Introduction
Renal cell carcinoma (RCC) is a common urological
neoplasm of the adult kidney. The diagnostic modalities and
therapeutic techniques for RCC continue to improve and the overall
incidence and mortality of RCC has increased in the last 20 years
(1). The 5-year survival rate is
∼98% for stage I disease and ∼50% for stage III disease (2), which underscore the importance of
early detection and treatment of RCC. However, early detection of
often difficult, because early-stage renal tumors are often
asymptomatic and non-palpable. Therefore, the identification of
non-invasive biomarkers for early-stage RCC would be of benefit.
However, no accurate biomarker for RCC currently exists (3).
Recent studies suggest that microRNAs (miRNAs),
which are non-protein-coding small RNAs, are involved in cancer
progression and metastasis. MicroRNAs are ∼22 nucleotides in length
and regulate gene expression at the post-transcriptional level by
binding to the untranslated region (3′UTR) of target mRNAs, leading
to translational inhibition and/or mRNA degradation (4). Specific expression profiles of miRNAs
in tissue have been reported in a variety of cancers, including RCC
(5). Early studies suggested that
miRNAs were strictly intracellular molecules, but more recent
studies suggest that miRNAs are highly stable and abundant in the
serum, urine and other body fluids. This is because exosomes likely
protect miRNAs against degradation by RNase (6,7).
Interestingly, serum miRNA levels are similar in men and women and
do not vary with patient age (8).
Thus, circulating miRNAs might be good non-invasive
biomarkers for diagnostic and prognostic considerations in a
variety of cancers. Indeed, several studies have reported that
specific circulating miRNAs were useful for distinguishing patients
with cancer (e.g., colorectal cancer, breast cancer, prostate
cancer) from healthy controls (HCs) (6,9,10).
However, the number of studies regarding circulating miRNAs in
patients with RCC is small.
Several studies using miRNA microarray analysis
demonstrated that miR-210 expression in clear cell carcinoma (CCC),
which is the largest subtype of RCC, was significantly upregulated
in tumor tissues and cell lines (11). In addition, some groups reported
that miR-210 upregulation played an important role in tumorigenesis
in various types of human cancers (12,13).
The goal of this study was to determine whether
circulating miR-210 was a useful diagnostic biomarker for
distinguishing CCC patients from HCs.
Materials and methods
Patients and sample collection
The study was approved by the ethics committee of
Tottori University Hospital, Japan and all the patients provided
written informed consent. We prospectively collected tissue and
serum samples from patients undergoing radical nephrectomy or
nephron-sparing surgery for renal tumors. Sample collection was
performed between 2011 and 2013 at the Department of Urology,
Tottori University Hospital. Thirty-four patients with
histologically confirmed CCC and 23 HCs with no previous history of
any cancer were included in this analysis. Detailed
clinicopathological parameters of patients are summarized in
Table I.
 | Table I.Clinicopathological parameters. |
Table I.
Clinicopathological parameters.
| Serum samples
|
|---|
| CCC (n=34) | HC (n=23) |
|---|
| Sex | | |
| Male | 26 | 11 |
| Female | 8 | 12 |
| Age (years) | | |
| Mean | 66.5 | 53.5 |
| Range | 29–86 | 36–84 |
| Pathological
stage | | |
| pT1a | 17 | - |
| pT1b | 8 | - |
| pT2a | 3 | - |
| pT2b | 1 | - |
| pT3a | 4 | - |
| pT3b | 1 | - |
| pT4 | 0 | - |
| Lymph nodes
metastasis | 2 | - |
| Distant
metastasis | 5 | - |
| Grade | | |
| G1 | 9 | - |
| G2 | 24 | - |
| G3 | 1 | - |
Tissue samples were obtained from tumor tissues and
matched normal tissues from the same kidney specimen in patients
with CCC. Immediately after resection, tissue samples were frozen
in liquid nitrogen and stored at −80°C.
Blood samples were obtained prior to
surgery
Serum was separated after centrifugation (3,000 rpm,
10 min) and stored at −80°C.
