Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most common cancer affecting the head and neck region, causing
~42,440 new cases and 8,390 deaths annually in the United States.
In spite of the many advances in surgical treatment, radiotherapy
and chemotherapy used in OSCC, the 5-year survival rate after
diagnosis for OSCC remains at ~50% (1). In addition, the limited survival of
OSCC patients is likely due to a high proportion of patients with
advanced disease stages, lack of suitable markers for early
detection, failure to respond to available drugs and incomplete
knowledge of the mechanisms of the poor survival risk factors
(2–5). Studies on the mechanisms underlying
the malignant progression and finding a therapeutic target of OSCC
might be an effective way to improve the treatment efficacy of this
cancer. Recent discoveries have dramatically improved our
understanding of the mechanisms of cell development and progression
in OSCC. However, most studies are focusing on the protein-coding
genes and the studies on non-coding RNA are limited, especially on
long non-coding RNAs.
Long non-coding RNAs (lncRNAs) are transcripts
longer than 200 nts, and without a functional open reading frame
(ORF) in most cases, but they might play widespread roles in gene
regulation and other cellular processes, including their
involvement in epigenetic regulation, gene transcription, and
post-transcription regulation (6).
In addition, recent studies suggests that lncRNAs are involved in
the development of human diseases, particularly in cancer (7,8).
Among the lncRNAs, HOX transcript antisense RNA (HOTAIR), a
paradigm for long non-coding RNA function in cancer, is transcribed
from the HOXC cluster in an antisense manner, functions as a
molecular scaffold to link the polycomb repressive complexes 2
(PRC2) and the lysine specific demethylase 1 (LSD1) complexes, then
regulates gene expression by mediating the modulation of chromatin
structures in trans across the 40-kb HOXD locus. Evidence
has shown that HOTAIR is pervasively overexpressed in many cancers
and involved in tumor invasion, progression, metastasis and poor
prognosis (9–11). However, the functions and the
molecular mechanisms of HOTAIR in OSCC are still unknown. In the
present studies, we measured the expression of HOTAIR and the
relationship with clinicopathological factors and survival in OSCC
patients. We investigated the functions of HOTAIR involvement in
OSCC cell proliferation, invasion and apoptosis in vitro.
Interestingly, we found that HOTAIR recruits EZH2 and represses
E-cadherin in OSCC. In addition, the study results may assist to
reveal the mechanism of long non-coding RNA HOTAIR in OSCC and to
find the molecular targets to inhibit invasion and metastasis in
OSCC.
Materials and methods
OSCC patients and cell lines
OSCC samples and adjacent histological normal
tissues were obtained from 76 patients who were admitted to the
Department of Maxillofacial Surgery and and Otorhinolaryngology
Head and Neck Surgery of Tianjin Medical University between January
2001 and March 2009. None of the patients received treatment prior
to radical surgical treatment. Tumor tissues and non-malignant
tissues that were ≥1.5 cm distal to the tumor margins were
snap-frozen in liquid nitrogen and then stored at −80°C until use.
The clinicopathological characteristics of patients are summarized
in Table I. The study was approved
by the Institutional Ethics Committee of Tianjin Medical University
Cancer Institute and Hospital, and patients signed a statement of
informed consent before entry into the study.
 | Table IHOTAIR expression and
clinicopathological factors in OSCC. |
Table I
HOTAIR expression and
clinicopathological factors in OSCC.
| | HOTAIR
expression | |
|---|
| |
| |
|---|
| Clinicopathological
factors | n (%) | High | Low | P-value |
|---|
| Gender | | | | 0.479 |
| Male | 47 (62) | 22 | 25 | |
| Famale | 29 (38) | 16 | 13 | |
| Age (years) | | | | 0.243 |
| ≤60 | 45 (59) | 20 | 25 | |
| >60 | 31 (41) | 18 | 13 | |
| Smoking and/or
alcohol | | | | 0.238 |
| Yes | 47 (62) | 26 | 21 | |
| No | 29 (38) | 12 | 17 | |
| Location | | | | 0.065 |
| OTSCCa | 42 (55) | 25 | 17 | |
| OSCC | 34 (45) | 13 | 21 | |
| OTSCC
(excepted) |
| Tumor size
(cm) | | | | 0.758 |
| ≤2 | 21 (28) | 11 | 10 | |
| 2–4 | 41 (54) | 19 | 22 | |
| >4 | 14 (18) | 8 | 6 | |
| Histological
differentiation | | | | 0.019 |
| High | 29 (38) | 9 | 20 | |
| Median | 35 (46) | 20 | 15 | |
| Poor | 12 (16) | 9 | 3 | |
| Stage | | | | 0.001 |
| I–II | 32 (42) | 9 | 23 | |
| III–IV | 44 (58) | 29 | 15 | |
| Lymph node
metastasis | | | | 0.000 |
| N1 | 34 (45) | 26 | 8 | |
| N0 | 42 (55) | 12 | 30 | |
Two OSCC cell lines (TSCCA, Tca8223) were purchased
from the Institute of Basic Medical Sciences of Chinese Academy of
Medical Sciences (Beijing, China). CAL-27 cell line was purchased
from Cell Resource Center, IBMS, CAMS/PUMC (Beijing, China), and
Tb3.1 cell line was obtained from Shanghai Ninth People’s Hospital
(Shanghai, China). Cells were cultured in RPMI-1640, DMEM or MEM
(Invitrogen, CA, USA) medium supplemented with 10% fetal bovine
serum (FBS), (Hyclone, UT, USA) in humidified air with 5%
CO2 at 37°C.
RNA preparation, reverse transcription,
and quantitative real-time PCR
Total RNA was extracted from tissues or cultured
cells with TRIzol reagent (Invitrogen, NY, USA) according to the
manufacturer’s protocol. qRT-PCR assays were performed to detect
HOTAIR expression using the PrimeScript RT reagent kit and SYBR
Premix Ex Taq (Takara, Dalian, China) according to the
manufacturer’s instructions. Results were normalized to the
expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The
primers used were as follows: HOTAIR F:
5′-AAATATGGCGGCGTCTACACGGA-3′, R: 5′-TCCAGAACCCTCTGACATTTGCCT-3′;
E-cadherin F: 5′-TGCCCAGAAAATGAAAAAGG-3′, R:
5′-GTGTATGTGGCAATGCGTTC-3′; GAPDH F: 5′-GAAGGTGAAGGTCGGAGTC-3′, R:
5′-GAAGATGGTGATGGGATTTC-3′; EZH2 F: 5′-AATCAGAGTACATGCGACTGAGA-3′,
R: 5′-GCTGTATCCTTCGCTGTTTCC-3′. qRT-PCR and data collection were
performed on an ABI 7500 (Applied Biosystems, CA, USA). qRT-PCR
results were analyzed and expressed relative to CT (threshold
cycle) values, and then converted to fold changes.
