Introduction
The increasing age of the world’s population has
been the most dramatic demographic change since the 20th century
(1). As the fifth leading cause of
death in women and the first cause of gynecologic cancer death,
ovarian cancer incidence and mortality rate increase continuously
with advancing age and peaks in the 7th to 8th decades of life
(2–6). Due to lack of symptoms in early-stage
of this disease, in China, indeed the long-term survival for
elderly patients with advanced disease does not exceed 20%.
Therefore, the poor prognosis of ovarian cancer in the elderly has
been recently recognized (4). A
prior report showed that, 28,082 women were diagnosed with
epithelial ovarian cancer (EOC) from 1988 to 2001. The largest
histology subgroup, 49.3% (13,835) of patients had ovarian
papillary serous carcinoma (OPSC) (7). OPSC increases steadily after
menopause, and the major patients of OPSC with advanced-stage (FIGO
stages III or IV) diagnoses have a <30% 5-year survival rate
(7,8). Thus, age should be considered as an
important prognostic variable. In addition, ovarian clear cell
carcinoma (OCC) was frequently present at early stage (9), and has a distinct, aggressive
biologic behavior with poor survival rates compared to other
epithelial counterparts (7,10–12).
Given that elderly advanced OPSC and OCC all carry poor prognosis,
it is advantageous to study them together, find the essential
differences of cancer epidemiology and biology, and then improve
the understanding of the molecular basis which plays an important
part in pathogenesis of elderly advanced OPCS or OCC cases.
MicroRNAs (miRNAs) are small non-coding RNA
molecules of ~22 nucleotides that act post-transcriptionally to
regulate gene expression (8). They
show great potential in discovering new biological pathways; could
be used as a diagnostic and prognostic tool for cancer patients and
even serve as molecular targets for therapy.
Considering the above evidence, the OPSC patients
aged ≥50 years with advanced-stage and OCC patients, who all carry
poor prognosis, were studied. The goal was to identify the
differences of key miRNA and possible regulators through miRNA
microarray chip, functional target prediction, and clinical outcome
between the elderly advanced OPSC and OCC patients to find the
pathogenic basis, and to provide insight into clinical diagnosis
and therapy for advanced OPSC, especially in elderly patients.
Materials and methods
Patients and samples
Patients, who were surgically treated for ovarian
cancer at the Obstetrics and Gynecology Hospital, Dalian, China
between July 2004 and November 2010 were identified. The study was
approved by a central and by institutional ethics committees.
Corresponding 58 tumor specimens; OPSC (n=30); OCC (n=28) were
dissected during the operations, and formalin fixed paraffin
embedded (FFPE) blocks were further obtained after surgery. All
pathological specimens from primary surgery were reviewed by two
independent pathologists with no knowledge of patients’ clinical
data. Cases were classified according to the FIGO staging system.
All cases were newly diagnosed, and only serous papillary (≥50
years and FIGO stages III or IV tumors) and OCC histology were
included.
MicroRNA array and data analysis
A microarray platform optimized for the analysis of
a panel of 768 human miRNAs was used to analyze and compare the
pattern of miRNA expression between OCC (n=9) and elderly advanced
OPSC (n=8). Total RNA that enriched miRNAs was extracted from the
FFPE tissue by using the Ambion mirVana microRNA isolation kit
(Ambion, Austin, TX). The quality of total RNA was assessed using
the Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA).
Individual quantitative real-time polymerase chain reaction assays
were formatted into a TaqMan low-density array (TLDA; Applied
Biosystems), which was performed at the Shannon McCormack Advanced
Molecular Diagnostics Laboratory Research Services, Dana Farber
Cancer Institute, Harvard Clinic and Translational Science Center.
The normalized microarray data were managed and analyzed by
Statminer version 3.0 (Integromics™).
MiRNA targets prediction and pathway
analysis
The analysis of miRNA predicted targets was
determined using several computational approaches, the MicroCosm
Targets version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
and miRBase (http://www.mirbase.org/). The target
prediction algorithm used here is estimated to have a 20–30% false
positive rate. This level of false discovery is unlikely to affect
the overall network findings obviously, although the number of top
predicted gene targets is large. Functional analysis of these
predicted gene targets identified biologic pathways with
significant involvement for gene expression. In order to retrieve
only the most relevant targets, we listed only genes targeted by
the miRNAs that were differentially expressed in the patient with
OPSC and OCC. To further understand and interpret literature
information of our unique miRNAs, an analysis of biologic pathway
relationships was performed using commercially available software
(Ingenuity Systems, Redwood City, CA).
Quantitative real-time RT-PCR
To validate key microarray results, quantitative
reverse transcription was performed using the TaqMan MicroRNA
Reverse Transcription Kit [Applied Biosystems (ABI), Foster City,
CA] with ABI miRNA specific primers and primer kits on an Agilent
Technologies Stratagent Mx3000P. Specific kits used were as
follows: hsa-miR-505*: ABI#4395198;
hsa-miR-30a*: ABI#4373062; hsa-miR-30e*:
ABI#4427975. Relative expression levels were calculated using the
comparative Ct (2−ΔΔCt) method with U6 small nuclear RNA
as the endogenous control. Samples were analyzed in triplicate.
Technical validations were carried out using a subset of the
original samples that were used in the discovery phase of the study
with miRNA array. Biological validation was performed using
additional FFPE cases of OPSC and OCC (n=21 in total) (13).
qRT-PCR was performed to validate target prediction
results. Total RNA (0.5 μg) in 1 μl of RNase-free water was used in
20 μl of RT mix. Primer pairs were as follows: activating
transcription factor 3 (ATF3) forward 5′-CTGCAGAAAGAGTCGGAG-3′ and
reverse 5′-TGAGCCCGGACAATACAC-3′; β-actin forward
5′-GACTACCTCATGAAGATC-3′ and reverse 5′-GATCCACA TCTGCTGGAA-3′
(Invitrogen). Real-time PCR was done using the LightCycler system
and Roche fast-Start Light Cycler-Master Hybridization Probes
master mix (Roche Diagnostics), and the product was detected with
SYBR-Green I. Samples from at least three independent experiments,
each measured in duplicate, were analyzed and the data expressed as
the averages ± SE.
Immunohistochemistry (IHC)
The IHC staining procedure was described previously
(14) with modification. Briefly,
after dewaxing in xylene and descending concentrations of ethanol,
the sections were incubated in 3% H2O2 for 30
min to suppress endogenous peroxidase activity. The sections were
further blocked with a mixed solution [10% goat serum and 3%
albumin bovine (BSA) in PBS] for 1 h and incubated with the primary
antibodies of ATF3 (1:100 dilution; Bioworld, cat#BS1245); Stmn1
(1:100 dilution; Bioworld, cat#BS3615); or MYC (1:100 dilution;
Bioworld, cat#BS2261) overnight at 4°C. On the second day, after
rinsing for 10 min × 3, the sections were incubated with
streptavidin-peroxidase staining system kit according to the
manufacturer’s instructions (SP-9001, Zhongshan Golden Bridge
Biotechnology Company, Beijing, China). Finally,
3,3′-diaminobenzidine (DAB) liquid chromogen substrate kit
(ZLI-9032, Zhongshan Golden Bridge Biotechnology Company) offered a
clean detection performance and subsequent hematoxylin staining
provided a light blue counter stain contrasting with DAB. For
negative controls, all the conditions were performed at the same
procedures except that primary antibody was eliminated.
