Introduction
Normal haematopoiesis during adult life in animals
like in humans is a multistep complex process occurring within the
bone marrow microenvironment (1,2).
Haematopoietic stem cells (HSCs) besides their self-renewal
potential can be triggered to give rise to different blood cells
(red blood cells, white blood cells, platelets) via
lineage-restricted cell pathways (3–5).
HSCs interact with bone marrow meschenchymal (stroma) cells as well
as with growth factors and differentiation signals needed for
maturation (6). During
erythropoiesis, HSCs give rise to pro-erythroid progenitors which
are then converted into orthochromatophilic normoblasts,
subsequently reticulocytes and finally red blood cells entering the
peripheral blood stream (7–10).
Impairment of normal erythropoiesis is directly related to the
pathophysiology of haematological disorders of metabolic or genetic
nature such as porphyria, anemia, leukemia and myelodysplastic
syndromes (MDS) (11,12).
The induced erythroid maturation program of murine
erythroleukemia (MEL) cells in culture by chemical inducers is
accompanied by orchestrated gene expression patterns that involve
upregulation of developmentally regulated genes and downregulation
of those controlling potential for cell proliferation (13,14).
It has been previously reported that down-regulation of genes
encoding ribosomal RNAs (rRNAs) and specific ribosomal proteins
(RPs), (e.g., RPS5 and RPL35a), occurs very early in
MEL erythroid maturation program (15–17).
At this early period (latent period; <24 h) of induction, the
overall number of ribosomes also declines substantially and cells
synthesize far less protein (15,18).
Simultaneously, a salt-labile translationally inactive form of
ribosomes has been shown to exist (19). The precise molecular mechanisms
underlying such progressive reduction of ribosome biogenesis and
protein synthesis in mature MEL cells are still elusive; however,
the assessment of ribosomal dysfunction has been considered
essential in understanding the pathophysiology of reticulocytes
disorders (20) and
ribosomopathies (21). Besides,
overexpression of RPS5 gene in stably transfected MEL cells (e.g.,
MEL-C14) has been associated with a delay in the initiation of MEL
erythroid differentiation program in vitro. Interestingly,
it has been demonstrated that RPS5 interferes with the ability of
MEL-C14 to differentiate through perturbation of cell entrance into
G0/G1 cell cycle arrest and CDK2, CDK4 and
CDK6 levels (22).
In this study, we established two additional MEL
cell cultures with altered RPS5 gene expression. One culture in
which the cells were stably transfected with the full length of
RPS5 antisense cDNA (MEL-antisenseRPS5) and another in which
the cells were transiently transfected with siRNAs specific for
RPS5 gene silencing (MEL-RPS5siRNA). Parental MEL and MEL
cells transiently transfected with scrambled siRNAs
(MEL-scrambled siRNA) were used as control cultures.
Moreover, previously established MEL-C14 culture in which
overexpression of RPS5 exists was also included in this study. The
data obtained thus far indicate that: a) MEL-antisenseRPS5
exhibited a more pronounced delay in the initiation of
differentiation program as compared to MEL-C14; b) RNAi-mediated
transient silencing of RPS5 gene expression resulted in
complete inability of MEL cells to differentiate in vitro;
however, when these cells were permitted to restore normal RPS5
gene expression levels, their maximum differentiation capacity has
been regained; c) interestingly, differentiation of
MEL-antisenseRPS5 and MEL-RPS5siRNA cells was
accompanied by cellular morphology changes, altered gene expression
profiles and impairment of their potential for proliferation and
even cell death. Overall, these findings support the concept for
the first time that genetic manipulation of RPS5 gene
expression level (up- and/or downregulation) critically affects the
potential of MEL cells to fully complete their erythroid maturation
program in vitro.
Materials and methods
Chemicals and antibodies
Dimethylsulfoxide (DMSO),
hexamethylene-bis-acetamide (HMBA), benzidine dihydrocloride,
vanadyl ribonucleotide complexes (VRC) and proteinase K was
purchased from Sigma (St. Louis, MO, USA). [γ-32P]-dCTP
(111 Tbq/mmol) was obtained from Izotop, Institute of Isotopes Co.,
Ltd., Budapest, Hungary, whereas the DNA 32P-labeling
system kit was purchased from Invitrogen (Red Prime DNA Labeling
System). The rabbit anti-RPS5 polyclonal antibody (pAb) raised
against C-terminal oligopeptide of RPS5 was kindly provided by Dr
Shuetsu Fukushi (R&D Center, BioMedical Laboratories, Matoba,
Kawagoe, Saitama, Japan). The rat anti-mouse MYC, CDK2, CDK4, CDK6
and Gata-1 mAbs were purchased from Invitrogen, Cell Signaling,
Transduction Bioscience (BD) and Santa Cruz. Also, goat anti-mouse
IgG and goat anti-rabbit IgG were obtained from Santa Cruz,
respectively.
Assessment of MEL cell differentiation
and the cellular content of haemoglobin
Parental MEL cells were maintained in Dulbecco’s
modified Eagle’s medium as previously published (22). MEL cell cultures were exposed to
DMSO (1.5% v/v) and/or HMBA (5 mM) as indicated under individual
figure. At certain time intervals upon exposure, the accumulation
of differentiated (haemoglobin-producing; Bz+ cells)
cells was assessed cytochemically with
benzidine-H2O2 solution as previously
described (16). Also the
haemoglobin content within the cells was determined
spectrophotometrically as described elsewhere (23).
Stable transfection of MEL cells with the
full length mouse RPS5 cDNA either in sense or antisense
orientation
MEL-C14 cells generated by stable transfection of
MEL cells with the full length mouse RPS5 cDNA to express
recombinant RPS5-Myc-His protein has been previously described
(22). Stable transfection of MEL
cells with the full length mouse antisense RPS5 cDNA was
carried out as briefly described below: logarithmically growing MEL
cells were transfected with the recombinant vector pcDNA3.1(+)
carrying the full-length mouse RPS5 cDNA (715 bp in length).
Such construct was generated through the ligation of the
EcoRI/XhoI fragment derived from the original
pBluescript SK +/− plasmid that carry the full length of
RPS5 cDNA cloned in our laboratory (16) (GenBank accession no. Y12431) into
the respective EcoRI/XhoI sites of pcDNA3.1(+)
vector. The RPS5 DNA fragment was inserted into the
recombinant pcDNA3.1(+) vector (Invitrogen Life Technologies, USA)
in antisense orientation with respect to CMV promoter to generate
the construct pcDNA3.1(+)-anti-RPS5. Subsequent stable
transfection of MEL cells was performed by using
Lipofectamine-2000™ reagent (Invitrogen Life Technologies) and 1 μg
plasmid DNA of the recombinant construct
pcDNA3.1(+)-anti-RPS5 according to the accompanying
manufacturer’s protocol. Stably transfected cells were then
selected with G418, (Gibco BRL, Gaithersburg, MD, USA; 0.6 mg/ml)
added in the culture medium. G418-resistant cells outgrown from the
transfected pcDNA3.1(+)-anti-RPS5 MEL culture allowed us to
establish a culture designated as MEL-antisenseRPS5 and used
then throughout this study. After the establishment of
MEL-antisenseRPS5 culture, G418 (0.2–0.25 mg/ml) was used
throughout all experiments described in this study.
Isolation of total cytoplasmic RNA,
northern blot hybridization and RT-PCR analysis
Total cytoplasmic RNA isolated from control and/or
inducer-treated cells at various time intervals upon incubation in
culture was subjected to northern blot hybridization analysis to
assess the steady-state levels of RNA transcripts encoded the
RPS5 and βmajor globin genes, as previously
described (22,29).
For RT-PCR analysis, total cytoplasmic RNA (0.2–0.5
μg) isolated from various MEL cell cultures was used for RT-PCR.
The PCR experiments were performed by using the RobusT™ I kit
(Finnzymes). In detail, the primers used were:
5′-GCGGGATCCATGACTGAGTGGGAAGCA-3′ (forward) and
5′-GCGGAATTCTCAGCGGTTAGACTTGGC-3′ (reverse) specifically designed
to allow the detection of mRNA level of endogenous RPS5 (441
bp) as previously published (22);
5′-GGACTTCGAGCAAGAGATGG-3′ (forward) and 5′-AGC
ACTGTGTTGGCGTACAG-3′ (reverse) for β-actin (234 bp);
5′-GACCTCGACTACGACTCCGTAC-3′ (forward) and
5′-CCACTGAGGGGTCAATGCAC-3′ (reverse) for c-myc (547 bp);
5′-CTGCTGGTTGTCTACCCTTGG-3′ (forward) and
5′-CCTGAAGTTCTCAGGATCCAC-3′ (reverse) for
βmajor globin (222 bp).
Transfection with scrambled and RPS5
specific siRNAs
All siRNAs used including a negative control siRNA
[negative control siRNA (#1022076)] designed with no significant
homology to any sequence in the mouse genome (scrambled siRNA)
along with two predesigned siRNAs directed against mouse
RPS5 (NM_009095) (Mm_Rps5_4 #SI01407322; Mm_Rps5_4
#SI01407336) were obtained from Qiagen. Based on extensive
preliminary data (not shown), however, only Mm_Rps5_4 was capable
of specifically downregulating the expression of RPS5 gene
in parental MEL cells and, therefore, was used throughout this
study. Scrambled and Mm_Rps5_4 siRNAs were transfected in MEL cells
using HiPerFect reagent (Qiagen). The oligo-nucleotide sequence for
siRNAs included in Mm_Rps5_4 were: sense,
r(GCGCUUCCGCAAAGCACAA)dTdT; antisense,
r(UUGUGCUUUGCGGAAGCGC)dTdT), whereas in scrambled siRNA were
sense, UUCUCCGAACGUGUCACGUdTdT; antisense,
ACGUGACACGUUCGGAGAAdTdT. The procedure briefly was as follows. The
day before transfection, MEL cells were seeded at a density of
0.4–1.6×106 cells in 60-mm dishes with 4 ml of DMEM
containing serum and antibiotics. On the day of transfection either
RPS5 specific or scrambled siRNA were diluted in culture to
give a final concentration of 120 nM. The transfection of siRNAs
was facilitated by adding 20 μl of HiPerFect reagent buffer
according to the manufacturer’s protocol and the cells were further
incubated under normal growth conditions before silencing of the
RPS5 gene to be monitored by RT-PCR analysis.
