Introduction
Multiple myeloma (MM) is a refractory hematopoietic
malignancy showing clonal plasma cell accumulation. A major
breakthrough in MM treatment has been the introduction of the
first-in-class proteasome inhibitor bortezomib (BZ) (1,2).
Moreover, the treatment of relapsed and refractory MM is now
possible with carfilzomib, a second-generation proteasome
inhibitor, and immunomodulatory agents. This has offered new
alternatives for vulnerable patients (3,4).
However, patients with relapsed and refractory MM are still urgent
issues and require treatment combinations (5,6).
Increasing lines of evidence indicate that proteasome inhibition
induces misfolded protein accumulation in the endoplasmic reticulum
(ER) (7,8). This evokes ER stress followed by the
unfolded protein response (UPR) (9,10).
UPR mainly functions: i) to decrease protein entry into the ER by
suppressing translational rate; and ii) to increase the folding
capacity of the ER via chaperon protein translational activations.
Following incorrect folding in the ER, proteins are
retro-translocated for degradation in the cytoplasm via the
ubiquitin (Ub)-proteasome pathway [i.e., ER-associated degradation
(ERAD)]. A failure in all the adaptation strategies triggers
apoptosis and induces C/EBP homologous protein (CHOP) (GADD153), a
pro-apoptotic transcription factor and other pathways (7–10).
Since MM is characterized by the uncontrolled cell growth of
monoclonal antibody (mAb)-producing plasma cells, production of
large quantities of unfolded or misfolded immunoglobulin triggers
ER stress. Thus, therapeutic manipulation of the UPR pathway
appears to disrupt cellular mechanisms for processing high protein
loads and cellular stress, and further leads to death of MM
cells.
Macroautophagy (hereafter, ‘autophagy’) occurs when
cellular proteins and organelles (e.g., ER) are enveloped in an
autophagosome and degraded in lysosomes by lysosomal hydrolases
(11,12). Although autophagy is considered a
bulk non-selective degradation of long-lived proteins and
organelles, recent reports revealed the selective degradation
pathway of ubiquitinated protein via autophagy using docking
proteins (e.g., p62) and related proteins (e.g., NBR1), having both
a microtubule-associated protein 1 light chain 3 (LC3)-interacting
region and a Ub-associated domain (13,14).
Thus, overflowed ubiquitinated proteins are bound to p62 and
subsequently engulfed into an autophagosome via the LC3-interacting
region in p62. This indicates that autophagy also acts as a
compensatory degradation system when the proteasome system is
impaired (14). We previously
reported that the inhibition of autophagy using the autophagy
inhibitor bafilomycin A1 (BAF) enhanced BZ-induced
apoptosis by burdening ER stress in MM cell lines (15). We also reported that macrolide
antibiotics [e.g., clarithromycin (CAM) and azithromycin (AZM)]
attenuated or blocked autophagy flux, possibly mediated through the
inhibition of the lysosomal function, and that ER stress loading
was enhanced by both the BZ-induced inhibition of the Ub-proteasome
system and the CAM- or AZM-induced inhibition of the
autophagy-lysosome system. This is followed by CHOP transcriptional
activation and the induction of apoptosis in MM and breast cancer
cells (16,17). Therefore, concomitant blocking of
proteasome and autophagy appears to be a promising combination
therapy.
Moreover, misfolded/unfolded proteins are
sequestrated into aggregates and transported. They are then
discarded from the cytoplasm by dynein motors through the
microtubule network to the aggresome (18). The class II histone deacetylase 6
(HDAC6), which is a microtubule-associated deacetylase and an
aggresome component, has the capacity to bind both
polyubiquitinated misfolded proteins and dynein motors (19). By acting as an adaptor between
ubiquitinated protein aggregates and dynein, HDAC6 enables
aggregated protein loading onto the dynein motor protein complex
(20). Thus, in the formation of
aggresome at the microtubule-organizing center (MTOC), HDAC6,
dynein, and polyubiquitinated proteins functionally interact with
each other (19,20). If HDAC6 is lacking, cells will not
be able to clear cytoplasmic unfolded protein aggregates; cells
cannot appropriately form aggresomes; and cells become
hypersensitive to unfolded protein accumulation (19). Therefore, HDAC6 appears to be
another critical factor in the management of unfolded
protein-induced stress at the cellular level. Moreover, some parts
of the aggresome are reported to be degraded via the
autophagy-lysosome system (21,22).
A recent study also revealed that p62 regulates the accumulation
and autophagic clearance of protein aggregates by directly binding
to HDAC6, and this interaction appears to regulate HDAC6 activity
(23).
All these lines of evidence suggest the integrated
intracellular networks of proteasome, autophagy, and aggresome for
unfolded protein processing. ER stress loading may be further
enhanced by targeting both intracellular proteolytic pathways and
aggresome formation (24). To
prove this hypothesis aimed for clinical application, we
intentionally attempted to use well-approved drugs such as CAM, BZ,
and vorinostat [suberoylanilide hydroxamic acid (SAHA)], an orally
bioavailable inhibitor of HDAC with a half maximal inhibitory
concentration (IC50) of 37 nM and which has passed the
assessment of the Food and Drug Administration for cutaneous T-cell
lymphoma treatment (25,26). In the present investigation, we
clearly demonstrated that the SAHA/CAM/BZ combination treatment
induced marked ER stress-mediated MM cell death. This provides a
promising treatment for MM patients in the form of ‘ER
stress-loading therapy’.
Materials and methods
Reagents
BZ was purchased from Selleck Chemicals (Houston,
TX, USA). CAM was purchased from Tokyo Chemical Industry Co., Ltd.
(Tokyo, Japan), SAHA was from Cayman Chemical Co. (Ann Arbor, MI,
USA), and tubacin was from Sigma-Aldrich (St. Louis, MO, USA). BZ,
SAHA and tubacin were dissolved in dimethyl sulfoxide to make stock
solutions at concentrations of 1, 10 and 1 mM, respectively. CAM
was dissolved in ethanol to prepare stock solutions of 5 mg/ml.
Cell lines and culture conditions
For this study, the MM cell lines IM-9 and
RPMI-8226, and the lung carcinoma cell line H226 were obtained from
the American Type Culture Collection (ATCC) (Manassas, VA, USA).
The human MM cell line KMS-12-PE was obtained from the Japanese
Collection of Research Bioresources (JCRB) (Osaka, Japan). A
CHOP−/− murine embryonic fibroblast (MEF) cell line
(CHOP-KO-DR) established from a 13.5-day-old CHOP−/−
mouse embryo by SV-40 immortalization and a CHOP+/+ MEF
cell line (DR-wild-type) established by SV-40 immortalization as a
control cell line for CHOP-KO-DR were also obtained from the ATCC.
These authorized cell lines were expanded and frozen in aliquots
within 1 month after obtaining from the cell banks. Each aliquot
was thawed and the cells were used for the experiments within 2
months after thawing. IM-9, H226, RPMI-8226, and KMS-12-PE cells
were cultured in RPMI-1640 medium (Sigma-Aldrich) supplemented with
10% fetal bovine serum (FBS) (Biowest SAS, Nuaillé, France), 2 mM
L-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml)
(Wako Pure Chemicals Industries, Tokyo, Japan). CHOP-KO-DR and
DR-wild-type cells were maintained in Dulbecco’s modified Eagle’s
medium (Sigma-Aldrich) supplemented with 10% FBS, 2 mM L-glutamine,
penicillin (100 U/ml), and streptomycin (100 μg/ml). All cell lines
were cultured in a humidified incubator containing 5%
CO2 and 95% air at 37°C.
Assessment of viable number of cells
The number of viable cells was assessed using
CellTiter-Blue Cell Viability Assay kit (Promega Corp., Madison,
WI, USA) according to the manufacturer’s instructions as previously
described in detail (16).
Immunoblotting
Immunoblotting was performed as previously described
(15,16). In brief, cells were lysed with RIPA
lysis buffer supplemented with a protease and phosphatase inhibitor
cocktail (both from Nacalai Tesque, Kyoto, Japan). Cellular
proteins were quantified using a DC Protein Assay kit (Bio-Rad,
Hercules, CA, USA). Equal amounts of proteins were loaded onto the
gels, separated by sodium dodecyl sulfate polyacrylamide gel
electrophoresis (SDS-PAGE), and transferred onto Immobilon-P
membrane (Millipore, Billerica, MA, USA). The membranes were probed
with primary antibodies (Abs) such as anti-acetylated-α-tubulin
(6–11B-1) mAb, anti-α-tubulin (B-7) mAb, anti-GAPDH (6C5) mAb,
anti-HDAC6 (H-300) Ab, and anti-Ub (P4D1) mAb, which were all
purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA)
and anti-PARP Ab (Cell Signaling Technology, Inc., Danvers, MA,
USA). Immunoreactive proteins were detected with horseradish
peroxidase-conjugated secondary Abs (Cell Signaling Technology,
Inc.) and an enhanced chemiluminescence reagent (Millipore).