Statistical analysis
The results were analyzed by SPSS 15.0 (Chicago,
IL). Data are expressed as arithmetic means ± SEM of the number (n)
of experiments. Samples were analyzed with repeated measures
analysis of variance; differences in the incidence were analyzed
using ANOVA. The differences of the positive area and integrate
optical density (IOD) per vision-field of ×400 immunohistochemistry
photographs were taken with Image-Pro plus vision 6.0. Overall
survival was defined as the time from initial cytoreductive surgery
to date of last follow-up or death. Survival time course was
studied using the Kaplan-Meier method, and groups were compared
using the log-rank test. P<0.05 was considered statistically
significant.
Results
Patient characteristics
Patient characteristics are shown in Table I. Fifty-eight patients were
identified to fit the study criteria, including 30 OPSC patients
(age ≥50 years with advanced-stage) and 28 OCC patients. The
percentage of patients with stage III disease and the presence of
positive lymph nodes were all significantly higher in those
patients with elderly advanced OPSC compared with those patients
with elderly OCC (8.3 vs. 93.3%).
 | Table IClinicopathological characteristics
of the patients. |
Table I
Clinicopathological characteristics
of the patients.
|
Characteristics | Total
(n=58)
No. (%) | Advance OPSC ≥50
(n=30)
No. (%) | OCC (n=28) | P-valuea |
|---|
|
|---|
<50
(n=16)
No. (%) | ≥50
(n=12)
No. (%) | Total OCC
(n=28)
No. (%) |
|---|
| Age at
diagnosis |
| Median | 54.1 | 59.9 | 38.6 | 54.7 | 45.5 | 0.035 |
| Range | (29–74) | (50–74) | (29–48) | (50–62) | (29–62) | |
| FIGO stage at
diagnosis | | | | | | <0.001c |
| I | 23 (39.7) | 0 (0.0) | 12 (75.0) | 11 (91.7) | 23 (82.1) | |
| IA | 13 (22.4) | 0 (0.0) | 6 (37.5) | 7 (58.3) | 13 (46.4) | |
| IC | 10 (17.2) | 0 (0.0) | 6 (37.5) | 4 (33.3) | 10 (35.7) | |
| II | 2 (3.4) | 0 (0.0) | 2 (12.5) | 0 (0.0) | 2 (7.1) | |
| IIC | 2 (3.4) | 0 (0.0) | 2 (12.5) | 0 (0.0) | 2 (7.1) | |
| IIIb | 31 (53.4) | 28 (93.3) | 2 (12.5) | 1 (8.3) | 3 (10.7) | |
| IIIB | 3 (5.2) | 1 (3.3) | 1 (6.3) | 1 (8.3) | 2 (7.1) | |
| IIIC | 27 (46.6) | 26 (86.7) | 1 (6.3) | 0 (0.0) | 1 (3.6) | |
| IV | 2 (3.4) | 2 (6.7) | 0 (0.0) | 0 (0.0) | 0 (0.0) | |
| Graded |
| 1 | 2 (3.4) | 1 (3.3) | 0 (0.0) | 1 (8.3) | 1 (3.6) | |
| 2 | 9 (15.5) | 6 (20.0) | 1 (6.3) | 2 (16.7) | 3 (10.7) | |
| 3 | 29 (50.0) | 22 (73.3) | 4 (25) | 3 (25.0) | 7 (25.0) | |
| Unknown | 18 (31.0) | 1 (3.3) | 11 (68.8) | 6 (50.0) | 17 (63.0) | |
|
Lymphadenectomy |
| Yes | 44 (75.9) | 20 (66.7) | 15 (93.8) | 9 (75) | 24 (85.7) | |
| No | 12 (20.7) | 9 (30.0) | 1 (6.2) | 2 (16.7) | 3 (10.7) | |
| Unknown | 2 (3.5) | 1 (3.3) | 0 (0.0) | 1 (8.3) | 1 (3.6) | |
| Median no. nodes
resected | 20 | 19 | 18 | 23 | 20 | |
| Presence of
positive nodes | | | | | | 0.002 |
| Yes | 14 (24.1) | 12 (40.0) | 1 (6.3) | 1 (8.3) | 2 (7.1) | |
| No | 30 (51.7) | 8 (26.7) | 14 (87.5) | 8 (66.7) | 22 (78.6) | |
| Unknown | 14 (24.1) | 10 (33.3) | 1 (6.3) | 3 (25.0) | 4 (14.3) | |
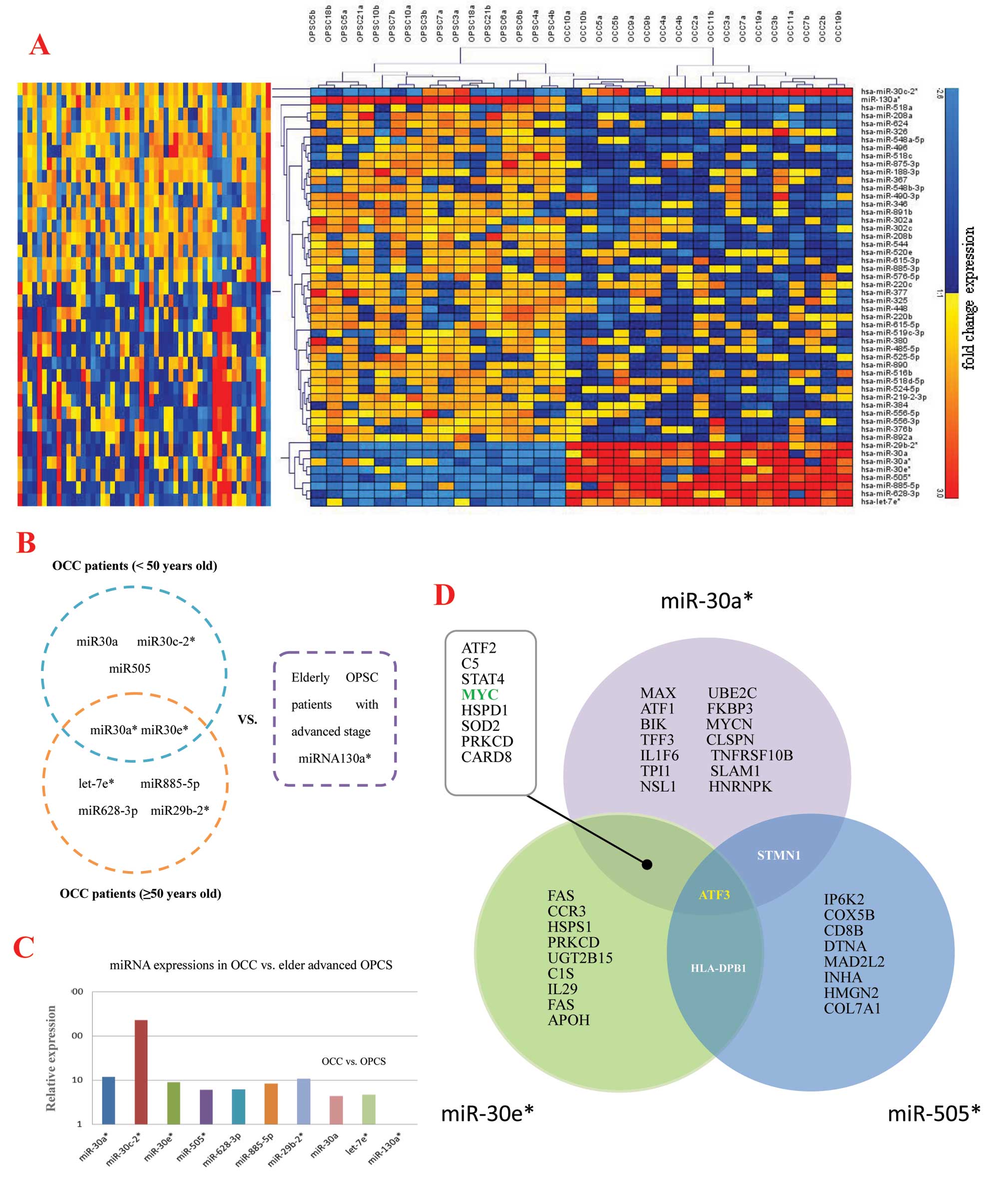
miRNA expression pattern of OPSC and
OCC
miRNAs have been reported in different types of
tumors derived from different organs, including ovarian cancer.