Flow cytometry DNA analysis and
assessment of apoptosis
Samples of parental MEL cells as well from those
exposed either to specific RPS5 (MEL-RPS5siRNA) or
scrambled (MEL-scrambled-siRNA) siRNAs before harvested from
culture at time intervals as shown in each figure, were subjected
to flow cytometry DNA analysis and assessment of apoptosis, as
previously published (22,24).
Results
Stable transfection of mouse antisense
RPS5 cDNA affects the commitment and decreases the onset of MEL
cells to erythroid maturation in vitro
Stable transfection of MEL cells with the mouse
antisense RPS5 cDNA allowed us to assess the proliferation
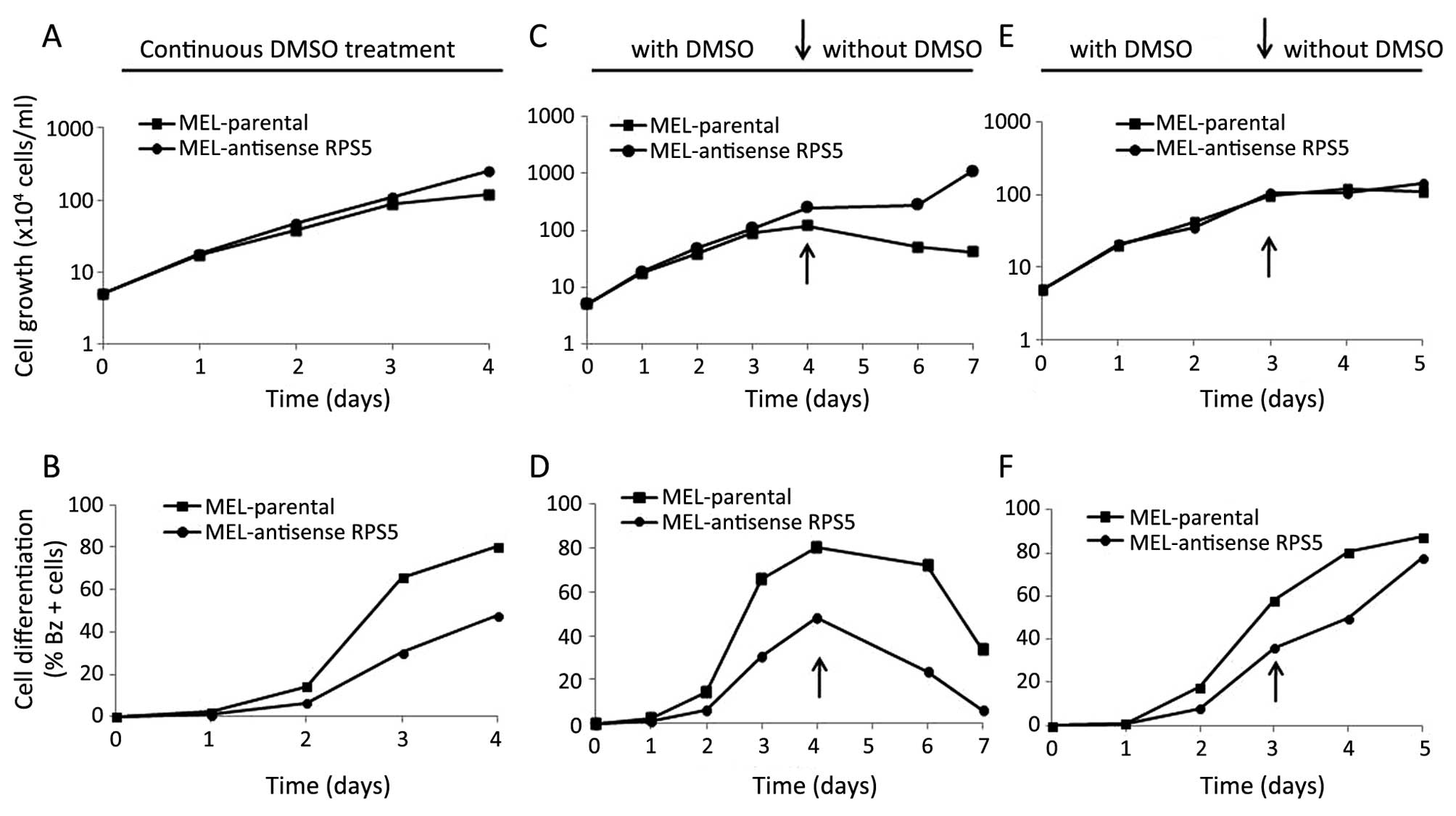
and differentiation behavior of such cells in culture. As shown in
Fig. 1B, MEL-antisenseRPS5
cells failed to reach the maximum differentiation level upon
exposure to chemical inducer DMSO as compared to parental MEL
cells, although comparatively no significant change on their cell
growth capacity was observed (Fig.
1A). In particular, parental MEL cells exhibited high levels of
Bz+ (haemoglobin-producing) cells after 96-h exposure to
DMSO (>80%) as expected (22).
On the contrary, MEL-antisenseRPS5 culture has shown a delay
of 18–24 h in the initiation of differentiation and the proportion
of Bz+ cells was ~50% (Fig.
1B). These data suggest that stable transfection of MEL cells
with antisense RPS5 cDNA delayed the onset of
differentiation by reducing the number of terminally differentiated
cells accumulated in culture.
By being aware on how the commitment to erythroid
maturation proceeds in MEL cells, a more thorough kinetic analysis
was performed in cultures to better understand the behavior of
MEL-antisenseRPS5. In particular it has been reported
earlier that in parallel to haemoglobin synthesis, commitment of
inducer-treated cells to terminal erythroid maturation leads to
limitation of proliferation potential of MEL cells up to 4–5
cellular divisions (13,14). This event becomes quite clear upon
dilution of cultures of inducer-treated cells. Such dilution
approach has been successfully applied in our work in MEL-C14
culture overexpressing the recombinant RPS5-Myc-His protein
(22). Inducer-treated cells, fail
to grow and divide continuously in contrast to
uncommitted-undifferentiated MEL cells, which exhibit an unlimited
proliferation potential. To this end, MEL-antisenseRPS5
cells were exposed continuously to inducer DMSO for either 96 or 72
h, before the dilution of cultures with fresh medium to permit
cells to proliferate for further 3 days in the absence (Fig. 1C and D) or 2 days in the presence
of DMSO (Fig. 1E and F),
respectively. Under these conditions, cells are capable of
exhibiting their maximum proliferation potential by executing the
erythroid maturation program in the absence of the inducer. As
shown in Fig. 1C, parental MEL
cells initially growing for 96 h in the presence of DMSO continued
in the absence of DMSO for additional 72 h, they reached their
plateau phase of growth and maintained at that level despite the
dilution (1:10) of the culture with fresh medium. The latter result
is attributed to the differentiation-dependent restriction of
proliferation potential of DMSO-treated cells (13,14,22),
since they achieved high level of differentiated cells (>80%)
after 96 h in culture (Fig. 1D).
However, the proportion of differentiated cells decreased
thereafter upon dilution as expected, since the committed cells can
go on only for 4–5 divisions, as mentioned above. On the contrary,
MEL-antisenseRPS5 cells did not exceed the level of 50% of
differentiated cells under the same culture conditions (Fig. 1D). Moreover in this culture,
cellular growth increased after the 6th day (Fig. 1C), since the higher number of
non-committed cells (~50%) continued to grow in the absence of
DMSO, as expected. These data suggest that in
MEL-antisenseRPS5 culture fewer cells enter the
differentiation program at a slower rate. Such observation proposes
that the plateau phase of growth seen in antisense-RPS5-transfected
MEL cells before dilution has to be attributed to cell-cell contact
inhibition rather than differentiation-dependent restriction of
cell proliferation potential, as seen in parental MEL cells. To
confirm this observation, the same experiment was repeated but
allowing cells to grow in the presence of DMSO even after dilution
of cultures. Interestingly enough, in that case the
MEL-antisenseRPS5 cells reached the same high level of
differentiation (>80%) after 5 days in culture compared to
parental MEL, as illustrated in Fig.
1E and F. These results suggest that the onset of
differentiation has been significantly delayed by the transfection
of antisense-RPS5 cDNA and the presence of inducer is required
throughout the entire exposure period for MEL cells to complete
their erythroid maturation program. At this point it has to be
noted that such behavior is unique for MEL-antisenseRPS5
culture, since both parental MEL and MEL-C14 did not require the
presence of inducer after initiation of commitment to execute the
differentiation program, as previously published (22).
