Introduction
ARD1 was originally identified in yeast as an
N-acetyltransferase that catalyzes N-terminal acetylation of newly
synthesized proteins. In yeast, ARD1 is required for entry into the
stationary phase and sporulation during nitrogen deprivation
(1). Subsequently, mammalian ARD1
was identified and found to catalyze not only N-terminal
acetylation but also lysine acetylation of several proteins
including hypoxiainducible factor-1 α (HIF-1α), β-catenin, myosin
light chain kinase, the androgen receptor, tuberous sclerosis 2
(TSC2) and the tubulin complex (2–7). In
mammalian cells, ARD1 regulates diverse cellular activities
including growth, apoptosis, autophagy and differentiation
(7–12). In particular, ARD1 garnered
attention as a molecule that plays a critical role in cancer
progression (13–15). ARD1 expression is elevated in
various human cancers such as lung, breast, prostate, thyroid, and
colorectal cancer (16–20). Furthermore, depletion of ARD1 leads
to impaired proliferation or induces apoptosis in human cancer
cells (3,21). Thus, emerging evidence suggests
that ARD1 could be a potential target for cancer therapy.
There are several isoforms of ARD1 derived from
alternative splicing of mRNA. Alternative splicing of ARD1 mRNA is
a species-specific event, thus isoform compositions differ between
humans and mice (22). Previously,
we identified three mouse (mARD1198,
mARD1225, mARD1235) and two human
(hARD1131, hARD1235) ARD1 variants (23). Among these, mARD1225,
mARD1235 and hARD1235 have been well
characterized and were found to have different cellular
localizations with distinct roles in tumor angiogenesis (2,22–27).
However, the cellular expression profiles and biological functions
of mARD1198 and hARD1131 remain
unelucidated.
The current study was designed to characterize the
hARD1 variant, hARD1131 and to investigate how its
cellular functions differ from hARD1235, the most common
form of hARD1. Our results demonstrate that the human ARD1 (hARD1)
variants, hARD1131 and hARD1235, have
different subcellular localizations and play distinct roles in the
regulation of cell proliferation.
Materials and methods
Reagents and antibodies
Anti-GFP and cyclin D1 antibodies were purchased
from Santa Cruz Biotechnology (Santa Cruz, CA, USA).
Anti-acetyl-lysine antibody was purchased from Cell Signaling
Technology (Danvers, MA, USA). Anti-tubulin was purchased from
Sigma-Aldrich (St. Louis, MO, USA).
Cell culture
HeLa and 293T cells were maintained in Dulbecco’s
modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) at 37°C and 5% CO2 in a humid
atmosphere.
Plasmid constructions and
transfection
To construct expression vectors for human ARD1
variants, ARD1 cDNA was amplified by PCR and sub-cloned into a
GFP-tagged pCS2+ vector for cell expression, and a pGEX-4T vector
for bacterial induction of the recombinant protein. Transfection
was carried out using Lipofectamine (Life Technology, Carlsbad, CA,
USA) or Polyfect (Qiagen, Valencia, CA, USA), according to the
manufacturer’s instructions.
Immunoblotting and
immunoprecipitation
Cells were harvested and proteins were extracted
using protein lysis buffer (10 mM HEPES at pH 7.9, 40 mM NaCl, 0.1
mM EDTA, 5% glycerol, 1 mM DTT and protease inhibitors). The
concentration of extracted protein was measured using a BCA assay.
Total cell lysates were resolved by SDS-PAGE and transferred onto
nitrocellulose membranes (Amersham Pharmacia Bioscience,
Piscataway, NJ, USA). The membrane was probed with a primary
antibody followed by a secondary antibody conjugated to horseradish
peroxidase, and protein was visualized using the ECL system (Intron
Biotechnology, Gyeonggi-do, Korea).
In vitro acetylation assay
Recombinants of GST-hARD1 variants were freshly
prepared as previously described (21). ARD1 recombinants were incubated in
reaction mixture (50 mM Tris-HCl at pH 8.0, 0.1 mM EDTA, 1 mM DTT,
10% glycerol and 10 mM acetyl-CoA) at 37°C for 1 h.
Immunofluorescence staining and
microscopy
Cells were placed on cover slips then incubated with
Hoechst 33342 (Molecular Probes, Eugene, OR, USA) for nucleus
staining. Axiovert M200 microscopes (Carl Zeiss, Jena, Germany)
were used for immunofluorescence imaging.
