Introduction
Esophageal cancer ranks sixth among all cancers in
mortality worldwide because of its extremely aggressive nature
(1,2). The predominant histological types of
esophageal cancer are adenocarcinoma and squamous cell carcinoma
(3). Adenocarcinoma of the distal
esophagus predominates in Western countries, whereas esophageal
squamous cell carcinoma (ESCC) predominates in Asia (3). The mechanism of carcinogenesis of
ESCC differs from that of adenocarcinoma, which has been widely
studied in North America and Europe (3). Further, exogenous factors such as
smoking, drinking, nitrosamines, and consumption of hot beverages
correlate significantly with the development of ESCC but not with
adenocarcinoma of the esophagus (4,5).
Recent advances in our understanding of the molecular biology of
ESCC document the role of genetic alterations in tumorigenesis
(6,7). Therefore, a better understanding of
the molecular mechanisms of progression and recurrence is of
paramount importance, and identification of the genes that mediate
ESCC pathogenesis will increase our understanding of the molecular
and cellular processes involved and provide new biomarkers that may
facilitate diagnosis, risk stratification, and monitoring
recurrences of ESCC (8,9).
The transmembrane adherens junctions-associated
protein-1 (AJAP1) targets the basolateral membrane of polarized
epithelial cells and interacts with E-cadherin-catenin complexes of
adherens junctions (10). The
locus encoding AJAP1 resides in chromosome 1p36 and is frequently
deleted from the genomes of various tumor cells or is
epigenetically silenced, indicating that AJAP1 acts as a tumor
suppressor (11–13). Although AJAP1 is involved in
cell-cell and cell-extracellular matrix interactions potentially
involved in the motility, migration, and invasion of glioblastoma
cells (11,14), little evidence is available on its
role in oncogenesis. To our knowledge, there are no studies of the
expression and regulatory mechanisms of AJAP1 transcription
in gastrointestinal cancers, including ESCC. To address these
issues, we analyzed ESCC cell lines and tumor tissues to evaluate
AJAP1 expression and its regulatory mechanisms. Our results
indicate that AJAP1 expression levels provide a potential
clinical biomarker of the progression and recurrence of ESCC.
Materials and methods
Sample collection
Nine ESCC cell lines (TE1, TE2, TE3, NUEC1, NUEC2,
NUEC3, TT, TTn and WSSC) and a control nontumorigenic epithelial
cell line (FHs74) were obtained from the American Type Culture
Collection (ATCC; Manassas, VA, USA), the Japanese Collection of
Research Bioresources Cell Bank (Osaka, Japan), or were established
in our institute. Cells were stored at −80°C using cell
preservative solution (Cell Banker; Mitsubishi Chemical Medience
Co., Tokyo, Japan) and cultured in RPMI-1640 (Sigma-Aldrich, St.
Louis, MO, USA) supplemented with 10% fetal bovine serum and in an
atmosphere containing 5% CO2 at 37°C (15,16).
Seventy-eight primary ESCC tissues and adjacent normal tissues were
acquired from patients who underwent radical esophageal resection
at Nagoya University Hospital between December 2001 and October
2013. All tissue samples were diagnosed histologically as ESCC,
frozen immediately after resection, and stored at −80°C. None of
the patients underwent preoperative treatment such as chemotherapy
and radiation. Specimens were classified histologically using the
seventh edition of the UICC staging system for esophageal
cancer.
Patients were questioned to determine their levels
of alcohol consumption, and excessive alcohol consumption was
defined as >210 g/week for ≥3 years (2,17).
The mean age of the 78 patients was 64.9±8.2 years (mean ± standard
deviation; range, 44–82 years). The male-to-female ratio was 62:16.
Nine, 29, 30 and 10 patients were in stages I, II, III and IV,
respectively, according to the UICC staging system (seventh
edition). The median duration of patient follow-up was 73.7 months
(range, 5.3–149 months) or until death. Postoperative follow-up
examinations included physical examination, measurement of serum
tumor markers every 3 months, and enhanced computed tomography of
the chest and abdominal cavity every 6 months. Adjuvant
chemotherapy was administered to selected patients according to the
patient's condition and the physician's discretion.
The present study conforms to the ethical guidelines
of the World Medical Association Declaration of Helsinki: Ethical
Principles for Medical Research Involving Human Subjects. Written
informed consent for use of clinical samples and data was required
by the Institutional Review Board at Nagoya University, Japan and
was obtained from all patients (18).
Analysis of the nucleotide sequences
flanking the AJAP1 transcription initiation site
Nucleotide sequence analysis was conducted to
determine the presence of CpG islands around the promoter region of
AJAP1. CpG islands were defined as follows: ≥200-bp region
with a GC content >50% and CpG:expected CpG ≥0.6 identified
using CpG Island Searcher software (http://cpgislands.usc.edu/) (19–21).
Methylation-specific polymerase chain
reaction (MSP-PCR) and 5-aza-2′-deoxycytidine (5-aza-dC)
treatment
Genomic DNA samples from 10 cell lines were
subjected to bisulfite treatment, and MSP-PCR was conducted to
determine the presence or absence of hypermethylation of the
AJAP1 promoter. To evaluate the influence of promoter
hypermethylation on AJAP1 transcription, ESCC cells
(1.5×106) were treated with 10 μM of the DNA
methylation inhibitor 5-aza-dC (Sigma-Aldrich) and cultured for 6
days with medium changes on days 1, 3 and 5.
Bisulfite sequence analysis
Genomic DNAs of ESCC cell lines treated with
bisulfite were sequenced to verify the accuracy of the MSP-PCR
results. After PCR amplification using specific primers (Table I), PCR products were subcloned into
a TA cloning vector (Invitrogen, Carlsbad, CA, USA). The DNAs were
mixed with 3 ml of a specific primer (M13) and 4 ml of Cycle
Sequence mix (ABI PRISM Terminator v1.1 Cycle Sequencing kit;
Applied Biosystems, Foster City, CA, USA). Sequence analysis was
conducted using an Applied Biosystems ABI310, and sequence
electropherograms were generated using ABI Sequence Analysis 3.0
software (Applied Biosystems) (22).
 | Table IPrimers and annealing
temperature. |
Table I
Primers and annealing
temperature.
