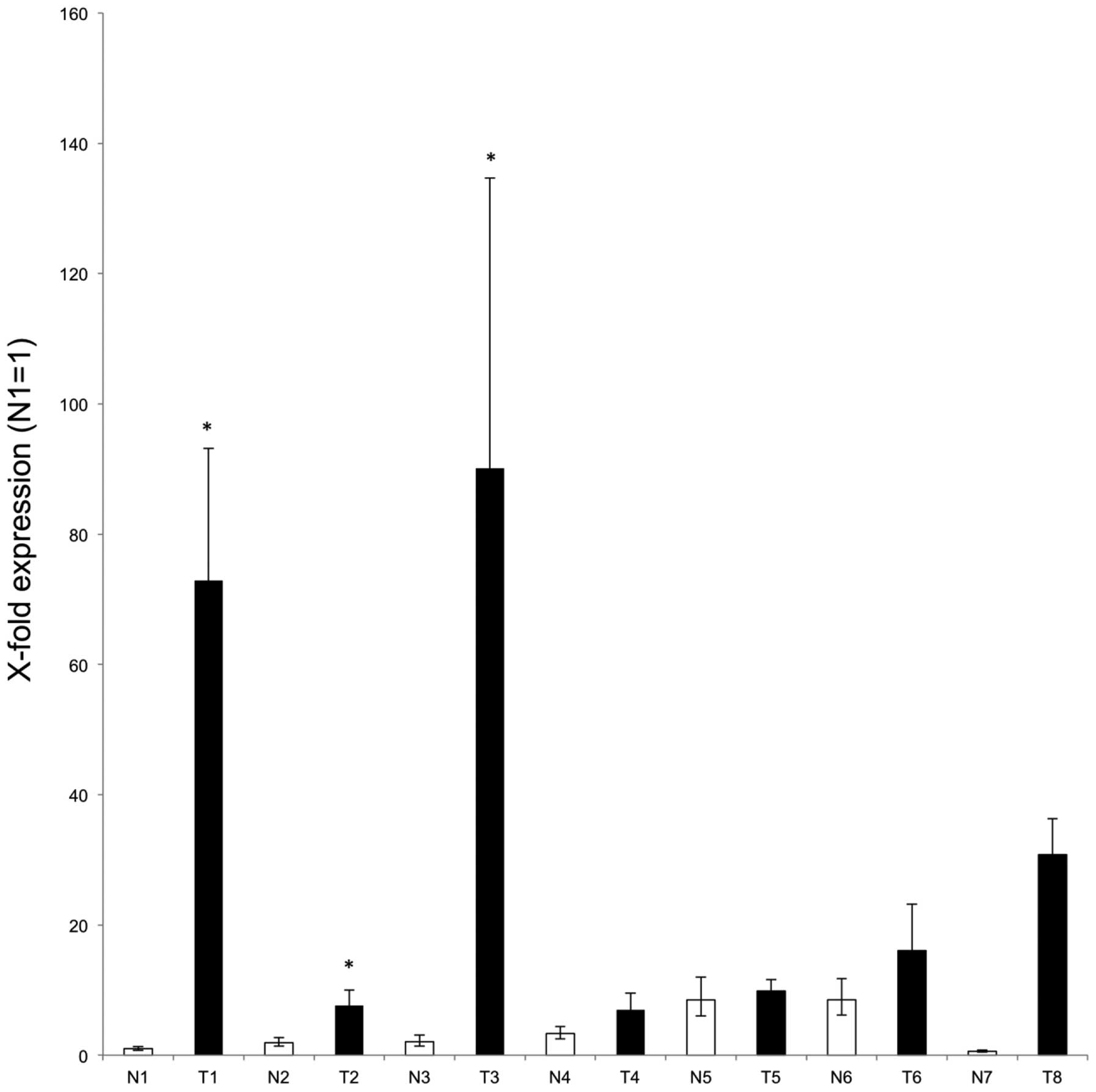
Finally, quantitative real-time RT-PCR was performed using tumour
and matched adjacent normal tissue from six patients with NSCLC
lung cancer, as well as two non-matched samples (for clinical
details, see Table I). A higher
level of GHSROS expression was observed in each of the
tumour samples compared to their matched adjacent normal tissue
with samples 1, 2 and 3 being statistically significant (Fig. 4).