Correlation of RPS5 gene expression with
modulation of CDKs in DMSO-treated MEL-antisenseRPS5
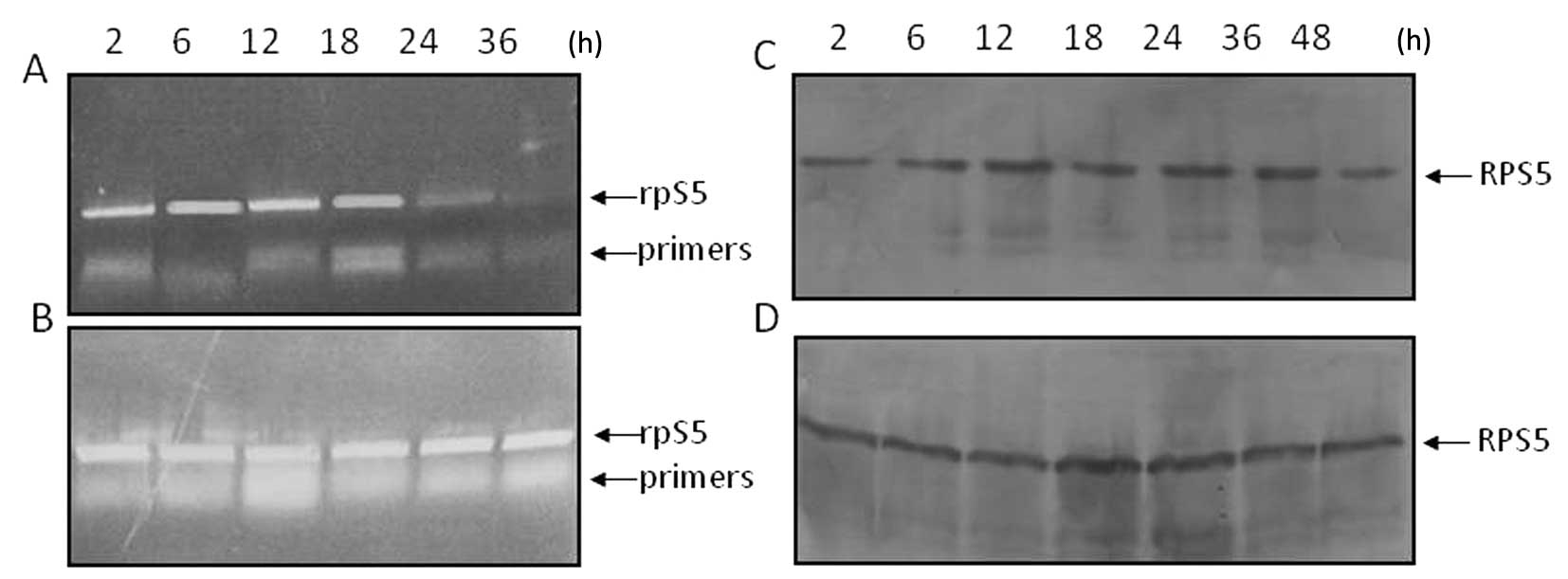
To assess any changes in the steady-state level of
RPS5 RNA transcripts in MEL-antisenseRPS5 cells in
proliferating and/or differentiating cultures, isolated cytoplasmic
RNA was subjected to northern blot hybridization analysis using
βmajor globin gene as internal control. As shown in
Fig. 2A, the level of RPS5
RNA transcripts in the cytoplasm remained constant in
MEL-antisenseRPS5 cells growing in culture in the absence of
DMSO. Similar data were obtained even after 96-h exposure to DMSO
(Fig. 2C). On the contrary, the
gradual cytoplasmic accumulation of βmajor globin RNA
transcripts was evident in differentiating cells only after 48–72 h
(Fig. 2D). The latter, represents
a difference in what was seen in differentiating parental MEL cells
where a substantial decrease of RPS5 mRNA after 24 h and a
significant accumulation of βmajor mRNA after 36 h has
been detected (16,22). These marginal differences seen in
the expression level of both RPS5 and βmajor
globin genes in differentiating MEL-antisenseRPS5 cells
seems more likely to be related to their limited commitment
potential to erythroid maturation, as shown in Fig. 1.
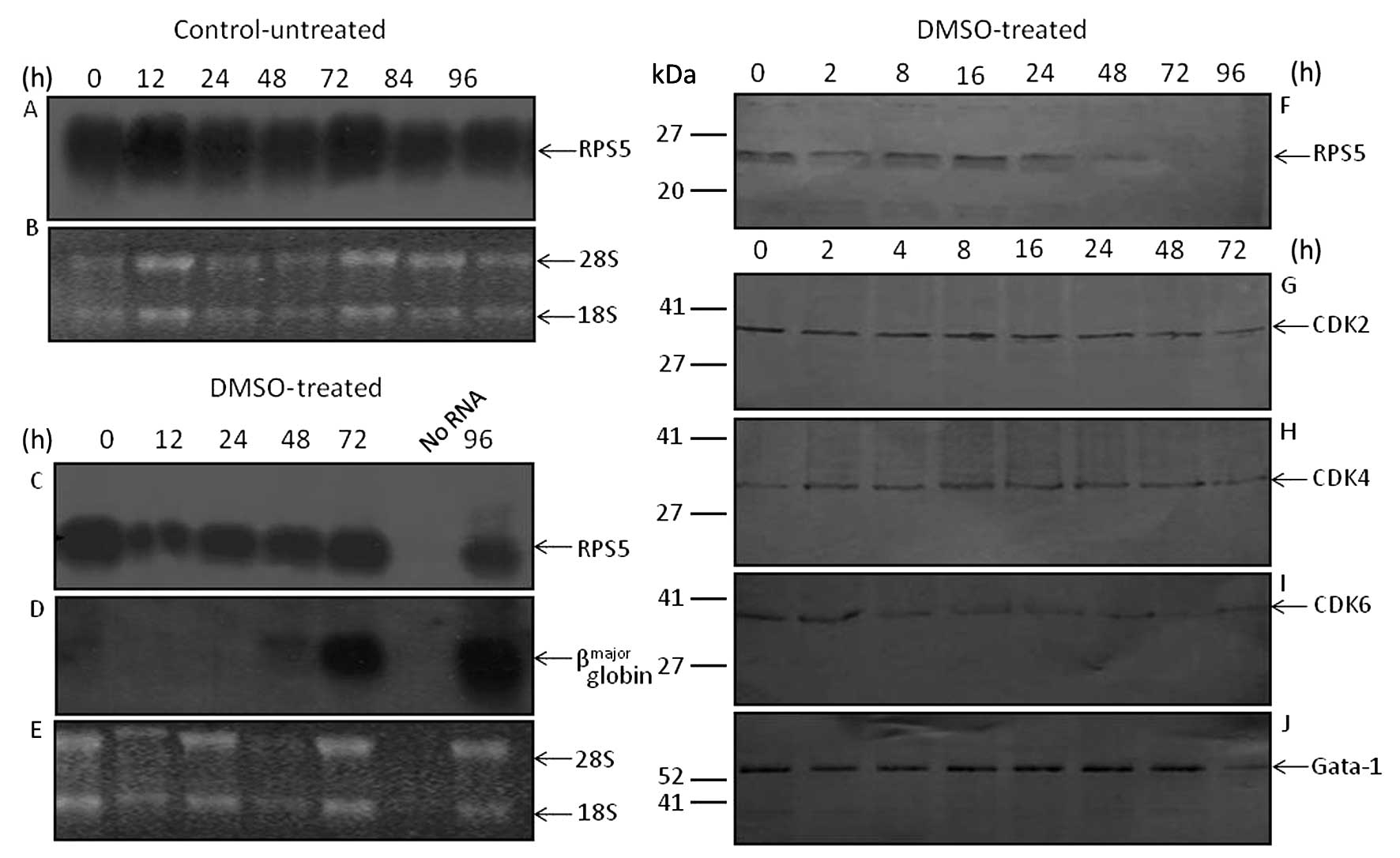 | Figure 2Assessment by northern blot
hybridization of the steady-state level of RPS5 and
βmajor globin RNA transcripts as well as RPS5 and CDKs
protein level in MEL-antisenseRPS5 cell cultures grown in
the presence of DMSO. MEL-antisenseRPS5 cell cultures were
grown in DMEM supplemented with 10% v/v FBS and G418 in the absence
(control-untreated) or presence (DMSO-treated) of DMSO (1.5% v/v).
At times indicated, cells were harvested from culture and total
cytoplasmic RNA and cellular protein extracts were isolated, as
described in Materials and methods. (A, C and D) Cytoplasmic RNA
(10 μg) was electrophoretically separated on 1% agarose gel,
transferred onto a nylon membrane, and hybridized at 65°C with
[32P]-labeled DNA fragments coding for mouse RPS5
mRNA (715 bp) (A and C) and/or βmajor globin gene
(7,304-bp genomic DNA fragment containing the entire gene) (D). The
ethidium-bromide staining pattern of the isolated cytoplasmic RNA
transcripts from control-untreated (B) and DMSO-treated (E)
MEL-antisenseRPS5 cells is shown. The position of the 28S
and 18S rRNAs is indicated by the arrows. (F–J) Western blot
analysis using 30 μg protein extracts from each culture preparation
was carried out for the assessment of RPS5 (F), CDK-2 (G), CDK-4
(H), CDK-6 (I) and Gata-1 (J) protein levels. Abs for RPS5
(C-terminal), CDK2, CDK-4 and CDK-6 (1:1,000 dilution) and for
Gata-1 (1:400 dilution) was used for immunoblotting, as shown in
Materials and methods. |
By knowing that commitment of MEL cells to erythroid
maturation is correlated with cell cycle changes, the assessment of
the expression profile of endogenous RPS5 and CDKs (CDK2, CDK4 and
CDK6) upon differentiation of MEL-antisenseRPS5 cells was
investigated. During the course of differentiation, the Gata-1
protein level was also assessed as an internal control for MEL
cells, as previously described (22). Cell lysates prepared from
differentiating MEL-antisenseRPS5 cultures and subjected to
immunoblotting revealed that the RPS5 protein level decreased quite
early after 24 h (Fig. 2F). This
is interesting, since in parental MEL cells the decrease of RPS5
protein is detected only after 72 h (22). Regarding the assessment of CDKs,
the level of CDK2 and CDK4 proteins remained almost constant,
whereas that of CDK6 decreased very early ~2 h after the induction
of differentiation (Fig. 2G–I).
