Introduction
In developed countries, endometrial cancer (EC) is
the most common malignancy among women, accounting for ~25% of all
deaths related to cancer of the female genital tract (1). Unopposed estrogen therapy, obesity,
nulliparity, diabetes mellitus and arterial hypertension have been
linked to an increased risk of ECs (2). ECs are clinicohistologically
classified into two subgroups: type I and type II. Type I tumors,
which account for ~80% of all cases, are estrogen-dependent,
low-grade tumors, while type II tumors are more aggressive and
exhibit invasion into the myometrium (3,4).
Currently, there is a lack of effective treatments for patients
with advanced stage and recurrent EC (5); thus, more effective treatment
strategies based on genomic data are needed.
In the post-genome sequencing era, the discovery of
non-coding RNA (ncRNA) has been a conceptual breakthrough in cancer
research fields (6). For example,
microRNAs (miRNAs) are small ncRNA molecules (19–22 bases in
length) that function to regulate the expression of multiple
protein-coding genes by repressing translation or cleaving RNA
transcripts in a sequence-specific manner (7,8).
Bioinformatic predictions indicate that miRNAs regulate 30–60% (or
more) of the protein-coding genes in the human genome. Numerous
studies have reported that various miRNAs are aberrantly expressed
in many types of human cancers, affecting the development and
metastasis of cancers through oncogenic or tumor-suppressive
functions (9,10).
Elucidation of cancer-related miRNA networks has
provided important new information about human cancers. In our
previous studies, we used our miRNA expression signatures to
investigate several tumor-suppressive miRNAs and their regulated
cancer pathways. We recently showed that miR-1/133a
clustered miRNAs are significantly down-regulated in several cancer
tissues (11–14). From our miRNA signatures, we have
sequentially reported functional roles of the miR-1/133a
cluster and the molecular targets/pathways regulated by these
miRNAs. However, the contributions of these miRNAs in EC cells have
not been fully elucidated.
The aim of the present study was to investigate the
functional significance of the miR-1/133a cluster and to
identify the molecular targets regulated by these miRNAs in EC
cells. We found that restoration of mature miR-1 or
miR-133a in EC cells significantly inhibited cell migration
and invasion. Gene expression data and in silico analysis
demonstrated that phosphodiesterase 7A (PDE7A), an enzyme
that hydrolyzes intracellular cAMP, was a potential target of the
miR-1/133a cluster. Elucidation of the cancer-related
signaling pathways and targets regulated by the tumor-suppressive
miR-1/133a cluster will provide new insights into the
potential mechanisms of EC oncogenesis and metastasis.
Materials and methods
Clinical specimens
A total of 27 primary EC specimens were collected
from patients who had undergone surgical treatment at Chiba
University Hospital. Eight non-cancer endometrial specimens were
obtained from patients who underwent total hysterectomy because of
other gynecologic diseases (Table
I). The samples were processed and stored in liquid nitrogen
until RNA extraction. Our study was approved by the Bioethics
Committee of Chiba University; prior written informed consent and
approval was given by each patient.
 | Table ICharacteristics of endometrial cancer
specimens and non-cancer specimens. |
Table I
Characteristics of endometrial cancer
specimens and non-cancer specimens.
| Sample no. |
|---|
| (a) Endometrial
cancer |
| Total no. | 27 |
| Median age, years
(range) | 58 (39–80) |
| Pathological tumor
stage (UICC7th) |
| 1A | 13 (48.1%) |
| 1B | 5 (18.5%) |
| 2 | 1 (3.7%) |
| 3A | 2 (7.4%) |
| 3B | 1 (3.7%) |
| 3C1 | 2 (7.4%) |
| 3C2 | 2 (7.4%) |
| 4 | 1 (3.7%) |
|
Differentiation |
| G1 | 7 (25.9%) |
| G2 | 10 (37.0%) |
| G3 | 10 (37.0%) |
| Lymphatic
metastasis |
| (+) | 4 (14.8%) |
| (−) | 20 (74.1%) |
| Unknown | 3 (11.1%) |
| (b) Normal
endometrium |
| Total no. | 8 |
| Median age, years
(range) | 41 (34–76) |
Cell lines and cell culture
Hec1B cells (derived from endometrioid
adenocarcinoma G1) and Hec265 cells (derived from endometrioid
adenocarcinoma G2) were used in this analysis. Hec1B cells were
grown in E-MEM medium supplemented with 10% fetal bovine serum.
Hec265 cells were grown in E-MEM medium supplemented with 15% fetal
bovine serum. All cells were cultured at 37°C in a humidified
atmosphere containing 5% CO2.
RNA isolation
Total RNA was isolated using TRIzol reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
protocol. RNA concentrations were determined
spectrophotometrically. RNA quality was confirmed using a NanoDrop
1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA,
USA).
Quantitative real-time reverse
transcription polymerase chain reaction (qRT-PCR)
First-strand cDNA was synthesized from 1 μg of total
RNA using a High Capacity cDNA Reverse Transcription kit (Applied
Biosystems, Foster City, CA, USA). Gene-specific PCR products were
assayed continuously using a 7300-HT Real-Time PCR system according
to the manufacturer’s protocol. The initial PCR step consisted of a
10- min hold at 95°C, followed by 40 cycles consisting of
denaturation for 15 sec at 95°C and annealing/extension for 1 min
at 60°C.
The expression levels of miR-1 (assay ID:
002222) and miR-133a (assay ID: 0002246) were analyzed by
TaqMan qPCR (TaqMan MicroRNA assay; Applied Biosystems) and
normalized to RNU48 (assay ID: 001006). All reactions were
performed in duplicate. TaqMan probes and primers for PDE7A
(P/N: Hs00300285_m1), DDX3X (P/N: Hs00606179_m1),
CORO1C (P/N: Hs00170938_m1), SPTBN1 (P/N:
Hs00162271_m1) and GUSB (P/N: Hs00939627_m1; used as an
internal control) were obtained from Applied Biosystems
(Assay-On-Demand Gene Expression Products). All reactions were
performed in triplicate and included negative control reactions
that lacked cDNA. The ΔΔCt method was adopted and applied to
calculate the relative quantities of target genes.
Transfections with mature miRNA and
small-interfering RNA (siRNA)
Cells were transfected with 10 nM mature miRNA or
siRNA molecules using Lipofectamine RNAiMAX transfection reagent
(Invitrogen) and Opti-MEM (Invitrogen). The following RNA species
were used in this study: mature miRNA, Pre-miR™ miRNA Precursors
(hsa-miR-1; P/N: PM10617, hsa-miR-133a; P/N: PM10413;
Applied Biosystems), negative control miRNA (P/N: AM17111; Applied
Biosystems), siRNA (Stealth siRNAs, si-PDE7A, P/N: HSS107737
and HSS107739; Invitrogen) and negative control siRNA (Stealth RNAi
Negative Control Med GC, P/N: 12935-300; Invitrogen).
Cell proliferation, migration and
invasion assays
For cell proliferation assays, cells were
transfected with 10 nM miRNA or siRNA by reverse transfection and
plated in 96-well plates at 3×103 cells per well. After
72 h, cell proliferation was determined with XTT assays using a
Cell Proliferation Kit II (Roche Applied Science, Tokyo,
Japan).