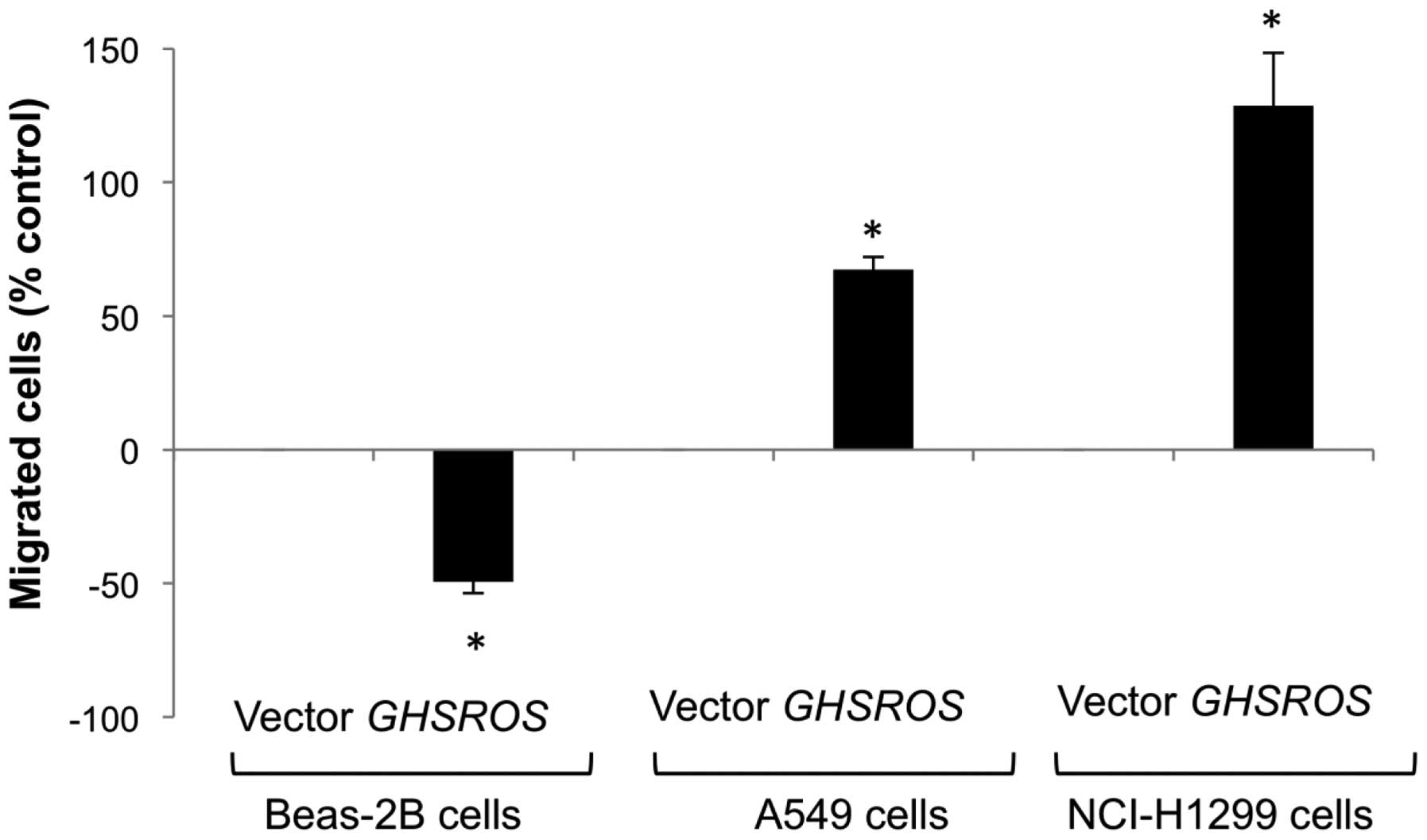
GHSROS overexpression increases the
migration of A549 and NCI-H1299 NSCLC cell lines, but reduces
migration in the Beas-2B cell line
The functional significance of GHSROS in the
lung was studied by creating stable transfectants in the A549,
NCI-H1299 and Beas-2B cell lines. Migration was significantly
decreased in GHSROS overexpressing Beas-2B cells over 24 h
(49% below vector-only control, P<0.05) (Fig. 5, lanes 1 and 2). In contrast,
migration was significantly increased in the GHSROS
overexpressing NSCLC cell lines examined, with an increase of 67%
above control (p<0.05) in A549 cells (Fig. 5, lanes 3 and 4) and 129% above
control (p<0.05) in NCI-H1299 cells after 6 h (Fig. 5, lanes 5 and 6). The observed
differences in cell migration were not due to changes in cell
number, as overexpression of GHSROS did not significantly
alter cell proliferation in the A549, NCI-N1299, or Beas-2B cell
lines at these time points compared to cells expressing the vector
alone (data not shown).
Discussion
We demonstrate that the intronic region of the
ghrelin receptor gene, GHSR, encodes a long non-coding RNA,
termed GHSROS, which is expressed in lung cancer and
promotes cell migration in lung cancer cell lines. Research into
the role of ncRNAs in normal development and disease processes has
predominantly focused on microRNAs (miRNAs), however, a number of
long ncRNAs (lncRNAs), including MALAT-1, H19,
B2 and lincRNA-p21 are known to play a role in lung
cancer progression (11). Long
ncRNAs are greater than 200 nucleotides in length, lack significant
open reading frames and often harbour protein-coding mRNA-like
features such as transcription by RNA polymerase II,
polyadenylation and alternative splice variants. They control gene
expression via the regulation of a broad range of processes