Transfection of siRNA
Three individual FAM-siRNAs (siHOTAIR1, siHOTAIR2
and siHOTAIR3) siSUZ12 and siEZH2, negative control FAM-siRNA
(silencer negative control siRNA) were purchased from GenePharma
(Shanghai, China). siRNA oligonucleotides (10 nmol/l) in Opti-MEM
(Invitrogen, NY, USA) were transfected into cells using
Lipofectamine 2000 (Invitrogen, NY, USA) following the
manufacturer’s protocol. Transfection efficiencies were detected by
fluorescence microscope 6 and 48 h post-transfection, HOTAIR
expression levels were measured. Target sequences for HOTAIR siRNAs
were as follows: siHOTAIR1 sense, 5′-GCACAGAGCAACUCUAUAATT-3′;
antisense, 5′-UUAUAGAGUUGCUCUGUGCTT-3′. siHOTAIR2 sense,
5′-GCCUUUGGAAGCUCUUGAATT-3′; antisense, 5′-UUCAAGAGCUUCCAAGGCTT-3′.
siCT sense, 5′-UUCUCCGAACGUGUCACGUTT-3′; antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. siEZH2 sense,
5′-TTCATGCAACACCCAACACT-3′; siEZH2 antisense,
5′-GAGAGCAGCAGCAAACTCCT-3′.
Wound healing scratch assay
Cells were seeded in 6-well plates in normal cell
growth media and incubated to confluence. A 20-μl tip was used to
make a straight scratch, simulating a wound. The medium was changed
to MEM or 1640 containing 2% FBS. After 48-h incubation, the area
occupied by migrated cells in the scratch was evaluated.
Matrigel invasion assay
The Matrigel invasion assay was done using the BD
Biocoat Matrigel Invasion Chamber (pore size, 8 mm, 24-well; BD
Biosciences, CA, USA) following the manufacturer’s protocol. Cells
were plated in the upper chamber in serum-free medium. The bottom
chamber contained medium with 10% FBS. After 48 h, the bottom of
the chamber insert was stained with methanol and 0.1% crystal
violet, imaged, and counted using an IX70 inverted microscope
(Olympus, Tokyo, Japan). Each Matrigel invasion assay was conducted
in at least 3 replicates.
Flow cytometric analysis of
apoptosis
TSCCA and Tca8113 cells, transiently transfected
with siHOTAIR or siCT, were harvested 48 h after transfection by
trypsinization. After double staining with FITC-Annexin V and
propidium iodide, cells were analyzed by flow cytometry using
CellQuest software (BD Biosciences). Cells were discriminated into
viable cells, dead cells, early apoptotic cells, and apoptotic
cells, and then the relative ratio of apoptotic cells was compared
to the control from each experiment. All samples were assayed in
triplicate.
Colony formation assay
A total of 500 HOTAIR siRNA-transfected Tscca and
Tca8113 cells were placed in a fresh 6-well plate and maintained in
media containing 10% FBS, replacing the medium every 3 days. After
14 days, cells were fixed with methanol and stained with 0.1%
crystal violet. Visible colonies were manually counted. For each
treatment group wells were measured in triplicate.
Western blot analysis and antibodies
Cells were lysed using RIPA protein extraction
reagent (Beyotime, Shanghai, China) supplemented with a protease
inhibitor cocktail (Roche, Basel, Switzerland). Protein
concentration was measured using the Bio-Rad protein assay kit
(Bio-Rad, Beijing, China). Approximately 30 μg of protein extract
was separated by 10% SDS polyacrylamide gel electrophoresis, then
transferred to nitrocellulose membrane (Sigma, MO, USA) and
incubated with mouse anti-human E-cadherin, EZH2, H3K27me3 (Abcam,
MA, USA). ECL chromogenic substrate was used to visualize the bands
and the density of the band on the western blotting was measured
using the ChemiDoc XRS imaging system and QuantityOne software
(Bio-Rad). β-actin was used as a control.
Chromatin immunoprecipitation (ChIP)
assay
ChIP assay was performed with the EZ-Magna ChIP kit
(Millipore) according to the manufacturer’s instructions. For each
ChIP assay, 2 μg of antibodies were used: EZH2 (Cell Signaling
Technology), H3K27me3 (Abcam) and H3 (Cell Signaling Technology).
The percentage of the bound DNA was quantified against the original
DNA input by qRT-PCR analysis. Primer sequences used are as
follows: E-cadherin-a sense, 5′-TGTCCGCCCCGACTTGTCTCTC-3′;
antisense, 5′-GTCCTCTGGCCCCAGCCTCTCT-3′. E-cadherin-b sense,
5′-AGACCCCATCTCCAAAACGAACAAA-3′;antisense,5′-GCATAGACGCGGTGACCCTCTAGCC-3′.
E-cadherin-c sense, 5′-TGTCCGCCCCGACTTGTCTCTC-3′; antisense,
CGGTCCTCTGGCCCCAGCCTCT-3′.
Statistical analysis
Statistical tests for data analysis included the
Wilcoxon test, χ2 test, Fisher’s exact test and
Student’s two-tailed t-test. Kaplan-Meier analysis and log-rank
test were applied to evaluate the prognostic significance of HOTAIR
expression level in terms of patient survival. The statistical
analyses were performed using SPSS 19.0 statistical software
package (SPSS, Chicago, IL, USA). The data represent the mean ± SD.
P-values of P<0.05 were considered to be statistically
significant.
