Introduction
As one of the most common malignancies in the world,
hepatocellular carcinoma (HCC) has a very high morbidity and
mortality (1). Risk factors of HCC
have been well established, and chronic hepatitis B virus (HBV)
infection is a major cause in China. The smallest open reading
frame of the HBV genome, HBX, which encodes the hepatitis B virus X
protein (HBx), has been implicated in hepatocarcinogenesis and
considered to be oncogenic (2).
HBx interacts with many signal pathways in the initiation of
hepatocarcinogenesis, including AKT/PKB, ERK1/2, SAPK, NF-κB signal
transduction pathway (3). Recent
studies have indicated the role of Notch pathway in HBx-related HCC
(4–8). According to the conventional model,
Notch itself is a cell-surface receptor that transduces short-range
signals by interacting with transmembrane ligands such as Delta
(termed Delta-like in humans) and Serrate (termed Jagged in humans)
on neighboring cells. Proteolytic cleavage within the transmembrane
subunit of the Notch receptor results in translocation of the
intracellular domain of Notch (ICN) to the nucleus where it
interacts with the transcriptional repressor CSL, also known as
CBF-1 or RBP-Jκ. Binding of ICN displaces co-repressor complexes,
thereby activates transcription by promoters with CSL binding
elements and then modulates the expression of many genes, such as
Hes1 (9–11). Notch signaling may generate
opposing effect in different steps of carcinogenesis, depending on
the tumor cell type and the status of other signaling pathways,
such as Wnt signaling pathway (12).
The effect of Notch in HCC is highly controversial.
Some studies have described a direct role of Notch in promoting
HCC. Giovannini et al (13)
reported aberrant nuclear expression of Notch1 and Notch3 in HCC
tissues compared with the surrounding cirrhotic tissues.
Furthermore, silencing of Notch3 in HCC cells enhanced sensitivity
to doxorubicin-induced cell death via a p53-dependent mechanism. In
keeping with the view, Villanueva et al (14) reported that Notch signaling was
activated in human HCC samples and promoted formation of liver
tumors in mice. Other groups, however, have pointed out the
different role of Notch signaling in HCC. Qi et al (15) demonstrated that Notch1 induced
apoptosis of HCC cells by altering the balance between p53 and
Bcl-2. Consistent with the observation, upregulation of p53 induced
by Notch1 sensitized HCC cells to tumor necrosis factor-related
apoptosis-inducing ligand-induced apoptosis (16). These discoveries were further
confirmed in a mouse model of HCC generated by genetic inactivation
of the retino blastoma pathway, activation of the Notch signaling
reduced HCC cell proliferation and tumor growth (17). In a word, contradictory data exist
on the effect of Notch signaling in HCC, and its role in
HBx-related HCC is even less documented. Our previous studies have
shown that activated Notch1 signaling is required for HBx to
promote proliferation and survival of L02 (4,7).
However, the exact mechanisms remain elusive.
To address this question, we used L02/HBx cell lines
as a cell model to study the relationship between Notch and
Wnt/β-catenin pathways in promoting proliferation. These data
suggested that Wnt pathway might be involved in Notch1-mediated
effects.
The Wnt signaling pathway has long been recognized
for its role in regulating embryonic development, and has recently
been linked to cancer in adults (18). Canonical Wnt signaling is the most
well-known pathway, which is centered around β-catenin. At the cell
membrane, Wnt binds to Frizzled receptors and co-receptors such as
low density lipoprotein receptor-related proteins (LRP). This
results in stabilization of β-catenin through disheveled-mediated
inhibition of the destruction complex. Soluble β-catenin then
translocates to the nuclei and displaces groucho-related repressor
from T cell factor (TCF)/lymphoid enhancer factor (LEF)
transcription factor. TCF/LEF is then able to regulate expression
of target genes, such as cyclin D1 (19).
There is now emerging evidence of cross-talk between
Notch and Wnt pathways in cancer. Camps et al (20) identified LNX2, a gene that was not
associated with cancer before, as mediating cross-talk between WNT
and NOTCH signaling cascades in colorectal cancer. Li et al
(21) found evidence of cross-talk
in non-small cell lung cancer. However, little is known about
cross-talk between Notch and Wnt signaling pathways in HCC, and in
HBx-related HCC cross-talk and the potential role of Wnt signaling
are even less documented.
In this study, we address the question of how Notch
signaling can modulate HBx-related HCC and discover novel
observations that close cooperation between Notch and Wnt pathways
is important in HBx-related HCC.
Materials and methods
Cell culture
The human non-tumor hepatic cell line L02 was a
generous gift from Dr Xinyuan Liu (Shanghai Institutes for
Biological Sciences, Shanghai, China). This cell line was
originated histologically from normal human liver tissue
immortalized by stable transfection with the hTERT gene, which has
been used previously (22).
L02/HBx and L02/pcDNA3.1 cell lines, which derived from L02 cells
by transfection with HBx expression plasmid or its empty plasmid
(pcDNA3.1(+)/V5-HisB), respectively, were successfully established
previously (23). All cell lines
were cultured in DMEM (Gibco, Carlsbad, CA, USA) containing 10%
fetal bovine serum (FBS; Gibco, Grand Island, NY, USA) and
maintained in humidified incubator at 37°C in a 5% CO2
atmosphere, L02/pcDNA3.1 and L02/HBx cell lines were supplemented
with 250 μg/ml G418 (Invitrogen, Carlsbad, CA, USA).
RNA isolation, quantitative real-time PCR
(qRT-PCR)
Total RNA was isolated from cultured cells using
TRIzol reagent (Invitrogen) and cDNA was synthesized from 100 ng
RNA using PrimeScript RT reagent kit (Takara, Japan). Real-time
quantitative PCR using SYBRP remix (Takara) was performed as
described previously (8).
Amplifications were performed in a Step One Real-Time PCR system
(Applied Biosystems, USA) following the manufacturer’s
instructions. The expression of RNA was determined from the
threshold cycle (Ct) and the relative expression levels were
calculated by the 2−ΔΔCt method. All samples were
assayed in triplicate. The primer sequences used to amplify
specific target genes are listed in Table I.
 | Table IPrimer sequences for quantitative
RT-PCR analysis. |
Table I
Primer sequences for quantitative
RT-PCR analysis.
| Gene | Sequences | PCR product
(bp) | GenBank accession
no. |
|---|
| Notch1 |
| Sense |
5′-CCGCAGTTGTGCTCCTGAA-3′ | 109 | NM_017617 |
| Antisense |
5′-ACCTTGGCGGTCTCGTAGCT-3′ | | |
| Hes1 |
| Sense |
5′-GCTAAGGTGTTTGGAGGCT-3′ | 122 | NM_005524 |
| Antisense |
5′-CCGCTGTTGCTGGTGTA-3′ | | |
| Fzd7 | | | |
| Sense |
5′-TTCTCGGACGATGGCTACC-3′ | 132 | NM_003507 |
| Antisense |
5′-GAACCAAGTGAGAGACAGAATGACC-3′ | | |
| Fzd10 | | | |
| Sense |
5′-CCCGATTATGGAGCAGTTCA-3′ | 117 | NM_007197 |
| Antisense |
5′-TCGTCCGAGCCGTTGTT-3′ | | |
| cyclin D1 |
| Sense |
5′-GGCTGAAGTCACCTCTTGGTTACAG-3′ | 177 | NM_053056 |
| Antisense |
5′-TAGCGTATCGTAGGAGTGGGACAG-3′ | | |
| Actin |
| Sense |
5′-GTTGCGTTACACCCTTTCTTG-3′ | 157 | NM_001101 |
| Antisense |
5′-GACTGCTGTCACCTTCACCGT-3′ | | |
Western blot analysis
Cells were lysed as described (8). Protein (40 μg) from each sample was
examined by SDS-12% PAGE and then electrotransferred to
nitrocellulose membranes using a semidry transfer apparatus
(Bio-Rad). Nitrocellulose membranes were subsequently blocked with
5% BSA in TBST for 2 h at room temperature, and incubated with each
primary antibody overnight. Rabbit anti-Notch1, anti-Fzd7, anti-
Fzd10, anti-β-catenin, anti-cyclin D1 were purchased from
Proteintech Group (Proteintech Group, Chicago, IL, USA), and rabbit
anti-Hes1, anti-actin were obtained from Santa Cruz Biotechnology
(Santa Cruz, CA, USA). The membranes were washed and incubated with
horseradish peroxidase-labeled secondary antibody (1:4,000; Santa
Cruz Biotechnology), visualization was performed by an enhanced
chemiluminescence kit (Pierce) and exposure to X-ray film (Kodak,
Rochester, NY, USA). Immunoblotting with anti-actin antibody was
used as an internal control to confirme quivalent protein loading.