including gene expression at the transcriptional and
post-transcriptional levels (splicing, transcript degradation,
epigenetic modification, chromatin remodelling, and sub-cellular
transport) and some are precursors for small RNAs (11,30).
The GHSROS gene encodes a transcript ∼1.1 kb
in size and is a putative mRNA-like non-coding RNA, as it is likely
to be derived from a classical TATA-box promoter and is 5′ capped
and 3′ polyadenylated. Although we predict that GHSROS is a
non-coding gene, we cannot currently dismiss the possibility that
GHSROS encodes short, bioactive peptides. For example, the
assumed ncRNA gene pri in Drosophila has been shown
to encode functional peptides of 11 and 32 amino acids (31,32).
It has recently been recognised that short open reading
frame-encoded polypeptides (SEPs) may be very abundant in the
proteome and are also derived from ncRNAs (33). Proteomics studies, for example
using assays such as multiple reaction monitoring mass spectrometry
(34), would be most useful in
assessing whether any of the 13 short open reading frames of
GHSROS are indeed translated and have independent
functions.
GHSROS overlaps a MER5B DNA transposable
element. Repeat elements in non-coding RNAs have been reported by a
number of investigators (35–40).
Accumulating evidence suggests that transposable elements often
harbour promoters for natural antisense transcripts (41) and these elements lead to the
transcription of novel, species-specific, non-coding RNA
transcripts involved in gene regulation (42). It has been hypothesised that
non-coding RNAs in introns may regulate the abundance, or splicing
of their overlapping protein-coding transcripts (22,23,43–46).
It is equally likely, however, that many intronic RNAs regulate a
large number of genes in trans, at sites that are distant to
their host loci (44,47).