Results
HOTAIR is significantly upregulated in
human OSCC tissues
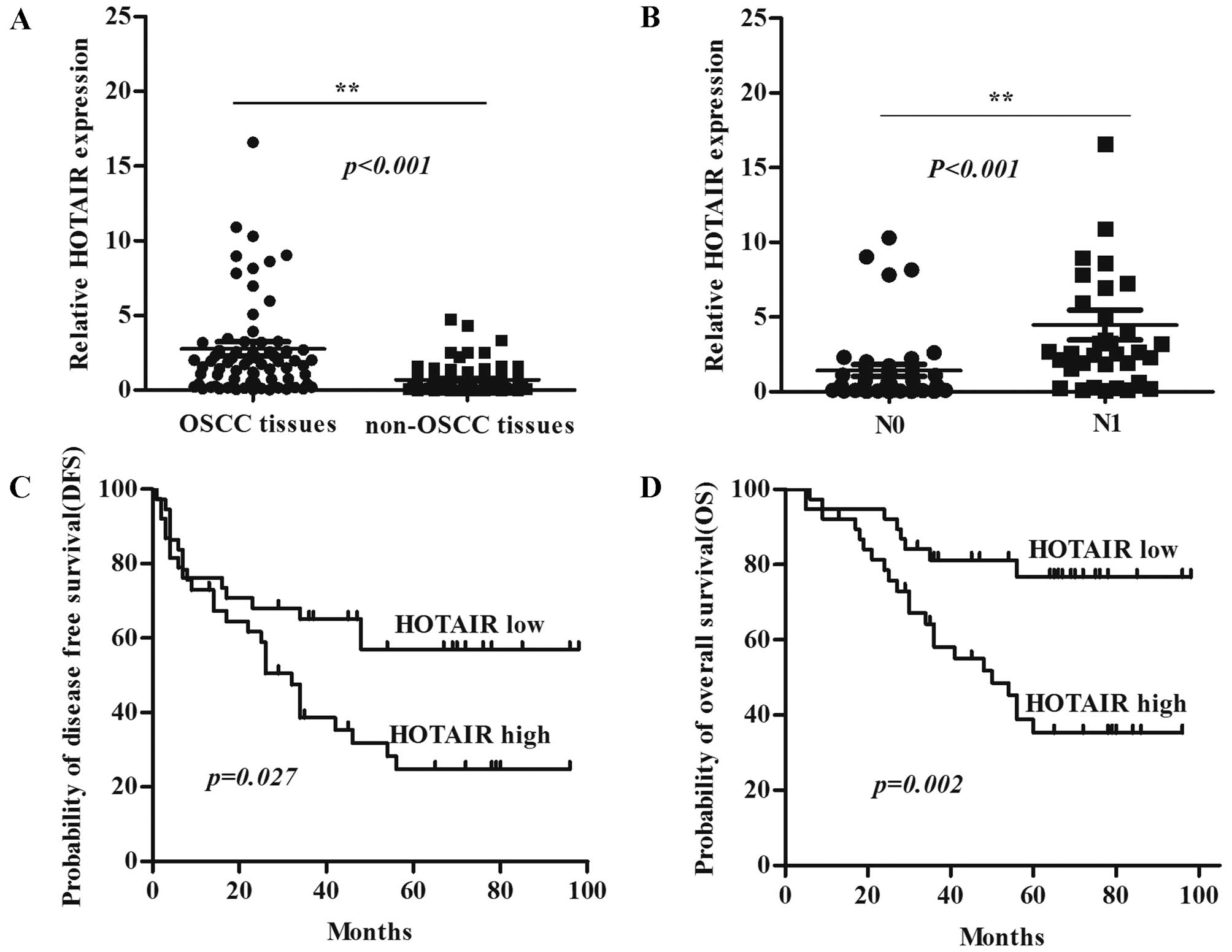
Expression level of HOTAIR was detected in 76 OSCC
samples and adjacent normal tissues by qRT-PCR and normalized to
GAPDH. HOTAIR was more highly expressed in OSCC tumor compared with
non-tumor tissues (P<0.001) (Fig.
1A), and HOTAIR was more highly expressed in tumors with
regional lymph node metastasis (N1) compared with tumors without
lymph node metastasis (N0) (P<0.001) (Fig. 1B). The univariate analysis revealed
that HOTAIR expression strongly correlated with the clinical stage,
lymph node metastasis, and histological differentiation in patients
with OSCC (Table I). Kaplan-Meier
survival analysis showed that patients with high HOTAIR expression
had significantly decreased disease-free survival (DFS) (Fig. 1C) and overall survival (OS)
(Fig. 1D) compared with patients
with low HOTAIR expression. These results suggest that the HOTAIR
is overexpressed in OSCC and is associated with development and
progression of OSCC patients. Moreover, HOTAIR may be a potential
predictive biomarker for disease outcome and prognosis in OSCC
patients.
HOTAIR promotes the malignancy of OSCC
cells in vivo
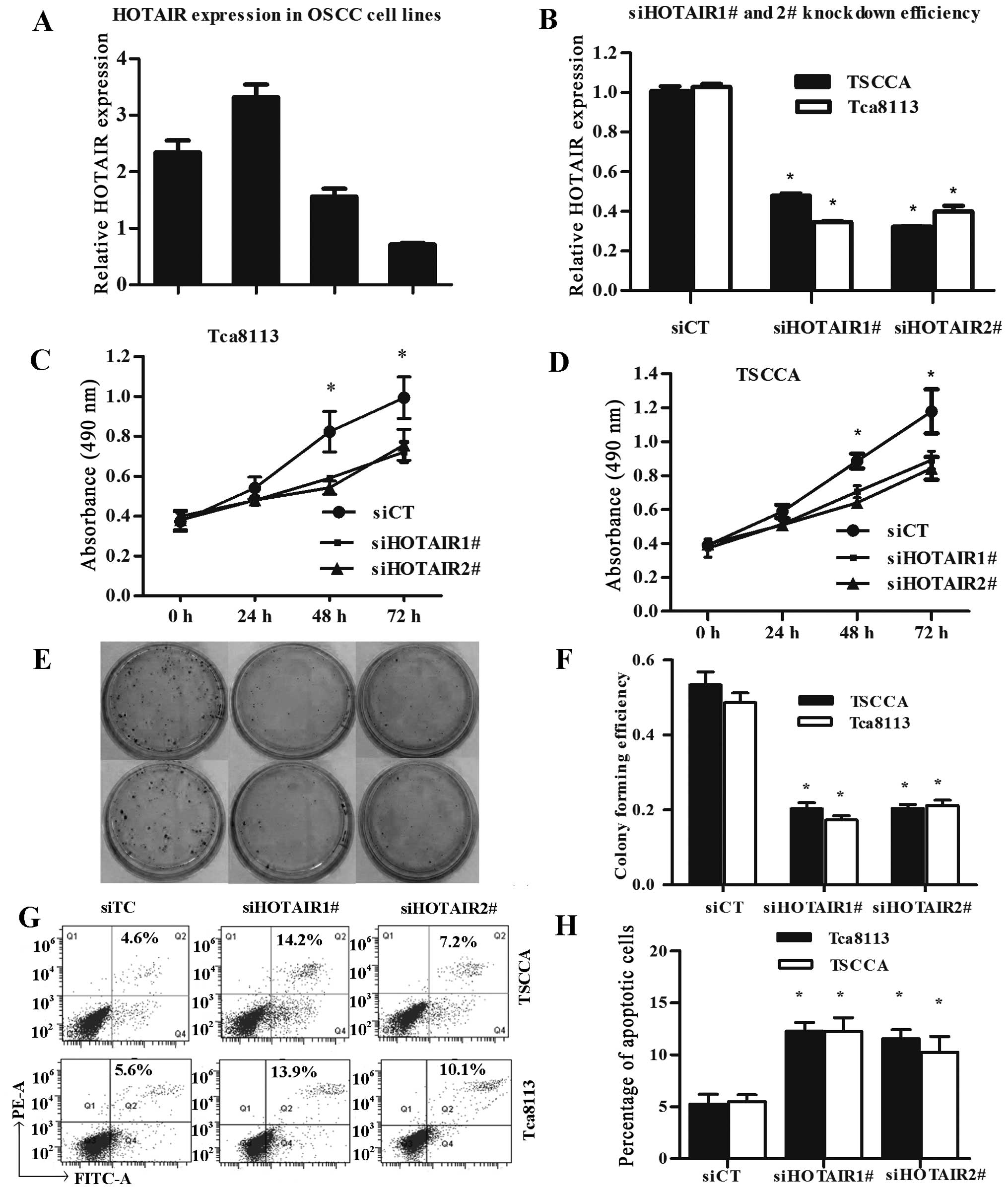
Next, we performed qRT-PCR analysis to examine the
expression level of HOTAIR in human OSCC cell lines. Of these 4
OSCC cell lines, TSCCA and Tca8113 expressed significantly higher
levels of HOTAIR (Fig. 2A). In
TSCCA and Tca8113 cells, HOTAIR knockdown was performed using two
different siRNAs (siHOTAIR1# and siHOTAIR2#), and siCT was
performed as control. Lipofectamine 2000 transfection reagent was
used for siRNA transfection. Forty-eight hours after treatment,
HOTAIR expression was effectively knocked down in TSCCA and Tca8113
cells (Fig. 2B).