Cell migration assay
Modified Boyden Chambers (Trans-wells, no. 3422;
Corning, NY, USA) were used. Cells were transfected with 10 nM
miRNA or siRNA by reverse transfection and plated in 10-cm dishes
at 8×105 cells/dish. After 48 h, 1×105 cells
were added to the upper chamber of each migration well and were
allowed to migrate for 48 h. After gentle removal of the
non-migratory cells from the filter surface of the upper chamber,
the cells that migrated to the lower side were fixed and stained
with Diff-Quick (no. 16920; Sysmex Corp., Japan). The number of
cells migrating to the lower surface was determined microscopically
by counting four areas of constant size per well. Cell invasion
assays were carried out using modified Boyden chambers in 24-well
tissue culture plates at 1×105 cells per well (Matrigel
invasion chamber, no. 354480; BD Biocoat, USA). All experiments
were performed in duplicate.
Search for miR-1 and miR-133a target
genes
To identify putative miR-1- and
miR-133a-regulated genes, we searched the TargetScan
database (http://www.targetscan.org) for genes
having conserved sites for both miR-1 and miR-133a.
Then, we analyzed gene expression using the GEO database. Gene
expression data for clinical EC specimens were entered into the GEO
database (accession no. GSE17025). The procedure used for the
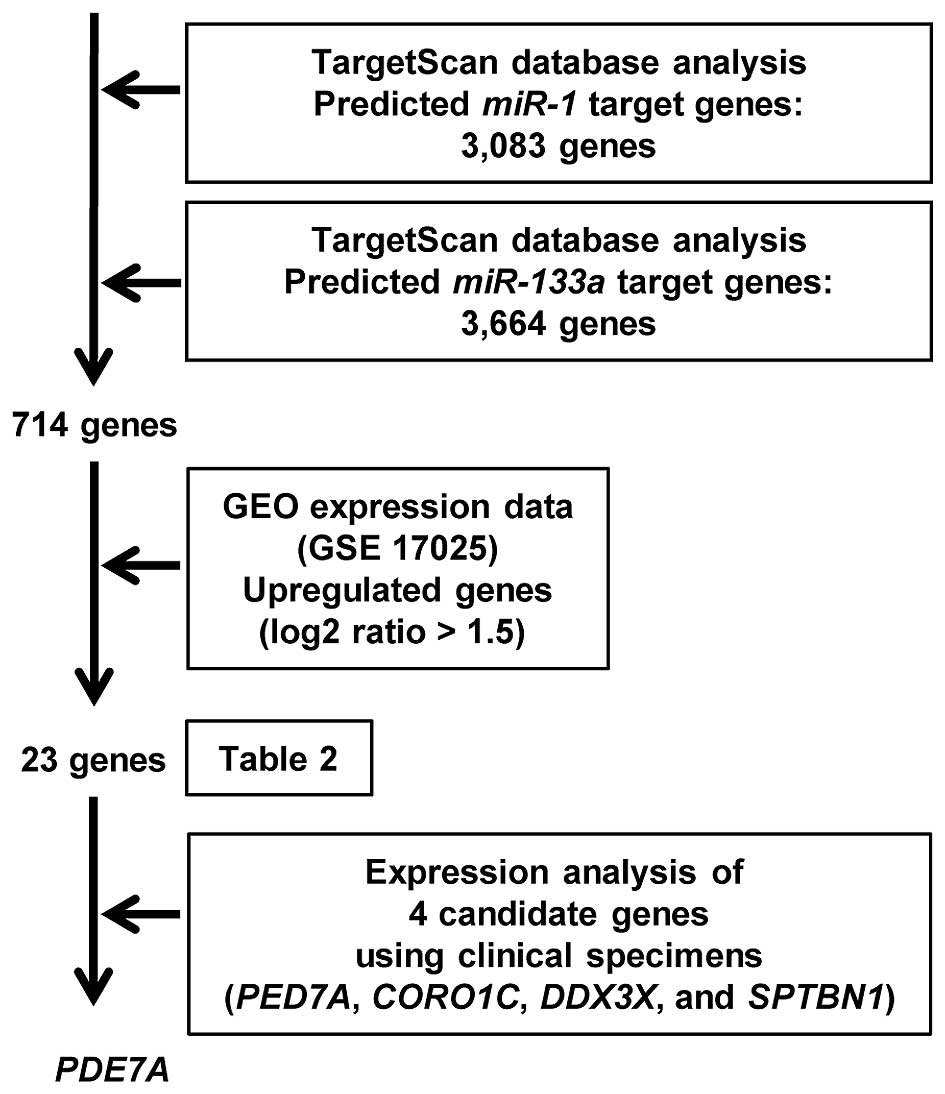
selection of miR-1 and miR-133a genes is shown in Fig. 3.
Western blot analysis
Cells were harvested and lysed 72 h after
transfection. Cell lysates (50 μg of protein each) were separated
using Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA), followed
by subsequent transfer to PVDF membranes. Immunoblotting was
performed with polyclonal anti-PDE7A antibodies (ab154857; Abcam,
Cambridge, UK). Anti-GAPDH antibodies (ab8245; Abcam) were used as
an internal control.
Plasmid construction and dual-luciferase
reporter assays
Partial sequences of the PDE7A 3′
untranslated region (3′UTR) containing the miR-1 and
miR-133a target sites were inserted between the XhoI
and PmeI restriction sites in the 3′UTR of the hRluc
gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA).
Hec265 cells were then transfected with 5 ng vector or 10 nM mature
miRNA.
Statistical analysis
The relationships between 2 variables and numerical
values were analyzed using the Mann-Whitney U test, and the
relationships between 3 variables and numerical values were
analyzed using the Bonferroni-adjusted Mann-Whitney U test. Expert
StatView analysis software (ver. 4; SAS Institute Inc., Cary, NC,
USA) was used in both analyses. In the comparison of 3 variables,
an unadjusted statistical level of significance of P<0.05
corresponded to the Bonferroni-adjusted level of P<0.0083.
