Introduction
Breast cancer is the most commonly diagnosed cancer
in women, and the second leading cause of cancer deaths in the
developed world. Although many advanced treatments have emerged
following improvement in clinical instruments and methods,
metastasis still leads to cancer mortality and poor prognosis
(1). Almost 30% of early breast
cancers eventually develop recurrence and metastasis (2). As a result, research and development
of treatment targeting breast cancer are of great importance.
miRNAs are endogenous, noncoding small RNAs with
20–25 nucleotides in length (3).
They play an important regulatory role through complimentary
binding of the 3′ untranslated regions (UTRs) of target genes thus
resulting in the degradation of the target mRNA and inhibition of
translation (4). Since the initial
discovery of miRNAs in 1993 (5),
they have been shown to affect multiple cellular processes
(6), and in particular, have been
shown to play significant roles in cancer development and
progression (7,8). Abnormal patterns of miRNA expression
have been observed in various cancer types, including breast
cancers (6,9–11),
colon (12), lymphomas and
leukemias (13), head and neck
(14) and hepatocellular carcinoma
(15). Functionally, abnormal
miRNA expression can affect tumor cell proliferation (16), apoptosis (8), development of metastases (17), invasion (16,17),
chemo- and radiation-sensitivity (18). Previous studies have indicated that
miRNAs can be useful for cancer diagnosis and therapy (19). Let-7 was the first identified miRNA
and its downregulation has a prognostic impact on the survival of
surgically treated lung cancer patients (20). Let-7a expression increases after
differentiation and in mature tissue, but is nearly undetectable in
the embryonic stage (21).
Although let-7 family members can all function as tumor suppressors
(22–24), let-7a is the one that is the most
reported to downregulate c-myc (24–26).
In this study, we investigated function of let-7a in
human breast cancer. First, total RNAs of breast cancer cells
(MDA-MB-231, MCF-7), breast cancer tissues and corresponding
adjacent normal tissues were extracted and used to detect let-7a
expression by qRT-PCR. Secondly, the effects of let-7a on
proliferation, colony formation, migration and invasion of breast
cancer cells were assessed by in vitro cell culture
experiments, which further clarified the role of let-7a in breast
cancer development. Finally, western blotting demonstrated that
let-7a negatively regulated HMGA1 protein expression which in turn
contributed to tumor formation.
Materials and methods
Breast cancer tissues and normal
tissues
Documented informed consent was obtained from all
subjects and the Ethics Committee of Jiangsu University approved
all aspects of the study. Breast cancer tissues and corresponding
adjacent normal tissues were obtained at the Department of Surgery,
the Second People’s Hospital of Kunshan, China. Both tumor tissues
and corresponding adjacent normal tissues were histologically
confirmed. The tissues obtained were immediately stored at
−80°C.
Cell culture
The human breast cancer cell lines MCF-7, and
MDA-MB-231 were provided from Nanjing University and cultured in
Dulbecco’s modified Eagle’s medium with low glucose (L-DMEM, Gibco)
supplemented with 10% fetal bovine serum (FBS, ExCell Biology,
China) at 37°C in humidified 5% CO2.
miRNA transfection
Let-7a mimics or mimics negative control (mimics NC)
and let-7a inhibitor or inhibitor negative control (inhibitor NC)
were synthesized and purified by GenePharma (Shanghai, China). All
of the sequences are listed in Table
I. Transfection was performed with lipofectamine 2000
(Invitrogen). Let-7a mimics and let-7a inhibitors were used at a
concentration of 100 nM. The same concentrations of mimics and
inhibitor negative controls were used.
 | Table IThe sequence of let-7a mimics, let-7a
inhibitor and negative control. |
Table I
The sequence of let-7a mimics, let-7a
inhibitor and negative control.
| Name | Sequence |
|---|
| Hsa-let-7a
mimic |
| Sense |
5′-UGAGGUAGUAGGUUGUAUAGUU-3′ |
| Anti-sense |
5′-CUAUACAACCUACUACCUCAUU-3′ |
| Mimic-negative
control |
| Sense |
5′-UUCUCCGAACGUGUCACGUTT-3′ |
| Anti-sense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| Hsa-let-7a
inhibitor |
5′-AACUAUACAACCUACUACCUCA-3′ |
| Inhibitor negative
control |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
RNA isolation and qRT-PCR
The total RNA of tissues was isolated with the
TRIzol reagent (Invitrogen) according to the manufacturer’s
instructions. The RNA was also isolated with the TRIzol reagent
from cell lines which were transfected with let-7a mimic and
negative control or let-7a inhibitor and inhibitor negative control
for 24 h. For measurement of let-7a RNA expression, qRT-PCR was
performed using a SYBR green-containing PCR kit (GenePharma). For
the detection of HMGA1 mRNA, cDNA was synthesized from 1 μg of
total RNA using the reverse reaction kit in accordance with the
manufacturer’s instructions (Thermo). After that, qRT-PCR was
performed using UltraSYBR Mixture (with ROX) Assay kits (CWBio,
China) according to the manufacturer’s instructions. The CFX-96
real-time fluorescence thermal cycler (Bio-Rad) was used for
quantitative miRNA and mRNA detection. The relative expression
levels of miRNA and mRNA were normalized to the expression of U6
snRNA and β-actin mRNA, respectively. The expression of each gene
was quantified by measuring cycle threshold (Ct) values and
normalized using the 2−ΔCt or 2−ΔΔCt Ct
method relative to U6 snRNA or β-actin mRNA. All the primer
sequences are listed in Table
II.
 | Table IISpecific primers for target and
control genes. |
Table II
Specific primers for target and
control genes.
| Name | Sequence |
|---|
| Let-7a primer | F:
5′-CGATTCAGTGAGGTAGTAGGTTGT-3′
R: 5′-TATGGTTGTTCTGCTCTCTGTCTC-3′ |
| U6snRNA primer | F:
5′-ATTGGAACGATACAGAGAAGATT-3′
R: 5′-GGAACGCTTCACGAATTTG-3′ |
| HMGA1 primer | F:
5′-CAGCGAAGTGCCAACACCTA-3′
R: 5′-AGGAAGCTGCTCCTCCAGTG-3′ |
| β-actin primer | F:
5′-TGGCACCCAGCACAATGAA-3′
R: 5′-CTAAGTCATAGTCCGCCTAGAAGCA-3′ |
Cell proliferation assay
Cell proliferation was measured 3 days after
transfection with let-7a mimic and negative control or let-7a
inhibitor and inhibitor negative control by Thiazolyl Blue
Tetrazolium Bromide (MTT, Amresco). The results are represented as
proliferating cells quantified at OD 490 nm by FLx800 Fluorescence
Microplate Reader (BioTek). The experiment was performed in
triplicate.
Colony forming assay
The tumor cells were transfected with let-7a mimic
and negative control or let-7a inhibitor and inhibitor negative
control for 6 h. Then, 1×103 cells were seeded into
6-well plates and incubated for 10 days, fixed and stained,
followed by colony counting. The experiment was performed in
triplicate.