Given that overexpression of HOTAIR was
significantly associated with progression in patients with OSCC we
examined whether HOTAIR regulated the proliferation of OSCC cells.
MTT assay was used to examine the effect of HOTAIR silence on the
proliferation of TSCCA and Tca8113 cells in vitro. We
estimated the proliferation rate of OSCC cells at 0, 24, 48 and 72
h after transfection, compared with the negative control, knockdown
of HOTAIR notably repressed the growth of OSCC cells (Fig. 2C and D). Furthermore, colony
formation assay indicated that the silence of HOTAIR expression
significantly reduced colonies of the OSCC cells (Fig. 2E and F).
To further assess the role of HOTAIR in apoptosis
and cell death of OSCC cells, we analyzed the HOTAIR knocked down
cells using flow cytometry. In addition, the results indicated that
knockdown of HOTAIR promoted both early apoptosis and later
apoptosis of OSCC cells Tca8113and TSCCA (Fig. 2G and H).
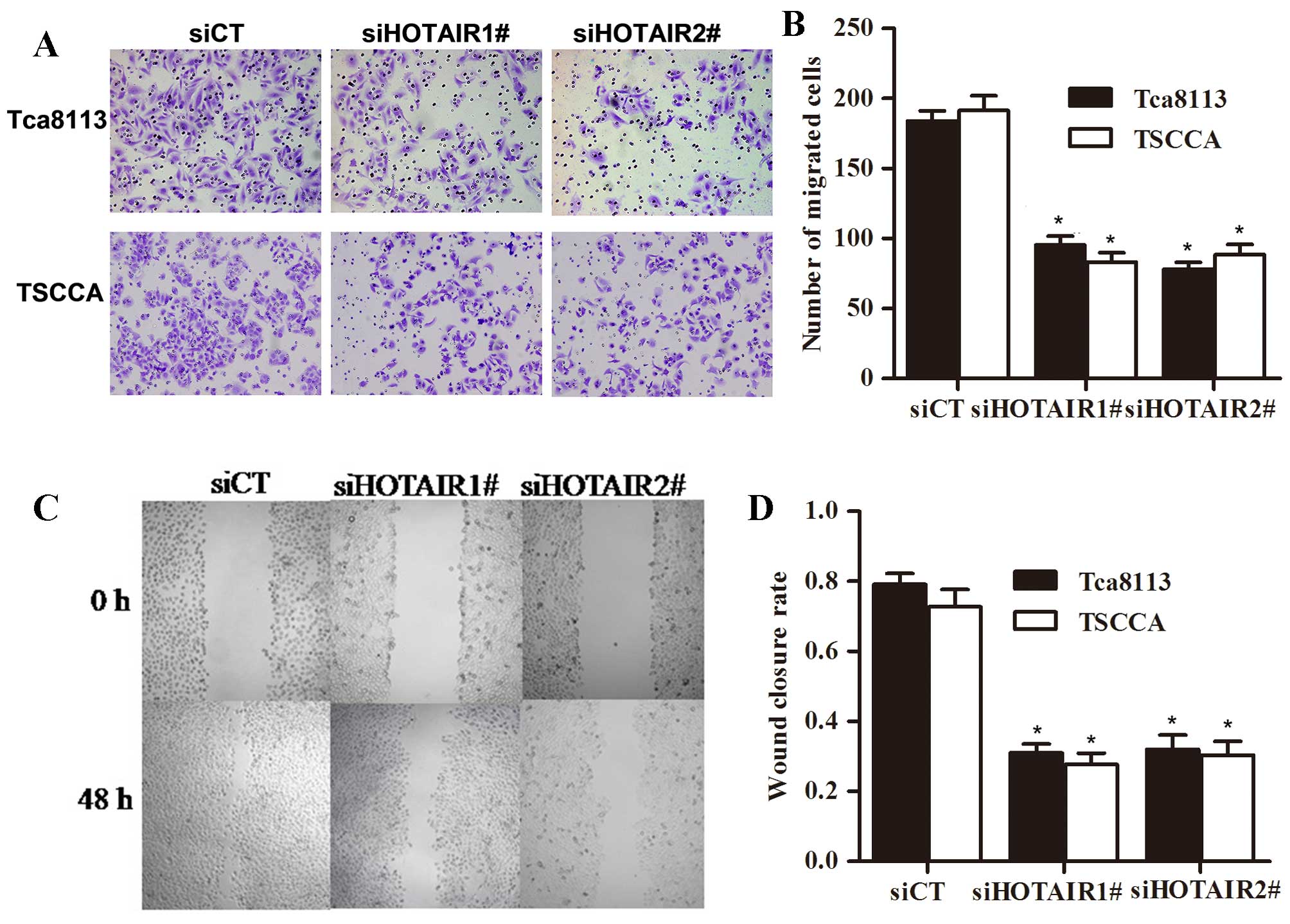
We use wound healing scratch assay and Matrigel
invasion assay to evaluate whether HOTAIR alters invasion and
migration of OSCC cells. As shown, silence of HOTAIR decreased
their migration and invasion ability. Conversely, the 2 OSCC cell
lines Tca8113 and TSCCA treated with siCT did not affect the cell
invasion and migration. Based on our results, expression of HOTAIR
promoted migration and invasion of OSCC cells in vitro
(Fig. 3).