We report that GHSROS is expressed in
clinical samples from lung tissues and lung cell lines, with a
similar, but not statistically significant, trend towards higher
GHSROS expression in lung tumours. We hypothesise that this
observation could be due to adjacent normal tissue samples with
elevated GHSROS levels exhibiting pre-neoplastic alterations
in gene expression, as described in other studies (48,49),
or that tumour cells have spread into the normal tissue. Moreover,
lung tissue is highly heterogeneous, consisting of a range of cell
types, including cartilage, smooth muscle, epithelial and
endothelial cells (50). Laser
capture microdissection would be useful to isolate specific cell
types for future experiments (50,51).
As was the case for the lncRNA, HOTAIR, that is
overexpressed in metastatic breast tumours (52), it is also feasible that the
heterogeneous GHSROS expression pattern observed in our
limited panel of primary tumours can be resolved by measuring the
levels of the transcript in a larger and more diverse patient
tissue panel. This will determine whether GHSROS could be a
useful biomarker for predicting metastatic progression and patient
survival.
The most well-studied long non-coding RNA in lung
cancer, MALAT-1 (Metastasis Associated in Lung
Adeno-carcinoma Transcript-1), plays a role in cancer progression
through a number of mechanisms including the regulation of gene
transcription (53).
MALAT-1 is overexpressed in NSCLC, in NSCLC cell lines and
in a number of other tumour types (5,54–56).
It has an important role in cancer cell motility and migration, and
regulating genes related to these processes, and may be involved in
other cancer related processes (11,55).
Furthermore, knockdown of MALAT-1 expression in NSCLC
xenograft mouse models reduces tumour growth and prevents lung
cancer cell metastasis (6,53). MALAT-1 is, therefore, a
potential therapeutic target for lung cancer (53) and also has potential as a
prognostic biomarker for NSCLC, breast, prostate, pancreatic,
colon, liver and endometrial cancers (6,57–60).
H19 is an imprinted lncRNA which is highly
expressed in the embryo and is oncogenic in some cell types
including lung cancer cell lines, while it has tumour-suppressing
activity in other cell types (61–65).
It is upregulated by a number of carcinogens, and expression is
greatly increased in the airway epithelium of smokers (66). Long non-coding RNAs work through a
number of different mechanisms and H19 is a precursor for at
least one miRNA (67,68). This may allow it to play
contrasting roles in different tissues and at different stages of
development. The lncRNA lincRNA-p21 is a global repressor of
the p53 tumor suppressor pathway (69) and acts a post-transcriptional
inhibitor of the translation of target genes through an interaction
with the RNA binding protein HuR (70).
A hallmark of tumour cell behaviour is the ability
to migrate and ultimately to metastasise to secondary sites in the
body (12,13). Engineered GHSROS
overexpression resulted in a decreased rate of migration in the
normal lung-derived Beas-2B cell line, while it stimulated cell
migration in the two NSCLC cell lines. Such cell-type and
context-specific effects are also observed for miRNAs, which can
regulate a large number of genes and function as either oncogenes
or tumour suppressors (71,72).
Indeed, evidence is emerging that many short and long RNA
transcripts may have dual functions, and their ultimate biological
effects may be dependent on complex ncRNA-DNA-protein interactions
(73,74). Interestingly, it has recently been
reported that lncRNAs are able to deplete miRNA, acting as miRNA
sponges (75–77). This could explain the global
changes in gene expression observed with lncRNA overexpression
and/or knockdown. Conversely, lncRNAs may also exert global effects
as precursors for miRNAs, as observed for H19 (67). Further studies are underway in our
laboratory to explore the mechanisms by which GHSROS
promotes migration in lung cancer cells.