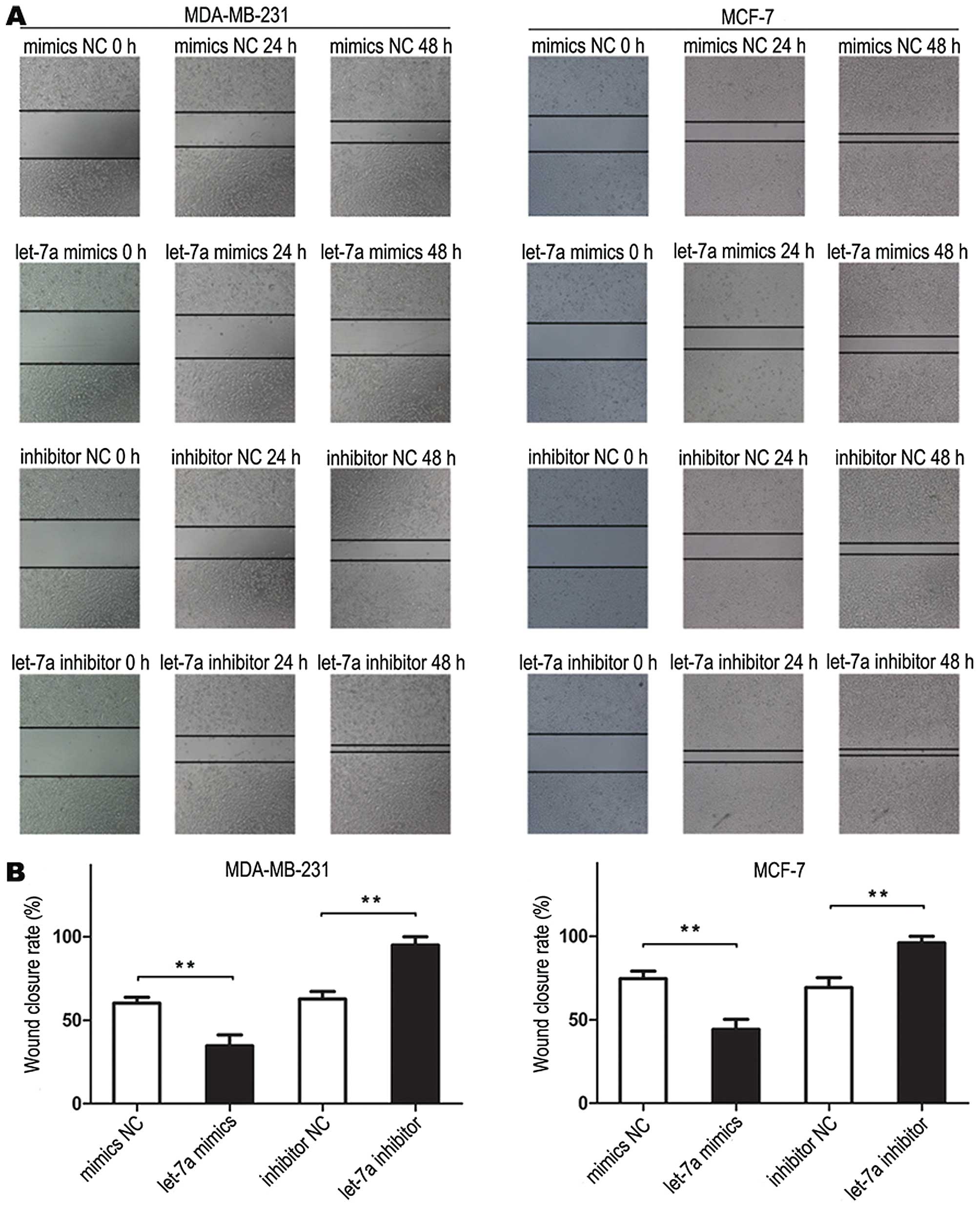
Wound healing assay
The tumor cells were seeded into 6-well plates and
transfected with let-7a mimic and negative control or let-7a
inhibitor and inhibitor negative control and allowed to grow until
100% confluency. Next, the cell layer was scratched through the
central axis using a sterile plastic tip and loose cells were
washed away by PBS. The wound healing was observed and photographed
at three pre-selected time points (0, 24, and 48 h) in three
randomly selected microscopic fields for each condition and time
point. The experiment was performed in triplicate.
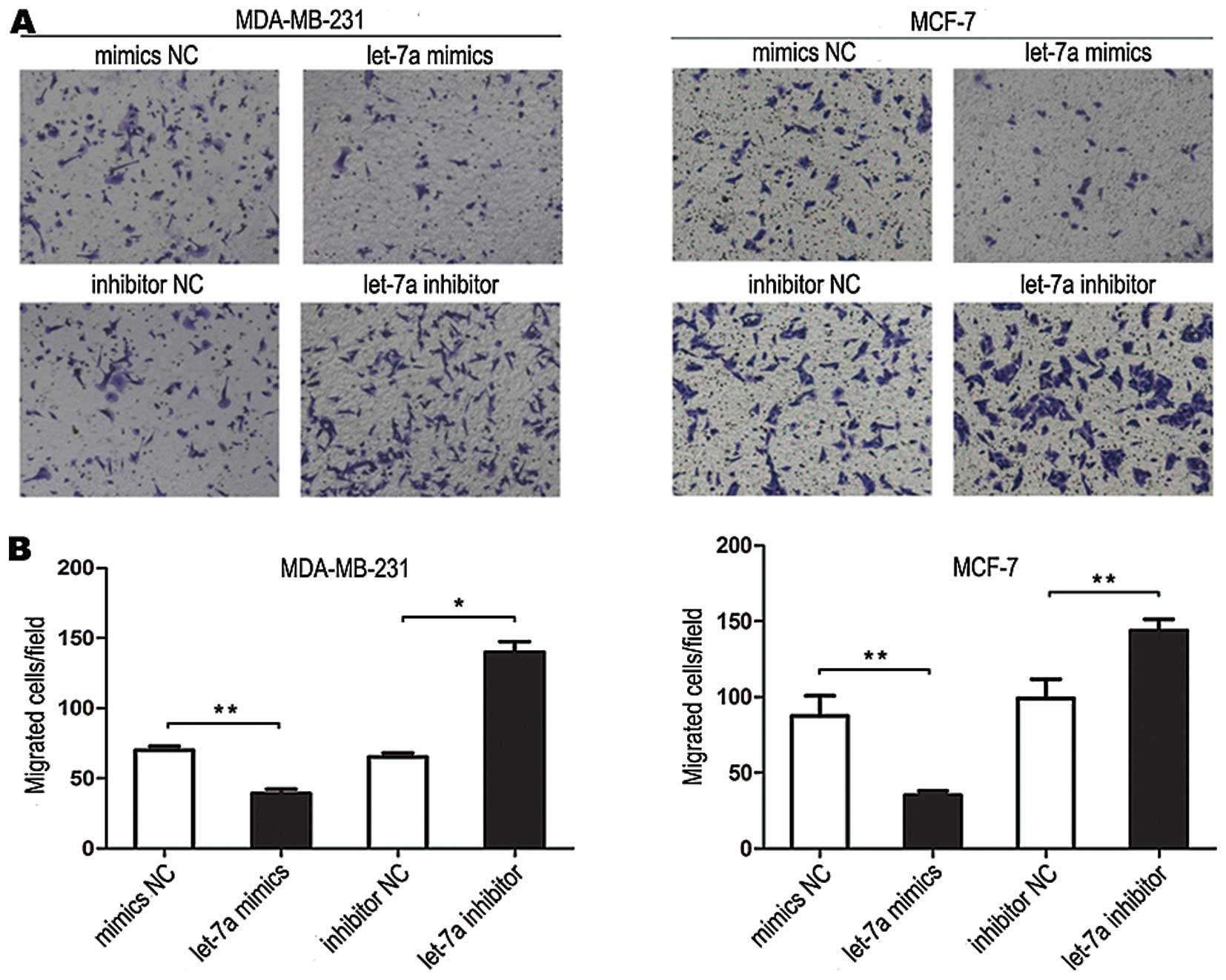
Cell invasion assay
The tumor cell invasion was evaluated using a
Transwell insert (8 μm, Corning). Cells were transfected with
let-7a mimic and negative control or let-7a inhibitor and inhibitor
negative control. After 24 h, the cells were starved in L-DMEM
without fetal bovine serum overnight, and then 4×104
cells resuspended in 0.2 ml serum-free L-DMEM were added to the
upper chamber and L-DMEM containing 10% fetal bovine serum was
added to the lower chamber as a chemoattractant at 37°C in
humidified 5% CO2 for 14 h (MDA-MB-231) or 16 h (MCF-7).