Total RNA isolation
Total RNA was extracted from tissue using the
mirVana™ miRNA isolation kit (Ambion, USA) and from 200 μl
of serum using the microRNA extractor SP kit (Wako, Japan)
according to the manufacturer’s recommendation (final elution
volume, 50 μl). RNA quantity and purity were determined
using a NanoDrop Spectrophotometer ND-1000 (Thermo Scientific,
USA). The RNA samples were stored at −80°C until reverse
transcription (RT) reaction.
Quantitative real-time PCR
Complementary DNA (cDNA) was synthetized from total
RNA using the TaqMan MicroRNA RT kit (Applied Biosystems) and using
miRNA-specific RT primers from the TaqMan MicroRNA assay (Applied
Biosystems). RT reaction mixtures were incubated for 30 min at
16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C.
Quantitative real-time PCR was performed using the TaqMan MicroRNA
assay on the ABI PRISM 7900HT system (Applied Biosystems). All
experiments were performed as specified in the manufacturer’s
protocols.
Statistical methods
Analysis of the real-time PCR data was done using
SDS software, version 2.4 (Applied Biosystems). MicroRNA levels in
tissue were normalized against miR-145 and microRNA levels in serum
were normalized against miR-16. We confirmed that the expression of
miR-145 in tissue and miR-16 in serum were not significantly
different when comparing patients and HCs. The relative expression
levels of miR-210 were determined by the equation: 2−ΔCT
(ΔCT=CT miR-210 - CT miR-145 or 16).
Statistical analyses were performed using PASW statistics 18 (SPSS,
Chicago, IL, USA). Sensitivity, specificity and area under curve
(AUC) for serum microRNA levels were determined using receiver
operator characteristic (ROC) analysis. The relationship between
clinicopathologic parameters and microRNA levels was examined using
the Mann-Whitney U or Kruskal-Wallis test, as appropriate. P-values
of <0.05 were considered to represent statistical
significance.
Results
Validation in tissue samples
Using real-time PCR, we first assessed tissue
miR-210 levels normalized against miR-145 in 34 pairs of tumor
tissues and matched normal tissues obtained from patients with CCC.
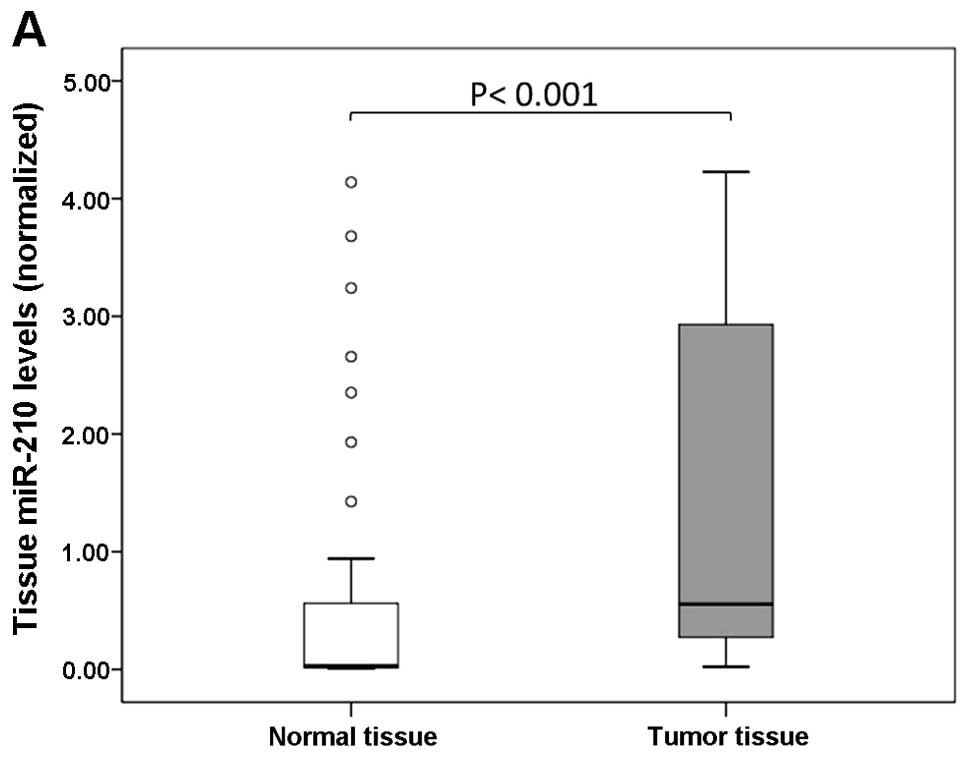
Tissue miR-210 levels were significantly higher in tumor tissues
than in normal tissues (P<0.001; Fig. 1A). There was no difference in the
CT values for miR-145 when comparing tumor tissues and normal
tissues (P=0.236; Fig. 1B). In 31
cases (92%), the miR-210 level in tumor tissues was increased by
>2-fold when compared with that in normal tissues.
