Introduction
Lung and colon cancers are among the main causes of
death in developed countries. The life expectancy of patients is
very limited, especially in metastatic disease. Nevertheless, in
recent years, the development of targeted therapies (Tyrosine
Kinase inhibitors or inhibitors of receptors which hyperactivate
survival pathways) has shown great therapeutic promise. For
example, patients with a mutated EGFR gene and wild-type
KRAS gene lung tumor are eligible for gefitinib therapy
(1). In colon cancer, patients
with wild-type KRAS and BRAF could be treated with
panitumumab or cetuximab (2). In
skin cancer, vemurafenib (3) or
imatinib (4) can be used to treat
mutated BRAF (V600E) or c-KIT melanoma, respectively.
As the efficacy of these targeted therapies depends on specific
genetic abnormalities, molecular diagnosis has become essential for
the treatment of cancers. Since 2008, molecular biology platforms
have screened for genetic alterations in EGFR (exons 18–21),
KRAS (codons 12 and 13) and BRAF (codon 600). At the
beginning, the gold standard was Sanger sequencing, but the
technique has low sensitivity and is expensive. With the increasing
number of samples and gene alterations to screen for,
alternon-amplified techniques were developed. For example, allelic
discrimination (5) was developed
to screen for a specific mutation quickly at a relatively low
price. In parallel, screening technologies, such as High Resolution
Melting (6) or fragment analysis
(for indel alterations), were developed to use with Sanger
sequencing, but only in the presence of potential mutations in the
region of interest. Over the years, the number of genes with
genetic alterations that could be targeted by therapies has
increased rapidly. This medical progress has led to the need for
more and more molecular diagnoses. This need has now been met by
the recent development of Next Generation Sequencing (NGS), which
has revolutionized molecular diagnosis. Indeed, the sequencing
capacity allows the analysis of dozens of genes on multiplexed
samples. In this paper, we describe the results we obtained with
the Truseq Cancer Panel. In addition to the routinely detected
mutations, NGS analysis, thanks to its high sensitivity, revealed
new mutations in routinely analyzed genes.
Materials and methods
Patients and DNA samples
Eighteen tissue samples with >400 ng gDNA
(Table I) from patients treated at
the Centre Georges-François Leclerc between 2009 and 2013 were
randomly chosen. Genomic DNA was extracted from FFPE tissues with
either the QIAamp DNA mini kit (Qiagen, Heidelberg, Germany) or the
Maxwell 16 FFPE Plus LEV DNA purification kit (Promega, Madison,
USA). The samples had already been genotyped by allelic
discrimination, fragment analysis and Sanger sequencing. Written
consent was provided by all patients, and the researchers obtained
authorization from the diagnostic centers to use the tumor
samples.
 | Table IClinical details of studied
patients. |
Table I
Clinical details of studied
patients.
| Patients | Organ of
origin | Histology | Age (years) |
|---|
| L1 | Lung | Keratinizing poorly
differentiated squamous carcinoma | 61 |
| L2 | Lung | Moderately
differentiated adenocarcinoma | 67 |
| L3 | Lung | Poorly
differentiated adenocarcinoma | 62 |
| L4 | Lung | Adenocarcinoma | 50 |
| L5 | Lung | Adenocarcinoma | 69 |
| L6 | Lung | Adenocarcinoma | 55 |
| L7 | Lung | Mucus-secreting
adenocarcinoma | 68 |
| L8 | Lung | Acinar
differentiated mucus-secreting adenocarcinoma | 86 |
| C1 | Colon | Moderately
differentiated adenocarcinoma | 55 |
| C2 | Colon | Adenocarcinoma | 64 |
| C3 | Colon | Moderately
differentiated adenocarcinoma | 59 |
| C4 | Colon | Adenocarcinoma | 72 |
| C5 | Colon | Adenosquamous
adenocarcinoma | 70 |
| C6 | Colon | Poorly
differentiated adenocarcinoma | 55 |
| C7 | Colon | Adenocarcinoma | 80 |
| C8 | Colon | Poorly
differentiated adenocarcinoma | 72 |
| C9 | Colon | Well differentiated
infiltrating lieberkunien adenocarcinoma | 58 |
| C10 | Colon | Colloidal
adenocarcinoma | 67 |
Whole genome amplification
The Repli-g FFPE kit (Qiagen) was used to amplify
300 ng of gDNA from patients L7 and L8: 10 μl of gDNA solution were
mixed with 8 μl of FFPE buffer, 1 μl of ligation enzyme and 1 μl of
FFPE enzyme. The solution was then incubated at 24°C for 30 min, at
95°C for 5 min and then kept at 4°C. The Repli-g master mix was
prepared by mixing, per sample, 29 μl of Repli-g Midi reaction
buffer and 1 μl of Repli-g Midi DNA Polymerase. This second mixture
(30 μl) was added to the gDNA solution. This solution was then
incubated at 30°C for 2 h, 95°C for 10 min and kept at 4°C.
Amplified DNA was stored at −20°C. Thanks to this protocol, we
obtained 6900 and 6400 ng of amplified gDNA.
Preparation of libraries
Libraries were prepared with the Truseq Cancer Panel
(Illumina, San Diego, USA) by following the manufacturer protocol.
Briefly, 400–1250 ng of gDNA in 5 μl water was hybridized with an
oligo pool. Then, unbound oligos were removed, and
extension-ligation of bound oligos was followed by PCR
amplification. PCR products were cleaned and checked for quality
using Tapestation analysis (Agilent). The PCR product size had to
be around 350 bp. Before sequencing, the libraries were normalized
by the normalization process of the Truseq Cancer Panel.
Sequencing with MiSeq device
As each library possessed a specific primer index
combination (i5 and i7), the libraries were pooled for 2 sequencing
runs (pool no. 1, 10 libraries; pool no. 2, 9 libraries). For the
MiSeq sample sheet, each sample was identified by its specific
index combination. Libraries were paired-end sequenced with 2×151
bp cycles.