Results
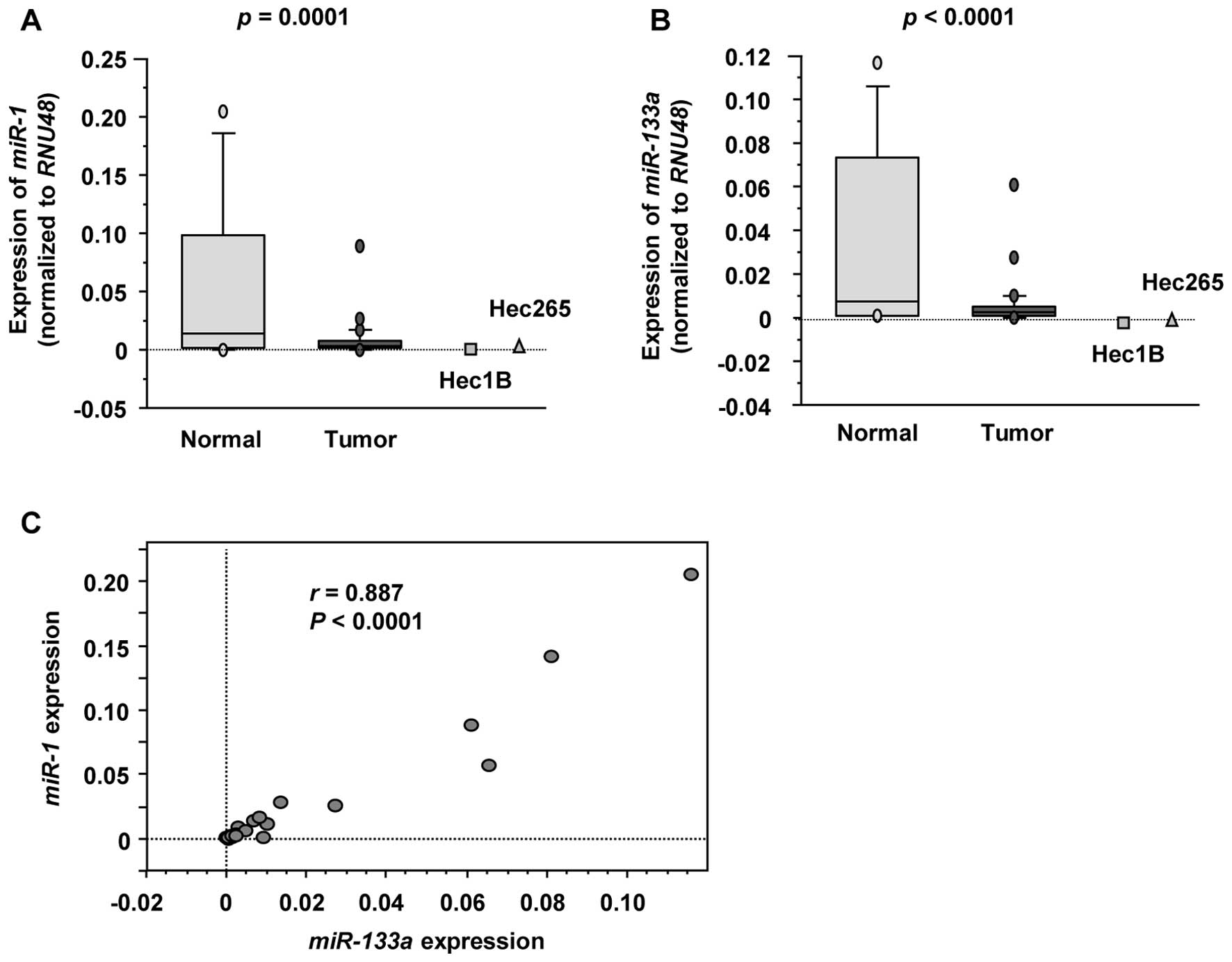
Expression levels of miR-1 and miR-133a
in EC specimens and cell lines
To validate our previous miRNA expression
signatures, we evaluated the expression levels of miR-1 and
miR-133a in 27 EC specimens and 8 non-cancer endometrial
specimens. The backgrounds and clinicopathological characteristics
of patients are summarized in Table
I. Quantitative stem-loop RT-PCR demonstrated that miR-1
and miR-133a expression levels were significantly lower in
cancer specimens compared with non-cancer specimens (P<0.0001;
Fig. 1A and B, respectively). The
expression levels of miR-1 and miR-133a were also
reduced in 2 EC cell lines (Hec1B and Hec265). Spearman’s rank test
showed a positive correlation between the expression of
miR-1 and that of miR-133a (r=0.887, P<0.0001;
Fig. 1C).
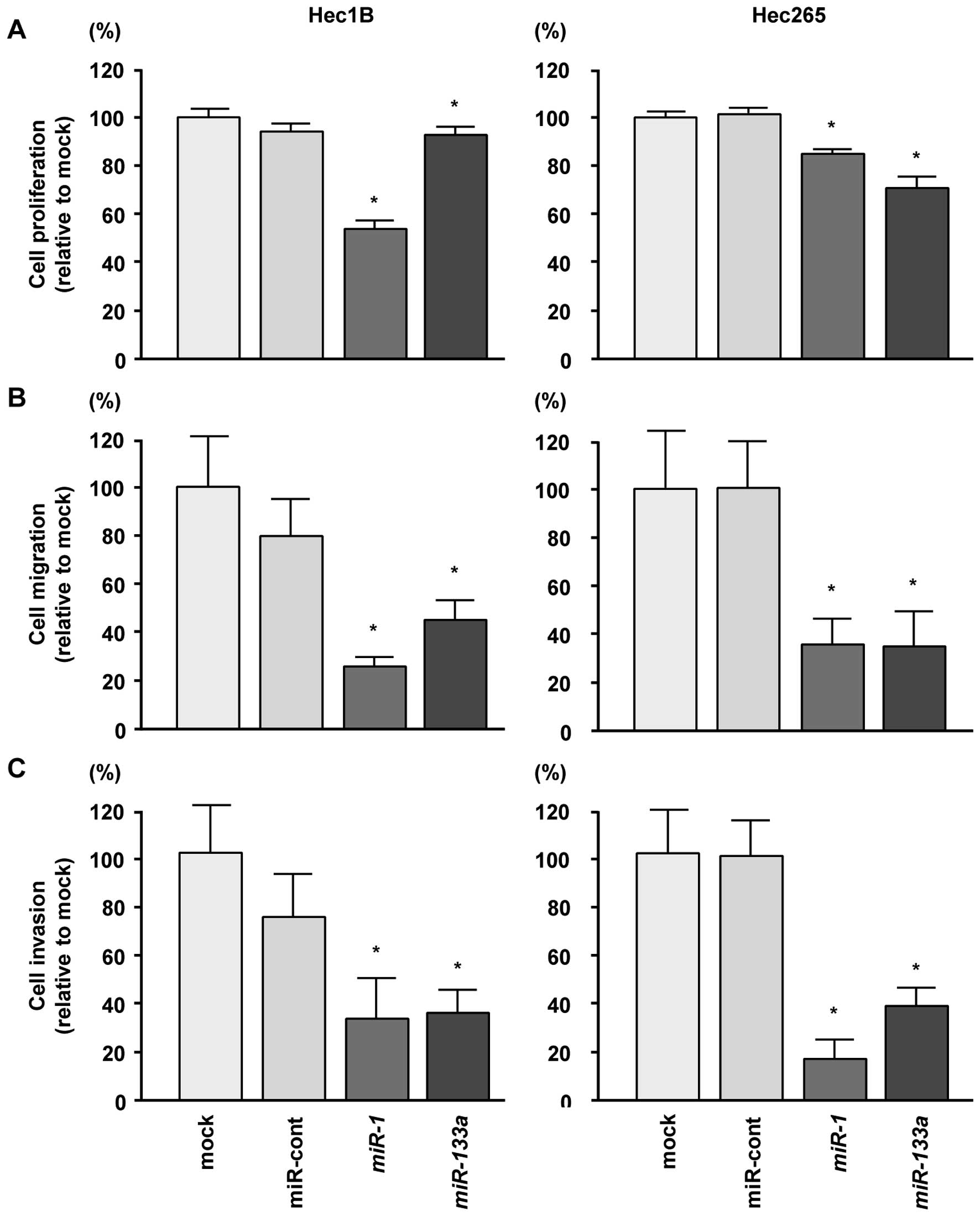
Effects of transfection with miR-1 and
miR-133a on cell proliferation, migration and invasion in EC cell
lines
To examine the functional roles of miR-1 and
miR-133a, we performed gain-of-function assays by
transfecting mature miRNAs into Hec1B and Hec265 cells. XTT assays
showed that cell proliferation was inhibited by transfection with
miR-1 and miR-133a in both Hec1B and Hec265 cells
compared with mock and miRNA-control transfections (P<0.0083,
Fig. 2A).