| Gene | Experiment | Type | Sequence
(5′-3′) | Product size
(bp) | Annealing
temperature (°C) |
|---|
| AJAP1 | qRT-PCR | Forward |
GTTAGCACAACGGAGCCTTC | 105 | 60 |
| | Reverse |
GATGATCTGATGGACAGCCA | | |
| MSP | Forward |
GGTCGCGAGTTTCGCGTTTC | 184 | 64 |
| | Reverse |
CCGATCTCCGACTCTCGATC | | |
| U-MSP | Forward |
GTGTTGATTGGTGGTGGAGT | 152 | 64 |
| | Reverse |
TCCCAACACACAACTCTTAC | | |
| Bisulfite
sequencing | Forward |
GTTTTTAGGATTTAGGTGAG | 316 | 60 |
| | Reverse |
CTACTAACTCCTAAAACTAC | | |
| SRC | qRT-PCR | Forward |
CTGACCGCATGGACCGT | 107 | 58 |
| | Reverse |
AAGCCAACCTGTCACTTGGTA | | |
| EZR | qRT-PCR | Forward |
GATAGTCGTGTTTTCGGGGA | 91 | 60 |
| | Reverse |
CTCTGCATCCATGGTGGTAA | | |
| FAK | qRT-PCR | Forward |
GCCAAAAGGATTTCTAAACCAG | 110 | 64 |
| | Reverse |
CCTGGTCCACTTGATCAGCTA | | |
| DPYSL3 | qRT-PCR | Forward |
AGAAGAAGGAGGGAGGGAGC | 110 | 60 |
| | Reverse |
CTCCCTTGATAAGGAGACGG | | |
| GAPDH | qRT-PCR | Forward |
GAAGGTGAAGGTCGGAGTC | 226 | 60 |
| | Probe |
CAAGCTTCCCGTTCTCAGCC | | |
| | Reverse |
GAAGATGGTGATGGGATTTC | | |
Quantitative real-time reverse
transcription-PCR (qRT-PCR)
The levels of AJAP1 mRNA were determined
using qRT-PCR. Total RNA (10 μg) isolated from cell lines,
78 primary ESCCs, and adjacent normal tissues were used as
templates for cDNA synthesis. Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) mRNA levels (TaqMan, GAPDH Control
Reagents; Applied Biosystems) were quantified to normalize
expression levels. qRT-PCR was performed using the SYBR-Green PCR
Core Reagents kit (Applied Biosystems) as follows: one cycle at
95°C for 10 min, 40 cycles at 95°C for 5 sec, and 60°C for 60 sec.
All samples were tested in triplicate, and samples without template
were included in each PCR plate as negative controls. Real-time
detection of SYBR-Green fluorescence was conducted using an ABI
StepOnePlus Real-Time PCR system (Applied Biosystems). The
expression level of each sample is shown as the value of the
AJAP1 amplicon divided by that of GAPDH (23,24).
To identify cell adhesion molecules that may interact with AJAP1,
10 cell lines were analyzed using qRT-PCR to determine the
expression levels of the ezrin (EZR), focal adhesion kinase
(FAK), SRC (SRC), and dihydropyrimidinase-like 3
(DPYSL3) genes (18,25,26).
Primers specific for AJAP1, GAPDH, EZR,
FAK, SRC, and DPYSL3 are listed in Table I.
Copy number analysis
AJAP1 copy numbers of 10 cell lines were
determined using TaqMan Copy Number Assays (Applied Biosystems).
Twenty nanograms of genomic DNA was amplified using specific primer
pairs according to the manufacturer's instructions. Two assays were
employed as follows: upstream (assay ID Hs04540488_cn, chromosome
1, map position 4798221 in AJAP1 intron 2) and downstream
(assay ID Hs01575789_cn, chromosome 1, map position 4834502 in the
intron 4 to exon 5 of AJAP1 gene). Data were analyzed using
CopyCaller software (Invitrogen Life Technologies, Carlsbad, CA,
USA). Copy number loss was defined as the copy number value equal
to 1 determined in the regions upstream, downstream, or both of the
AJAP1 loci.
Statistical analysis
Correlations between the levels of AJAP1 mRNA
with those of EZR, FAK, SRC, and DPYSL3
were analyzed using the Spearman's rank correlation test.
Differences in the levels of AJAP1 mRNA between ESCC and
adjacent normal tissues were analyzed using the Mann-Whitney test.
The χ2 test was used to analyze the significance of the
association between the expression levels of AJAP1 and
clinicopathological parameters. Overall and disease-free survival
rates were calculated using the Kaplan-Meier method, and the
difference in survival curves was analyzed using the log-rank test.
We performed multivariate regression analysis using the Cox
proportional hazards model to identify prognostic factors, and
variables with P-values <0.05 were entered into the final model.
Statistical analyses were performed using JMP 10 software (SAS
Institute, Inc., Cary, NC, USA). P<0.05 was considered
statistically significant.
