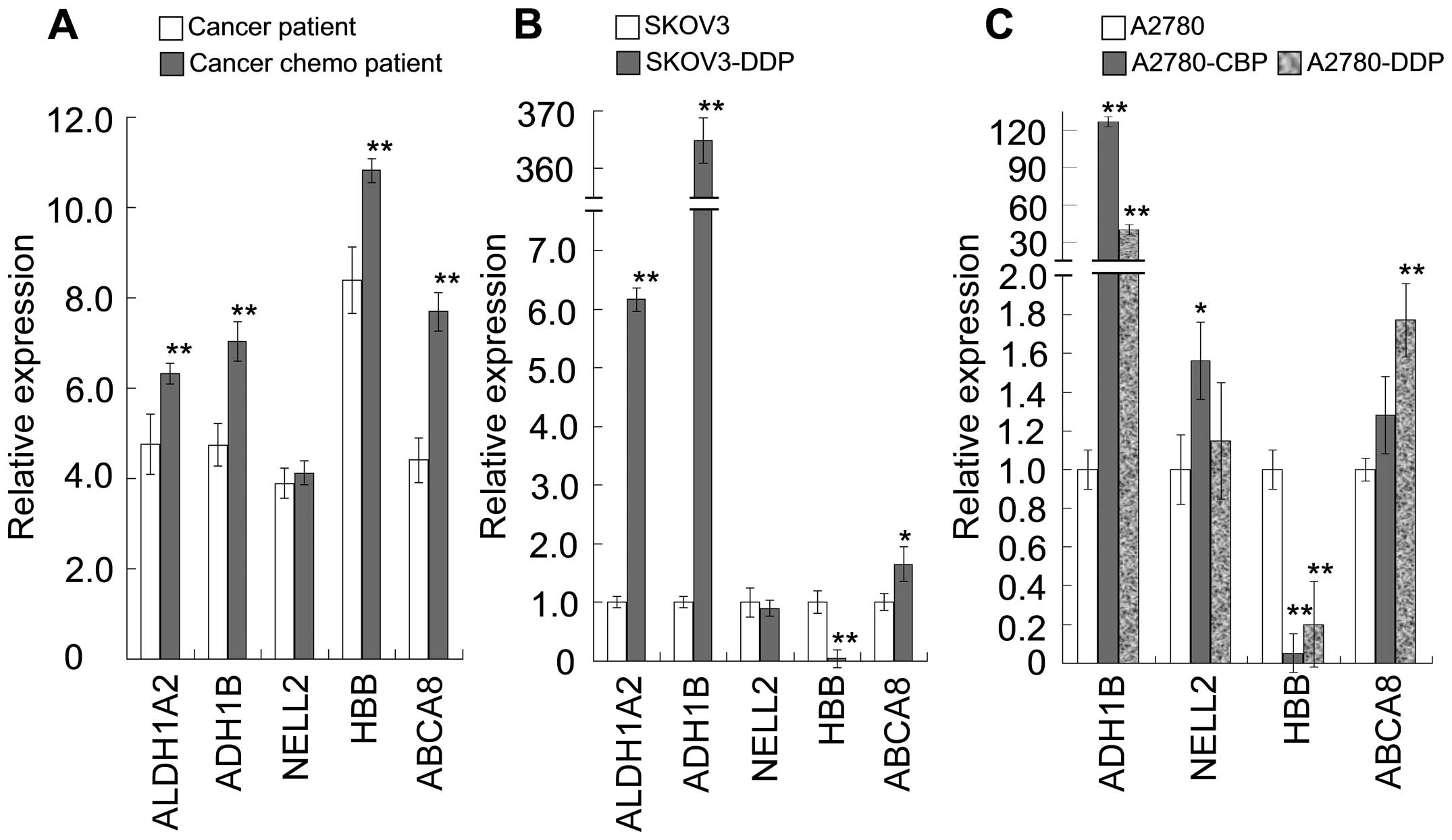
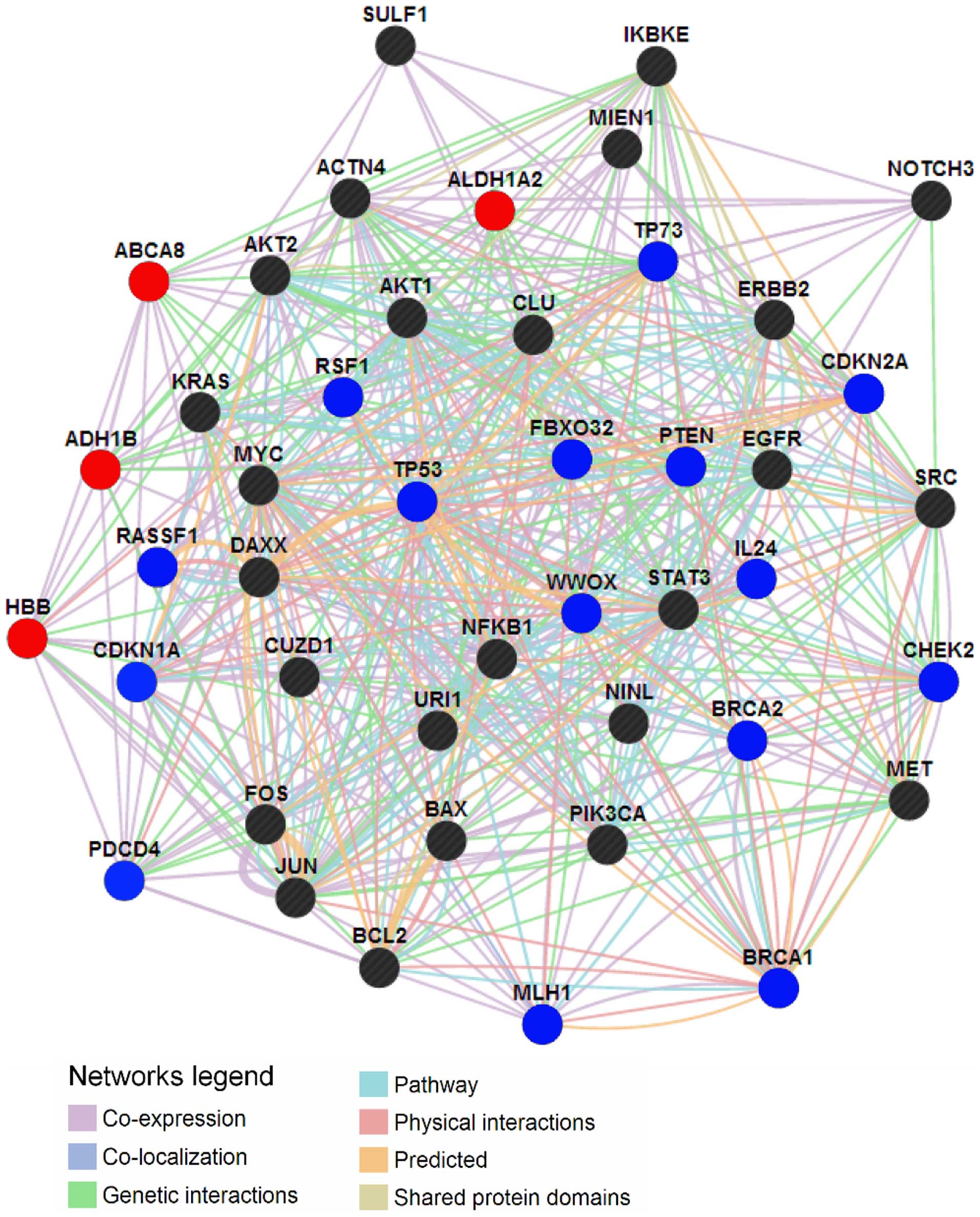