Cell migration assays demonstrated that cell
migration was significantly inhibited in miRNA-transfected cells
compared with mock- or miRNA-control-transfected cells
(P<0.0083, Fig. 2B).
Moreover, in Matrigel invasion assays, transfection
with miR-1 and miR-133a significantly inhibited cell
invasion as compared with mock or miRNA-control transfection
(P<0.0083, Fig. 2C). These
results suggested that the miR-1/133a cluster could
represent a putative tumor suppressor in EC cells.
Identification of common targets of miR-1
and miR-133a by in silico analysis and gene expression data
To identify putative genes regulated by the
miR-1/133a cluster (i.e., both miR-1 and
miR-133a), we searched the TargetScan database (Release 6.2,
http://www.targetscan.org/) and analyzed
expression data of EC clinical specimens using the Gene Expression
Omnibus (GEO accession no. GSE 17025). Our strategy for
identification of target genes of the miR-1/133a cluster is
shown in Fig. 3. We found that 23
genes were upregulated in EC specimens and had putative target
sites for miR-1 and miR-133a in their 3′UTRs.
Therefore, these genes were annotated as putative targets of the
miR-1/133a cluster (Table
II). Among 23 genes, we evaluated the expression of 4 genes
(PDE7A, DDX3X, CORO1C and SPTBN1) in clinical
specimens. As a result, the expression of PDE7A mRNA was
significantly higher in clinical EC specimens.
 | Table IIPutative target genes regulated by
the miR-1/133a cluster in endometrial cancer cells. |
Table II
Putative target genes regulated by
the miR-1/133a cluster in endometrial cancer cells.
| | | | | Putative target
site |
|---|
| | | | |
|
|---|
| | | | | miR-1 |
miR-133a |
|---|
| | | | |
|
|
|---|
| Entrez Gene ID | Gene symbol | Gene name | Expression (log2
ratio) | P-value | Conserved | Poorly | Conserved | Poorly |
|---|
| 5150 | PDE7A | Phosphodiesterase
7A | 1.79 | 1.2E-07 | 1 | | 1 | |
| 6711 | SPTBN1 | Spectrin, β,
non-erythrocytic 1 | 1.71 | 1.6E-03 | 1 | | 1 | 2 |
| 23603 | CORO1C | Coronin, actin
binding protein, 1C | 1.67 | 2.0E-06 | 2 | | 1 | |
| 1654 | DDX3X | DEAD
(Asp-Glu-Ala-Asp) box polypeptide 3, X-linked | 1.51 | 1.9E-03 | 1 | | 1 | |
| 377 | ARF3 | ADP-ribosylation
factor 3 | 2.62 | 2.5E-05 | 1 | 2 | | 1 |
| 10888 | GPR83 | G protein-coupled
receptor 83 | 2.07 | 2.5E-05 | 1 | | | 1 |
| 6373 | CXCL11 | Chemokine (C-X-C
motif) ligand 11 | 2.04 | 6.6E-04 | 1 | | | 1 |
| 7267 | TTC3 | Tetratricopeptide
repeat domain 3 | 1.99 | 2.0E-03 | 1 | | | 1 |
| 7705 | ZNF146 | Zinc finger protein
146 | 1.66 | 3.4E-04 | 1 | | | 1 |
| 4804 | NGFR | Nerve growth factor
receptor | 1.58 | 3.8E-03 | 1 | | | 1 |
| 93685 | ENTPD7 | Ectonucleoside
triphosphate diphosphohydrolase 7 | 1.75 | 5.7E-07 | 1 | 1 | | 1 |
| 2321 | FLT1 | fms-related