Analysis of obtained sequences
At the end of the run, sequences were aligned to the
human genome reference hg19. Generated BAM files were analyzed with
the Genome Golden Helix software (Golden Helix, Bozeman, USA). A
genetic variation was defined by a Q-score above 30 (except for
indel alteration).
Results
All mutations detected with standard
methods were detected with NGS
In the routine diagnosis of lung or colon
carcinomas, mutations in KRAS (exon 2), EGFR (exon
18–21), BRAF (exon 15) and HER2 (exon 20) genes are
analyzed using three different methods: allelic discrimination for
targeted mutations, fragment analysis for the screening of indel
variations and Sanger sequencing for non-targeted mutations or
characterization of indel abnormalities detected by fragment
analysis. All mutation hotspots are analyzed one by one. Over the
years, more and more genes and mutation hotspots will need to be
explored. For example, exons 3 and 4 of KRAS, and exons 2–4
of NRAS and HRAS need to be analyzed before anti-EGFR
antibody can be prescribed for colon cancer (7,8). In
the first step of our study, we used the Truseq Cancer Panel kit to
sequence samples that had already been analyzed in routine
diagnosis. We then compared the results obtained by NGS with the
results of the routine diagnosis at the same mutational hotspots.
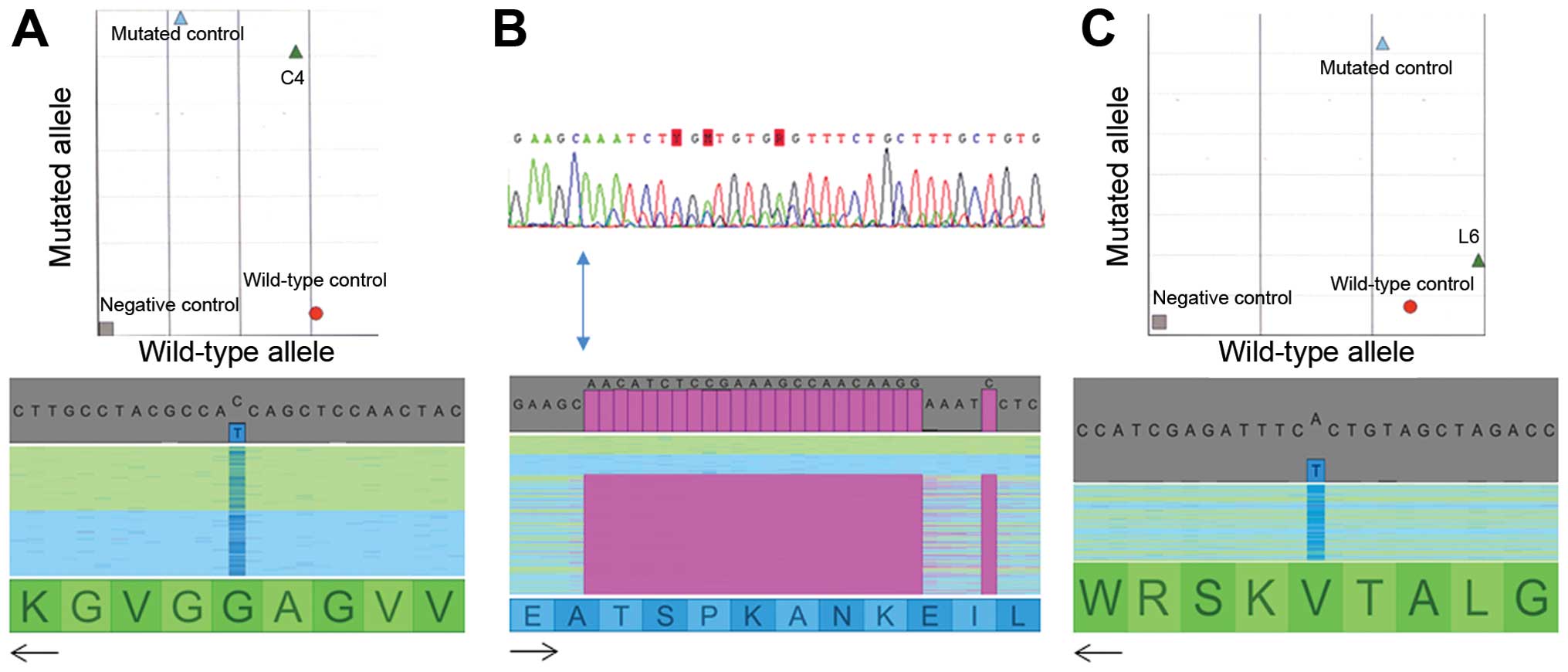
All the mutations detected in the routine diagnosis were also
detected by NGS (Fig. 1 and
Table II). Moreover, mutations
not found in routine diagnosis were detected by NGS. These included
a 15-nt deletion (c.2235_2249delGGAATTAAGAGAAGC) in two lung
carcinomas classified as wild-type using routine methods (patients
L1 and L6). In the L1 sample, another mutation in the KRAS gene
(G12D) was also identified. In patient L6, this 15-nt deletion in
EGFR was concomitant with a V600E BRAF mutation.
 | Table IIComparison of results obtained
routinely and with NGS. |
Table II
Comparison of results obtained
routinely and with NGS.