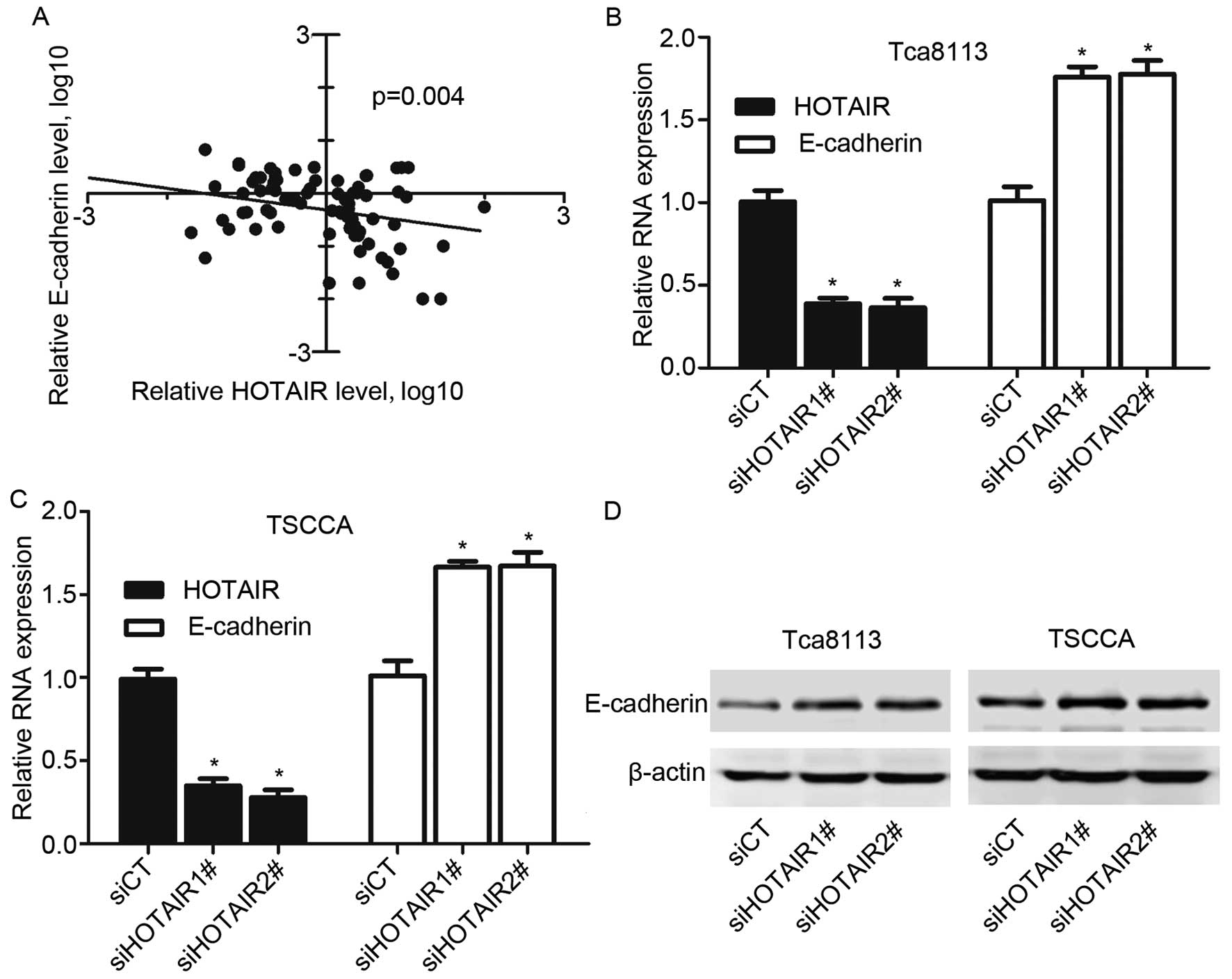
HOTAIR negatively correlates to
E-cadherin expression in OSCC tissues and cell lines
To define functional correlation between HOTAIR and
epithelial-mesenchymal transition (EMT), we examined the effects of
HOTAIR knockdown on E-cadherin expression. We first assayed the
expression level of HOTAIR and E-cadherin in OSCC tissues. A
significant negative correlation is observed between the E-cadherin
mRNA levels and the HOTAIR expression levels in OSCC tissues
(r2=−0.327, P=0.004) (Fig.
4A). Then we investigated whether HOTAIR regulates E-cadherin
expression in OSCC cells. Silence of HOTAIR significantly increased
E-cadherin expression at both transcript and protein levels in
TSCCA and Tca8113 cells (Fig. 4B and
C). These results demonstrated that HOTAIR promotes EMT in OSCC
tissues and cell lines.
