Introduction
Peptide nucleic acids (PNAs) are DNA analogues in
which the sugar-phosphate backbone has been replaced by
N-(2-aminoethyl)glycine units (1–5).
These very interesting molecules were described for the first time
by Nielsen et al (1) and,
despite a radical structural change with respect to DNA and RNA,
they are capable of sequence-specific and efficient hybridization
with complementary DNA and RNA, forming Watson-Crick double helices
(1). In addition, they are able to
generate triple helix formation with double stranded DNA and
perform strand invasion (2–4).
Accordingly, they have been used as very efficient tools for
pharmacologic alteration of gene expression, both in vitro
and in vivo (1–5). PNAs and PNA-based analogues were
proposed as antisense molecules targeting mRNAs, triple-helix
forming molecules targeting eukaryotic gene promoters, artificial
promoters, decoy molecules targeting transcription factors
(4–10). Recently, PNAs have been shown to be
able to alter biological functions of microRNAs, both in
vitro and in vivo (11–18).
Cheng et al, for instance, demonstrated that attachment of a
peptide-(anti-miR) PNA construct is able to target the tumor
microenvironment and to transport the anti-miR PNA across plasma
membranes under acidic conditions such as those found in solid
tumors. This treatment led to an efficient inhibition of the target
oncomiR in a tumor mouse model (18).
MicroRNAs (miRNAs or miRs) are short non-coding RNA
molecules, which act as gene regulators by repressing translation
or by inducing the cleavage of target RNA transcripts (19). Emerging evidence suggests that the
altered expression of miRNA may be involved in the pathogenesis of
cancer (20–25). Therefore, in the case of oncomiRNAs
and metastamiRNAs, approaches based on the targeting of microRNAs
can be designed to inhibit tumor cell growth and metastasis, and to
counteract the resistance of tumor cells to anticancer drugs. Chan
et al demonstrated that sequence-specific functional
inhibition of oncomiR-138 in malignant gliomas prevents tumor
sphere formation in vitro and impedes tumorigenesis in
vivo (26). In addition,
Wagenaar et al were able to deregulate the transcriptional
network in hepatocellular carcinomas by targeting miR-21 with
sequence-specific antagomiRs, thus inducing a significant
de-repression of direct targets of miR-21 which led to loss of
viability in the majority of hepatocarcinoma cell lines tested
(27). In another example Ma et
al reported that therapeutic silencing of the oncogenic miR-10b
inhibits metastasis in a mouse model of mammary tumor (28). In this context, the use of PNAs
directed against oncomiRNAs might be of great interest (20).
Among the possible microRNA targets involved in
cancer, the cluster miR-221/222 plays a very important role
(29–34). Lee et al found that high
levels of miR-221/222 cause increased cell invasion and poor
prognosis in glioblastomas (33).
Accordingly, co-suppression of miR-221/222 is associated with
inhibition of cell growth, increased expression of the miR-221/222
targets, in particular of the p53-upregulated modulator of
apoptosis (PUMA), and activation of apoptosis (35). Enforced expression of miR-221/222
induces cell survival whereas knockdown of miR-221/222 stimulates
cellular apoptosis. Interestingly, the miR-221/222 cluster is also
involved in sensitization of glioma cells to anticancer drugs
(34). Accordingly, downregulation
of miR-221/222 sensitizes glioma cells to temozolomide, regulating
apoptosis (34).
We have recently designed and studied a peptide
nucleic acid targeting miR-221 (R8-PNA-a221) (36), bearing an oligoarginine peptide
(R8) to facilitate uptake by glioma cells (11,12).
The effects of the R8-PNA-a221 were analyzed in U251, U373 and T98G
glioma cells and found to strongly inhibit miR-221. In addition,
the effects of R8-PNA-a221 on p27Kip1 (a target of
miR-221) were analyzed in U251 and T98G cells by RT-qPCR and by
western blotting. We found an increase of p27Kip1 mRNA
and of p27Kip1 protein in cells treated with R8-PNA-a221
(36).
The present study was aimed at determining the
biological activity of a combined treatment of glioma cell lines
with two PNAs, directed against miR-221 and miR-222 and conjugated
to octaarginine for cellular delivery. Apoptosis was analyzed to
determine whether co-administration of the two PNAs is important to
obtain the highest effects. The effects of the combined treatment
of glioma cells on the reversion of drug-resistance phenotype were
assessed in the temozolomide-resistant T98G glioma cell line.
Materials and methods
Synthesis and characterization of
PNAs
The synthesis and characterization of anti-miR-221
PNAs have been previously reported (36), the new anti-miR-222 PNAs synthesis
was performed using standard Fmoc-based automate peptide
synthesizer (Syro II, MultiSynTech GmbH, Witten, Germany), using a
ChemMatrix-RinkAmide resin loaded with Fmoc-Gly-OH (0.2 mmol/g) as
first monomer and using commercially available monomers (Link
Technologies, Bellshill, UK) with HBTU/DIPEA coupling. After
purification the PNAs were characterized by UPLC-MS on a Water
Acquity System equipped with Acquity UPLC BEH C18 (1.7 μm, 2.1×50
mm). Gradient: 100% A for 0.9 min, then from 0 to 50% B in 5.7 min
at 0.25 ml/min flow (A, water + 0.2% formic acid; B, acetonitrile +
0.2% formic acid).