These differences seen in the expression level of CDKs seems to be
correlated with the differentiation potential of
MEL-antisenseRPS5 cells, since the expression profile of
Gata-1 remained almost constant throughout the course of the
experiment (Fig. 2J). The
expression profile of CDKs detected at late stages of
differentiating MEL cells coincide with previous observations
(22,25–28).
In an effort to rule out the possibility that the
delay observed in the onset of differentiation of
MEL-antisenseRPS5 cells could be attributed to inducer DMSO,
the chemical inducer hexamethylene-bis-acetamide (HMBA) has been
also included for further analysis (13,14).
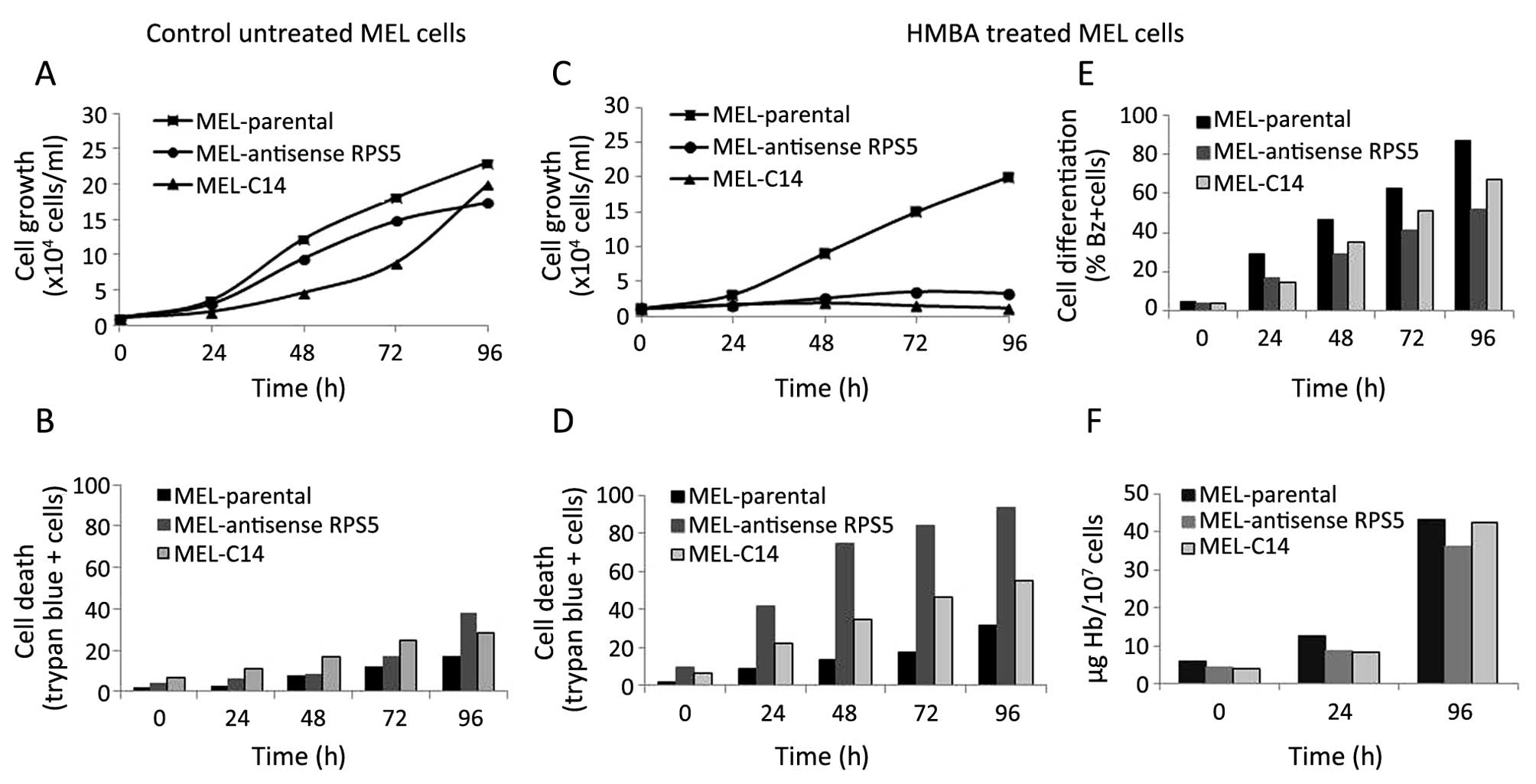
Based on the experimental data obtained, a decrease in the onset of
differentiation was also recorded since the number of
differentiating MEL-antisenseRPS5 cells accumulated in
culture after 96 h is less (~40%) as compared to that seen in
parental MEL cells (>80%) (Fig.
3E). Interestingly, this effect was more pronounced even by
comparing the number of differentiating cells seen in MEL-C14 cells
(~60%). The latter culture, as reported earlier, also exhibits a
delay in the initiation and the onset of differentiation upon
treatment with HMBA attributed to the overexpression of recombinant
RPS5-Myc-His protein (22).
Moreover, the amount of haemoglobin accumulated in differentiating
MEL-antisenseRPS5 is lower than that measured in parental
MEL and MEL-C14 cultures (Fig.
3F). In addition, cell growth is decreased dramatically in
MEL-antisenseRPS5 culture upon exposure to HMBA similarly to
MEL-C14 (Fig. 3C), whereas the
number of non-viable cells is much higher in
MEL-antisenseRPS5 (~85%) compared to both parental MEL
(~25%) and MEL-C14 (~45%) cells (Fig.
3D). However, these differences in cell growth capacity and
viability recorded in cultures is more likely to be attributed to
their differentiation potential, since the untreated-control cells
behave similarly and achieve comparable high numbers of
proliferating and viable cells after 96 h (Fig. 3A and B). This notion is further
supported by examining the morphology of cells in culture as well
as assessing the gene expression profile of β-actin (housekeeping
gene), RPS5 (gene of interest) and βmajor globin (a
developmentally regulated gene) upon induction of differentiation
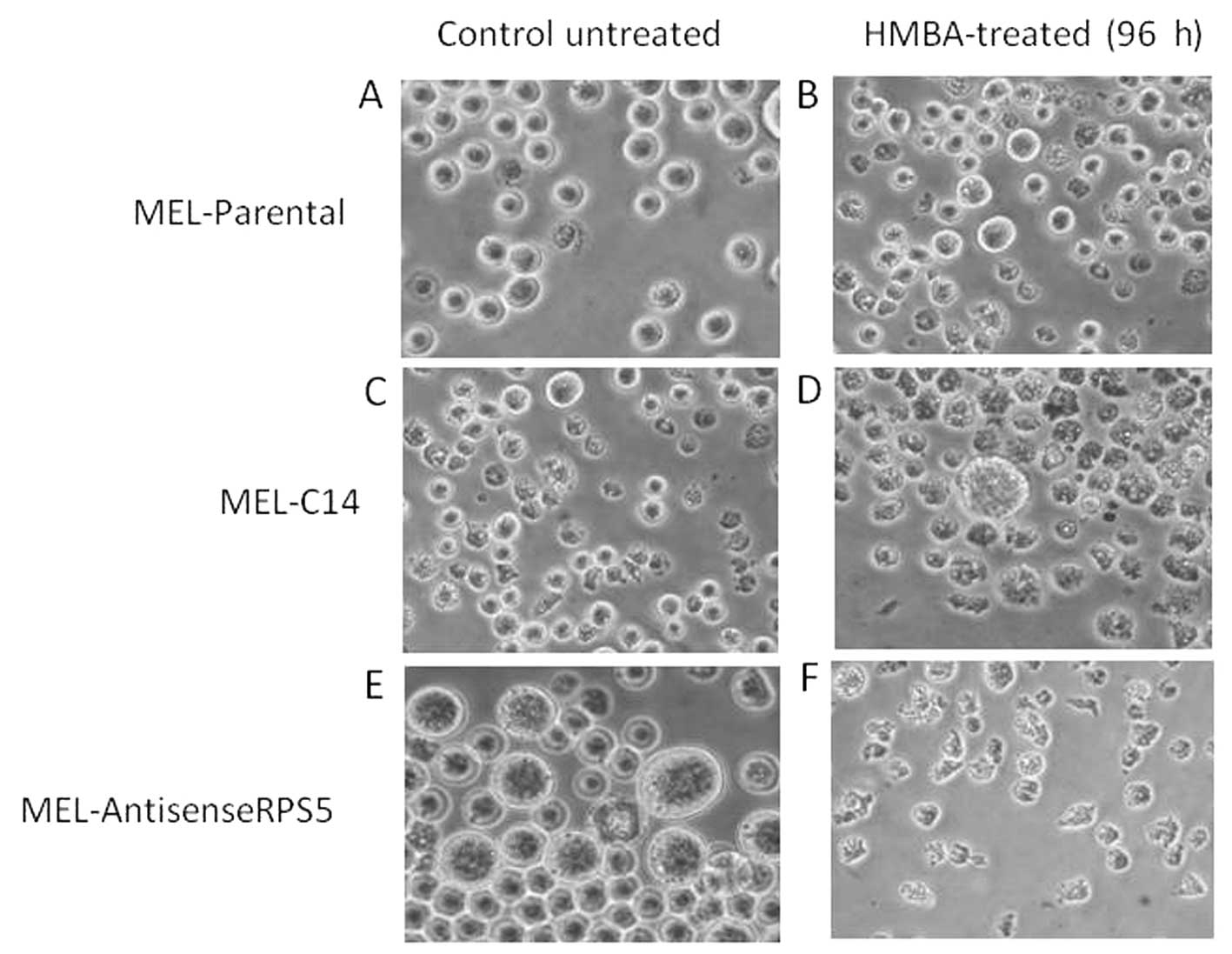
of MEL-antisenseRPS5. Indeed, the morphology of untreated
MEL-antisenseRPS5 exhibit an altered phenotype with a larger
size that, however, upon 96-h exposure to inducing agent HMBA is
further exacerbated by giving smaller although abnormally
differentiated cells (distorted nucleolus and nucleus) (Fig. 4). Moreover, a dramatic decrease in
mRNA levels for all three genes was observed after 48–72-h exposure
of cells to HMBA (data not shown), whereas no significant
alteration was seen for RNA transcripts of β-actin,
βmajor globin and c-myc (an oncogene with crucial role
in the differentiation potential of MEL cells) in control-untreated
cells continuously growing in culture even for 120 h. Such data
collectively propose a correlation between antisense-RPS5
transfection and the decrease in the onset of differentiation seen
in MEL-antisenseRPS5 cell culture, since dismantling of the
differentiation program is observed very early regardless of the
nature of the chemical inducer (DMSO and/or HMBA) employed.