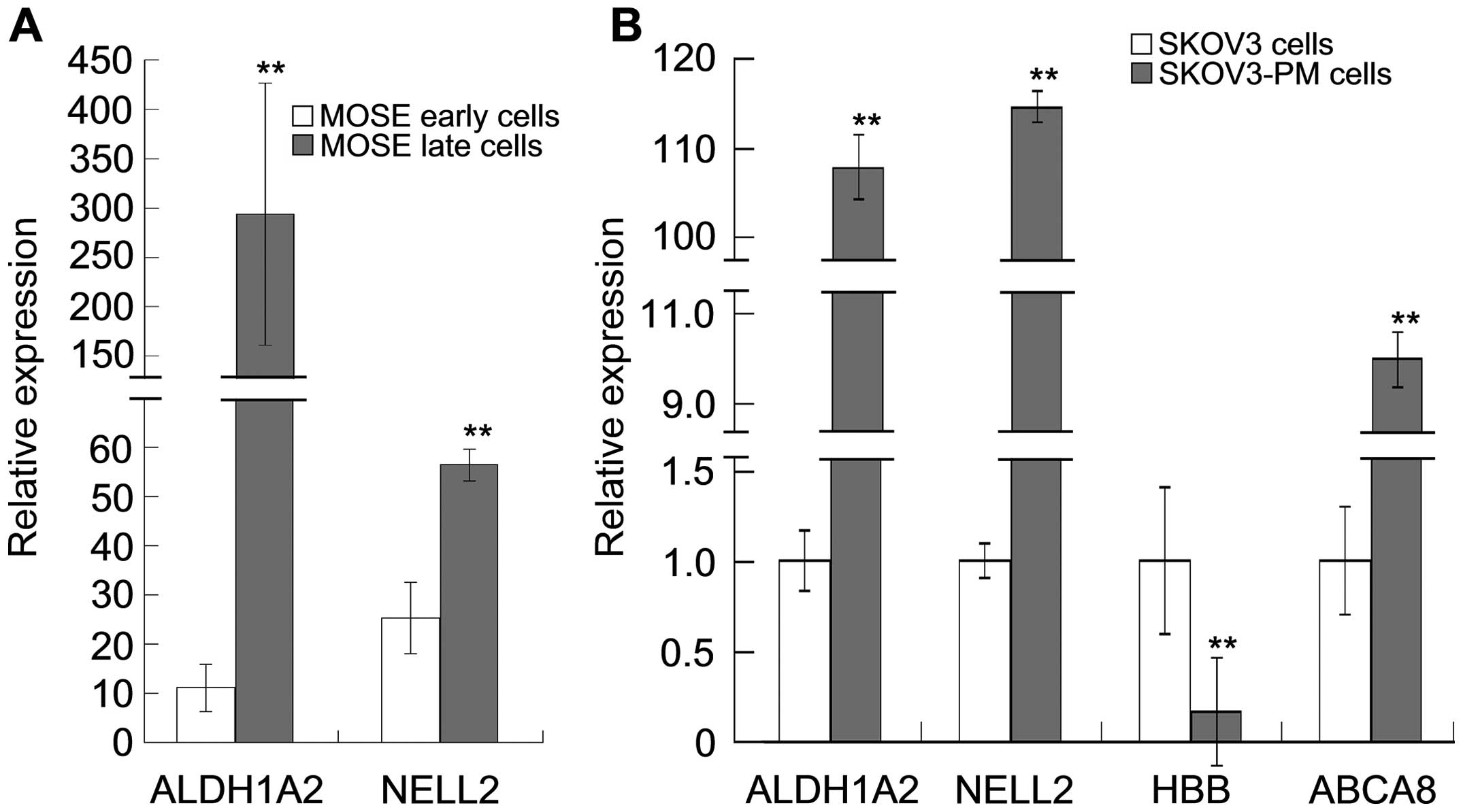
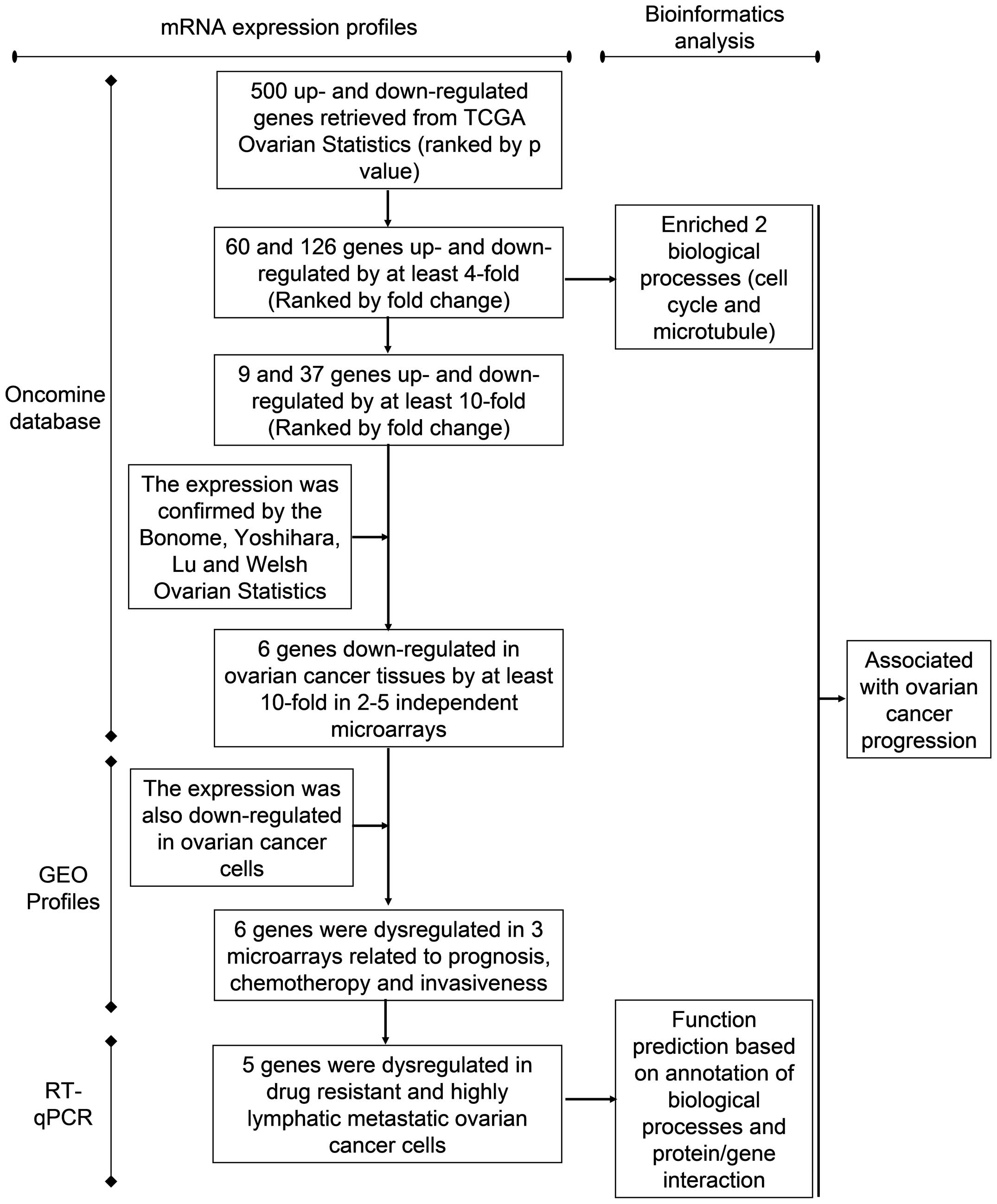
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sharan R, Ulitsky I and Shamir R:
Network-based prediction of protein function. Mol Syst Biol.