In conclusion, we have identified a novel, long
non-coding RNA gene in the intron of the ghrelin receptor gene,
GHSR, that exhibits high levels of expression in lung tumour
tissue and regulates cell migration in cultured cells of lung
origin. These observations suggest that GHSROS may be
significant in cancer progression and could be a useful therapeutic
target for inhibiting tumour migration. Further studies on the role
of GHSROS in normal physiology and cancer progression are
required to dissect the function and mechanism of action of this
long non-coding RNA gene. The identification of novel stimulators
of NSCLC progression such as GHSROS will lead to earlier
detection, better prognostic biomarkers and therapeutic approaches
for patients diagnosed with NSCLC in the future.
Acknowledgements
This study was supported by grants
from the National Health and Medical Research Council (NHMRC), the
National Breast Cancer Foundation, The Cancer Council Queensland
(to L.K.C. and A.C.H.), the Queensland University of Technology
(QUT) Early Career Researcher grants (to I.S. and E.J.W.). We thank
Professor Kwun Fong, Professor Ian Yang and Professor Rayleen
Bowman, and Santiyagu Mary Savarimuthu Francis (Department of
Thoracic Medicine, the Prince Charles Hospital, Brisbane,
Australia) for the Beas-2B normal bronchoepithelial lung cell line.
We also thank the staff at the Ontario Tumour Bank (OTC), Canada,
for the recruitment, retention and dispersion of the lung-derived
tissue samples.
References
|
1.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2.
|
Molina JR, Yang P, Cassivi SD, Schild SE
and Adjei AA: Non-small cell lung cancer: epidemiology, risk
factors, treatment, and survivorship. Mayo Clin Proc. 83:584–594.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Nagamachi Y, Tani M, Shimizu K, Tsuda H,
Niitsu Y and Yokota J: Orthotopic growth and metastasis of human
non-small cell lung carcinoma cell injected into the pleural cavity
of nude mice. Cancer Lett. 127:203–209. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Wang Y, Yang H, Liu H, Huang J and Song X:
Effect of staurosporine on the mobility and invasiveness of lung
adenocarcinoma A549 cells: an in vitro study. BMC Cancer.
9:1742009. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Ji P, Diederichs S, Wang W, et al:
MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Schmidt LH, Spieker T, Koschmieder S, et
al: The long noncoding MALAT-1 RNA indicates a poor prognosis in
non-small cell lung cancer and induces migration and tumor growth.
J Thorac Oncol. 6:1984–1992. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Lee J, Yoo J, Yoo H, et al: The novel
miRNA hc-smR-S2-5 decrease the proliferation and migration of human
lung cancer cells by targeting c-Met. Mol Cancer Res. 11:43–53.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liu J, Lu KH, Liu ZL, Sun M, De W and Wang
ZX: MicroRNA-100 is a potential molecular marker of non-small cell
lung cancer and functions as a tumor suppressor by targeting
polo-like kinase 1. BMC Cancer. 12:5192012. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Enfield KS, Pikor LA, Martinez VD and Lam
WL: Mechanistic roles of noncoding RNAs in lung cancer biology and
their clinical implications. Genet Res Int.
2012:7374162012.PubMed/NCBI
|
|
10.
|
Prasanth KV and Spector DL: Eukaryotic
regulatory RNAs: an answer to the ‘genome complexity’ conundrum.
Genes Dev. 21:11–42. 2007.
|
|
11.
|
Gutschner T and Diederichs S: The
hallmarks of cancer: a long non-coding RNA point of view. RNA Biol.
9:703–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Chiang AC and Massague J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar
|
|
13.
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: the next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Ovcharenko I, Nobrega MA, Loots GG and
Stubbs L: ECR Browser: a tool for visualizing and accessing data
from comparisons of multiple vertebrate genomes. Nucleic Acids Res.
32:W280–W286. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Larkin MA, Blackshields G, Brown NP, et
al: Clustal W and Clustal X version 2.0. Bioinformatics.