Results
Expression, methylation, and copy number
analysis of cell lines. AJAP1
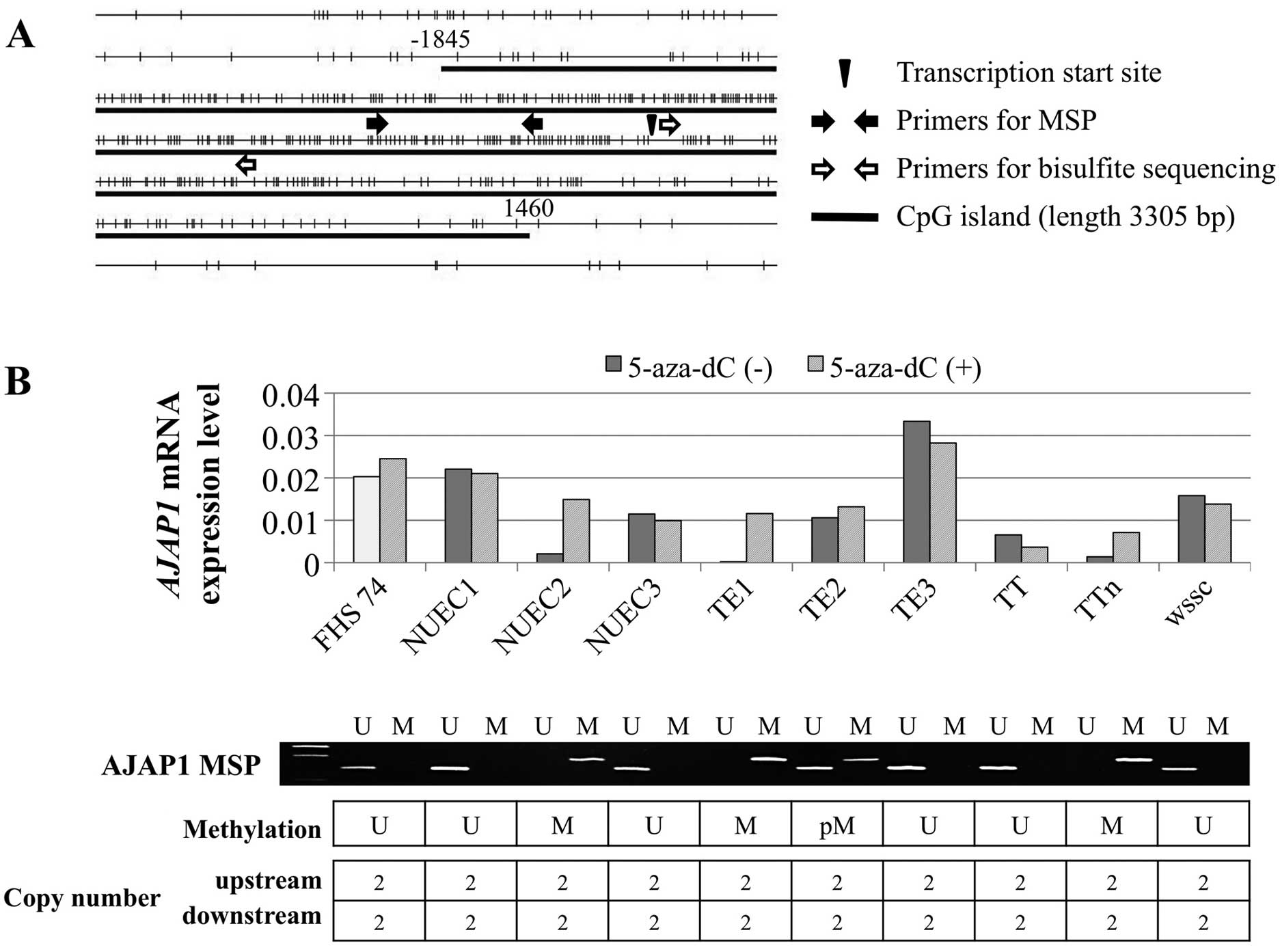
harbors a CpG island (length 3305 bases, 70.3% GC,
and Observed CpG:Expected CpG=0.883) flanking the transcription
initiation site (Fig. 1A).
AJAP1 mRNA expression levels differed among the nine ESCC
cell lines, and five expressed levels <50% of that of the
control FHs74 cells (Fig. 1B).
MSP-PCR detected methylation of the DNAs of NUEC2, TE1, TE2, and
TTn cells, which expressed reduced levels of AJAP1 mRNA.
When we compared the levels of AJAP1 mRNA in ESCC cell lines
before and after demethylation, reactivation of AJAP1
transcription was detected in cells with complete methylation of
AJAP1 before treatment with 5-aza-dC. Moreover, there was no
detectable loss of copy number in ESCC cell lines and FHs74 cells
(Fig. 1B).
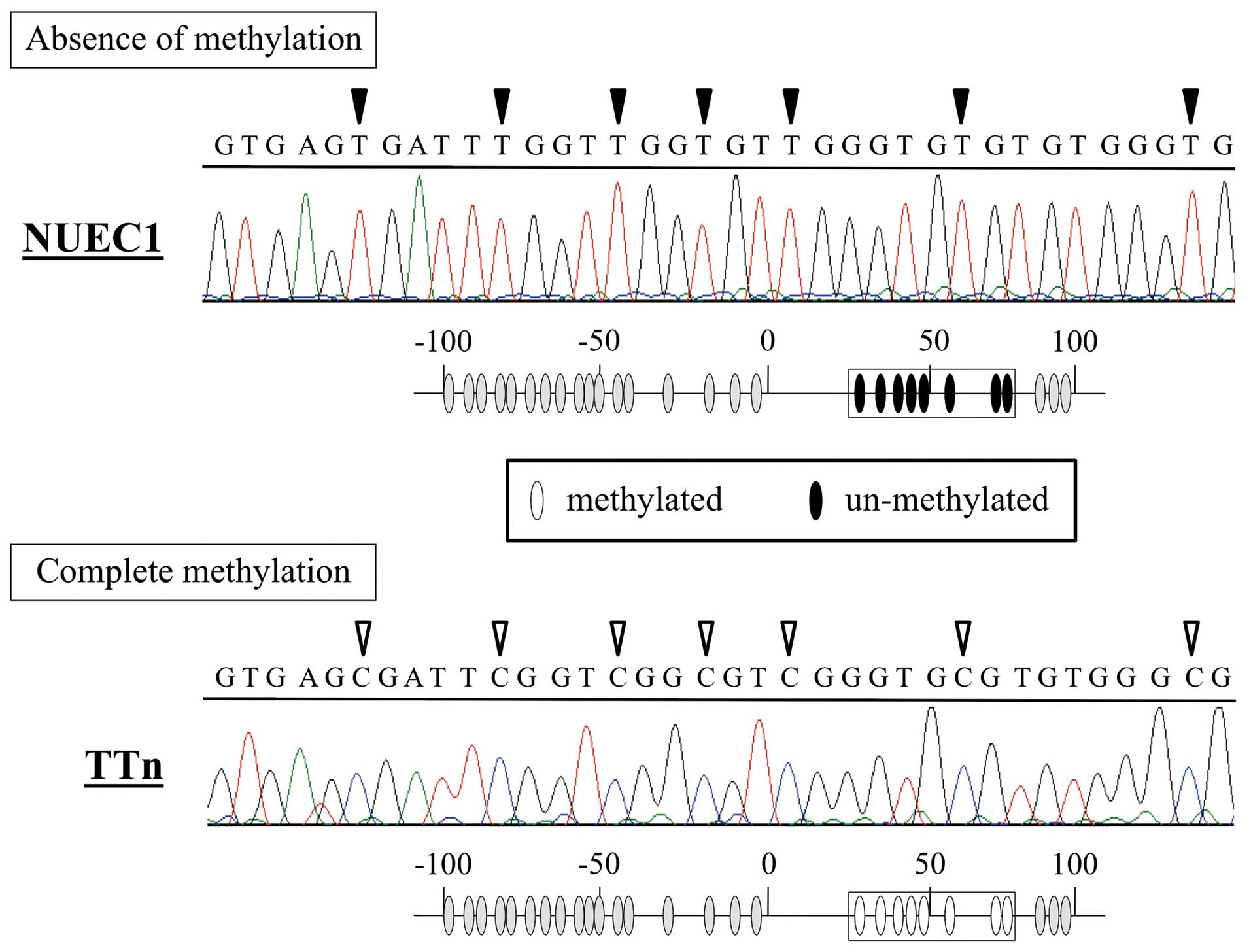
Bisulfite sequence analysis
Sequence analysis revealed that all CpG sites in TTn
DNA (complete methylation) were CG (cytosine and guanine) and that
the corresponding positions in NUEC1 DNA (absence of methylation)
were TG (thymine and guanine) (Fig.