However, the pathobiological significances of aberrant miRNA
expression in human ovarian cancer have not been well documented.
To further characterize the unique miRNAs in EOC development, we
initially analyzed miRNA expression in tumors of 8 elderly OPSC
patients with advanced stage and 9 OCC patients for microarray
analysis. According to the background-subtracted and normalized
fluorescent intensities of array sample results, 52 unique miRNAs
were detected with significant (p<0.00001 for all), fold-change
(FC) in expression level, of which 9 were upregulated, whereas 43
miRNAs were downregulated in OCC patients compared to elderly OPSC
patients with advanced stage (Table
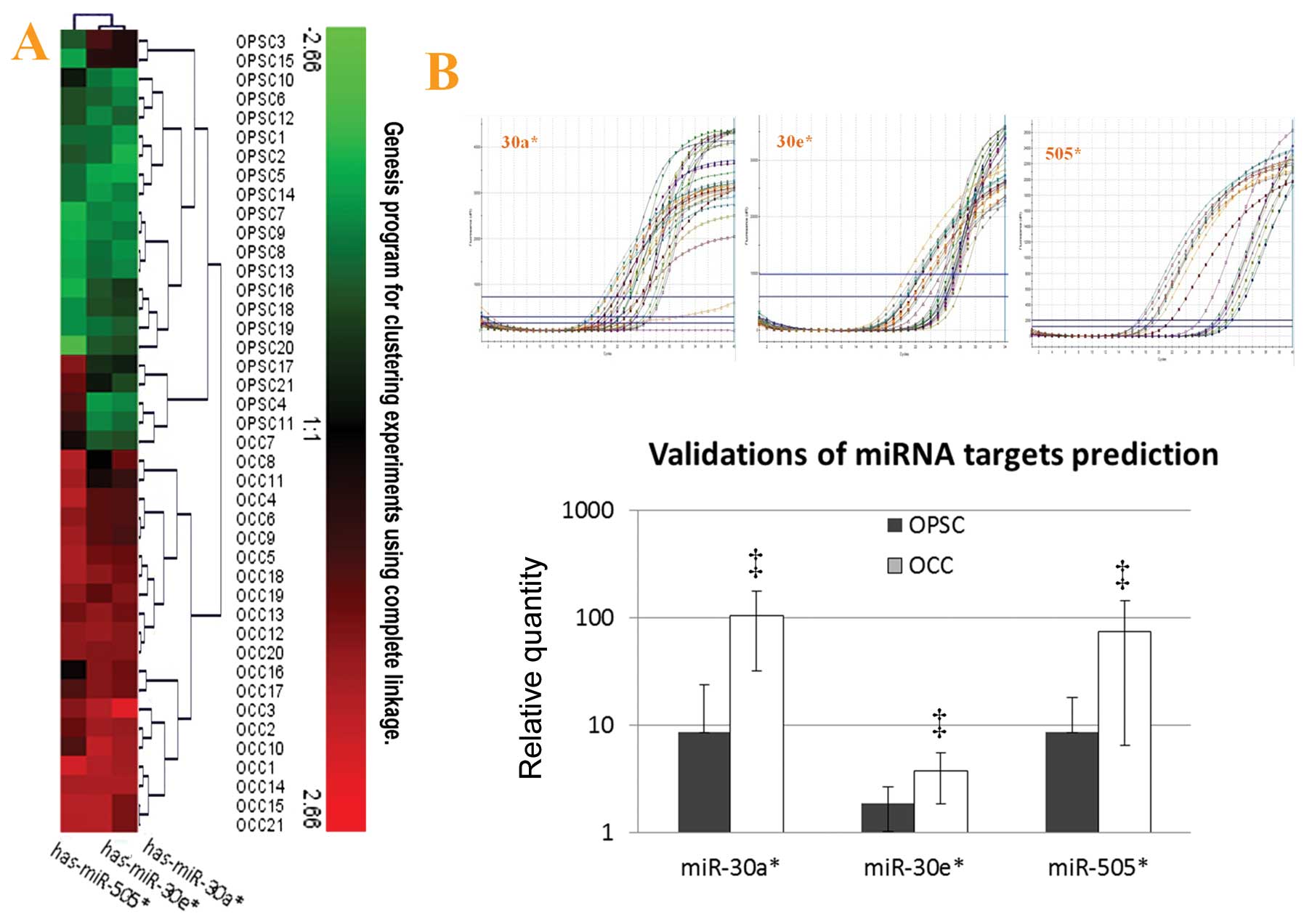
II and Fig. 1A). Moreover,
among the top significant unique miRNAs (FC >4), 9 were
preferentially expressed in the OCC patients, whereas only 1 miRNA
was most highly expressed in the elderly OPSC with advanced stage
(Table II). The prediction
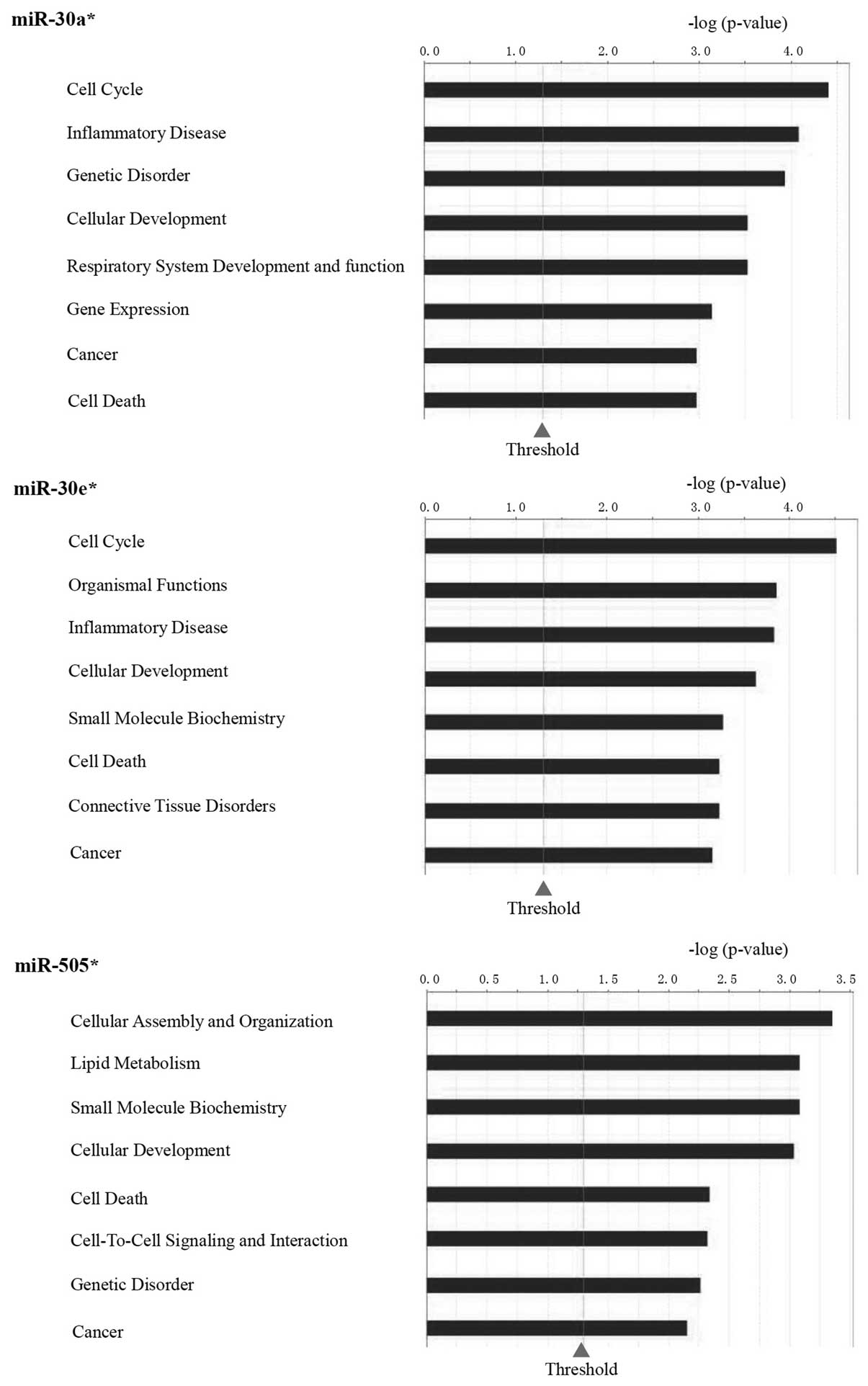
targets of these 9 miRNAs are shown in Fig. 1B and C.
 | Figure 1Target predictions of miRNAs through
microarray analysis. (A) Results of the unsupervised hierarchical
clustering of microarray assay by qRT-PCR. Among the top
significantly unique miRNAs (fold-change >4, FDR <0.01, B and
C) using microarray platform to predict target miRNAs, 9 miRNAs are
preferentially expressed in the OCC patients when compared to the
elderly OPSC with advanced stage, and their relative expressions
are shown in the column map. Only miR-130a*, is highly
expressed in OPSC (its FC value is 0.0035528). These 9 miRNAs are
distributed in different ages of OCC patients, and
miR-30a* and miR-30e* are the co-prediction
targets of younger (<50 years) and elderly (≥50 years) OCC
patients. (D) Venn diagram of selected miRNAs and their putative
gene targets. The MicroCosm Targets version 5 and miRBase were used
to analyze predicted targets for the 10 top significantly unique
miRNAs which were highly expressed in OPSC vs. OCC with
p<0.00001, and change >4-fold. Three miRNAs were found with
some common targets, including miR-30a*,
miR-30e* and miR-505*. These events suggest
they might have an important role in ovarian cancer differences
between OCC and elder advanced OPSC together or partly, and ATF3 is
the co-target of these relevant miRNAs. |
 | Table IIThe miRNA expression in elderly OPSC
and OCC patients with advanced stage. |
Table II
The miRNA expression in elderly OPSC
and OCC patients with advanced stage.
| miR name | P-value | FDR (%)a | Fold-change
(FC) | Fold-change
grade | Higher expression
associated with | Cancer
involvementb |
|---|
|
hsa-miR-130a* | 1.40E-08 | <0.01 | 0.003552833 | IV | OPSC | Brain, liver |
| hsa-miR-188-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-miR-29b-2* | 3.33E-07 | <0.01 | 10.83658116 | III | OCC | Thyroid, uterus,
AML |
| hsa-miR-208a | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-208b | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-miR-219-2-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-220b | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-220c | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-30a | 1.47E-06 | <0.01 | 4.376533535 | I | OPSC | Lung, colon,
myeloid leukemia, renal |
|
hsa-miR-30a* | 1.30E-08 | <0.01 | 11.97926536 | II | OCC | Colon, breast,
esophagus, bladder |
|
hsa-miR-30c-2* | 2.72E-07 | <0.01 | 226.4780393 | IV | OCC | Endometrial,
stomach, ovarian, breast, colon |
|
hsa-miR-30e* | 5.97E-09 | <0.01 | 8.945290166 | II | OCC | Ovarian, brain,
epithelium, esophagus, breast, lung, colon |
| hsa-miR-302a | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Skin, germ
cell |
| hsa-miR-302c | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Breast |