The invasive cells were fixed and stained with 0.1% crystal violet.
Three low-magnification areas (x100) were randomly selected and
counted for the cell numbers. The experiment was performed in
triplicate.
Protein extraction and western blot
analysis
Cells were transfected with let-7a mimic and
negative control or let-7a inhibitor and inhibitor negative
control. After 48 h, the total cellular protein of the cells was
extracted using a modified radioimmunoprecipitation assay (RIPA,
Vazyme Biotech, China) lysis buffer and phenylmethanesulfonyl
fluoride (PMSF, Beyotime, China). The protein concentration was
then determined by NanoDrop 1000 spectrophotometer (Thermo) and
equal amounts of protein lysates (100 μg) were separated by 12%
sodium dodecyl sulfate polyacrylamide gel electrophoresis
(SDS-PAGE, Beyotime) and then transferred to polyvinylidene
fluoride (PVDF) membrane (Beyotime). The membranes were blocked
with 5% defatted milk/TBST (20 mM Tris-HCl (pH 7.4), 150 mM NaCl,
and 0.1% Tween-20) at room temperature for 1 h and incubated with
primary antibodies at 4°C overnight. The next day, the membranes
were washed with TBST and then incubated with HRP-linked secondary
antibodies (anti-rabbit IgG, diluted at 1:1000; Cell Signaling
Technology). The protein bands were developed with
chemiluminescence (ECL) reagents (Beyotime). The antibodies were
anti-HMGA1 (diluted at 1:500; Abgent), anti-GAPDH (diluted at
1:1000; Cell Signaling Technology).
Statistical analysis
For statistical analyses, mean values with standard
deviation are shown in the graphs that were generated from several
repeats of biological experiments. P-values were obtained from
t-tests with paired or unpaired samples and <0.05 were
considered significant.
Results
Let-7a is decreased in breast cancer
tissues and breast cancer cells
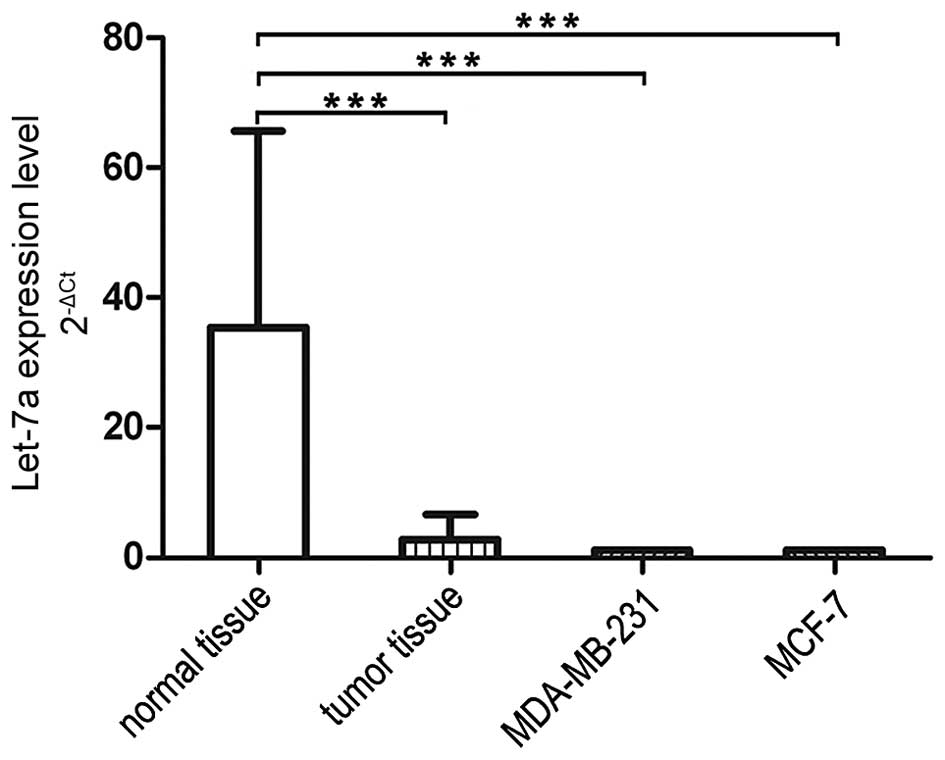
We performed qRT-PCR to determine let-7a levels in
breast cancer cells, 27 breast cancer tissues and adjacent normal
breast tissues. As shown in Fig.