23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Kong L, Zhang Y, Ye ZQ, et al: CPC: assess
the protein-coding potential of transcripts using sequence features
and support vector machine. Nucleic Acids Res. 35:W345–W349. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Kohany O, Gentles AJ, Hankus L and Jurka
J: Annotation, submission and screening of repetitive elements in
Repbase: RepbaseSubmitter and Censor. BMC Bioinformatics.
7:4742006. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Lai J, Lehman ML, Dinger ME, et al: A
variant of the KLK4 gene is expressed as a cis sense-antisense
chimeric transcript in prostate cancer cells. RNA. 16:1156–1166.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Fung JN, Seim I, Wang D, Obermair A,
Chopin LK and Chen C: Expression and in vitro functions of the
ghrelin axis in endometrial cancer. Horm Cancer. 1:245–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
|
|
21.
|
Mituyama T, Yamada K, Hattori E, et al:
The functional RNA database 3.0: databases to support mining and
annotation of functional RNAs. Nucleic Acids Res. 37:D89–D92. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Bertino EM, Confer PD, Colonna JE, Ross P
and Otterson GA: Pulmonary neuroendocrine/carcinoid tumors: a
review article. Cancer. 115:4434–4441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Nakaya HI, Amaral PP, Louro R, et al:
Genome mapping and expression analyses of human intronic noncoding
RNAs reveal tissue-specific patterns and enrichment in genes
related to regulation of transcription. Genome Biol. 8:R432007.
View Article : Google Scholar
|
|
24.
|
Wobbe CR and Struhl K: Yeast and human
TATA-binding proteins have nearly identical DNA sequence
requirements for transcription in vitro. Mol Cell Biol.
10:3859–3867. 1990.PubMed/NCBI
|
|
25.
|
Jurka J: Novel families of interspersed
repetitive elements from the human genome. Nucleic Acids Res.
18:137–141. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Tsuritani K, Irie T, Yamashita R, et al:
Distinct class of putative ‘non-conserved’ promoters in humans:
comparative studies of alternative promoters of human and mouse
genes. Genome Res. 17:1005–1014. 2007.
|
|
27.
|
Segal E and Widom J: Poly(dA:dT) tracts:
major determinants of nucleosome organization. Curr Opin Struct
Biol. 19:65–71. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Dinger ME, Pang KC, Mercer TR and Mattick
JS: Differentiating protein-coding and noncoding RNA: challenges
and ambiguities. PLoS Comput Biol. 4:e10001762008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Kozak M: An analysis of 5′-noncoding
sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res.
15:8125–8148. 1987.
|
|
30.
|
Taft R, Pang K, Mercer T, Dinger M and
Mattick J: Non-coding RNAs: regulators of disease. J Pathol.
220:126–139. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Kondo T, Hashimoto Y, Kato K, Inagaki S,
Hayashi S and Kageyama Y: Small peptide regulators of actin-based
cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol.
9:660–665. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Kondo T, Plaza S, Zanet J, et al: Small
peptides switch the transcriptional activity of Shavenbaby during
Drosophila embryogenesis. Science. 329:336–339. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Slavoff SA, Mitchell AJ, Schwaid AG, et
al: Peptidomic discovery of short open reading frame-encoded
peptides in human cells. Nat Chem Biol. 9:59–64. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Makawita S and Diamandis EP: The
bottleneck in the cancer biomarker pipeline and protein
quantification through mass spectrometry-based approaches: current
strategies for candidate verification. Clin Chem. 56:212–222. 2010.