Reverse transcription-PCR analysis
Total RNA was extracted using an RNA extraction kit
(Invitrogen, Carlsbad, CA, USA). cDNA was synthesized from 2 μg of
RNA using an oligo(dt) primer. Primers used for PCR reactions were
as follows: human ARD1, 5′-ATGAACATCCGCAATGCGAG-3′ (forward) and
5′-CTCATATCATGGCTCGAGAGG-3′ (reverse); cyclin D1,
5′-CTGGCCATGAACTACCTGGA-3′ (forward) and 5′-GTC
ACACTTGATCACTCTGG-3′ (reverse); GAPDH, 5′-ACCAC AGTCCATGCCATCAC-3′
(forward) and 5′-TCCACCACCCT GTTGCTGTA-3′ (reverse). The PCR
reaction was performed for 25 cycles to allow ARD1, cyclin D1 and
GAPDH amplification.
Cell proliferation assay
The rate of cell proliferation was measured using a
Non-Radioactive Proliferation Assay kit (Promega) according to the
manufacturer’s instructions. Briefly, cells were seeded into
96-well plates and cultured for three days. Subsequently, 20 μl of
substrate solution was added and the cells were incubated for 1 h
to allow color development. The absorbance at 492 nm was measured
to determine the number of proliferating cells.
Statistical analysis
Results are presented as means ± SD and P-values
were calculated by applying the two-tailed Student’s t-test to data
derived from three independent experiments. Differences were
considered statistically significant when P<0.05.
Results
Expression of hARD1 variants
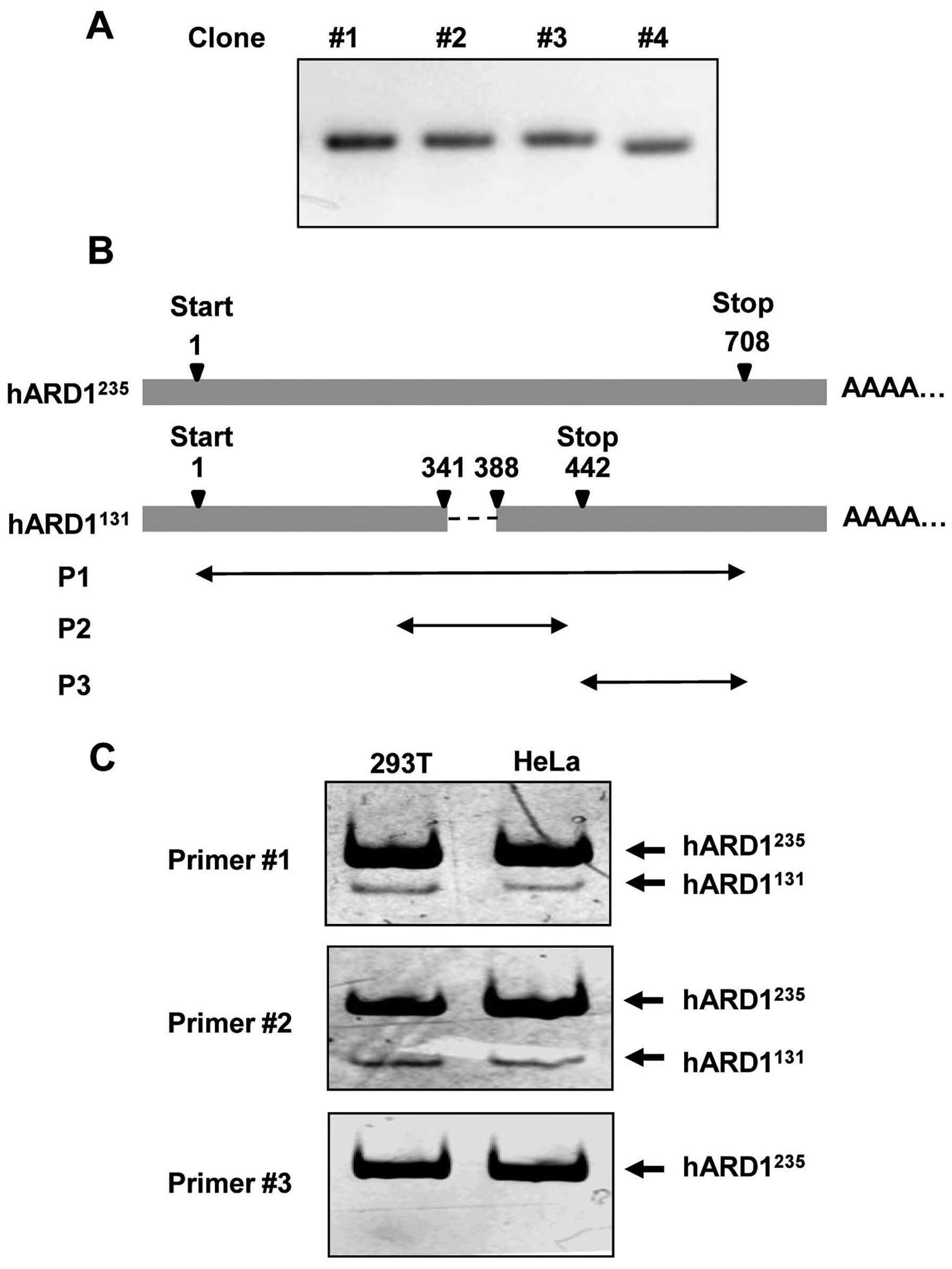
During the cloning of human ARD1 using mRNA prepared
from HeLa cells, we observed that cDNA from one clone (no. 4) was
shorter than cDNA from other clones (Fig. 1A). Sequence analysis revealed that
the clone with the shorter cDNA sequence (clone no. 4) was
hARD1131 (GenBank accession no. BC063377), whereas the
other clones were all hARD1235 (GenBank accession no.
