Introduction
Lung cancer remains the most frequent cause of
cancer-related death in developed countries, and an estimated 1.8
million new cases of lung cancer occurred in 2012 (1). Approximately 80% of lung cancers are
classified histopathologically as non-small cell lung cancer
(NSCLC), and NSCLC are subdivided into four major histological
subtypes: adenocarcinoma, squamous cell carcinoma (SCC), large cell
carcinoma, and neuroendocrine cancer (2). NSCLC, as compared to small cell lung
cancer, is less sensitive to anticancer drugs and radiation
therapy. Recently, molecular target therapies for adenocarcinoma
(gefitinib, erlotinib and crizotinib) have shown remarkable
therapeutic efficacy; however, no targeted therapeutics are
currently approved for treatment of lung SCC (3). Therefore, the lung squamous cell
carcinomas need a new treatment option.
The human genome sequence era and the discovery of
microRNAs (miRNAs) in human genomes have brought great changes to
the study of human cancers. miRNAs are endogenous small non-coding
RNA molecules (19–22 bases in length) that regulate protein-coding
gene expression by repressing translation or cleaving RNA
transcripts in a sequence-specific manner (4,5).
Substantial evidence suggests that miRNAs are aberrantly expressed
in many human cancers and play significant roles in human
oncogenesis and metastasis (6,7). The
nature of miRNAs is unique in that one miRNA has the ability to
regulate multiple protein-coding RNAs. Bioinformatic predictions
indicate that miRNAs regulate 30–60% of the protein-coding genes in
the human genome (5,6). Therefore, identification of
tumor-suppressive or oncogenic miRNAs and the miRNAs-mediated novel
cancer networks are the first step toward elucidating the molecular
mechanisms of human cancers.
Based on this foregoing discussion, we sequentially
identified tumor-suppressive miRNAs and the miRNA-regulated
oncogenic genes in various types of cancers (8–11). A
recent study of lung SCC showed that miRNA-1/133a-clustered
miRNAs inhibit cancer cell migration and invasion by targeting the
CORO1C gene encoding a member of the WD repeat protein family and
is involved in a variety of cellular processes (12). Moreover, tumor-suppressive
miR-206 inhibited dual signaling networks activated by MET
and EGFR in lung SCC cells (13).
Our miRNA expression signatures of human cancers
demonstrated that the miR-29 family (miR-29a,
miR-29b, and miR-29c) was downregulated in cancer
tissues compared to normal tissues (8,9,14–17),
suggesting that these miRNAs function as tumor suppressors in lung
SCC cells. However, miR-29s-regulating molecular networks
have not been sufficiently analyzed in this disease. The aim of
this study is to investigate the functional significance of
miR-29s in lung SCC and to identify
miR-29s-regulating molecular targets in lung SCC cells.
In this study, we found that the restoration of all
miR-29s inhibited cancer cell migration and invasion,
directly targeting the lysyl oxidase-like 2 (LOXL2) gene.
Moreover, overexpression of LOXL2 was detected in lung SCC clinical
specimens, and silencing of LOXL2 significantly inhibited
cell migration and invasion by cancer cells. The tumor-suppressive
miR-29-LOXL2 axis may provide new insights into the
potential mechanisms of lung SCC oncogenesis and metastasis.
Materials and methods
Clinical specimens and RNA
extraction
A total of 32 lung SCCs and 22 normal lung specimens
were collected from patients who underwent pneumonectomy at
Kagoshima University Hospital from 2010 to 2013. The patient
backgrounds and clinical characteristics are summarized in Table I. Archival formalin-fixed
paraffin-embedded (FFPE) samples were used for qRT-PCR analysis and
immunohistochemistry.
 | Table ICharacteristics of the lung cancer
and normal lung cases. |
Table I
Characteristics of the lung cancer
and normal lung cases.
| A, Characteristics
of the lung cancer cases |
|---|
|
|---|
| Lung cancer | n | (%) |
|---|
| Total number | 32 | |
| Median age
(range) | 71 | (50–88) |
| Gender |
| Male | 30 | (93.7) |
| Female | 2 | (6.3) |
| Pathological
stage |
| IA | 4 | (12.5) |
| IB | 8 | (25.0) |
|
IIA | 4 | (12.5) |
|
IIB | 5 | (15.6) |
|
IIIA | 8 | (25.0) |
|
IIIB | 1 | (3.1) |
| Unknown | 2 | (6.3) |
|
Differentiation |
| Well | 8 | (25.0) |
| Moderately | 19 | (59.4) |
| Poorly | 3 | (9.4) |
| Unknown | 2 | (6.3) |
| Pleural
invasion |
| (+) | 15 | (46.9) |
| (−) | 17 | (53.1) |
| Venous
invasion |
| (+) | 16 | (50.0) |
| ( −) | 16 | (50.0) |
| Lymphatic
invasion |
| (+) | 16 | (50.0) |
| ( −) | 16 | (50.0) |
| Recurrence |
| (+) | 9 | (28.1) |
| ( −) | 20 | (62.5) |
| Unknown | 3 | (9.4) |
|
| B, Characteristics
of the normal lung cases |
|
| Normal lung | n | (%) |
|
| Total number | 22 | |
| Median age
(range) | 71 | (50–88) |
| Gender |
| Male | 22 | |
| Female | 0 | |
Samples were staged according to the International
Association for the Study of Lung Cancer TNM classification, and
they were histologically graded (18). This study was approved by the
Institutional Review Board for Clinical Research of Kagoshima
University School of Medicine. Prior written informed consent and
approval were provided by each patient.
FFPE tissues were sectioned to a thickness of 10 μm,
and 8 tissue sections were used for RNA extraction. Total RNA
(including miRNA) was extracted using Recover All™ Total Nucleic
Acid Isolation kit (Ambion, Austin, TX, USA) using the
manufacturer's protocol. The integrity of the RNA was checked with
an RNA 6000 Nano assay kit and a 2100 Bioanalyzer (Agilent
Technologies, Santa Clara, CA, USA).
Cell culture and RNA extraction
We used human lung SCC cell lines (EBC-1 and
SK-MES-1) obtained from the Japanese Cancer Research Resources Bank
(JCRB) and the American Type Culture Collection (Manassas, VA,
USA), respectively. Cells were grown in RPMI-1640 medium
supplemented with 10% fetal bovine serum (FBS) and maintained in a
humidified incubator (5% CO2) at 37°C.
Total RNA was isolated using Isogen (Nippon Gene,
Tokyo, Japan) according to the manufacturer's protocol. The
integrity of the RNA was checked with an RNA 6000 Nano assay kit
and a 2100 Bioanalyzer (Agilent Technologies).
Quantitative reverse transcription PCR
(qRT-PCR)
The procedure for PCR quantification was described
previously (9–11). TaqMan probes and primers for
LOXL2 (P/N: Hs00158757_m1; Applied Biosystems, Foster City,
CA, USA) were assay-on-demand gene expression products. Stem-loop
RT-PCRs for miR-29a (P/N: 002112; Applied Biosystems),
miR-29b (P/N: 000413), and miR-29c (P/N: 000587) were
used to quantify the expression levels of miRNAs according to the
manufacturer's protocol. To normalize the data for quantification
of mRNA and miRNAs, we used human GUSB (P/N: Hs99999908_m1;
Applied Biosystems) and RNU48 (P/N: 001006; Applied
Biosystems), respectively.
Transfections with mature miRNA and small
interfering RNA (siRNA) into cell lines
The following mature miRNA species were used in the
present study: Pre-miR™ miRNA precursors (hsa-miR-29a-3p,
P/N: AM 12499; hsa-miR-29b-3p, P/N: AM 10103;
hsa-miR-29c-3p, P/N: AM 10518; and negative control miRNA,
P/N: AM 17111), Stealth Select RNAi siRNA, si-LOXL2 (P/N:
HSS106124, P/N: HSS106125 and P/N: HSS180848; Invitrogen, Carlsbad,
CA, USA), and negative-control siRNA (D-001810-10; Thermo Fisher
Scientific, Waltham, MA, USA). RNAs were incubated with OPTI-MEM
and Lipofectamine RNAiMAX Reagent (both from Invitrogen) as
described previously (9–11).
Cell proliferation, migration, and
invasion assays
Cells were transfected with 10 nM miRNAs by reverse
transfection and plated in 96-well plates at 8×103
cells/well. After 96 h, cell proliferation was determined with the
XTT assay using the Cell Proliferation kit II (Roche Molecular
Biochemicals, Mannheim, Germany) as described previously (9–11).
Cell migration activity was evaluated with wound
healing assays. Cells were plated in 6-well plates at
8×105 cells/well, and after 48 h of transfection, the
cell monolayer was scraped using a P-20 micropipette tip. The
initial gap length (0 h) and the residual gap length 24 h after
wounding were calculated from photomicrographs as described
previously (9–11).
Cell invasion assays were performed using modified
Boyden chambers, consisting of Transwell-precoated Matrigel
membrane filter inserts with 8 μm pores in 24-well tissue culture
plates (BD Biosciences, Bedford, MA, USA). After 72 h of
transfection, cells were plated in 24-well plates at
1×105 cells/well. Minimum essential medium containing
10% FBS in the lower chamber served as the chemoattractant, as
described previously (9–11). All experiments were performed in
triplicate.