2). These results confirm the MSP-PCR results.
Analysis of the levels of AJAP1 mRNA and
those representing potentially interacting molecules
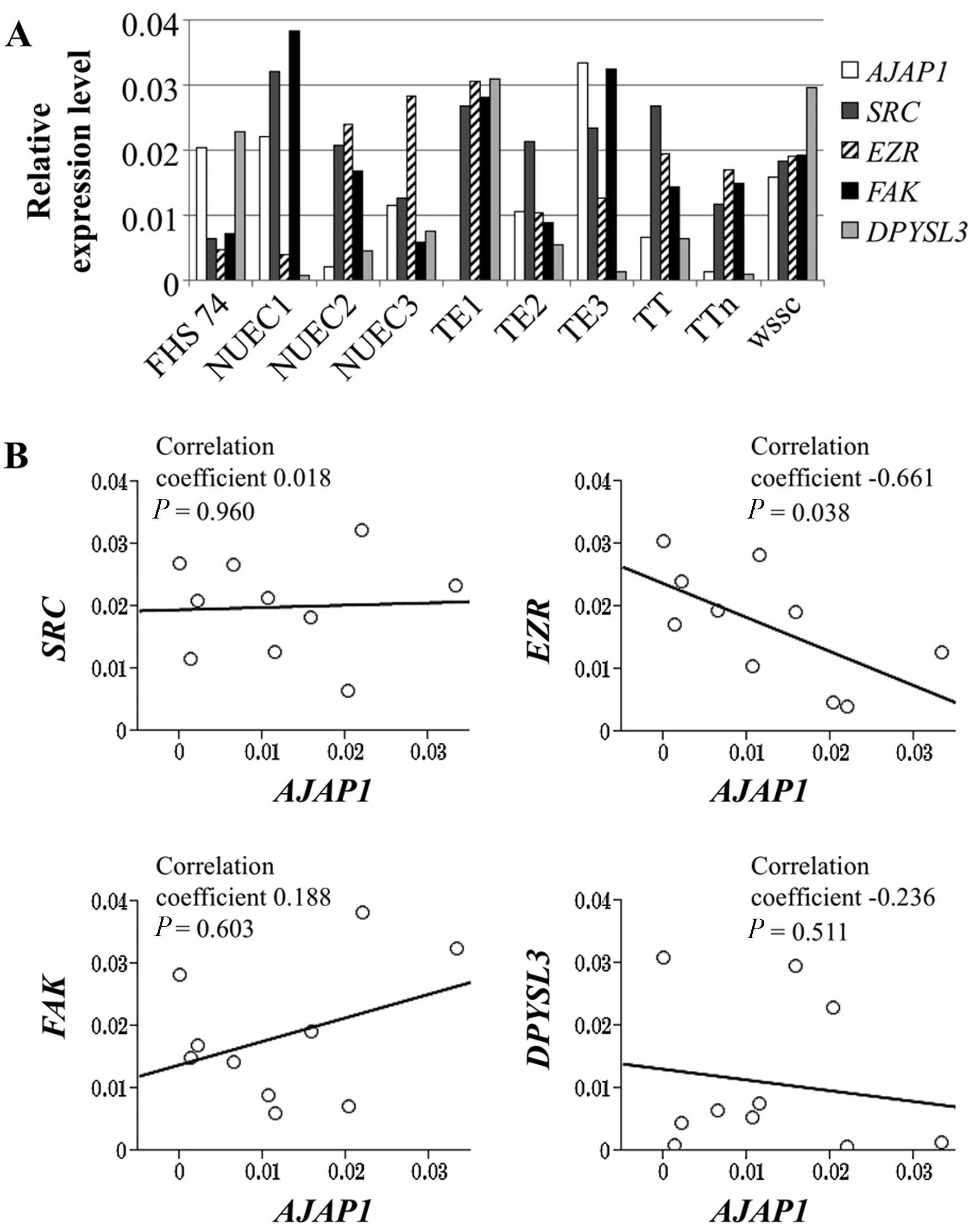
We evaluated the expression levels of genes encoding
other cell adhesion molecules that may functionally interact with
AJAP1. The relative expression levels of EZR,
FAK, SRC, DPYSL3, and AJAP1 mRNAs in
the ESCC and FHs74 cell lines are shown in Fig. 3A. The AJAP1 mRNA levels
correlated inversely with those of EZR (correlation
coefficient −0.661, P=0.038), and there was no significant
correlation with the levels of FAK, SRC and
DPYSL3 mRNAs (Fig. 3B).