NM_003491). As shown in Fig. 1B,
the nucleotide sequence from residue 342 to 387, which is located
in exon 6 and exon 7 of ARD1 mRNA, was deleted in
hARD1131. Therefore, the nucleotide sequence of
hARD1131 is 46 bp shorter than that of
hARD1235.
To confirm the expression of hARD1131 in
human cells, we performed RT-PCR using mRNA isolated from 293T and
HeLa cells. As described in Table
I and Fig. 1B, three kinds of
primer sets named P1, P2 and P3 were designed for the PCR analysis:
P1 and P2 contained the deleted mRNA region, whereas P3 did not. As
predicted, when P1 and P2 were used in PCR, two bands corresponding
to hARD1131 and hARD1235 were detected by
polyacrylamide gel electrophoresis. However, only one band,
corresponding to hARD1235, was detected following PCR
with the P3 primer set. These data confirm the expression of the
hARD1131 splice variant in human cells (Fig. 1C).
 | Table IDesign of the primer sets for
RT-PCR. |
Table I
Design of the primer sets for
RT-PCR.
| Primer | Sequence
(5′-3′) | Region
(nucleotide) |
|---|
| P1 sense |
ATGAACATCCGCAATGCCAGG | 1–21 |
| P1 antisense |
CTAGGAGGCTGAGTCGGAGGC | 688–708 |
| P2 sense |
AACTTCAATGCCAAATATGTC | 301–321 |
| P2 antisense |
TCATGGCATAGGCGTCCTCCC | 422–442 |
| P3 sense |
AGCGGGACCTCACTCAGATGG | 443–463 |
| P3 antisense |
CTAGGAGGCTGAGTCGGAGGC | 688–708 |
Sequence comparison of hARD1
variants
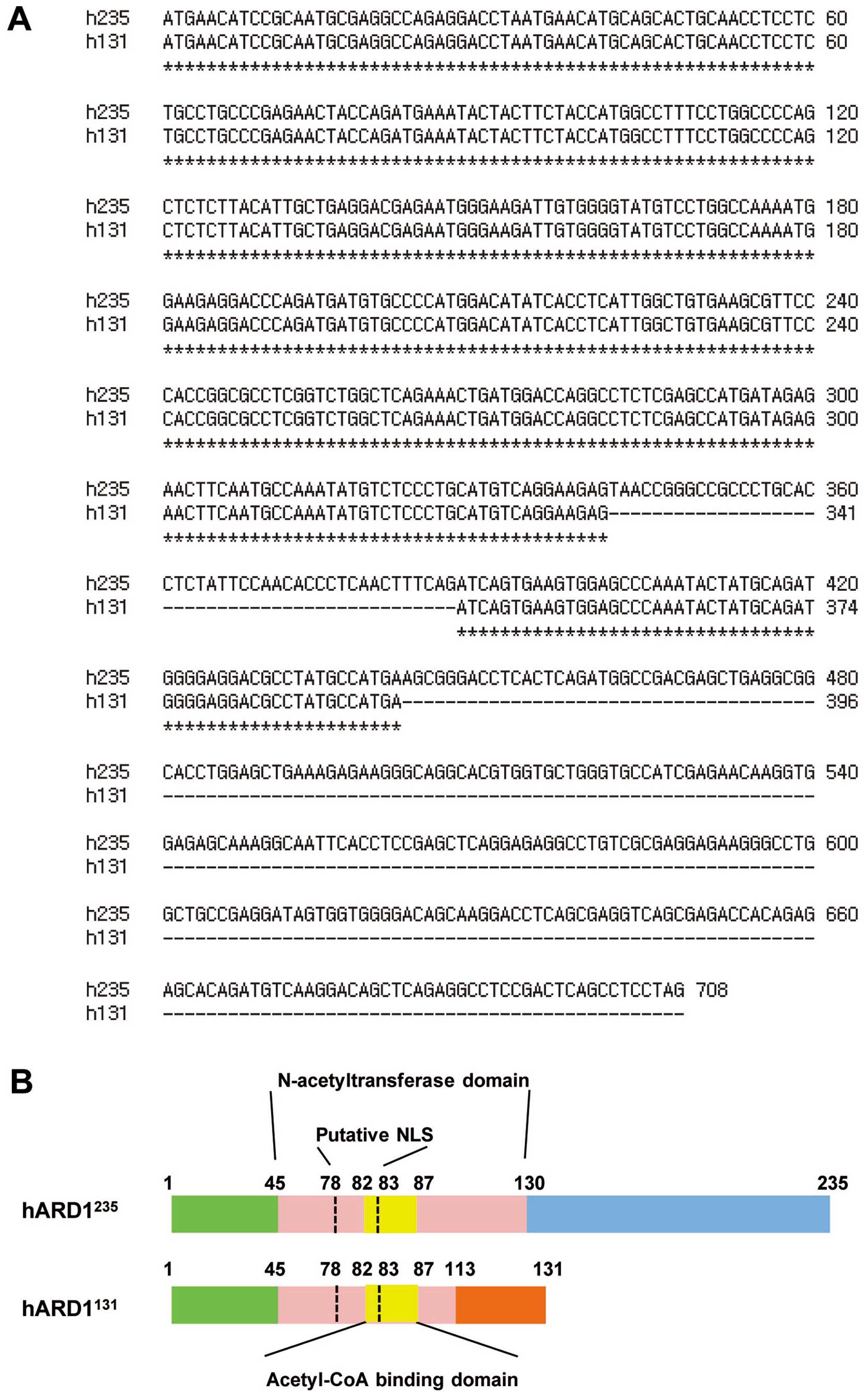
Next, we compared the coding sequences of
hARD1131 and hARD1235. The coding sequence of
hARD1235 terminates at amino acid 708, thus it encodes a
235 amino acid proteins. However, in hARD1131, deletion
of 46 bp in hARD1131 results in a frame shift after
amino acid 113, resulting in premature termination at amino acid
131 (Fig. 2A and B). ARD1 is
predicted to have an acetyltransferase domain located between amino
acid residues 45 and 130, in which an acetyl-CoA binding domain is
positioned between amino acid residues 82 and 87. Compared with
hARD1235, hARD1131 protein possesses a
conserved acetyl-CoA binding domain. However, 20% of the
acetyltransferase domain was deleted in hARD131 and it