Western blotting
After 96 h of transfection, protein lysates (50 μg)
were separated on NuPAGE on 4–12% Bis-Tris gels (Invitrogen) and
transferred to polyvinylidene fluoride membranes. Immunoblotting
was performed with diluted anti-LOXL2 antibodies (1:1,000; ab96233;
Abcam, Cambridge, UK) and anti-GAPDH antibodies (1:5,000; MAB374;
Chemicon, Temecula, CA, USA). The membrane was washed and then
incubated with an anti-rabbit-IgG, HRP-linked antibody (#7074; Cell
Signaling Technology, Danvers, MA, USA). Specific complexes were
visualized with an echochemiluminescence (ECL) detection system (GE
Healthcare, Little Chalfont, UK) as described previously (9–11).
Plasmid construction and dual-luciferase
reporter assay
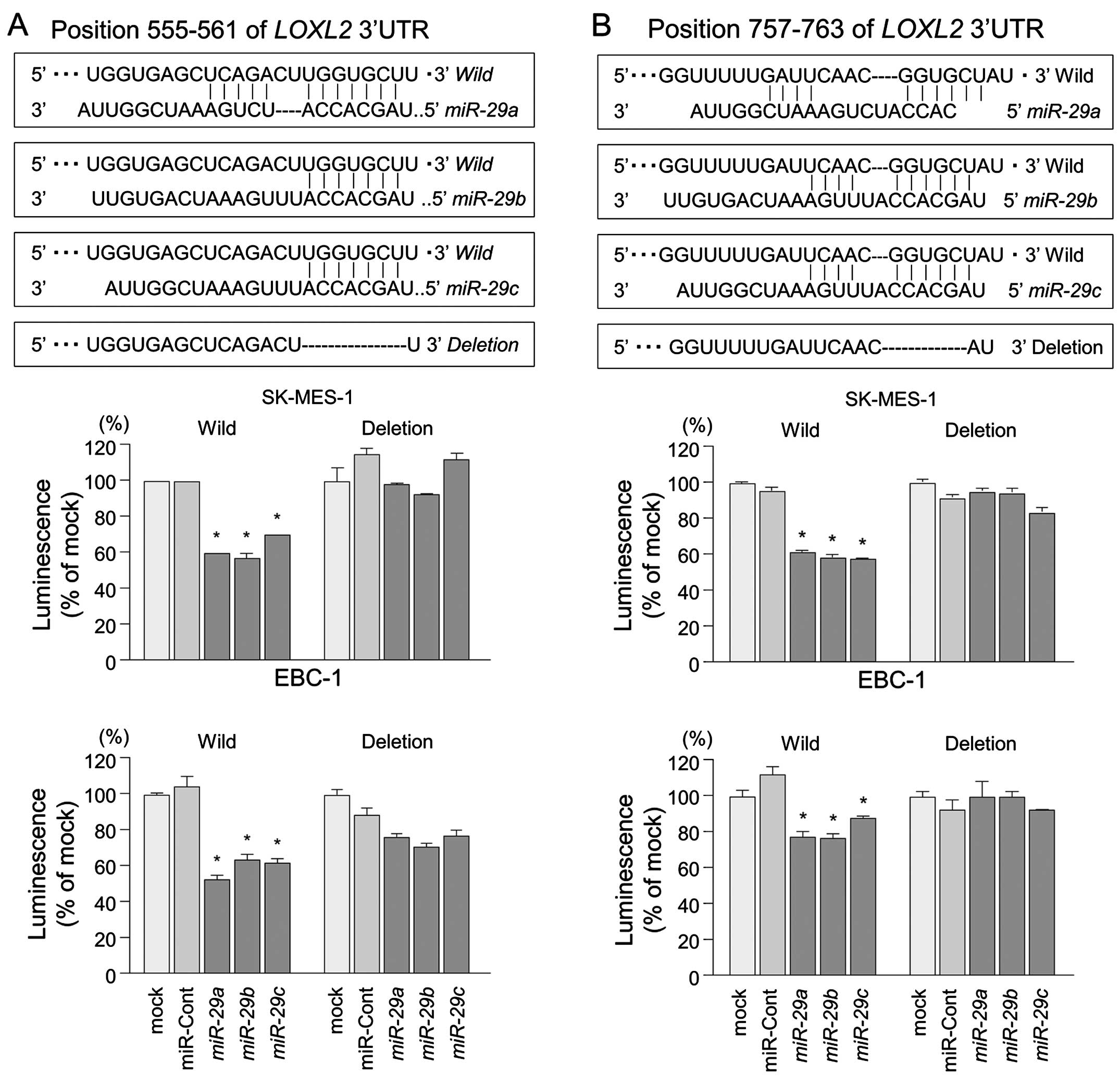
A partial wild-type sequence of the LOXL2
3′-UTR or those with a deleted miR-29 target site (position
555–561 or position 757–763 of the LOXL2 3′-UTR) was
inserted between the XhoI-PmeI restriction sites in
the 3′-UTR of the hRluc gene in the psiCHECK-2 vector (C8021;
Promega, Madison, WI, USA).
The synthesized DNA was cloned into the psiCHECK-2
vector. EBC-1 cells and SK-MES-1 cells were transfected with a 50
ng vector, 10 nM miRNAs, and 1 μl of Lipofectamine 2000 in 100 μl
of Opti-MEM (both from Invitrogen). The activities of Firefly and
Renilla luciferases in cell lysates were determined with a
dual-luciferase assay system (E1910; Promega). Normalized data were
calculated as the quotient of Renilla/Firefly luciferase
activities.
Immunohistochemistry
We stained the tissue array (LC2083; US Biomax,
Rockville, MD, USA). The tissues were immunostained following the
manufacturer's protocol with an UltraVision Detection System
(Thermo Fisher Scientific). The primary rabbit polyclonal
antibodies against LOXL2 (ab96233) were diluted 1:1,000. The slides
were treated with biotinylated goat anti-rabbit antibodies.
Diaminobenzidine hydrogen peroxidase was the chromogen, and
counterstaining was done with 0.5% hematoxylin.
Identification of putative miR-29 target
genes
To identify putative miR-29-regulated genes,
we used the TargetScan database (http://www.targetscan.org/). We investigated the
expression status of putative targets of miR-29 using lung
SCC clinical expression data from the GEO database (accession no.
GSE 11117). Additionally, we performed gene expression analysis
using miR-29a transfected EBC-1 cells. Oligo-microarray
procedures and data mining methods were described in previous
studies (9,10).
Statistical analysis
Relationships between two or three variables and
numerical values were analyzed using the Mann-Whitney U test or
Bonferroni-adjusted Mann-Whitney U test. Expert StatView version 4
was used in these analyses.