Clinical significance of AJAP1 mRNA
levels
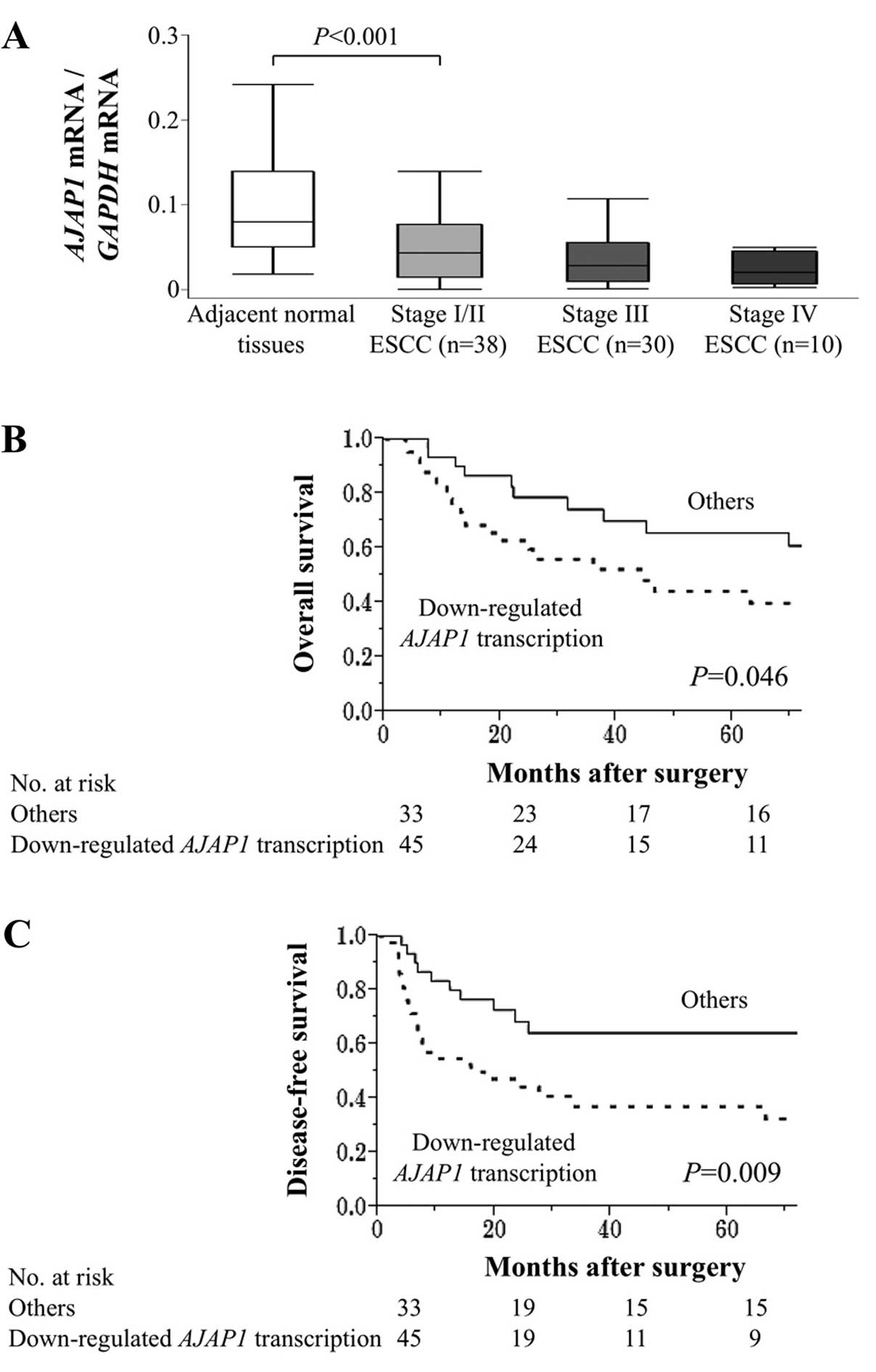
In resected samples, AJAP1 mRNA levels were
lower in ESCC tissues compared with those of adjacent normal
tissues in 67 (86%) of 78 patients. AJAP1 mRNA levels
gradually decreased according to the UICC stage (Fig. 4A). The AJAP1 mRNA levels of
45 patients with ESCC were less than half of those of adjacent
normal tissues, and these patients were designated as the
‘downregulated AJAP1 transcription’ group in the following
analyses. Downregulation of AJAP1 transcription associated
significantly with male individuals but not with
clinicopathological factors (Table
II). Patients with downregulated AJAP1 transcription
were more likely to experience shorter overall survival compared
with that of other patients (5-year survival rates were 40 and 66%,
respectively; P=0.046) (Fig. 4B).
Moreover, the disease-free survival of patients with downregulated
AJAP1 transcription was significantly shorter compared with
those of other patients (3-year survival rates were 37 and 64%,
respectively; P=0.009) (Fig. 4C).
Multivariate analysis of disease-free survival identified
downregulated AJAP1 transcription as an independent
prognostic factor (hazard ratio 2.04, 95% confidence interval (CI),
1.11–3.90; P=0.022) (Table
III).
 | Table IIAssociation between the expression of
AJAP1 mRNA and clinicopathological parameters of 78 patients
with squamous cell carcinoma of the esophagus. |
Table II
Association between the expression of
AJAP1 mRNA and clinicopathological parameters of 78 patients
with squamous cell carcinoma of the esophagus.
| Clinicopathological
parameters | Downregulated
AJAP1 transcription (n) | Other (n) | P-value |
|---|
| Age (years) | | | 0.874 |
| <65 | 24 | 17 | |
| ≥65 | 21 | 16 | |
| Gender | | | 0.016a |
| Male | 40 | 22 | |
| Female | 5 | 11 | |
| Preoperative
symptoms | | | 0.705 |
| Absent | 8 | 7 | |
| Present | 37 | 26 | |
| Brinkman index | | | 0.167 |
| <1,000 | 23 | 22 | |
| ≥1,000 | 22 | 11 | |
| Excessive alcohol
consumption | | | 0.453 |
| Absent | 9 | 9 | |
| Present | 36 | 24 | |
| CEA (ng/ml) | | | 0.225 |
| ≤5 | 42 | 28 | |
| >5 | 3 | 5 | |
| SCC (ng/ml) | | | 0.240 |
| ≤1.5 | 27 | 24 | |
| >1.5 | 18 | 9 | |
| Tumor size
(cm) | | | 0.724 |
| <5.0 | 20 | 16 | |
| ≥5.0 | 25 | 17 | |
| UICC T factor | | | 0.838 |
| T1–2 | 16 | 11 | |
| T3–4 | 29 | 22 | |
|
Differentiation | | | 0.961 |
| Moderate to
well | 38 | 28 | |
| Poor | 7 | 5 | |
| Lymphatic
involvement | | | 0.421 |
| Absent | 10 | 10 | |
| Present | 35 | 23 | |
| Vessel
invasion | | | 0.898 |
| Absent | 28 | 21 | |
| Present | 17 | 12 | |
| Intraepithelial
spread | | | 0.875 |
| Absent | 12 | 20 | |
| Present | 13 | 20 | |
| Lymph node
metastasis | | | 0.522 |
| Absent | 24 | 20 | |
| Present | 21 | 13 | |
 | Table IIIPrognostic factors for disease-free
survival of 78 patients with squamous cell carcinoma of the
esophagus. |
Table III
Prognostic factors for disease-free
survival of 78 patients with squamous cell carcinoma of the
esophagus.