Validation in serum samples
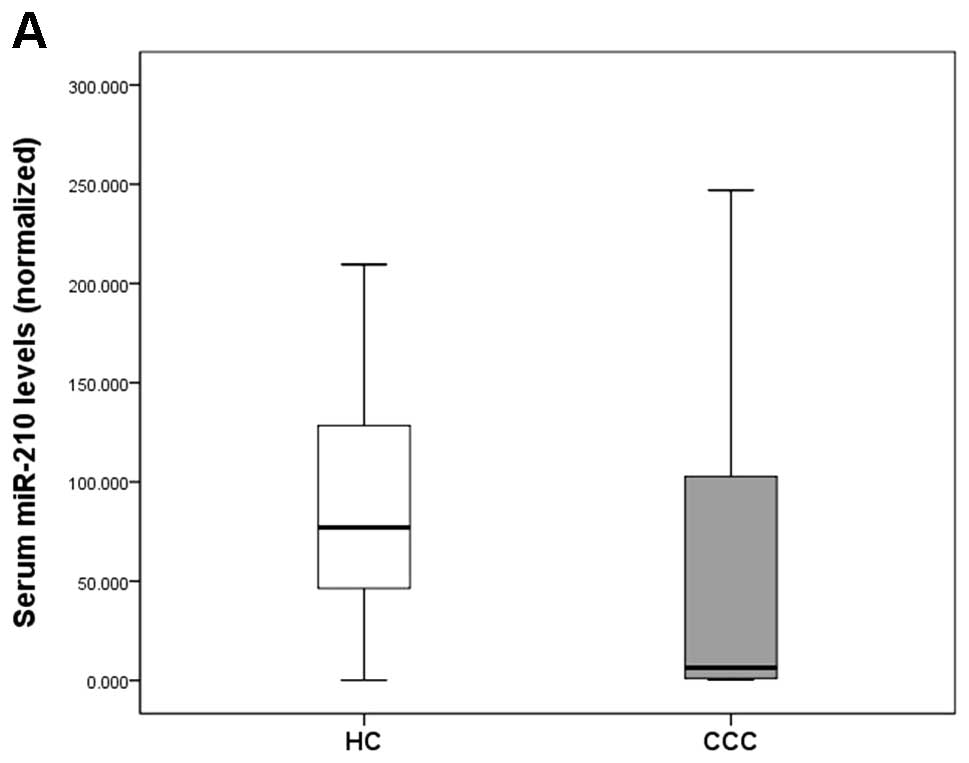
We assessed serum miR-210 levels normalized against
miR-16 in 34 CCC patients and 23 HCs. The 34 serum samples from CCC
patients perfectly matched up with tissue samples described above.