HOTAIR represses E-cadherin expression by
associating with EZH2
HOTAIR is thought to regulate transcription by
directing the action of PRC2 complexes to control the epigenetic
state of the cell and subsequently regulate gene expression. We
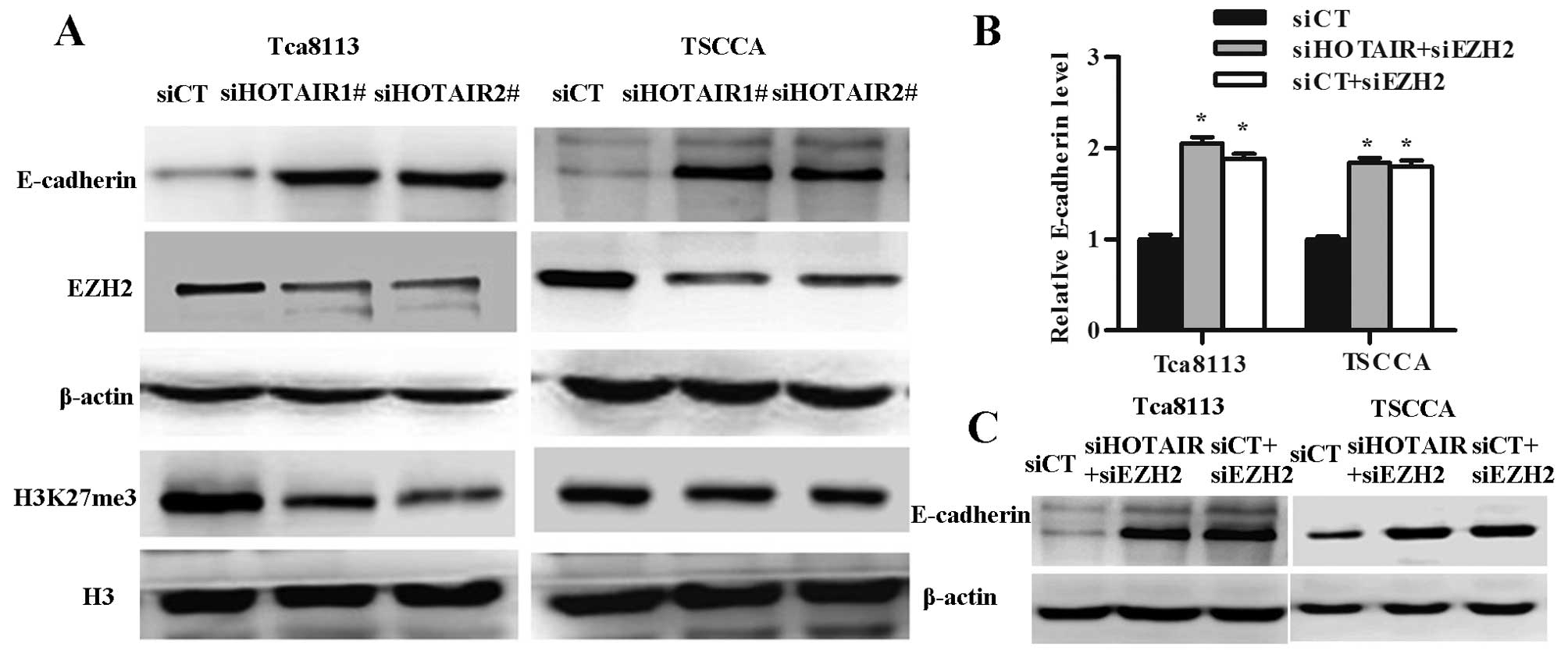
performed western blotting to test the enrichment of EZH2 and
H3K27me3, and expression of E-cadherin in OSCC cell lines treated
with HOTAIR siRNA. We observed enrichment of EZH2 and H3K27me3
significantly decreased and with the expression of E-cadherin
significantly increased in the HOTAIR knockdown OSCC cells
(Fig. 5A).
To check whether expression of the E-cadherin was
controlled by EZH2, we analyzed the E-cadherin expression after
EZH2 and/or HOTAIR knockdown. E-cadherin mRNA and protein levels
increased in Tca8113 and TSCCA cells treated with siEZH2 and
siEZH2+siHOTAIR groups (Fig. 5B and
C).
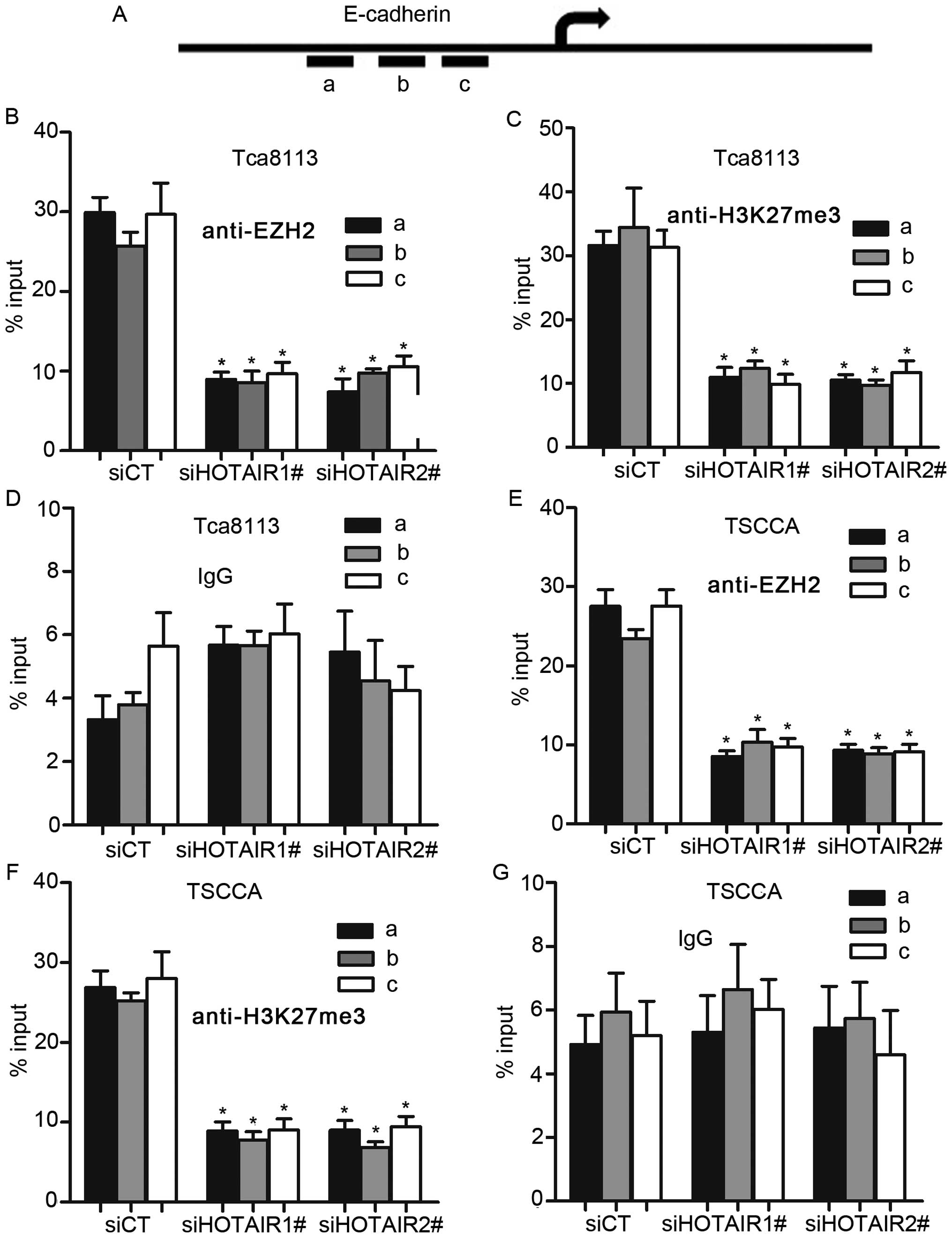
To further address how HOTAIR regulated E-cadherin
through enrichment of EZH2, we performed ChIP-arrays in OSCC cell
lines with HOTAIR knockdown. In addition, results showed that
HOTAIR silence decreased the binding of EZH2 and H3K27me3 with the
E-cadherin promoter in OSCC cells, whereas, we did not detect the
binding of IgG with E-cadherin promoter after HOTAIR inhibition
(Fig. 6B–G). These results suggest
that HOTAIR represses E-cadherin expression partly by associating
with EZH2.