tyrosine kinase 1 | 2.95 | 7.2E-06 | | 1 | 1 | |
| 55143 | CDCA8 | Cell division cycle
associated 8 | 2.67 | 5.5E-07 | | 1 | 1 | |
| 6789 | STK4 | Serine/threonine
kinase 4 | 2.32 | 3.4E-06 | | 1 | 1 | |
| 5451 | POU2F1 | POU class 2
homeobox 1 | 1.87 | 1.3E-02 | | 1 | 2 | 1 |
| 2043 | EPHA4 | EPH receptor
A4 | 3.01 | 1.5E-04 | | 1 | | 1 |
| 7545 | ZIC1 | Zic family member
1 | 2.99 | 4.9E-02 | | 1 | | 1 |
| 57823 | SLAMF7 | SLAM family member
7 | 2.07 | 1.9E-03 | | 1 | | 1 |
| 57522 | SRGAP1 | SLIT-ROBO Rho
GTPase activating protein 1 | 1.99 | 1.2E-06 | | 1 | | 1 |
| 85645 | EPT1 |
Ethanolaminephosphotransferase 1
(CDP-ethanolamine-specific) | 1.99 | 2.5E-09 | | 1 | | 2 |
| 13869 | Erbb4 | v-erb-a
erythroblastic leukemia viral oncogene homolog 4 (avian) | 1.69 | 3.0E-02 | | 1 | | 1 |
| 72555 | Shisa9 | Shisa homolog 9
(Xenopus laevis) | 1.56 | 7.0E-03 | | 1 | | 1 |
| 10640 | EXOC5 | Exocyst complex
component 5 | 1.51 | 1.8E-02 | | 1 | | 1 |
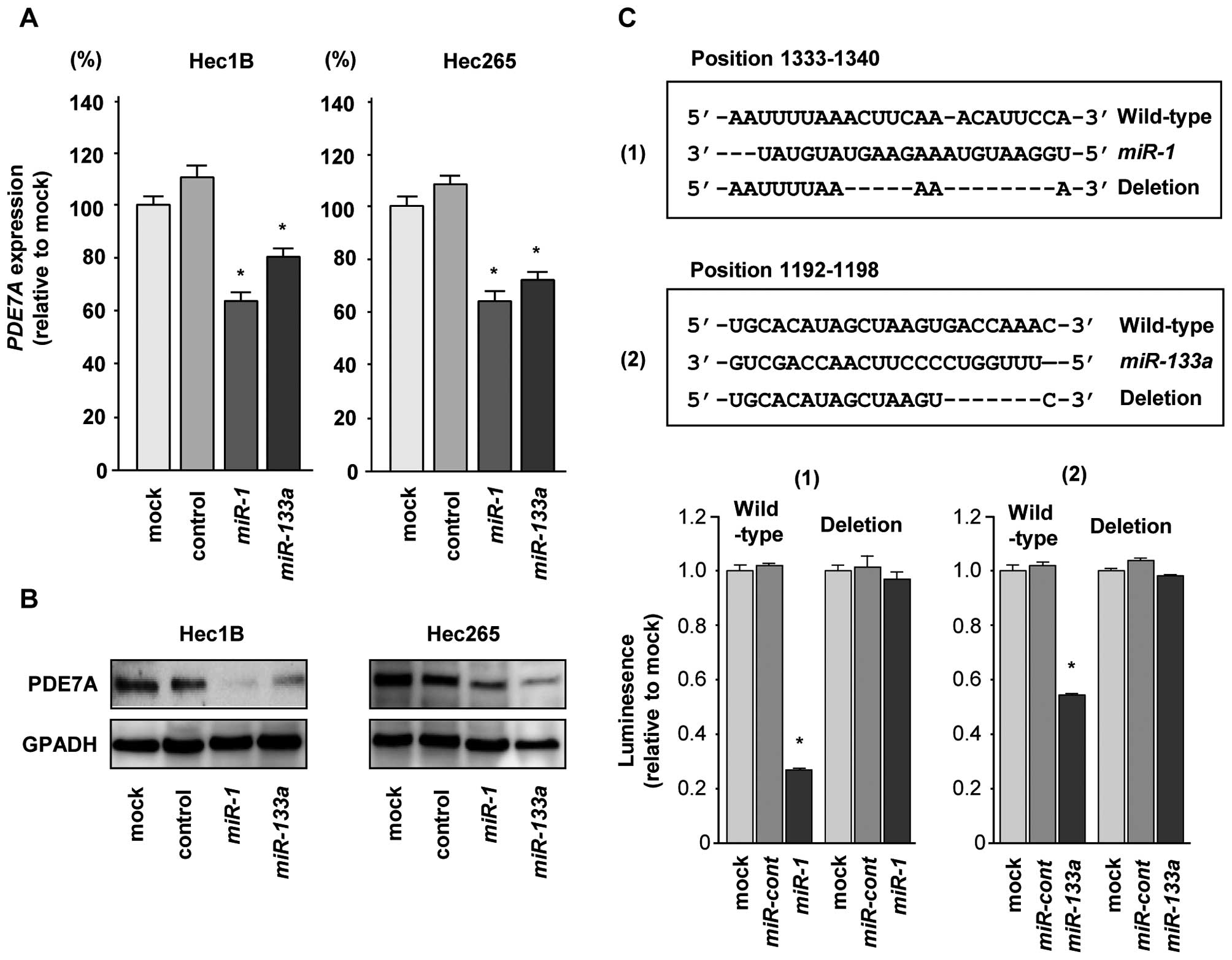
PDE7A was a direct target of the
miR-1/133a cluster in EC cells
Next, we performed qRT-PCR and western blotting to
confirm downregulation of PDE7A mRNA and protein following
restoration of miR-1 or miR-133a in Hec1B and Hec265
EC cells. The mRNA and protein expression levels of PDE7A were
significantly repressed in miR-1 and miR-133a
transfectants in comparison with mock or miR-control transfectants
(P<0.0083, Fig. 4A and B).
We then performed luciferase reporter assays in EC
cells to determine whether PDE7A was directly regulated by
miR-1 and miR-133a. The TargetScan database predicted
that there was one binding site for miR-1 in the 3′UTR of
PDE7A (positions 1333–1340; Fig. 4C) and one binding site for
miR-133a in the 3′UTR of PDE7A (positions 1192–1198;
Fig. 4C). We then used vectors
encoding the partial wild-type sequence of the 3′UTR of
PDE7A mRNA, including the predicted miR-1 or
miR-133a target sites. We found that the luminescence
intensity was significantly reduced by cotransfection with
miR-1 or miR-133a and the vector carrying the
wild-type 3′UTR of PDE7A. In contrast, transfection with the
mutant vector, in which the sequence within positions 1333–1340 or
1192–1198 had been changed, blocked the decrease in luminescence
(P<0.0001, Fig. 4C). These data
suggested that miR-1 and miR-133a bound directly to
specific sites in the 3′UTR of PDE7A mRNA.
Expression levels of PDE7A in EC clinical
specimens
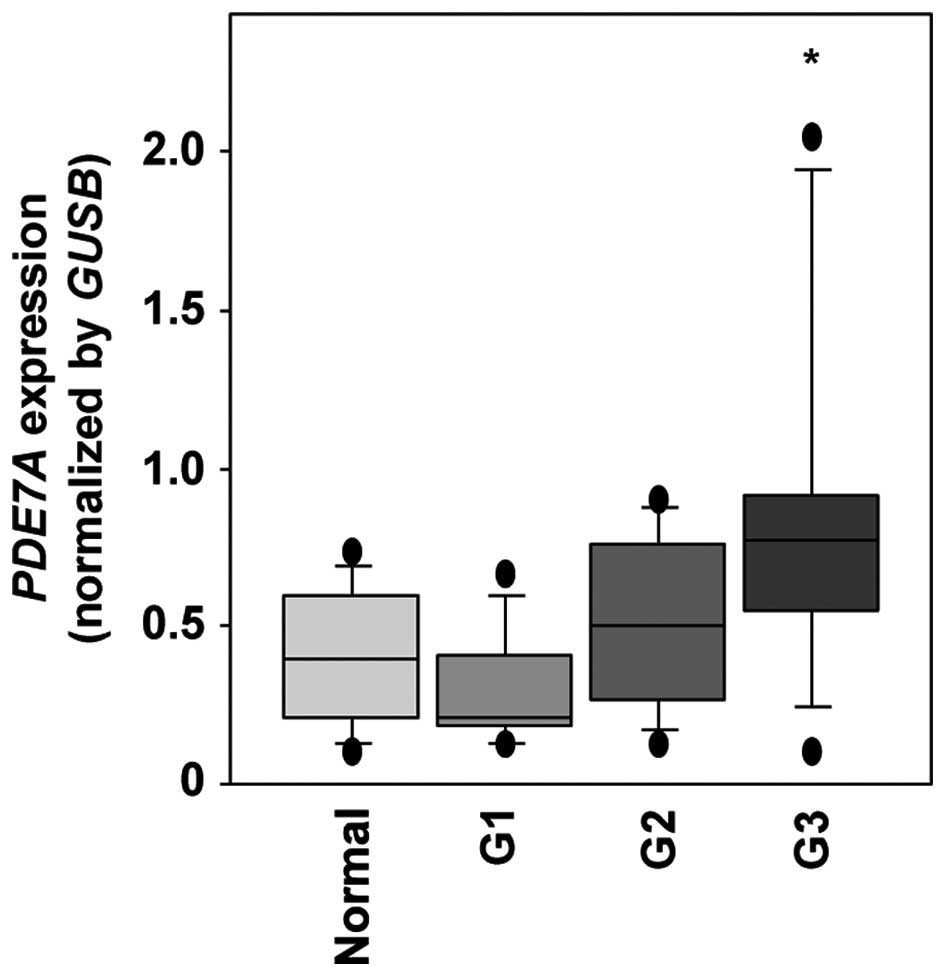
Twenty-seven EC and 8 normal endometrium specimens
were subjected to PDE7A mRNA expression analysis in this
study. qRT-PCR analysis showed that the expression of PDE7A
mRNA was significantly higher in clinical EC (differentiation G3)
specimens than in normal specimens (P=0.0022, Fig. 5).
