Introduction
Tetrahydrolipstatin (orlistat), a well-known
irreversible inhibitor of pancreatic and gastric lipases, is
extensively used as anti-obesity drug (1). This agent, that is administered by
oral route, is minimally absorbed by the gastrointestinal tract and
is able to prevent the absorption of a large percentage of lipids,
thereby reducing lipid supply from outside sources.
Of particular interest is the finding that orlistat
is also a potent inhibitor of fatty acid synthase (FASN), an enzyme
that plays an important role in tumor growth and progression
(2) and is considered a metabolic
oncogene (3). Accordingly, the
agent shows antiproliferative activity against cancer cells both
in vitro and in animal models (2).
The antitumor activity of orlistat, however, does
not seem to be entirely dependent on its activity against FASN. A
large amount of experimental data indicates that the antineoplastic
effects of the agent may also be due to the inhibition of fatty
acid synthesis through different metabolic pathways without
altering FASN activity (4) or
impairing mitochondrial respiration (5). In any case, orlistat produces
cytotoxic effects through activation of apoptotic pathways preceded
by endoplasmic reticulum stress (6), although it is not clear whether this
mechanism is preferentially involved in the suppression of
neoplastic cells.
It has been previously shown that saturated fatty
acids downregulate cell response to DNA damage, thus favoring
malignant transformation (7).
According to this mechanism, one would expect that FASN inhibition
could be of value not only as a device for attaining antitumor
effects, but also as an anticancer treatment modality. In addition,
orlistat has been found to augment pro-apoptotic NOXA protein
(8), thus reinforcing its possible
cancer protective activity. However, preclinical studies performed
on rats exposed to predisposing factors for colon cancer (i.e.,
receiving high fat diet alone or combined with the carcinogen
compound methyl hydrazine), showed that long-term treatment with
orlistat lead to severe crypt alterations of colonic mucosa, that
are considered colon cancer biomarkers (9,10).
These results do not seem to be in line with the
expected cancer prevention activity of orlistat. Therefore, we
decided to obtain further insight into the pharmacodynamic
properties of orlistat by evaluating its activity on other
cell-associated biochemical functions. In view of the extensive
protective role against malignant transformation played by several
DNA repair enzymes, we elected to dedicate our attention in
particular to O6-methylguanine-DNA
methyltransferase (MGMT). This protein plays a primary role in the
defense against chemical carcinogenesis (reviewed in ref. 11) since it removes methyl adducts at
O6-guanine in DNA, which are produced by
several DNA mono-methylating agents, including environmental
carcinogenic compounds such as N-nitroso derivatives involved in
colorectal cancer (12). Moreover,
it has been found that MGMT loss could be responsible of PIK3CA
mutations present in human colon cancer (13).
The results of the present study pointed out that
orlistat is able to downregulate MGMT protein expression at a
concentration that could be reasonably attainable at the level of
intestinal epithelial cells under oral long-term treatment with the
drug (i.e., 120 mg three times a day). It follows that our findings
support the indication that long-term use (≥1 year) of high doses
of orlistat in overweight subjects would require appropriate
surveillance for possible gastrointestinal carcinogenesis
threats.
Materials and methods
Cell lines
The human colon cancer cell line HCT116, kindly
provided by Dr G. Marra (Institute of Molecular Cancer Research,
University of Zurich, Zurich, Switzerland), was maintained in
McCoy’s 5A medium (Sigma-Aldrich, Milan, Italy) supplemented with
10% heat-inactivated (56°C, 30 min) fetal calf serum (FCS,
Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), and 50 μg/ml
gentamicin (Euroclone, Milan, Italy).
The human colon cancer cell line HT-29, obtained
from the American Type Culture Collection (ATCC, Rockville, MD,
USA), was routinely grown in Dulbecco’s modified Eagle’s medium
(DMEM, Sigma-Aldrich), supplemented with 2 mM L-glutamine, 60 μM
gentamicin and 10% heat-inactivated FCS (Sigma-Aldrich), hereafter
referred to as complete medium D (CMD). Both HCT116 and HT-29 cells
growing as plastic adherent cells were removed using a solution of
0.05% trypsin and 0.02% EDTA in phosphate-buffered saline (PBS)
without calcium and magnesium.
The human Jurkat CD4+ T cell leukemia
cell line and the human promyelocytic leukemia cell line HL-60 were
obtained from the ATCC and were cultured at 37°C in a 5%
CO2 humidified atmosphere in RPMI-1640 (Sigma-Aldrich),
supplemented with 10% heat-inactivated FCS (Sigma-Aldrich), 2 mM
L-glutamine (Sigma-Aldrich), and 50 μg/ml gentamicin (Euroclone)
hereafter referred to as complete medium (CM).
The human melanoma cell line M10, kindly donated by
Dr D. Del Bufalo (Regina Elena Cancer Institute, Rome, Italy), was
originally established from a cutaneous metastasis of advanced
melanoma. The line, regularly cultured in CM in our laboratory,
shows fairly high levels of MGMT activity (14). M10 cells were removed from
continuous culture as described for the colon cancer cell
lines.
Peripheral blood mononuclear cells were obtained by
centrifugation on a Ficoll-Hypaque (Lymphoprep™, Axis-Shield, Oslo,
Norway) gradient of buffy coats from healthy blood donors, and
washed twice in RPMI-1640 medium. Non adherent mononuclear cells
(NAMNC) were separated by plastic adherence at 37°C for 1 h in CM.
Informed consent was obtained from blood donors according to our
institutional guidelines.
Drugs
Orlistat was purchased from Sigma-Aldrich, dissolved
in sterile DMSO (Sigma-Aldrich) at the concentration of 20 mM,
aliquoted and stored at −70°C until use. Lomeguatrib (LM) was a
generous gift from Professor G. Margison (Centre for Occupational
and Environmental Health, University of Manchester, Manchester,
UK). The compound was dissolved in DMSO at the concentration of 10
mM, aliquoted and stored at −70°C until use. Temozolomide (TMZ) was
supplied by Schering-Plough Co. (Milan, Italy) and was always
prepared freshly in culture medium and added immediately to cell
suspensions, because the drug readily decomposes in aqueous
solution.
MGMT activity assay
MGMT activity was determined by measuring the
transfer of 3H-methyl groups from a DNA substrate to the
MGMT protein (15). Briefly, cell
pellets (1×106 cells) were re-suspended in 1 ml of lysis
buffer (0.5% CHAPS, 50 mM Tris-HCl pH 8.0, 1 mM EDTA, 3 mM
dithiothreitol, 100 mM NaCl, 10% glycerol) supplemented with a
cocktail of protease inhibitors (Roche, Mannheim, Germany) and
incubated for 30 min at 4°C. Cell lysates were then centrifuged at
18,000 × g for 10 min at 4°C. Aliquots of supernatants were then
diluted in 50 mM Tris-HCl buffer, pH 8.3, containing 1 mM EDTA, and
3 mM dithiothreitol, and incubated with 10 μg of
3H-methylated-DNA at 37°C for 1 h. DNA was then
hydrolyzed by heating samples at 75°C for 45 min, in the presence
of 1 N perchloric acid, and protein precipitated using 1 mg bovine
serum albumin as carrier. Pellets were washed with 1 N perchloric
acid, re-suspended in 0.01 N NaOH, and radioactivity measured in a
liquid scintillation counter (TRI-CARB 1900, Packard Instruments
Co., Meriden, CT, USA), after addition of scintillation liquid
(Ultima Gold, Packard Instruments Chemical Operation, Groningen,
The Netherlands). Protein concentration in supernatants was
evaluated according to the method of Bradford using the Bio-Rad
Protein Assay Dye reagent (Bio-Rad Laboratories Inc., Hercules, CA,
USA) and bovine serum albumin as standard. MGMT activity was
expressed in terms of fmoles of 3H-methyl groups
transferred per mg of protein in cell extract.