Results
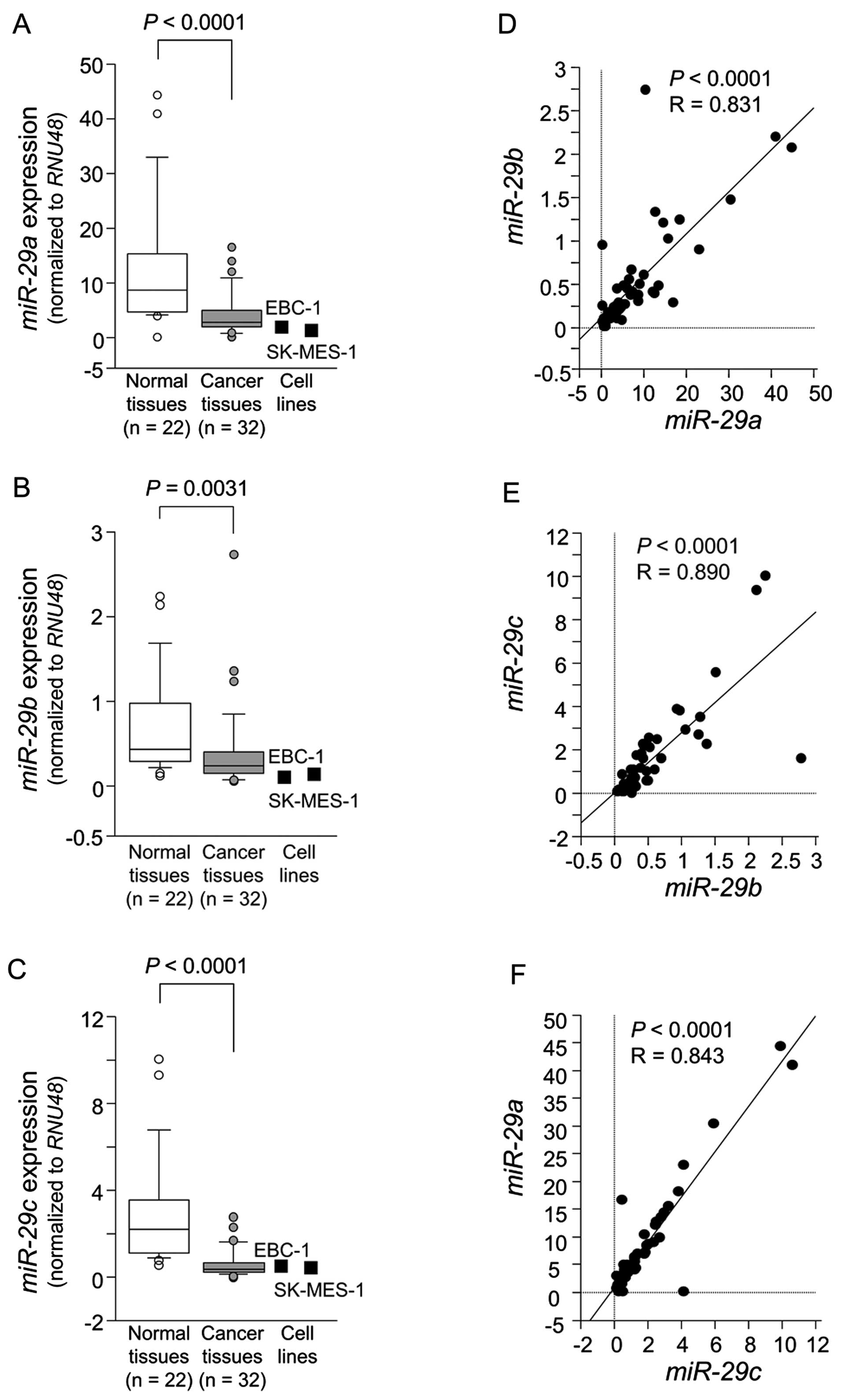
Expression levels of miR-29a, miR-29b and
miR-29c in lung SCC clinical specimens
The expression levels of miR-29a,
miR-29b and miR-29c were significantly reduced in
tumor tissues compared to corresponding noncancerous tissues
(P<0.0001, P=0.0031 and P<0.0001, respectively) (Fig. 1A–C). Spearman's rank test showed a
positive correlation between the expression of miR-29a and
that of miR-29b (R=0.836 and P<0.0001) (Fig. 1D). The expression of miR-29a
was positively correlated with that of miR-29c (R=0.878 and
P<0.0001) (Fig. 1E). Similarly,
the expression of miR-29b was positively correlated with
that of miR-29c (R=0.744 and P<0.0001) (Fig. 1F).
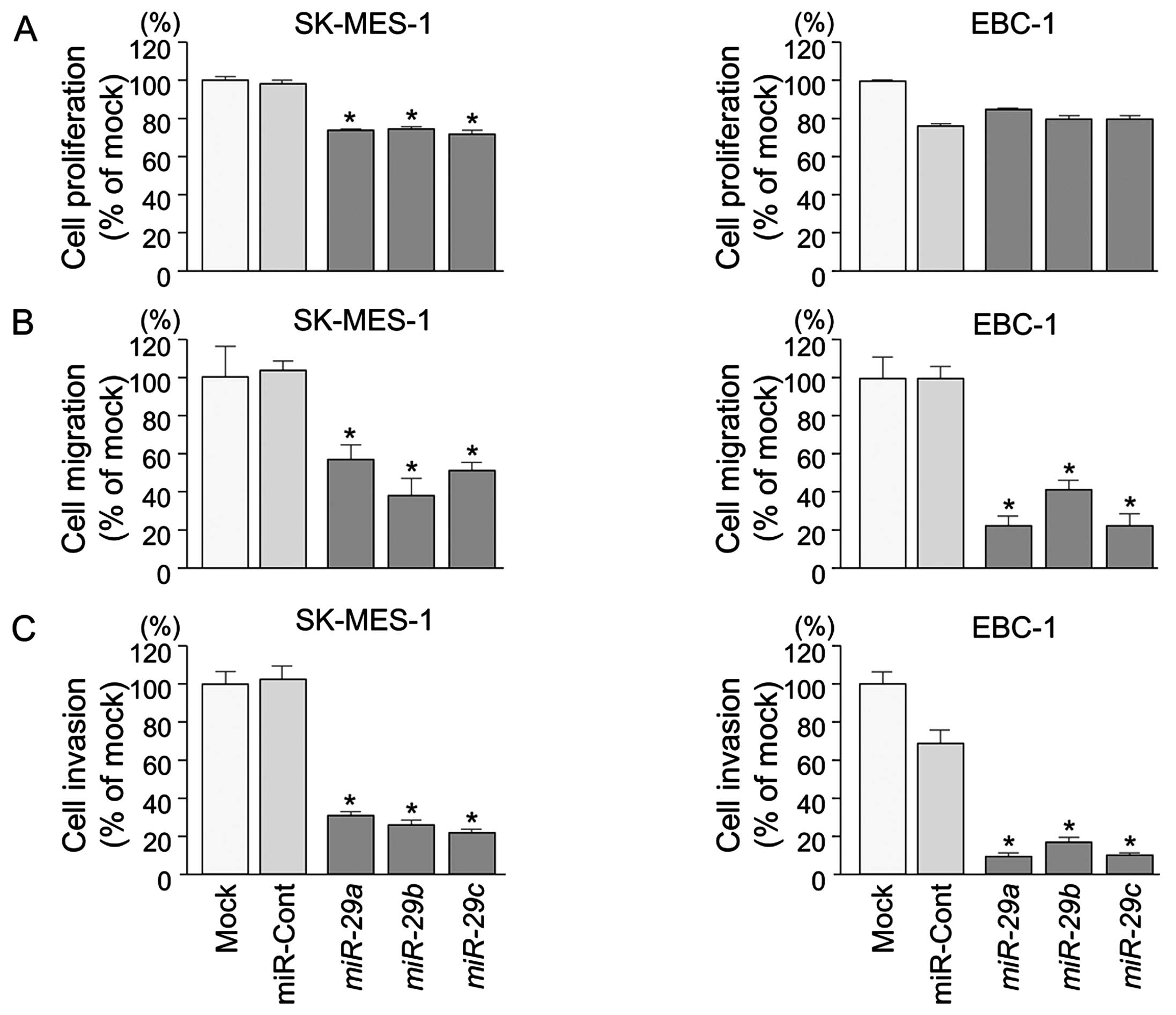
Effects of miR-29a, miR-29b and miR-29c
restoration on the proliferation, migration and invasion in lung
SCC cell lines
To examine the functional roles of the miR-29
family (miR-29a, miR-29b and miR-29c), we
performed gain-of-function studies using miRNA transfection into
lung SCC cell lines (EBC-1 and SK-MES-1).
XTT assays revealed significant inhibition of cell
proliferation in SK-MES-1 cells transfected with miR-29s in
comparison with mock or control transfectants (P<0.0001)
(Fig. 2A). Otherwise, in EBC-1
cells transfected with miR-29s, there was no significant
inhibition of cell proliferation in comparison with control
transfectants (Fig. 2A).
Wound healing assays showed significant inhibition
of cell migration activity after transfection with miR-29s
(P<0.0001) (Fig. 2B).
Similarly, Matrigel invasion assays revealed that
transfection with miR-29 reduced cell invasion activities
(P<0.0001) (Fig. 2C).