Densitometry was performed using a Molecular Imager ChemiDoc XRS
System (Bio-Rad).
RNA interference
For the gene silencing of HDAC6 in RPMI-8226 and
H226 cells, HDAC6 siRNA and control siRNA were purchased from Life
Technologies (Grand Island, NY, USA) and whose sequences are
described as follows: HDAC6 sense, CCAGCACAGUCUUAUGGAUGCUAU and
antisense, AUAGCCAUCCAUAAGACUGUGCUGG. siRNAs were diluted to a
final concentration of 33 nM in Opti-MEM I (Life Technologies).
Transfection was performed with the cells at 40% confluency using
Lipofectamine RNAiMAX transfection reagent (Life Technologies)
according to the manufacturer’s instructions. Knockdown efficiency
was assessed by immunoblotting.
Gene expression analysis
Total RNA was isolated from cell pellets using
Isogen (Wako Pure Chemicals Industries) and genomic DNA was removed
using RQ1 RNase-Free DNase (Promega Corp.) at 37°C for 30 min,
followed by extraction with phenol chloroform and ethanol
precipitation. Reverse-transcription using a PrimeScript RT Master
Mix (Takara Bio, Inc., Shiga, Japan) was performed according to the
manufacturer’s instructions. Real-time polymerase chain reaction
(PCR) was performed on 3 ng of cDNA using validated SYBR-Green gene
expression assays for human ER stress-related genes (CHOP,
BAX, BIM, DR5, GADD34 and GRP78)
in combination with SYBR Premix Ex Taq II (Takara Bio, Inc.). The
sequences of primers and reaction conditions were previously
described (16). Quantitative
real-time PCR was performed in duplicates in a Thermal Cycler Dice
Real-Time System TP800 (Takara Bio, Inc.). The data were analyzed
using Thermal Cycler Dice Real-Time System Software version 5.00
(Takara Bio, Inc.), and the comparative Ct method
(2−ΔΔCt) was used for the relative quantification of
gene expression. The data of real-time PCR products were
standardized to GAPDH as an internal control. To confirm the
specific amplification of target genes, each gene product was
further separated by using 1.5% agarose gel after real-time PCR to
detect a single band at the theoretical product size, as well as by
analysis of the dissociation curve for detecting a single peak.
Immunocytochemistry and confocal
microscopy
Cells were spread on slide glasses using Cytospin 4
Centrifuge (Thermo Fisher Scientific, Inc., Rockford, IL, USA) to
make slide glass preparations. Cells were fixed for 20 min in
ice-cold methanol and permeabilized with 0.1% Triton X-100 for 20
min, followed by blocking with 2% bovine serum albumin in TBST (25
mM Tris, 137 mM NaCl, 2.7 mM KCl, 0.05% Tween-20, pH 7.4) for 1 h.
Cells were immunostained with primary Abs such as mouse
anti-vimentin (V9) mAb, mouse anti-Ub mAb, and lysosomal-associated
membrane protein-1 (LAMP-1) (H4A3) mAbs (all from Santa Cruz
Biotechnology, Inc.). The secondary Abs used for fluorescence
detection were Alexa Fluor® 488 F(ab′)2 fragment of goat
anti-mouse IgG (H+L) Ab (Life Technologies). Nuclei were stained
with 4′,6-diamidino-2-phenylindole (DAPI) (Wako Pure Chemicals
Industries). Slides were mounted with SlowFade Gold antifade
reagent (Life Technologies). Analysis by confocal microscopy was
performed using the confocal laser scanning fluorescence microscope
FV10i-DOC.(Olympus Corp., Tokyo, Japan).
Assessment of aggresome by fractionation
of detergent-soluble and -insoluble proteins
Cells were lysed with Triton X-100 lysis buffer (10
mM Tris-HCl, 150 mM NaCl, 2% Triton X-100, pH 7.8) supplemented
with a protease inhibitor cocktail (Nacalai Tesque). The lysates
were centrifuged at 12,000 g for 30 min at 4°C. The supernatant was
then collected as the soluble fraction. The pellets (which contain
the insoluble protein) were then resuspended in sodium dodecyl
sulfate (SDS) lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 2% SDS, pH
7.8) and sonicated for 30 sec with a tip sonicator VP-5S (Taitec,
Saitama, Japan) to prepare the insoluble fraction. Equal volumes of
each pellet and supernatants were boiled for 5 min in SDS-PAGE
sample buffer (125 mM Tris-HCl, 4% SDS, 20% glycerol, 0.002% BPB,
pH 6.8) and analyzed by SDS-PAGE.
Electron microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1 M
phosphate buffer (pH 7.4) for 1 h. The samples were further fixed
in 1% osmium tetroxide for 1 h, dehydrated in graded ethanol
(30–100%), and embedded in Quetol 812 epoxy resin (Nisshin EM Co.,
Ltd., Tokyo, Japan). Ultrathin sections were cut with an Ultracut J
microtome (Reichert-Jung, Vienna, Austria). The sections were
stained with lead nitrate and uranium acetate, and subjected to
electron microscopic analysis using the scanning electron
microscope JEM-1200 EXII (JEOL, Tokyo, Japan).
Statistical analysis
All data are expressed as mean ± SD. Statistical
analysis was performed using Mann-Whitney U test (two-tailed).
Results
SAHA, BZ, and CAM combination treatment
potently enhanced MM cell apoptosis
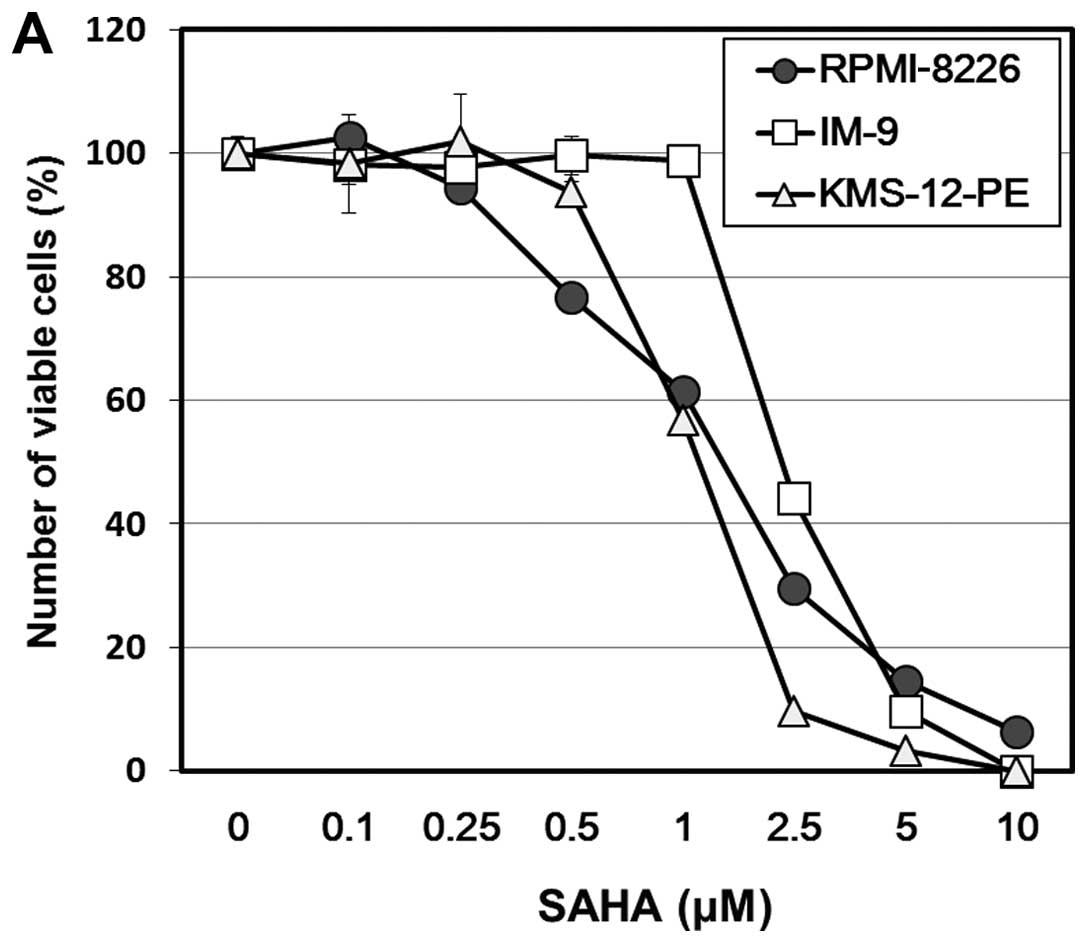
Treatment with SAHA for 48 h resulted in a
dose-dependent inhibition of cellular growth in all MM cell lines
(Fig. 1A). The IC50 was
1.2 μM in KMS-12-PE, 1.5 μM in RPMI-8226, and 2.1 μM in IM-9 cells.