3:882007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stuart JM, Segal E, Koller D and Kim SK: A
gene-coexpression network for global discovery of conserved genetic
modules. Science. 302:249–255. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Uetz P, Giot L, Cagney G, Mansfield TA,
Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart
P, et al: A comprehensive analysis of protein-protein interactions
in Saccharomyces cerevisiae. Nature. 403:623–627. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Phuong T and Nhung N: Predicting gene
function using similarity learning. BMC Genomics. 14(Suppl 4):
S42013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hsu FH, Serpedin E, Hsiao TH, Bishop AJ,
Dougherty ER and Chen Y: Reducing confounding and suppression
effects in TCGA data: An integrated analysis of chemotherapy
response in ovarian cancer. BMC Genomics. 13(Suppl 6): S132012.
View Article : Google Scholar :
|
|
8
|
Liu X, Gao Y, Lu Y, Zhang J, Li L and Yin
F: Upregulation of NEK2 is associated with drug resistance in
ovarian cancer. Oncol Rep. 31:745–754. 2014.
|
|
9
|
Liu X, Gao Y, Lu Y, Zhang J, Li L and Yin
F: Downregulation of NEK11 is associated with drug resistance in
ovarian cancer. Int J Oncol. 45:1266–1274. 2014.PubMed/NCBI
|
|
10
|
Yin F, Liu L, Liu X, Li G, Zheng L, Li D,
Wang Q, Zhang W and Li L: Downregulation of tumor suppressor gene
ribonuclease T2 and gametogenetin binding protein 2 is associated
with drug resistance in ovarian cancer. Oncol Rep. 32:362–372.
2014.PubMed/NCBI
|
|
11
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Bioinformatic analysis of chemokine (C-C motif) ligand 21 and
SPARC-like protein 1 revealing their associations with drug
resistance in ovarian cancer. Int J Oncol. 42:1305–1316.
2013.PubMed/NCBI
|
|
12
|
Ruan HY, Li DR, Li L, Guan X and Zhang W:
Establishment of human ovarian carcinoma cell lines with
directional highly lymphatic metastasis and study of their
biological characteristics. Zhonghua Fu Chan Ke Za Zhi. 42:482–486.
2007.(In Chinese). PubMed/NCBI
|
|
13
|
Rhodes DR, Yu J, Shanker K, Deshpande N,
Varambally R, Ghosh D, Barrette T, Pandey A and Chinnaiyan AM:
ONCOMINE: A cancer microarray database and integrated data-mining
platform. Neoplasia. 6:1–6. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Edgar R, Domrachev M and Lash AE: Gene
expression omnibus: NCBI gene expression and hybridization array
data repository. Nucleic Acids Res. 30:207–210. 2002. View Article : Google Scholar :
|
|
15
|
Barrett T and Edgar R: Mining microarray
data at NCBI’s gene expression omnibus (GEO)*. Methods Mol Biol.
338:175–190. 2006.
|
|
16
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:pl12013. View Article : Google Scholar
|
|
18
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
19
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
20
|
de Leeuw N, Dijkhuizen T, Hehir-Kwa JY,
Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van
Ravenswaaij-Arts CMA, et al: Diagnostic interpretation of array
data using public databases and internet sources. Hum Mutat.