RNAi-mediated silencing of RPS5 gene
expression blocked MEL cell differentiation in vitro
In order to more thoroughly investigate the
correlation of RPS5 gene expression with the capacity of MEL
cells to initiate their erythroid maturation program in
vitro, we applied the RNA interference (RNAi) methodology to
specifically downregulate the expression of RPS5 in this
culture. As shown in Fig. 5, by
using specific siRNAs the silencing of RPS5 gene in MEL cells was
achieved in both mRNA and protein level after 24 and 36 h,
respectively (Fig. 5A and C).
Interestingly, this effect was evident even after 72–96-h
continuous exposure to specific siRNAs for the RPS5 gene and
not for the scrambled siRNAs used as control (data not shown). The
successful RNAi-mediated silencing of RPS5 gene in MEL cells
prompted us to assess the capacity of such culture to differentiate
in vitro upon exposure to the chemical inducer HMBA. The
experimental design shown in Fig.
6 allowed us to verify for each experiment the successful RPS5
gene silencing and also to assess the differentiation capacity of
such a culture (Fig. 6A). In
particular, the cells were initially exposed to suitable siRNAs
(scrambled and/or RPS5-specific) for 24 h before the addition of
HMBA in culture. After an additional period of 24 h, the silencing
of RPS5 gene expression was verified by RT-PCR, as shown in
Fig. 6B. The RPS5 protein level
was not detectable after 96-h exposure to RPS5-specific siRNAs, in
contrast to cultures exposed to scrambled siRNAs (Fig. 6D). Importantly however, the
monitoring of differentiation potential of these cultures for the
entire 96-h exposure period to inducer HMBA indicated that only MEL
cells exposed to RPS5-specific siRNAs failed to initiate their
erythroid maturation program in vitro (Fig. 6F). The number of differentiating
cells (cells producing haemoglobin) accumulated after 96 h in
culture exposed to specific RPS5 siRNAs is very low (~15%),
whereas MEL cells exposed to scrambled siRNAs reach a level
comparable to parental ones (>85%). In parallel, assessment of
trypan-blue positive cells in these cultures revealed that in both
conditions the number of dead cells marginally exceed 20% after
72-h exposure to HMBA; however, after 96 h in culture only MEL
cells exposed to RPS5-specific siRNAs exhibited a high proportion
(80%) of dead cells (Fig. 6G).
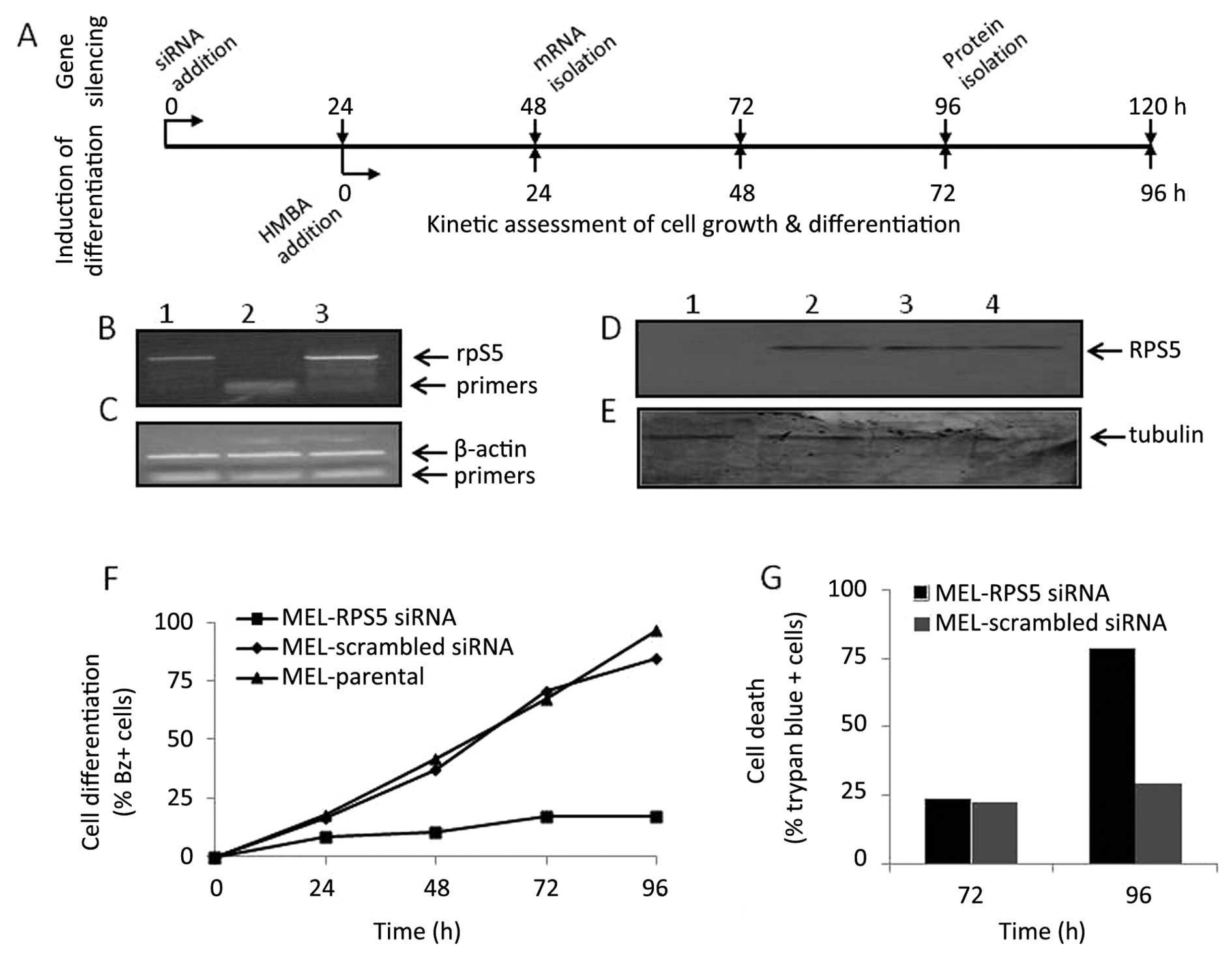 | Figure 6Assessment of differentiation
potential of parental MEL cells treated with RPS5 specific siRNAs.
Parental MEL cells were treated with either scrambled
(MEL-scrambled siRNA) or RPS5 specific (MEL-RPS5 siRNA) siRNAs as
indicated in Fig. 4 for 24 h,
before the addition of inducer HMBA (5 mM). Note that parental MEL
cells treated without siRNA molecules (MEL parental) exposed to
HMBA were also included in this experimental design as control.
Afterwards, the number of cells producing haemoglobin
(benzidine-positive cells; differentiated cells) accumulated in
culture was assessed kinetically for a 96-h period (F), as
described in Materials and methods. After 48- and 96-h continuous
exposure to siRNAs in culture, as shown diagrammatically (A),
isolation of total cytoplasmic RNA and protein extracts was carried
out to assess the expression RPS5 gene and protein level by RT-PCR
[(B) 0.5 μg RNA] and western blotting [(D) 20 μg protein],
respectively. The assessment of β-actin mRNA (C) and tubulin
protein (E) was also carried out and used as controls. (G) The
accumulation of trypan-blue (dead) staining cells was measured
after 72- and 96-h exposure of cells to the inducer HMBA (A). (B
and C) Cultures treated with HMBA for 24 h; lane 1, M_L parental;
lane 2, M_L-PRS5 siRNA; lane 3, M_L-scrambled siRNA. (D and E)
Cultures treated with (lanes 1–3) or without (lane 4) the presence
of HMBA for 72 h; lane 1, MEL-RPS5 siRNA; lane 2, MEL-scrambled
siRNA; lanes 3 and 4, parental MEL. The data shown are from a
representative experiment repeated at least three times and the
data represent the mean of two independent measurements. |
MEL cultures exposed to RPS5-specific siRNAs and
being induced to differentiate by HMBA exhibit different cellular
morphology. In particular, parental and scrambled siRNA-treated MEL
cells showed the typical erythroid maturation phenotype as expected
after 72–96 h. However, differentiating cells exposed to
RPS5-specific siRNAs present a phenotype more resembling that of
undifferentiated cells where nucleolus and nucleus are distorted
(data not shown). The latter, is in accordance with the inability
of such a culture to initiate commitment of erythroid maturation
and accumulate high proportion of haemoglobin-producing cells, as
presented above. In order to clarify such differentiation behavior
of cells, the assessment of the expression level of RPS5,
c-myc, βmajor globin and
β-actin genes was further investigated. Upon induction of
MEL cell differentiation with HMBA, parental MEL, MEL-C14 and
MEL-scrambled siRNA-treated cells exhibited decreased level
of RPS5 (data not shown). On the contrary, the steady-state level
of RPS5 RNA transcripts in MEL-siRNA RPS5 culture was much
lower from the beginning and throughout the entire exposure period,
as expected. The steady-state mRNA level of c-myc in
parental MEL showed an early initial increase at 6 h before a final
decline after 48 h, in agreement with a previous report (29). In MEL-C14 culture, the steady-state
c-myc RNA transcripts indicated a biphasic profile, where an
initial downregulation was observed very early (3 h) followed by an
increase after 24 h before the final decrease afterwards (data not
shown). Moreover, in both MEL-scrambled siRNA and MEL-siRNA
RPS5 cultures, the low expression level of c-myc at 24–48 h was
followed by a significant increase at 72 h before a final decrease
later on. In parallel, the level of βmajor globin gene
expression in differentiating parental MEL, MEL-C14 and
MEL-scrambled siRNAs cells showed a time-dependent increase,
as expected, with the higher level seen after 96 h in all cultures.