Downregulation of PDE7A expression in EC
cells affected cell proliferation, migration and invasion
activities
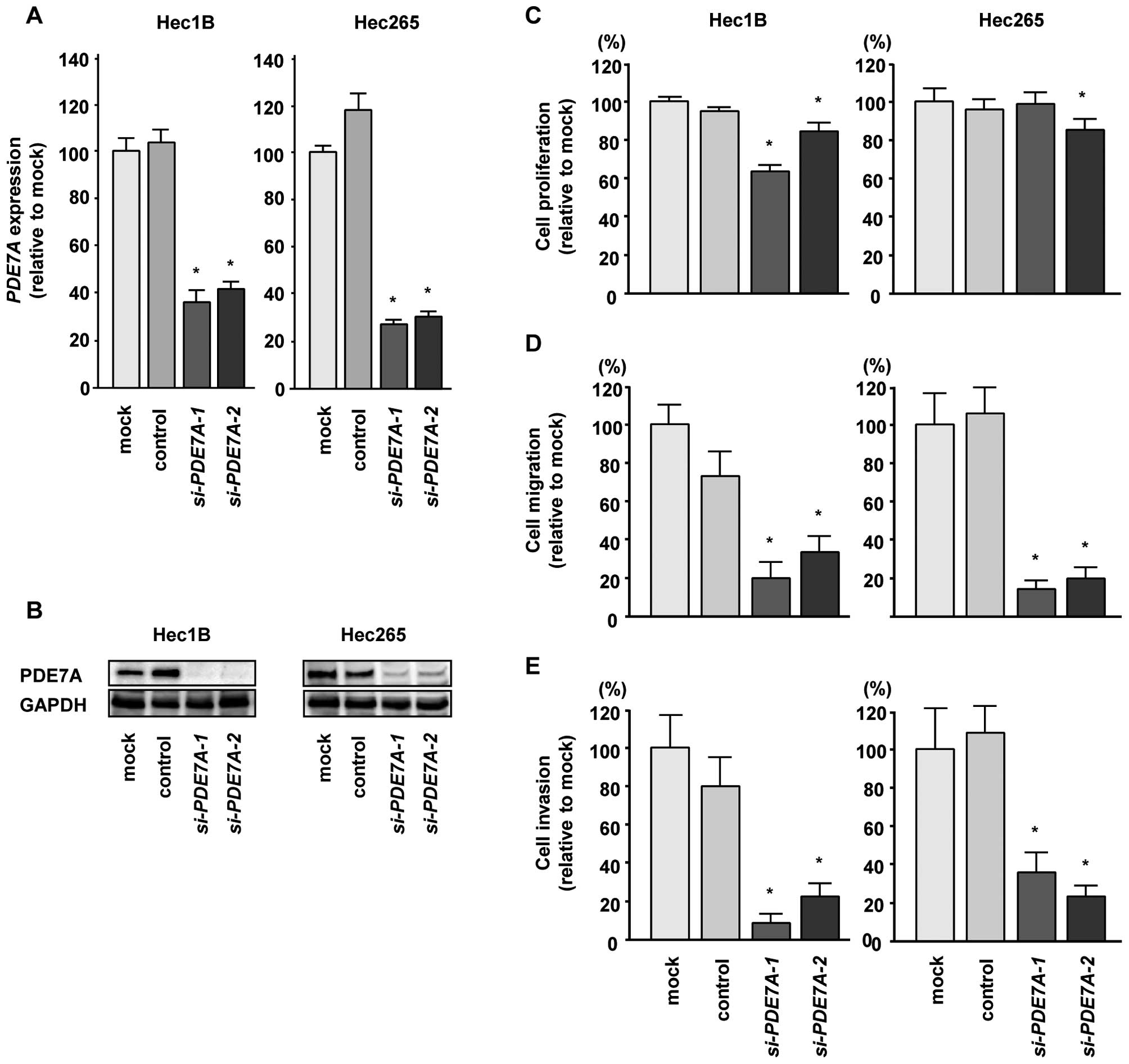
To investigate the functional role of PDE7A,
we performed loss-of-function studies using si-PDE7A
transfectants. First, we evaluated the knockdown efficiency of
si-PDE7A transfection in Hec1B and Hec265 EC cells. Western
blotting and qRT-PCR indicated that si-PDE7A effectively
downregulated PDE7A expression in EC cells (P<0.0083, Fig. 6A and B).
Next, we analyzed the functional effects of
PDE7A knockdown in EC cells. XTT assays demonstrated that
cell proliferation was significantly inhibited in si-PDE7A
transfectants in comparison with mock or si-control transfectants
(P<0.005, Fig. 6C). Moreover,
cell migration assays revealed significant inhibition of cell
migration in si-PDE7A transfectants in comparison with mock
or si-control transfectants (P<0.0001, Fig. 6D). Similarly, Matrigel invasion
assays revealed that the number of invading cells was significantly
decreased when EC cells were transfected with si-PDE7A
(P<0.0001, Fig. 6E). These
findings suggested that PDE7A acted as an oncogene in EC
cells.
Discussion
The 5-year survival rate of patients with stage I EC
is >90%; however, that in patients with stages III or IV EC is
much lower, ranging from 40 to 80% (3,4).
Previous studies have demonstrated that mutation of K-ras or PTEN
is common in low-grade EC, while high-grade EC is associated with
P53 mutation (15). These data
suggest that differences in expression of cancer-related genes have
substantial effects on disease progression. However, EC is a
complex disease and cannot be explained only by mutations in these
few genes; thus, elucidation of the involvement of other unknown
genetic abnormalities and signaling pathways, including ncRNAs, is
critical.
miRNAs are unique in their ability to regulate
multiple protein-coding genes. Recent bioinformatic predictions
have shown that miRNAs regulate >30–60% of the protein-coding
genes in the human genome (9,10).
Accumulating evidence has suggested that aberrantly expressed
miRNAs disrupt tightly regulated RNA networks in cancer cells.
These events are believed to initiate cancer cell development and
metastasis. Therefore, identification of key miRNAs and the
networks regulated by these miRNAs will provide new insights into
the potential mechanisms of cancer initiation, development and
metastasis. Recent studies have reported the differential
expression of miRNAs in EC cells; for examples, miR-205,
miR-210, miR-429 and miR-449 are upregulated in EC
tissues, whereas let-7e, miR-30c, miR-204 and miR-221
are downregulated in EC tissues (16). Upregulation of miR-205 is
significantly correlated with disease survival in EC, and thus,
miR-205 is considered a potential prognostic marker in EC.
Interestingly, miR-205 directly regulates the expression of
PTEN and inhibits apoptosis in EC cells (17).