Preparation of cell extracts
Cells were washed with PBS. The cell pellet was
suspended in 100 μl extraction buffer [(12.5 mM
Na2HPO4, pH 7.2; 94 mM NaCl; 50 mM NaF; 1%
di-Triton X-100; 2 mM EGTA, 1% protease inhibitor cocktail
(Sigma-Aldrich), 0.5% saponin (Sigma-Aldrich) and 1 mM
Na3VO4: (Sigma-Aldrich)] kept on ice for 30
min, and centrifuged for 30 min at 15,000 × g at 4°C in an
Eppendorf microcentrifuge. The protein concentrations were
determined using Bio-Rad Protein Assay Dye. The samples were stored
at −80°C until use.
Electrophoresis and immunoblotting
The proteins were heated in a boiling water bath for
2 min and separated by running cell extracts on 10% polyacrylamide
pre-cast gels (NuPAGE® Novex Bis-Tris, Invitrogen, Life
Technologies, Grand Island, NY, USA), using XCell SureLock™
Mini-Cell apparatus (Invitrogen) following the instructions of the
producer. At the end of the electrophoretic separation, proteins
were transferred to Hybond-ECL nitrocellulose filters (GE
Healthcare Life Sciences, Pittsburg, PA, USA) by electrotransfer
with miniblot apparatus (Invitrogen). The transfer was carried out
at 25 V overnight. Thereafter, the membranes were incubated with 3%
non-fat dry milk (Bio-Rad) in Tris-buffered saline (TBS) at pH 7.5,
for 60 min at room temperature and then incubated with an
anti-actin rabbit polyclonal antibody (Sigma-Aldrich) diluted
1:3,000 in TBS containing 0.05% Tween-20 (TBST) and mouse
monoclonal antibody against MGMT (Millipore, Billerica, MA, USA)
diluted 1:500 in TBST for 60 min. The membranes were washed twice
with TBST and incubated for 45 min with the secondary alkaline
phosphate-conjugated antibody. Bands were developed using
westernBreeze chemiluminescent immunodetection kit (Invitrogen),
according to the manufacturer’s instructions. The film was scanned
on a GS-710 Calibrated Imaging Densitometer and analyzed by means
of Quantity One software, version 4.1.1 (Bio-Rad Laboratories). The
optical density (OD) of MGMT and actin bands was expressed as
arbitrary units. MGMT expression was evaluated on the basis
MGMT:actin ratio (OD-R).
RNA isolation and RT-qPCR
Total RNA was extracted, from 2×106
viable cells using 1 ml of TRI Reagent solution (Ambion, Life
Technologies, Monza, Italy) according to the manufacturer’s
instructions. RNA purity and concentration were checked with a
NanoQuant Infinite M200 instrument (Tecan Group Ltd., Mannedorf,
Switzerland). Two microliters of RNA was purified by clearance of
DNA traces using Turbo DNA-free kit (Applied Biosystems, Life
Technologies, Monza, Italy). cDNA was synthesized from 2 μg of RNA
by reverse transcription using TaqMan RT kit (Applied Biosystems),
according to the manufacturer’s instructions. Five microliters
(i.e., 2 μg) of cDNA/sample was amplified, according to the
manufacturer’s instructions, by the Stratagene Mx3005P qPCR System
(La Jolla, CA, USA) using a TaqMan gene expression assay kit
(Applied Biosystems, Assay-On-Demand: code no. Hs.00172470_m1 for
MGMT). Levels of MGMT mRNA were normalized against glyceraldehyde
3-phosphate dehydrogenase (GAPDH) housekeeping gene expression
(Applied Biosystems, code no. 4326317E). All RT-qPCR reactions were
performed in triplicate.
The relative quantification of MGMT was performed
using the comparative threshold cycle (CT) method (as
described by Applied Biosystems protocol) that uses an arithmetic
formula (2−ΔΔCT). ΔΔCT is the difference
between the ΔCT of the sample under investigation and
the ΔCT of the calibrator sample. The RNA obtained from
Jurkat cells was chosen as the calibrator sample.
Statistical analysis
Statistical significance among different mean values
was assessed using two-tailed Student’s t-test analysis.
Results
Effect of orlistat treatment on MGMT
expression in tumor cells or in normal NAMNC
In order to determine the concentration range of
orlistat to be utilized in the experiments illustrated in the
present report, a concentration/effect study was performed using
Jurkat target cells. Leukemic cells were cultured in the presence
of graded concentrations of orlistat for two days. Control cultures
were exposed to DMSO alone at the concentration corresponding to
that utilized for orlistat 40 μM. At the end of the in vitro
treatment, leukemic cells were lysed and subjected to western blot
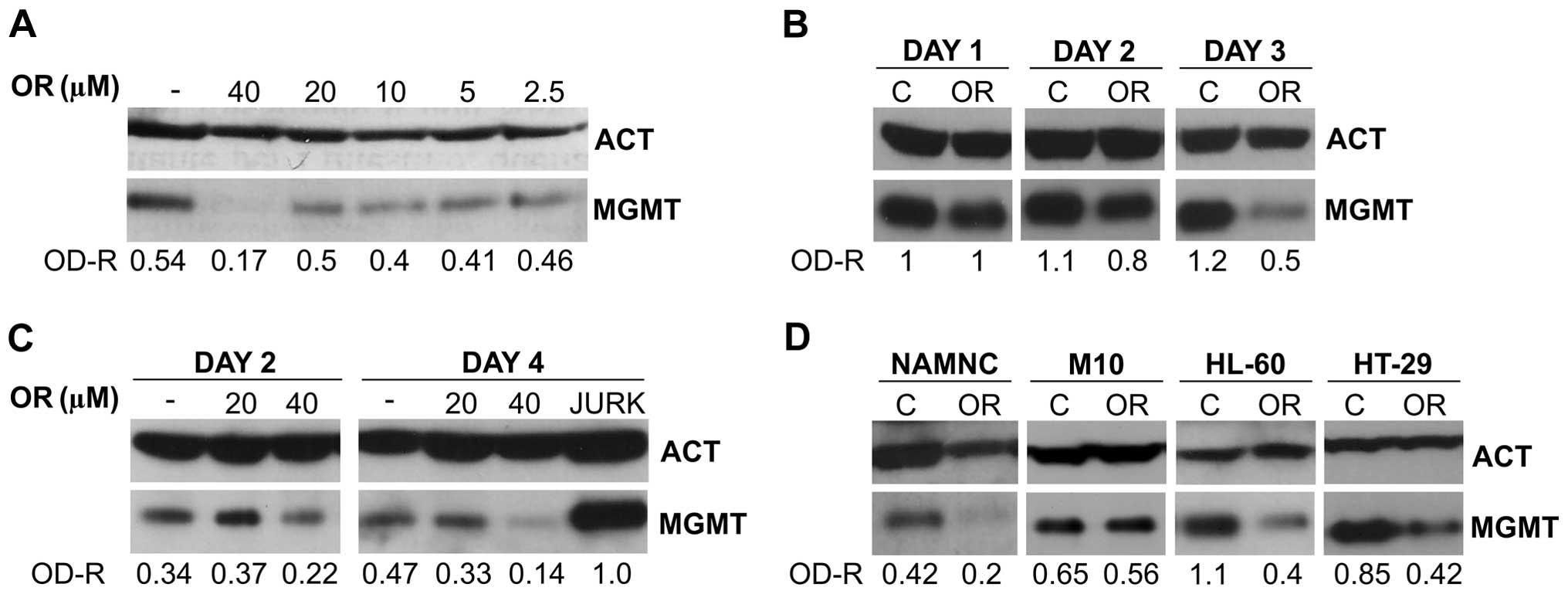
(WB) analysis. The results, illustrated in Fig. 1A, indicate that orlistat, at the
concentration of 40 μM, was able to reduce by >50% the MGMT
level, whereas little or no effect was found when lower
concentrations were used. These findings were qualitatively
confirmed by four additional experiments conducted with Jurkat
cells (data not shown), although downregulation of MGMT protein
expression appears to be noticeably variable (i.e., inhibition
among different experiments ranging from ~30 to 70%). Based on
these results, we decided to utilize the concentration of 40 μM as
the standard concentration of the agent in the majority of the
experimental procedures herein described.