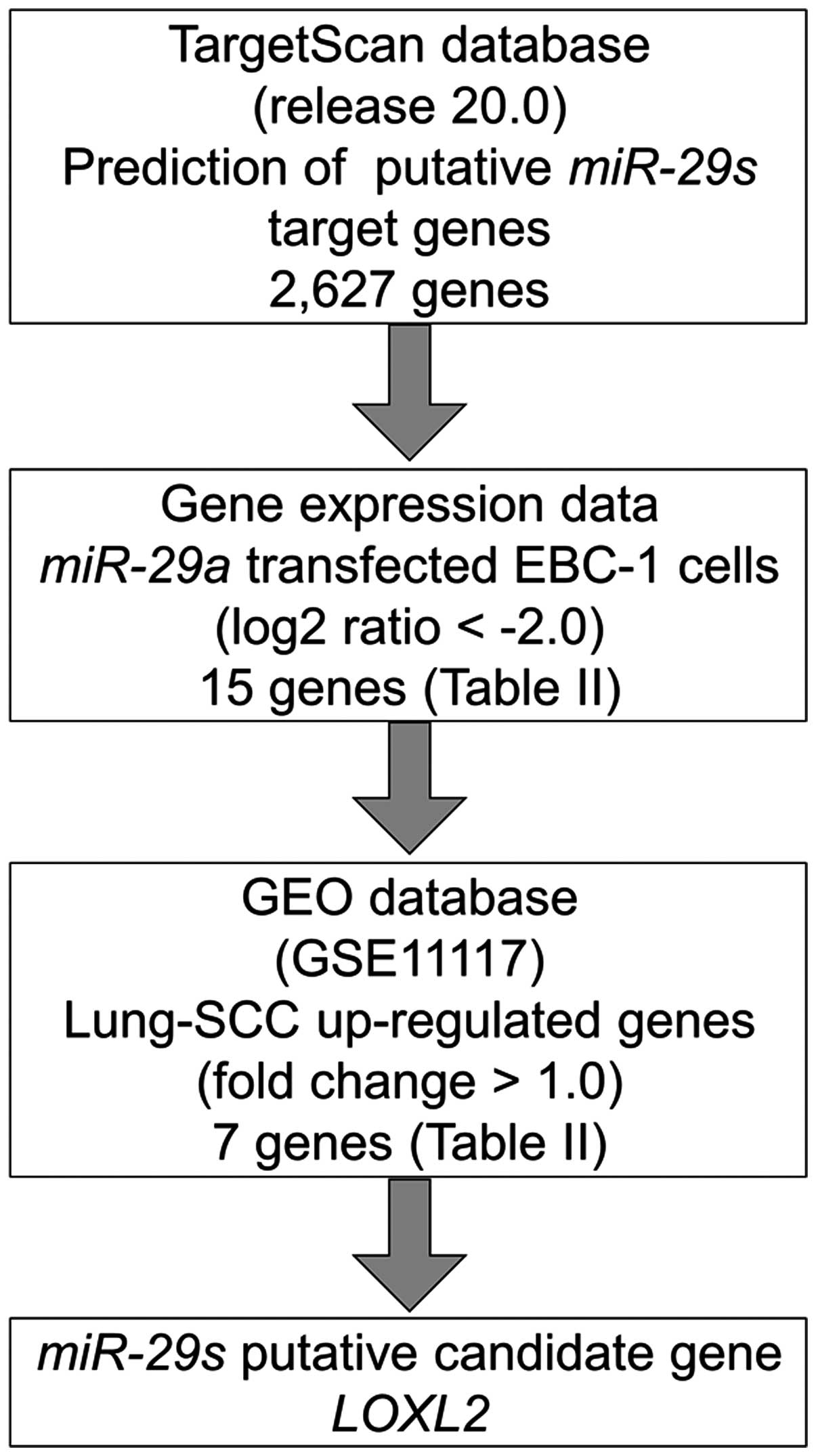
Identification of candidate target genes
of miR-29s in lung SCC
To identify molecular targets of miR-29s, we
used a combination of in silico analysis and lung SCC gene
expression data. In total, 2,627 genes were putative targets of
miR-29s according to the TargetScan database. Next, we pared
down the list of genes based on two kinds of gene expression data:
downregulated genes (Log2 ratio <-2.0) following
miR-29a-transfected EBC-1 cells and upregulated genes
determined by the gene expression data set of lung SCC clinical
specimens according to the GEO database (accession no. GSE 11117).
From this selection, 7 candidate genes were identified as targets
of the miR-29s (Table II).
Among these genes, we focused on the LOXL2 gene and examined
the LOXL2 function and characteristics in further analyses.
Our strategy for selecting miR-29 target genes is shown in
Fig. 3.
 | Table IIDownregulated genes in miR-29a
transfectant. |
Table II
Downregulated genes in miR-29a
transfectant.
| Entrez gene ID | Gene symbol | Description | EBC-1 miR-29
transfectant Log2 ratio | miR-29a
conserved site | miR-29a
poorly conserved site | GSE1117
fold-change |
|---|
| 4017 | LOXL2 | Lysyl oxidase-like
2 | −4.05 | 1 | 1 | 2.10 |
| 3038 | HAS3 | Hyaluronan synthase
3 | −3.37 | 3 | | 4.19 |
| 9535 | GMFG | Glia maturation
factor γ | −3.10 | 2 | | ND |
| 3655 | ITGA6 | Integrin α6 | −2.62 | 1 | | 2.78 |
| 634 | CEACAM1 | Carcinoembryonic
antigen-related cell adhesion molecule 1 (biliary
glycoprotein) | −2.58 | 1 | 1 | 1.41 |
| 871 |
SERPINH1 | Serpin peptidase
inhibitor, clade H (heat shock protein 47), member 1 (collagen
binding protein 1) | −2.57 | 1 | | ND |
| 22801 | ITGA11 | Integrin α11 | −2.42 | 1 | | 1.50 |
| 80381 | CD276 | CD276 molecule | −2.35 | 1 | | ND |
| 50848 | F11R | F11 receptor | −2.26 | 1 | 1 | ND |
| 8894 | EIF2S2 | Eukaryotic
translation initiation factor 2, subunit 2β, 38 kDa | −2.17 | 1 | 1 | 1.70 |
| 91584 | PLXNA4 | Plexin A4 | −2.15 | 1 | 1 | ND |
| 55920 | RCC2 | Regulator of