33:930–940. 2012. View Article : Google Scholar
|
|
21
|
Mostafavi S, Ray D, Warde-Farley D,
Grouios C and Morris Q: GeneMANIA: A real-time multiple association
network integration algorithm for predicting gene function. Genome
Biol. 9(Suppl 1): S42008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zuberi K, Franz M, Rodriguez H, Montojo J,
Lopes CT, Bader GD and Morris Q: GeneMANIA prediction server 2013
update. Nucleic Acids Res. 41:W115–22. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hedditch EL, Gao B, Russell AJ, Lu Y,
Emmanuel C, Beesley J, Johnatty SE, Chen X, Harnett P, George J, et
al; Australian Ovarian Cancer Study Group. ABCA transporter gene
expression and poor outcome in epithelial ovarian cancer. J Natl
Cancer Inst. 106:1062014. View Article : Google Scholar
|
|
24
|
Bowen NJ, Walker LD, Matyunina LV, Logani
S, Totten KA, Benigno BB and McDonald JF: Gene expression profiling
supports the hypothesis that human ovarian surface epithelia are
multipotent and capable of serving as ovarian cancer initiating
cells. BMC Med Genomics. 2:712009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Partheen K, Levan K, Osterberg L and
Horvath G: Expression analysis of stage III serous ovarian
adenocarcinoma distinguishes a sub-group of survivors. Eur J
Cancer. 42:2846–2854. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Moreno CS, Matyunina L, Dickerson EB,
Schubert N, Bowen NJ, Logani S, Benigno BB and McDonald JF:
Evidence that p53-mediated cell-cycle-arrest inhibits
chemotherapeutic treatment of ovarian carcinomas. PLoS One.
2:e4412007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yin F, Liu X, Li D, Wang Q, Zhang W and Li
L: Tumor suppressor genes associated with drug resistance in
ovarian cancer (Review). Oncol Rep. 30:3–10. 2013.PubMed/NCBI
|
|
28
|
Liu X, Gao Y, Lu Y, Zhang J, Li L and Yin
F: Oncogenes associated with drug resistance in ovarian cancer. J
Cancer Res Clin Oncol. 141:381–395. 2015. View Article : Google Scholar
|
|
29
|
Zhang X, Wang X, Song X, Liu C, Shi Y,
Wang Y, Afonja O, Ma C, Chen YH and Zhang L: Programmed cell death
4 enhances chemosensitivity of ovarian cancer cells by activating
death receptor pathway in vitro and in vivo. Cancer Sci.
101:2163–2170. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Creekmore AL, Silkworth WT, Cimini D,
Jensen RV, Roberts PC and Schmelz EM: Changes in gene expression
and cellular architecture in an ovarian cancer progression model.
PLoS One. 6:e176762011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cancer Genome Atlas Research Network.
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Braun R, Finney R, Yan C, Chen QR, Hu Y,
Edmonson M, Meerzaman D and Buetow K: Discovery analysis of TCGA
data reveals association between germline genotype and survival in
ovarian cancer patients. PLoS One. 8:e550372013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ying H, Lv J, Ying T, Jin S, Shao J, Wang
L, Xu H, Yuan B and Yang Q: Gene-gene interaction network analysis
of ovarian cancer using TCGA data. J Ovarian Res. 6:882013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wan YW, Mach CM, Allen GI, Anderson ML and
Liu Z: On the reproducibility of TCGA ovarian cancer microRNA
profiles. PLoS One. 9:e877822014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets - update. Nucleic Acids Res. 41:D991–D995. 2013.
View Article : Google Scholar
|
|
36
|
Brazma A, Hingamp P, Quackenbush J,
Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA,
Causton HC, et al: Minimum information about a microarray
experiment (MIAME)-toward standards for microarray data. Nat Genet.
29:365–371. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
D’Andrilli G, Kumar C, Scambia G and
Giordano A: Cell cycle genes in ovarian cancer: Steps toward
earlier diagnosis and novel therapies. Clin Cancer Res.