Each experiment was repeated three times.
Transfection and RNA interference
L02/HBx cells were seeded in 6-well plates. The next
day the cells (30–50% confluence) were treated with Notch1 siRNAs
or Fzd10 siRNAs, and control siRNA (which does not match any known
mammalian GenBank sequences). The siRNA sequences are listed in
Tables II and III. All siRNAs were purchased from
RiboBio (Guangzhou, China). Cells were transiently transfected with
Notch1 siRNAs or Fzd10 siRNAs using Lipofectamine™ 2000
(Invitrogen). Media were replaced 6 h after transfection. Cells
were allowed to grow for 48 h and harvested to choose the sequence
of maximum inhibition effect on Notch1 and Fzd10, based on which
recombinant plasmids pH1-MCS-CMV-GFP-Hygro- Notch1 shRNA and -Fzd10
shRNA were constructed for further stable transfection. After 48 h,
transfected cells were selected for 3 weeks with 100 μg/ml
hygromycin B (Sigma, St. Louis, MO, USA) to obtain stable
Notch1-knockdown cell lines (Notch1-shRNA), Fzd10-knockdown cell
lines (Fzd10-shRNA) and negative control cells (NC). Individual
colonies were picked and then the stable cells were expanded with
50 μg/ml hygromycin B.
 | Table IIThree siRNA sequences against Notch1
sequence (NM_017617). |
Table II
Three siRNA sequences against Notch1
sequence (NM_017617).
| Target
sequences | | siRNA
sequences |
|---|
| siRNA1 |
|
GGTGTCTTCCAGATCCTGA | Sense |
5′-GGUGUCUUCCAGAUCCUGA dTdT-3′ |
| Antisense | 3′-dTdT
CCACAGAAGGUCUAGGACU-5′ |
| siRNA2 |
|
TGGCGGGAAGTGTGAAGCG | Sense |
5′-UGGCGGGAAGUGUGAAGCG dTdT-3′ |
| Antisense | 3′-dTdT
ACCGCCCUUCACACUUCGC-5′ |
| siRNA3 |
|
GGACCAACTGTGACATCAA | Sense |
5′-GGACCAACUGUGACAUCAA dTdT-3′ |
| Antisense | 3′-dTdT
CCUGGUUGACACUGUAGUU-5′ |
 | Table IIIThree siRNA sequences against Fzd10
sequence (NM_007197). |
Table III
Three siRNA sequences against Fzd10
sequence (NM_007197).
| Target
sequences | | siRNA
sequences |
|---|
| siRNA1 |
|
CGATTATGGAGCAGTTCAA | Sense |
5′-CGAUUAUGGAGCAGUUCAA dTdT-3′ |
| Antisense | 3′-dTdT
GCUAAUACCUCGUCAAGUU-5′ |
| siRNA2 |
|
GCTACAACATGACTCGTAT | Sense |
5′-GCUACAACAUGACUCGUAU dTdT-3′ |
| Antisense | 3′-dTdT
CGAUGUUGUACUGAGCAUA-5′ |
| siRNA3 |
|
CCATCCAGTTGCACGAGTT | Sense |
5′-CCAUCCAGUUGCACGAGUU dTdT-3′ |
| Antisense | 3′-dTdT
GGUAGGUCAACGUGCUCAA-5′ |
Construction of Fzd10-expression vector
and transfection
Fzd10 gene was obtained from cDNA library via PCR,
and then the Fzd10 gene was cloned into XhoI and KpnI
sites of pCMV-MCS-3FLAG-SV40-Puromycin (pCV061) expression vector
using T4 DNA ligase at 16°C overnight. The resulting construct was
confirmed by DNA sequencing. cDNA library and pCV061 were purchased
from GeneChem (Shanghai, China). The primerse quences used to
amplify Fzd10 gene: sense 5′-TCCGCTCGAGATGCAGCGCCCGGGCCCCC
GCCTGT-3′, antisense 5′-ATGGGGTACCGACACGCAGG TGGGCGACTGGGCAG-3′.
Ninety percent-confluent Notch1- shRNA cells were transfected with
pCV061-Fzd10 plasmids or pCV061 control vector using Lipofectamine
2000, respectively (Invitrogen). After 6 h of transfection, cells
were washed and allowed to recover in DMEM medium supplemented with
10% fetal calf serum. After 48 h, transfected cells were selected
for 3 weeks with 2 μg/ml puromycin (Sigma) to obtain stable
Fzd10-overexpressed cell lines (pCV061-Fzd10) and control cells
(pCV061). Individual colonies were picked and then the stable cells
were expanded with 1 μg/ml puromycin.
Cell proliferation and viability
assay
Cells were seeded in a 96-well plate with
104 cells/well and Cell Counting kit-8 (Dojindo
Laboratories, Kumamoto, Japan) was used to test cell proliferation
at the indicated times (24, 48, 72 and 96 h). The absorbance value
at 450 nm was measured using a spectrophotometer after incubation
with WST-8 reagent (10 μl) for 2 h; 630 nm was the reference
wavelength. All experiments were repeated eight times.
Analysis of colony formation
For clonogenicity analysis, 1000 viable cells were
placed in 6-well plates and maintained in complete medium for 2
weeks. Colonies were fixed with methanol and stained with methylene
blue.
Xenograft tumor model
Twelve male BALB/c-nu/nu mice (BW, approximately 19
g; age, 4 weeks) were purchased from the animal centre of Tongji
Medical College, Huazhong University of Science and Technology
(Wuhan, China). All animal experiments were approved by the
Institutional Animal Care and Use Committee of the Huazhong
University of Science and Technology. The mice were randomized into
two groups (n=6 in each group), and housed in the Animal Institute
of Tongji Medical College in laminar flow cabinets. Cells
(2×106) (NC and Fzd10-shRNA) in 0.2 ml each were
injected subcutaneously into the posterior neck of each nude mouse.
The length (L) and width (W) of the tumors were measured externally
with a vernier caliper every 3 days. Tumor volume was calculated
with the formula: V = (L × W2)/2. After 20 days,
tumor-bearing mice and controls were sacrificed, and the tumors
were excised and measured.
Histology and immunohistochemistry
For histological analysis, tissues were fixed in 4%
paraformaldehyde at 4°C, embedded in paraffin, cut into 5-μM
sections and transferred to silicon-coated slides. Tissue sections
were then stained with hematoxylin and eosin. For
immunohistochemical analysis, staining for β-catenin was carried
out, using rabbit polyclonal antibodies against β-catenin
(Proteintech Group) at a dilution of 1:100. Visualization was
performed using the 3,3′-diaminobenzidine tetrahydrochloride (DAB;
Vector Laboratories, Burlingame, CA), followed by counterstaining
with Mayer’s hematoxylin (Merck, Darmstadt, Germany).
Immunofluorescence analysis
Cells seeded on 24-well plates were fixed in 4%
paraformaldehyde for 30 min at 37°C. Then cells were permeabilized
with 0.5% Triton-X 100 for 20 min, and blocked with 5% BSA for 30
min. Cells were incubated with anti-β-catenin (1:100, Proteintech
Group) at 4°C overnight. Primary antibody was visualized with
Cy3-conjugated secondary antibodies (1:100, Boster, China) for 1 h
in the dark. Nuclei were stained with DAPI (Boster) for 5 min.
Images were collected using an LSM410 confocal laser scanning
microscope (Carl Zeiss, Jena, Germany).
Statistics
Each experiment was repeated at least three times.
The data are presented as mean ± SEM. SPSS version 17.0 software
(SPSS for Windows, Inc., Chicago, IL, USA) was used for all
statistical analyses. Statistical analysis of data was performed
using standard One-way ANOVA or One-way ANOVA for repeated
measures, followed by Bonferroni post-hoc test. A two-tailed
Student’s paired t-test was also used to compare the difference in
values between 2 groups. A P-value of <0.05 is considered as
statistically significant.