View Article : Google Scholar
|
|
35.
|
Jacquot C, Carbonnelle D, Tomasoni C,
Papaconstadinou A, Roussis V and Roussakis C: Identification of a
novel putative non-coding RNA involved in proliferation arrest of a
non-small cell lung carcinoma cell line treated with an original
chemical substance, methyl-4-methoxy-3-(3-methyl-2-butanoyl)
benzoate. Int J Oncol. 25:519–527. 2004.
|
|
36.
|
Chen LL and Carmichael GG: Long noncoding
RNAs in mammalian cells: what, where, and why? Wiley Interdiscip
Rev RNA. 1:2–21. 2010.PubMed/NCBI
|
|
37.
|
Moh MC, Lee LH, Yang X and Shen S:
Identification of a novel gene HEPT3 that is overexpressed in human
hepatocellular carcinoma and may function through its noncoding
RNA. Int J Oncol. 31:293–301. 2007.PubMed/NCBI
|
|
38.
|
Panzitt K, Tschernatsch MM, Guelly C, et
al: Characterization of HULC, a novel gene with striking
up-regulation in hepatocellular carcinoma, as noncoding RNA.
Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Sonkoly E, Bata-Csorgo Z, Pivarcsi A, et
al: Identification and characterization of a novel, psoriasis
susceptibility-related noncoding RNA gene, PRINS. J Biol Chem.
280:24159–24167. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Amaral PP, Neyt C, Wilkins SJ, et al:
Complex architecture and regulated expression of the Sox2ot locus
during vertebrate development. RNA. 15:2013–2027. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Conley AB, Miller WJ and Jordan IK: Human
cis natural antisense transcripts initiated by transposable
elements. Trends Genet. 24:53–56. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Pheasant M and Mattick JS: Raising the
estimate of functional human sequences. Genome Res. 17:1245–1253.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Reis EM, Louro R, Nakaya HI and
Verjovski-Almeida S: As antisense RNA gets intronic. Omics. 9:2–12.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Louro R, Smirnova AS and Verjovski-Almeida
S: Long intronic noncoding RNA transcription: expression noise or
expression choice? Genomics. 93:291–298. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Louro R, Nakaya HI, Amaral PP, et al:
Androgen responsive intronic non-coding RNAs. BMC Biol. 5:42007.
View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Michael DR, Phillips AO, Krupa A, et al:
The human hyaluronan synthase 2 (HAS2) gene and its natural
antisense RNA exhibit coordinated expression in the renal proximal
tubular epithelial cell. J Biol Chem. 286:19523–19532. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Khaitan D, Dinger ME, Mazar J, et al: The
melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates
apoptosis and invasion. Cancer Res. 71:3852–3862. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Lonergan KM, Chari R, Coe BP, et al:
Transcriptome profiles of carcinoma-in-situ and invasive non-small
cell lung cancer as revealed by SAGE. PLoS One. 5:e91622010.
View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Dempsey EC, Cool CD and Littler CM: Lung
disease and PKCs. Pharmacol Res. 55:545–559. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Espina V, Wulfkuhle JD, Calvert VS, et al:
Laser-capture microdissection. Nat Protoc. 1:586–603. 2006.
View Article : Google Scholar
|
|
51.
|
Edwards RA: Laser capture microdissection
of mammalian tissue. J Vis Exp. 2007:3092007.PubMed/NCBI
|
|
52.
|
Gupta RA, Shah N, Wang KC, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Gutschner T, Hammerle M, Eissmann M, et
al: The non-coding RNA MALAT1 is a critical regulator of the
metastasis phenotype of lung cancer cells. Cancer Res.
73:1180–1189. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Xu C, Yang M, Tian J, Wang X and Li Z:
MALAT-1: a long non-coding RNA and its important 3′ end functional
motif in colorectal cancer metastasis. Int J Oncol. 39:169–175.
2011.
|
|
55.
|
Ying L, Chen Q, Wang Y, Zhou Z, Huang Y
and Qiu F: Upregulated MALAT-1 contributes to bladder cancer cell
migration by inducing epithelial-to-mesenchymal transition. Mol
Biosyst. 8:2289–2294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Tano K, Mizuno R, Okada T, et al: MALAT-1
enhances cell motility of lung adenocarcinoma cells by influencing
the expression of motility-related genes. FEBS Lett. 584:4575–4580.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Guffanti A, Iacono M, Pelucchi P, et al: A
transcriptional sketch of a primary human breast cancer by 454 deep
sequencing. BMC Genomics. 10:1632009. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Lai MC, Yang Z, Zhou L, et al: Long
non-coding RNA MALAT-1 overexpression predicts tumor recurrence of
hepatocellular carcinoma after liver transplantation. Med Oncol.