R8-PNA-a222: Rt = 2.70 min;
calculated mw, 6,226.3 g/mol; m/z found: 1,246.5
[M+5H]5+, 1,039.0 [M+6H]6+, 890.4
[M+7H]7+, 779.5 [M+8H]8+, 693.0
[M+9H]9+, 623.8 [M+10H]10+, 567.2
[M+11H]11+.
R8-PNA-a222-MUT: Rt = 2.70 min;
calculated mw, 6,226.34 g/mol; m/z found: 1,246.2
[M+5H]5+, 1,038.7 [M+6H]6+, 890.3
[M+7H]7+, 779.1 [M+8H]8+, 692.8
[M+9H]9+.
Bioinformatic tools for molecular
interaction studies
Predicted interactions between microRNAs and target
mRNA sequences were determined using the following tools: a) the
ViennaRNA Web Services (Institute of Theoretical Chemistry, Vienna
University), webRNAfold and WebServer (http://rna.tbi.univie.ac.at/), which predict minimum
free energy secondary structures and base pair probabilities from
single stranded RNA sequences; b) the UCSC (University of
California Santa Cruz) Genome Browser (https://genome.ucsc.edu/) Gene Sorter, which shows
expression, homology and other information on genes, and it was
used for the 3′-UTR base sequence; c) the microRNA database miRBase
(University of Manchester, http://www.mirbase.org/), for published miRNA
sequences and annotation.
Biospecific interaction analysis (BIA)
with the Biacore X100
All procedures were performed at 25°C and at a
5-μl/min flow rate, by using HBS-EP (0.01 M HEPES pH 7.4, 0.15 M
NaCl, 3 mM EDTA, 0.005% v/v Surfactant P20) (GE Healthcare) as
running buffer. CM5 sensor chips (GE Healthcare) containing PNAs
were obtained by the amine coupling chemistry, exploiting the
terminal amine group of the PNA. To this aim, the Amine Coupling
kit (GE Healthcare) was used: briefly, carboxyl groups on the
sensor chip surface were first activated with a 1:1 mixture of 0.4
M 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide (EDC) and 0.1 M
N-hydroxysuccinimide (NHS) to give reactive succinimide esters; the
R8-PNA-a221, R8-PNA-a222, R8-PNA-a221-MUT and R8-PNA-a222-MUT were
then passed over the surface at the concentration of 50 μg/ml for
10 min, in order to form a covalent bond; finally, 1 M
ethanolamine-HCl at pH 8.5 was injected to quench the excess of
reactive groups. Hybridization to pre-miR-221 and pre-miR-222 were
performed in 10% HBS buffer, followed by washing with HBS and a
1-min pulse of 50 mM NaOH for regeneration of flow cells. Biacore
X100 Control Software and Biacore X100 Evaluation Software, version
2.0.1 (GE Healthcare) were used for operation and data analysis,
respectively (37,38).
Glioma cell lines and culture
conditions
U251, U373 and T98G cells (39–41)
were cultured in humidified atmosphere of 5% CO2/air in
RPMI-1640 medium (Life Technologies, Monza, Italy) supplemented
with 10% fetal bovine serum (FBS, Celbio, Milan, Italy), 100 U/ml
penicillin and 100 mg/ml streptomycin. To verify the effect on
proliferation, cell growth was monitored by determining the cell
number/ml using a Z2 Coulter Counter (Coulter Electronics, Hialeah,
FL, USA).
RNA extraction
Cultured cells were tripsinized and collected by
centrifugation at 1,500 rpm for 10 min at 4°C, washed with PBS,
lysed with Tri-reagent (Sigma-Aldrich, St. Louis, MO, USA),
according to the manufacturer's instructions. The isolated RNA was
washed once with cold 75% ethanol, dried and dissolved in nuclease
free pure water before use.
Quantitative analyses of miRNAs
For miRNA quantification using real-time RT-qPCR
reagents, the primers and probes (hsa-miR-221, TM:000524,
hsa-miR-222, TM:000525) were obtained from Applied Biosystems.
Reverse transcriptase (RT) reactions were performed using the
TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems,
Foster City, CA, USA); real-time PCR was performed according to the
manufacturer's protocols (36).
Twenty nanograms per sample were used for the assays. All RT
reactions, including no-template controls and RT-minus controls,
were performed in duplicate using the CFX96 Touch Real-Time PCR
Detection System (Bio-Rad, Hercules, CA, USA). The relative
expression was calculated using the comparative cycle threshold
method and U6 snRNA (TM:001973) and hsa-let-7c (TM:000379) were
used as references to normalize all RNA samples, since they remain
constant in the assayed samples by miR-profiling and quantitative
RT-PCR analysis, as previously reported (36).