Results
HBx activates Notch1 and Wnt/β-catenin
signaling pathways in L02 cells, and increase Fzd10 expression
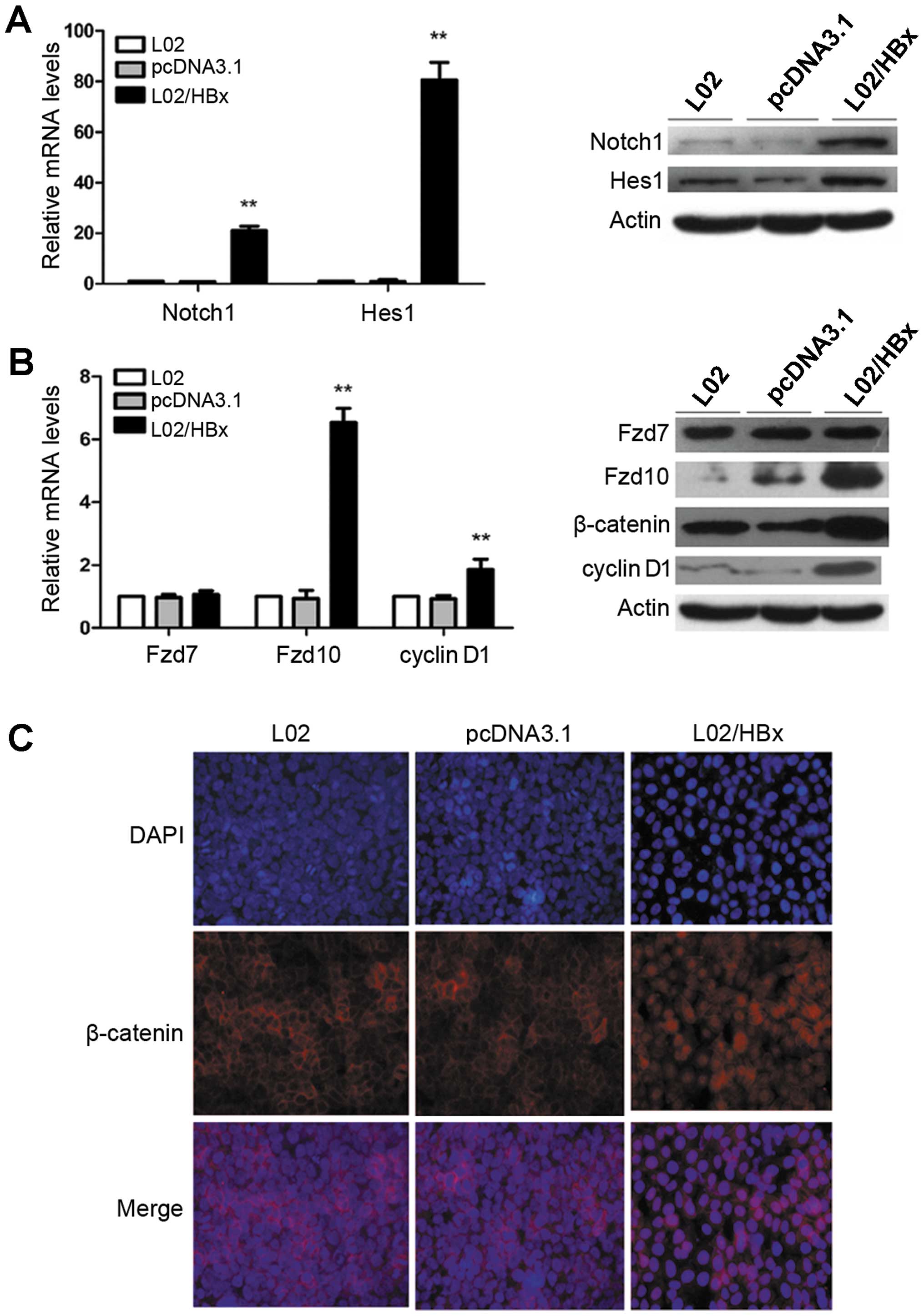
We first used qRT-PCR and Western blot analysis in
L02, L02/pcDNA3.1 and L02/HBx cells. As shown in Fig. 1, the mRNA and protein expression
levels of Notch1, Hes1 in L02/HBx cells were elevated relative to
controls (Fig. 1A;
**P<0.01). In addition, we analyzed the mRNA and
protein expression levels of Fzd7, Fzd10 and cyclin D1, along with
the protein expression of β-catenin. We found that their expression
levels were increased in L02/HBx cells as compared with controls
except Fzd7 (Fig. 1B;
**P<0.01). Moreover, β-catenin translocated from the
membrane and cytoplasm to the nuclei (Fig. 1C). Taken together, these results
suggest that HBx may activate Notch1 and Wnt/β-catenin signaling
pathways upregulating Fzd10 expression.
Inhibition of Notch1 signaling pathway
decreases the activity of Wnt/β-catenin signaling pathway, and
reduces the Fzd10 expression in L02/HBx cells
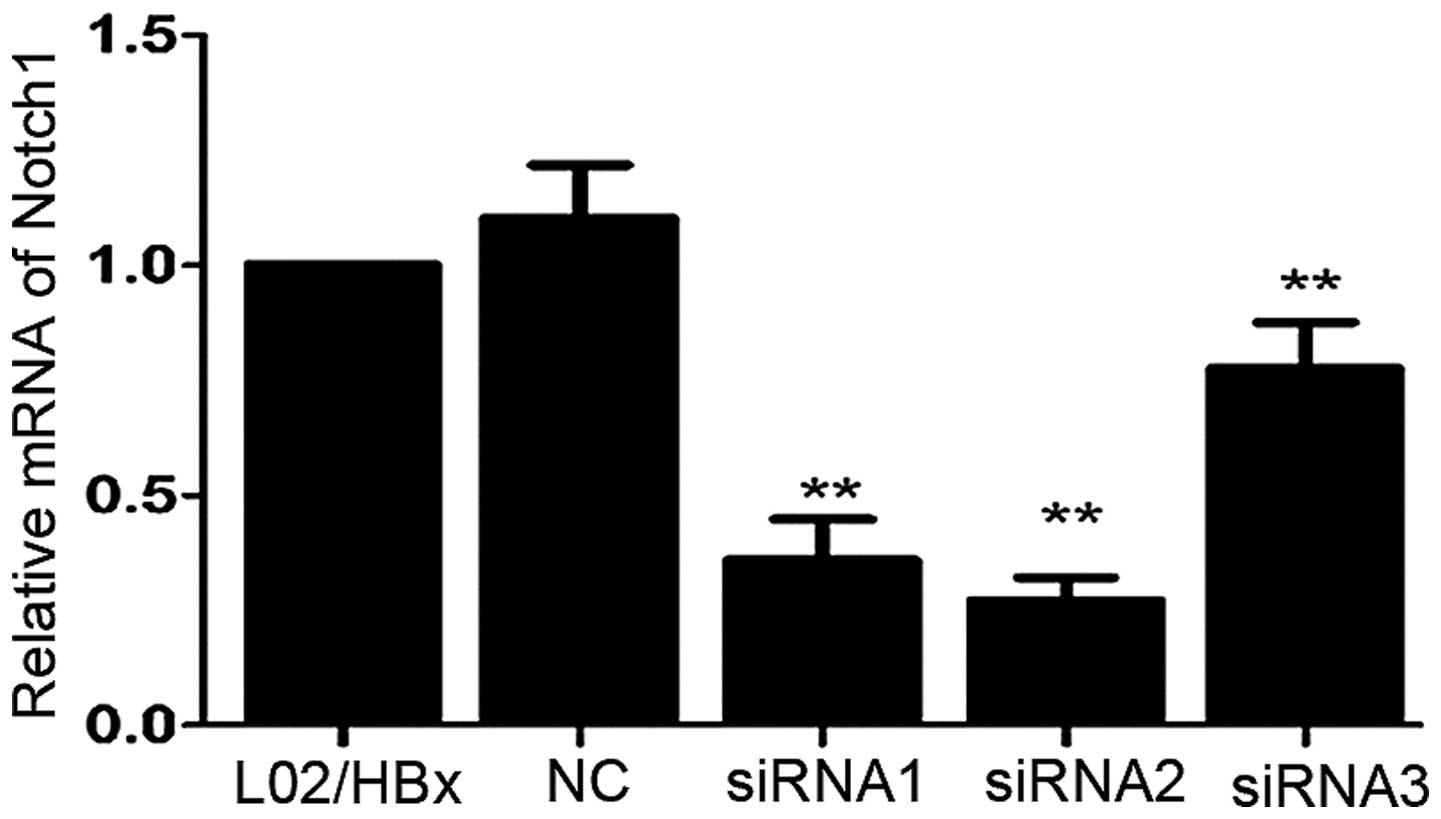
In order to identify the effective siRNA target
gene, the mRNA expression of Notch1 in L02/HBx cells was determined
by qRT-PCR 48 h after transfection with siRNAs. Comparing to the
non-transfected cells, transfection of Notch1-siRNA1, 2 or 3
(siRNA1, 2, 3) into L02/HBx cells resulted in downregulating of
Notch1 mRNA expression up to 64.2, 72.9 and 22.4%, respectively
(Fig. 2; **P<0.01).