Discussion
Evidence has shown that HOTAIR can act as a key
regulation factor in the molecular mechanisms underlying the
development and progression of cancer (12,13),
therefore potentially leading to new strategies for cancer therapy.
In addition, in the clinic, it is necessary to solve the problems
to improve the treatment outcome in patients of OSCC, therefore
exploring new strategies for molecular targeted therapy is urgent
needed. As a novel lncRNA, HOTAIR initially become well-known for
its involvement in cancer invasiveness and metastasis (10,14–17),
and is expected to be the most potential treatment target in
OSCC.
Several reports have suggested that HOTAIR is
upregulated in breast cancer, colorectal cancer, esophageal
squamous cell carcinoma (ESCC) and promotes cancer invasion and
metastasis and poor survival (10,14,16–21).
However, this study is the first to demonstrate the regulation of
HOTAIR in OSCC and revealed that HOTAIR expression is correlated
with the aggressive biological behavior of OSCC. In the present
study, HOTAIR is overexpressed in OSCC tissues and cell lines. In
addition, the patients with high HOTAIR expression had more
cervical lymph node metastasis, more advanced stage and poorer
histological differentiation than those with low HOTAIR expression.
However, other clinicopathological factors, such as tumor size,
age, gender and smoking and/or alcohol abuse, were not correlated
with the expression of HOTAIR. In addition, it is known that
patients with oral tongue squamous cell carcinoma (OTSCC) have a
significantly worse prognosis than those with similar lesions of
the other oral cavity sites (22),
however, the expression of HOTAIR did not show a significant
difference between the two groups. Besides, we found that the
patients with high level of HOTAIR had lower OS rate and DFS rate
which suggested that overexpression of HOTAIR might be an indicator
of poor outcome in OSCC patients.
Emerging evidence suggests that HOTAIR regulates
proliferation, invasion and apoptosis of a variety of tumor cells
in vitro (15,18,23,24).
Enforced expression of HOTAIR in breast carcinoma cells promoted
cell invasion (10), knockdown of
HOTAIR reduced pancreatic cancer cell invasion, inhibited cell
growth, modulated cell cycle progression and induced apoptosis
(14), and the results were
confirmed in colorectal cancer (16), non-small cell lung cancer (17,25,26),
gastric cancer (27–29) and other cancers (15,20,30–32).
To further understand the biological function of HOTAIR in OSCC
progression, in vitro assays were performed in the OSCC cell
lines TSCCA and Tca8113. Our data showed that knockdown of HOTAIR
mediated by siRNA led to significant decrease in proliferation,
colony forming efficiency and invasive ability and significant
increase in apoptosis in OSCC cells.
Determination of the targeted proteins associated
with HOTAIR and the genes regulated by HOTAIR would reveal the
molecular mechanisms underlying cancer development and progression
by HOTAIR. HOTAIR functions in the recruitment and binding of the
PRC2 and LSD1 complexes to the HOXD locus on chromosome 2 where
genes involved in metastasis are regulated through H3K27
methylation and H3K4 demethylation (9,33).
Reports have revealed that HOTAIR led to decreased expression of
some mesenchymal markers, while the expression of epithelial
markers (such as E-cadherin) were increased in various cancers
(20,34,35).
EZH2, a unit of the PRC2 complex, has been reported to downregulate
the level of E-cadherin gene expression through H3K27me3 in the
E-cadherin promoter (36–38). Thus, we speculate HOTAIR induces
EMT related markers through EZH2. In our study, we found a
significantly negative correlation between HOTAIR levels and
E-cadherin levels in OSCC tissues and cell lines. In addition,
knocking down of EZH2, but not changing the HOTAIR expression level
could also upregulate E-cadherin expression in vivo.
Furthermore, we performed ChIP analysis in HOTAIR-silenced OSCC
cell lines, and results showed that HOTAIR contributes to the
binding of EZH2 and H3K27me3 with the E-cadherin promoter.
In conclusion, this study has shown that HOTAIR,
defined as an oncogene is overexpressed in OSCC patients and
indicates poor prognosis, and poor outcome of patients with high
HOTAIR expression due to the ability of HOTAIR to promote cancer
development and progression by inducing cancer cell proliferation
and invasion and reducing cancer cell apoptosis. Moreover, HOTAIR
regulates EMT related marker E-cadherin through recruitment of EZH2
and H3K27me3. Therefore, these findings indicate that HOTAIR plays
a vital role in the development and progression of OSCC and
downregulation of such lncRNAs could be a valuable predictive
marker as well as novel therapeutic target in future OSCC
treatment.
Acknowledgements
This study was supported by National Basic Research
Program of China (973 Program), no. 2010CB529301 (to Xishan Hao)
and supported in part by the China National Natural Scientific Fund
(81172573) (to Lun Zhang).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Molinolo AA, Amornphimoltham P, Squarize
CH, Castilho RM, Patel V and Gutkind JS: Dysregulated molecular
networks in head and neck carcinogenesis. Oral Oncol. 45:324–334.
2009. View Article : Google Scholar :
|
|
3
|
Sabour S and Moezizadeh M: Prediction of
OSCC using biomarkers: Methodological mistake. Oral Oncol.
48:e512012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Raj LSM, Boaz K and Natarajan S:
Prognostic significance of lymph node pattern in oral squamous cell
carcinoma (OSCC). J Clin Diagn Res. 8:232–235. 2014.
|
|
5
|
Grimm M: Prognostic value of
clinicopathological parameters and outcome in 484 patients with
oral squamous cell carcinoma: Microvascular invasion (V+) is an
independent prognostic factor for OSCC. Clin Transl Oncol.
14:870–880. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee JT: Epigenetic regulation by long
noncoding RNAs. Science. 338:1435–1439. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Woo CJ and Kingston RE: HOTAIR lifts
noncoding RNAs to new levels. Cell. 129:1257–1259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun L and Fang J: Writer meets eraser in
HOTAIR. Acta Biochim Biophys Sin (Shanghai). 43:1–3. 2011.