Time-course studies were performed on Jurkat cells
in order to explore the effect of orlistat on MGMT expression when
the leukemic blasts were exposed to the drug for a total of 72 h.
Incubation of target cells with orlistat (40 μM) started on day 0,
and MGMT levels were tested by WB analysis on days 1, 2 and 3. The
results (Fig. 1B) show that
downregulation of MGMT by orlistat is not detectable on day 1,
starts on day 2 and persists up to day 3.
Similar concentration/effect and time-course studies
were conducted with the epithelial colon cancer HCT116 cells. The
results, illustrated in Fig. 1C,
confirm that downregulation of MGMT expression is produced by
orlistat in these cells at the concentration of 40 μM. This effect
was even more pronounced after 4-day exposure to the agent, showing
a decline of MGMT protein concentration >70%.
Analysis of the effect of continuous exposure to
orlistat was extended to normal NAMNC and to other tumor cell
types, such as melanoma M10, promyelocytic leukemia HL-60, and
colon cancer HT-29 cell lines. After 2-day incubation with the
agent, the results, illustrated in Fig. 1D, confirm that the drug provoked an
~50% reduction of MGMT level in all target cells except for
melanoma M10 cells that showed no downregulation of the
protein.
Comparative studies on the effect of
orlistat and LM on MGMT expression
Among well established MGMT inhibitors, LM
represents one of the best molecule able to rapidly and potently
inhibit MGMT functional activity (16). This agent behaves as a false
substrate that binds MGMT irreversibly and promotes its degradation
through the ubiquitin-proteasome pathway (17). This mechanism implies that MGMT
function is almost immediately suppressed by LM and that the
inactive protein undergoes a fairly rapid degradation within the
cell. Thereafter, the cell starts to synthesize de novo the
MGMT protein in order to restore its DNA repair functional
activity. These observations prompted us to compare the effect of
orlistat with that of LM on the kinetics of MGMT protein level
decline in a short-term assay. Previous studies pointed out that 10
μM LM was able to downregulate markedly the MGMT function in target
cells (18). Therefore, we
employed this inhibitor concentration in all experiments described
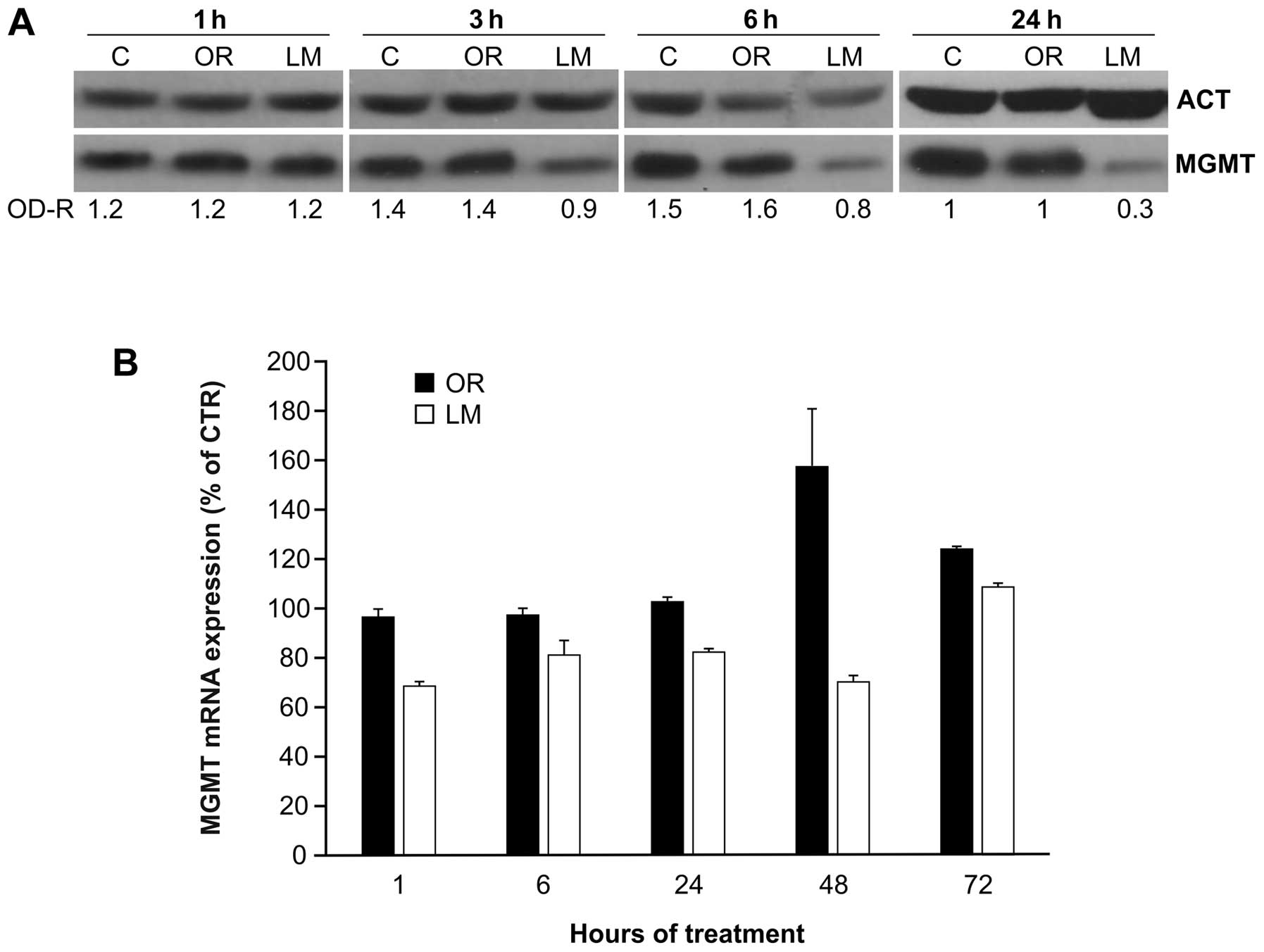
in the present report. Jurkat cells were incubated with orlistat
(40 μM) alone, or with LM alone. At different time intervals (i.e.,
1, 3, 6 and 24 h) the cells were collected and tested for MGMT
protein expression by WB analysis. The results, illustrated in
Fig. 2A, confirmed that no
noticeable downregulation of MGMT protein level was found at all
time intervals explored when target cells were treated with
orlistat. In contrast, a manifest decline of MGMT protein was
detected in LM-treated cells as early as 3 h after start of
treatment, followed by a further reduction of ~70% at 24 h.