SAHA-treated MM cells exhibited apoptotic morphologic features such
as fragmentation of the nucleus and formation of an apoptotic body
along with cleavage of PARP and caspase-3 (data not shown). The
regulation of the stability and function of a microtubule has been
associated with α-tubulin reversible acetylation, and HDAC6
functions as α-tubulin deacetylase (27). Immunoblotting using a specific Ab
for the acetylated α-tubulin revealed that, in response to SAHA (at
0.5 μM for KMS-12-PE and RPMI-8226, at 1 μM for IM-9), the
acetylation of α-tubulin was detectable within 16 h and persisted
for at least 48 h in all three cell lines tested (Fig. 1B). In our previous studies,
autophagy flux was shown to be blocked by CAM, and that the BZ and
CAM combination treatment resulted in ER-stress overloading
followed by enhanced induction of apoptosis in MM and breast cancer
cells (16,17). Since HDAC6 has been implicated in
aggresome formation that results in sequestering overabundant
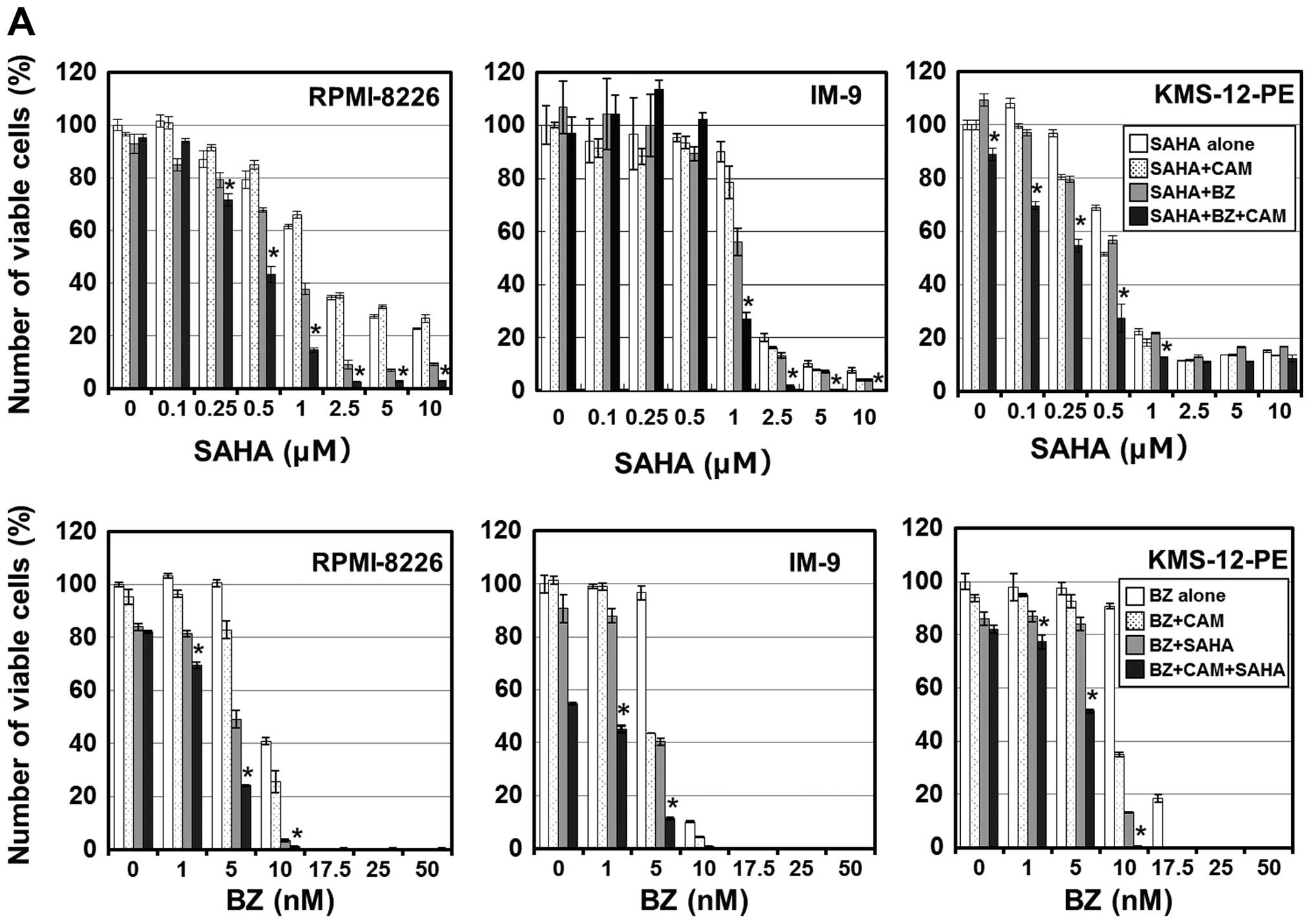
intracellular unfolded proteins (18), we examined whether the SAHA plus BZ
and/or CAM combination treatment further increased cytotoxicity via
ER stress loading. As shown in Fig.
2A, the combined treatment with SAHA/BZ, but not with SAHA/CAM,
potentiated cell growth inhibition. Notably, although CAM treatment
alone at 50 μg/ml did not inhibit the growth of cells, the combined
SAHA/BZ/CAM treatment showed a clearly pronounced cytotoxicity
compared with the SAHA/BZ or BZ/CAM treatment in all MM cell lines.
This enhanced cytotoxicity was mediated through apoptosis induction
since the pronounced expression of the cleaved PARP was observed in
response to the suboptimal concentrations of the SAHA/BZ/CAM
combination in IM-9 cells (Fig.
2B).
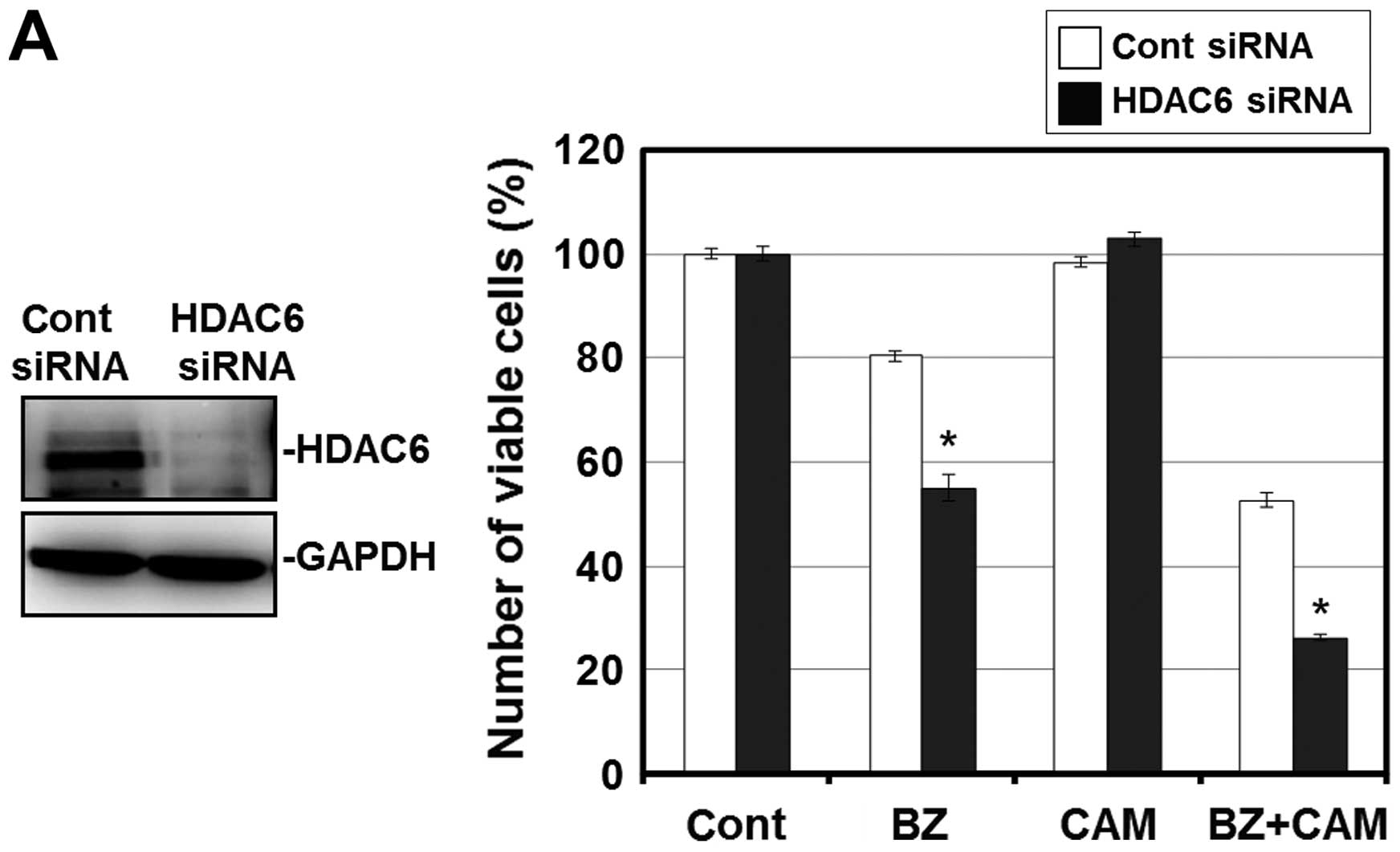
To confirm that this enhanced cytotoxicity is
mediated through HDAC6 inhibition, we next attempted to knock down
HDAC6 with siRNA. Pre-treatment with HDAC6 siRNA effectively
suppressed the expression level of HDAC6 in RPMI-8226 cells. Under
this condition, the cell growth inhibition by the BZ/CAM treatment
was pronounced compared with the cells pre-treated with control
siRNA. In addition, tubacin, a specific HDAC6 inhibitor, reproduced
the pronounced cytotoxicity when combined with BZ and BZ/CAM
(Fig. 3A and B) (28). These results suggest that HDAC6
inhibition appears to be involved in the enhancement of
cytotoxicity induced by the addition of SAHA to BZ/CAM-containing
cell culture medium.