| hsa-miR-325 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-326 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Brain |
| hsa-miR-346 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Follicular
thyroid |
| hsa-miR-367 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Ependymomas |
| hsa-miR-376b | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-377 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Breast |
| hsa-miR-380 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-384 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Laryngeal,
breast |
| hsa-miR-448 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Prostate,
breast |
| hsa-miR-485-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Ovarian |
| hsa-miR-490-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Colon |
| hsa-miR-496 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Ovary, AML |
|
hsa-miR-505* | 4.98E-06 | <0.01 | 6.122042066 | II | OCC | Bladder |
| hsa-miR-516b | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-518a | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Follicular
lymphoma |
| hsa-miR-518c | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Colon, bladder |
|
hsa-miR-518d-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-miR-519c-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Lung |
| hsa-miR-520e | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Cholangio,
liver |
| hsa-miR-524-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-525-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-544 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-miR-548a-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-miR-548b-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-556-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Prostate |
| hsa-miR-556-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Prostate |
| hsa-miR-576-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Ovary |
| hsa-miR-615-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Breast, colon,
prostate, blood, ovary |
| hsa-miR-615-5p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Breast, colon,
prostate, blood, ovary |
| hsa-miR-628-3p | 2.43E-07 | <0.01 | 6.140440621 | II | OCC | Brain, breast,
thyroid |
| hsa-miR-624 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-875-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Pancreas |
| hsa-miR-885-3p | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Lung, brain |
| hsa-miR-885-5p | 5.11E-06 | <0.01 | 8.409273981 | II | OCC | Renal, brain,
ovary |
| hsa-miR-890 | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | Nasopharyngeal |
| hsa-miR-891b | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
| hsa-miR-892a | 8.87E-06 | <0.01 | 0.62304526 | I | OPSC | |
|
hsa-let-7e* | 5.99E-06 | <0.01 | 4.703680442 | I | OPSC | Head and neck,
retinoblastoma, pleural |
Unique miRNAs and their co-target
prediction
The Micro Cosm Targets version 5 and miRBase were
used to analyze predicted targets for the 10 top significantly
unique miRNAs highly expressed in OPSC vs. OCC with p-values
<0.00001, and FC >4-fold. The target prediction algorithm
used here is estimated to have a 20–30% false positive rate. This
level of false discovery is unlikely to affect the overall network
findings obviously, although the number of top predicted gene
targets is large. Functional analysis of these predicted gene
targets identified biologic pathways with significant involvement
for gene expression. In order to retrieve only the most relevant
targets, we listed only genes targeted by miR-30a*,
miR-30e* and miR-505* that were found having
some targets in common suggesting they might play important roles
in pathogenesis between OCC and elderly advanced OPSC together or
partly (Figs. 1D and 2). Fig.