Importantly, however, the level of βmajor globin gene
expression in differentiating MEL-RPS5 siRNA exhibited no
substantial change by being kept at low levels even after 96 h as
compared to all other cultures (data not shown). Furthermore, the
level of β-actin mRNA did not show any significant change either
between the four cultures or during the course of
differentiation.
Restoration of normal RPS5 expression
levels in RNAi-mediated RPS5 gene silencing MEL cells leads the
culture to regain maximum erythroid maturation capacity in
vitro
The commitment of MEL cells to erythroid maturation
is associated with early accumulation of cells in
G0/G1 cell cycle phase arrest. In order to
assess that propensity of MEL cells in both RPS5-specific
siRNAs- and/or scrambled siRNAs-treated cultures, the accumulation
of cells to G0/G1 was evaluated upon
differentiation in vitro. No substantial difference between
the two cultures was recorded in the 24–72-h period tested (data
not shown). However, a difference in the differentiation potential
in the two cell cultures was previously seen and discussed
regarding the inability to commit only in cells treated with
RPS5-specific siRNAs. We reasoned additionally to assess the
accumulation of apoptotic cells upon induction of differentiation
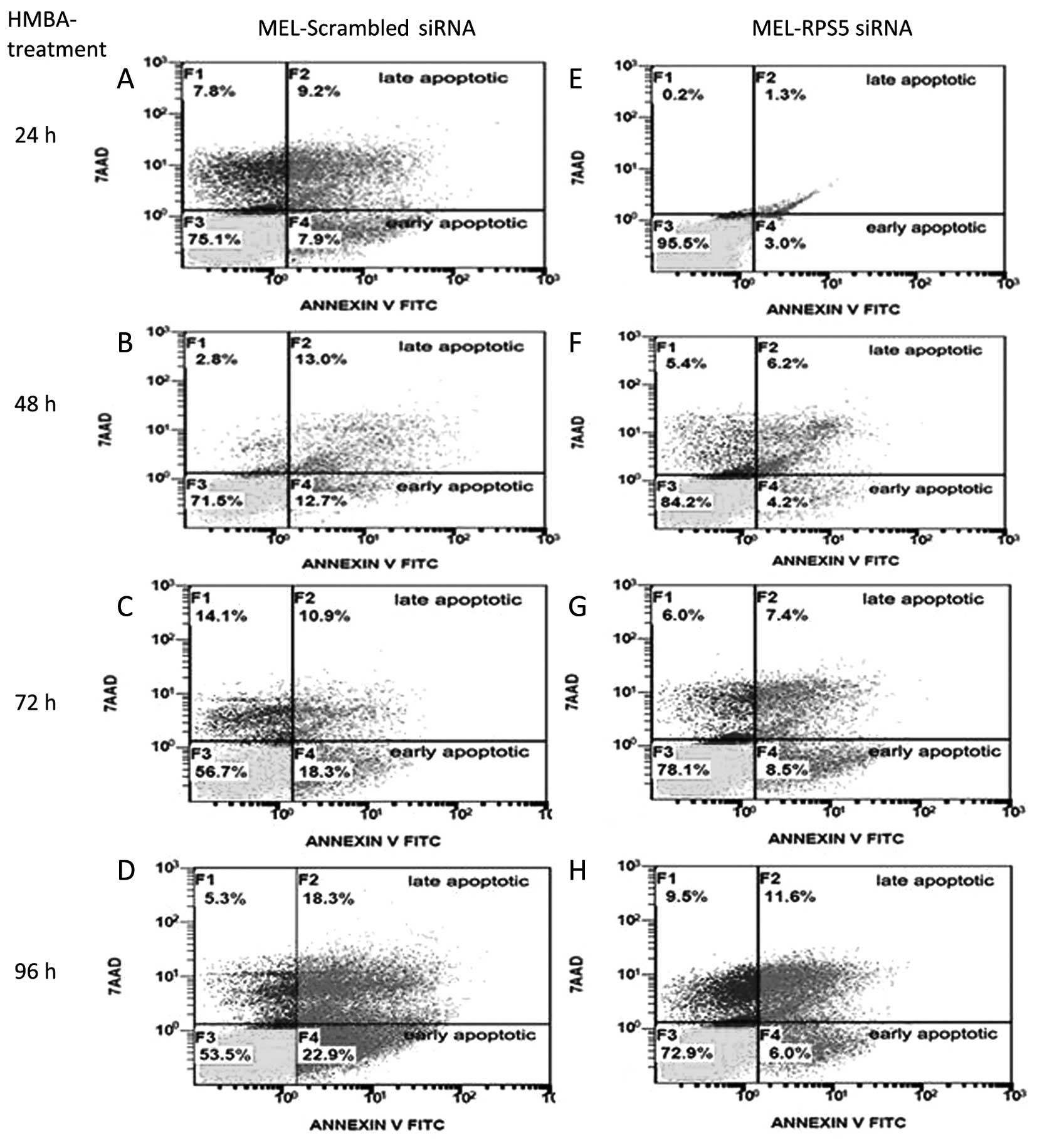
with HMBA. Indeed, as shown in Fig.
7, by using flow cytometry to assess Annexin V-positive cells,
we were able to evaluate the kinetics of apoptotic cells
accumulated in these cultures for the entire 24–96-h period.
MEL-RPS5siRNA culture from the beginning of the induction of
differentiation showed much lower proportion of cells exhibiting a
pre-apoptotic or apoptotic phenotype. Interestingly, MEL cells
treated with scrambled siRNAs accumulate ~32% of both pre-apoptotic
and apoptotic cells after 96 h, whereas specific RPS5
siRNAs-treated cells only ~17% during the same period.
The fact that the modulated gene expression level of
RPS5 is associated with an altered differentiation capacity of MEL
cell cultures in vitro (MEL-C14, MEL-antisenseRPS5
and MEL-RPS5 siRNA treated cells) has clearly proposed a
correlation between MEL cell differentiation potential and
RPS5 gene expression. In order to more precisely
inter-correlate such connection, an attempt was made to restore the
normal RPS5 expression levels in MEL cultures that previously
exhibited an RNAi-mediated RPS5 gene silencing and then reversely
assess their capacity to commit to erythroid maturation. In doing
so, the experiment shown in Fig. 8
was designed. MEL cells were exposed for 48 h with either
scrambled- or RPS5-specific siRNAs, before the replenishment
of cultures with fresh medium containing only the inducer HMBA, no
siRNAs (shown in Fig. 8A). MEL
cells cultured in the presence of specific RPS5 siRNAs for
48 h, were washed out and changed into fresh medium with the
addition of inducer, restored their RPS5 gene expression
levels within 24 h (Fig. 8B),
whereas the level of β-actin mRNA used as control remained
unchanged (Fig. 8C). However, such
MEL cells exhibit their maximum differentiation potential
comparable to that seen for both parental MEL and scrambled-siRNAs
treated MEL cultures (Fig. 8F).
Similarly, the kinetics of cellular proliferation in both terms of
growth (Fig. 8D) and trypan-blue
positive (dead) cells (Fig. 8E)
are indistinguishable between the three differentiating MEL
cultures. Overall, such data propose a close relationship between
the recorded RPS5 gene expression level in MEL cells and
their capacity to initiate the in vitro differentiation
program to erythroid maturation.
Discussion
In this study and in order to more thoroughly
investigate the function of RPS5 in the initiation of commitment to
erythroid maturation and the onset of MEL differentiation program
to accumulate haemoglobin-containing cells in culture, we stably
transfected MEL cells with the full length mouse anti-sense
RPS5 cDNA (MEL-antisenseRPS5) and also applied RNAi
methodology to transiently silence the expression of RPS5
gene (MEL-RPS5siRNA). The fact that overexpression of
recombinant RPS5 in MEL-C14 cells abrogated their reprogramming to
differentiation in vitro has shown a novel potential
extraribosomal function of this protein in erythropoiesis (22). Indeed, this conclusion has been
further supported through the establishment of
MEL-antisenseRPS5 culture and the induction of
differentiation upon exposure to chemical inducers. In particular,
and comparatively to MEL-C14, these cells achieved lower onset of
differentiation in culture, whereas they exhibited a delay in
completing their erythroid maturation program in full (haemoglobin
synthesis and limitation of proliferation potential). The latter
effect would be attributed to the fact that although these cells do
not produce full recombinant RPS5 RNA transcripts, they
might accumulate, however, truncated antisense RPS5 molecules in
the cytoplasm interfere with endogenous RPS5 mRNA to affect its
function. A similar effect has been reported in the case of Notch,
where the transfection of MEL cells with recombinant constructs of
Notch antisense cDNA modulated the endogenous Notch mRNA level and
function within the cells (30).
The comprehensive analysis of the experimental data presented in
this study indicated that although the cytoplasmic steady-state
level of endogenous RPS5 RNA transcripts remains unchanged in
differentiating MEL-antisenseRPS5 cells, the synthesis of
the corresponding RPS5 protein dramatically decreases very early,
soon after induction of differentiation as shown in this study, in
contrast to MEL-C14 cells where this effect happened at latter
stages of differentiation (22).