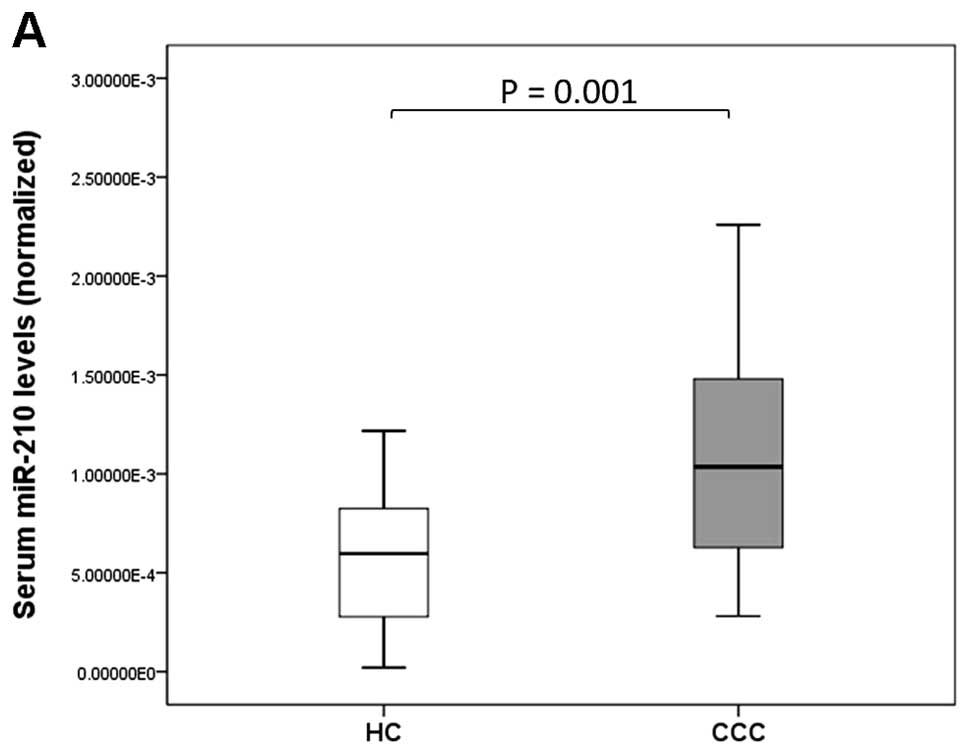
Serum miR-210 levels were significantly higher in CCC patients than
in HCs (P=0.001; Fig. 2A). There
was no significant difference in the CT values of miR-16 when
comparing CCC patients and HCs (P=0.614; Fig. 2B).
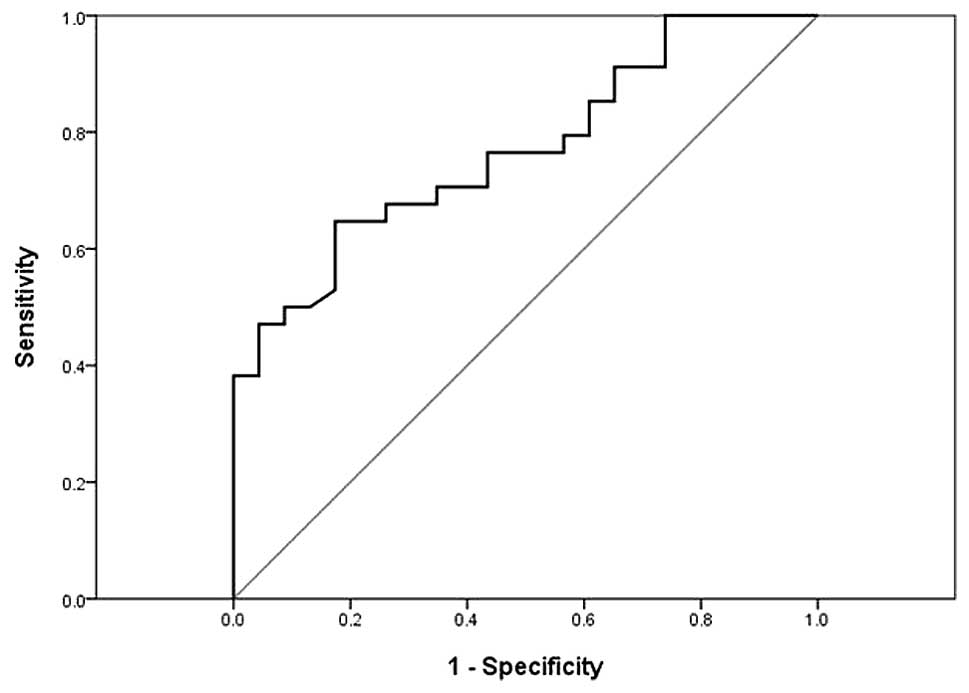
ROC curve analysis indicated that the serum miR-210
level might serve as a useful biomarker for differentiating
patients with CCC from those with HCs; the AUC was 0.77 (95%
confidence interval, 0.65–0.89) and the sensitivity and specificity
was 65 and 83%, respectively (Fig.