3 show the function analysis of miR-30a*,
miR-30e* and miR-505*.
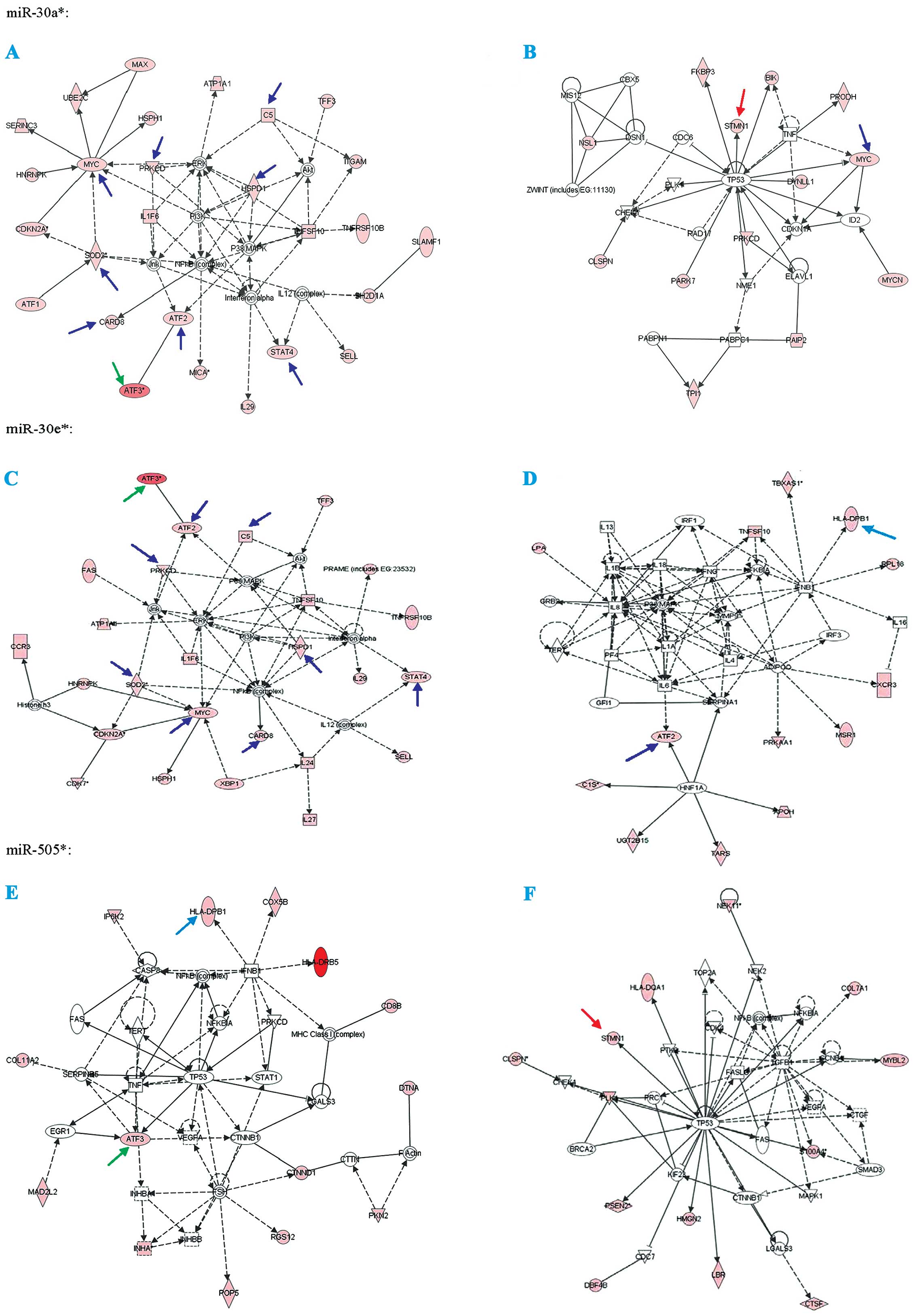 | Figure 2The analysis of prediction protein
targets for miR-30a*, miR-30e* and
miR-505* were determined through using the MicroCosm
Targets version 5 (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/)
and miRBase (http://www.mirbase.org/). Green
arrows, co-targets of miR-30a*, miR-30e* and
miR-505*; light blue arrows, co-targets of
miR-30e* and miR-505*; blue arrows,
co-targets of miR-30a* and miR-30e*; and red
arrows, co-targets of miR-30a* and miR-505*.
Associated network functions are: (A) DNA replication,
recombination and repair, cancer, cell death of
miR-30a*; (B) cell cycle, cancer, cell death of
miR-30a*; (C) cell morphology, cancer, cell death of
miR-30e*; (D) dermatological diseases and conditions,
cellular movement, hematological system development and function of
miR-30e*; (E) cancer, cellular growth and proliferation,
cell death of miR-505*; and (F) cell cycle, cancer,
renal and urological disease of miR-505*. |
qRT-PCR validation for microarray
results
In order to confirm microarray results, RNA was
isolated from a new set of FFPE tissues as described above to
increase the likelihood that the observed differences in miRNA
expression profiles represent biologically significant changes.
MiR-30a*, miR-30e* and miR-505*
were the upregulated miRNAs in OCC with different fold-changes
(from 6–12) when compared with OPSC by using microarray analysis.
Validation of miRNA expression analysis was repeated with qRT-PCR
(miR-30a*, miR-30e* and miR-505*)
and representative analyses are shown in Fig. 4A (unsupervised hierarchical
clustering of validation) and Fig.
4B. Through this additional analysis, the expression patterns
found in the arrays could be confirmed. In keeping with microarray
results, miR-30a*, miR-30e* and
miR-505* were all highly expressed in OCC with
statistical significance.
Validations of miRNA target prediction
and their top co-targets
In target prediction experiment using multiple
software, ATF3 was predicted as a potential co-target of
miR-30a*, miR-30e* and miR-505*,
and presented as a regulator in the different pathways, which
include cancer and cell death. At the same time, we found that MYC
is the co-target of miR-30a* and miR-30e*;
stathmin1 (STMN1) is co-target of miR-30a* and
miR-505*; HLA-DPB1 is co-target of miR-30e*
and miR-505*. To examine the biological significance of
these miRNAs in ovarian cancer, we focus on the ATF3, STMN1 and
MYC, which were already proven to be cancer markers.
Immunohistochemical assay for the ATF3, STMN1 and
MYC proteins showed a relevant upregulation in OPSC cells compared
to OCC cells (Fig. 5A). It is
clear that these co-targets were all extensively distributed in the
cytoplasm of cancer cells in the tissue of OPSC samples (Fig. 5A1–3) comparing with the OCC
sections (Fig. 5A4–6). Through
analysis with Image-Pro plus vision 6.0, positive area and IOD per
vision-field of ×400 immunohistochemistry photographs were
detected. These results also support the conclusions of the
immunohistochemical assay (Fig. 5B and
C).