chromosome condensation 2 | −2.11 | 1 | | ND |
| 9076 | CLDN1 | Claudin 1 | −2.10 | 1 | | ND |
| 2118 | ETV4 | Ets variant 4 | −2.09 | 1 | | 4.84 |
| 284119 | PTRF | Polymerase I and
transcript release factor | −2.05 | 1 | | −1.86 |
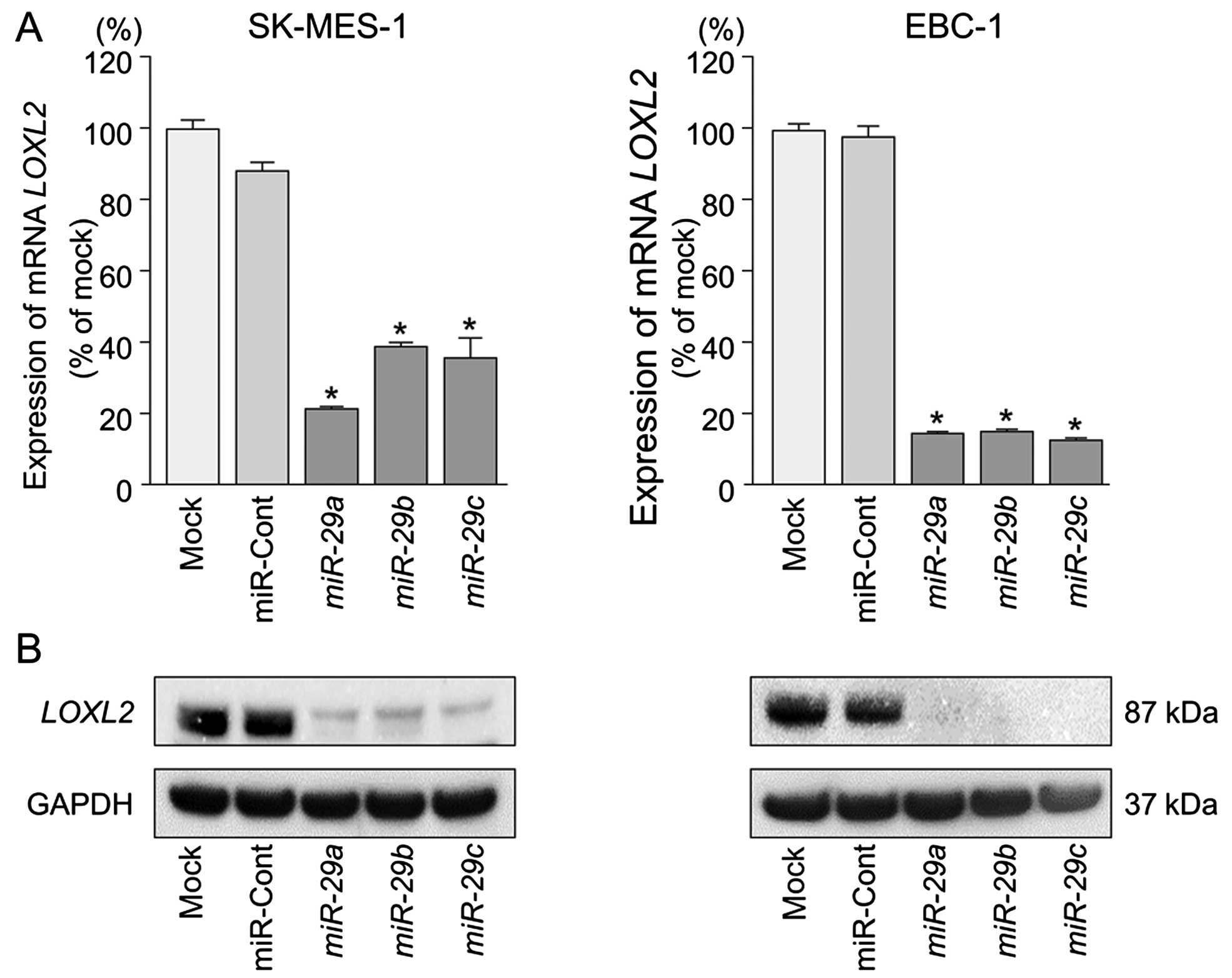
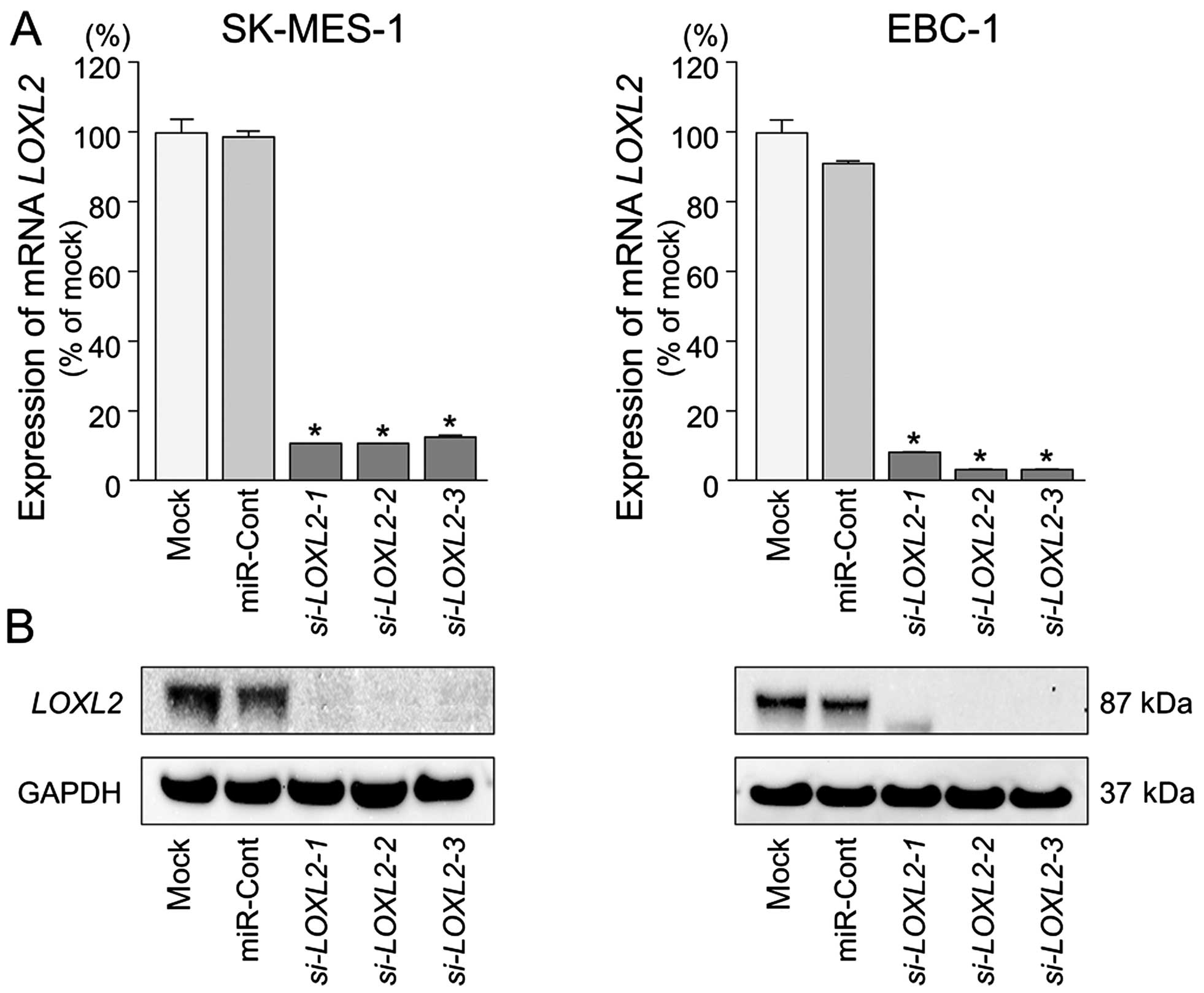
LOXL2 is directly regulated by miR-29s in
lung SCC cells
We performed qRT-PCR and western blotting to confirm
LOXL2 downregulation following restoration of miR-29s
expression in lung SCC cell lines. The mRNA and protein expression
levels of LOXL2 were significantly repressed in miR-29s
transfectants in comparison with mock or miR-control transfectants
(P<0.001) (Fig. 4).
The TargetScan database identified two putative
target sites in the 3′-UTR of LOXL2 (Fig. 5, upper part). A lucif-erase
reporter assay confirmed that the 3′-UTR of LOXL2 was indeed
an actual target of miR-29s. Luciferase activity was
significantly decreased in two miR-29 target sites (positions
555–561 and 757–763 in the 3′-UTR of LOXL2) (Fig. 5, lower part).
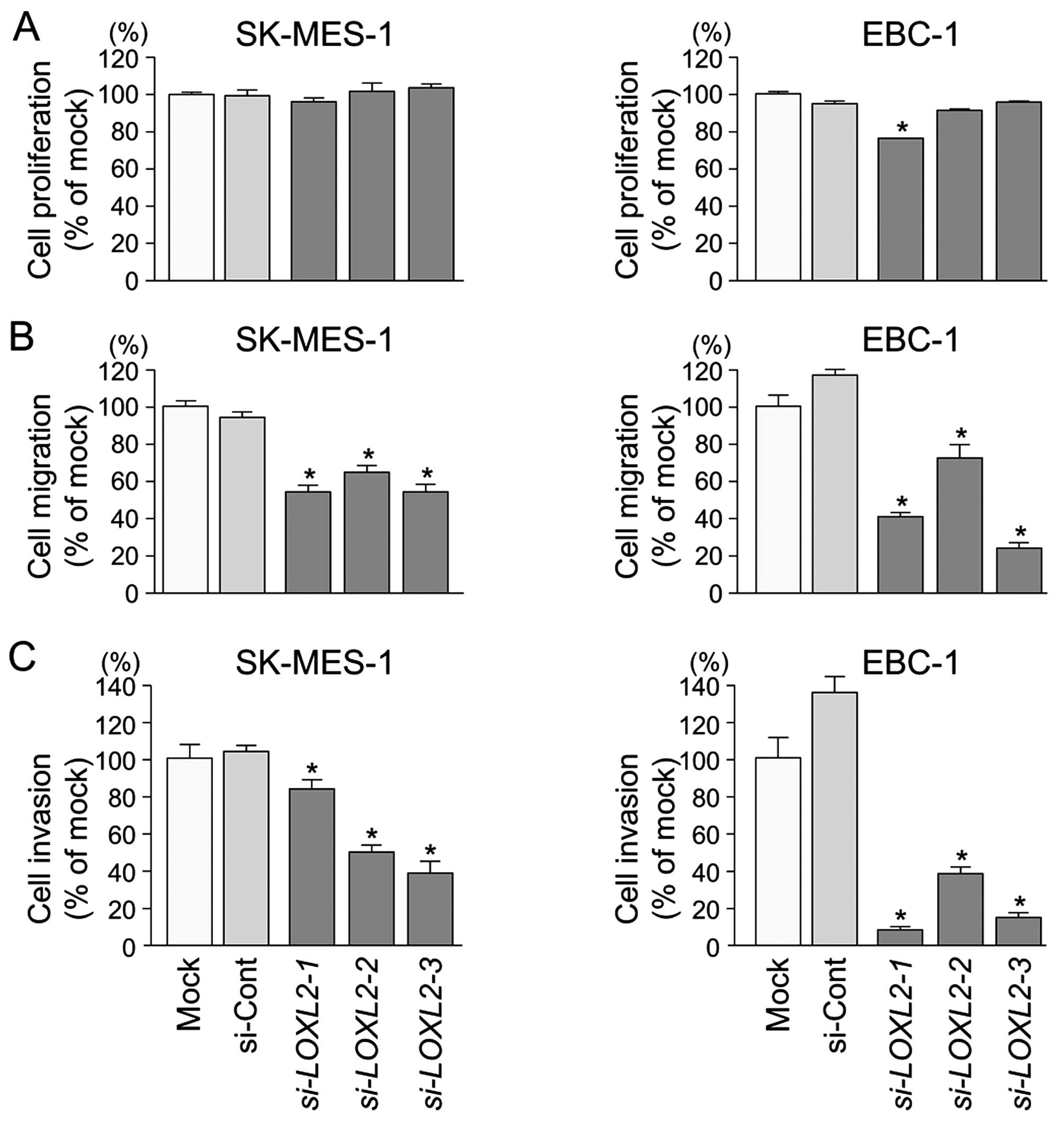
Effects of downregulating LOXL2 on cell
proliferation, migration, and invasion in lung SCC cell lines
To investigate the functional role of LOXL2
in lung SCC cells, we performed loss-of-function studies using
si-LOXL2 transfectants. First, we evaluated the knockdown
efficiency of si-LOXL2 transfection in lung SCC cells.
Western blotting and qRT-PCR indicated that si-LOXL2
effectively downregulated LOXL2 expression in lung SCC cells
(P<0.0001) (Fig. 6).
XTT assays demonstrated that cell proliferation was
not inhibited in si-LOXL2 transfectants in comparison with
mock or si-control transfectants in lung SCC cells (Fig. 7A).
Wound healing assays showed significant inhibition
of cell migration in si-LOXL2 transfectants in comparison
with mock or si-control transfectants in lung SCC cells
(P<0.0001) (Fig. 7B).
Similarly, Matrigel invasion assays revealed that
the number of invading cells was significantly decreased when lung
SCC cells were transfected with si-LOXL2 (P<0.0001)
(Fig. 7C).