So the target sequence of Notch1-siRNA2 was chosen for construction
of Notch1-shRNA.
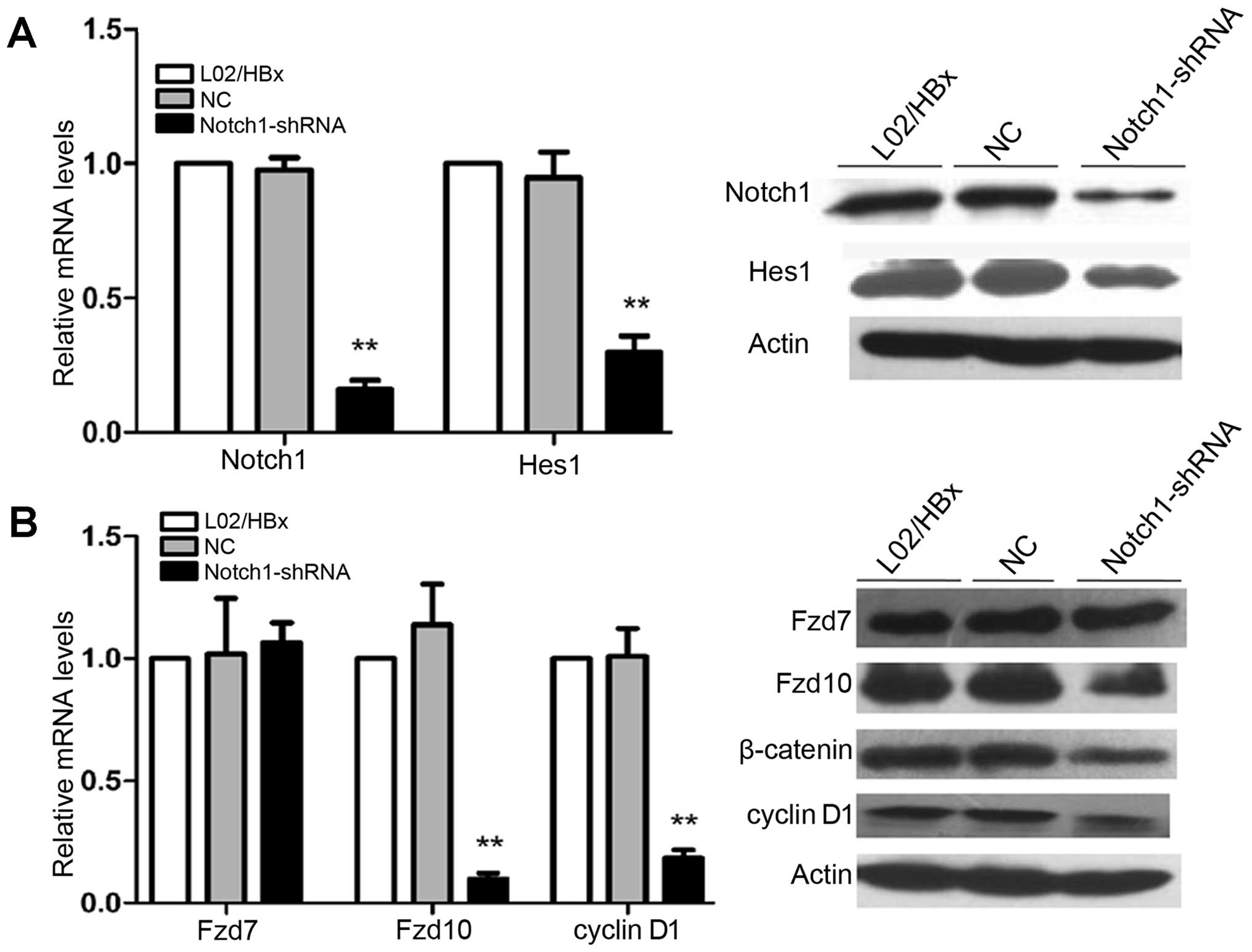
We successfully constructed the pH1-MCS-CMVGFP-
Hygro-Notch1 shRNA (Notch1-shRNA) and the pH1-MCS-CMV-GFP-Hygro
negative control vector (NC). Comparing to the non-transfected
cells, transfection of Notch1-shRNA into L02/HBx cells resulted in
dramatically downregulating of Notch1 mRNA and protein expression,
but there was no significant change in NC-transfected group. Hes1,
a target gene of Notch signaling, was downregulated after
transfected with Notch1-shRNA (Fig.
3A; **P<0.01). These results indicated that
Notch1 signaling pathway in L02/HBx cells had been partially
blocked by Notch1-shRNA. Then, we tested the mRNA and protein
expression levels of Fzd7, Fzd10 and cyclin D1, along with the
protein expression of β-catenin. We found that their expression
levels except Fzd7 were decreased in L02/HBx cells as compared with
controls (Fig. 3B;
**P<0.01). These results suggested that
downregulation of Notch1 expression by shRNA decreases the
activation of Wnt/β-catenin pathway in L02/HBx cells.
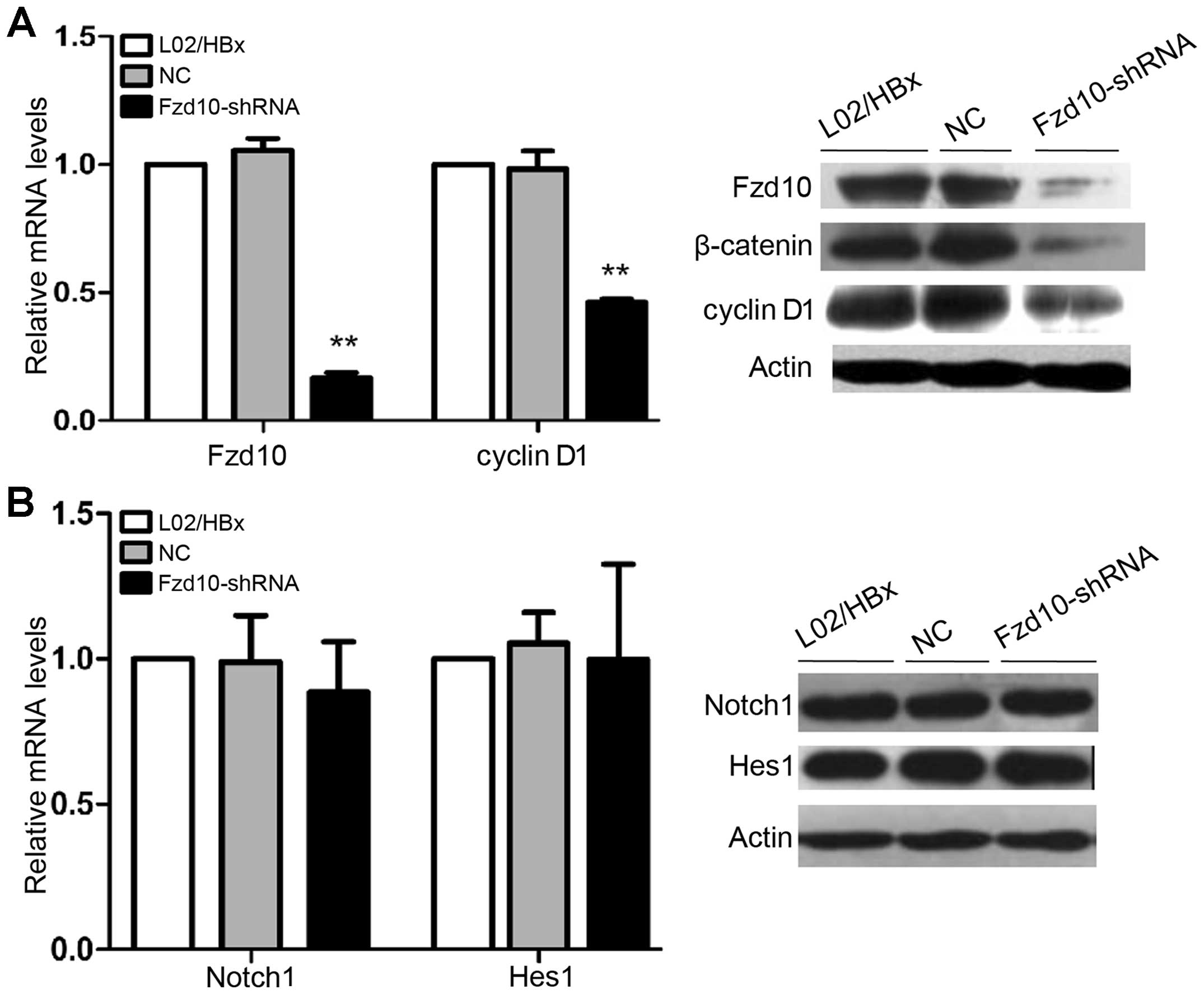
Inhibition of Wnt/β-catenin signaling
pathway via Fzd10 shRNA does not affect the activity of Notch1
signaling pathway in L02/HBx cells
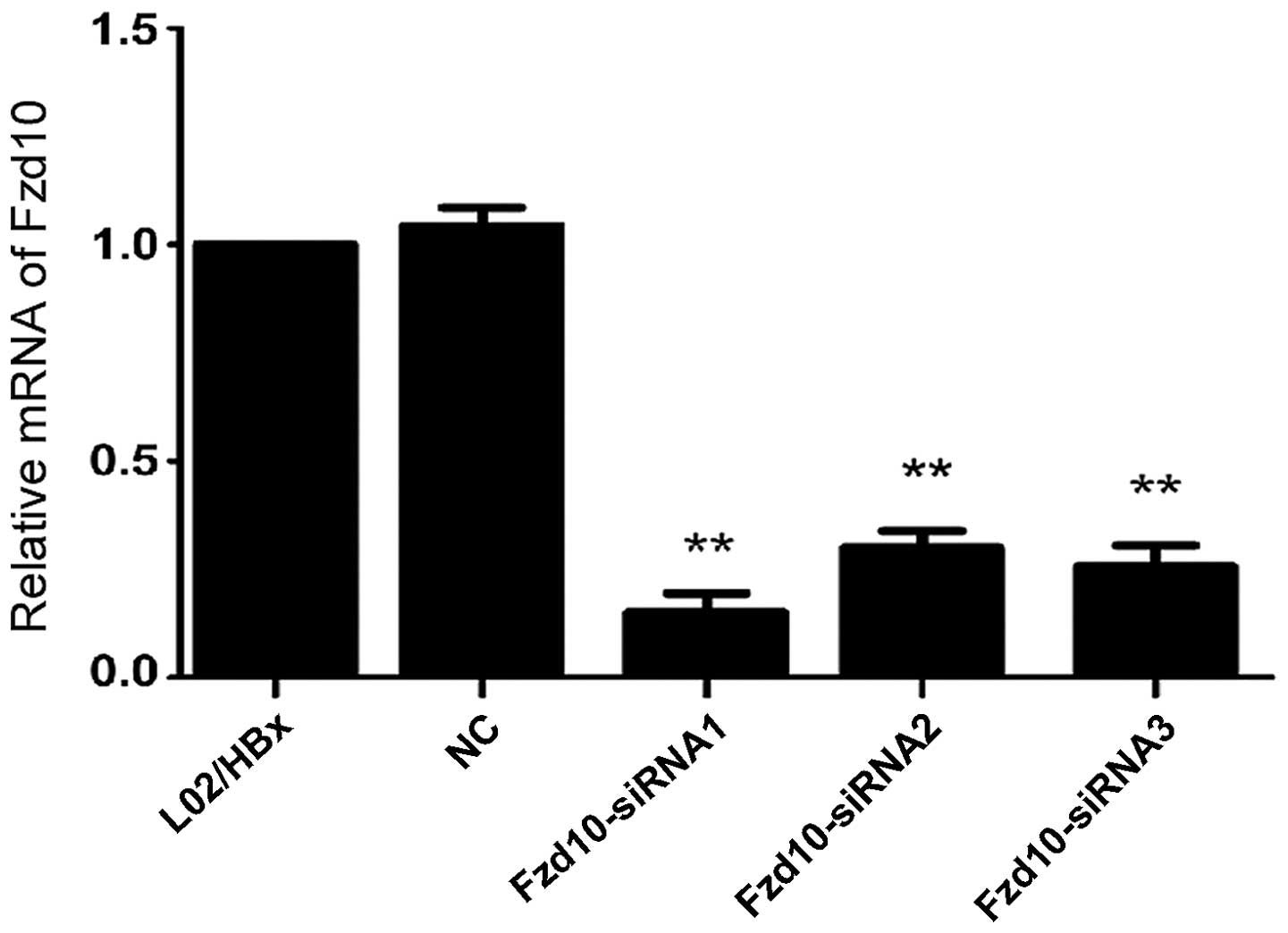