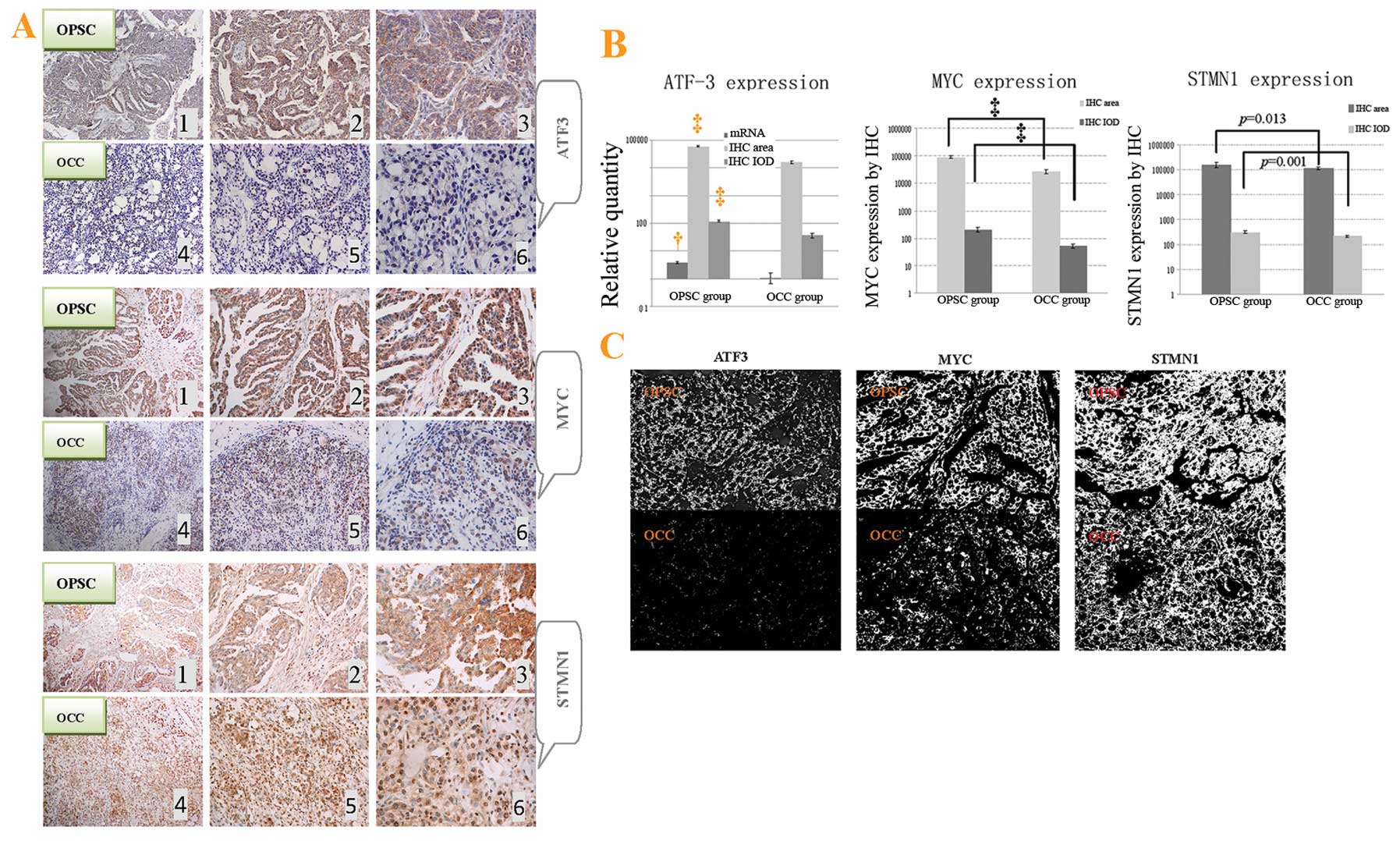 | Figure 5ATF3, STMN1 and MYC protein
expressions in paraffin-embedded tissues in the immunohistochemical
assay, and overall survival of ovarian cancer patients with elderly
advanced OPSC and OCC. (A) Different expression of ATF3, STMN1 and
MYC protein between OPSC and OCC cells by immunohistochemistry
(IHC) with hematoxylin counter staining. ATF3, STMN1 and MYC, are
extensively expressed in the cytoplasm of OPSC tumor cells (A1, 2
and 3) and poorly expressed in OCC tissues (A4, 5 and 6).
Magnifications from left to right are ×100, ×200 and ×400,
respectively. (B) As the only co-target of these miRNAs, ATF3 mRNA
expression was also detected through real-time PCR. Different
expressions for positive area and integrate optical density (IOD)
of ATF3, STMN1 and MYC (per vision-field of ×400 photograph taken
with Image-Pro plus vision 6.0) between OPSC and OCC tumor through
immunohistochemistry staining, respectively, are shown. (C) Image
(x400) showing positive staining area of above proteins by
Image-Pro plus vision 6.0, respectively. Compared with the OCC,
†p<0.05, and ‡p<0.01. |
Overall survival analysis of elderly
advanced OPSC and OCC patients
We next compared the prognosis of elderly advanced
OPSC patients in groups stratified according the expression levels
of individual miRNAs. For each miRNA, we divided the samples into
two sub-sections according to high and low expression level of the
miR-30a*, miR-30e* and miR-505*
(Fig. 6A–C). The association of
these three miRNAs with survival indicated that lower expression of
miR-30a*, miR-30e* and miR-505*,
all associated with poorer prognosis. We also studied the overall
survival of ovarian cancer patients with OPSC and OCC (Fig. 6D). They were associated with
significant differences (OPSC was lower, log-rank p<0.01) in
overall survival.