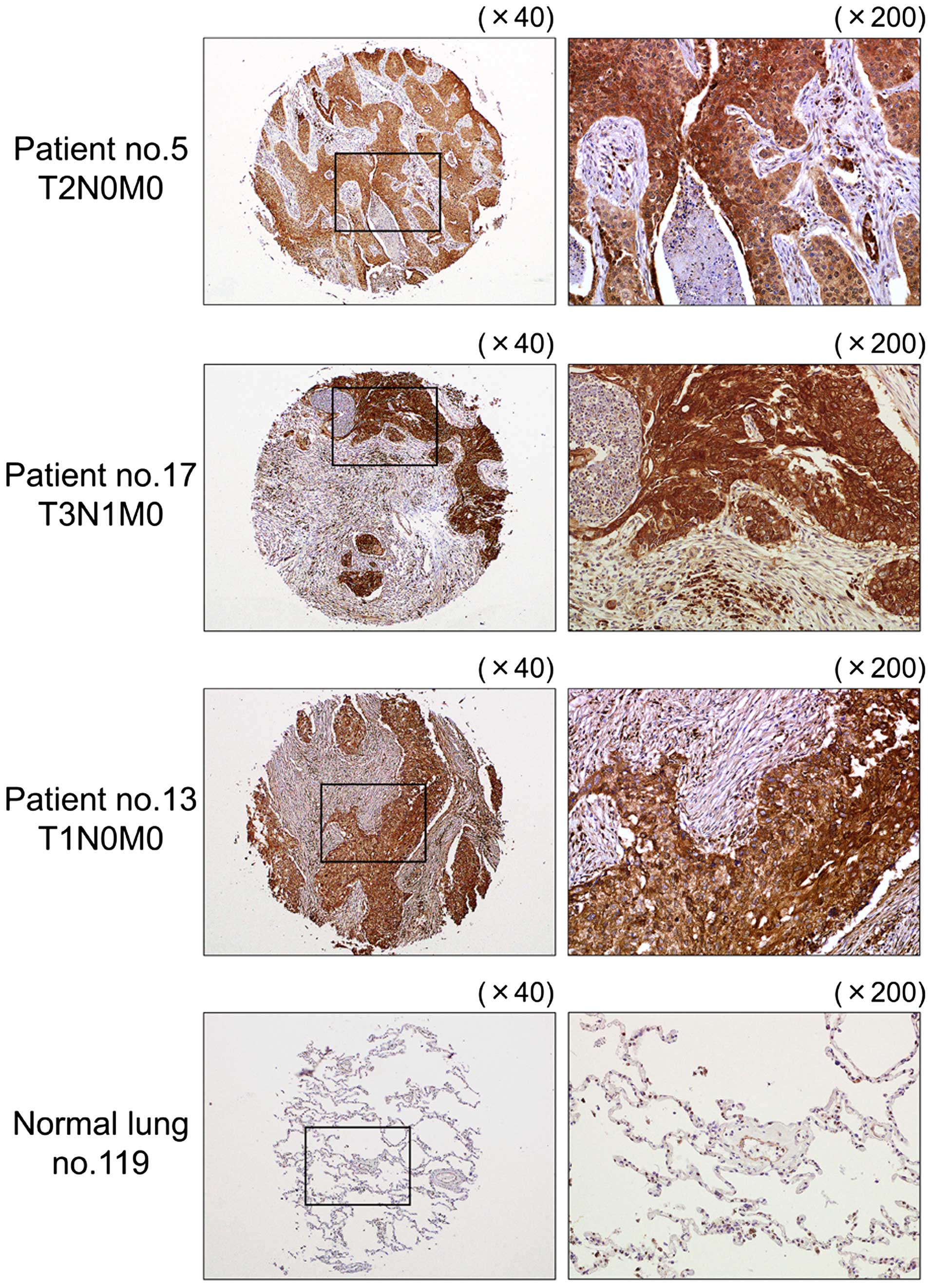
Immunohistochemical staining of LOXL2 in
lung SCC clinical specimens
We confirmed the expression status of LOXL2
in lung SCC clinical specimens using immunohistochemical staining.
Fifty specimens were checked in this study: 32 of 40 lung SCC
specimens stained moderately or strongly, and 9 of 10 normal lung
specimens stained weakly or negatively for LOXL2 (Table III and Fig. 8).
 | Table IIIImmunohistochemistry status and
characteristics of the lung cancer and normal lung cases. |
Table III
Immunohistochemistry status and
characteristics of the lung cancer and normal lung cases.
| A,
Immunohistochemistry status and characteristic of the lung cancer
cases |
|---|
|
|---|
| Patient no. | Grade | T | N | M | Pathological
stage |
Immunohistochemistry |
|---|
| 1 | 1 | 3 | 1 | 0 |
IIIA | (++) |
| 2 | 2 | 2 | 1 | 0 | IIA | (++) |
| 3 | 2 | 2 | 0 | 0 | IB | (+) |
| 4 | 2 | 2 | 0 | 0 | IB | (+++) |
| 5 | 2 | 2 | 0 | 0 | IB | (+++) |
| 6 | 2 | 2 | 1 | 0 | IIB | (++) |
| 7 | 2 | 3 | 2 | 0 |
IIIA | (+++) |
| 8 | 2 | 2 | 0 | 0 | I | (++) |
| 9 | 2 | 2 | 2 | 0 |
IIIA | (+) |
| 10 | 2 | 2 | 2 | 0 |
IIIA | (+) |
| 11 | 2 | 2 | 2 | 0 |
IIIA | (+) |
| 12 | 2 | 3 | 0 | 0 | IIB | (+) |
| 13 | 2 | 1 | 0 | 0 | IA | (+++) |
| 14 | 2 | 2 | 0 | 0 | I | (++) |
| 15 | 2 | 2 | 1 | 0 | IIB | (++) |
| 16 | 2 | 2 | 1 | 0 | IB | (++) |
| 17 | 2 | 3 | 1 | 0 |
IIIA | (+++) |
| 18 | 2 | 2 | 0 | 0 | IB | (++) |
| 19 | 2 | 3 | 1 | 0 |
IIIA | (+++) |
| 20 | 2 | 2 | 0 | 0 | IB | (++) |
| 21 | 3 | 2 | 0 | 0 | IB | (+) |
| 22 | 3 | 2 | 1 | 0 | II | (++) |
| 23 | 3 | 2 | 0 | 0 | IB | (++) |
| 24 | 3 | 2 | 0 | 0 | IB | (++) |
| 25 | 3 | 2 | 0 | 0 | IB | (+++) |
| 27 | 3 | 3 | 2 | 0 |
IIIA | (++) |
| 28 | 3 | 2 | 0 | 0 | IB | (+++) |
| 29 | 2 | 3 | 1 | 0 |
IIIA | (+++) |
| 30 | 3 | 3 | 1 | 0 | IIIA | (++) |
| 31 | 3 | 2 | 0 | 0 | IB | (++) |
| 32 | 3 | 3 | 1 | 0 |
IIIA | (++) |
| 33 | 3 | 2 | 1 | 0 | IIA | (++) |
| 34 | 3 | 1 | 2 | 0 |
IIIA | (++) |
| 35 | 3 | 2 | 2 | 0 |
IIIA | (+) |
| 36 | 3 | 2 | 0 | 0 | I | (+) |
| 37 | | 2 | 0 | 0 | IB | (+++) |
| 38 | 3 | 2 | 0 | 0 | IB | (++) |
| 39 | 3 | 2 | 0 | 0 | IB | (+++) |
| 40 | 3 | 1 | 0 | 0 | IA | (+++) |
| 41 | 3 | 2 | 0 | 0 | IB | (++) |
|
| B,
Immunohistochemistry status of normal lung cases |
|
| Patient no. |
Immunohistochemistry |
|
| 111 | (+) |
| 112 | (+) |
| 113 | (−) |
| 114 | (++) |
| 115 | (−) |
| 116 | (+) |
| 117 | (+) |
| 118 | (−) |
| 119 | (−) |
| 120 | (−) |
Discussion
Recent studies have suggested that the interaction
of cancer cells with their microenvironment has influenced the
initiation, development, and metastasis of tumors (19,20).
Overexpression of extracellular matrix (ECM) components has
frequently been observed in cancer lesions and aberrantly expressed
ECM-mediated signals have triggered cancer cell metastasis
(21,22). Our past studies demonstrated that
miR-29s and miR-218 directly regulated
laminin-integrin signaling and thereby activated cancer cell
migration and invasion (23–25).
Other studies indicated that miR-29s modulated ECM
components such as collagen, laminin and elastin (26). Therefore, the identification of
ECM-regulated tumor-suppressive miRNAs may provide a better
appreciation of novel pathways and how their interrelations are
involved in cancer metastasis.