In order to identify the effective siRNA target
gene, the mRNA expression of Fzd10 in L02/HBx cells was determined
by qRT-PCR 48 h after transfection with siRNAs. Comparing to the
non-transfected cells, transfection of Fzd10-siRNA1, 2 or 3 into
L02/HBx cells resulted in downregulating of Fzd10 mRNA expression
up to 85.0, 70.0 and 74.3%, respectively (Fig. 4; **P<0.01). So the
target sequences of Fzd10-siRNA1 were chosen for construction of
Fzd10-shRNA.
We successfully constructed the pH1-MCS-CMV-GFP-
Hygro-Fzd10 shRNA (Fzd10-shRNA). Comparing to the non-transfected
cells, transfection of Fzd10-shRNA into L02/HBx cells resulted in
dramatic downregulation of Fzd10 mRNA and protein expressions, but
there was no significant change in NC-transfected group. β-catenin,
a dominant effector and cyclin D1, a target gene of Wnt/β-catenin
signaling, were downregulated after transfected with Fzd10- shRNA
(Fig. 5A; **P<0.01).
These results indicated that Wnt/β-catenin pathway in L02/HBx cells
had been partially blocked by Fzd10-shRNA. Then, we tested the mRNA
and protein expression levels of Notch1 and Hes1. We found that
their expression levels were not changed in L02/HBx cells as
compared with controls (Fig. 5B).
These results indicated that downregulation of Fzd10 expression by
shRNA does not significantly affect the Notch signaling pathway in
L02/HBx cells.