3).
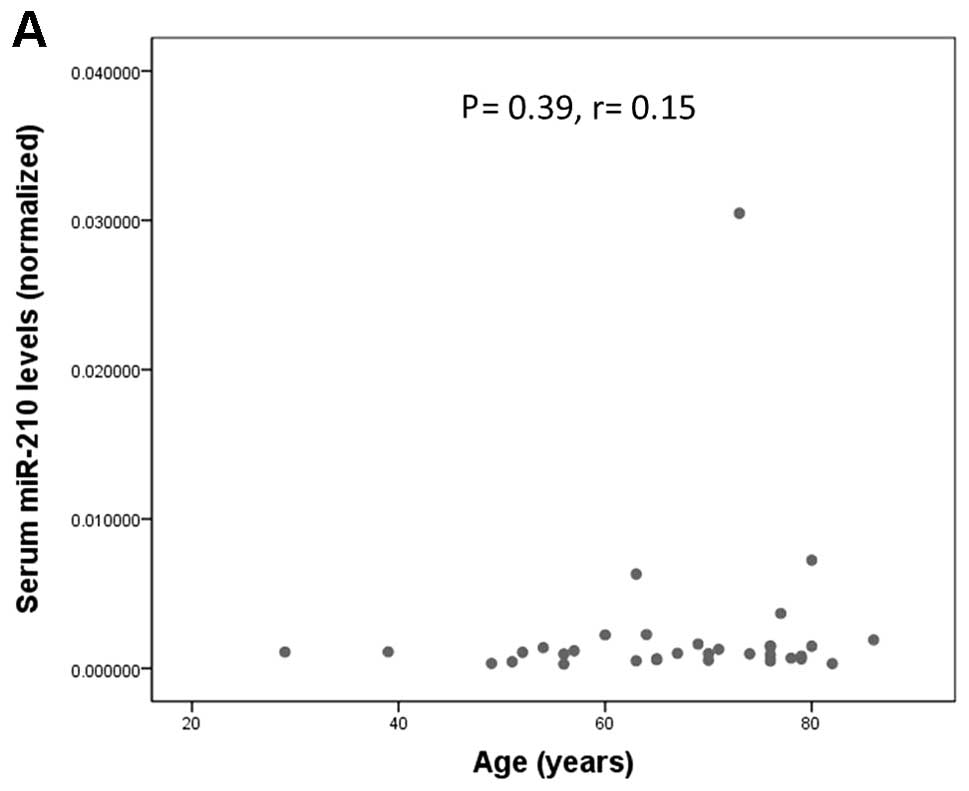
We analyzed the relationship between serum miR-210
levels and clinicopathological parameters. There was no significant
association between serum miR-210 levels and age, sex, tumor size,
or existence of metastasis at diagnosis (Fig. 4). Although serum miR-210 level
tended to be higher in patients with metastasis at diagnosis when
compared with patients without metastasis (P=0.067; Fig. 4D), this difference did not reach
the level of statistical significance.
Discussion
Previous studies have described the potential use of
circulating microRNA as a non-invasive biomarker for various
cancers [e.g., miR-29a and miR-92 in colorectal cancer (9); miR-195 in breast cancer (14); miR-17-5p, miR-21, miR-106a and
miR-106b in gastric cancer (15);
and miR-141 and miR-26a in prostate cancer (6,16)].
In the case of RCC, several recent studies using miRNA microarray
analysis showed different microRNA expression profiles when
comparing tumor tissues and matched normal tissues. However, some
of the data regarding the number and type of up-/downregulated
microRNAs is conflicting (17–19).
Regardless, several recent studies have described the use of
circulating microRNA as a new biomarker for RCC.
The present study showed that serum miR-210 levels
were significantly higher in CCC patients than in HCs. Furthermore,
there was no correlation between serum miR-210 levels and
clinicopathological parameters. These results indicate that
upregulation of serum miR-210 may occur in the early stage of CCC
and can serve as a potential biomarker of early diagnosis in
CCC.