SAHA suppressed the aggresome formation
induced by simultaneous inhibition of proteasome and autophagy in
MM cells
To further confirm that the pronounced cytotoxicity
by SAHA shown in Fig. 2A was due
to the inhibition of HDAC6 activity involved in aggresome
formation, we assessed aggresome formation in the presence or
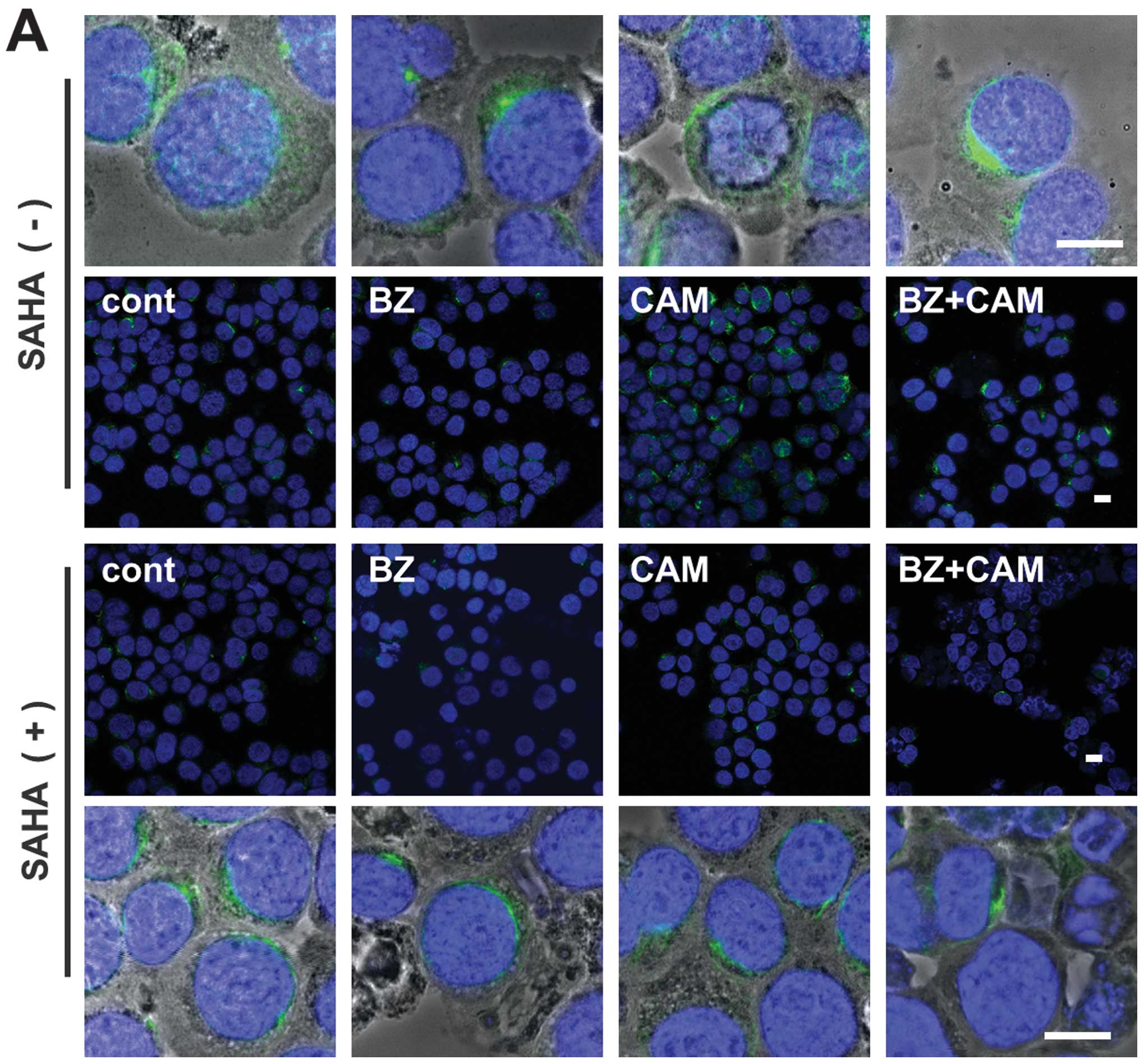
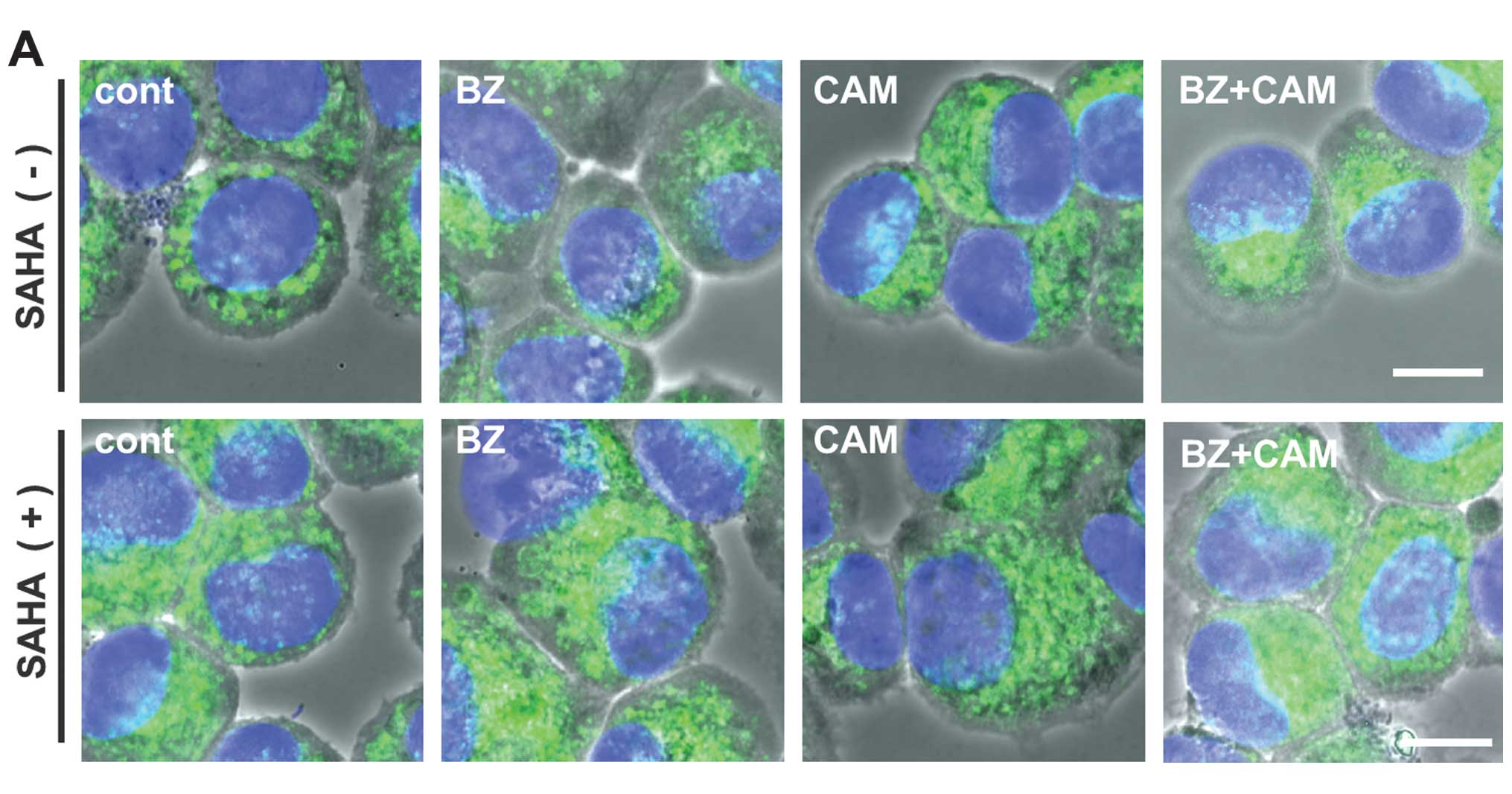
absence of SAHA. In RPMI-8226 cells, immunocytochemistry using
anti-vimentin mAb showed that the combined treatment with BZ/CAM
induced a dense deposit of vimentin in the perinuclear region,
which is a well-known marker for aggresome (29). This vimentin deposit was clearly
suppressed in the presence of SAHA (Fig. 4A). However, since the MM cell lines
including RPMI-8226 were very sensitive to the SAHA/BZ/CAM
treatment, in which apoptosis was induced within 18 h of exposure
to these reagents (data not shown), precise analysis of aggresome
formation was practically difficult using MM cells. We therefore
used the lung squamous carcinoma cell line H226 which is less
sensitive, but shows pronounced cytotoxicity in response to the
SAHA/BZ/CAM combination similarly to MM cell lines (Fig. 4B). As shown in Fig. 4C, the prominent vimentin deposit in
the perinuclear region was observed after combined treatment with
BZ/CAM for 24 h. The vimentin deposit with high fluorescent
intensity was evidently suppressed in the presence of SAHA,
indicating that SAHA inhibits BZ/CAM-induced aggresome formation
(Fig. 4C). Similarly to SAHA,
treatment with HDAC6 siRNA suppressed the vimentin condensation in
the perinuclear region in H226 cells (Fig. 4D–F).