Additional experiments were conducted with the
intent to establish whether orlistat could be able to influence the
functional activity of MGMT with a mechanism conceivably similar to
that of LM. Therefore, HCT116 and Jurkat cells were treated with 40
μM orlistat or 10 μM LM, and MGMT activity was tested as early as 2
h after start of treatment. As expected, and in line with previous
reports (16,18), LM provoked a drastic decrease of
MGMT function in both cell lines, whereas minimal or no significant
downregulation of the enzymatic activity was detected in Jurkat and
HCT116 samples, respectively, exposed to orlistat (Table I).
 | Table IEffect of orlistat or LM on MGMT
enzymatic activity of Jurkat and HCT116 cells. |
Table I
Effect of orlistat or LM on MGMT
enzymatic activity of Jurkat and HCT116 cells.
| Control
(DMSO)a | Orlistat (40
μM)a | | LM (10 μM)a |
|---|
|
|
| |
|
|---|
| Cell line | Exp 1 | Exp 2 | Mean (SE)b | Exp 1 | Exp 2 | Mean (SE)b | P-valuec | Exp 1 | Exp 2 |
|---|
| Jurkat | 641 | 592 | 616.5 (24.5) | 527 | 543 | 535 (8) | <0.05 | <1 | <1 |
| HCT116 | 102 | 108 | 105 (2.8) | 102 | 105 | 103.5 (1.8) | NS | <1 | <1 |
Further studies were performed to evaluate the
possible influence of orlistat and LM on MGMT gene transcription.
RT-qPCR performed on Jurkat cells at different times after exposure
to orlistat (40 μM) or to LM (10 μM) pointed out that no remarkable
changes of MGMT transcription respect to DMSO-treated controls were
detectable in orlistat-treated cells, for ≤24 h after start of
treatment (Fig. 2B). However, a
transient increase of MGMT mRNA was found at 48 and 72 h. Limited
downregulation of MGMT expression was found instead, shortly after
exposure to LM. In this case, mRNA values returned to normal levels
at 72 h of incubation with the MGMT inhibitor.
Effects of orlistat combined with LM on
MGMT protein expression
The results illustrated in the previous section
favor the hypothesis that orlistat inhibits MGMT expression with a
mechanism distinct from that involved in LM activity. Therefore it
was of interest to explore the combined effects of the two agents
on MGMT protein levels of normal or leukemic cells.
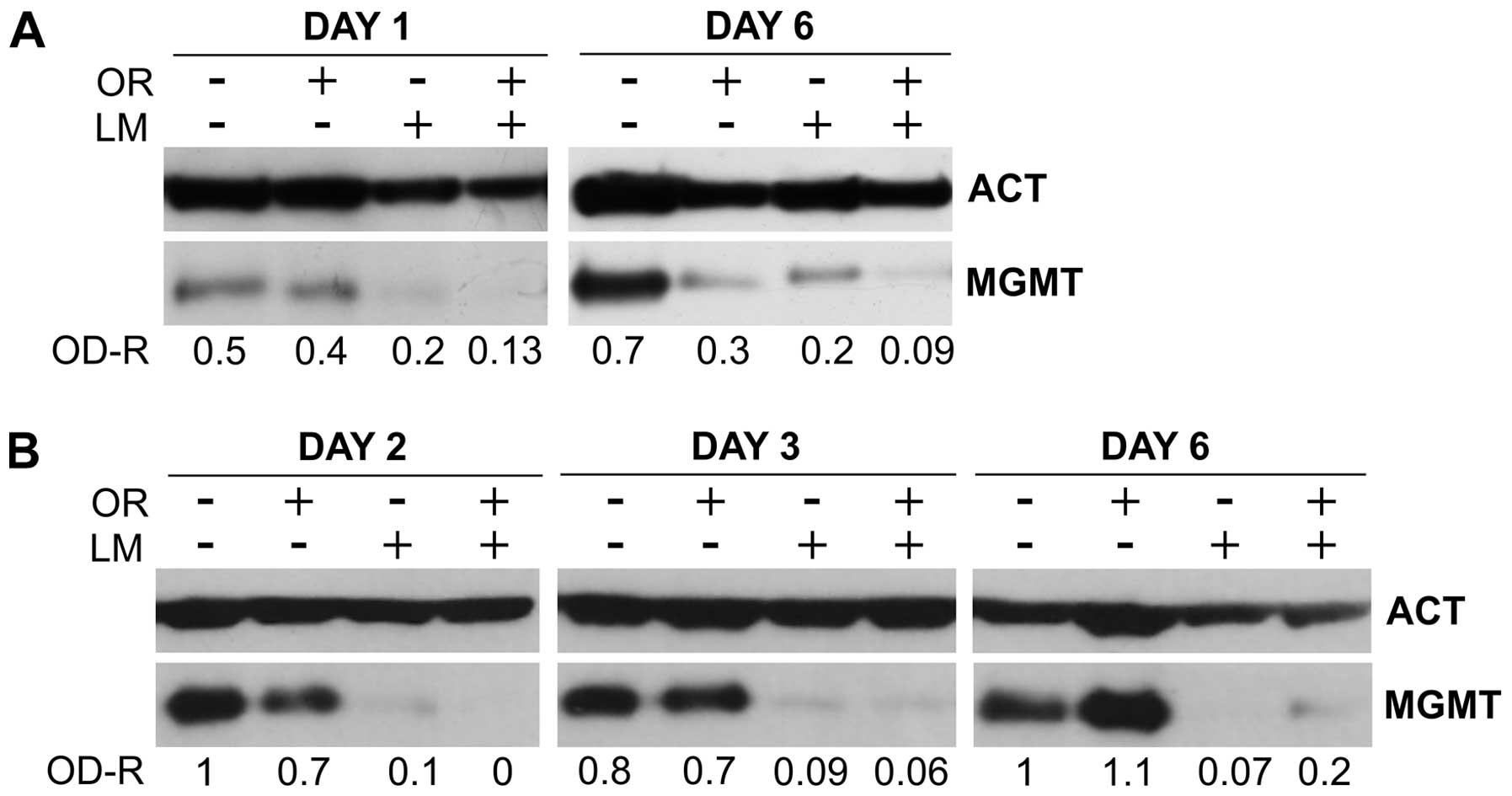
Normal NAMNC were exposed to 40 μM orlistat alone,
to 10 μM LM alone, or to orlistat + LM for 6 consecutive days. WB
analysis was performed on cells collected shortly (i.e., on day 1),
or late (i.e., on day 6) from the onset of drug treatment. The
results, shown in Fig. 3A,
demonstrate that, in line with the previous results, orlistat alone
had marginal effect on MGMT protein levels on day 1, whereas LM
reduced MGMT expression by >50%. Combination of the two agents
was slightly additive, as confirmed by other two experiments that
gave similar results. When the WB assay was performed on day 6,
both orlistat and LM showed remarkable inhibitory effects, that
were higher when the two drugs were used in combination.