was found to have a different C-terminal region (Fig. 2B).
Subcellular localization of hARD1
variants
In a previous study, we reported that splice
variants of mouse ARD1 have a different subcellular localization
(22). Therefore, we speculated
that hARD1131 might have a distinct localization
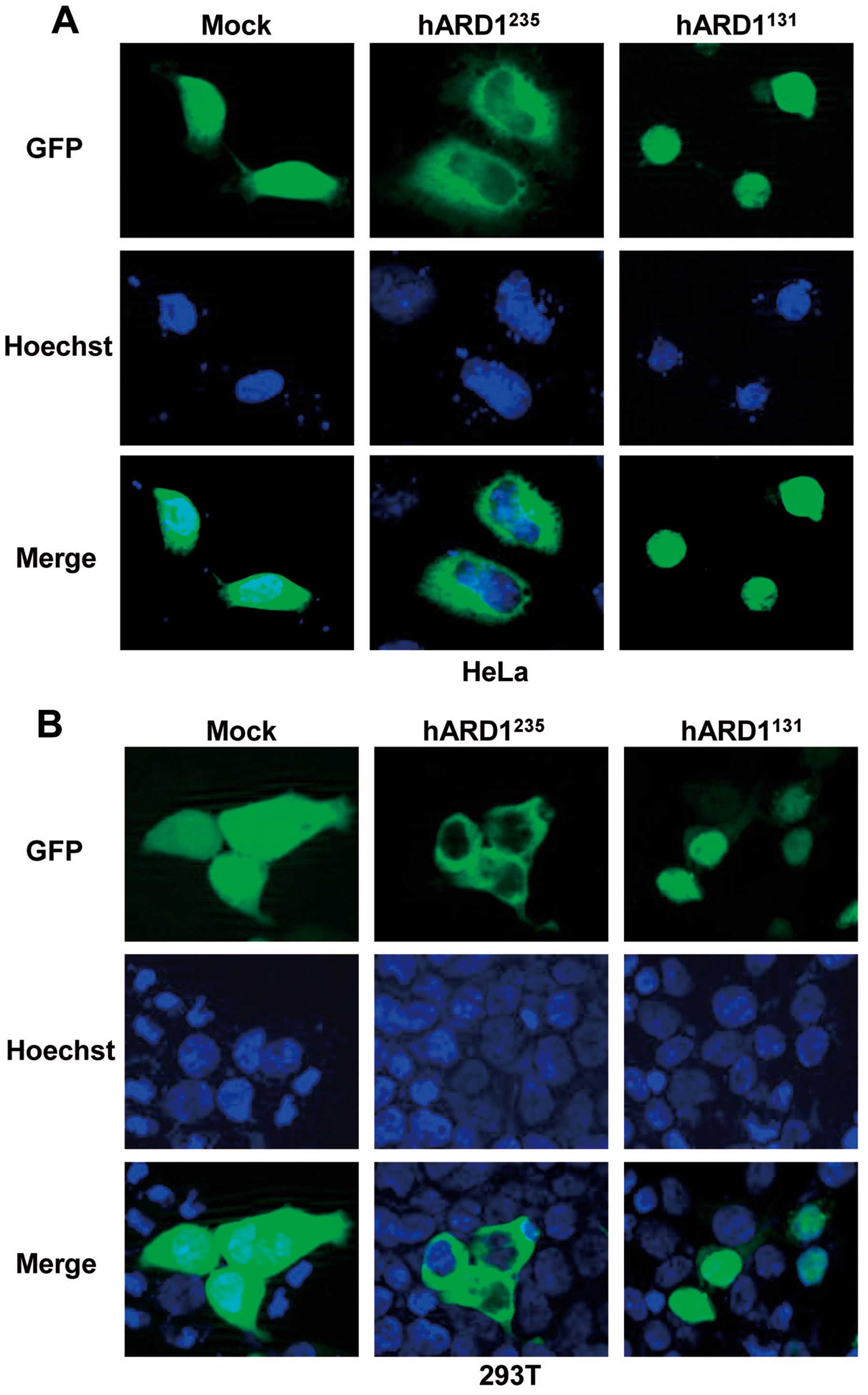
compared to hARD1235. To investigate ARD1 location in
human cells, we constructed GFP-tagged plasmids containing
hARD1131 and hARD1235, which were
subsequently transfected into HeLa and 293T cells. The localization
of GFP-hARD1 variants and control GFP protein was analyzed by
fluorescence microscopy. As shown in Fig. 3, GFP-hARD1235 was
observed predominantly in the cytoplasm, despite hARD1 containing a
putative nuclear localization sequence (NLS) (Fig. 2B). however, hARD1131 was
specifically located in the cell nucleus. These results demonstrate
that hARD1 variants have distinct subcellular localizations, and
suggest that the roles of hARD1131 and
hARD1235 might differ within the cell.