Four studies have investigated the utility of
circulating microRNAs as a diagnostic biomarker for RCC. Wulfken
et al were the first to report that the serum miR-1233 level
was increased in 84 patients with RCC from a multicenter cohort
(AUC, 0.588; sensitivity, 77.4%; specificity, 37.6%). Moreover,
they investigated 13 samples from patients with angiomyolipoma or
oncocytoma whose serum miR-1233 levels were similar to those of
patients with RCC (20). Redova
et al also demonstrated that serum miR-378 and miR-451 were
potential biomarkers for RCC. When the utility of miR-378 and
miR-451 was evaluated in an independent cohort of 90 patients with
RCC and 35 HCs, the combination of serum miR-378 and miR-451
enabled identification of RCC with relatively high accuracy rate
(AUC 0.86; sensitivity, 81%; specificity, 83%) (21). Hauser et al confirmed that
serum miR-378 was significantly increased in 25 CCC patients
(P=0.006), but they did not detect a difference in the level of
this biomarker when comparing 117 patients with RCC versus 123 HCs
(22). Zhao et al reported
that tissue miR-210 levels in 33 CCC patients were significantly
higher in tumor tissue than in adjacent non-tumoral renal
parenchyma (P= 0.004). Serum miR-210 levels were also significantly
higher in 68 CCC patients than in 42 HCs (P<0.001) (AUC, 0.87;
sensitivity, 81.0%; specificity, 79.4%). Furthermore, serum miR-210
levels in patients with CCC decreased by 1 week after surgical
resection of the tumor (23).
The present study normalized the assessed RNA values
in tissue and serum against those of miR-145 and miR-16,
respectively, which is a different approach from that used by Zhao
et al. There is no consensus regarding the optimal
normalization gene to use for quantitative real-time polymerase
chain reaction (qRT-PCR) analysis of circulating mRNAs (24). The present study did not utilize
RNU6B or 5s rRNA for normalization of qRT-PCR data because some
studies have reported that RNU6B and 5s rRNA were not stable in
human tissues or body fluids. In their study targeting serum miRNA
in gastric cancer patients, Song et al demonstrated that
RNU6B could not be detected in almost half of the serum samples; in
the other half of patients, it was detected with Ct values of
>40. Moreover, RNU6B was less stably expressed than let-7a and
miR-16 in normal and cancerous human solid tissues (25). Although we actually examined the
same study using RNU6B for normalization, there was marked
variation in Ct values when comparing CCC patients and HCs
(Fig. 5). Therefore, we selected
other miRNAs (miR-16, miR-103a, miR-122 and miR-145) for the
purposes of normalization, based on previous reports (21,26).
Then, we determined which of these miRNAs was expressed in similar
amounts when comparing CCC patients and HCs. As a result of these
investigations, miR-145 and miR-16 were selected for use as
normalizing genes in the present study.
MiR-210 is upregulated in various types of human
cancers, suggesting its important role in tumorigenesis (27). Jung et al reported that
plasma miR-210 levels in human breast cancer were associated with
trastuzumab sensitivity, tumor presence and lymph node metastasis
(28). Although the mechanism of
upregulation of miR-210 is still unclear, some groups have reported
that miR-210 is induced under-hypoxic conditions via
hypoxia-inducible factors (HIFs) in various cancer cell lines
(28). It is well known that HIF1α
and HIF2α accumulate in CCC as a result of abrogated
ubiquitin-mediated degradation due to loss or deficiency of the
von Hippel-Lindau tumor suppressor (VHL) tumor suppressor
gene (29). Nakada et al
demonstrated that miR-210 was highly expressed in RCC cell lines
and that its expression clearly correlated with the accumulation of
hypoxia-inducible factor 1α (HIF1α) under normoxia as well as under
hypoxia, suggesting that miR-210 upregulation in CCC was most
likely due to accumulation of HIF1α. Further, they confirmed that
restoration of VHL expression in the VHL-deficient cell line led to
the degradation of HIF1α and suppressed the expression of miR-210,
suggesting that miR-210 expression is regulated via the VHL-HIF1α
pathway (11). Since VHL
inactivation is observed in >70% of CCC patients, its
inactivation may occur in the early stage of tumorigenesis. This
raises the possibility that the expression of miR-210 is
upregulated and plays a role in the early stage of tumorigenesis in
CCC tissue and that serum miR-210 is upregulated in the human
peripheral blood in the early stages of CCC.