In addition, immunocytochemistry using anti-Ub Ab
exhibited the clustering of Ub-positive dots in the perinuclear
region by BZ/CAM, whereas they were dispersed in the presence of
SAHA in H226 cells (data not shown). As an alternative assessment
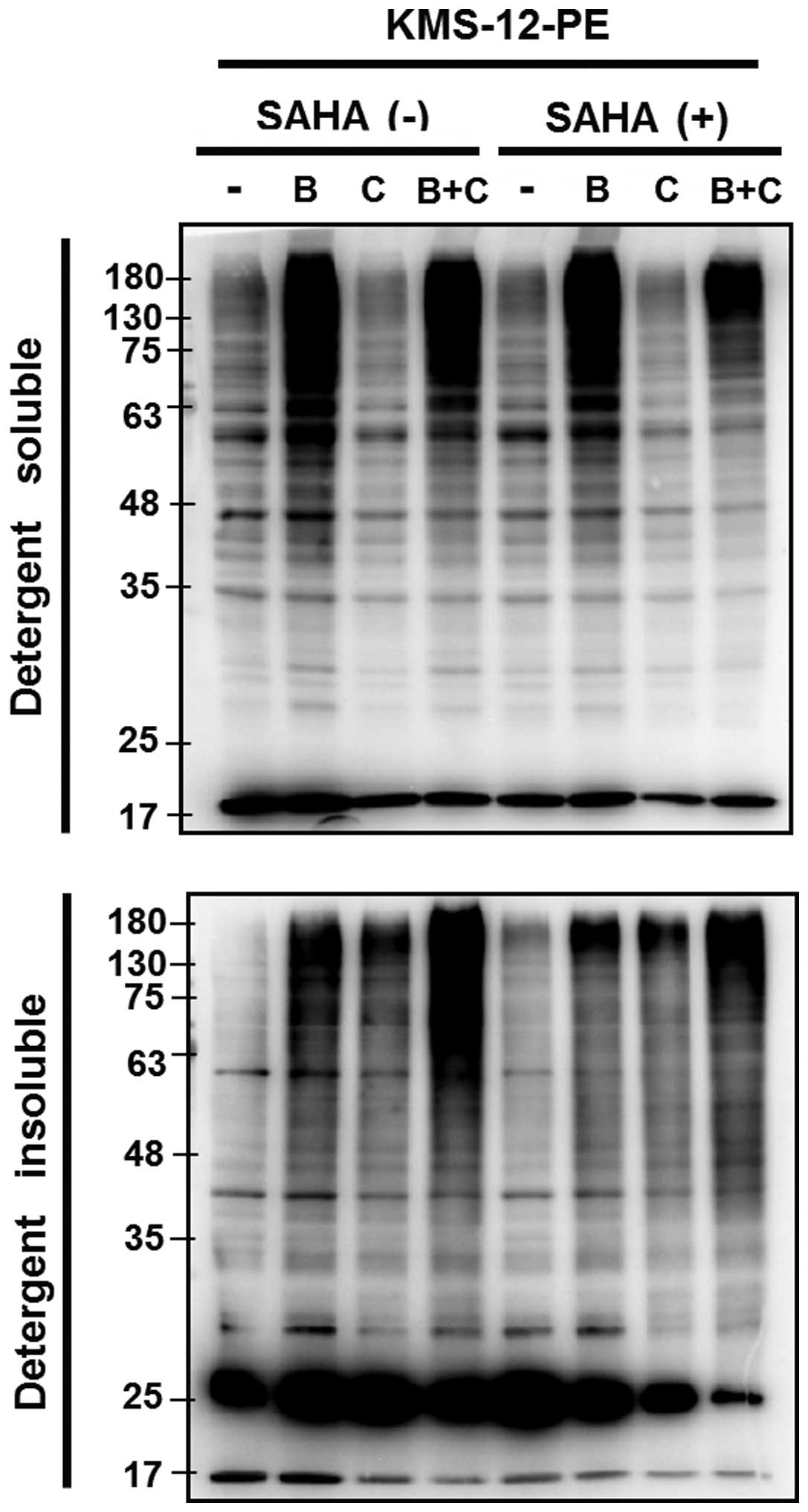
of aggresome, we treated KMS-12-PE with BZ, CAM, or BZ/CAM in the
presence or absence of SAHA. The cells were then treated with lysis
buffer containing 2% Triton X-100, and cellular proteins were
subsequently fractionated into the detergent-soluble and the
-insoluble fraction. The detergent-insoluble fraction (cell
pellets) was sonicated for 30 sec in the presence of 2% SDS and
boiled for 5 min. Thereafter, the samples were loaded on the gels,
separated by 11.25% SDS-PAGE, and immunoblotted with anti-Ub Ab.
The samples derived from the detergent-soluble fractions were
processed similarly. As shown in Fig.
5, the increased intracellular polyubiquitinated proteins were
detectable after treatment with BZ and BZ/CAM, but not with CAM or
SAHA in the detergent-soluble fraction (upper panel). In the
detergent-insoluble fraction (lower panel) at higher molecular
weights, prominent polyubiquitinated proteins were detected as
aggresome contents in the cells treated with BZ/CAM. Treatment with
BZ alone also induced accumulation of the detergent-insoluble
polyubiquitinated proteins but to a lesser extent than that induced
by treatment with BZ/CAM. It was noteworthy that the increased
ubiquitinated proteins in the detergent-insoluble fraction were
suppressed in the presence of SAHA. This also indicates the
inhibition of aggresome in response to SAHA.
Immunocytochemistry using anti-LAMP-1 mAb showed
lysosome accumulation in the perinuclear region along with
aggresome formation after BZ/CAM treatment. As is the case of
vimentin-positive aggresome formation, the clustering of
LAMP-1-positive dots was suppressed in the presence of SAHA
(Fig. 6A). We therefore performed
electron microscopy for precise analysis of the cytosolic area
strongly stained with anti-vimentin Ab. However, contrary to what
we had expected, electron microscopy exhibited no typical features
of the aggresome structure such as massive accumulation of closely
packed electron-dense particles surrounded by a cage of bundles of
intermediated filaments in the region of the centrosome, which was
first described by Johnston et al in 1998 (30). Instead, there was the clustering of
a number of lysosomes, autophagosomes, autolysosomes, and
mitochondria with increased filaments in response to the BZ/CAM
treatment (Fig. 6B). A few numbers
of unstructured particles with a rather high density, which might
be the aggresome precursor, were detectable in this area (Fig. 6C). It was noteworthy that many
lysosomal fusions or lysosomal aggregates in various sizes were
observed. These might represent the phenomenon termed ‘lysophagy’
which has recently been reported by others, that is, the lysosome
engulfed into an autophagosome or autolysosome (Fig. 6D) (31). In the presence of SAHA, the
clustering of these organelles in the perinuclear region was
suppressed to some extent along with the decreased filamentous
structures. However, an increased number of lysosomes including
autolysosomes was still observed compared with the cells treated
with either CAM or BZ alone (Fig.