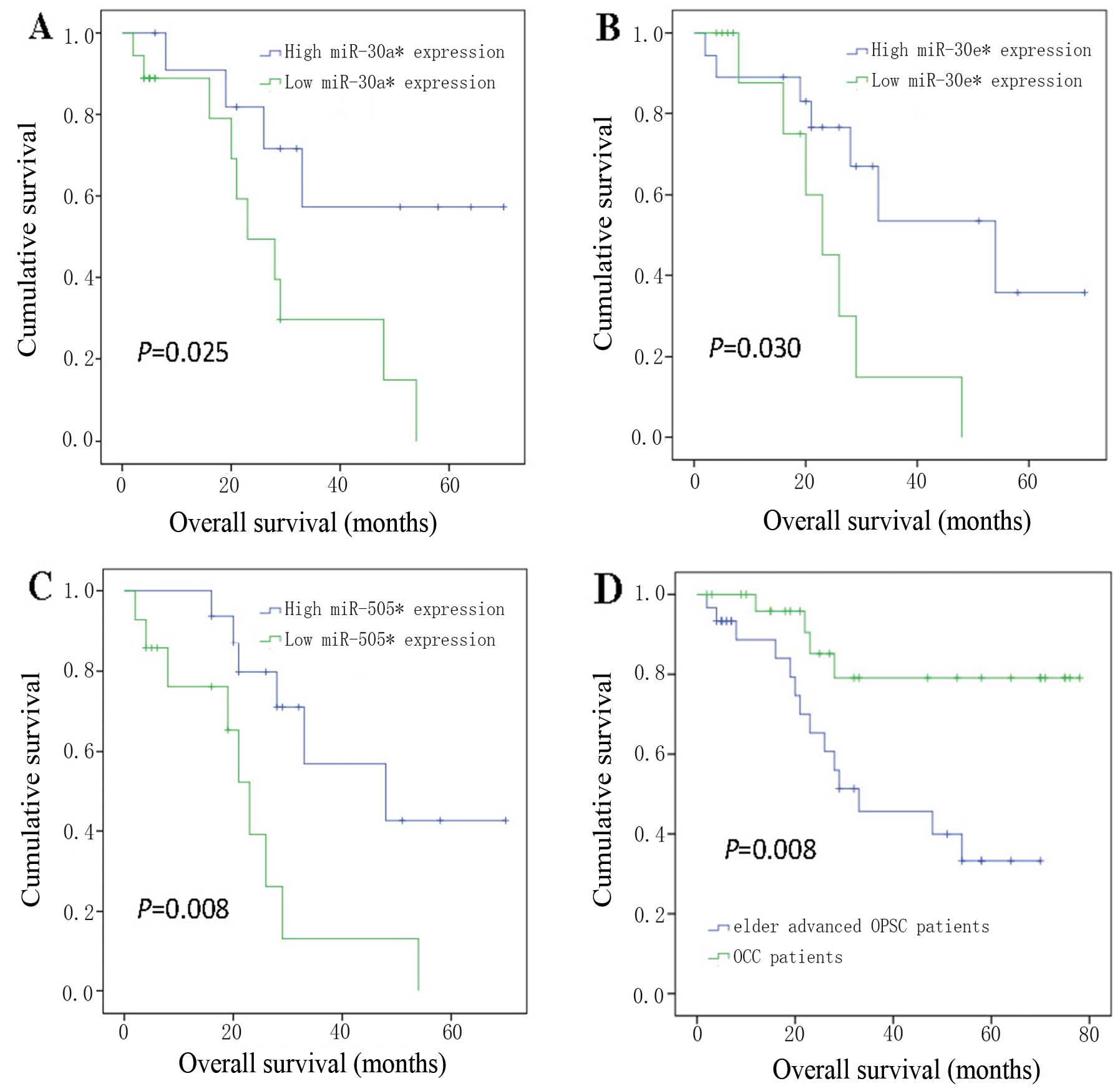 | Figure 6Kaplan-Meier curves showing overall
survival for groups of elderly OPSC patients with advanced disease
(A, B and C), stratified by expression levels of (A)
hsa-miR-30a*, (B) hsa-miR-30e* and (C)
hsa-miR-505*; (D) overall survival of ovarian cancer
patients with OPSC and OCC. The samples of elder advanced OPSC
patient were divided into two groups with high expression levels
(blue line) and low expression levels (green line) of (A)
hsa-miR-30a* (p=0.0025), (B) hsa-miR-30e*
(p=0.030) and (C) hsa-miR-505* (p=0.008), n=30 for all.
Lower expressions of miR-30a*, miR-30e* and
miR-505*, all associated with poorer prognosis. (D)
Overall survival associated with the histological type, elder
advanced OPSC and OCC, was also tested (n=58, p=0.008). The overall
survival of elder advanced OPSC is obviously lower than OCC
patients. P-values are calculated by log-rank test comparing the
low and high expression groups. Censoring events are marked by
vertical lines. |
Discussion
EOC is the most important cause of gynecologic
malignancy-related mortality in women (15) rising continuously with advancing
age. Some recent reports show that cancer incidence is 11-fold
greater for the older population (4,16).
Therefore, it is necessary to study and understand cancer
epidemiology, biology and therapy to the elderly patient. The
incidence of OPSC increases steadily after menopause, and this
histologic subgroup makes up the largest part of EOC patients.
Considering that for most women menopause is after
the age of 50 years, OPSC patients aged ≥50 years with
advanced-stage as well as OCC patients were studied. The purpose
was to apply microarray analysis to investigate the difference,
especially molecular mechanism between elderly OPSC with advanced
stage and OCC, which both carry poor prognosis (8,9).
Through miRNA microarray and target prediction analysis, 10 miRNAs
were found to be differentially expressed in tumor from OPSC vs.
OCC (p<0.00001 and FC >4). Moreover, in order to retrieve
only the most relevant targets, we only investiged three miRNAs
(miR-30a*, miR-30e* and miR-505*)
which were found with some common cancer associated pathways. We
confirmed these predictions and relations. Biological pathways were
predicted using multiple software, and ATF3 was indicated as an
only potential co-target of these three miRNAs.
Advanced OPSC has its own specific pathogenic
factors, especially in elderly patients. Lower expressions of
miR-30a*, miR-30e* and miR-505*
were validated in elderly advanced OPSC patients consistent with
microarray analysis. The survival analysis was investigated for
elderly advanced OPSC patients in groups stratified according the
expression levels of individual miRNAs, and the results revealed
that lower expressions of miR-30a*, miR-30e*
and miR-505*, all associated with significantly poorer
prognosis. The results strongly suggested that they could be
critical oncogenes and take important roles in ovarian cancer
etiology with advantaged stage.
In order to validate our above hypothesis, we
selected some typical downstream prediction targets which have
associated with cancer as supporting evidence. ATF3 is a member of
the ATF/cyclic adenosine monophosphate response element binding
family of transcription factors (17,18).
It is expressed at low levels in normal and quiescent cells, but
can rapidly and significantly increase in various cancers (19). Previous research supports that
overexpression of ATF3 could play an oncogenic role in
carcinogenesis. These studies provide correlative evidence that
ATF3 expression contributes to the successful propagation of human
cancer (17,20–22),
and also could promote metastasis, cell adhesion and invasion in
vitro and in vivo(23,24).
Furthermore, STMN1 overexpression is associated with polyploidy,
tumor-cell invasion, early recurrence and poor prognosis in human
hepatoma and ovarian cancer (25–27).
MYC was also proven to have a pivotal role as a regulator of
tumorigenesis in numerous human cancers of diverse origin (28). In our results, they all increased
more in elderly advanced OPSC group, strongly supporting our
pathogenic hypothesis for miR-30a*, miR-30e*
and miR-505*, and suggesting that OPSC also has
aggressive biologic behavior when presented with advanced-stage.
Epidemiology results show that incidence and mortality rate of
advanced OPSC rise continuously with advancing age. Previous
reports have shown that women with OCC have a poorer prognosis
compared to serous ovarian cancer (7,29).
However, most of these studies originate from limited research with
different age and stage thus yielding different prognosis. In the
current study, we found the survival rate of elderly advanced OPSC
was significantly shorter than that for patients with OCC.
Although, there are no data supporting the concept that elderly
women with cancer should receive differential treatment based on
age alone, the actual condition is that cancer risk increases with
age (4). Pignata and Vermorken
(1) already demonstrated that
ageing is associated with important changes which can affect
pharmacologic properties of cytotoxic agents including
pharmacokinetic, pharmacodynamic and toxicity profiles. Therefore,
age should be regarded as an important prognostic variable in the
pathogenesis and treatment of advanced OPSC. Major questions about
ovarian cancer in older-aged women need urgent attention from the
research community since the incidence and the prognosis of this
population is continuously worsening (1).