10:8132–8141. 2004. View Article : Google Scholar
|
|
38
|
D’Andrilli G, Giordano A and Bovicelli A:
Epithelial ovarian cancer: The role of cell cycle genes in the
different histotypes. Open Clin Cancer J. 2:7–12. 2008. View Article : Google Scholar
|
|
39
|
Huang Y, Zhang X, Jiang W, Wang Y, Jin H,
Liu X and Xu C: Discovery of serum biomarkers implicated in the
onset and progression of serous ovarian cancer in a rat model using
iTRAQ technique. Eur J Obstet Gynecol Reprod Biol. 165:96–103.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giardina B, Messana I, Scatena R and
Castagnola M: The multiple functions of hemoglobin. Crit Rev
Biochem Mol Biol. 30:165–196. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Onda M, Akaishi J, Asaka S, Okamoto J,
Miyamoto S, Mizutani K, Yoshida A, Ito K and Emi M: Decreased
expression of haemoglobin beta (HBB) gene in anaplastic thyroid
cancer and recovery of its expression inhibits cell growth. Br J
Cancer. 92:2216–2224. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Suresh A, Vannan M, Kumaran D, Gümüs ZH,
Sivadas P, Murugaian EE, Kekatpure V, Iyer S, Thangaraj K and
Kuriakose MA: Resistance/response molecular signature for oral
tongue squamous cell carcinoma. Dis Markers. 32:51–64. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Annilo T, Chen ZQ, Shulenin S and Dean M:
Evolutionary analysis of a cluster of ATP-binding cassette (ABC)
genes. Mamm Genome. 14:7–20. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nagase T, Ishikawa K, Suyama M, Kikuno R,
Hirosawa M, Miyajima N, Tanaka A, Kotani H, Nomura N and Ohara O:
Prediction of the coding sequences of unidentified human genes. XII
The complete sequences of 100 new cDNA clones from brain which code
for large proteins in vitro. DNA Res. 5:355–364. 1998. View Article : Google Scholar
|
|
45
|
Tucker SL, Gharpure K, Herbrich SM, Unruh
AK, Nick AM, Crane EK, Coleman RL, Guenthoer J, Dalton HJ, Wu SY,
et al: Molecular biomarkers of residual disease after surgical
debulking of high-grade serous ovarian cancer. Clin Cancer Res.
20:3280–3288. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Januchowski R, Zawierucha P, Andrzejewska
M, Ruciński M and Zabel M: Microarray-based detection and
expression analysis of ABC and SLC transporters in drug-resistant
ovarian cancer cell lines. Biomed Pharmacother. 67:240–245. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Seitz HK and Stickel F: Molecular
mechanisms of alcohol-mediated carcinogenesis. Nat Rev Cancer.
7:599–612. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tsai ST, Wong TY, Ou CY, Fang SY, Chen KC,
Hsiao JR, Huang CC, Lee WT, Lo HI, Huang JS, et al: The interplay
between alcohol consumption, oral hygiene, ALDH2 and ADH1B in the
risk of head and neck cancer. Int J Cancer. 135:2424–2436. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu M, Chang SC, Kampman E, Yang J, Wang
XS, Gu XP, Han RQ, Liu AM, Wallar G, Zhou JY, et al: Single
nucleotide polymorphisms of ADH1B, ADH1C and ALDH2 genes and
esophageal cancer: A population-based case-control study in China.
Int J Cancer. 132:1868–1877. 2013. View Article : Google Scholar
|
|
50
|
Duell EJ, Sala N, Travier N, Muñoz X,
Boutron-Ruault MC, Clavel-Chapelon F, Barricarte A, Arriola L,
Navarro C, Sánchez-Cantalejo E, et al: Genetic variation in alcohol
dehydrogenase (ADH1A, ADH1B, ADH1C, ADH7) and aldehyde
dehydrogenase (ALDH2), alcohol consumption and gastric cancer risk
in the European Prospective Investigation into Cancer and Nutrition
(EPIC) cohort. Carcinogenesis. 33:361–367. 2012. View Article : Google Scholar
|
|
51
|
Leclerc J, Courcot-Ngoubo Ngangue E,
Cauffiez C, Allorge D, Pottier N, Lafitte JJ, Debaert M, Jaillard
S, Broly F and Lo-Guidice JM: Xenobiotic metabolism and disposition
in human lung: Transcript profiling in non-tumoral and tumoral
tissues. Biochimie. 93:1012–1027. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Murray GI, Weaver RJ, Paterson PJ, Ewen
SW, Melvin WT and Burke MD: Expression of xenobiotic metabolizing
enzymes in breast cancer. J Pathol. 169:347–353. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Akman SA, Forrest G, Chu FF, Esworthy RS
and Doroshow JH: Antioxidant and xenobiotic-metabolizing enzyme
gene expression in doxorubicin-resistant MCF-7 breast cancer cells.