| | Univariate | Multivariate |
|---|
| |
|
|
|---|
| Variable | n | Hazard ratio | 95% CI | P-value | Hazard ratio | 95% CI | P-value |
|---|
| Age (≥65) | 37 | 1.70 | 0.87–3.35 | 0.119 | | | |
| Gender (male) | 62 | 2.69 | 1.06–9.06 | 0.035 | 1.86 | 0.70–6.45 | 0.227 |
| Preoperative
symptoms | 63 | 1.29 | 0.60–3.20 | 0.541 | | | |
| Brinkman index
(≥1,000) | 33 | 1.61 | 0.83–3.12 | 0.153 | | | |
| Excessive alcohol
consumption | 60 | 0.99 | 0.47–2.32 | 0.975 | | | |
| CEA (>5
ng/ml) | 8 | 1.38 | 0.47–3.25 | 0.521 | | | |
| SCC (>1.5
ng/ml) | 27 | 0.80 | 0.37–1.61 | 0.539 | | | |
| Tumor size (≥5.0
cm) | 42 | 1.20 | 0.62–2.31 | 0.591 | | | |
| UICC T factor
(T3–4) | 51 | 1.60 | 0.79–3.48 | 0.197 | | | |
| Tumor
differentiation (poor) | 12 | 1.75 | 0.74–3.67 | 0.186 | | | |
| Lymphatic
involvement | 58 | 5.18 | 1.85–21.5 | <0.001 | 4.37 | 1.38–19.4 | 0.011a |
| Vessel
invasion | 29 | 1.89 | 0.97–3.66 | 0.062 | 1.39 | 0.70–2.76 | 0.346 |
| Intraepithelial
spread | 33 | 1.02 | 0.52–1.97 | 0.951 | | | |
| Lymph node
metastasis | 34 | 1.92 | 0.95–4.18 | 0.069 | 1.01 | 0.43–2.08 | 0.973 |
| Downregulated
AJAP1 transcription | 45 | 2.53 | 1.26–5.51 | 0.009 | 2.19 | 1.07–4.90 | 0.032a |
Discussion
Despite numerous and intensive recent studies
devoted to improving the treatment of esophageal cancer, clinical
outcomes remain unsatisfactory as indicated dramatically by 5-year
survival rates of 49.3 and 2.8% for localized and metastatic
disease, respectively (1,27). To develop novel treatment options
for ESCC, molecular biological approaches were applied to identify
specific molecular diagnostic markers and therapeutic targets
(28). We decided to study
AJAP1 for this purpose, because it encodes a transmembrane
protein of adherens junctions in epithelial cells that plays
pivotal roles in cell growth and migration and is involved in the
pathogenesis of glioblastoma (11,13).
Here, we determined the levels of AJAP1
expression in patients with ESCC to determine the underlying
regulatory mechanism. We detected reduced levels of AJAP1
mRNA in 78 and 86% of ESCC cell lines and resected ESCC tissues,
respectively, and the loss of AJAP1 expression correlated
with methylation of the AJAP1 promoter region without loss
of copy number. Moreover, AJAP1 transcription was restored
when ESCC cell lines were treated with 5-aza-dC. Downregulation of
AJAP1 was associated with worse patient outcomes,
particularly postoperative recurrence. The present study shows that
the levels of AJAP1 mRNA were frequently decreased in ESCC
cell lines and tissues, indicating that AJAP1 plays a role in the
pathogenesis of ESCC.
In the ESCC cell lines, differentially expressed
AJAP1 mRNA, and AJAP1 promoter hypermethylation were
detected only in cells with significantly decreased AJAP1
mRNA levels. Further, AJAP1 mRNA levels were increased in
cells treated with a DNA methylation inhibitor, indicating that
promoter hypermethylation is a pivotal regulatory mechanism of
AJAP1 transcription, which is consistent with studies of
patients with glioma (11,14,29,30).
In contrast, we identified some ESCC cells with reduced expression
of AJAP1 mRNA without DNA methylation, leading us to assume
that other mechanisms regulate AJAP1 transcription, such as
loss of heterozygosity (LOH), because AJAP1 resides within
chromosome 1p36, a known hotspot of chromosomal alterations
(13,31,32).
However, we did not detect a loss of AJAP1 copy number in
ESCC cell lines. Because copy number analysis addressed a limited
region of the AJAP1 locus, further investigations, including
detection of LOH and epigenetic modifications other than DNA
methylation are required.
In the present study, we determined the relative
levels of mRNAs encoding selected cell adhesion molecules to
identify novel proteins that may interact with AJAP1 in ESCC cells.
Thus, cell adhesion molecules act coordinately to mediate the
migration and invasion of tumor cells (33). We found that the levels of
AJAP1 mRNA correlated significantly with those of EZR
in ESCC and FHs74 cell lines. EZR is a member of the
ezrin-radixin-moesin family and acts as a cross-linker between the
plasma membrane and the actin cytoskeleton (34). Inactive EZR is located in the
cytoplasm, and its C-terminal domain, an F-actin-binding site, is
masked by the EZR N-terminal domain or those of other ERM proteins
(35). Moreover, EZR is a key
signaling molecule that is involved in a wide variety of cellular
processes such as cell adhesion, survival, and motility as well as
signal transduction (36,37). Moreover, EZR contributes to
tumorigenesis, development, invasion, and metastasis, likely
through regulation of adhesion molecules and signaling to other
cell membrane channels in tumors, including ESCC (38–40).
Further, the AKT/EZR/NF-κB signaling pathway
regulates the epidermal growth factor-induced
epithelial-mesenchymal transition (EMT) in squamous cell carcinoma
of the tongue (41). Our present
results indicate a possible interaction between AJAP1 and EZR that
may provide the first step required to understand the role of AJAP1
in oncogenesis. Further investigation of the correlation between
the expression of AJAP1 and EMT-associated molecules are
required.
We show here that AJAP1 mRNA levels gradually
decreased as a function of UICC tumor stage, highlighting the
diagnostic implications of analyzing AJAP1 expression in
esophageal tissues. Downregulation of AJAP1 mRNA in ESCC
tissues associated significantly with worse prognosis after
curative esophagectomy, particularly with disease-free survival.
This finding emphasizes that AJAP1 expression is a potential
biomarker for patients with ESCC who are susceptible to
recurrence.