In a second set of experiments Jurkat cells were
incubated with either 40 μM orlistat or 10 μM LM or with the
combination of the two drugs. In this case, however, target cells
were incubated with the agents for 24 h only. Thereafter, the cells
were washed carefully and all cultures were reconstitute in culture
medium alone. MGMT protein levels were determined on days 2, 3 and
6 of total culture time. The results, illustrated in Fig. 3B, show that progressive decline of
MGMT levels occurred during the time-course study in all groups
treated with LM, alone or in combination with orlistat, with the
combination being particularly active on day 2. However, since this
type of experiment was conducted with target cells exposed to drugs
for 24 h only, it was possible to detect a complete recovery of
MGMT levels in the group treated with orlistat alone. On the
contrary, the effect of LM, alone or in combination was fully
detectable up to 6 days, as confirmed by other two experiments
(data not shown).
Effects of orlistat combined with TMZ on
MGMT expression
It is known that methylation of oxygen 6 of DNA
guanine by endogenous or exogenous DNA methylating agents is an
important step in chemical carcinogenesis leading to
mutation-mediated malignant transformation (19,20).
MGMT is crucially involved in the physiological defenses against
carcinogenesis consequent to methylation of
O6-guanine and undergoes inactivation and
proteasome-mediated degradation after removing, in a stoichiometric
reaction, methyl adducts from O6-guanine
(21). We therefore decided to
investigate the effects of a combined treatment with low
concentrations of a mono-methylating agent followed by orlistat on
MGMT protein levels.
We directed our attention to TMZ, a triazene
compound that is activated in vitro and is capable of
inducing DNA O6-guanine methyl adducts (20). This drug has been found recently to
increase and accelerate tumorigenesis in intestinal cells of mice
in a murine model of Lynch syndrome (22). Removal of TMZ-induced methyl
adducts operated by MGMT is necessarily followed by depletion of
the enzyme. Therefore, it is not surprising that TMZ downregulates
the enzymatic activity (23) and
the cellular levels of MGMT as a result of the DNA repair
process.
In the first set of experiments, the ability of the
triazene compound to downregulate the level of MGMT protein in a
short-term assay was tested using HCT116 target cells that were
incubated with graded concentrations of TMZ (40–320 μM) for 24 h.
No influence of TMZ was detectable on MGMT levels when drug
concentrations were lower than 160 μM (data not shown). At the
concentration of 160 or 320 μM the drug provoked a marginal or a
strong (close to 70%) downregulation of MGMT, respectively
(Fig. 4A), thus confirming that
TMZ was able to produce the expected enzymatic depletion in our
model.
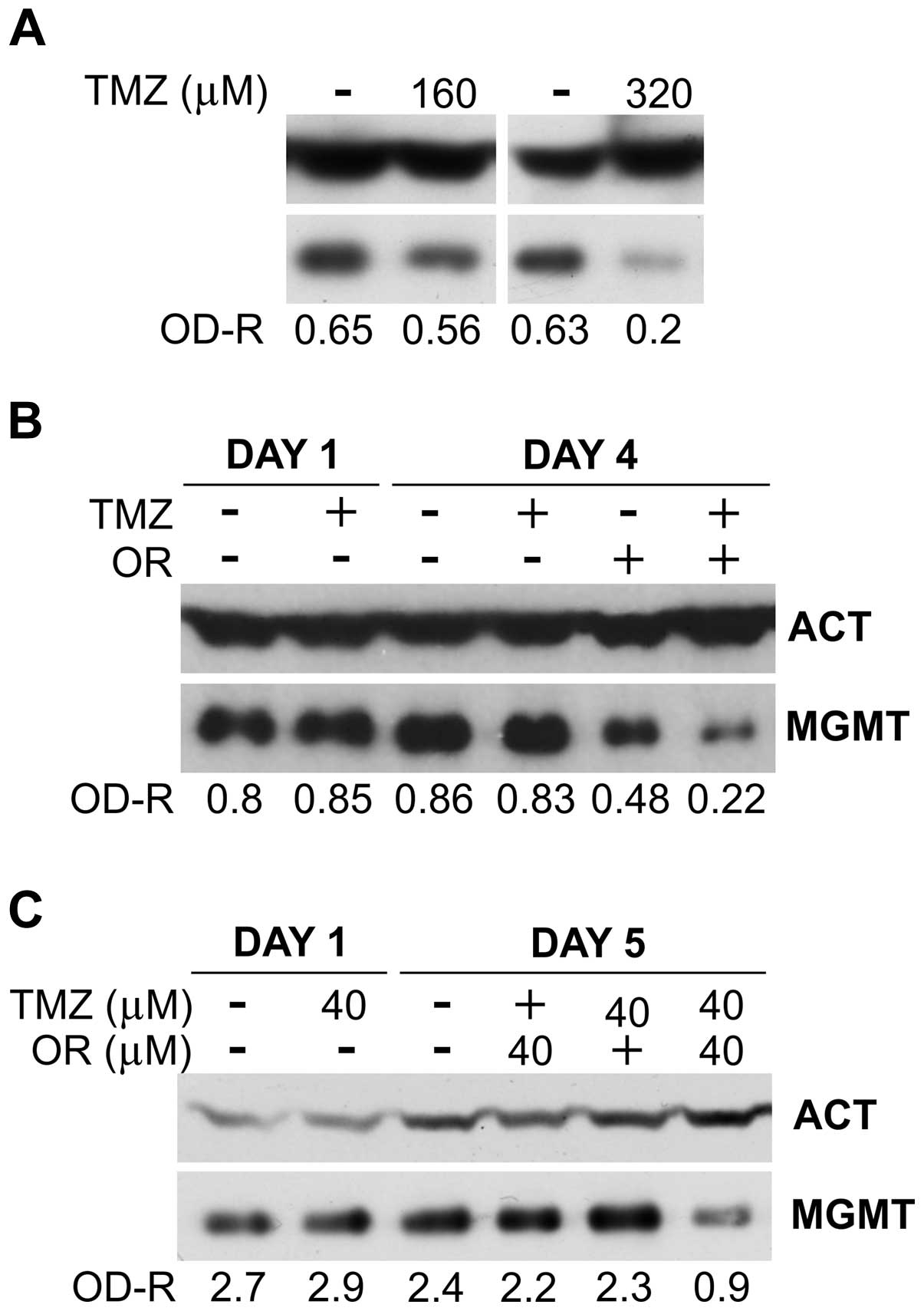 | Figure 4Effect of temozolomide (TMZ) or
orlistat (OR, 40 μM), alone or in combination, on MGMT protein
expression in colon cancer cells (for abbreviations, see legend of
Fig. 1). (A) MGMT levels in HCT116
cells after 1 day of culture with TMZ alone. (B) Effects of TMZ and
orlistat, alone or in combination, on MGMT protein levels in HCT116
cells. Target cells were cultured with medium alone or with TMZ (40
μM) for 24 h. Thereafter the cells were washed twice with medium
alone, and cultured for additional 3 days with medium alone or with
orlistat (40 μM). The results were confirmed in two additional
independent experiments. (C) Effects of TMZ and orlistat, alone or
in combination, on MGMT protein levels in HT-29 cells. Target cells
were cultured in medium alone or with 40 μM TMZ for 24 h.