To identify novel miRNA-mediated RNA networks in
cancer cells, we have constructed miRNA expression signatures in
several types of cancers and investigated the roles of miRNAs in
oncogenesis and metastasis using differentially expressed miRNAs
(12,18,19).
These miRNA signatures have revealed that the miR-1/133a
cluster is frequently down-regulated in several types of cancers,
including head and neck squamous cell carcinoma, prostate cancer,
bladder cancer and lung cancer (11–14).
Our present study demonstrated that miR-1 and
miR-133a were significantly downregulated in EC specimens
and cell lines. Moreover, restoration of these miRNAs significantly
inhibited cancer cell migration and invasion, suggesting that this
miRNA cluster may function as a tumor suppressor in EC cells,
similar to its function in other cancers (20–22).
A full understanding of the targets in EC cells that are regulated
by the miR-1/133a cluster may contribute to our knowledge on
EC oncogenesis and metastasis. Recently, we established a strategy
for identification of pathways and genes regulated by
tumor-suppressive miRNAs (14,22).
In the present study, we used this strategy and found 23 putative
candidate genes potentially regulated by the miR-1/133a
cluster in EC cells. This is the first report demonstrating that
PDE7A is directly regulated by the miR-1/133a cluster
in EC cells.
PDEs are enzymes that regulate the cellular levels
of the secondary messengers cAMP and cGMP by controlling their
rates of degradation. PDEs can be categorized into 11 families
(PDE1-11), which are structurally related but functionally distinct
(23). PDE7, including isoforms
PDE7A and PDE7B, is a high-affinity cAMP-specific PDE (24–26).
Three variant forms of PDE7A have been annotated: PDE7A1 and PDE7A2
are N-terminal variants, and PDE7A3 is a C-terminal variant
(25,27). The expression of PDE7A is elevated
in pro-inflammatory and immune cells, supporting the role of PDE7A
as a therapeutic target for inflammation disorders (28). A recent study indicated that the
PDE7A-specific inhibitor ASB16165 suppresses keratinocyte
proliferation on TPA-induced skin inflammation (29).
Many tumor cells exhibit significantly decreased
cAMP levels as a consequence of overexpression of PDEs in chronic
lymphocytic leukemia (CLL) and malignant carcinoma cells (30–32).
PDE7A is overexpressed in CLL, and stimulation of the cAMP
signaling pathway has been shown to induce apoptosis and augment
the effects of glucocorticoids in inducing apoptosis in CLL cells
(33). Increasing intracellular
concentrations of cAMP may arrest growth, induce apoptosis and
attenuate cancer cell migration in various cancers (34–37).
The effects of cAMP are mediated by two ubiquitously expressed
intracellular cAMP receptors, protein kinase A (PKA) and exchange
protein directly activated by cAMP (Epac) (38). The cAMP/PKA signaling pathway may
have an important role in tumor migration. Indeed, activation of
the cAMP/PKA pathway inhibits cancer cell migration in various
cancers by targeting matrix metalloproteinase (MMP)2, actin,
integrin, MMP9 and MMP4 (39–42).
Interestingly, PDE7A contains a PKA pseudosubstrate site within 2
repeated sequences at the N-terminal region of PDE7A. The PDE7A1
N-terminal repeat region inhibits the C subunit of PKA (C) activity
and suppresses C-dependent, cAMP-independent, physiological
responses. These observations demonstrate that PDE7A1 can inhibit
cAMP signaling via direct binding to C (43).
In conclusion, downregulation of the
miR-1/133a cluster was a frequent event in EC. Moreover, the
tumor-suppressive miR-1/133a cluster directly regulated
PDE7A, a high-affinity cAMP-specific enzyme. Restoration of
miR-1/miR-133a or silencing of PDE7A inhibited cancer cell
migration and invasion, suggesting that the
miR-1/miR-133a-PDE7A pathway contributes to the metastasis
of EC. Identification of molecular targets regulated by
tumor-suppressive miRNAs will provide insights into the potential
mechanisms of EC oncogenesis and metastasis, facilitating the
development of novel therapeutic strategies for the treatment of
this disease.
Acknowledgements
This study was supported by the KAKENHI (C),
24592590.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rose PG: Endometrial carcinoma. N Engl J
Med. 335:640–649. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Creasman WT, Odicino F, Maisonneuve P,
Quinn MA, Beller U, Benedet JL, Heintz AP, Ngan HY and Pecorelli S:
Carcinoma of the corpus uteri. FIGO 26th Annual Report on the
Results of Treatment in Gynecological Cancer. Int J Gynaecol
Obstet. 95(Suppl 1): S105–S143. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Murali R, Soslow RA and Weigelt B:
Classification of endometrial carcinoma: More than two types.
Lancet Oncol. 15:e268–e278. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Makker V, Hensley ML, Zhou Q, Iasonos A
and Aghajanian CA: Treatment of advanced or recurrent endometrial
carcinoma with doxorubicin in patients progressing after
paclitaxel/carboplatin: Memorial Sloan-Kettering Cancer Center
experience from 1995 to 2009. Int J Gynecol Cancer. 23:929–934.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Friedman JM and Jones PA: MicroRNAs:
Critical mediators of differentiation, development and disease.
Swiss Med Wkly. 139:466–472. 2009.PubMed/NCBI
|
|
11
|
Nohata N, Hanazawa T, Kikkawa N, Sakurai
D, Sasaki K, Chiyomaru T, Kawakami K, Yoshino H, Enokida H,
Nakagawa M, et al: Identification of novel molecular targets
regulated by tumor suppressive miR-1/miR-133a in maxillary sinus
squamous cell carcinoma. Int J Oncol. 39:1099–1107. 2011.PubMed/NCBI
|
|
12
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kojima S, Chiyomaru T, Kawakami K, Yoshino
H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, et
al: Tumour suppressors miR-1 and miR-133a target the oncogenic
function of purine nucleoside phosphorylase (PNP) in prostate
cancer. Br J Cancer. 106:405–413. 2012. View Article : Google Scholar :
|
|
14
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar
|
|
15
|
Berg A, Hoivik EA, Mjøs S, Holst F, Werner
HM, Tangen IL, Taylor-Weiner A, Gibson WJ, Kusonmano K, Wik E, et
al: Molecular profiling of endometrial carcinoma precursor, primary
and metastatic lesions suggests different targets for treatment in
obese compared to non-obese patients. Oncotarget. 6:1327–1339.
2015.
|
|
16
|
Banno K, Yanokura M, Kisu I, Yamagami W,
Susumu N and Aoki D: MicroRNAs in endometrial cancer. Int J Clin
Oncol. 18:186–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Karaayvaz M, Zhang C, Liang S, Shroyer KR
and Ju J: Prognostic significance of miR-205 in endometrial cancer.