View Article : Google Scholar
|
|
14
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
15
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion, and
metastatic potential of metastatic melanoma. BioMed Res Int.
2013:2510982013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu XH, Liu ZL, Sun M, Liu J, Wang ZX and
De W: The long non-coding RNA HOTAIR indicates a poor prognosis and
promotes metastasis in non-small cell lung cancer. BMC Cancer.
13:4642013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ono H, Motoi N, Nagano H, Miyauchi E,
Ushijima M, Matsuura M, Okumura S, Nishio M, Hirose T, Inase N, et
al: Long noncoding RNA HOTAIR is relevant to cellular
proliferation, invasiveness, and clinical relapse in small-cell
lung cancer. Cancer Med. 3:632–642. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li X, Wu Z, Mei Q, Li X, Guo M, Fu X and
Han W: Long non-coding RNA HOTAIR, a driver of malignancy, predicts
negative prognosis and exhibits oncogenic activity in oesophageal
squamous cell carcinoma. Br J Cancer. 109:2266–2278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Qiu JJ, Lin YY, Ye LC, Ding JX, Feng WW,
Jin HY, Zhang Y, Li Q and Hua KQ: Overexpression of long non-coding
RNA HOTAIR predicts poor patient prognosis and promotes tumor
metastasis in epithelial ovarian cancer. Gynecol Oncol.
134:121–128. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen FJ, Sun M, Li SQ, Wu QQ, Ji L, Liu
ZL, Zhou GZ, Cao G, Jin L, Xie HW, et al: Upregulation of the long
non-coding RNA HOTAIR promotes esophageal squamous cell carcinoma
metastasis and poor prognosis. Mol Carcinog. 52:908–915. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bello IO, Soini Y and Salo T: Prognostic
evaluation of oral tongue cancer: Means, markers and perspectives
(I). Oral Oncol. 46:630–635. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bhan A, Hussain I, Ansari KI, Bobzean SA,
Perrotti LI and Mandal SS: Bisphenol-A and diethylstilbestrol
exposure induces the expression of breast cancer associated long
noncoding RNA HOTAIR in vitro and in vivo. J Steroid Biochem Mol
Biol. 141:160–170. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
He X, Bao W, Li X, Chen Z, Che Q, Wang H
and Wan XP: The long non-coding RNA HOTAIR is upregulated in
endometrial carcinoma and correlates with poor prognosis. Int J Mol
Med. 33:325–332. 2014.
|
|
25
|
Zhuang Y, Wang X, Nguyen HT, Zhuo Y, Cui
X, Fewell C, Flemington EK and Shan B: Induction of long intergenic
non-coding RNA HOTAIR in lung cancer cells by type I collagen. J
Hematol Oncol. 6:352013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nakagawa T, Endo H, Yokoyama M, Abe J,
Tamai K, Tanaka N, Sato I, Takahashi S, Kondo T and Satoh K: Large
noncoding RNA HOTAIR enhances aggressive biological behavior and is
associated with short disease-free survival in human non-small cell
lung cancer. Biochem Biophys Res Commun. 436:319–324. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Endo H, Shiroki T, Nakagawa T, Yokoyama M,
Tamai K, Yamanami H, Fujiya T, Sato I, Yamaguchi K, Tanaka N, et
al: Enhanced expression of long non-coding RNA HOTAIR is associated
with the development of gastric cancer. PLoS One. 8:e770702013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ,
Huang L, Yu PF and Cheng XD: Knockdown of long non-coding RNA
HOTAIR suppresses tumor invasion and reverses
epithelial-mesenchymal transition in gastric cancer. Int J Biol
Sci. 9:587–597. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hajjari M, Behmanesh M, Sadeghizadeh M and
Zeinoddini M: Up-regulation of HOTAIR long non-coding RNA in human
gastric adenocarcinoma tissues. Med Oncol. 30:6702013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ge XS, Ma HJ, Zheng XH, Ruan HL, Liao XY,
Xue WQ, Chen YB, Zhang Y and Jia WH: HOTAIR, a prognostic factor in
esophageal squamous cell carcinoma, inhibits WIF-1 expression and
activates Wnt pathway. Cancer Sci. 104:1675–1682. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J
and Liu M: Long intergenic noncoding RNA HOTAIR is overexpressed
and regulates PTEN methylation in laryngeal squamous cell
carcinoma. Am J Pathol. 182:64–70. 2013. View Article : Google Scholar
|
|
32
|
Niinuma T, Suzuki H, Nojima M, Nosho K,
Yamamoto H, Takamaru H, Yamamoto E, Maruyama R, Nobuoka T, Miyazaki
Y, et al: Upregulation of miR-196a and HOTAIR drive malignant
character in gastrointestinal stromal tumors. Cancer Res.
72:1126–1136. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu L, Murat P, Matak-Vinkovic D, Murrell A
and Balasubramanian S: Binding interactions between long noncoding
RNA HOTAIR and PRC2 proteins. Biochemistry. 52:9519–9527. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Cai K, Wang J, Wang X, Cheng K,
Shi F, Jiang L, Zhang Y and Dou J: MiR-7, inhibited indirectly by
lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of
breast cancer stem cells by downregulating the STAT3 pathway. Stem
Cells. 32:2858–2868. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pádua Alves C, Fonseca AS, Muys BR, de
Barros E, Lima Bueno R, Bürger MC, de Souza JE, Valente V, Zago MA
and Silva WA Jr: Brief report: The lincRNA Hotair is required for
epithelial-to-mesenchymal transition and stemness maintenance of
cancer cell lines. Stem Cells. 31:2827–2832. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang C, Liu X, Chen Z, Huang H, Jin Y,
Kolokythas A, Wang A, Dai Y, Wong DT and Zhou X: Polycomb group
protein EZH2-mediated E-cadherin repression promotes metastasis of
oral tongue squamous cell carcinoma. Mol Carcinog. 52:229–236.
2013. View Article : Google Scholar
|
|
37
|
Liu L, Xu Z, Zhong L, Wang H, Jiang S,
Long Q, Xu J and Guo J: EZH2 promotes tumor cell migration and
invasion via epigenetic repression of E-cadherin in renal cell
carcinoma. BJU Int. Feb 25–2014.(Epub ahead of print). View Article : Google Scholar
|
|
38
|
Sun NX, Ye C, Zhao Q, Zhang Q, Xu C, Wang
SB, Jin ZJ, Sun SH, Wang F and Li W: Long noncoding RNA-EBIC
promotes tumor cell invasion by binding to EZH2 and repressing
E-cadherin in cervical cancer. PLoS One. 9:e1003402014. View Article : Google Scholar : PubMed/NCBI
|