Taken together, our analyses of AJAP1 promise
to improve clinical management of ESCC as follows: i) The
expression levels of AJAP1 in biopsy tissue obtained using
endoscopic surveillance may identify patients requiring intensive
systemic treatment or neoadjuvant therapy; ii) the expression
levels of AJAP1 in surgical specimens may predict recurrence
and subsequent adverse prognosis, leading to the design of
appropriate therapeutic strategies; and iii) demethylating agents
targeting AJAP1 may serve as therapeutics. However, this
study is limited by its lack of direct functional analysis of
AJAP1, and we are unable to conclude that AJAP1 acts a suppressor
of ESCC. Further, the mechanisms that regulate AJAP1
expression, other than promoter hypermethylation, remain to be
determined. Further studies are therefore necessary to identify the
molecular mechanisms underlying the phenotypes of ESCC cells.
In summary, our data suggest that AJAP1
expression is frequently suppressed in ESCC and that
hypermethylation of the AJAP1 promoter region is a pivotal
regulatory mechanism of AJAP1 expression in ESCC.
Downregulation of AJAP1 in ESCC tissues may represent a
promising biomarker for predicting ESCC recurrence.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oya H, Kanda M, Takami H, Hibino S,
Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, et al:
Overexpression of melanoma-associated antigen D4 is an independent
prognostic factor in squamous cell carcinoma of the esophagus. Dis
Esophagus. 28:188–195. 2015. View Article : Google Scholar
|
|
3
|
Enzinger PC and Mayer RJ: Esophageal
cancer. N Engl J Med. 349:2241–2252. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mayne ST and Navarro SA: Diet, obesity and
reflux in the etiology of adenocarcinomas of the esophagus and
gastric cardia in humans. J Nutr. 132(Suppl): 3467S–3470S.
2002.PubMed/NCBI
|
|
5
|
Kamangar F, Chow WH, Abnet CC and Dawsey
SM: Environmental causes of esophageal cancer. Gastroenterol Clin
North Am. 38:27–57. vii2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Song Y, Li L, Ou Y, Gao Z, Li E, Li X,
Zhang W, Wang J, Xu L, Zhou Y, et al: Identification of genomic
alterations in oesophageal squamous cell cancer. Nature. 509:91–95.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang JC, Lam KY, Law S, Wong J and
Srivastava G: Detection of genetic alterations in esophageal
squamous cell carcinomas and adjacent normal epithelia by
comparative DNA fingerprinting using inter-simple sequence repeat
PCR. Clin Cancer Res. 7:1539–1545. 2001.PubMed/NCBI
|
|
8
|
Hibino S, Kanda M, Oya H, Takami H,
Shimizu D, Nomoto S, Hishida M, Niwa Y, Koike M, Yamada S, et al:
Reduced expression of DENND2D through promoter hypermethylation is
an adverse prognostic factor in squamous cell carcinoma of the
esophagus. Oncol Rep. 31:693–700. 2014.
|
|
9
|
Oya H, Kanda M, Koike M, Iwata N, Niwa Y,
Shimizu D, Takami H, Sueoka S, Hashimoto R, Ezaka K, et al:
Detection of serum melanoma-associated antigen D4 in patients with
squamous cell carcinoma of the esophagus. Dis Esophagus. May
8–2015.(Epub ahead of print). View Article : Google Scholar
|
|
10
|
Schreiner A, Ruonala M, Jakob V, Suthaus
J, Boles E, Wouters F and Starzinski-Powitz A: Junction protein
shrew-1 influences cell invasion and interacts with
invasion-promoting protein CD147. Mol Biol Cell. 18:1272–1281.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han L, Zhang KL, Zhang JX, Zeng L, Di CH,
Fee BE, Rivas M, Bao ZS, Jiang T, Bigner D, et al: AJAP1 is
dysregulated at an early stage of gliomagenesis and suppresses
invasion through cytoskeleton reorganization. CNS Neurosci Ther.
20:429–437. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
McDonald JM, Dunlap S, Cogdell D, Dunmire
V, Wei Q, Starzinski-Powitz A, Sawaya R, Bruner J, Fuller GN,
Aldape K, et al: The SHREW1 gene, frequently deleted in
oligodendrogliomas, functions to inhibit cell adhesion and
migration. Cancer Biol Ther. 5:300–304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin N, Di C, Bortoff K, Fu J, Truszkowski
P, Killela P, Duncan C, McLendon R, Bigner D, Gregory S, et al:
Deletion or epigenetic silencing of AJAP1 on 1p36 in glioblastoma.
Mol Cancer Res. 10:208–217. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zeng L, Fee BE, Rivas MV, Lin J and
Adamson DC: Adherens junctional associated protein-1: A novel 1p36
tumor suppressor candidate in gliomas (Review). Int J Oncol.
45:13–17. 2014.PubMed/NCBI
|
|
15
|
Kanda M, Shimizu D, Nomoto S, Takami H,
Hibino S, Oya H, Hashimoto R, Suenaga M, Inokawa Y, Kobayashi D, et
al: Prognostic impact of expression and methylation status of
DENN/MADD domain-containing protein 2D in gastric cancer. Gastric
Cancer. 18:288–296. 2015. View Article : Google Scholar
|
|
16
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Hibino S, Shimizu D, Takami H, Hashimoto R, Okamura Y, Yamada S, et
al: B cell translocation gene 1 serves as a novel prognostic
indicator of hepatocellular carcinoma. Int J Oncol. 46:641–648.
2015.
|
|
17
|
Long MJ, Jiang CQ, Lam TH, Lin JM, Chan
YH, Zhang WS, Jin YL, Liu B, Thomas GN and Cheng KK: Alcohol
consumption and electrocardiographic left ventricular hypertrophy
and mediation by elevated blood pressure in older Chinese men: The
Guangzhou Biobank Cohort Study. Alcohol. 47:473–480. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tanaka Y, Kanda M, Sugimoto H, Shimizu D,
Sueoka S, Takami H, Ezaka K, Hashimoto R, Okamura Y, Iwata N, et
al: Translational implication of Kallmann syndrome-1 gene
expression in hepatocellular carcinoma. Int J Oncol. 46:2546–2554.
2015.PubMed/NCBI
|
|
19
|
Kanda M, Shimizu D, Nomoto S, Hibino S,
Oya H, Takami H, Kobayashi D, Yamada S, Inokawa Y, Tanaka C, et al:
Clinical significance of expression and epigenetic profiling of
TUSC1 in gastric cancer. J Surg Oncol. 110:136–144. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takai D and Jones PA: The CpG island
searcher: A new WWW resource. In Silico Biol. 3:235–240.