1, the expression levels of the let-7a were downregulated in
cancer tissues compared to the adjacent normal tissues. We compared
let-7a expression in breast cancer cell lines MDA-MB-231 and MCF-7
which was in the similar range of that in breast cancer tissues,
thus we used these two breast cancer cell lines for further
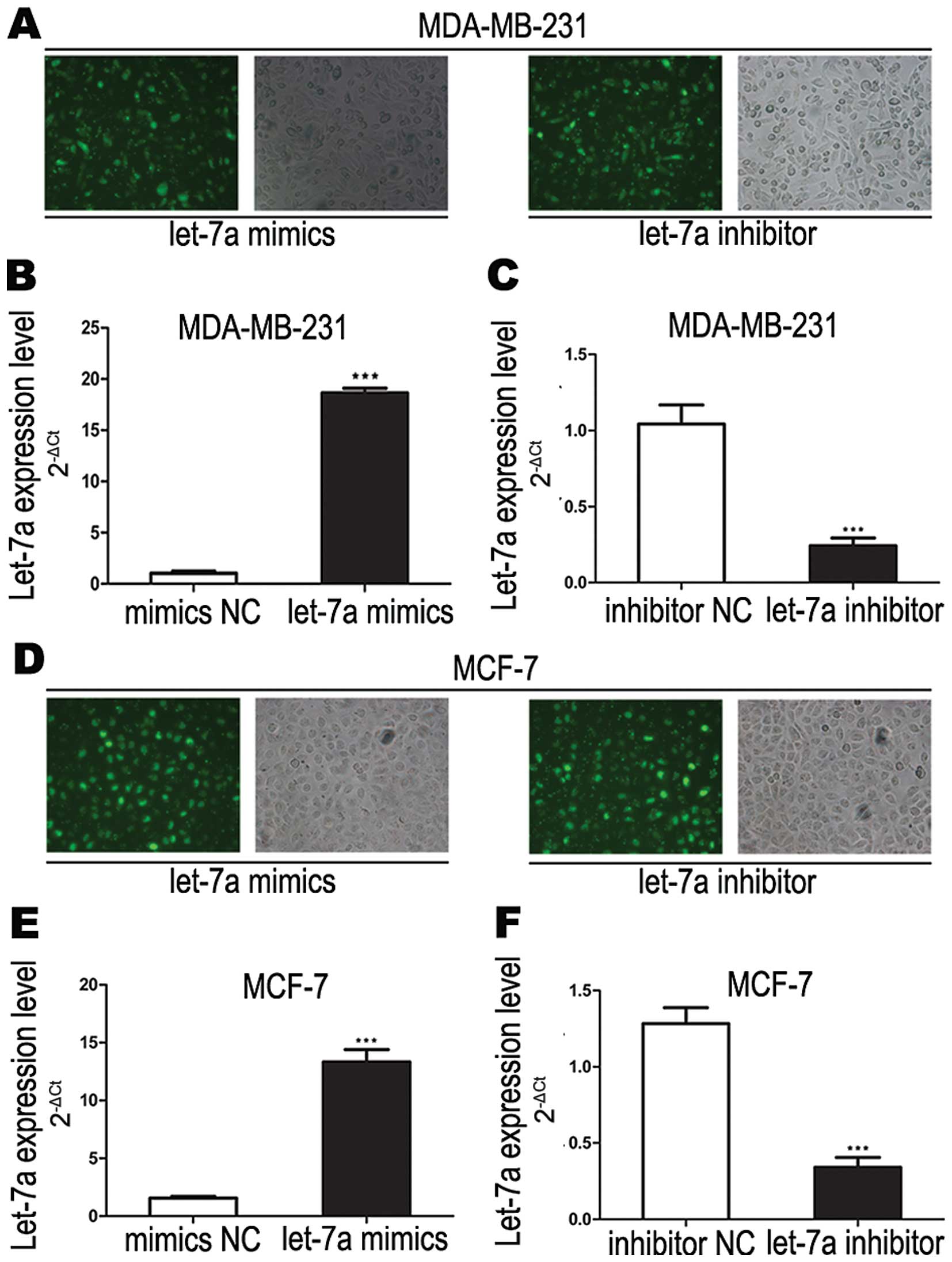
experiments in this study. To confirm the function of let-7a, we
transfected let-7a mimics and inhibitor into breast cancer cell
lines (MDA-MB-231 and MCF-7). At 6 h after transfection of let-7a
mimics and inhibitor into breast cancer cells, the transfection
efficiency were estimated by fluorescence microscopy and the let-7a
expression level was verified by real-time PCR (Fig. 2). We found that let-7a mimics
significantly increased let-7a RNA expression while let-7a
inhibitor significantly decreased let-7a RNA expression in both
breast cancer cell lines.
Let-7a inhibits breast cancer cell
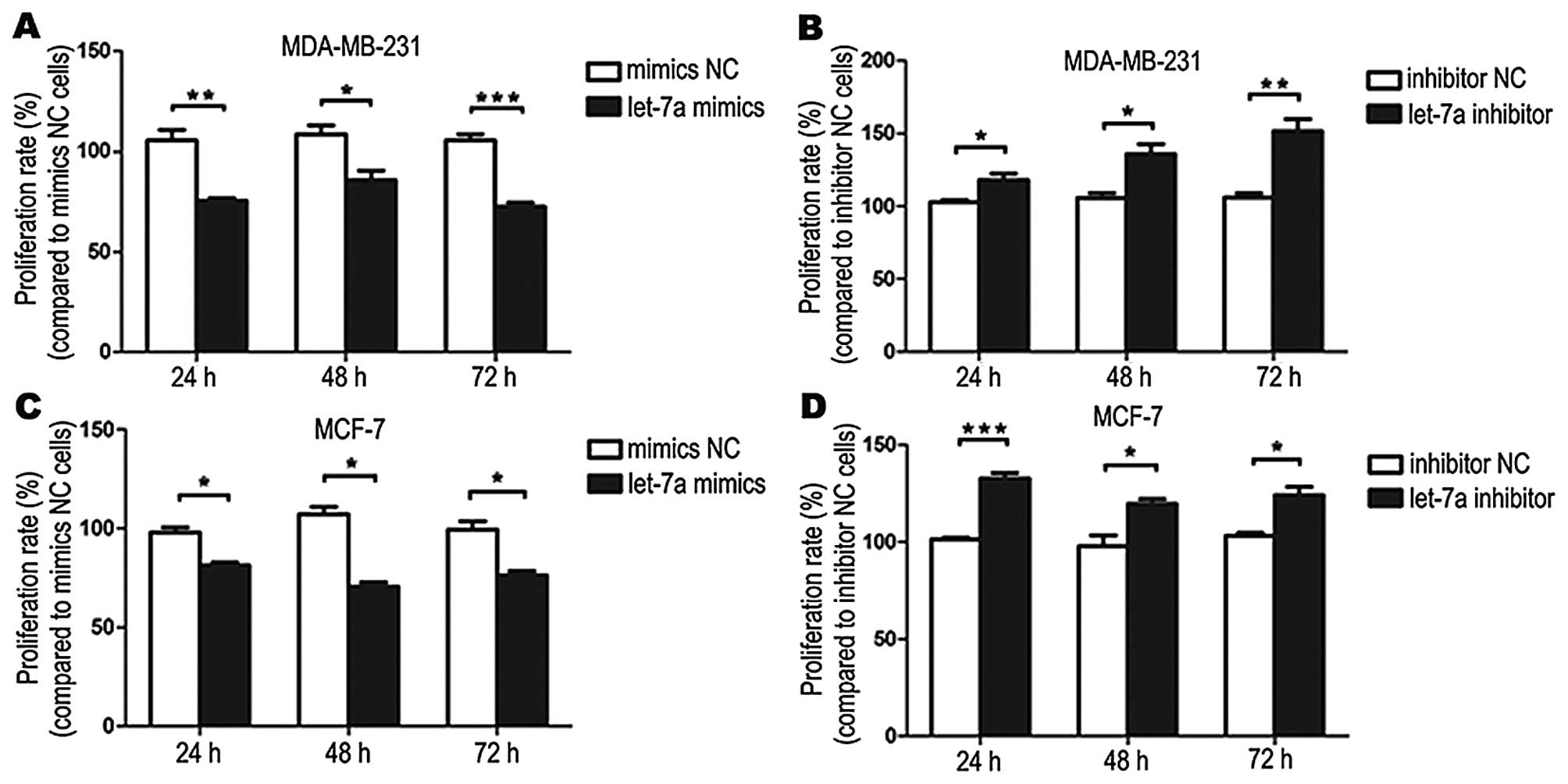
proliferation
Breast cancer cells were treated with let-7a mimics
or mimics NC and let-7a inhibitor or inhibitor NC for 24, 48, 72 h
and measured the absorbance at 490 nm. Compared with the treatment
with the mimics NC, cell proliferation was inhibited when cells
were treated with let-7a mimics. Compared with the inhibitor NC
treatment, cell proliferation was promoted when cells were treated
with let-7a inhibitor (Fig.