6A).
Concomitant aggresome, proteasome, and
autophagy targeting enhanced ER stress loading
Since aggresome is reported to form as an adaption
process against overabundant intracellular unfolded or misfolded
proteins, we hypothesized that HDAC6 inhibition with SAHA further
enhances BZ/CAM-induced ER stress loading to the cells. As we
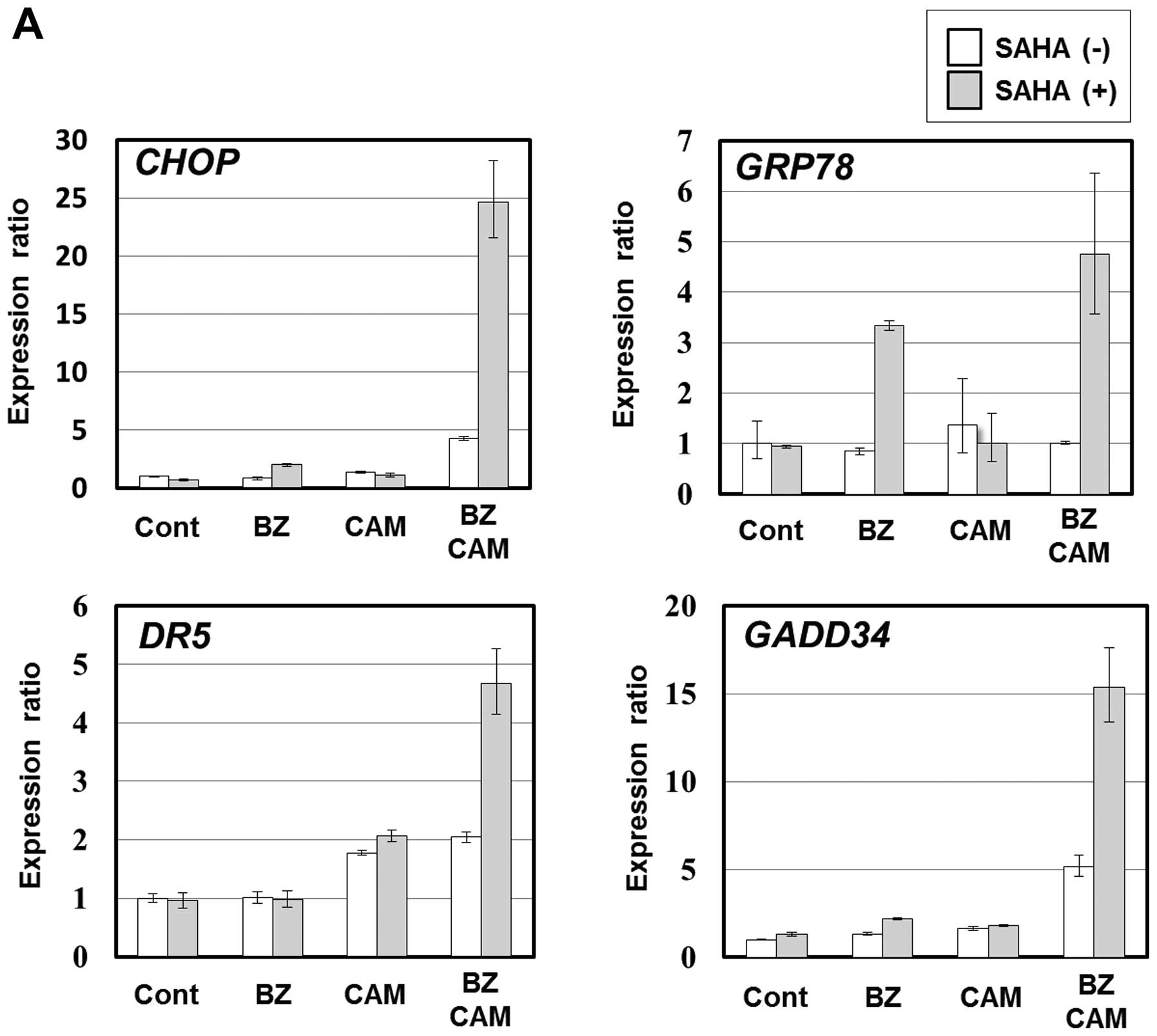
expected, the expression levels of ER stress-related genes
including CHOP were markedly pronounced in the presence of
SAHA; the expression ratios of CHOP to untreated control cells
increased to 5-fold by BZ/CAM treatment, and further increased to
25-fold by BZ/CAM/SAHA treatment in IM-9 cells (Fig. 7A). In H226 cells, similarly to
SAHA, HDAC6 siRNA treatment enhanced BZ/CAM-induced CHOP expression
compared with the cells treated with control siRNA (Fig. 7B). CHOP is an ER stress-related
pro-apoptotic transcription factor which upregulates pro-apoptotic
genes (e.g., BIM, BAX, DR5) and one of the
critical molecules playing a role in the induction of apoptosis in
response to ER stress (9,10). In addition, enhanced cytotoxicity
was shown by a wild-type MEF cell line along with CHOP upregulation
by the combination of the three reagents, whereas the
CHOP−/− MEF cell line derived from a
CHOP-deficient mouse almost completely canceled this
pronounced cytotoxicity (Fig. 7C).
Therefore, the pronounced cytotoxicity and apoptosis in response to
the SAHA/BZ/CAM combination treatment shown in Fig. 2A is strongly suggested to be due to
the upregulation of CHOP in response to ER stress loading.
Discussion
We showed that ER stress-mediated apoptosis in MM
cells was potently enhanced by the simultaneous targeting of
aggresome formation, proteasome, and the autophagy-lysosome system.
Concomitant SAHA, BZ, and CAM treatment markedly enhanced CHOP
induction via ER stress loading in MM cells (Fig. 7A), whereas a CHOP−/− MEF
cell line nearly completely cancelled the pronounced cytotoxicity
in response to the SAHA/BZ/CAM combination treatment (Fig. 7C). These results strongly suggest
that CHOP upregulation appears to be involved, at least in part, in
the pronounced cytotoxicity shown in Fig. 2A. Our recent report also showed
that combined treatment with SAHA/BZ/CAM exhibited potent
cytotoxicity along with upregulation of CHOP in breast cancer cell
lines (24). Since this enhanced
cytotoxicity by ER stress loading is associated with inhibition of
the BZ/CAM-induced aggresome formation (Figs. 4 and 5), induction of aggresome itself appears
to function as cytoprotective against intracellular overloading of
unfolded proteins as previously demonstrated in various
neurodegenerative disorders (32,33).
In this study, we intentionally attempted to use the
clinically approved drugs for blocking aggresome formation and the
proteolytic pathways. Although exhibiting a potent inhibitory
effect on HDAC6, SAHA (vorinostat) is a pan-HDAC inhibitor;
therefore, the inhibitory effects on other HDACs cannot be
completely excluded for the pronounced ER stress loading and
apoptosis induction shown in Figs.
2A and 7A. However, tubacin,
which is a specific inhibitor of HDAC6, also exhibited pronounced
cytotoxicity (Fig. 3B), and
knockdown of HDAC6 superimposed the effects of SAHA, such as
suppression of vimentin-positive aggresome formation induced by
BZ/CAM treatment (Fig. 4C and E),
and further enhanced the ER stress-related gene upregulation along
with the enhanced BZ/CAM treatment cytotoxicity (Figs. 3A and 7B). Therefore, HDAC6 appears to be a
pivotal target for efficient ER stress loading under
proteasome/autophagy inhibition. From the standpoint of ER stress
loading, intracellular active protein synthesis appears to be
theoretically an important factor for determining sensitivity. MM
cell sensitivity to BZ/SAHA-induced cell death is regulated by Myc
(34). Intracellular ER content,
protein synthesis rates, percentage of aggresome-positive cells,
and sensitivity to BZ/SAHA-induced cell death directly correlated
with Myc expression. We previously reported that cycloheximide
suppressed BZ/CAM-induced ER stress loading and cytotoxicity in MM
cells (16). Thus, the potent
effects shown in this study should depend in part on de novo
protein synthesis. This may explain some differences among the MM
cell lines, H226 cells, and MEF cell lines in terms of cytotoxicity
and ER stress loading in response to these reagents shown in this
study.