| KRAS | BRAF | EGFR | HER2 |
|---|
|
|
|
|
|
|---|
| Patients | Routinea | NGS | Routineb | NGS | Routinec | NGS | Routined | NGS |
|---|
| L1 | WT | G12D | WT | WT | WT | 15-nt
E19 | WT | WT |
| L2 | WT | WT | WT | WT | 15-nt E19 | 15-nt E19 | WT | WT |
| L3 | WT | WT | WT | WT | 24-nt E19 | 24-nt E19 | WT | WT |
| L4 | WT | WT | WT | WT | L858R | L858R | WT | WT |
| L5 | WT | WT | WT | WT | 15-nt E19
T790M | 15-nt E19
T790M | WT | WT |
| L6 | WT | WT | V600E | V600E | WT | 15-nt
E19 | WT | WT |
| L7 | G12C | G12C | WT | WT | WT | WT | WT | WT |
| L8 | WT | WT | WT | WT | WT | WT | WT | WT |
| C1 | WT | WT | WT | V600R | ND | | ND | |
| C2 | WT | WT | WT | WT | ND | | ND | |
| C3 | G13D | G13D | WT | WT | ND | | ND | |
| C4 | G12D | G12D | WT | WT | ND | | ND | |
| C5 | WT | WT | WT | WT | ND | | ND | |
| C6 | WT | WT | WT | WT | ND | | ND | |
| C7 | G13D | G13D | WT | WT | ND | | ND | |
| C8 | WT | WT | WT | WT | ND | | ND | |
| C9 | WT | WT | WT | WT | ND | | ND | |
| C10 | WT | WT | WT | WT | ND | | ND | |
In colon cancer, a ‘common’ 15-nt
deletion in the EGFR gene was detected only with NGS
Up to now, rare mutations of the KRAS gene
have not been routinely analyzed in lung carcinomas. In our small
population, a Q61H mutation in the KRAS gene was found in
the sample L8. This mutation was localized in exon 3, which is not
routinely analyzed in lung cancer. No other alteration was found in
the routinely analyzed genes (HRAS and NRAS).
In colon cancer, only the genes KRAS,
BRAF and very recently NRAS and HRAS are
studied. Concerning rare mutations of KRAS, NRAS and
HRAS, we detected a Q61K mutation in the NRAS gene in
patient C5. As numerous genes were sequenced by the Cancer Panel
kit, we analyzed the results obtained for the PIK3CA,
HER2 and EGFR genes, which are routinely analyzed in
lung carcinomas. No mutations were detected in exon 20 of
HER2, or in exons 18, 20 or 21 of EGFR. In exon 20 of
PIK3CA, an H1047L mutation was detected in patient C9.
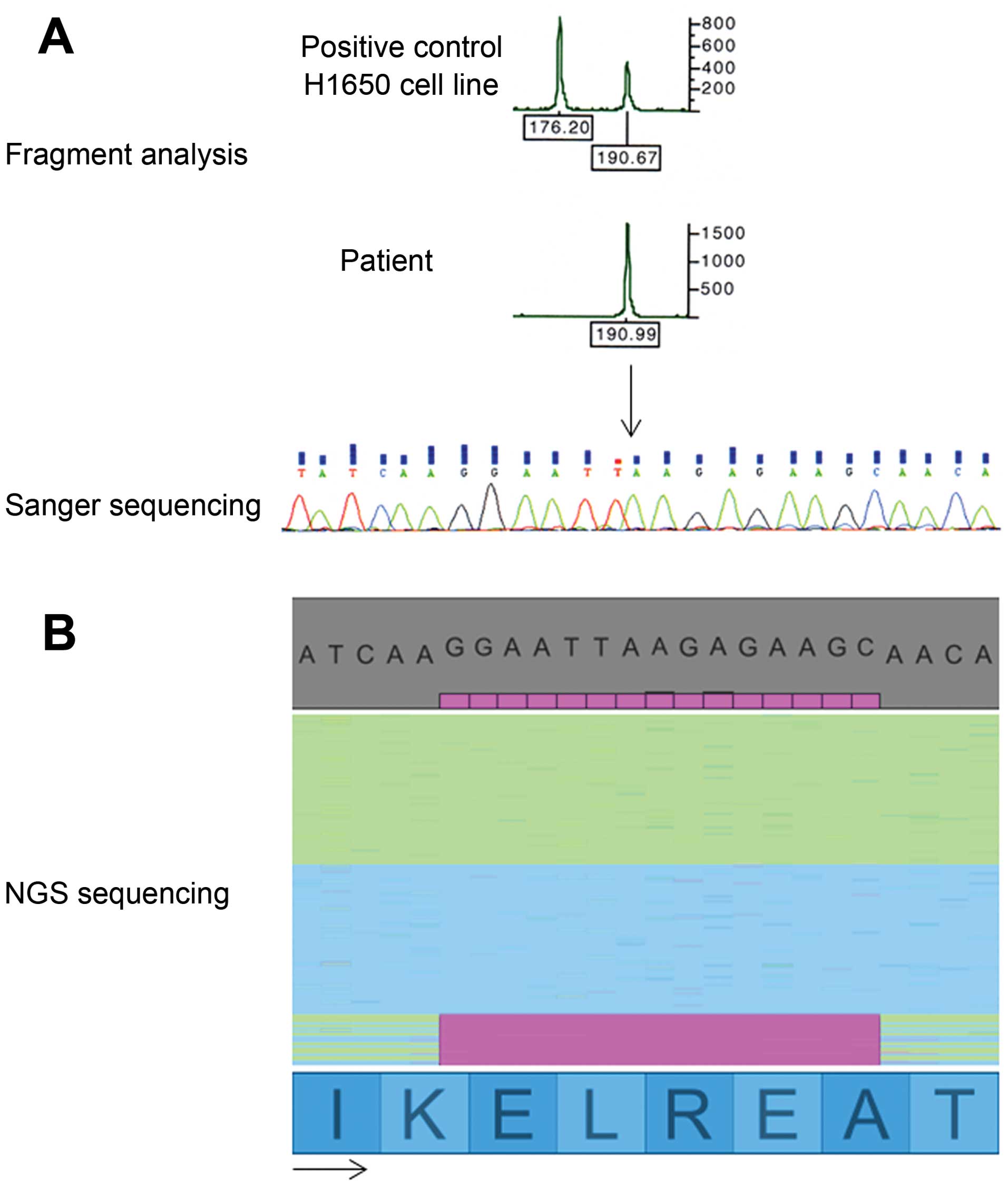
Concerning exon 19 of EGFR, a 15-nt deletion (the same as
that observed in lung carcinomas) was detected in three patients
(C4, C6 and C7). As this region was not routinely analyzed for
colon cancer, we decided to perform both fragment analysis and
Sanger sequencing. Neither fragment analysis, nor Sanger sequencing
was able to detect the 15-nt deletion in exon 19 of EGFR in
colon cancer (Fig. 2A). In
contrast, NGS sequencing detected the deletion in >8% of
sequenced fragments (Fig. 2B) for
one patient. The two other patients harbored the mutation in
approximately 4% of read sequences. Among these three patients,
only one did not present a concomitant KRAS mutation.