3).
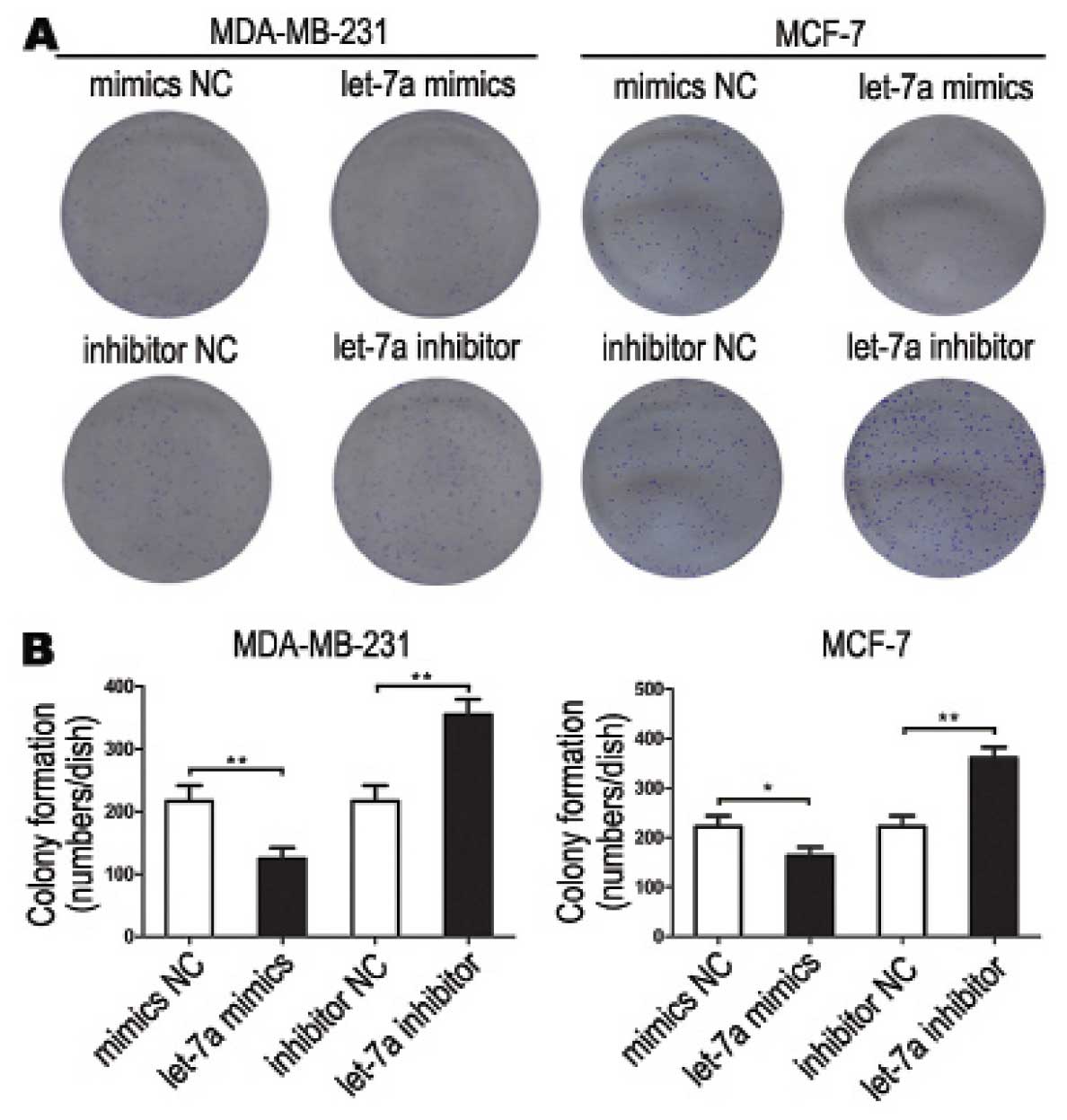
Let-7a decreases breast cancer cell
colony formation
Colonies formed from let-7a mimic-transfected cells
were significantly less than that of mimics NC transfected cells.
The let-7a inhibitor transfected cells formed significantly more
colonies than that of inhibitor NC transfected cells (Fig. 4). These data demonstrated that
let-7a inhibited breast cancer cell colony formation.
Let-7a inhibits breast cancer cells
migration
Cell scratch assay showed that cells transfected
with let-7a mimics migrated slowly. The scratch in mimics NC
treated cells was almost healed 48 h after the scratch had been
made, but not in let-7a mimics treated cells. The cells transfected
with let-7a inhibitor migrated more rapidly than cells transfected
with inhibitor NC (Fig. 5). These
data demonstrated that let-7a inhibited breast cancer cell
migration.
Let-7a inhibited breast cancer cell
invasion
Transwell invasion assays showed that the number of
tumor cells invading out of the chamber after treatment with let-7a
mimics was significantly less than that after treatment with mimics
NC. The number of tumor cells invading from the chamber after
treatment of let-7a inhibitor was significantly more than that
after treatment with inhibitor NC, demonstrating that let-7a
inhibited tumor cell invasion (Fig.
6).