Such a result implies that antisense-RPS5 RNA molecules may
cause a translational inhibition of endogenous RPS5 mRNA, as seen
in the case of antisense-Notch cDNA transfection in MEL cells,
leading to modulated RPS5 function in the timed reprogramming of
these cells to differentiate. More importantly, the causal
relationship between the RPS5 gene expression and the
differentiation potential of MEL cells is undoubtedly supported by
the experiments presented in this study based on the selective
RNAi-mediated silencing of RPS5 gene. By exhibiting silencing of
RPS5 gene, the inability of such MEL culture to initiate
commitment to erythroid maturation in vitro has been
strikingly reversed by allowing cells to restore normal RPS5 gene
expression level. The latter, confirms previous observations
showing that imbalance in differentiation potential and
differentiation-dependent apoptosis is correlated with the capacity
of cells to continue proliferation in culture (13,14).
On the other hand, the cellular morphology showing a distortion of
nucleus and nucleolus, coincides with an imbalance between the need
for either proliferation (neoplasm phenotype), or
differentiation/apoptosis decisions (erythroid maturation
phenotype). Such a conclusion is in accordance with previously
published data showing that MEL cell differentiation in
vitro is associated very early with a noticeable decrease in
the expression of RPS5 gene, as well as a sharp inactivation
of the transcription of rRNAs genes (15,16).
Being also aware on the highly coordinated nature of the MEL
erythroid maturation program regarding the transcriptional
activation and/or repression of crucial genes, it is reasonable to
assume that somehow the deregulation of RPS5 contributes to an
imbalance in ribosomal biogenesis and function. It has been
reported that the blockade of MEL erythroid differentiation program
by methylation inhibitors (neplanocin A, 3-deaza-neplanocin A, and
cycloleucine), caused constitutive expression of RPS5, thus
implying a differentiation-dependent regulation for RPS5 gene in
these cells (31).
The ribosomal proteins are thought to have mainly
structural role to facilitate the proper configuration of rRNAs as
integral moieties of ribosomal subunits, thereby promoting the
speed and accuracy of the translation process (32). This conservative concept, however,
is changing, by knowing that besides their profound structural and
functional role in ribosome integrity and protein synthesis,
ribosomal proteins exert extraribosomal roles in gene
transcription, cell proliferation, apoptosis and differentiation
(33). Also recently, an intense
interest in delineating the role of ribosome function in the
pathogenesis of certain blood disorders has been emerged. Indeed,
ribosomal proteins have unexpectedly been found to contribute in a
series of pathological conditions such as Diamond-Blackfan anemia
(DBA) or 5q− syndrome. The latter happened at a rate
that led to the term ‘ribosomopathies’ which has been coined to
describe these and other syndromes characterized by abrogated
ribosome biogenesis (21).
Interestingly, the molecular pathogenesis of DBA depends on
mutations in a number of different ribosomal protein genes with
that of RPS19 accounting for ~25% of DBA patients (34). The striking specificity of the
defect manifestation mainly on red blood cell precursors is still
elusive, while the list of ribosomal protein genes found mutated is
growing to include members of the large as well the small ribosomal
subunit such as RPS24, RPS17, RPS10, RPS26, RPL35a, RPL5 and
RPL11 (35–39). On the other hand, RPS14 is
directly linked to 5q− syndrome pathogenesis, which
presents many similarities to the clinical symptoms of DBA
(40). Haploinsufficiency of
certain ribosomal protein genes in Drosophila has been
associated with a growth restricted phenotype called ‘minute’,
while in Zebrafish it leads to tumorigenesis by an unknown
mechanism (41). One of the major
events during erythroid differentiation is the modulation of the
number of ribosomes thus ensuring the synthesis of large amounts of
haemoglobin needed for red blood cell function (13). The pathophysiology at the molecular
level correlating to the selective dysfunction of the erythroid
cell lineage with disorders of ribosome biogenesis is still elusive
besides the fact that the p53 activation and p21 accumulation has
been shown to cause cell cycle arrest in such ribosome abnormal
erythroid progenitor cells. The latter, proposes that the erythroid
lineage has a low threshold for the induction of p53, thus
providing a basis for the failure of erythropoiesis in disorders
like 5q− syndrome and DBA (42). A clear connection between
erythropoiesis, ribosome biogenesis and specific ribosomal protein
function is still elusive. We do not know precisely how ribosomal
proteins contribute to the formation of the 40S and 60S ribosomal
subunits and to which extent they contribute to the protein
synthesis. In MEL cells for example, it has been proposed that
ribosomal proteins are added sequentially during the formation of
the small (40S) ribosomal subunit (43). In particular, RPS5 together with
RPS4 and RPS12 have been implicated in the signaling step for the
formation of a peptide bond (after the binding of tRNA in the
ribosome) during the translation process (44). For example, the targeted disruption
of mouse PRS19 gene that was lethal prior to implantation, thus
exhibiting a potential role in erythropoiesis and development
(45). Moreover, it is also
noteworthy that RPS3 can act as a potential receptor for chemical
inducers that initiate MEL cell differentiation in vitro,
thus enriching our knowledge regarding an important extraribosomal
functional role of this specific protein within cancer cells
(46).
Initiation of MEL cell differentiation has been
reported to occur within the G1/S interphase and that
terminal erythroid cell maturation is associated with
G0/G1 cell cycle arrest and the coordinated
function of CDK2, CDK4 and CDK6 along with their inhibitors (CDKIs)
(13,14,25–28).
As reported earlier, MEL-C14 cells delayed the initiation of the
onset of differentiation upon exposure to chemical inducers,
whereas at the same time a slower entrance of cells in
G0/G1 phase and significant changes in the
profile of CDK2, CDK4 and CDK6 have been recorded (22). Such a conclusion also applies for
differentiating MEL-antisenseRPS5 cells as reported in this
study, where a delay in the onset of initiation of differentiation
has been uncovered with simultaneous significant alterations in the
protein levels of individual CDKs. Alternatively, the deregulation
in cell cycle and differentiation seen upon modulating RPS5
gene expression in MEL cells could be attributed to the altered
c-myc expression profile seen in this study. Indeed, recent studies
have clearly indicated the existence of a molecular link connecting
cell growth control and ribosome biogenesis, as well as ribosome
dysfunction and cell cycle regulation (47,48).
In particular, c-myc plays a major role in balancing
ribosome component production, whereas impairment of ribosome
biogenesis (e.g., deregulation of RPL11 and RPL5)
leads to p53 induction thus hindering cell cycle progression.
Whether such a connection already exists for RPS5 in MEL cells is
still elusive. As shown in this study, the extra-ribosomal function
of RPS5 related to the modulation of the onset of MEL cell
differentiation program must be attributed to the latent period
before the initiation of commitment. This is an interesting, and
intriguing possibility requiring further experimental verification
and validation to provide insights into how RPS5 exerts its roles
for crucial cellular decisions, such as proliferation,
differentiation and apoptosis. In a previous study, the selective
repression of RPS19 gene expression in human CD34+ cells
caused abnormal ribosomal biogenesis that consequently has been
clearly correlated with irregular cell cycle arrest and
lineage-specific defects of erythroid progenitors (49). The data presented in this study
contribute to the better understanding of the role of RPS5 in the
initiation of commitment of MEL cells to terminal erythroid
maturation. Moreover, such results provide new knowledge on the
borderlines of erythropoiesis on how the pathogenesis of
ribosomopathies and the modulated gene expression of RPS5 could
impair red blood cell production and the function of haemoglobin.
This is considered very attractive especially in light of evidence
(50) showing that individual
components of the translation machinery are deregulated in cancer
cells, an event that also suggests the therapeutic exploitation of
specific ribosome-related molecules as candidate targets for cancer
therapy.
Acknowledgements
We would like to thank Elsa P. Amanatiadou (Ph.D.
Fellow, Laboratory of Pharmacology, Department of Pharmaceutical
Sciences at Aristotle University of Thessaloniki) for her
substantial help both in experimentation and upon the preparation
of this manuscript. This study was supported by interdepartmental
funds and the Research Committee of the Aristotle University of
Thessaloniki (no. 88074 to I.S.V.).
References
|
1
|
Adams GB and Scadden DT: The hematopoietic
stem cell in its place. Nat Immunol. 7:333–337. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Moore KA and Lemischka IR: Stem cells and
their niches. Science. 311:1880–1885. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Stamatoyannopoulos G: Control of globin
gene expression during development and erythroid differentiation.
Exp Hematol. 33:259–271. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Orkin SH and Zon LI: Hematopoiesis: An
evolving paradigm for stem cell biology. Cell. 132:631–644. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Palis J: Ontogeny of erythropoiesis. Curr
Opin Hematol. 15:155–161. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsiftsoglou AS, Vizirianakis IS and
Strouboulis J: Erythropoiesis: Model systems, molecular regulators,
and developmental programs. IUBMB Life. 61:800–830. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tsiftsoglou AS, Bonovolias ID and
Tsiftsoglou SA: Multilevel targeting of hematopoietic stem cell
self-renewal, differentiation and apoptosis for leukemia therapy.
Pharmacol Ther. 122:264–280. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Edling CE and Hallberg B: c-Kit - a
hematopoietic cell essential receptor tyrosine kinase. Int J
Biochem Cell Biol. 39:1995–1998. 2007. View Article : Google Scholar
|
|
9
|
Elliott S, Pham E and Macdougall IC:
Erythropoietins: A common mechanism of action. Exp Hematol.
36:1573–1584. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang CC and Lodish HF: Cytokines
regulating hematopoietic stem cell function. Curr Opin Hematol.