Cancer Res. 50:1397–1402. 1990.PubMed/NCBI
|
|
54
|
Tsuruoka S, Ishibashi K, Yamamoto H,
Wakaumi M, Suzuki M, Schwartz GJ, Imai M and Fujimura A: Functional
analysis of ABCA8, a new drug transporter. Biochem Biophys Res
Commun. 298:41–45. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Januchowski R, Zawierucha P, Ruciński M,
Andrzejewska M, Wojtowicz K, Nowicki M and Zabel M: Drug
transporter expression profiling in chemoresistant variants of the
A2780 ovarian cancer cell line. Biomed Pharmacother. 68:447–453.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Auersperg N: The stem-cell profile of
ovarian surface epithelium is reproduced in the oviductal fimbriae,
with increased stem-cell marker density in distal parts of the
fimbriae. Int J Gynecol Pathol. 32:444–453. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Kim H, Lapointe J, Kaygusuz G, Ong DE, Li
C, van de Rijn M, Brooks JD and Pollack JR: The retinoic acid
synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate
cancer. Cancer Res. 65:8118–8124. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ju ZH, Ying MG, Ai X, Shi TP, Wang BJ,
Wang C, Zhang GX and Zhang X: Effects of 5-Aza-2′-deoxycitydine and
trichostatin A on expression and apoptosis of ALDH1a2 gene in human
bladder cancer cell lines. Zhonghua Wai Ke Za Zhi. 48:378–382.
2010.(In Chinese). PubMed/NCBI
|
|
59
|
Moreb JS, Ucar D, Han S, Amory JK,
Goldstein AS, Ostmark B and Chang LJ: The enzymatic activity of
human aldehyde dehydrogenases 1A2 and 2 (ALDH1A2 and ALDH2) is
detected by Aldefluor, inhibited by diethylaminobenzaldehyde and
has significant effects on cell proliferation and drug resistance.
Chem Biol Interact. 195:52–60. 2012. View Article : Google Scholar :
|
|
60
|
Kawasoe M, Yamamoto Y, Okawa K, Funato T,
Takeda M, Hara T, Tsurumi H, Moriwaki H, Arioka Y, Takemura M,
Matsunami H, Markey SP and Saito K: Acquired resistance of leukemic
cells to AraC is associated with the upregulation of aldehyde
dehydrogenase 1 family member A2. Exp Hematol. 41:597–603 e2. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Cortes-Dericks L, Froment L, Boesch R,
Schmid RA and Karoubi G: Cisplatin-resistant cells in malignant
pleural mesothelioma cell lines show ALDH(high)CD44(+) phenotype
and sphere-forming capacity. BMC Cancer. 14:3042014. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Kim DH, Roh YG, Lee HH, Lee SY, Kim SI,
Lee BJ and Leem SH: The E2F1 oncogene transcriptionally regulates
NELL2 in cancer cells. DNA Cell Biol. 32:517–523. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Maeda K, Matsuhashi S, Tabuchi K, Watanabe
T, Katagiri T, Oyasu M, Saito N and Kuroda S: Brain specific human
genes, NELL1 and NELL2, are predominantly expressed in
neuroblastoma and other embryonal neuroepithelial tumors. Neurol
Med Chir (Tokyo). 41:582–588; discussion 589. 2001. View Article : Google Scholar
|
|
64
|
Kuroda S, Oyasu M, Kawakami M, Kanayama N,
Tanizawa K, Saito N, Abe T, Matsuhashi S and Ting K: Biochemical
characterization and expression analysis of neural
thrombospondin-1-like proteins NELL1 and NELL2. Biochem Biophys Res
Commun. 265:79–86. 1999. View Article : Google Scholar : PubMed/NCBI
|