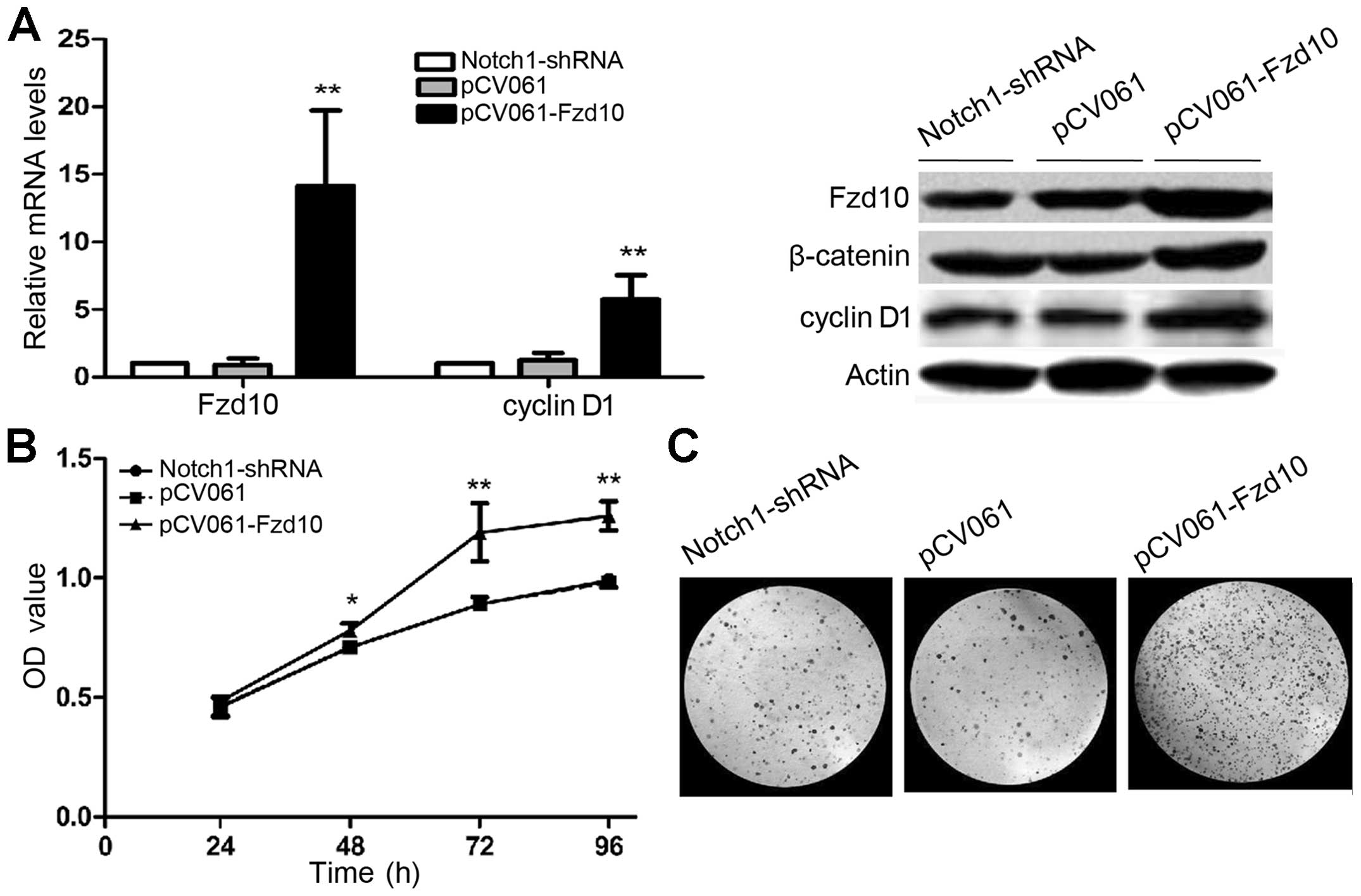
Activated Wnt/β-catenin pathway is
required for L02/HBx cell proliferation in vivo and in vitro
L02/HBx cells lacking Notch1 have impaired ability
of proliferation and survival (7).
We asked whether activation of Wnt signaling can overcome this
defect. To address this question, we constructed pCV061-Fzd10.
After transfection of pCV061-Fzd10, L02/HBx-Notch1 shRNA cells that
previously expressed low levels of Fzd10, cyclin D1 and β-catenin,
showed significantly increased levels of mRNA and protein compared
to non-transfected cells (Fig. 6A;
**P<0.01). There was no remarkable change in the
pCV061 empty vector-transfected group, suggesting that the
transfection was successful and Wnt signaling was activated with
Fzd10-expression vector. Moreover, CCK-8 assay (Fig. 6B; *P<0.05,
**P<0.01) and colony formation assay (Fig. 6C) showed that activation of Wnt
signaling restored proliferation that was inhibited in
L02/HBx-Notch1 shRNA cells. These data suggest that Wnt signaling
is downstream of Notch in regulation of cell proliferation.
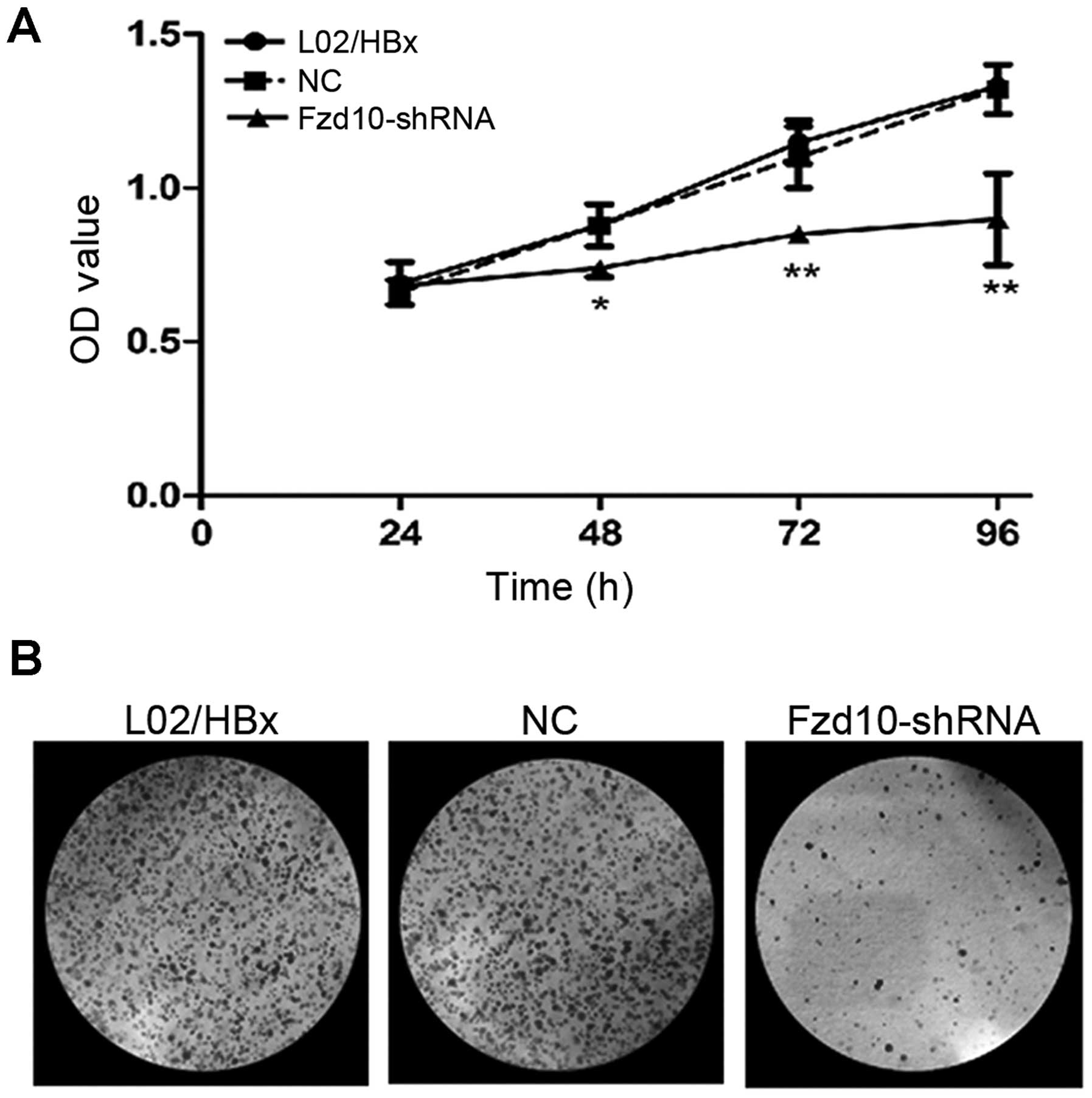
To confirm the above observations, we used an
alternative experimental system where activated Notch1 signaling
exists in L02/HBx cells, and is required for HBx to promote
proliferation and survival of L02 cells (4,7). To
verify that the Notch effect on cell proliferation was mediated via
Wnt pathway, L02/HBx cells were transfected with either NC or
Fzd10-shRNA and then carried out CCK-8 (Fig. 7A; *P<0.05,
**P<0.01) and colony formation assays (Fig. 7B). Notch1 may induce a significant
increase in the proliferation of L02 (7).
This effect was absent when L02/HBx cells were
transfected with Fzd10-shRNA. Fzd10-shRNA abrogated the effect of
Notch1 on proliferation. These data were consistent with the
results of experiments with L02/HBx-Notch1 shRNA cells and indicate
that Wnt pathway is downstream of Notch signaling in proliferation
of L02/HBx cells, and Wnt pathway is required for Notch1 to promote
proliferation of L02/HBx cells.