WGA does not alter the NGS sequencing
results
An important limitation in routine diagnosis is the
quantity of gDNA extracted from FFPE samples and another paraffin
block cannot be obtained in most cases. To counteract this
limitation, we tested the impact of Whole Genome Amplification
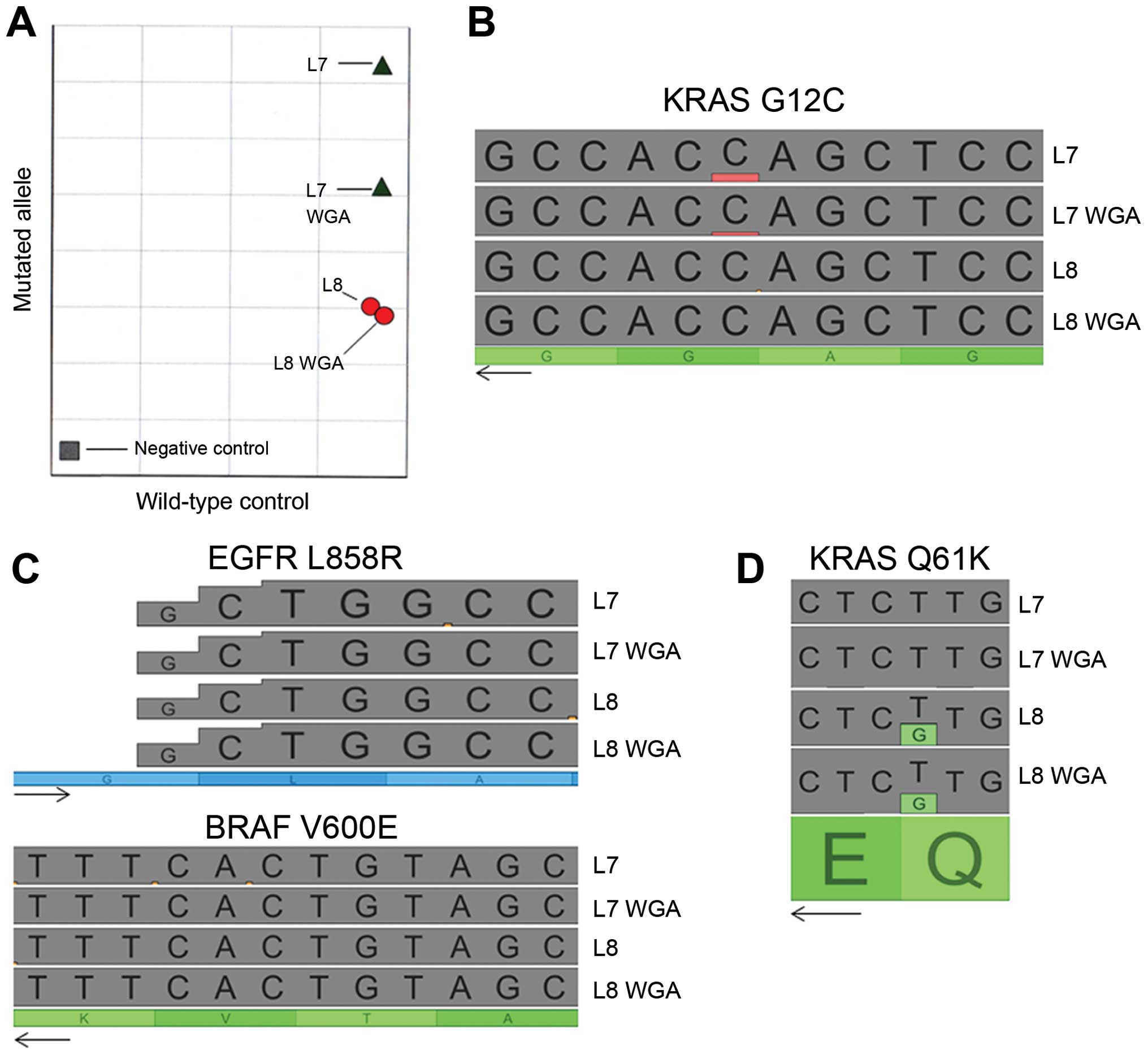
(WGA) on two samples of gDNA obtained from FFPE tissues. We then
performed allelic discrimination on non-amplified gDNA and
amplified gDNA (Fig. 3A). A
KRAS G12C mutation was detected in both the amplified and
non-amplified sample from patient L7. For patient L8, no
KRAS G12C mutation was observed in either sample. In NGS
analysis, the KRAS G12C mutation was also observed in
patient L7 (non-amplified and WGA) but not in patient L8 (Fig. 3B). We also analyzed other routinely
studied genes to compare the sequences before and after WGA.
Whatever the gene analyzed, no point mutation was induced by the
WGA (e.g., with the V600E BRAF and L858R EGFR
hotpoint mutations in Fig. 3C).
Even the rare mutation Q61H of KRAS was detected in both
non-amplified and WGA gDNA from patient L8 (Fig. 3D). Moreover, the variant allele
fraction was not modified after amplification.