Our present data showed that all members of the
miR-29 family were significantly reduced in lung SCC
specimens. Our previous studies also showed the downregulation of
miR-29s in renal cell carcinoma, cervical cell carcinoma,
and head and neck squamous cell carcinoma (24,25,27,28)
and are consistent with present data on lung SCC. However, the
molecular mechanisms underlying the dysregulated expression of the
miR-29s in lung SCC cells are still unclear. The genomic
structure of the miR-29 family consists of two clusters in
the human genome: miR-29b-1 and miR-29a in 7q32 and
miR-29b-2 and miR-29c in 1q32 (26). Several studies indicated the
molecular mechanisms of the silencing of miR-29s (26). Previous studies demonstrated that
the promoter regions of miR-29b-1/miR-29a showed that
two putative E-box (MYC-binding) sites, a Gli-binding site and four
NF-κB-binding sites, were contained within this region, and c-Myc
and NF-κB suppressed miR-29 expression at transcriptional levels
(29). A recent study showed that
cancer cells with high c-Myc, low miR-29b, and low
FHIT expression had a shorter overall survival and
relapse-free survival in NSCLC patients (30). In breast cancer, GATA3 is a
transcription factor that specifies and maintains luminal
epithelial cell differentiation in the mammary gland (31). Loss of GATA3 is involved in breast
cancer pathogenesis (31). The
miR-29a/miR-29b-1 promoter region contains three
GATA3-binding sites, and GATA3 induced miR-29b
expression, which inhibits metastasis by targeting metastatically
involved genes (31). Moreover,
recent data have suggested that TGF-β inhibited the expression of
miR-29s and promoted the expression of ECM components
(32,33). However, the silencing of molecular
mechanisms of miR-29s in lung SCC are still unclear;
detailed examination will be necessary to better understand these
processes.
In this study, the restoration of miR-29s
into cancer cells significantly inhibited migration and invasion;
thus miR-29-mediated novel targets deeply contribute to
meta-static pathways. To better understand lung SCC metastasis, we
searched putative miR-29-regulated genes by using gene
expression analysis combined with in silico analysis.
Finally, 15 putative candidate genes were listed in this analysis.
Among these genes, integrin α6 (ITGA6) and serpin peptidase
inhibitor, clade H, member 1 (SERPINH1) have already been
reported by our group as miR-29-regulated genes in head and
neck squamous cell carcinoma and cervical cancer (24,28).
Moreover, another group showed that miR-29c may be involved
in the regulation of cell proliferation through targeting regulator
of chromosome condensation 2 (RCC2) in gastric carcinoma (34). The target gene list provided by
this analysis is effective for miR-29-regulated target
analysis.
Here, we focused on the LOXL2 gene and
validated the direct regulation of miR-29s in lung SCC
cells. Furthermore, overexpression of LOXL2 was detected in lung
SCC clinical specimens, and silencing of LOXL2 expression in
lung SCC cells inhibited cancer cell migration and invasion,
indicating that LOXL2 acts as an oncogene in the disease.
Interestingly, our latest data on renal cell carcinoma also showed
that LOXL2 was a direct regulator of tumor-suppressive
miR-29s (27). The lysyl
oxidase (LOX) protein family comprises LOX and four LOX-like
proteins (LOXL1-LOXL4). These proteins are copper- and
quinone-dependent amino oxidases (35). Basically, the function of the LOX
family is the covalent cross-linking of collagen and/or elastin in
the ECM (36–38). Several studies suggested that
LOXL2-mediated cancer progression cause ECM modification and
increased ECM deposition, and subsequent tissue stiffness derives
malignant progression through activation of ECM-integrin or
ECM-growth factor signaling (39–42).
It has been reported that overexpression of LOXL2 in a number of
cancers and high expression levels of LOXL2 are correlated with
cancer cell invasion, lymph node metastasis, and poor overall
patient survival in patients with gastric cancer, breast cancer and
squamous cell carcinomas (43–45).
It is well known that metastasis is associated with
the aberrant activation of epithelial-mesenchymal transition
(EMT)-related transcriptional factors and TGF-β signaling, which
endows cancer cells with elevated capabilities to invade and
disseminate to distant sites (46). Previous studies have shown that
LOXL2 is a direct transcriptional target of HIF1 (46). Moreover, nuclear LOXL2 interacts
with transcription factor SNAIL1 and represses E-cadherin as well
as inducing EMT (47,48). These findings suggest that hypoxia
conditions and overexpression of LOXL2 trigger the ability for
metastasis acquisition of the cancer cells. Several studies
indicated that targeting LOXL2 with antibodies inhibited primary
and metastatic xenograft models of cancers via suppression of
SRC/FAK signaling or the production of growth factors and cytokines
and TGF-β pathways (49,50). Interestingly, recent data indicated
that expression of the LOX family was induced by TGF-β (51,52).
In contrast, TGF-β inhibited the expression of miR-29s and
promoted the expression of ECM components (53,54).
The present data suggest that the miR-29-LOXL2 axis
regulates the cancer cell microenvironment and activates metastatic
pathways. Therefore, the miR-29-regulated meta-static
pathway is a potential target in the development of novel therapies
to treat pathological lung SCC.
In conclusion, downregulation of miR-29s was
frequently observed in lung SCC clinical specimens, and all members
of the miR-29 family act as tumor-suppressive miRNAs in this
disease. LOXL2 was a direct regulator of miR-29s in
lung SCC cells. Overexpression of LOXL2 was detected in lung SCC
clinical specimens, and functional assays showed that LOXL2
promoted cancer cell invasion and migration, indicating this gene
as an oncogene in lung SCC cells. The identification of novel
molecular pathways mediated by the miR-29-LOXL2 axis
may lead to a better understanding of lung SCC and the development
of new therapeutic strategies to treat this disease.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Iorio MV and Croce CM: MicroRNAs in
cancer: Small molecules with a huge impact. J Clin Oncol.
27:5848–5856. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fukumoto I, Hanazawa T, Kinoshita T,
Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida
H, Nakagawa M, et al: MicroRNA expression signature of oral
squamous cell carcinoma: Functional role of microRNA-26a/b in the
modulation of novel cancer pathways. Br J Cancer. 112:891–900.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fukumoto I, Kinoshita T, Hanazawa T,
Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R,
Nakagawa M, et al: Identification of tumour suppressive
microRNA-451a in hypopharyngeal squamous cell carcinoma based on
microRNA expression signature. Br J Cancer. 111:386–394. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Matsushita R, Seki N, Chiyomaru T,
Inoguchi S, Ishihara T, Goto Y, Nishikawa R, Mataki H, Tatarano S,
Itesako T, et al: Tumour-suppressive microRNA-144-5p directly
targets CCNE1/2 as potential prognostic markers in bladder cancer.
Br J Cancer. 113:282–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goto Y, Kojima S, Nishikawa R, Enokida H,
Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T and Seki
N: The microRNA-23b/27b/24-1 cluster is a disease progression
marker and tumor suppressor in prostate cancer. Oncotarget.
5:7748–7759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar
|
|
13
|
Mataki H, Seki N, Chiyomaru T, Enokida H,
Goto Y, Kumamoto T, Machida K, Mizuno K, Nakagawa M and Inoue H:
Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR
oncogenic signaling in lung squamous cell carcinoma. Int J Oncol.
46:1039–1050. 2015.
|
|
14
|
Goto Y, Kurozumi A, Enokida H, Ichikawa T
and Seki N: Functional significance of aberrantly expressed
microRNAs in prostate cancer. Int J Urol. 22:242–252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yoshino H, Seki N, Itesako T, Chiyomaru T,
Nakagawa M and Enokida H: Aberrant expression of microRNAs in
bladder cancer. Nat Rev Urol. 10:396–404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kikkawa N, Hanazawa T, Fujimura L, Nohata
N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y and Seki
N: miR-489 is a tumour-suppressive miRNA target PTPN11 in
hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer.
103:877–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fuse M, Kojima S, Enokida H, Chiyomaru T,
Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M,
et al: Tumor suppressive microRNAs (miR-222 and miR-31) regulate
molecular pathways based on microRNA expression signature in
prostate cancer. J Hum Genet. 57:691–699. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shepherd FA, Crowley J, Van Houtte P,
Postmus PE, Carney D, Chansky K, Shaikh Z and Goldstraw P:
International Association for the Study of Lung Cancer
International Staging Committee and Participating Institutions: The
International Association for the Study of Lung Cancer lung cancer
staging project: Proposals regarding the clinical staging of small
cell lung cancer in the forthcoming (seventh) edition of the tumor,
node, metastasis classification for lung cancer. J Thorac Oncol.