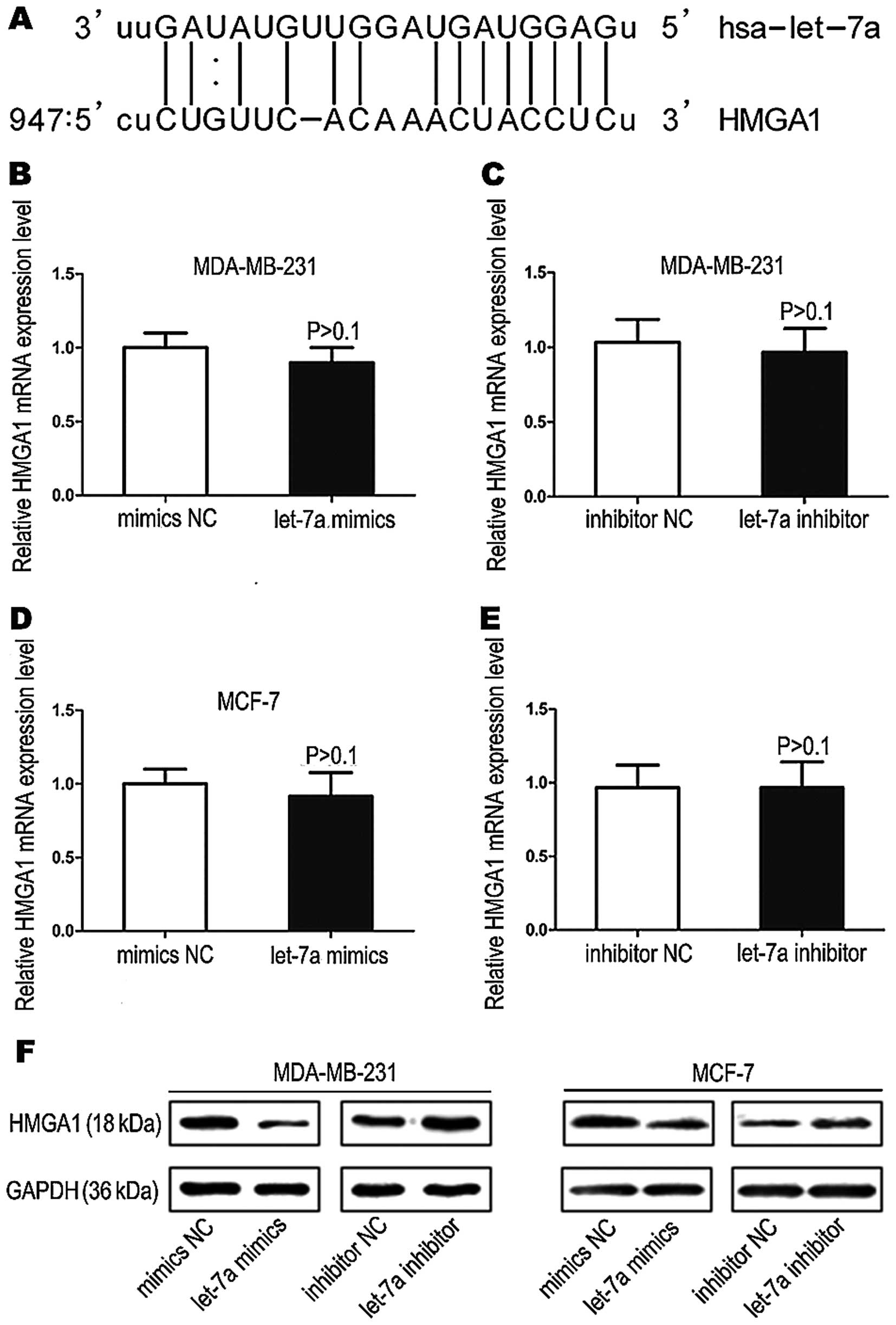
HMGA1 is a target gene of let-7a
In silico analyses of potential let-7a
targets (www.microrna.org and www.targetscan.org) indicated that the proteins of the
high mobility group A1 (HMGA1) is a possible target of let-7a.
HMGA1 mRNA has one potential complimentary binding site with let-7a
within its 3′ UTR (Fig. 7A). Based
on these results, we performed qRT-PCR assays and western blot
analysis to assess the impact of let-7a on HMGA1 expression.
Although we did not find apparent effects on HMGA1 mRNA expression
in the breast cancer cells after treatment of let-7a mimics or
mimics NC and let-7a inhibitor or inhibitor NC (Fig. 7B–E), western blot analysis showed
that HMGA1 protein expression was significantly decreased or
increased in let-7a mimics or let-7a inhibitor transfected cells
(Fig. 7F). These data suggest that
let-7a may target HMGA1 mRNA, and inhibits its translation into
proteins.
Discussion
Breast cancer is the most common type of malignant
tumor and its metastatic progression is a complex process (27–30).
At present, surgery and chemotherapy are the primary treatments for
breast cancer, but many patients under chemotherapy have early
tumor recurrence and metastasis, which results in poor prognosis.
Consequently, it is especially important to explore targeted
treatment for breast cancer. Over the last decades, miRNA has
become a hotspot for research. Previous studies suggest that
dysregulation of miRNAs is a common event in breast cancer
(31) and that they may thus act
as key regulators of tumorigenesis of breast cancer. Based on these
findings, it has been proposed that more effective targeted drugs
for treatment of breast cancer may involve miRNAs.
Among human cancer-related miRNAs, the let-7 family
has attracted significant attention because its family members are
expressed aberrantly in many cancers, such as lung carcinoma, and
colon carcinoma (20,22). As a member of the let-7 miRNA
family, let-7a has been reported to be expressed at lower than
normal levels in a variety of cancer (32–34).
However, the suppressive role of let-7a in tumorigenesis is still
poorly understood. In this study, we examined the expression levels
of let-7a in breast cancer cells (MDA-MB-231 and MCF-7), breast
cancer tissues and corresponding adjacent normal tissues. We found
that the expression of let-7a was significantly lower in breast
cancer cells and breast cancer tissues than corresponding adjacent
normal tissues, which suggested that downregulation of let-7a was
associated with the development of breast cancer. Thus, we
hypothesized that let-7a may function as a tumor suppressor. To
prove the role of let-7a in breast cancer, we transfected let-7a
mimics or let-7a inhibitor into breast cancer cells to induce
overexpression or low expression of let-7a. Exogenous
overexpression of let-7a significantly inhibited the cell growth as
indicated by MTT and colony formation assays whereas low expression
of let-7a significantly promote cell growth. Moreover, cell
migration and invasion were also significantly decreased or
increased by overexpression or low expression of let-7a in breast
cancer cells as shown by wound healing and Transwell assays. Recent
evidence indicated that let-7a plays a role in the progression of
human tumors such as renal cell carcinoma, gastric carcinoma and
hepatocellular carcinoma (35–37).