The above data of this research supported our
hypothesis and strongly suggest that miR-30a*,
miR-30e* and miR-505* may be the important
pathogenic factors for elderly OPSC patients with advanced stage.
This is the first report indicating and validating the differences
and significance of miR-30a*, miR-30a* and
miR-505*, and their targets (ATF3, STMN1 and MYC) in
elderly OPSC with advanced stage. We hope this can improve
understanding of molecular underpinnings during EOC development and
progression, especially in elderly advanced OPSC patients; and to
identify putative targets, including mRNA and proteins, which may
open a new field for the understanding of this disease and
providing improved diagnostic, prognostic and therapeutic
approaches to individual patients, especially to the elderly.
Acknowledgements
This study was supported through the Chinese
Ministry of science and technology projects (no. 2008DFA30720).
References
|
1
|
Pignata S and Vermorken JB: Ovarian cancer
in the elderly. Crit Rev Oncol Hematol. 49:77–86. 2004. View Article : Google Scholar
|
|
2
|
Boren T, Xiong Y, Hakam A, et al:
MicroRNAs and their target messenger RNAs associated with ovarian
cancer response to chemotherapy. Gynecol Oncol. 113:249–255. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu JF, Hirsch MS, Lee H and Matulonis UA:
Prognosis and hormone receptor status in older and younger patients
with advanced-stage papillary serous ovarian carcinoma. Gynecol
Oncol. 115:401–406. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moore DH, Kauderer JT, Bell J, Curtin JP
and Van Le L: An assessment of age and other factors influencing
protocol versus alternative treatments for patients with epithelial
ovarian cancer referred to member institutions: a Gynecologic
Oncology Group study. Gynecol Oncol. 94:368–374. 2004. View Article : Google Scholar
|
|
5
|
Shih KK, Qin LX, Tanner EJ, et al: A
microRNA survival signature (MiSS) for advanced ovarian cancer.
Gynecol Oncol. 121:444–450. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yancik R, Ries LG and Yates JW: Ovarian
cancer in the elderly: an analysis of Surveillance, Epidemiology,
and End Results Program data. Am J Obstet Gynecol. 154:639–647.
1986. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chan JK, Teoh D, Hu JM, Shin JY, Osann K
and Kapp DS: Do clear cell ovarian carcinomas have poorer prognosis
compared to other epithelial cell types? A study of 1411 clear cell
ovarian cancers. Gynecol Oncol. 109:370–376. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wyman SK, Parkin RK, Mitchell PS, et al:
Repertoire of microRNAs in epithelial ovarian cancer as determined
by next generation sequencing of small RNA cDNA libraries. PLoS
One. 4:e53112009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Jaarsveld MT, Helleman J, Berns EM and
Wiemer EA: MicroRNAs in ovarian cancer biology and therapy
resistance. Int J Biochem Cell Biol. 42:1282–1290. 2010.PubMed/NCBI
|
|
10
|
Sugiyama T, Kamura T, Kigawa J, et al:
Clinical characteristics of clear cell carcinoma of the ovary: a
distinct histologic type with poor prognosis and resistance to
platinum-based chemotherapy. Cancer. 88:2584–2589. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yoshida S, Furukawa N, Haruta S, et al:
Theoretical model of treatment strategies for clear cell carcinoma
of the ovary: focus on perspectives. Cancer Treat Rev. 35:608–615.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Pectasides D, Pectasides E, Psyrri A and
Economopoulos T: Treatment issues in clear cell carcinoma of the
ovary: a different entity? Oncologist. 11:1089–1094. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Snowdon J, Zhang X, Childs T, Tron VA and
Feilotter H: The microRNA-200 family is upregulated in endometrial
carcinoma. PLoS One. 6:e228282011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cui S, Li C, Ema M, Weinstein J and
Quaggin SE: Rapid isolation of glomeruli coupled with gene
expression profiling identifies downstream targets in Pod1 knockout
mice. J Am Soc Nephrol. 16:3247–3255. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
American Cancer Society. Global Cancer
Facts and Figures 2007. American Cancer Society Inc; Atlanta, GA:
2007
|
|
16
|
Howlader N, Noone AM, Krapcho M, Neyman N,
Aminou R, Altekruse SF, Kosary CL, Ruhl J, Tatalovich Z, Cho H,
Mariotto A, Eisner MP, Lewis DR, Chen HS, Feuer EJ and Cronin KA:
SEER Cancer Statistics Review: 1975–2008. National Institutes of
Health; Bethesda, MD: 2011
|
|
17
|
Kim MS, In SG, Park OJ, et al: Increased
expression of activating transcription factor 3 is related to the
biologic behavior of cutaneous squamous cell carcinomas. Hum
Pathol. 42:954–959. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li ZD, Hu XW, Wang YT and Fang J: Apigenin
inhibits proliferation of ovarian cancer A2780 cells through Id1.
FEBS Lett. 583:1999–2003. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hai T and Hartman MG: The molecular
biology and nomenclature of the activating transcription
factor/cAMP responsive element binding family of transcription
factors: activating transcription factor proteins and homeostasis.
Gene. 273:1–11. 2001. View Article : Google Scholar
|
|
20
|
Thompson MR, Xu D and Williams BR: ATF3
transcription factor and its emerging roles in immunity and cancer.
J Mol Med (Berl). 87:1053–1060. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pelzer AE, Bektic J, Haag P, et al: The
expression of transcription factor activating transcription factor
3 in the human prostate and its regulation by androgen in prostate
cancer. J Urol. 175:1517–1522. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Janz M, Hummel M, Truss M, et al:
Classical Hodgkin lymphoma is characterized by high constitutive
expression of activating transcription factor 3 (ATF3), which
promotes viability of Hodgkin/Reed-Sternberg cells. Blood.
107:2536–2539. 2006. View Article : Google Scholar
|
|
23
|
Bandyopadhyay S, Wang Y, Zhan R, et al:
The tumor metastasis suppressor gene Drg-1 down-regulates the
expression of activating transcription factor 3 in prostate cancer.
Cancer Res. 66:11983–11990. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ishiguro T, Nagawa H, Naito M and Tsuruo
T: Inhibitory effect of ATF3 antisense oligonucleotide on ectopic
growth of HT29 human colon cancer cells. Jpn J Cancer Res.
91:833–836. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hsieh SY, Huang SF, Yu MC, et al:
Stathmin1 overexpression associated with polyploidy, tumor-cell
invasion, early recurrence, and poor prognosis in human hepatoma.
Mol Carcinog. 49:476–487. 2010.PubMed/NCBI
|
|
26
|
Lee HS, Lee DC, Park MH, et al: STMN2 is a
novel target of beta-catenin/TCF-mediated transcription in human
hepatoma cells. Biochem Biophys Res Commun. 345:1059–1067. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su D, Smith SM, Preti M, et al: Stathmin
and tubulin expression and survival of ovarian cancer patients
receiving platinum treatment with and without paclitaxel. Cancer.
115:2453–2463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ponzielli R, Katz S, Barsyte-Lovejoy D and
Penn LZ: Cancer therapeutics: targeting the dark side of Myc. Eur J
Cancer. 41:2485–2501. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee YY, Kim TJ, Kim MJ, et al: Prognosis
of ovarian clear cell carcinoma compared to other histological
subtypes: a meta-analysis. Gynecol Oncol. 122:541–547. 2011.
View Article : Google Scholar : PubMed/NCBI
|