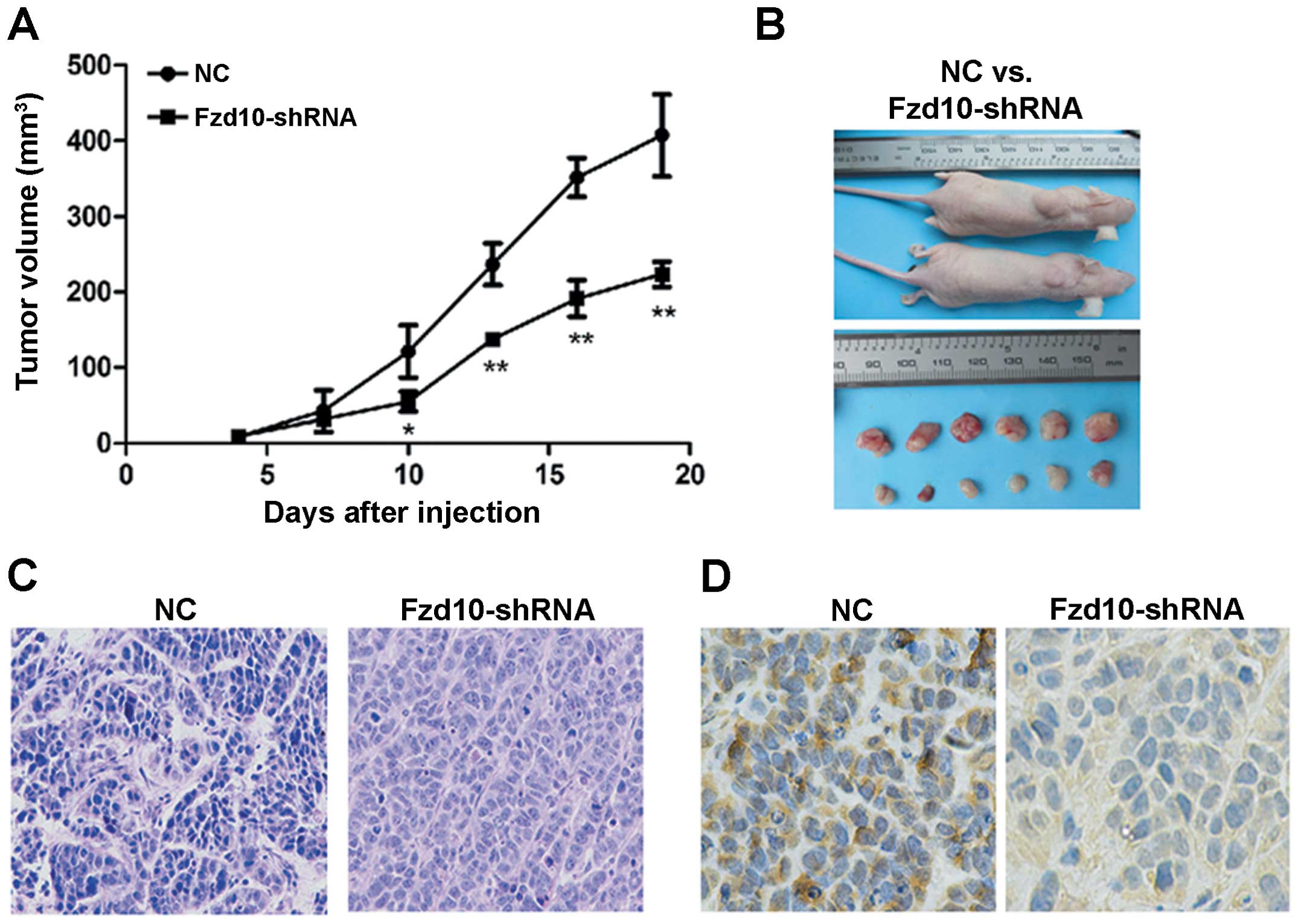
Coincidentally, the tumorigenicity was remarkably
suppressed in nude mice injected with L02/HBx cells pre-treated
with Fzd10-shRNA in vivo (Fig.
8A and B; *P<0.05, **P<0.01).
Pathological examination of the primary tumors revealed malignant
phenotypes in both groups by hematoxylin and eosin (H&E)
staining (Fig. 8C). The effective
inhibition of Wnt signaling pathway was confirmed by
immunoreactivity analysis with β-catenin in mouse tumor models
in vivo (Fig. 8D).
Discussion
Notch signaling was previously reported to be
implicated in the development of HCC. However, the available data
were controversial. Some groups found aberrant nuclear expression
of Notch1 and Notch3 in human liver cancer tissue compared to the
surrounding cirrhotic tissue, and Notch signaling promoted
formation of liver tumors in mice. Utilizing experiments with ‘loss
of function’ based on knockdown of Notch3 in HCC cells showed
enhanced sensitivity to doxorubicin-induced cell death (13,14).
On the contrary, other studies demonstrated that the Notch1
signaling results in significant growth inhibition of HCC cells
both in vitro and in vivo (15–17).
So contradictory data exists in respect to the effect of Notch
signaling on HCC, and scarce data exist on its effect on
HBx-related HCC. Our research may be helpful to resolve some of
those contradictions and clarify the role of Notch signaling in
HBx-related HCC.
We have reported that activated Notch1 signaling is
required for HBx to promote malignant transformation of L02 cells
(4,7). In this study, we showed that Notch1
may exert its effect on HBx-related HCC primarily via activation
the Wnt pathway. Activation of Wnt signaling restored the
proportion of proliferation inhibited by L02/HBx-Notch1 shRNA, and
inhibition of Wnt signaling impaired the effect of Notch1 on
proliferation and tumor formation of L02/HBx cells in BALB/c nude
mice.
Our data provide a potential new model for
explaining the machanism of HBx-related HCC. It appears that Notch
signaling may affect Wnt signaling in L02/HBx via regulating
expression level of Fzd10 instead of Fzd7. Several lines of
evidence have demonstrated that Notch modulates Wnt/β-catenin
pathway by regulating Fzd10 expression in HBx-expressing L02 cells:
HBx upregulated Notch signaling pathway and Wnt/β-catenin pathway,
along with Fzd10 rather than Fzd7; inhibition of Notch pathways
resulted in the decreased expression of Fzd10 and the activation of
Wnt pathway; Fzd10-shRNA significantly reduced the activation of
Wnt pathway, but did not apparently affect the Notch signaling
pathway.
Human FZD10 has been found to be expressed in adult
normal placenta, skeletal muscle, brain, heart, lung, pancreas,
spleen, prostate and fetal kidney, lung and brain (24). Our data are in agreement with
observations from some groups that FZD10 expression was higher in
some cancer tissues (colon, lung squamous cell carcinoma) (25–27).
Moreover, the effectiveness of anti-FZD10 antibody therapy was
reported in synovial sarcomas since FZD10 increases cell growth in
synovial sarcoma (28,29). However, it was previously
demonstrated that FZD7 activated Wnt/β-catenin signaling in several
cancer types including hepatocellular carcinoma (24,30),
and the Fzd7 steady-state mRNA levels were upregulated in hepatitis
B, C and non-viral-induced HCC cell lines and mouse models
(30–33), and inhibition of Fzd7 with small
interfering peptides displayed antitumor properties in
hepatocellular carcinoma (34).
Given that our data are inconsistent with these reports, we
speculate Fzd10 is related to the initial progression of
HBx-related HCC, and Fzd7 is linked to the development of HCC.
Our results demonstrate a critical role of the
canonical Wnt pathway for malignant transformation of human hepatic
cells induced by HBx. Downregulation of Wnt pathway in L02/HBx
cells inhibited proliferation and tumor formation of L02/HBx cells
in BALB/c nude mice. Our results are consistent with the recent
observations that HBx competitively binds adenomatous polyposis
coli (APC) to displace β-catenin from its degradation complex,
which results in the activation of Wnt signaling (35); ectopic expression of HBx along with
Wnt-1 activated Wnt/β-catenin signaling in hepatoma cells, which
was achieved by suppressing glycogen synthase kinase3 activity via
the activation of Src kinase, suggesting that HBx could contribute
to hepatocarcinogenesis via the activation of Wnt/β-catenin
signaling (36).
Our data suggest that the Wnt pathway can be
downstream of Notch in malignant transformation of human hepatic
cells induced by HBx. This model may also explain the discrepancy
in reports of Notch effects on HCC. On one hand, activation of the
Notch pathway by itself is not sufficient for HCC and requires the
Wnt pathway. However, activation of the Wnt pathway may overcome
the absence of Notch signaling. This cooperation between Notch and
Wnt signaling may provide a novel field of vision for research on
the pathogenesis of HBx-related HCC.
Acknowledgements
This study was supported by the Graduate student
innovation foundation of Huazhong University of Science and
Technology, HF-11-29-2013 and the National Science Foundation of
China, No. 81172063 and No. 81372352.