2:1067–1077. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Weaver VM, Petersen OW, Wang F, Larabell
CA, Briand P, Damsky C and Bissell MJ: Reversion of the malignant
phenotype of human breast cells in three-dimensional culture and in
vivo by integrin blocking antibodies. J Cell Biol. 137:231–245.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bissell MJ and Radisky D: Putting tumours
in context. Nat Rev Cancer. 1:46–54. 2001. View Article : Google Scholar
|
|
21
|
Paszek MJ and Weaver VM: The tension
mounts: Mechanics meets morphogenesis and malignancy. J Mammary
Gland Biol Neoplasia. 9:325–342. 2004. View Article : Google Scholar
|
|
22
|
Paszek MJ, Zahir N, Johnson KR, Lakins JN,
Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M,
Boettiger D, et al: Tensional homeostasis and the malignant
phenotype. Cancer Cell. 8:241–254. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kinoshita T, Hanazawa T, Nohata N, Kikkawa
N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto
Y, et al: Tumor suppressive microRNA-218 inhibits cancer cell
migration and invasion through targeting laminin-332 in head and
neck squamous cell carcinoma. Oncotarget. 3:1386–1400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kinoshita T, Nohata N, Hanazawa T, Kikkawa
N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto
Y, et al: Tumour-suppressive microRNA-29s inhibit cancer cell
migration and invasion by targeting laminin-integrin signalling in
head and neck squamous cell carcinoma. Br J Cancer. 109:2636–2645.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Nishikawa R, Goto Y, Kojima S, Enokida H,
Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y,
et al: Tumor-suppressive microRNA-29s inhibit cancer cell migration
and invasion via targeting LAMC1 in prostate cancer. Int J Oncol.
45:401–410. 2014.PubMed/NCBI
|
|
26
|
Wang Y, Zhang X, Li H, Yu J and Ren X: The
role of miRNA-29 family in cancer. Eur J Cell Biol. 92:123–128.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nishikawa R, Chiyomaru T, Enokida H,
Inoguchi S, Ishihara T, Matsushita R, Goto Y, Fukumoto I, Nakagawa
M and Seki N: Tumour-suppressive microRNA-29s directly regulate
LOXL2 expression and inhibit cancer cell migration and invasion in
renal cell carcinoma. FEBS Lett. 589:2136–2145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yamamoto N, Kinoshita T, Nohata N, Yoshino
H, Itesako T, Fujimura L, Mitsuhashi A, Usui H, Enokida H, Nakagawa
M, et al: Tumor-suppressive microRNA-29a inhibits cancer cell
migration and invasion via targeting HSP47 in cervical squamous
cell carcinoma. Int J Oncol. 43:1855–1863. 2013.PubMed/NCBI
|
|
29
|
Mott JL, Kurita S, Cazanave SC, Bronk SF,
Werneburg NW and Fernandez-Zapico ME: Transcriptional suppression
of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J
Cell Biochem. 110:1155–1164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wu DW, Hsu NY, Wang YC, Lee MC, Cheng YW,
Chen CY and Lee H: c-Myc suppresses microRNA-29b to promote tumor
aggressiveness and poor outcomes in non-small cell lung cancer by
targeting FHIT. Oncogene. 34:2072–2082. 2015. View Article : Google Scholar
|
|
31
|
Chou J, Lin JH, Brenot A, Kim JW, Provot S
and Werb Z: GATA3 suppresses metastasis and modulates the tumour
micro-environment by regulating microRNA-29b expression. Nat Cell
Biol. 15:201–213. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Maurer B, Stanczyk J, Jüngel A,
Akhmetshina A, Trenkmann M, Brock M, Kowal-Bielecka O, Gay RE,
Michel BA, Distler JH, et al: MicroRNA-29, a key regulator of
collagen expression in systemic sclerosis. Arthritis Rheum.
62:1733–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roderburg C, Urban GW, Bettermann K, Vucur
M, Zimmermann H, Schmidt S, Janssen J, Koppe C, Knolle P, Castoldi
M, et al: Micro-RNA profiling reveals a role for miR-29 in human
and murine liver fibrosis. Hepatology. 53:209–218. 2011. View Article : Google Scholar
|
|
34
|
Matsuo M, Nakada C, Tsukamoto Y, Noguchi
T, Uchida T, Hijiya N, Matsuura K and Moriyama M: MiR-29c is
down-regulated in gastric carcinomas and regulates cell
proliferation by targeting RCC2. Mol Cancer. 12:152013. View Article : Google Scholar
|
|
35
|
Barker HE, Cox TR and Erler JT: The
rationale for targeting the LOX family in cancer. Nat Rev Cancer.
12:540–552. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kagan HM and Li W: Lysyl oxidase:
Properties, specificity, and biological roles inside and outside of
the cell. J Cell Biochem. 88:660–672. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vadasz Z, Kessler O, Akiri G,
Gengrinovitch S, Kagan HM, Baruch Y, Izhak OB and Neufeld G:
Abnormal deposition of collagen around hepatocytes in Wilson's
disease is associated with hepatocyte specific expression of lysyl
oxidase and lysyl oxidase like protein-2. J Hepatol. 43:499–507.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim YM, Kim EC and Kim Y: The human lysyl
oxidase-like 2 protein functions as an amine oxidase toward
collagen and elastin. Mol Biol Rep. 38:145–149. 2011. View Article : Google Scholar
|
|
39
|
Erler JT and Weaver VM: Three-dimensional
context regulation of metastasis. Clin Exp Metastasis. 26:35–49.
2009. View Article : Google Scholar :
|
|
40
|
Kauppila S, Stenbäck F, Risteli J, Jukkola
A and Risteli L: Aberrant type I and type III collagen gene
expression in human breast cancer in vivo. J Pathol. 186:262–268.
1998. View Article : Google Scholar
|
|
41
|
Mackie EJ, Chiquet-Ehrismann R, Pearson
CA, Inaguma Y, Taya K, Kawarada Y and Sakakura T: Tenascin is a
stromal marker for epithelial malignancy in the mammary gland. Proc
Natl Acad Sci USA. 84:4621–4625. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhu GG, Risteli L, Mäkinen M, Risteli J,
Kauppila A and Stenbäck F: Immunohistochemical study of type I
collagen and type I pN-collagen in benign and malignant ovarian
neoplasms. Cancer. 75:1010–1017. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kasashima H, Yashiro M, Kinoshita H,
Fukuoka T, Morisaki T, Masuda G, Sakurai K, Kubo N, Ohira M and
Hirakawa K: Lysyl oxidase-like 2 (LOXL2) from stromal fibroblasts
stimulates the progression of gastric cancer. Cancer Lett.
354:438–446. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ahn SG, Dong SM, Oshima A, Kim WH, Lee HM,
Lee SA, Kwon SH, Lee JH, Lee JM, Jeong J, et al: LOXL2 expression
is associated with invasiveness and negatively influences survival
in breast cancer patients. Breast Cancer Res Treat. 141:89–99.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Peinado H, Moreno-Bueno G, Hardisson D,
Pérez-Gómez E, Santos V, Mendiola M, de Diego JI, Nistal M,
Quintanilla M, Portillo F, et al: Lysyl oxidase-like 2 as a new
poor prognosis marker of squamous cell carcinomas. Cancer Res.
68:4541–4550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Schietke R, Warnecke C, Wacker I, Schödel
J, Mole DR, Campean V, Amann K, Goppelt-Struebe M, Behrens J,
Eckardt KU, et al: The lysyl oxidases LOX and LOXL2 are necessary
and sufficient to repress E-cadherin in hypoxia: Insights into
cellular transformation processes mediated by HIF-1. J Biol Chem.
285:6658–6669. 2010. View Article : Google Scholar :
|
|
48
|
Moon HJ, Finney J, Xu L, Moore D, Welch DR
and Mure M: MCF-7 cells expressing nuclear associated lysyl
oxidase-like 2 (LOXL2) exhibit an epithelial-to-mesenchymal
transition (EMT) phenotype and are highly invasive in vitro. J Biol
Chem. 288:30000–30008. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Peng L, Ran YL, Hu H, Yu L, Liu Q, Zhou Z,
Sun YM, Sun LC, Pan J, Sun LX, et al: Secreted LOXL2 is a novel
therapeutic target that promotes gastric cancer metastasis via the
Src/FAK pathway. Carcinogenesis. 30:1660–1669. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Barry-Hamilton V, Spangler R, Marshall D,
McCauley S, Rodriguez HM, Oyasu M, Mikels A, Vaysberg M, Ghermazien
H, Wai C, et al: Allosteric inhibition of lysyl oxidase-like-2
impedes the development of a pathologic microenvironment. Nat Med.
16:1009–1017. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Voloshenyuk TG, Landesman ES, Khoutorova
E, Hart AD and Gardner JD: Induction of cardiac fibroblast lysyl
oxidase by TGF-β1 requires PI3K/Akt, Smad3, and MAPK signaling.
Cytokine. 55:90–97. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Roy R, Polgar P, Wang Y, Goldstein RH,
Taylor L and Kagan HM: Regulation of lysyl oxidase and
cyclooxygenase expression in human lung fibroblasts: Interactions
among TGF-beta, IL-1 beta, and prostaglandin E. J Cell Biochem.
62:411–417. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tan J, Tong BD, Wu YJ and Xiong W:
MicroRNA-29 mediates TGFβ1-induced extracellular matrix synthesis
by targeting wnt/β-catenin pathway in human orbital fibroblasts.
Int J Clin Exp Pathol. 7:7571–7577. 2014.
|
|
54
|
Yang T, Liang Y, Lin Q, Liu J, Luo F, Li
X, Zhou H, Zhuang S and Zhang H: miR-29 mediates TGFβ1-induced
extracellular matrix synthesis through activation of PI3K-AKT
pathway in human lung fibroblasts. J Cell Biochem. 114:1336–1342.
2013. View Article : Google Scholar
|