For these reasons, serum miR-210, which can be
easily measured on an outpatient basis, could be a useful biomarker
for early diagnosis and facilitate earlier treatment and, hence,
improves outcomes. Further studies with a larger number of patients
are warranted to validate these results.
In conclusion, early detection and treatment of RCC
is critical to improve outcomes. Serum miR-210 is a useful
biomarker for early CCC.
References
|
1.
|
Hollingsworth JM, Miller DC, Daignault S
and Hollenbeck BK: Rising incidence of small renal masses: a need
to reassess treatment effect. J Natl Cancer Inst. 98:1331–1334.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Devita VT Jr, Hellman S and Rosenberg SA:
Cancer Principles and Practice of Oncology. 8th edition. Lippincott
Williams & Wilkins; 2008
|
|
3.
|
Ljungberg B, Cowan NC, Hanbury DC, Hora M,
Kuczyk MA, Merseburger AS, Patard JJ, Mulders PF and Sinescu IC;
European Association of Urology Guideline Group: EAU guidelines on
renal cell carcinoma: the 2010 update. Eur Urol. 58:398–406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Slaby O, Jancovicova J, Lakomy R, Svoboda
M, Poprach A, Fabian P, Kren L, Michalek J and Vyzula R: Expression
of miRNA-106b in conventional renal cell carcinoma is a potential
marker for prediction of early metastasis after nephrectomy. J Exp
Clin Cancer Res. 29:902010. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M,
Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M,
Harris CC and Croce CM: A microRNA expression signature of human
solid tumors defines cancer gene targets. Proc Natl Acad Sci USA.
103:2257–2261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant
KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt
DL, Gentleman R, Vessella RL, Nelson PS, Martin DB and Tewari M:
Circulating microRNAs as stable blood-based markers for cancer
detection. Proc Natl Acad Sci USA. 105:10513–10518. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Chim SS, Shing TK, Hung EC, Leung TY, Lau
TK, Chiu RW and Lo YM: Detection and characterization of placental
microRNAs in maternal plasma. Clin Chem. 54:482–490. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Hunter MP, Ismail N, Zhang X, Aguda BD,
Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, Nana-Sinkam
SP, Jarjoura D and Marsh CB: Detection of microRNA expression in
human peripheral blood microvesicles. PLoS One. 3:e36942008.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Huang Z, Huang D, Ni S, Peng Z, Sheng W
and Du X: Plasma microRNAs are promising novel biomarkers for early
detection of colorectal cancer. Int J Cancer. 127:118–126. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Roth C, Rack B, Müller V, Janni W, Pantel
K and Schwarzenbach H: Circulating microRNAs as blood-based markers
for patients with primary and metastatic breast cancer. Breast
Cancer Res. 12:R902010. View
Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Nakada C, Tsukamoto Y, Matsuura K, Nguyen
TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M and Moriyama M:
Overexpression of miR-210, a downstream target of HIF1α, causes
centrosome amplification in renal carcinoma cells. J Pathol.
224:280–288. 2011.PubMed/NCBI
|
|
12.
|
Crosby ME, Kulshreshtha R, Ivan M and
Glazer PM: MicroRNA regulation of DNA repair gene expression in
hypoxic stress. Cancer Res. 69:1221–1229. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Fasanaro P, D’Alessandra Y, Di Stefano V,
Melchionna R, Romani S, Pompilio G, Capogrossi MC and Martelli F:
MicroRNA-210 modulates endothelial cell response to hypoxia and
inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol
Chem. 283:15878–15883. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Heneghan HM, Miller N, Lowery AJ, Sweeney
KJ, Newell J and Kerin MJ: Circulating microRNAs as novel minimally
invasive biomarkers for breast cancer. Ann Surg. 251:499–505. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Tsujiura M, Ichikawa D, Komatsu S,
Shiozaki A, Takeshita H, Kosuga T, Konishi H, Morimura R, Deguchi
K, Fujiwara H, Okamoto K and Otsuji E: Circulating microRNAs in
plasma of patients with gastric cancers. Br J Cancer.