Different functions of hARD1 variants in
the regulation of cell proliferation
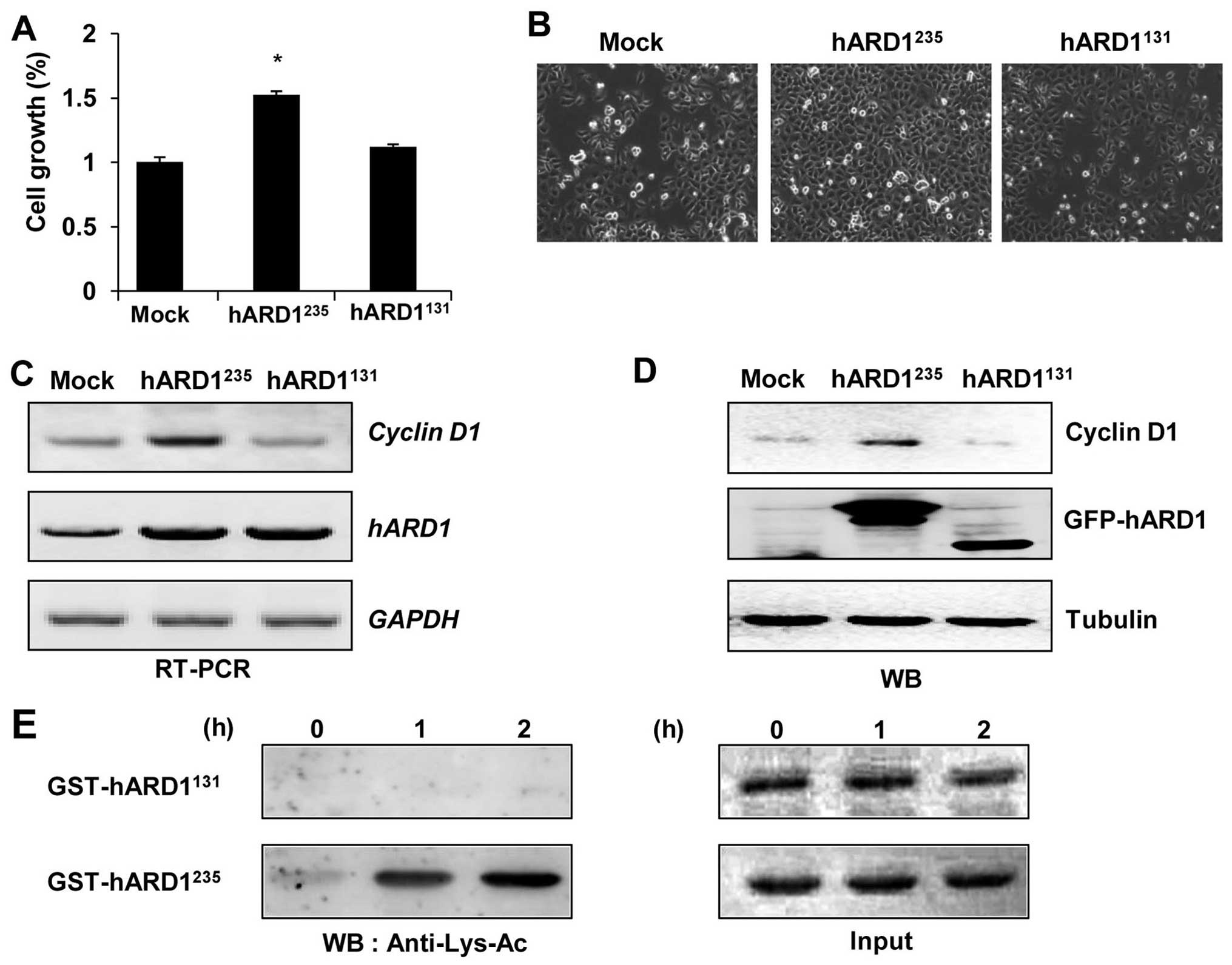
Several studies have reported that
hARD1235 regulates the cell cycle and stimulates cell
proliferation (3,21). Thus, we aimed to determine whether
hARD1131 could promote cellular growth in a similar way
to hARD1235. Consistent with previous studies, compared
to control cells, cell proliferation was significantly accelerated
in hARD1235-expressing HeLa cells. However,
hARD1131 had no effect on cell growth, suggesting that
hARD1 variants have different roles in the regulation of cell
proliferation (Fig. 4A and B). In
a previous study, cyclin D1 was found to mediate ARD1-induced cell
growth, therefore we examined the expression levels of cyclin D1
mRNA and protein in hARD1235 and hARD1235
transfected cells (3,21). Consistent with enhanced cellular
growth, cyclin D1 mRNA and protein expression were significantly
increased in hARD1235, but not hARD1131
transfected cells (Fig. 4C and D).
These results suggest that the diverse roles of hARD1 variants in
the regulation of cell proliferation may be due to their different
effects on cyclin D1 expression.
Previously, we showed that the autoacetylation
activity of hARD1235 was required for enhanced cell
proliferation (21). Thus, we
compared the autoacetylation activities of hARD1131 and
hARD1235 using an in vitro acetylation assay.
While the hARD1235 recombinant acetylated itself,
hARD1131 was not acetylated in vitro, indicating
a lack of autoacetylation activity in this splice variant (Fig. 4E). These results suggest that,
unlike hARD1235, hARD1131 has no
autoacetylation activity, and is therefore unable to upregulate
cyclin D1 expression and promote cell proliferation.
Discussion
Alternative mRNA splicing is a common process in the
regulation of gene expression by which a single gene codes for
multiple proteins. This process contributes to protein diversity,
and different proteins produced from alternative splicing often
have distinct cellular functions (28,29).