In addition, it has been shown that let-7a is downregulated in
Burkitt’s lymphoma and acted as an anticancer miRNA repressing
C-MYC expression at the translational level (38). These previous studies are
consistent with our current data, suggesting that let-7a plays a
role as a tumor suppressor in regulation of breast cancer
progression.
In this study, we first predicted by online
biological software that HMGA1 is a potential target gene of
let-7a. HMGA family members have previously been reported to be
involved in breast carcinogenesis (39). They are non-histone and DNA-binding
proteins, which are often referred to as architectural
transcription factors. They contain basic A-T hook domains which
mediate binding to the minor groove of AT-rich regions of
chromosomal DNA. Upon binding to DNA, HMGA proteins regulate gene
expression by forming the transcriptional complex through
protein-protein and protein-DNA interactions (40–42).
The HMGA family includes the products of the HMGA1 and HMGA2 genes.
HMGA1 has been found to be abnormally expressed in several types of
malignant tumors, including breast (43–45),
ovarian (46), leukemia (47), colon (48), pancreatic (49), thyroid (50), lung (51), prostate (52), endometrial (53), and head and neck malignant tumor
(54).
Next, we tested whether HMGA1 is a target of let-7a
by qRT-PCR and western blotting. Noteworthy, we found that the mRNA
expression of HMGA1 did not alter in let-7a mimics or let-7a
inhibitor transfected breast cancer cells, but protein expression
was significantly decreased or increased in let-7a mimics or let-7a
inhibitor transfected cells. Previous studies indicated that one
miRNA might have multiple mRNA targets and that one mRNA might be
targeted by multiple miRNAs. When a miRNA is perfectly
complementary to its target, it can specifically degrade the target
mRNA (55). However, if it is not
perfectly complementary to its target, the miRNA will inhibit mRNA
translation (56). Most miRNAs are
involved in the regulation of the expression of target genes
through the two pathways discussed above (57). Therefore, the result indicates that
let-7a is not perfectly complementary to its target. In other
words, let-7a regulates HMGA1 only at protein level, but not at the
genetic level. The result also confirmed the prediction from
bioinformatics.
Collectively, these data strongly suggested that
let-7a might act as a tumor suppressor in breast cancer by
targeting HMGA1. The upregulation of let-7a targeting HMGA1 shows
promise as new strategy to treat breast cancer.
Acknowledgements
This study was supported by the foundation of the
Jiangsu University for senior talented man (Grant no. 11JDG0089),
the innovation project of Cultivating Graduate of Jiangsu Province
(Grant no. CXLX13_689) and the Science Foundation of Kunshan (Grant
no. KS1331).
References
|
1
|
Gonzalez-Angulo AM, Morales-Vasquez F and
Hortobagyi GN: Overview of resistance to systemic therapy in
patients with breast cancer. Adv Exp Med Biol. 608:1–22.
2007.PubMed/NCBI
|
|
2
|
Lv YG, Yu F, Yao Q, Chen JH and Wang L:
The role of survivin in diagnosis, prognosis and treatment of
breast cancer. J Thorac Dis. 2:100–110. 2010.PubMed/NCBI
|
|
3
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
4
|
de Moor CH, Meijer H and Lissenden S:
Mechanisms of translational control by the 3′ UTR in development
and differentiation. Semin Cell Dev Biol. 16:49–58. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Blenkiron C, Goldstein LD, Thorne NP,
Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE,
Green AR, Ellis IO, et al: MicroRNA expression profiling of human
breast cancer identifies new markers of tumor subtype. Genome Biol.
8:R2142007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Silveri L, Tilly G, Vilotte JL and Le
Provost F: MicroRNA involvement in mammary gland development and
breast cancer. Reprod Nutr Dev. 46:549–556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hui AB, Shi W, Boutros PC, Miller N,
Pintilie M, Fyles T, McCready D, Wong D, Gerster K, Waldron L, et
al: Robust global micro-RNA profiling with formalin-fixed
paraffin-embedded breast cancer tissues. Lab Invest. 89:597–606.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wan K, Wang G, Ba Y, et al: Role of miR-143 targeting
KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nicoloso MS, Spizzo R, Shimizu M, Rossi S
and Calin GA: MicroRNAs - the micro steering wheel of tumour
metastases. Nat Rev Cancer. 9:293–302. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hui AB, Lenarduzzi M, Krushel T, Waldron
L, Pintilie M, Shi W, Perez-Ordonez B, Jurisica I, O’Sullivan B,
Waldron J, et al: Comprehensive microRNA profiling for head and
neck squamous cell carcinomas. Clin Cancer Res. 16:1129–1139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gramantieri L, Ferracin M, Fornari F,
Veronese A, Sabbioni S, Liu CG, Calin GA, Giovannini C, Ferrazzi E,
Grazi GL, et al: Cyclin G1 is a target of miR-122a, a microRNA
frequently downregulated in human hepatocellular carcinoma. Cancer
Res. 67:6092–6099. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M,
et al: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ma J, Dong C and Ji C: MicroRNA and drug
resistance. Cancer Gene Ther. 17:523–531. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
et al: microRNAs exhibit high frequency genomic alterations in
human cancer. Proc Natl Acad Sci USA. 103:9136–9141. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takamizawa J, Konishi H, Yanagisawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Akao Y, Nakagawa Y and Naoe T: let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Park SM, Shell S, Radjabi AR, Schickel R,
Feig C, Boyerinas B, Dinulescu DM, Lengyel E and Peter ME: Let-7
prevents early cancer progression by suppressing expression of the
embryonic gene HMGA2. Cell Cycle. 6:2585–2590. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong TS, Man OY, Tsang CM, Tsao SW, Tsang
RK, Chan JY, Ho WK, Wei WI and To VS: MicroRNA let-7 suppresses
nasopharyngeal carcinoma cells proliferation through downregulating
c-Myc expression. J Cancer Res Clin Oncol. 137:415–422. 2011.
View Article : Google Scholar :
|
|
25
|
He XY, Chen JX, Zhang Z, Li CL, Peng QL
and Peng HM: The let-7a microRNA protects from growth of lung
carcinoma by suppression of K-Ras and c-Myc in nude mice. J Cancer
Res Clin Oncol. 136:1023–1028. 2010. View Article : Google Scholar
|
|
26
|
Long XB, Sun GB, Hu S, Liang GT, Wang N,
Zhang XH, Cao PP, Zhen HT, Cui YH and Liu Z: Let-7a microRNA
functions as a potential tumor suppressor in human laryngeal
cancer. Oncol Rep. 22:1189–1195. 2009.PubMed/NCBI
|
|
27
|
Chiang AC and Massagué J: Molecular basis
of metastasis. N Engl J Med. 359:2814–2823. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Talmadge JE and Fidler IJ: AACR centennial
series: the biology of cancer metastasis: historical perspective.