NGS analysis revealed cancer
susceptibility SNP and genetic alterations in some genes
The Illumina Cancer Panel kit studies exons with
mutation hotpoints of 48 genes. We therefore analyzed all covered
sequences for the 8 lung carcinomas and 10 colon carcinomas.
Twenty-eight genetic alterations and three SNP related to cancer
susceptibility or different protein activities were found (Table III). The most frequently altered
gene was TP53 with nine alterations detected in nine
patients. Double mutations in the colon cancer susceptibility genes
APC and SMAD4 were detected in two patients (C3 and
C8, respectively), suggesting a familial risk of colon cancer in
these patients. For the patient with the APC mutation, we
detected a concomitant c-MET activating mutation E168D.
Concerning patient C8, we found a large number of alterations in
different genes (c-KIT, c-MET, FBXW7,
FGFR3, FLT3, IDH1, KRAS, RB1,
SMAD4 and TP53), suggesting high genetic instability
in this SMAD4 mutated tumor. Moreover, thanks to the
non-targeted analysis, we detected two BRAF exon 15
mutations, N581S and V600R, which induce intermediate and strong
activation of the protein, respectively. Furthermore, two mutations
with unknown impact were detected in PIK3CA and PTEN.
Concerning SNP, two patients (L7 and C4) harbored the breast cancer
susceptibility ATM F858L SNP, and one patient (C10) had the
rare c-KIT M541L SNP, which may influence the response to
imatinib. Finally, 10 patients (8 heterozygotes and 2 homozygotes
H472H) harbored the KDR Q472H polymorphism, which has been
reported to increase tumor microvasculature.
 | Table IIIExonic SNP and genetic alterations in
other analyzed genes. |
Table III
Exonic SNP and genetic alterations in
other analyzed genes.
| Genes | Nucleotide
variation | Protein sequence
variation | Patients | Impact |
|---|
| APC | c.2626C→T | R876X | C3 | Loss of function
(familial mutation) |
| c.3944C→T | S1315X | C3 | Loss of function
(somatic mutation) (26) |
| ATM | c.2572T→C | F858L | L7, C4 | Breast cancer
susceptibility SNP (23) |
| BRAF | c.1742A→G | N581S | C2 | Intermediate
activated (27) |
|
c.1798_1799GT→AG | V600R | C1 | Strongly activated
(28) |
| c-KIT | c.1621A→C | M541L | C10 | SNP with a
potential effect on imatinib response (29) |
| c.2146G→A | D716N | C8 | Possible resistance
to imatinib (24) |
| c-MET | c.504G→T | E168D | C3 | Activated (19) |
| c.1156C→A | L386I | C8 | Unknown (never
observed) |
| FBXW7 | c.832C→T | R278X | C8 | Uncertain
significance (30) |
| FGFR3 |
c.1196_1197GC→AG | R399H | C8 | Unknown (31) |
| FLT3 | c.2039C→T | A680V | C8 | Activated (32) |
| IDH1 | c.290G→A | G97V | C8 | Loss of wild-type
function (33) |
| KDR | c.1416A→T | Q472H | H: L1-L3, L5,
L7-L8, C5-C6
O: C7, C9 | Increased activity
SNP (25) |
| KRAS | c.408T→A | S136K | C8 | Unknown |
| PIK3CA | c.2176G→A | E726K | C2 | Unknown (34) |
| PTEN | c.563A→T | D187V | L1 | Unknown |
| RB1 |
c.2074_2075insATGA | Y692FsX2 | L2, L5 | Loss of
function |
| c.2119T→C | S707P | C8 | Unknown |
| SMAD4 | c.1009G→A | E337K | C8 | Unknown |
| c.1082G→A | R361H | C8 | Loss of function
(35) |
| TP53 | c.310C→T | Q104X | L1 | Unknowna |
| c.523C→T | R175V | C8 | Unknown |
| c.527G→T | C176F | L5 | Partially
functional/deleteriousa |
| c.536A→G | H179R | C3 |
Non-functional/deleteriousa |
| IVS5+2T→G | G187Fs | C1 | Unknown |
| c.709A→C | M237L | C10 | Partially
functional/deleteriousa |
| c.743G→A | R248Q | C6 |
Non-functional/deleteriousa |
| c.742C→T | R248W | C5 |
Non-functional/deleteriousa |
| c.830G→T | C277F | C7 |
Non-functional/deleteriousa |
Discussion
Molecular diagnosis is the current challenge in
cancer management. Indeed, with the increased number of targeted
therapies and resistance mechanisms developed by cancer cells, the
molecular analysis of tumors is a very important task to achieve
optimal cancer therapy. Sanger sequencing, even when accompanied by
alternon-amplified technologies, such as allelic discrimination or
high resolution melting technology, has shown its limits. Today,
next-generation sequencing is providing exciting new perspectives.
In this study, we tested Truseq Amplicon technology for the
analysis of mutation hotspots of 48 genes in gDNA extracted from
FFPE samples. All of the mutations detected by routine Sanger
sequencing, allelic discrimination or fragment analysis (in
KRAS, BRAF, EGFR genes) were also identified
with NGS analysis. Moreover, other alterations at the mutation
hotspots of the routinely analyzed genes were also detected. These
additional alterations included a G12D KRAS, a V600R
BRAF and a 15-nt deletion in exon 19 of EGFR. The
additional deletion in the EGFR gene was concomitant with
the G12D KRAS mutation in one patient, and with a V600E
BRAF mutation in another patient. KRAS, BRAF and
EGFR mutations are normally exclusive (9) but concomitant KRAS and
EGFR mutations have already been described (10,11).