Along with vimentin-positive aggresome formation in
the perinuclear region, Fig. 6
shows a number of autophagosomes and autolysosomes after BZ/CAM
treatment. Electron microscopy demonstrated the clustering of
autolysosomes in this area. These findings are in agreement with a
recent study demonstrating that proteasome inhibition causes the
formation of a zone around the centrosome where microtubular
transport of lysosomes is suppressed, resulting in lysosome
entrapment and accumulation (35).
Interestingly, the authors of the previous study reported that the
microtubule-dependent transport of other organelles, including
autophagosomes, mitochondria and endosomes, is also blocked in this
entrapment zone. Therefore, the perinuclear region stained with
anti-vimentin Ab, where autolysosomes clustered along with the
increased number of mitochondria, may in part overlap to ‘the
entrapment zone’ (Figs. 4E and
6B) (35). It is also suggested that, upon
proteasome inhibition, the targeting of aggregated proteins to
aggresome is coordinated with lysosome positioning around the body
to facilitate degradation via autophagy. However, our data showed
that lysosomal clustering including autolysosome further became
prominent by the combined treatment with CAM/BZ compared with that
by the BZ treatment alone (Fig.
4F). Indeed, the addition of SAHA in the CAM/BZ culture medium
suppressed lysosomal clustering in the perinuclear region; however,
an increased number of cytosolic autolysosomes were still
detectable (data not shown). This was possibly because of the
suppression of lysosomal clearance upon autophagy inhibition by
CAM. We and others have reported that macrolide antibiotics inhibit
autophagy flux, although the precise molecular mechanism still
remains to be clarified (16,17,36).
The macrolide antibiotic BAF, which is a well-used autophagy
inhibitor for in vitro experiments, was initially evaluated
for its selective inhibition of a proton-pumping V-ATPase (37). BAF induces the disruption of
vesicular proton gradients and raises the pH of acidic vesicles at
nanomolar concentrations. This disruption of vesicular
acidification in response to BAF appears to prevent autophagosome
fusion with lysosomes, resulting in the inhibition of autophagy. It
was also reported that treatment with AZM increased lysosomal pH in
macrophages, which may lead to the inhibition of lysosomal
hydrolases having an optimal low pH for their enzymatic activities
(38). In this context, if CAM
exerts the same effect on lysosomes, accumulation of autolysosomes
by the suppression of lysosomal clearance itself appears to make
‘lysophagy’ apparent under proteasome inhibition. A similar
phenomenon regarding the selective sequestration of damaged
lysosome by autophagy was previously reported, in which under the
condition of lysosomal damage induced by silica, monosodium urate,
and a lysomotropic reagent, autophagy loss results in the in
vitro inhibition of lysosomal biogenesis and the in vivo
deterioration of acute kidney injury (39). Identification of the target
molecules of macrolide antibiotics involved in autophagy flux
inhibition, as well as the molecular mechanism of lysophagy
including the recognition process of impaired lysosomes appears to
be important issues that need to be clarified.
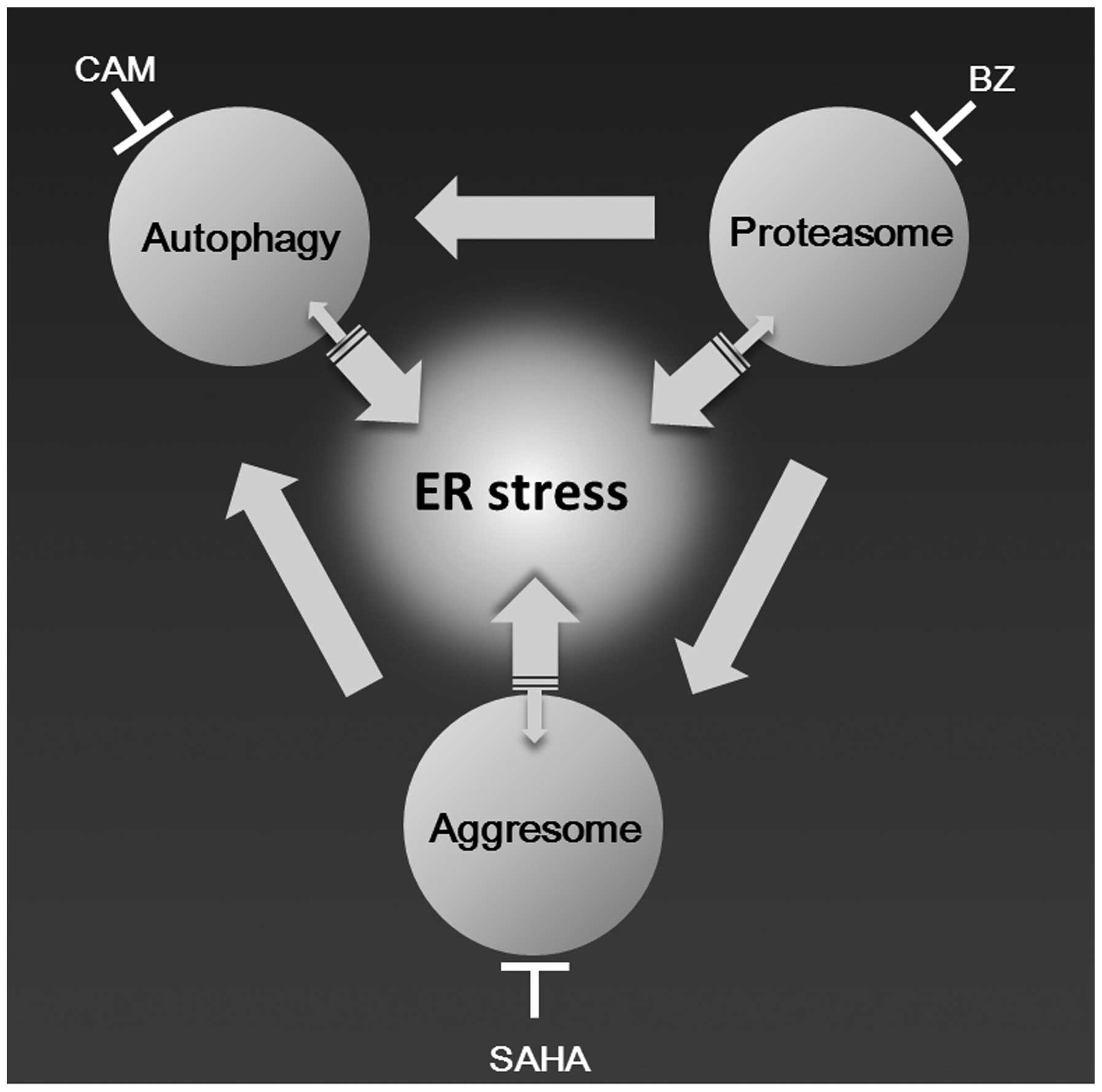
Taking our data and accumulating lines of evidence
together, we can draw an integrated network scheme among aggresome,
proteasome, and autophagy as shown in Fig. 8 (9,10,13,14).
Upon inhibition of proteasome and autophagy, overabundant
ubiquitinated protein aggregates are transported to MTOC to form
aggresomes. Under this condition, further inhibition of aggresome
formation most efficiently induces ER stress loading in cells with
a high protein synthesis rate such as MM cells. Thus, by combining
clinically available drugs such as vorinostat, BZ, and CAM, this
systematic strategy for blocking the processing of intracellular
unfolded proteins appears to be applicable to refractory/relapsed
MM patients.
Acknowledgements
The authors are indebted to Dr Edward F. Barroga,
Associate Professor and Senior Medical Editor of the Department of
International Medical Communications of Tokyo Medical University
for the editorial review of the report. This study was supported by
funds of a Grant-in-Aid for Scientific Research (C) (no. 26460478)
and MEXT-Supported Program for the Strategic Research Foundation at
Private Universities (S1411011, 2014–2018) from the Ministry of
Education, Culture, Sports, Science and Technology of Japan, a
Grant-in-Aid from Tokyo Medical University Cancer Research, and a
Grant-in-Aid from Taisho Toyama Pharmaceutical Co., Ltd. (Tokyo,
Japan) to K.M.; and a Grant-in-Aid for Young Scientist (B) from the
Ministry of Education, Culture, Sports, Science and Technology of
Japan (no. 25860398) and a Research Grant from Tokyo Medical
University to S.M.
References
|
1
|
Richardson PG, Barlogie B, Berenson J, et
al: A phase 2 study of bortezomib in relapsed, refractory myeloma.