Cancer Res. 70:5649–5669. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Valastyan S and Weinberg RA: MicroRNAs:
Crucial multitasking components in the complex circuitry of tumor
metastasis. Cell Cycle. 8:3506–3512. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou
X, Zhang ZJ, Peng YH, Yang YZ and Yun JP: miR-720 inhibits tumor
invasion and migration in breast cancer by targeting TWIST1.
Carcinogenesis. 35:469–478. 2014. View Article : Google Scholar
|
|
32
|
Johnson CD, Esquela-Kerscher A, Stefani G,
Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J,
Shingara J, et al: The let-7 microRNA represses cell proliferation
pathways in human cells. Cancer Res. 67:7713–7722. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meng F, Henson R, Wehbe-Janek H, Smith H,
Ueno Y and Patel T: The microRNA let-7a modulates
interleukin-6-dependent STAT-3 survival signaling in malignant
human cholangiocytes. J Biol Chem. 282:8256–8264. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mayr C, Hemann MT and Bartel DP:
Disrupting the pairing between let-7 and Hmga2 enhances oncogenic
transformation. Science. 315:1576–1579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liu Y, Yin B, Zhang C, Zhou L and Fan J:
Hsa-let-7a functions as a tumor suppressor in renal cell carcinoma
cell lines by targeting c-myc. Biochem Biophys Res Commun.
417:371–375. 2012. View Article : Google Scholar
|
|
36
|
Yang Q, Jie Z, Cao H, Greenlee AR, Yang C,
Zou F and Jiang Y: Low-level expression of let-7a in gastric cancer
and its involvement in tumorigenesis by targeting RAB40C.
Carcinogenesis. 32:713–722. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang Y, Lu Y, Toh ST, Sung WK, Tan P, Chow
P, Chung AY, Jooi LL and Lee CG: Lethal-7 is down-regulated by the
hepatitis B virus x protein and targets signal transducer and
activator of transcription 3. J Hepatol. 53:57–66. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sampson VB, Rong NH, Han J, Yang Q, Aris
V, Soteropoulos P, Petrelli NJ, Dunn SP and Krueger LJ: MicroRNA
let-7a downregulates MYC and reverts MYC-induced growth in Burkitt
lymphoma cells. Cancer Res. 67:9762–9770. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Peluso S and Chiappetta G: High-mobility
group A (HMGA) proteins and breast cancer. Breast Care (Basel).
5:81–85. 2010. View Article : Google Scholar
|
|
40
|
Reeves R and Beckerbauer L: HMGI/Y
proteins: Flexible regulators of transcription and chromatin
structure. Biochim Biophys Acta. 1519:13–29. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fedele M and Fusco A: HMGA and cancer.
Biochim Biophys Acta. 1799:48–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Resar LM: The high mobility group A1 gene:
Transforming inflammatory signals into cancer? Cancer Res.
70:436–439. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Dolde CE, Mukherjee M, Cho C and Resar LM:
HMG-I/Y in human breast cancer cell lines. Breast Cancer Res Treat.
71:181–191. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chiappetta G, Ottaiano A, Vuttariello E,
Monaco M, Galdiero F, Gallipoli A, Pilotti S, Jodice G, Siranoush
M, Colombo M, et al: HMGA1 protein expression in familial breast
carcinoma patients. Eur J Cancer. 46:332–339. 2010. View Article : Google Scholar
|
|
45
|
Flohr AM, Rogalla P, Bonk U, Puettmann B,
Buerger H, Gohla G, Packeisen J, Wosniok W, Loeschke S and
Bullerdiek J: High mobility group protein HMGA1 expression in
breast cancer reveals a positive correlation with tumour grade.
Histol Histopathol. 18:999–1004. 2003.PubMed/NCBI
|
|
46
|
Masciullo V, Baldassarre G, Pentimalli F,
Berlingieri MT, Boccia A, Chiappetta G, Palazzo J, Manfioletti G,
Giancotti V, Viglietto G, et al: HMGA1 protein over-expression is a
frequent feature of epithelial ovarian carcinomas. Carcinogenesis.
24:1191–1198. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Y, Sumter TF, Bhattacharya R, Tesfaye
A, Fuchs EJ, Wood LJ, Huso DL and Resar LM: The HMG-I oncogene
causes highly penetrant, aggressive lymphoid malignancy in
transgenic mice and is overexpressed in human leukemia. Cancer Res.
64:3371–3375. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Chiappetta G, Manfioletti G, Pentimalli F,
Abe N, Di Bonito M, Vento MT, Giuliano A, Fedele M, Viglietto G,
Santoro M, et al: High mobility group HMGI(Y) protein expression in
human colorectal hyperplastic and neoplastic diseases. Int J
Cancer. 91:147–151. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Abe N, Watanabe T, Masaki T, Mori T,
Sugiyama M, Uchimura H, Fujioka Y, Chiappetta G, Fusco A and Atomi
Y: Pancreatic duct cell carcinomas express high levels of high
mobility group I(Y) proteins. Cancer Res. 60:3117–3122.
2000.PubMed/NCBI
|
|
50
|
Chiappetta G, Tallini G, De Biasio MC,
Manfioletti G, Martinez-Tello FJ, Pentimalli F, de Nigris F, Mastro
A, Botti G, Fedele M, et al: Detection of high mobility group I
HMGI(Y) protein in the diagnosis of thyroid tumors: HMGI(Y)
expression represents a potential diagnostic indicator of
carcinoma. Cancer Res. 58:4193–4198. 1998.PubMed/NCBI
|
|
51
|
Hillion J, Wood LJ, Mukherjee M,
Bhattacharya R, Di Cello F, Kowalski J, Elbahloul O, Segal J,
Poirier J, Rudin CM, et al: Upregulation of MMP-2 by HMGA1 promotes
transformation in undifferentiated, large-cell lung cancer. Mol
Cancer Res. 7:1803–1812. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Tamimi Y, van der Poel HG, Denyn MM, Umbas
R, Karthaus HF, Debruyne FM and Schalken JA: Increased expression
of high mobility group protein I(Y) in high grade prostatic cancer
determined by in situ hybridization. Cancer Res. 53:5512–5516.
1993.PubMed/NCBI
|
|
53
|
Tesfaye A, Di Cello F, Hillion J, Ronnett
BM, Elbahloul O, Ashfaq R, Dhara S, Prochownik E, Tworkoski K,
Reeves R, et al: The high-mobility group A1 gene up-regulates
cyclooxygenase 2 expression in uterine tumorigenesis. Cancer Res.
67:3998–4004. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Rho YS, Lim YC, Park IS, Kim JH, Ahn HY,
Cho SJ and Shin HS: High mobility group HMGI(Y) protein expression
in head and neck squamous cell carcinoma. Acta Otolaryngol.
127:76–81. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wightman B, Ha I and Ruvkun G:
Posttranscriptional regulation of the heterochronic gene lin-14 by
lin-4 mediates temporal pattern formation in C. elegans. Cell.
75:855–862. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|