The identification of concomitant mutations should increase with
the higher sensitivity of NGS technologies. Nevertheless, as it may
be impossible to confirm these ‘new’ mutations using routine
techniques, their clinical relevance and even their existence may
be debatable (12). In the same
way, some mutations are detected by NGS in very few of the read
sequences. For example, the 15-nt deletion found in three colon
carcinomas was detected in less than 8% of the read sequences, and
among these three colon carcinomas, one had no KRAS/BRAF
mutation, one had a concomitant G12D KRAS mutation, and one
also had a G13D KRAS mutation. This observation is quite
disturbing, and raises two questions: was the 15-nt deletion true,
and if so, was this alteration clinically relevant given the small
number detected. Today, the only way to have an answer would be to
treat these patients with EGFR tyrosine kinase inhibitors or to
observe anti-EGFR antibody resistance in these patients. To date,
only patients with KRAS (13), HRAS or NRAS mutations
(7,8) can be diagnosed as immediately
resistant. Concerning our three colon carcinomas, two may benefit
from treatment with EGFR TKI as the G13D KRAS mutation does
not seem to interfere with the inhibition of the EGFR pathway
(14).
With the increase in the number of genes to be
analyzed for molecular diagnosis, the quantity of gDNA obtained
from FFPE tissues will rapidly become a major problem, especially
for lung carcinomas. In this work, we tested Whole Genome
Amplification in two lung carcinomas and analyzed the resulting
samples by NGS. The genetic profile obtained before and after WGA
was qualitatively the same and quantitatively close. Indeed, only
the intensity of the G12C KRAS mutation in patient L7 was
slightly lower in the amplified sample. The strong similarity
between amplified and non-amplified samples is in accordance in
very recent studies, which showed that WGA can be safely used for
diagnosis (15,16). Moreover, through this experiment,
we showed that Truseq Amplicon technology is compatible with
samples treated by WGA.
Among the genes or codons studied in the panel but
not analyzed in routine molecular diagnosis, we detected 27
different alterations in 14 genes. Of these, 9 were detected in the
TP53 gene, which is the most frequently altered gene in
cancer (17). Eight mutations were
in the DNA binding domain of the protein, indicating that these
mutations are deleterious. One mutation occurred in a splice site,
inducing a frameshift that may not be deleterious (18). We detected 2 genes with a double
mutation, APC and SMAD4. The presence of two
mutations in these two colon cancer predisposition genes indicated
that these patients could have been members of families with a high
risk of colon cancer. Both patients harbored mutations in the c-MET
genes. The APC mutated patient had the activating mutation E168D
(19), making him/her eligible for
crizotinib therapy, which is generally used in lung cancer
(20). Concerning the SMAD4
mutated tumor, we detected eight other altered genes, suggesting
high genetic instability in this tumor type (21,22).
The impact of most of these alterations is unknown. Nevertheless,
the activating A680V mutation of FLT3 may be targeted by anti-FLT3
therapies currently in clinical development for the treatment of
leukemia.
Three SNP modifying protein sequences were found.
ATM F858L polymorphism, detected in two patients, is
associated with an increased risk of breast cancer (23), but the small number of patients in
our study does not allow us to draw any conclusions with regard to
the predisposition for colon and lung cancer. Then, c-KIT
M541L polymorphism was found in only one patient. The impact of
this polymorphism in not known, but it has been suggested that it
may affect the response to imatinib (24). Finally, KDR Q472H polymorphism was
the most interesting alteration. Indeed, tumors with histidine show
higher vascularization than do tumors with glutamine (25). In the Caucasian population, the
frequency of each genotype is 58, 36 and 6% for Q472Q, Q472H and
H472H, respectively. In our small population of tumors (n=18), we
found enrichment of the histidine allele in 55.5% of our tumors
(44.5% Q/Q, 44.5% Q/H and 11% H/H). In lung carcinomas we observed
an enrichment of heterozygous tumors (75%), whereas in colon, the
enrichment concerned homozygous H/H tumors. Moreover, during the
analysis, we found that the presence of each allele was not 50/50
in heterozygous tumors, but varied from 9 to 96% of the read
sequences. This raises the question of true polymorphism or a
selection of tumor cells with a high angiogenic capacity. To answer
this question, it would be necessary to analyze the constitutive
DNA of each patient. According to the study by Glubb et al,
patients with heterozygous or homozygous histidine tumors could be
more sensitive to inhibiting treatments of VEGFR2 or to
bevacizumab.
In conclusion, the analysis by NGS of FFPE lung and
colon carcinomas identified the alterations highlighted by routine
molecular diagnosis techniques. Thanks to its higher sensitivity,
NGS analysis revealed new mutations that were not detected
routinely. The impossibility to confirm the presence of these
mutations by another technology is problematic, and the only way to
answer this question is by conducting clinical trials that compare
treatments of patients diagnosed by routine techniques or by NGS.
Finally, the use of NGS in routine practice could revolutionize the
management of cancer patients. Indeed, simultaneous analysis of
numerous genes could identify drug-sensitive alterations generally
observed in other cancer types (for example a c-MET
alteration in a colon carcinoma that would be treated with
crizotinib in lung cancer). Nevertheless, high throughput studies
that combine NGS analysis and clinical trials need to be performed
before NGS analysis can be generalized in routine molecular
diagnosis.
Acknowledgements
We thank Philip Bastable for editing the
manuscript.
References
|
1
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J and
Haber DA: Activating mutations in the epidermal growth factor
receptor underlying responsiveness of non-small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Di Nicolantonio F, Martini M, Molinari F,
Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L,
Frattini M, Siena S and Bardelli A: Wild-type BRAF is required for
response to panitumumab or cetuximab in metastatic colorectal
cancer. J Clin Oncol. 26:5705–5712. 2008.
|
|
3
|
Chapman PB, Hauschild A, Robert C, Haanen
JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M,
Hogg D, Lorigan P, Lebbe C, Jouary T, Schadendorf D, Ribas A, O’Day
SJ, Sosman JA, Kirkwood JM, Eggermont AM, Dreno B, Nolop K, Li J,
Nelson B, Hou J, Lee RJ, Flaherty KT and McArthur GA; BRIM-3 Study
Group. Improved survival with vemurafenib in melanoma with BRAF
V600E mutation. N Engl J Med. 364:2507–2516. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carvajal RD, Antonescu CR, Wolchok JD,
Chapman PB, Roman RA, Teitcher J, Panageas KS, Busam KJ,
Chmielowski B, Lutzky J, Pavlick AC, Fusco A, Cane L, Takebe N,
Vemula S, Bouvier N, Bastian BC and Schwartz GK: KIT as a
therapeutic target in metastatic melanoma. JAMA. 305:2327–2334.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee LG, Connell CR and Bloch W: Allelic
discrimination by nick-translation PCR with fluorogenic probes.