N Engl J Med. 348:2609–2617. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
San Miguel JF, Schlag R, Khuageva NK, et
al: Bortezomib plus melphalan and prednisone for initial treatment
of multiple myeloma. N Engl J Med. 359:906–917. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Richardson PG, Xie W, Jagannath S, et al:
A phase 2 trial of lenalidomide, bortezomib, and dexamethasone in
patients with relapsed and relapsed/refractory myeloma. Blood.
123:1461–1469. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel DS, Martin T, Wang M, et al: A
phase 2 study of single-agent carfilzomib (PX-171–003-A1) in
patients with relapsed and refractory multiple myeloma. Blood.
120:2817–2825. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dimopoulos M, Siegel DS, Lonial S, et al:
Vorinostat or placebo in combination with bortezomib in patients
with multiple myeloma (VANTAGE 088): a multicentre, randomised,
double-blind study. Lancet Oncol. 14:1129–1140. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Orlowski RZ: Novel agents for multiple
myeloma to overcome resistance in phase III clinical trials. Semin
Oncol. 40:634–651. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Martinon F: Targeting endoplasmic
reticulum signaling pathways in cancer. Acta Oncol. 51:822–830.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hetz C, Chevet E and Harding HP: Targeting
the unfolded protein response in disease. Nat Rev Drug Discov.
12:703–719. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tabas I and Ron D: Integrating the
mechanisms of apoptosis induced by endoplasmic reticulum stress.
Nat Cell Biol. 13:184–190. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Verfaillie T, Salazar M, Velasco G and
Agostinis P: Linking ER stress to autophagy: potential implications
for cancer therapy. Int J Cell Biol. 2010:9305092010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mizushima N, Levine B, Cuervo AM and
Klionsky DJ: Autophagy fights disease through cellular
self-digestion. Nature. 451:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mizushima N: Autophagy in protein and
organelle turnover. Cold Spring Harb Symp Quant Biol. 76:397–402.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kirkin V, McEwan DG, Novak I and Dikic I:
A role for ubiquitin in selective autophagy. Mol Cell. 34:259–269.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Korolchuk VI, Menzies FM and Rubinsztein
DC: Mechanisms of cross-talk between the ubiquitin-proteasome and
autophagy-lysosome systems. FEBS Lett. 584:1393–1398. 2010.
View Article : Google Scholar
|
|
15
|
Kawaguchi T, Miyazawa K, Moriya S, Ohtomo
T, Che XF, Naito M, Itoh M and Tomoda A: Combined treatment with
bortezomib plus bafilomycin A1 enhances the cytocidal
effect and induces endoplasmic reticulum stress in U266 myeloma
cells: Crosstalk among proteasome, autophagy-lysosome and ER
stress. Int J Oncol. 38:643–654. 2011.
|
|
16
|
Moriya S, Che XF, Komatsu S, Abe A,
Kawaguchi T, Gotoh A, Inazu M, Tomoda A and Miyazawa K: Macrolide
antibiotics block autophagy flux and sensitize to bortezomib via
endoplasmic reticulum stress-mediated CHOP induction in myeloma
cells. Int J Oncol. 42:1541–1550. 2013.PubMed/NCBI
|
|
17
|
Komatsu S, Miyazawa K, Moriya S, Takase A,
Naito M, Inazu M, Kohno N, Itoh M and Tomoda A: Clarithromycin
enhances bortezomib-induced cytotoxicity via endoplasmic reticulum
stress-mediated CHOP (GADD153) induction and autophagy in breast
cancer cells. Int J Oncol. 40:1029–1039. 2012.
|
|
18
|
Simms-Waldrip T, Rodriguez-Gonzalez A, Lin
T, Ikeda AK, Fu C and Sakamoto KM: The aggresome pathway as a
target for therapy in hematologic malignancies. Mol Genet Metab.
94:283–286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kawaguchi Y, Kovacs JJ, McLaurin A, Vance
JM, Ito A and Yao TP: The deacetylase HDAC6 regulates aggresome
formation and cell viability in response to misfolded protein
stress. Cell. 115:727–738. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ouyang H, Ali YO, Ravichandran M, Dong A,
Qiu W, MacKenzie F, Dhe-Paganon S, Arrowsmith CH and Zhai RG:
Protein aggregates are recruited to aggresome by histone
deacetylase 6 via unanchored ubiquitin C termini. J Biol Chem.
287:2317–2327. 2012. View Article : Google Scholar :
|
|
21
|
Lee JY, Koga H, Kawaguchi Y, et al: HDAC6
controls autophagosome maturation essential for ubiquitin-selective
quality-control autophagy. EMBO J. 29:969–980. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Fusco C, Micale L, Egorov M, et al: The
E3-ubiquitin ligase TRIM50 interacts with HDAC6 and p62, and
promotes the sequestration and clearance of ubiquitinated proteins
into the aggresome. PLoS One. 7:e404402012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan J, Seibenhener ML, Calderilla-Barbosa
L, Diaz-Meco MT, Moscat J, Jiang J, Wooten MW and Wooten MC:
SQSTM1/p62 interacts with HDAC6 and regulates deacetylase activity.
PLoS One. 8:e760162013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Komatsu S, Moriya S, Che XF, Yokoyama T,
Kohno N and Miyazawa K: Combined treatment with SAHA, bortezomib,
and clarithromycin for concomitant targeting of aggresome formation
and intracellular proteolytic pathways enhances ER stress-mediated
cell death in breast cancer cells. Biochem Biophys Res Commun.
437:41–47. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Olsen EA, Kim YH, Kuzel TM, et al: Phase
IIb multicenter trial of vorinostat in patients with persistent,
progressive, or treatment refractory cutaneous T-cell lymphoma. J
Clin Oncol. 25:3109–3115. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kavanaugh SM, White LA and Kolesar JM:
Vorinostat: a novel therapy for the treatment of cutaneous T-cell
lymphoma. Am J Health Syst Pharm. 67:793–797. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hubbert C, Guardiola A, Shao R, Kawaguchi
Y, Ito A, Nixon A, Yoshida M, Wang XF and Yao TP: HDAC6 is a
microtubule-associated deacetylase. Nature. 417:455–458. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Haggarty SJ, Koeller KM, Wong JC,
Grozinger CM and Schreiber SL: Domain-selective small-molecule
inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin
deacetylation. Proc Natl Acad Sci USA. 100:4389–4394. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Garcia-Mata R, Bebök Z, Sorscher EJ and
Sztul ES: Characterization and dynamics of aggresome formation by a
cytosolic GFP-chimera. J Cell Biol. 146:1239–1254. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Johnston JA, Ward CL and Kopito RR:
Aggresomes: a cellular response to misfolded proteins. J Cell Biol.
143:1883–1898. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hung YH, Chen LM, Yang JY and Yang WY:
Spatiotemporally controlled induction of autophagy-mediated
lysosome turnover. Nat Commun. 4:21112013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Richter-Landsberg C and Leyk J: Inclusion
body formation, macroautophagy, and the role of HDAC6 in
neurodegeneration. Acta Neuropathol. 126:793–807. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hol EM, Fischer DF, Ovaa H and Scheper W:
Ubiquitin proteasome system as a pharmacological target in
neurodegeneration. Expert Rev Neurother. 6:1337–1347. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nawrocki ST, Carew JS, Maclean KH, Courage
JF, Huang P, Houghton JA, Cleveland JL, Giles FJ and McConkey DJ:
Myc regulates aggresome formation, the induction of Noxa, and
apoptosis in response to the combination of bortezomib and SAHA.
Blood. 112:2917–2926. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zaarur N, Meriin AB, Bejarano E, Xu X,
Gabai VL, Cuervo AM and Sherman MY: Proteasome failure promotes
positioning of lysosomes around the aggresome via local block of
microtubule-dependent transport. Mol Cell Biol. 34:1336–1348. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Nakamura M, Kikukawa Y, Takeya M, Mitsuya
H and Hata H: Clarithromycin attenuates autophagy in myeloma cells.
Int J Oncol. 37:815–820. 2010.PubMed/NCBI
|
|
37
|
Yamamoto A, Tagawa Y, Yoshimori T,
Moriyama Y, Masaki R and Tashiro Y: Bafilomycin A1
prevents maturation of autophagic vacuoles by inhibiting fusion
between autophagosomes and lysosomes in rat hepatoma cell line,
H-4-II-E cells. Cell Struct Funct. 23:33–42. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Renna M, Schaffner C, Brown K, et al:
Azithromycin blocks autophagy and may predispose cystic fibrosis
patients to mycobacterial infection. J Clin Invest. 121:3554–3563.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Maejima I, Takahashi A, Omori H, et al:
Autophagy sequesters damaged lysosomes to control lysosomal
biogenesis and kidney injury. EMBO J. 32:2336–2347. 2013.
View Article : Google Scholar : PubMed/NCBI
|