Thereafter, the cells were washed twice with medium alone and
cultured for additional 4 days with medium alone or with orlistat
(40 μM). Similar results were obtained in one additional
independent experiment. |
Drug combination experiments were performed on
HCT116 cells that were exposed to TMZ (40 μM) for 24 h. Thereafter,
the drug was removed by thorough washing and the cells were
incubated for additional three days with 40 μM orlistat. The
results confirm that MGMT protein levels were not affected by 40 μM
TMZ, either when the protein amounts were evaluate at the end of
drug treatment (Fig. 4B, day 1) or
after three additional days of culture in the absence of the drug
(Fig. 4B, day 4). Conversely,
3-day incubation with orlistat alone (i.e., not preceded by TMZ
treatment) reduced MGMT by ~50% respect to untreated controls. Of
particular interest is the finding that the group treated in
sequence with TMZ and orlistat showed an overall reduction of MGMT
protein level of approximately 75% with respect to untreated
control. A similar set of experiments were performed with the colon
cancer cell line HT-29 (Fig. 4C).
In these studies, cancer cells were exposed to TMZ (40 μM) for 24
h, washed and incubated with orlistat (40 μM) for four days (i.e.,
from days 1–5). The results of WB analysis performed on day 1 show
that TMZ, at the concentration of 40 μM, did not influence MGMT
level. On day 5 of culture, HT-29 cells treated with 40 μM TMZ for
24 h, followed by 4-day culture without drugs, did not display
changes of MGMT protein levels. Orlistat alone, at the
concentration of 40 μM produced marginal (~8%) inhibition, probably
due to lower susceptibility to orlistat of HT-29 cell line with
respect to that of HCT116 after 4-day incubation with the agent.
However, most importantly, orlistat combined with TMZ at the same
molar concentration (i.e., 40 μM), reduced MGMT levels by >60%
respect to control, suggesting that the combined effects of the two
agents on MGMT could be synergistic.
Discussion
The results of the present study reveal for the
first time that orlistat downregulates MGMT protein expression,
increases addictively the effects of a classical MGMT inhibitor
such as LM and amplifies the MGMT suppression induced by a DNA
monomethylating agent such as TMZ.
Alkylation and, in particular, methylation of
O6-guanine of DNA is a highly mutagenic
event produced by either endogenous (24) or exogenous chemical compounds. If
not repaired by MGMT, and in the absence of a functional mismatch
repair system (25), the
biochemical lesion is ignored and DNA synthesis proceeds leading to
frequent G-C→A-T transitions, as described by Ito et al
(26). The mismatch repair system
is estimated to be of great relevance in the control of mutational
events, being engaged in the rapid correction of the base
substitutions that compromise fidelity of the new DNA strand
generated during DNA synthesis (27). Therefore, failure of the functional
activity of this enzymatic system is connected with a number of
different pathologies including malignant transformation, that has
been documented extensively in colon rectal cancer development in
human subjects (28). It follows
that both MGMT and mismatch repair play a remarkable role in the
control of carcinogenesis affecting various target organs, with
particular regard to the colorectal segment of the digestive tract.
In any case, it is obvious that the presence of suitable levels of
MGMT ensures striking protection against mutagenesis whenever
O6-methylguanine
(O6-MeG) is induced on DNA molecules
(29), even in the absence of
adequate levels of mismatch repair function. The MGMT protein
behaves as an acceptor of the methyl adduct that is transferred in
a stoichiometric reaction from DNA O6-MeG
to a cysteine residue associated with the active site of MGMT. It
follows that the biochemical lesion is repaired, whereas MGMT is
inactivated according to a sort of suicide process (21) leading to ubiquitination followed by
proteasome-mediated degradation of the repair protein (17,30,31).
Affected cells restore their MGMT levels through an entirely de
novo synthesis of the protein. This mechanism helps in the
understanding why the functional activity of the MGMT is rapidly
suppressed by molecules that behave like pseudo substrates able to
compete with DNA O6-MeG for interaction
with the repair protein. This is in line with the results of the
present study that pointed out that LM, a potent competitor of
O6-MeG (32–34),
suppressed entirely MGMT activity of Jurkat or of HCT116 cells
within 2 h (Table I). On the other
hand, the MGMT protein of Jurkat cells started to decline at 3 h or
later, reaching low levels 24 h later, as shown by the WB assay
(Fig. 2A).
In oncology the biological significance of MGMT has
been considered from two opposite perspectives. The protein has
been considered responsible of malignant cell resistance against
alkylating agents such as triazenes, being capable of eliminating
drug-induced methyl adducts to DNA O6-G.
Consequently, agents able to suppress MGMT activity have been used
in association with triazene compounds in the clinic (16,32–35);
reviewed in ref. 36. On the
contrary, the role of MGMT as a protective protein against not only
endogenous, but also exogenous alkylating carcinogenic compounds
(37) has been firmly established
by a number of in vivo and in vitro investigations.
Of particular interest is the observation that MGMT protects
transgenic mice from N-methylnitrosourea-induced thymic lymphomas
(38), or nitrosamine-induced
hepatocarcinogenesis (39). This
issue has been adequately discussed in a relatively recent review
by Fahrer and Kaina (12) who
highlighted the crucial role that could be played by MGMT in colon
carcinogenesis secondary to nitrosamines. These molecules are well
known carcinogenic compounds particularly present in a variety of
preserved meat products (40).
The possibility that orlistat could be directly
involved in colorectal carcinogenesis has been matter of debate for
several years (9,10,41).
However, no conclusive results have been obtained so far, although
the finding that orlistat did not show carcinogenic potential
(41) does not support the
hypothesis that the agent is carcinogenic per se.
In the present study we found that in vitro
exposure to orlistat at the concentration of 40 μM for at least two
days is able to induce 30–70% reduction of MGMT protein in four
different human neoplastic cell lines and NAMNC. However,
particularly resistant to this biochemical effect of orlistat
appears to be the human melanoma M10, as shown in Fig. 1D. No data are presently available
to explain this differential behavior, that seems to deserve
further investigation. In all experiments, although orlistat at the
concentration of 40 μM produced a marked reduction of tumor cell
growth, cell viability was never found to be <85%. Moreover,
specificity of drug effect on MGMT protein expression was confirmed
by the finding that the same samples utilized for WB analysis did
not show any drug-induced downregulation of other cellular proteins
that have been tested (e.g., calreticulin or HSP-90, data not
shown).
At the present time we are not able to provide
sufficient experimental data to disclose the mechanism underlying
the effect of orlistat on MGMT protein expression. The finding that
the compound is unable to downregulate consistently MGMT function
within 2 h of treatment (Table I),
does not support the hypothesis that orlistat, similarly to LM,
could behave as pseudo-substrate able to bind and inactivate MGMT,
followed by a proteasome-dependent degradation process (17,30,31).
Several studies have pointed out that orlistat is a
potent FASN inhibitor (42).
Moreover, FASN inhibition is associated with p53 upregulation
(43–45), and p53 upregulation induces
inhibition of MGMT expression (46). However, if one considers the
defective p53 status of at least two tumor cell lines utilized in
the present study (i.e., Jurkat and HT-29 cells), the hypothesis
that orlistat acts on MGMT expression via p53 signaling does not
appear to be sufficiently supported.
Independently from the mechanism of action of
orlistat, its inhibitory effects on MGMT protein expression raise
the question of possible indirect carcinogenic effect of the
compound through a reduction of surveillance against endogenous or
exogenous chemical carcinogenesis. The finding that TMZ
pretreatment amplifies markedly the activity of orlistat on MGMT is
of particular interest. This in vitro active triazene
compound, able to induce large amounts of DNA
O6-guanine methyl adducts, engages
actively MGMT in its ‘suicidal’ repair function (21). However, in order to provoke a
detectable reduction of the repair protein, TMZ must be used at
very high concentrations (e.g., 320 μM to produce, an ~70% decline
of MGMT protein level, at least in the HCT116 tumor line, as shown
in Fig. 4A). Actually, at the
concentration of 40 μM the triazene compound was not able to show a
noticeable MGMT downregulation (Fig.