2003.PubMed/NCBI
|
|
21
|
Kanda M, Sugimoto H, Nomoto S, Oya H,
Shimizu D, Takami H, Hashimoto R, Sonohara F, Okamura Y, Yamada S,
et al: Clinical utility of PDSS2 expression to stratify patients at
risk for recurrence of hepatocellular carcinoma. Int J Oncol.
45:2005–2012. 2014.PubMed/NCBI
|
|
22
|
Kanda M, Nomoto S, Oya H, Hashimoto R,
Takami H, Shimizu D, Sonohara F, Kobayashi D, Tanaka C, Yamada S,
et al: Decreased expression of prenyl diphosphate synthase subunit
2 correlates with reduced survival of patients with gastric cancer.
J Exp Clin Cancer Res. 33:882014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kanda M, Nomoto S, Oya H, Takami H, Hibino
S, Hishida M, Suenaga M, Yamada S, Inokawa Y, Nishikawa Y, et al:
Downregulation of DENND2D by promoter hypermethylation is
associated with early recurrence of hepatocellular carcinoma. Int J
Oncol. 44:44–52. 2014.
|
|
24
|
Kanda M, Oya H, Nomoto S, Takami H,
Shimizu D, Hashimoto R, Sueoka S, Kobayashi D, Tanaka C, Yamada S,
et al: Diversity of Clinical Implication of B-cell translocation
gene 1 expression by histopathologic and anatomic subtypes of
gastric cancer. Dig Dis Sci. 60:1256–1264. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oya H, Kanda M, Sugimoto H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Okamura Y, Yamada S, Fujii T, et
al: Dihydropyrimidinase-like 3 is a putative hepatocellular
carcinoma tumor suppressor. J Gastroenterol. 50:590–600. 2015.
View Article : Google Scholar
|
|
26
|
Kanda M, Nomoto S, Oya H, Shimizu D,
Takami H, Hibino S, Hashimoto R, Kobayashi D, Tanaka C, Yamada S,
et al: Dihydropyrimidinase-like 3 facilitates malignant behavior of
gastric cancer. J Exp Clin Cancer Res. 33:662014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shim HJ, Shin MH, Kim HN, Kim JH, Hwang
JE, Bae WK, Chung IJ and Cho SH: The prognostic significance of
FGFR4 Gly388 polymorphism in esophageal squamous cell carcinoma
after concurrent chemoradiotherapy. Cancer Res Treat. May
14–2015.(Epub ahead of print). View Article : Google Scholar
|
|
28
|
Lee HW, Kwon J, Kang MC, Noh MK, Koh JS,
Kim JH and Park JH: Overexpression of HSP47 in esophageal squamous
cell carcinoma: Clinical implications and functional analysis. Dis
Esophagus. May 8–2015.(Epub ahead of print). View Article : Google Scholar
|
|
29
|
Cogdell D, Chung W, Liu Y, McDonald JM,
Aldape K, Issa JP, Fuller GN and Zhang W: Tumor-associated
methylation of the putative tumor suppressor AJAP1 gene and
association between decreased AJAP1 expression and shorter survival
in patients with glioma. Chin J Cancer. 30:247–253. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zeng L, Kang C, Di C, Fee BE, Rivas M, Lin
J and Adamson DC: The adherens junction-associated protein 1 is a
negative transcriptional regulator of MAGEA2, which potentiates
temozolomide-induced apoptosis in GBM. Int J Oncol. 44:1243–1251.
2014.PubMed/NCBI
|
|
31
|
Mori T, Nomoto S, Koshikawa K, Fujii T,
Sakai M, Nishikawa Y, Inoue S, Takeda S, Kaneko T and Nakao A:
Decreased expression and frequent allelic inactivation of the RUNX3
gene at 1p36 in human hepatocellular carcinoma. Liver Int.
25:380–388. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang H, Zhai Y, Hu Z, Wu C, Qian J, Jia
W, Ma F, Huang W, Yu L, Yue W, et al: Genome-wide association study
identifies 1p36.22 as a new susceptibility locus for hepatocellular
carcinoma in chronic hepatitis B virus carriers. Nat Genet.
42:755–758. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xin M, Dong XW and Guo XL: Role of the
interaction between galectin-3 and cell adhesion molecules in
cancer metastasis. Biomed Pharmacother. 69:179–185. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jin T, Jin J, Li X, Zhang S, Choi YH, Piao
Y, Shen X and Lin Z: Prognostic implications of ezrin and
phosphorylated ezrin expression in non-small cell lung cancer. BMC
Cancer. 14:1912014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bretscher A, Edwards K and Fehon RG: ERM
proteins and merlin: Integrators at the cell cortex. Nat Rev Mol
Cell Biol. 3:586–599. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Srivastava J, Elliott BE, Louvard D and
Arpin M: Src-dependent ezrin phosphorylation in adhesion-mediated
signaling. Mol Biol Cell. 16:1481–1490. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wang X, Liu M and Zhao CY: Expression of
ezrin and moesin related to invasion, metastasis and prognosis of
laryngeal squamous cell carcinoma. Genet Mol Res. 13:8002–8013.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie JJ, Xu LY, Xie YM, Zhang HH, Cai WJ,
Zhou F, Shen ZY and Li EM: Roles of ezrin in the growth and
invasiveness of esophageal squamous carcinoma cells. Int J Cancer.
124:2549–2558. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kong J, Li Y, Liu S, Jin H, Shang Y, Quan
C, Li Y and Lin Z: High expression of ezrin predicts poor prognosis
in uterine cervical cancer. BMC Cancer. 13:5202013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ghaffari A, Hoskin V, Szeto A, Hum M,
Liaghati N, Nakatsu K, LeBrun D, Madarnas Y, Sengupta S and Elliott
BE: A novel role for ezrin in breast cancer
angio/lymphangiogenesis. Breast Cancer Res. 16:4382014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wang Y, Lin Z, Sun L, Fan S, Huang Z,
Zhang D, Yang Z, Li J and Chen W: Akt/Ezrin Tyr353/NF-κB pathway
regulates EGF-induced EMT and metastasis in tongue squamous cell
carcinoma. Br J Cancer. 110:695–705. 2014. View Article : Google Scholar :
|