The current study characterized alternative splice variants of
human ARD1 and demonstrated differential biological functions and
cellular distributions of these variants.
Previously, we identified two hARD1 variants,
hARD1235 and hARD1131 (23). However, human cells dominantly
express hARD1235, and hARD1131 expression has
not been detected using RT-PCR or western blots in previous studies
(22,23). In this study, the expression of
hARD1131 was clearly detected using RT-PCR and
polyacrylamide gel electrophoresis, which can separate DNA well
beyond the resolving capabilities of an agarose gel (Fig. 1C). The basal expression level of
hARD1131 was relatively lower than that of
hARD1235. Therefore, we could not exclude the
possibility that the cellular activity of hARD1131 might
be small or performed preferentially by hARD1235.
Interestingly, hARD1131 has a unique
subcellular localization compared to hARD1235 (Fig. 3). Although the NLS is conserved at
amino acid residues 78–83 in all ARD1 variants, the different
subcellular localizations of hARD1 variants could be explained by
structural difference in C-terminal regions (Fig. 2).
The subcellular localization of a protein
corresponds to its biological function. As an N-acetyltransferase,
hARD1235 cooperates with NATH-1 for N-terminal
acetylation of newly synthesized proteins in the cytoplasm
(14). However, the nuclear
localization of hARD1131 suggests that it might have
functions other than N-acetylation. Indeed, N-terminal
acetyltransferase activity is associated with cellular growth.
However, hARD1131 had no effect on cellular growth
(Fig. 4A and B). Moreover,
autoacetylation activity, which is essential for the ability of
hARD1235 to stimulate cell proliferation, was also
absent in the hARD1131 variant (Fig. 4E). These results suggest novel
functions of hARD1131 that differ from that of
hARD1235, and suggest the necessity for further
experiments to investigate the diverse cellular functions of
hARD1131.
In summary, the present study revealed that hARD1
variants have different cell proliferative activities that might be
associated with their different subcellular localizations and
enzymatic activities. To further our understanding of hARD1
isoforms, future studies will focus on elucidating the specific
roles played by hARD1 variants and their relationships under
various physiological conditions.
Acknowledgements
This research was supported by the Global Research
Laboratory Program (2011-0021874), Global Core Research Center
(GCRC) Program (2011-0030001), National Research Foundation (NRF)
grant (2013-036038) funded by the Ministry of Science, ICT and
Future Planning (MSIP) and Basic Science Research Program through
the NRF of Korea funded by the Ministry of Education
(2013R1A1A2058956).
References
|
1
|
Driessen HP, de Jong WW, Tesser GI and
Bloemendal H: The mechanism of N-terminal acetylation of proteins.
CRC Crit Rev Biochem. 18:281–325. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jeong JW, Bae MK, Ahn MY, et al:
Regulation and destabilization of HIF-1alpha by ARD1-mediated
acetylation. Cell. 111:709–720. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lim JH, Park JW and Chun YS: Human arrest
defective 1 acetylates and activates beta-catenin, promoting lung
cancer cell proliferation. Cancer Res. 66:10677–10682. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ohkawa N, Sugisaki S, Tokunaga E, et al:
N-acetyltransferase ARD1-NAT1 regulates neuronal dendritic
development. Genes Cells. 13:1171–1183. 2008.PubMed/NCBI
|
|
5
|
Shin DH, Chun YS, Lee KH, Shin HW and Park
JW: Arrest defective-1 controls tumor cell behavior by acetylating
myosin light chain kinase. PLoS One. 4:e74512009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Wang Z, Guo J, et al: Inactivation
of androgen-induced regulator ARD1 inhibits androgen receptor
acetylation and prostate tumorigenesis. Proc Natl Acad Sci USA.
109:3053–3058. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kuo HP, Lee DF, Chen CT, et al: ARD1
stabilization of TSC2 suppresses tumorigenesis through the mTOR
signaling pathway. Sci Signal. 3:ra92010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xu H, Jiang B, Meng L, et al:
N-α-acetyltransferase 10 protein inhibits apoptosis through
RelA/p65-regulated MCL1 expression. Carcinogenesis. 33:1193–1202.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yi CH, Sogah DK, Boyce M, Degterev A,
Christofferson DE and Yuan J: A genome-wide RNAi screen reveals
multiple regulators of caspase activation. J Cell Biol.