Nucleic Acids Res. 21:3761–3766. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liew M, Pryor R, Palais R, Meadows C,
Erali M, Lyon E and Wittwer C: Genotyping of single-nucleotide
polymorphisms by high-resolution melting of small amplicons. Clin
Chem. 50:1156–1164. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Douillard JY, Oliner KS, Siena S,
Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham
D, Jassem J, Rivera F, Kocákova I, Ruff P, Błasińska-Morawiec M,
Šmakal M, Canon JL, Rother M, Williams R, Rong A, Wiezorek J, Sidhu
R and Patterson SD: Panitumumab-FOLFOX4 treatment and RAS mutations
in colorectal cancer. N Engl J Med. 369:1023–1034. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fornaro L, Lonardi S, Masi G, Loupakis F,
Bergamo F, Salvatore L, Cremolini C, Schirripa M, Vivaldi C, Aprile
G, Zaniboni A, Bracarda S, Fontanini G, Sensi E, Lupi C, Morvillo
M, Zagonel V and Falcone A: FOLFOXIRI in combination with
panitumumab as first-line treatment in quadruple wild-type (KRAS,
NRAS, HRAS, BRAF) metastatic colorectal cancer patients: a phase II
trial by the Gruppo Oncologico Nord Ovest (GONO). Ann Oncol.
24:2062–2067. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Reddi HV: Mutations in the EGFR pathway:
clinical utility and testing strategies. Clin Lab News. 39:14–16.
2013.
|
|
10
|
Schmid K, Oehl N, Wrba F, Pirker R, Pirker
C and Filipits M: EGFR/KRAS/BRAF mutations in primary lung
adenocarcinomas and corresponding locoregional lymph node
metastases. Clin Cancer Res. 15:4554–4560. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tuononen K, Mäki-Nevala S, Sarhadi VK,
Wirtanen A, Rönty M, Salmenkivi K, Andrews JM, Telaranta-Keerie AI,
Hannula S, Lagström S, Ellonen P, Knuuttila A and Knuutila S:
Comparison of targeted next-generation sequencing (NGS) and
real-time PCR in the detection of EGFR, KRAS, and BRAF mutations on
formalin-fixed, paraffin-embedded tumor material of non-small cell
lung carcinoma-superiority of NGS. Genes Chromosomes Cancer.
52:503–511. 2013. View Article : Google Scholar
|
|
12
|
McCourt CM, McArt DG, Mills K, Catherwood
MA, Maxwell P, Waugh DJ, Hamilton P, O’Sullivan JM and Salto-Tellez
M: Validation of next generation sequencing technologies in
comparison to current diagnostic gold standards for BRAF, EGFR and
KRAS mutational analysis. PLoS One. 8:e696042013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Misale S, Yaeger R, Hobor S, Scala E,
Janakiraman M, Liska D, Valtorta E, Schiavo R, Buscarino M,
Siravegna G, Bencardino K, Cercek A, Chen CT, Veronese S, Zanon C,
Sartore-Bianchi A, Gambacorta M, Gallicchio M, Vakiani E, Boscaro
V, Medico E, Weiser M, Siena S, Di Nicolantonio F, Solit D and
Bardelli A: Emergence of KRAS mutations and acquired resistance to
anti-EGFR therapy in colorectal cancer. Nature. 486:532–536.
2012.PubMed/NCBI
|
|
14
|
Tejpar S, Celik I, Schlichting M,
Sartorius U, Bokemeyer C and Van Cutsem E: Association of KRAS G13D
tumor mutations with outcome in patients with metastatic colorectal
cancer treated with first-line chemotherapy with or without
cetuximab. J Clin Oncol. 30:3570–3577. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Della Starza I, De Novi LA, Nunes V, Del
Giudice I, Ilari C, Marinelli M, Negulici AD, Vitale A, Chiaretti
S, Foà R and Guarini A: Whole-genome amplification for the
detection of molecular targets and minimal residual disease
monitoring in acute lymphoblastic leukaemia. Br J Haematol.
165:341–348. 2014.PubMed/NCBI
|
|
16
|
Hasmats J, Gréen H, Orear C, Validire P,
Huss M, Käller M and Lundeberg J: Assessment of whole genome
amplification for sequence capture and massively parallel
sequencing. PLoS One. 9:e847852014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vogelstein B, Sur S and Prives C: p53: the
most frequently altered gene in human cancers. Nat Educat.
3:62010.
|
|
18
|
Végran F, Rebucci M, Chevrier S, Cadouot
M, Boidot R and Lizard-Nacol S: Only missense mutations affecting
the DNA binding domain of p53 influence outcomes in patients with
breast carcinoma. PLoS One. 8:e551032013.PubMed/NCBI
|
|
19
|
Ma PC, Jagadeeswaran R, Jagadeesh S,
Tretiakova MS, Nallasura V, Fox EA, Hansen M, Schaefer E, Naoki K,
Lader A, Richards W, Sugarbaker D, Husain AN, Christensen JG and
Salgia R: Functional expression and mutations of c-Met and its
therapeutic inhibition with SU11274 and small interfering RNA in
non-small cell lung cancer. Cancer Res. 65:1479–1488. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tanizaki J, Okamoto I, Okamoto K, Takezawa
K, Kuwata K, Yamaguchi H and Nakagawa K: MET tyrosine kinase
inhibitor crizotinib (PF-02341066) shows differential antitumor
effects in non-small cell lung cancer according to MET alterations.