15:307–311. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rice KN and Jamieson CH: Molecular
pathways to CML stem cells. Int J Hematol. 91:748–752. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
List AF, Vardiman J, Issa JP and DeWitte
TM: Myelodysplastic syndromes. Hematology (Am Soc Hematol Educ
Program). 2004:297–317. 2004. View Article : Google Scholar
|
|
13
|
Tsiftsoglou AS, Pappas IS and Vizirianakis
IS: Mechanisms involved in the induced differentiation of leukemia
cells. Pharmacol Ther. 100:257–290. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsiftsoglou AS, Pappas IS and Vizirianakis
IS: The developmental program of murine erythroleukemia cells.
Oncol Res. 13:339–346. 2003.PubMed/NCBI
|
|
15
|
Tsiftsoglou AS, Wong W, Volloch V, Gusella
J and Housman D: Commitment of murine erythroleukemia (MEL) cells
to terminal differentiation is associated with coordinated
expression of globin and ribosomal genes. Prog Clin Biol Res.
102A:69–79. 1982.
|
|
16
|
Vizirianakis IS, Pappas IS, Gougoumas D
and Tsiftsoglou AS: Expression of ribosomal protein S5 cloned gene
during differentiation and apoptosis in murine erythroleukemia
(MEL) cells. Oncol Res. 11:409–419. 1999.
|
|
17
|
Pappas IS, Vizirianakis IS and Tsiftsoglou
AS: Cloning, sequencing and expression of a cDNA encoding the mouse
L35a ribosomal protein during differentiation of murine
erythroleukemia (MEL) cells. Cell Biol Int. 25:629–634. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Housman D, Volloch V, Tsiftsoglou AS,
Levenson R, Gusella JF, Kernen J and Mitrani A: Analysis of the
molecular basis of commitment in murine erythroleukemia (MEL)
cells. In Vivo and In Vitro Erythropoiesis: The Friend Cell System.
Rossi GB: Elsevier/North Holland Biomedical Press; Amsterdam: pp.
273–282. 1979
|
|
19
|
Hensold JO, Barth-Baus D and Stratton CA:
Inducers of erythroleukemic differentiation cause messenger RNAs
that lack poly(A)-binding protein to accumulate in translationally
inactive, salt-labile 80S ribosomal complexes. J Biol Chem.
271:23246–23254. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bessis M, Lessin LS and Beutler E:
Morphology of the erythron. Hematology. Williams WJ, Beutler E,
Erslev AJ and Lichtman MA: McGraw-Hill; New York, NY: pp. 257–279.
1983
|
|
21
|
Raiser DM, Narla A and Ebert BL: The
emerging importance of ribosomal dysfunction in the pathogenesis of
hematologic disorders. Leuk Lymphoma. 55:491–500. 2014. View Article : Google Scholar
|
|
22
|
Matragkou CN, Papachristou ET, Tezias SS,
Tsiftsoglou AS, Choli-Papadopoulou T and Vizirianakis IS: The
potential role of ribosomal protein S5 on cell cycle arrest and
initiation of murine erythroleukemia cell differentiation. J Cell
Biochem. 104:1477–1490. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Vizirianakis IS, Wong W and Tsiftsoglou
AS: Analysis of the inhibition of commitment of murine
erythroleukemia (MEL) cells to terminal maturation by
N6-methyladenosine. Biochem Pharmacol. 44:927–936. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gougoumas DD, Vizirianakis IS, Triviai IN
and Tsiftsoglou AS: Activation of Prn-p gene and stable
transfection of Prn-p cDNA in leukemia MEL and neuroblastoma N2a
cells increased production of PrP(C) but not prevented DNA
fragmentation initiated by serum deprivation. J Cell Physiol.
211:551–559. 2007. View Article : Google Scholar
|
|
25
|
Hsieh FF, Barnett LA, Green WF, Freedman
K, Matushansky I, Skoultchi AI and Kelley LL: Cell cycle exit
during terminal erythroid differentiation is associated with
accumulation of p27Kip1 and inactivation of cdk2 kinase.
Blood. 96:2746–2754. 2000.PubMed/NCBI
|
|
26
|
Matushansky I, Radparvar F and Skoultchi
AI: Manipulating the onset of cell cycle withdrawal in
differentiated erythroid cells with cyclin-dependent kinases and
inhibitors. Blood. 96:2755–2764. 2000.PubMed/NCBI
|
|
27
|
Matushansky I, Radparvar F and Skoultchi
AI: Reprogramming leukemic cells to terminal differentiation by
inhibiting specific cyclin-dependent kinases in G1. Proc Natl Acad
Sci USA. 97:14317–14322. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu L and Skoultchi AI: Coordinating cell
proliferation and differentiation. Curr Opin Genet Dev. 11:91–97.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Vizirianakis IS, Pappas IS and Tsiftsoglou
AS: Differentiation-dependent repression of c-myc, B22, COX II and
COX IV genes in murine erythroleukemia (MEL) cells. Biochem
Pharmacol. 63:1009–1017. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shelly LL, Fuchs C and Miele L: Notch-1
inhibits apoptosis in murine erythroleukemia cells and is necessary
for differentiation induced by hybrid polar compounds. J Cell
Biochem. 73:164–175. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vizirianakis IS and Tsiftsoglou AS:
Blockade of murine erythroleukemia cell differentiation by
hypomethylating agents causes accumulation of discrete small
poly(A)- RNAs hybridized to 3′-end flanking sequences of
beta(major) globin gene. Biochim Biophys Acta. 1743:101–114. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zimmermann RA: The double life of
ribosomal proteins. Cell. 115:130–132. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wool IG: Extraribosomal functions of
ribosomal proteins. Trends Biochem Sci. 21:164–165. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Draptchinskaia N, Gustavsson P, Andersson
B, Pettersson M, Willig TN, Dianzani I, Ball S, Tchernia G, Klar J,
Matsson H, et al: The gene encoding ribosomal protein S19 is
mutated in Diamond-Blackfan anaemia. Nat Genet. 21:169–175. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cmejla R, Cmejlova J, Handrkova H, Petrak
J and Pospisilova D: Ribosomal protein S17 gene (RPS17) is mutated
in Diamond-Blackfan anemia. Hum Mutat. 28:1178–1182. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gazda HT, Grabowska A, Merida-Long LB,
Latawiec E, Schneider HE, Lipton JM, Vlachos A, Atsidaftos E, Ball
SE, Orfali KA, et al: Ribosomal protein S24 gene is mutated in
Diamond-Blackfan anemia. Am J Hum Genet. 79:1110–1118. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Farrar JE, Nater M, Caywood E, McDevitt
MA, Kowalski J, Takemoto CM, Talbot CC Jr, Meltzer P, Esposito D,
Beggs AH, et al: Abnormalities of the large ribosomal subunit
protein, Rpl35a, in Diamond-Blackfan anemia. Blood. 112:1582–1592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gazda HT, Sheen MR, Vlachos A, Choesmel V,
O’Donohue MF, Schneider H, Darras N, Hasman C, Sieff CA, Newburger
PE, et al: Ribosomal protein L5 and L11 mutations are associated
with cleft palate and abnormal thumbs in Diamond-Blackfan anemia
patients. Am J Hum Genet. 83:769–780. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Doherty L, Sheen MR, Vlachos A, Choesmel
V, O’Donohue MF, Clinton C, Schneider HE, Sieff CA, Newburger PE,
Ball SE, et al: Ribosomal protein genes RPS10 and RPS26 are
commonly mutated in Diamond-Blackfan anemia. Am J Hum Genet.
86:222–228. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ebert BL, Pretz J, Bosco J, Chang CY,
Tamayo P, Galili N, Raza A, Root DE, Attar E, Ellis SR, et al:
Identification of RPS14 as a 5q-syndrome gene by RNA interference
screen. Nature. 451:335–339. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Perry RP: Balanced production of ribosomal
proteins. Gene. 401:1–3. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Dutt S, Narla A, Lin K, Mullally A,
Abayasekara N, Megerdichian C, Wilson FH, Currie T, Khanna-Gupta A,
Berliner N, et al: Haploinsufficiency for ribosomal protein genes
causes selective activation of p53 in human erythroid progenitor
cells. Blood. 117:2567–2576. 2011. View Article : Google Scholar :
|
|
43
|
Todorov IT, Noll F and Hadjiolov AA: The
sequential addition of ribosomal proteins during the formation of
the small ribosomal subunit in Friend erythroleukemia cells. Eur J
Biochem. 131:271–275. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Purohit P and Stern S: Interactions of a
small RNA with antibiotic and RNA ligands of the 30S subunit.
Nature. 370:659–662. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Matsson H, Davey EJ, Draptchinskaia N,
Hamaguchi I, Ooka A, Levéen P, Forsberg E, Karlsson S and Dahl N:
Targeted disruption of the ribosomal protein S19 gene is lethal
prior to implantation. Mol Cell Biol. 24:4032–4037. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Webb Y, Zhou X, Ngo L, Cornish V, Stahl J,
Erdjument-Bromage H, Tempst P, Rifkind RA, Marks PA, Breslow R, et
al: Photoaffinity labeling and mass spectrometry identify ribosomal
protein S3 as a potential target for hybrid polar
cytodifferentiation agents. J Biol Chem. 274:14280–14287. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lempiäinen H and Shore D: Growth control
and ribosome biogenesis. Curr Opin Cell Biol. 21:855–863. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Teng T, Thomas G and Mercer CA: Growth
control and ribosomopathies. Curr Opin Genet Dev. 23:63–71. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kuramitsu M, Hamaguchi I, Takuo M, Masumi
A, Momose H, Takizawa K, Mochizuki M, Naito S and Yamaguchi K:
Deficient RPS19 protein production induces cell cycle arrest in
erythroid progenitor cells. Br J Haematol. 140:348–359. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ruggero D and Pandolfi PP: Does the
ribosome translate cancer? Nat Rev Cancer. 3:179–192. 2003.
View Article : Google Scholar : PubMed/NCBI
|