179:619–626. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Arnesen T, Gromyko D, Pendino F, Ryningen
A, Varhaug JE and Lillehaug JR: Induction of apoptosis in human
cells by RNAi-mediated knockdown of hARD1 and NATH, components of
the protein N-alpha-acetyltransferase complex. Oncogene.
25:4350–4360. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asaumi M, Iijima K, Sumioka A, et al:
Interaction of N-terminal acetyltransferase with the cytoplasmic
domain of beta-amyloid precursor protein and its effect on A beta
secretion. J Biochem. 137:147–155. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Arnesen T, Anderson D, Baldersheim C,
Lanotte M, Varhaug JE and Lillehaug JR: Identification and
characterization of the human ARD1-NATH protein acetyltransferase
complex. Biochem J. 386:433–443. 2005. View Article : Google Scholar :
|
|
13
|
Kuo HP and Hung MC: Arrest-defective-1
protein (ARD1): tumor suppressor or oncoprotein? Am J Transl Res.
2:56–64. 2010.PubMed/NCBI
|
|
14
|
Kalvik TV and Arnesen T: Protein
N-terminal acetyltransferases in cancer. Oncogene. 32:269–276.
2013. View Article : Google Scholar
|
|
15
|
Arnesen T, Thompson PR, Varhaug JE and
Lillehaug JR: The protein acetyltransferase ARD1: a novel cancer
drug target? Curr Cancer Drug Targets. 8:545–553. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Shim JH, Chung YH, Kim JA, et al: Clinical
implications of arrest-defective protein 1 expression in
hepatocellular carcinoma: a novel predictor of microvascular
invasion. Dig Dis. 30:603–608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang ZH, Gong JL, Yu M, et al:
Up-regulation of human arrest-defective 1 protein is correlated
with metastatic phenotype and poor prognosis in breast cancer.
Asian Pac J Cancer Prev. 12:1973–1977. 2011.
|
|
18
|
Yu M, Gong J, Ma M, et al:
Immunohistochemical analysis of human arrest-defective-1 expressed
in cancers in vivo. Oncol Rep. 21:909–915. 2009.PubMed/NCBI
|
|
19
|
Ren T, Jiang B, Jin G, et al: Generation
of novel monoclonal antibodies and their application for detecting
ARD1 expression in colorectal cancer. Cancer Lett. 264:83–92. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Arnesen T, Gromyko D, Horvli O, Fluge O,
Lillehaug J and Varhaug JE: Expression of N-acetyl transferase
human and human arrest defective 1 proteins in thyroid neoplasms.
Thyroid. 15:1131–1136. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seo JH, Cha JH, Park JH, et al: Arrest
defective 1 autoacetylation is a critical step in its ability to
stimulate cancer cell proliferation. Cancer Res. 70:4422–4432.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chun KH, Cho SJ, Choi JS, Kim SH, Kim KW
and Lee SK: Differential regulation of splicing, localization and
stability of mammalian ARD1235 and ARD1225 isoforms. Biochem
Biophys Res Commun. 353:18–25. 2007. View Article : Google Scholar
|
|
23
|
Kim SH, Park JA, Kim JH, et al:
Characterization of ARD1 variants in mammalian cells. Biochem
Biophys Res Commun. 340:422–427. 2006. View Article : Google Scholar
|
|
24
|
Arnesen T, Kong X, Evjenth R, et al:
Interaction between HIF-1 alpha (ODD) and hARD1 does not induce
acetylation and destabilization of HIF-1 alpha. FEBS Lett.
579:6428–6432. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bilton R, Mazure N, Trottier E, et al:
Arrest-defective-1 protein, an acetyltransferase, does not alter
stability of hypoxia-inducible factor (HIF)-1alpha and is not
induced by hypoxia or HIF. J Biol Chem. 280:31132–31140. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fisher TS, Etages SD, Hayes L, Crimin K
and Li B: Analysis of ARD1 function in hypoxia response using
retroviral RNA interference. J Biol Chem. 280:17749–17757. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee MN, Lee SN, Kim SH, et al: Roles of
arrest-defective protein 1(225) and hypoxia-inducible factor 1alpha
in tumor growth and metastasis. J Natl Cancer Inst. 102:426–442.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brett D, Pospisil H, Valcarcel J, Reich J
and Bork P: Alternative splicing and genome complexity. Nat Genet.
30:29–30. 2002. View
Article : Google Scholar
|
|
29
|
Nilsen TW and Graveley BR: Expansion of
the eukaryotic proteome by alternative splicing. Nature.
463:457–463. 2010. View Article : Google Scholar : PubMed/NCBI
|