Abbreviations:
|
HCC
|
hepatocellular carcinoma
|
|
HBx
|
hepatitis B virus X protein
|
|
Hes
|
hairy and enhancer of split family
|
|
qRT-PCR
|
quantitative real-time RT-PCR
|
|
Fzd
|
Frizzled family of Wnt receptors
|
References
|
1
|
Block TM, Mehta AS, Fimmel CJ and Jordan
R: Molecular viral oncology of hepatocellular carcinoma. Oncogene.
22:5093–5107. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Martin-Vilchez S, Lara-Pezzi E,
Trapero-Marugan M, Moreno-Otero R and Sanz-Cameno P: The molecular
and pathophysiological implications of hepatitis B X antigen in
chronic hepatitis B virus infection. Rev Med Virol. 21:315–329.
2011.PubMed/NCBI
|
|
3
|
Diao J, Garces R and Richardson CD: X
protein of hepatitis B virus modulates cytokine and growth factor
related signal transduction pathways during the course of viral
infections and hepatocarcinogenesis. Cytokine Growth Factor Rev.
12:189–205. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang F, Zhou H, Xia X, Sun Q, Wang Y and
Cheng B: Activated Notch signaling is required for hepatitis B
virus X protein to promote proliferation and survival of human
hepatic cells. Cancer Lett. 298:64–73. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang F, Zhou H, Yang Y, Xia X, Sun Q, Luo
J and Cheng B: Hepatitis B virus X protein promotes the growth of
hepatocellular carcinoma by modulation of the Notch signaling
pathway. Oncol Rep. 27:1170–1176. 2012.PubMed/NCBI
|
|
6
|
Luo J, Zhou H, Wang F, Xia X, Sun Q, Wang
R and Cheng B: The hepatitis B virus X protein downregulates NF-κB
signaling pathways through decreasing the Notch signaling pathway
in HBx-transformed L02 cells. Int J Oncol. 42:1636–1643.
2013.PubMed/NCBI
|
|
7
|
Wang F, Xia X, Wang J, Sun Q, Luo J and
Cheng B: Notch1 signaling contributes to the oncogenic effect of
HBx on human hepatic cells. Biotechnol Lett. 35:29–37. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sun Q, Wang R, Wang Y, Luo J, Wang P and
Cheng B: Notch1 is a potential therapeutic target for the treatment
of human hepatitis B virus X protein-associated hepatocellular
carcinoma. Oncol Rep. 31:933–939. 2014.PubMed/NCBI
|
|
9
|
Kopan R and Ilagan MX: The canonical Notch
signaling pathway: unfolding the activation mechanism. Cell.
137:216–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kopan R: Notch signaling. Cold Spring Harb
Perspect Biol. 4:pii: a011213. 2012. View Article : Google Scholar
|
|
11
|
Kalaitzidis D and Armstrong SA: Cancer:
the flipside of Notch. Nature. 473:159–160. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: a little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Giovannini C, Gramantieri L, Chieco P, et
al: Selective ablation of Notch3 in HCC enhances doxorubicin’s
death promoting effect by a p53 dependent mechanism. J Hepatol.
50:969–979. 2009.PubMed/NCBI
|
|
14
|
Villanueva A, Alsinet C, Yanger K, et al:
Notch signaling is activated in human hepatocellular carcinoma and
induces tumor formation in mice. Gastroenterology. 143:1660–1669.
e16672012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qi R, An H, Yu Y, et al: Notch1 signaling
inhibits growth of human hepatocellular carcinoma through induction
of cell cycle arrest and apoptosis. Cancer Res. 63:8323–8329.
2003.PubMed/NCBI
|
|
16
|
Wang C, Qi R, Li N, et al: Notch1
signaling sensitizes tumor necrosis factor-related
apoptosis-inducing ligand-induced apoptosis in human hepatocellular
carcinoma cells by inhibiting Akt/Hdm2- mediated p53 degradation
and up-regulating p53-dependent DR5 expression. J Biol Chem.
284:16183–16190. 2009. View Article : Google Scholar
|
|
17
|
Viatour P, Ehmer U, Saddic LA, et al:
Notch signaling inhibits hepatocellular carcinoma following
inactivation of the RB pathway. J Exp Med. 208:1963–1976. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zimmerman ZF, Moon RT and Chien AJ:
Targeting Wnt pathways in disease. Cold Spring Harb Perspect Biol.
4:pii: a008086. 2012. View Article : Google Scholar
|
|
19
|
Zhou J, Cheng P, Youn JI, Cotter MJ and
Gabrilovich DI: Notch and wingless signaling cooperate in
regulation of dendritic cell differentiation. Immunity. 30:845–859.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Camps J, Pitt JJ, Emons G, et al: Genetic
amplification of the NOTCH modulator LNX2 upregulates the
WNT/beta-catenin pathway in colorectal cancer. Cancer Res.
73:2003–2013. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li C, Zhang Y, Lu Y, Cui Z, Yu M, Zhang S
and Xue X: Evidence of the cross talk between Wnt and Notch
signaling pathways in non-small-cell lung cancer (NSCLC):
Notch3-siRNA weakens the effect of LiCl on the cell cycle of NSCLC
cell lines. J Cancer Res Clin Oncol. 137:771–778. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao C, Zhao M, Song H, Uchida K, Yokoyama
KK and Li T: Identification of the gene for a novel liver-related
putative tumor suppressor at a high-frequency loss of
heterozygosity region of chromosome 8p23 in human hepatocellular
carcinoma. Hepatology. 32:721–727. 2000. View Article : Google Scholar
|
|
23
|
Cheng B, Zheng Y, Guo X, Wang Y and Liu C:
Hepatitis B viral X protein alters the biological features and
expressions of DNA repair enzymes in LO2 cells. Liver Int.
30:319–326. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ueno K, Hirata H, Hinoda Y and Dahiya R:
Frizzled homolog proteins, microRNAs and Wnt signaling in cancer.
Int J Cancer. 132:1731–1740. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Terasaki H, Saitoh T, Shiokawa K and Katoh
M: Frizzled-10, up-regulated in primary colorectal cancer, is a
positive regulator of the WNT - beta-catenin - TCF signaling
pathway. Int J Mol Med. 9:107–112. 2002.PubMed/NCBI
|
|
26
|
Nagayama S, Yamada E, Kohno Y, et al:
Inverse correlation of the up-regulation of FZD10 expression and
the activation of beta-catenin in synchronous colorectal tumors.
Cancer Sci. 100:405–412. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gugger M, White R, Song S, Waser B,
Cescato R, Riviere P and Reubi JC: GPR87 is an overexpressed
G-protein coupled receptor in squamous cell carcinoma of the lung.
Dis Markers. 24:41–50. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Nagayama S, Fukukawa C, Katagiri T, et al:
Therapeutic potential of antibodies against FZD 10, a cell-surface
protein, for synovial sarcomas. Oncogene. 24:6201–6212. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fukukawa C, Hanaoka H, Nagayama S, et al:
Radioimmunotherapy of human synovial sarcoma using a monoclonal
antibody against FZD10. Cancer Sci. 99:432–440. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Merle P, de la Monte S, Kim M, et al:
Functional consequences of frizzled-7 receptor overexpression in
human hepatocellular carcinoma. Gastroenterology. 127:1110–1122.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bengochea A, de Souza MM, Lefrancois L, et
al: Common dysregulation of Wnt/Frizzled receptor elements in human
hepatocellular carcinoma. Br J Cancer. 99:143–150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Merle P, Kim M, Herrmann M, et al:
Oncogenic role of the frizzled-7/beta-catenin pathway in
hepatocellular carcinoma. J Hepatol. 43:854–862. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
King TD, Zhang W, Suto MJ and Li Y:
Frizzled7 as an emerging target for cancer therapy. Cell Signal.
24:846–851. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nambotin SB, Lefrancois L, Sainsily X, et
al: Pharmacological inhibition of Frizzled-7 displays anti-tumor
properties in hepatocellular carcinoma. J Hepatol. 54:288–299.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hsieh A, Kim HS, Lim SO, Yu DY and Jung G:
Hepatitis B viral X protein interacts with tumor suppressor
adenomatous polyposis coli to activate Wnt/beta-catenin signaling.
Cancer Lett. 300:162–172. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cha MY, Kim CM, Park YM and Ryu WS:
Hepatitis B virus X protein is essential for the activation of
Wnt/beta-catenin signaling in hepatoma cells. Hepatology.
39:1683–1693. 2004. View Article : Google Scholar : PubMed/NCBI
|