J Thorac Oncol. 6:1624–1631. 2011. View Article : Google Scholar
|
|
21
|
Markowitz S, Wang J, Myeroff L, Parsons R,
Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein
B, et al: Inactivation of the type II TGF-beta receptor in colon
cancer cells with microsatellite instability. Science.
268:1336–1338. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Woodford-Richens KL, Rowan AJ, Gorman P,
Halford S, Bicknell DC, Wasan HS, Roylance RR, Bodmer WF and
Tomlinson IP: SMAD4 mutations in colorectal cancer probably occur
before chromosomal instability, but after divergence of the
microsatellite instability pathway. Proc Natl Acad Sci USA.
98:9719–9723. 2001. View Article : Google Scholar
|
|
23
|
Struewing JP: Genomic approaches to
identifying breast cancer susceptibility factors. Breast Dis.
19:3–9. 2004.PubMed/NCBI
|
|
24
|
Debiec-Rychter M, Cools J, Dumez H, Sciot
R, Stul M, Mentens N, Vranckx H, Wasag B, Prenen H, Roesel J,
Hagemeijer A, Van Oosterom A and Marynen P: Mechanisms of
resistance to imatinib mesylate in gastrointestinal stromal tumors
and activity of the PKC412 inhibitor against imatinib-resistant
mutants. Gastroenterology. 128:270–279. 2005. View Article : Google Scholar
|
|
25
|
Glubb DM, Cerri E, Giese A, Zhang W, Mirza
O, Thompson EE, Chen P, Das S, Jassem J, Rzyman W, Lingen MW,
Salgia R, Hirsch FR, Dziadziuszko R, Ballmer-Hofer K and Innocenti
F: Novel functional germline variants in the VEGF receptor 2 gene
and their effect on gene expression and microvessel density in lung
cancer. Clin Cancer Res. 17:5257–5267. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Albuquerque C, Breukel C, van der Luijt R,
Fidalgo P, Lage P, Slors FJ, Leitão CN, Fodde R and Smits R: The
‘just-right’ signaling model: APC somatic mutations are selected
based on a specific level of activation of the beta-catenin
signaling cascade. Hum Mol Genet. 11:1549–1560. 2012.
|
|
27
|
Fratev FF and Jónsdóttir SO: An in silico
study of the molecular basis of B-RAF activation and conformational
stability. BMC Struct Biol. 9:472009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Klein O, Clements A, Menzies AM, O’Toole
S, Kefford RF and Long GV: BRAF inhibitor activity in V600R
metastatic melanoma. Eur J Cancer. 49:1073–1079. 2013. View Article : Google Scholar
|
|
29
|
Grabellus F, Worm K, Sheu SY, Siffert W,
Schmid KW and Bachmann HS: The prevalence of the c-kit exon 10
variant, M541L, in aggressive fibromatosis does not differ from the
general population. J Clin Pathol. 64:1021–1024. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davis H, Lewis A, Behrens A and Tomlinson
I: Investigation of the atypical FBXW7 mutation spectrum in human
tumours by conditional expression of a heterozygous propellor tip
missense allele in the mouse intestines. Gut. 63:792–799. 2014.
View Article : Google Scholar
|
|
31
|
Imielinski M, Berger AH, Hammerman PS,
Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M,
Sivachenko A, Sougnez C, Auclair D, Lawrence MS, Stojanov P,
Cibulskis K, Choi K, de Waal L, Sharifnia T, Brooks A, Greulich H,
Banerji S, Zander T, Seidel D, Leenders F, Ansén S, Ludwig C,
Engel-Riedel W, Stoelben E, Wolf J, Goparju C, Thompson K, Winckler
W, Kwiatkowski D, Johnson BE, Jänne PA, Miller VA, Pao W, Travis
WD, Pass HI, Gabriel SB, Lander ES, Thomas RK, Garraway LA, Getz G
and Meyerson M: Mapping the hallmarks of lung adenocarcinoma with
massively parallel sequencing. Cell. 150:1107–1120. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Piccaluga PP, Bianchini M and Martinelli
G: Novel FLT3 point mutation in acute myeloid leukaemia. Lancet
Oncol. 4:6042003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ward PS, Cross JR, Lu C, Weigert O,
Abel-Wahab O, Levine RL, Weinstock DM, Sharp KA and Thompson CB:
Identification of additional IDH mutations associated with
oncometabolite R(−)-2-hydroxyglutarate production. Oncogene.
31:2491–2498. 2012.PubMed/NCBI
|
|
34
|
Jaiswal BS, Janakiraman V, Kljavin NM,
Chaudhuri S, Stern HM, Wang W, Kan Z, Dbouk HA, Peters BA, Waring
P, Dela Vega T, Kenski DM, Bowman KK, Lorenzo M, Li H, Wu J,
Modrusan Z, Stinson J, Eby M, Yue P, Kaminker JS, de Sauvage FJ,
Backer JM and Seshagiri S: Somatic mutations in p85alpha promote
tumorigenesis through class IA PI3K activation. Cancer Cell.
16:463–474. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miyaki M, Iijima T, Konishi M, Sakai K,
Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T,
Utsunomiya J, Kuroki T and Mori T: Higher frequency of Smad4 gene
mutation in human colorectal cancer with distant metastasis.
Oncogene. 18:3098–3103. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Petitjean A, Mathe E, Kato S, Ishioka C,
Tavtigian SV, Hainaut P and Olivier M: Impact of mutant p53
functional properties on TP53 mutation patterns and tumor
phenotype: lessons from recent developments in the IARC TP53
database. Hum Mutat. 28:622–629. 2007. View Article : Google Scholar
|