4B). Yet, when TMZ was followed by orlistat treatment, MGMT
inhibition was found to be double with respect to that operated by
orlistat alone suggesting a possible synergistic interaction. In
addition, when MGMT was directly downregulated by a potent
pseudo-substrate such as LM, addition of orlistat resulted in
additively increased suppressive activity (Fig. 3). This means that orlistat could
act in concert with other MGMT ‘consuming’ agents including those
that are endowed with carcinogenic potential such as TMZ (22).
In conclusion, the preliminary data illustrated in
this report appear to disclose an unexpected source of concern
about the clinical use of orlistat. Actually, the administration
modality of the drug in severely overweight subjects implies that
daily treatment with high doses of this agent is carried out for a
period of one or two years. At the present time, we are not able to
evaluate the impact that a chronic downregulation of a DNA repair
enzyme such as MGMT could have on host’s surveillance against
chemical carcinogenesis targeting digestive tract mucosa locally
exposed to high concentrations of orlistat. Therefore, the studies
illustrated in this report appear to provide the ground for casting
a note of caution in the long-term clinical use of orlistat
suggesting that appropriate control of the gastrointestinal
apparatus appears to be unreservedly advisable.
Acknowledgements
This study was supported in part by the Italian
Ministry of Health. The authors also thank Graziano Bonelli
(University of Rome ‘Tor Vergata’, School of Medicine, Rome, Italy)
for the artwork.
Abbreviations:
|
MGMT
|
O6-methylguanine-DNA methyltransferase
|
|
FASN
|
fatty acid synthase
|
|
NAMNC
|
non-adherent mononuclear cells
|
|
LM
|
lomeguatrib
|
|
TMZ
|
temozolomide
|
References
|
1
|
Yanovski SZ and Yanovski JA: Long-term
drug treatment for obesity: A systematic and clinical review. JAMA.
311:74–86. 2014. View Article : Google Scholar :
|
|
2
|
Flavin R, Peluso S, Nguyen PL and Loda M:
Fatty acid synthase as a potential therapeutic target in cancer.
Future Oncol. 6:551–562. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Santolla MF, Lappano R, De Marco P, Pupo
M, Vivacqua A, Sisci D, Abonante S, Iacopetta D, Cappello AR, Dolce
V, et al: G protein-coupled estrogen receptor mediates the
up-regulation of fatty acid synthase induced by 17β-estradiol in
cancer cells and cancer-associated fibroblasts. J Biol Chem.
287:43234–43245. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chuang HY, Chang YF and Hwang JJ:
Antitumor effect of orlistat, a fatty acid synthase inhibitor, is
via activation of caspase-3 on human colorectal carcinoma-bearing
animal. Biomed Pharmacother. 65:286–292. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rossato FA, Zecchin KG, La Guardia PG,
Ortega RM, Alberici LC, Costa RA, Catharino RR, Graner E, Castilho
RF and Vercesi AE: Fatty acid synthase inhibitors induce apoptosis
in non-tumorigenic melana cells associated with inhibition of
mitochondrial respiration. PLoS One. 9:e1010602014. View Article : Google Scholar
|
|
6
|
Little JL, Wheeler FB, Fels DR, Koumenis C
and Kridel SJ: Inhibition of fatty acid synthase induces
endoplasmic reticulum stress in tumor cells. Cancer Res.
67:1262–1269. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng L, Wu GZ, Goh KJ, Lee YM, Ng CC, You
AB, Wang J, Jia D, Hao A, Yu Q, et al: Saturated fatty acids
modulate cell response to DNA damage: Implication for their role in
tumorigenesis. PLoS One. 3:e23292008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dengler MA, Weilbacher A, Gutekunst M,
Staiger AM, Vöhringer MC, Horn H, Ott G, Aulitzky WE and van der
Kuip H: Discrepant NOXA (PMAIP1) transcript and NOXA protein
levels: A potential Achilles’ heel in mantle cell lymphoma. Cell
Death Dis. 5:e10132014. View Article : Google Scholar
|
|
9
|
Garcia SB, Barros LT, Turatti A,
Martinello F, Modiano P, Ribeiro-Silva A, Vespúcio MV and Uyemura
SA: The anti-obesity agent Orlistat is associated to increase in
colonic preneoplastic markers in rats treated with a chemical
carcinogen. Cancer Lett. 240:221–224. 2006. View Article : Google Scholar
|
|
10
|
Nairooz S, Ibrahim SH, Sahar MMO and Affan
M: Structural changes of the colonic mucosa induced by Orlistat:
Experimental study. Egypt J Histol. 33:635–648. 2010.
|
|
11
|
Christmann M, Verbeek B, Roos WP and Kaina
B: O(6)-Methylguanine-DNA methyltransferase (MGMT) in normal
tissues and tumors: Enzyme activity, promoter methylation and
immunohistochemistry. Biochim Biophys Acta. 1816:179–190.
2011.PubMed/NCBI
|
|
12
|
Fahrer J and Kaina B:
O6-methylguanine-DNA methyltransferase in the defense
against N-nitroso compounds and colorectal cancer. Carcinogenesis.
34:2435–2442. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nosho K, Kawasaki T, Ohnishi M, Suemoto Y,
Kirkner GJ, Zepf D, Yan L, Longtine JA, Fuchs CS and Ogino S:
PIK3CA mutation in colorectal cancer: Relationship with genetic and
epigenetic alterations. Neoplasia. 10:534–541. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pepponi R, Marra G, Fuggetta MP,
Falcinelli S, Pagani E, Bonmassar E, Jiricny J and D’Atri S: The
effect of O6-alkyl-guanine-DNA alkyltransferase and
mismatch repair activities on the sensitivity of human melanoma
cells to temozolomide, 1,3-bis(2-chloroethyl)1-nitrosourea, and
cisplatin. J Pharmacol Exp Ther. 304:661–668. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Watson AJ and Margison GP:
O(6)-Alkylguanine-DNA alkyl-transferase assay. Methods Mol Med.
28:167–178. 1999.
|
|
16
|
Ranson M, Middleton MR, Bridgewater J, Lee
SM, Dawson M, Jowle D, Halbert G, Waller S, McGrath H, Gumbrell L,
et al: Lomeguatrib, a potent inhibitor of
O6-alkylguanine-DNA-alkyltransferase: Phase I safety,
pharmacodynamic, and pharmacokinetic trial and evaluation in
combination with temozolomide in patients with advanced solid
tumors. Clin Cancer Res. 12:1577–1584. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Srivenugopal KS, Yuan XH, Friedman HS and
Ali-Osman F: Ubiquitination-dependent proteolysis of
O6-methylguanine-DNA methyltransferase in human and
murine tumor cells following inactivation with
O6-benzylguanine or
1,3-bis(2-chloroethyl)-1-nitrosourea. Biochemistry. 35:1328–1334.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Turriziani M, Caporaso P, Bonmassar L,
Buccisano F, Amadori S, Venditti A, Cantonetti M, D’Atri S and
Bonmassar E: O6-(4-bromothenyl)guanine (PaTrin-2), a
novel inhibitor of O6-alkylguanine DNA
alkyl-transferase, increases the inhibitory activity of
temozolomide against human acute leukaemia cells in vitro.