29:1810–1816. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Yamada K, Kano J, Tsunoda H, et al:
Phenotypic characterization of endometrial stromal sarcoma of the
uterus. Cancer Sci. 97:106–112. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Lin R, Maeda S, Liu C, Karin M and
Edgington T: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Lottin S, Adriaenssens E, Dupressoir T, et
al: Overexpression of an ectopic H19 gene enhances the tumorigenic
properties of breast cancer cells. Carcinogenesis. 23:1885–1895.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Matouk IJ, DeGroot N, Mezan S, et al: The
H19 non-coding RNA is essential for human tumor growth. PLoS One.
2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Yoshimizu T, Miroglio A, Ripoche MA, et
al: The H19 locus acts in vivo as a tumor suppressor. Proc Natl
Acad Sci USA. 105:12417–12422. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Barsyte-Lovejoy D, Lau SK, Boutros PC, et
al: The c-Myc oncogene directly induces the H19 noncoding RNA by
allele-specific binding to potentiate tumorigenesis. Cancer Res.
66:5330–5337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
65.
|
Yang F, Bi J, Xue X, et al: Up-regulated
long non-coding RNA H19 contributes to proliferation of gastric
cancer cells. FEBS J. 279:3159–3165. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66.
|
Kaplan R, Luettich K, Heguy A, Hackett NR,
Harvey BG and Crystal RG: Monoallelic up-regulation of the
imprinted H19 gene in airway epithelium of phenotypically normal
cigarette smokers. Cancer Res. 63:1475–1482. 2003.PubMed/NCBI
|
|
67.
|
Cai X and Cullen BR: The imprinted H19
noncoding RNA is a primary microRNA precursor. RNA. 13:313–316.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
68.
|
Keniry A, Oxley D, Monnier P, et al: The
H19 lincRNA is a developmental reservoir of miR-675 that suppresses
growth and Igf1r. Nat Cell Biol. 14:659–665. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Huarte M, Guttman M, Feldser D, et al: A
large intergenic noncoding RNA induced by p53 mediates global gene
repression in the p53 response. Cell. 142:409–419. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Yoon JH, Abdelmohsen K, Srikantan S, et
al: LincRNA-p21 suppresses target mRNA translation. Mol Cell.
47:648–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Gebeshuber CA, Zatloukal K and Martinez J:
miR-29a suppresses tristetraprolin, which is a regulator of
epithelial polarity and metastasis. EMBO Rep. 10:400–405. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Krol M, Polanska J, Pawlowski KM, et al:
Transcriptomic signature of cell lines isolated from canine mammary
adenocarcinoma metastases to lungs. J Appl Genet. 51:37–50. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
Mattick JS and Makunin IV: Non-coding RNA.
Hum Mol Genet 15 Spec No. 1:R17–R29. 2006. View Article : Google Scholar
|
|
74.
|
Ulveling D, Francastel C and Hube F: When
one is better than two: RNA with dual functions. Biochimie.
93:633–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Cesana M, Cacchiarelli D, Legnini I, et
al: A long noncoding RNA controls muscle differentiation by
functioning as a competing endogenous RNA. Cell. 147:358–369. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Wang J, Liu X, Wu H, et al: CREB
up-regulates long non-coding RNA, HULC expression through
interaction with microRNA-372 in liver cancer. Nucleic Acids Res.
38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Franco-Zorrilla JM, Valli A, Todesco M, et
al: Target mimicry provides a new mechanism for regulation of
microRNA activity. Nat Genet. 39:1033–1037. 2007. View Article : Google Scholar : PubMed/NCBI
|