PLoS One. 7:e351582012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Itesako T, Seki N, Yoshino H, Chiyomaru T,
Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa
M, et al: The microRNA expression signature of bladder cancer by
deep sequencing: The functional significance of the miR-195/497
cluster. PLoS One. 9:e843112014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
Yamada Y, Nohata N, Fuse M, Nakagawa M and Enokida H: Tumor
suppressive microRNA-1285 regulates novel molecular targets:
Aberrant expression and functional significance in renal cell
carcinoma. Oncotarget. 3:44–57. 2012.PubMed/NCBI
|
|
20
|
Kinoshita T, Hanazawa T, Nohata N, Kikkawa
N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto
Y, et al: Tumor suppressive microRNA-218 inhibits cancer cell
migration and invasion through targeting laminin-332 in head and
neck squamous cell carcinoma. Oncotarget. 3:1386–1400.
2012.PubMed/NCBI
|
|
21
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y, et al: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nishikawa R, Goto Y, Kojima S, Enokida H,
Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y,
et al: Tumor-suppressive microRNA-29s inhibit cancer cell migration
and invasion via targeting LAMC1 in prostate cancer. Int J Oncol.
45:401–410. 2014.PubMed/NCBI
|
|
23
|
Azevedo MF, Faucz FR, Bimpaki E, Horvath
A, Levy I, de Alexandre RB, Ahmad F, Manganiello V and Stratakis
CA: Clinical and molecular genetics of the phosphodiesterases
(PDEs). Endocr Rev. 35:195–233. 2014. View Article : Google Scholar :
|
|
24
|
Bloom TJ and Beavo JA: Identification and
tissue-specific expression of PDE7 phosphodiesterase splice
variants. Proc Natl Acad Sci USA. 93:14188–14192. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Han P, Zhu X and Michaeli T: Alternative
splicing of the high affinity cAMP-specific phosphodiesterase
(PDE7A) mRNA in human skeletal muscle and heart. J Biol Chem.
272:16152–16157. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Sasaki T, Kotera J, Yuasa K and Omori K:
Identification of human PDE7B, a cAMP-specific phosphodiesterase.
Biochem Biophys Res Commun. 271:575–583. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Glavas NA, Ostenson C, Schaefer JB, Vasta
V and Beavo JA: T cell activation up-regulates cyclic nucleotide
phosphodiesterases 8A1 and 7A3. Proc Natl Acad Sci USA.
98:6319–6324. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Safavi M, Baeeri M and Abdollahi M: New
methods for the discovery and synthesis of PDE7 inhibitors as new
drugs for neurological and inflammatory disorders. Expert Opin Drug
Discov. 8:733–751. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Goto M, Kadoshima-Yamaoka K, Murakawa M,
Yoshioka R, Tanaka Y, Inoue H, Murafuji H, Kanki S, Hayashi Y,
Nagahira K, et al: Phosphodiesterase 7A inhibitor ASB16165 impairs
proliferation of keratinocytes in vitro and in vivo. Eur J
Pharmacol. 633:93–97. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang L, Murray F, Zahno A, Kanter JR,
Chou D, Suda R, Fenlon M, Rassenti L, Cottam H, Kipps TJ, et al:
Cyclic nucleotide phosphodiesterase profiling reveals increased
expression of phosphodiesterase 7B in chronic lymphocytic leukemia.
Proc Natl Acad Sci USA. 105:19532–19537. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Marko D, Romanakis K, Zankl H,
Fürstenberger G, Steinbauer B and Eisenbrand G: Induction of
apoptosis by an inhibitor of cAMP-specific PDE in malignant murine
carcinoma cells overexpressing PDE activity in comparison to their
non-malignant counterparts. Cell Biochem Biophys. 28:75–101. 1998.
View Article : Google Scholar
|
|
32
|
Savai R, Pullamsetti SS, Banat GA,
Weissmann N, Ghofrani HA, Grimminger F and Schermuly RT: Targeting
cancer with phosphodiesterase inhibitors. Expert Opin Investig
Drugs. 19:117–131. 2010. View Article : Google Scholar
|
|
33
|
Dong H, Zitt C, Auriga C, Hatzelmann A and
Epstein PM: Inhibition of PDE3, PDE4 and PDE7 potentiates
glucocorticoid-induced apoptosis and overcomes glucocorticoid
resistance in CEM T leukemic cells. Biochem Pharmacol. 79:321–329.
2010. View Article : Google Scholar
|
|
34
|
Yamanaka Y, Mammoto T, Kirita T, Mukai M,
Mashimo T, Sugimura M, Kishi Y and Nakamura H: Epinephrine inhibits
invasion of oral squamous carcinoma cells by modulating
intra-cellular cAMP. Cancer Lett. 176:143–148. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Timoshenko AV, Xu G, Chak rabarti S, Lala
PK and Chakraborty C: Role of prostaglandin E2 receptors in
migration of murine and human breast cancer cells. Exp Cell Res.
289:265–274. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Murata K, Kameyama M, Fukui F, Ohigashi H,
Hiratsuka M, Sasaki Y, Kabuto T, Mukai M, Mammoto T, Akedo H, et
al: Phosphodiesterase type III inhibitor, cilostazol, inhibits
colon cancer cell motility. Clin Exp Metastasis. 17:525–530. 1999.
View Article : Google Scholar
|
|
37
|
McEwan DG, Brunton VG, Baillie GS, Leslie
NR, Houslay MD and Frame MC: Chemoresistant KM12C colon cancer
cells are addicted to low cyclic AMP levels in a phosphodiesterase
4-regulated compartment via effects on phosphoinositide 3-kinase.
Cancer Res. 67:5248–5257. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cheng X, Ji Z, Tsalkova T and Mei F: Epac
and PKA: A tale of two intracellular cAMP receptors. Acta Biochim
Biophys Sin (Shanghai). 40:651–662. 2008. View Article : Google Scholar
|
|
39
|
Dabizzi S, Noci I, Borri P, Borrani E,
Giachi M, Balzi M, Taddei GL, Marchionni M, Scarselli GF and
Arcangeli A: Luteinizing hormone increases human endometrial cancer
cells invasiveness through activation of protein kinase A. Cancer
Res. 63:4281–4286. 2003.PubMed/NCBI
|
|
40
|
Howe AK: Regulation of actin-based cell
migration by cAMP/PKA. Biochim Biophys Acta. 1692:159–174. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
McCawley LJ, Li S, Benavidez M, Halbleib
J, Wattenberg EV and Hudson LG: Elevation of intracellular cAMP
inhibits growth factor-mediated matrix metalloproteinase-9
induction and keratinocyte migration. Mol Pharmacol. 58:145–151.
2000.PubMed/NCBI
|
|
42
|
Ou Y, Zheng X, Gao Y, Shu M, Leng T, Li Y,
Yin W, Zhu W, Huang Y, Zhou Y, et al: Activation of cyclic AMP/PKA
pathway inhibits bladder cancer cell invasion by targeting
MAP4-dependent microtubule dynamics. Urol Oncol. 32:47.e21–47.e28.
2014. View Article : Google Scholar
|
|
43
|
Han P, Sonati P, Rubin C and Michaeli T:
PDE7A1, a cAMP-specific phosphodiesterase, inhibits cAMP-dependent
protein kinase by a direct interaction with C. J Biol Chem.
281:15050–15057. 2006. View Article : Google Scholar : PubMed/NCBI
|