Pharmacol Res. 53:317–323. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Pegg AE and Byers TL: Repair of DNA
containing O6-alkyl-guanine. FASEB J. 6:2302–2310.
1992.PubMed/NCBI
|
|
20
|
Kyrtopoulos SA, Anderson LM, Chhabra SK,
Souliotis VL, Pletsa V, Valavanis C and Georgiadis P: DNA adducts
and the mechanism of carcinogenesis and cytotoxicity of methylating
agents of environmental and clinical significance. Cancer Detect
Prev. 21:391–405. 1997.PubMed/NCBI
|
|
21
|
Gouws C and Pretorius PJ:
O6-methylguanine-DNA methyltransferase (MGMT): Can
function explain a suicidal mechanism? Med Hypotheses. 77:857–860.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wojciechowicz K, Cantelli E, Van Gerwen B,
Plug M, Van Der Wal A, Delzenne-Goette E, Song JY, De Vries S,
Dekker M and Te Riele H: Temozolomide increases the number of
mismatch repair-deficient intestinal crypts and accelerates
tumorigenesis in a mouse model of Lynch syndrome. Gastroenterology.
147:1064–1072.e5. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
D’Atri S, Graziani G, Lacal PM, Nisticò V,
Gilberti S, Faraoni I, Watson AJ, Bonmassar E and Margison GP:
Attenuation of O(6)-methylguanine-DNA methyltransferase activity
and mRNA levels by cisplatin and temozolomide in jurkat cells. J
Pharmacol Exp Ther. 294:664–671. 2000.
|
|
24
|
Xiao W and Samson L: In vivo evidence for
endogenous DNA alkylation damage as a source of spontaneous
mutation in eukaryotic cells. Proc Natl Acad Sci USA. 90:2117–2121.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iyama T and Wilson DM III: DNA repair
mechanisms in dividing and non-dividing cells. DNA Repair (Amst).
12:620–636. 2013. View Article : Google Scholar
|
|
26
|
Ito T, Nakamura T, Maki H and Sekiguchi M:
Roles of transcription and repair in alkylation mutagenesis. Mutat
Res. 314:273–285. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bak ST, Sakellariou D and Pena-Diaz J: The
dual nature of mismatch repair as antimutator and mutator: For
better or for worse. Front Genet. 5:2872014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cohen SA and Leininger A: The genetic
basis of Lynch syndrome and its implications for clinical practice
and risk management. Appl Clin Genet. 7:147–158. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Aquilina G, Biondo R, Dogliotti E, Meuth M
and Bignami M: Expression of the endogenous
O6-methylguanine-DNA-methyltransferase protects Chinese
hamster ovary cells from spontaneous G:C to A:T transitions. Cancer
Res. 52:6471–6475. 1992.PubMed/NCBI
|
|
30
|
Paranjpe A, Zhang R, Ali-Osman F, Bobustuc
GC and Srivenugopal KS: Disulfiram is a direct and potent inhibitor
of human O6-methylguanine-DNA methyltransferase (MGMT)
in brain tumor cells and mouse brain and markedly increases the
alkylating DNA damage. Carcinogenesis. 35:692–702. 2014. View Article : Google Scholar :
|
|
31
|
Xu-Welliver M and Pegg AE: Degradation of
the alkylated form of the DNA repair protein, O(6)-alkylguanine-DNA
alkyltransferase. Carcinogenesis. 23:823–830. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Khan O and Middleton MR: The therapeutic
potential of O6-alkylguanine DNA alkyltransferase
inhibitors. Expert Opin Investig Drugs. 16:1573–1584. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Marchesi F, Turriziani M, Tortorelli G,
Avvisati G, Torino F and De Vecchis L: Triazene compounds:
Mechanism of action and related DNA repair systems. Pharmacol Res.
56:275–287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bonmassar L, Marchesi F, Pascale E,
Franzese O, Margison GP, Bianchi A, D’Atri S, Bernardini S,
Lattuada D, Bonmassar E, et al: Triazene compounds in the treatment
of acute myeloid leukemia: A short review and a case report. Curr
Med Chem. 20:2389–2401. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Seiter K, Katragadda S, Ponce D, Rasul M
and Ahmed N: Temozolomide and cisplatin in relapsed/refractory
acute leukemia. J Hematol Oncol. 2:212009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang J, Stevens MF and Bradshaw TD:
Temozolomide: Mechanisms of action, repair and resistance. Curr Mol
Pharmacol. 5:102–114. 2012. View Article : Google Scholar
|
|
37
|
Niture SK, Velu CS, Smith QR, Bhat GJ and
Srivenugopal KS: Increased expression of the MGMT repair protein
mediated by cysteine prodrugs and chemopreventative natural
products in human lymphocytes and tumor cell lines. Carcinogenesis.
28:378–389. 2007. View Article : Google Scholar
|
|
38
|
Liu L, Allay E, Dumenco LL and Gerson SL:
Rapid repair of O6-methylguanine-DNA adducts protects
transgenic mice from N-methylnitrosourea-induced thymic lymphomas.
Cancer Res. 54:4648–4652. 1994.PubMed/NCBI
|
|
39
|
Nakatsuru Y, Matsukuma S, Nemoto N, Sugano
H, Sekiguchi M and Ishikawa T: O6-methylguanine-DNA
methyltransferase protects against nitrosamine-induced
hepatocarcinogenesis. Proc Natl Acad Sci USA. 90:6468–6472. 1993.
View Article : Google Scholar
|
|
40
|
Griesenbeck JS, Steck MD, Huber JC Jr,
Sharkey JR, Rene AA and Brender JD: Development of estimates of
dietary nitrates, nitrites, and nitrosamines for use with the Short
Willet Food Frequency Questionnaire. Nutr J. 8:162009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Orsolin PC, Silva-Oliveira RG and
Nepomuceno JC: Assessment of the mutagenic, recombinagenic and
carcinogenic potential of orlistat in somatic cells of Drosophila
melanogaster. Food Chem Toxicol. 50:2598–2604. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kridel SJ, Axelrod F, Rozenkrantz N and
Smith JW: Orlistat is a novel inhibitor of fatty acid synthase with
antitumor activity. Cancer Res. 64:2070–2075. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li JN, Gorospe M, Chrest FJ, Kumaravel TS,
Evans MK, Han WF and Pizer ES: Pharmacological inhibition of fatty
acid synthase activity produces both cytostatic and cytotoxic
effects modulated by p53. Cancer Res. 61:1493–1499. 2001.PubMed/NCBI
|
|
44
|
Zecchin KG, Rossato FA, Raposo HF, Melo
DR, Alberici LC, Oliveira HC, Castilho RF, Coletta RD, Vercesi AE
and Graner E: Inhibition of fatty acid synthase in melanoma cells
activates the intrinsic pathway of apoptosis. Lab Invest.
91:232–240. 2011. View Article : Google Scholar
|
|
45
|
Kant S, Kumar A and Singh SM: Tumor growth
retardation and chemosensitizing action of fatty acid synthase
inhibitor orlistat on T cell lymphoma: Implication of reconstituted
tumor microenvironment and multidrug resistance phenotype. Biochim
Biophys Acta. 1840:294–302. 2014. View Article : Google Scholar
|
|
46
|
Bocangel D, Sengupta S, Mitra S and Bhakat
KK: p53-mediated downregulation of the human DNA repair gene
O6-methyl-guanine-DNA methyltransferase (MGMT) via
interaction with Sp1 transcription factor. Anticancer Res.
29:3741–3750. 2009.PubMed/NCBI
|