102:1174–1179. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Mahn R, Heukamp LC, Rogenhofer S, von
Ruecker A, Müller SC and Ellinger J: Circulating microRNAs (miRNA)
in serum of patients with prostate cancer. Urology. 77:1265.e9–16.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Redova M, Svoboda M and Slaby O: MicroRNAs
and their target gene networks in renal cell carcinoma. Biochem
Biophys Res Commun. 405:153–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
White NM, Bao TT, Grigull J, Youssef YM,
Girgis A, Diamandis M, Fatoohi E, Metias M, Honey RJ, Stewart R,
Pace KT, Bjarnason GA and Yousef GM: miRNA profiling for clear cell
renal cell carcinoma: biomarker discovery and identification of
potential controls and consequences of miRNA dysregulation. J Urol.
186:1077–1083. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Wotschofsky Z, Busch J, Jung M,
Kempkensteffen C, Weikert S, Schaser KD, Melcher I, Kilic E, Miller
K, Kristiansen G, Erbersdobler A and Jung K: Diagnostic and
prognostic potential of differentially expressed miRNAs between
metastatic and nonmetastatic renal cell carcinoma at the time of
nephrectomy. Clin Chim Acta. 416:5–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Wulfken LM, Moritz R, Ohlmann C,
Holdenrieder S, Jung V, Becker F, Herrmann E, Walgenbach-Brünagel
G, von Ruecker A, Müller SC and Ellinger J: MicroRNAs in renal cell
carcinoma: diagnostic implications of serum miR-1233 levels. PLoS
One. 6:e257872011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Redova M, Poprach A, Nekvindova J, Iliev
R, Radova L, Lakomy R, Svoboda M, Vyzula R and Slaby O: Circulating
miR-378 and miR-451 in serum are potential biomarkers for renal
cell carcinoma. J Transl Med. 10:552012. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Hauser S, Wulfken LM, Holdenrieder S,
Moritz R, Ohlmann CH, Jung V, Becker F, Herrmann E,
Walgenbach-Brünagel G, von Ruecker A, Müller SC and Ellinger J:
Analysis of serum microRNAs (miR-26a-2*, miR-191,
miR-337-3p and miR-378) as potential biomarkers in renal cell
carcinoma. Cancer Epidemiol. 36:391–394. 2012.
|
|
23.
|
Zhao A, Li G, Péoc’h M, Genin C and
Gigante M: Serum miR-210 as a novel biomarker for molecular
diagnosis of clear cell renal cell carcinoma. Exp Mol Pathol.
94:115–120. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Kroh EM, Parkin RK, Mitchell PS and Tewari
M: Analysis of circulating microRNA biomarkers in plasma and serum
using quantitative reverse transcription-PCR (qRT-PCR). Methods.
50:298–301. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Song J, Bai Z, Han W, Zhang J, Meng H, Bi
J, Ma X, Han S and Zhang Z: Identification of suitable reference
genes for qPCR analysis of serum microRNA in gastric cancer
patients. Dig Dis Sci. 57:897–904. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Cheng Y, Wang X, Yang J, Duan X, Yao Y,
Shi X, Chen Z, Fan Z, Liu X, Qin S, Tang X and Zhang C: A
translational study of urine miRNAs in acute myocardial infarction.
J Mol Cell Cardiol. 53:668–676. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Camps C, Buffa FM, Colella S, Moore J,
Sotiriou C, Sheldon H, Harris AL, Gleadle JM and Ragoussis J:
hsa-miR-210 is induced by hypoxia and is an independent prognostic
factor in breast cancer. Clin Cancer Res. 14:1340–1348. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Jung EJ, Santarpia L, Kim J, Esteva FJ,
Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST, Pusztai
L and Calin GA: Plasma microRNA 210 levels correlate with
sensitivity to trastuzumab and tumor presence in breast cancer
patients. Cancer. 118:2603–2614. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Eble JN, Sauter G, Epstein JI and
Sesterhenn IA: World Health Organization Classification of Tumours.
Pathology and Genetics of Tumours of the Urinary System and Male
Genital Organs. IARC Press; Lyon: pp. 9–87. 2004
|