Analysis of apoptosis
Annexin V and Dead Cell assay on T98G cells,
untreated and treated with temozolomide (Sigma-Aldrich) (36) and different concentrations of PNAs,
were performed with Muse (Millipore Corp., Billerica, MA, USA)
method, according to the instructions supplied by the manufacturer.
This procedure utilizes Annexin V to detect PS (phosphatidylserine)
on the external membrane of apoptotic cells. A dead cell marker is
also used as an indicator of cell membrane structural integrity. It
is excluded from live, healthy cells, as well as early apoptotic
cells. Four populations of cells can be distinguished in this
assay. Cells were washed with sterile 1X PBS, trypsinized,
suspended and diluted (1:2) with the one step addition of the Muse
Annexin V & Dead Cell reagent. After incubation of 20 min at
room temperature in the dark, samples were analyzed. Data from
prepared samples are acquired and recorded utilizing the Annexin V
and Dead Cell Software Module (Millipore) (36).
Statistical analyses
Results are expressed as mean ± standard deviation
(SD). Comparisons between groups were made by using paired
Student's t test. Statistical significance was defined with
p<0.05 (significant) and p<0.01 (highly significant).
Results
Design of the PNA molecules targeting
miR-221 and miR-222
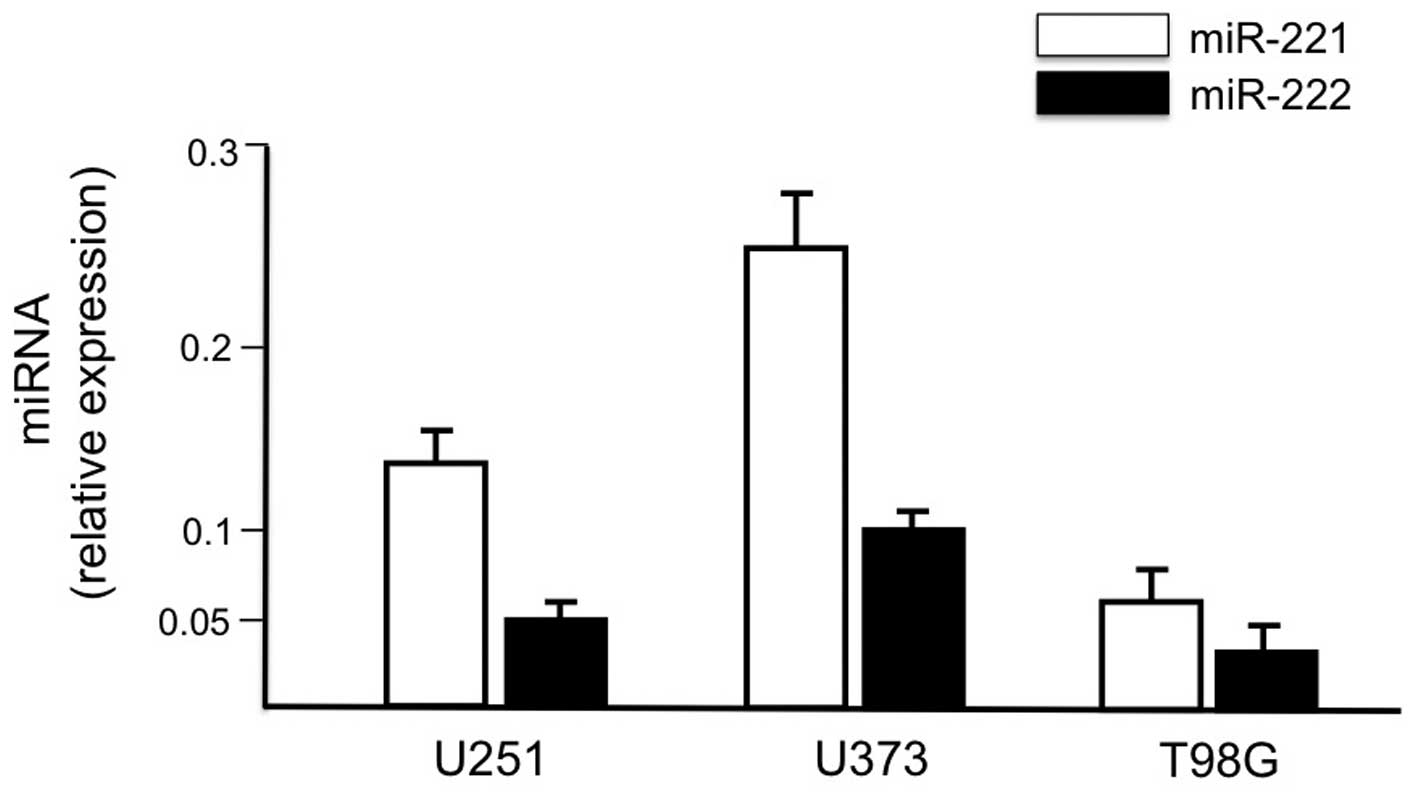
We first demonstrated that the human glioma cell
lines U251, U373 and T98G express at high levels both miR-221 and
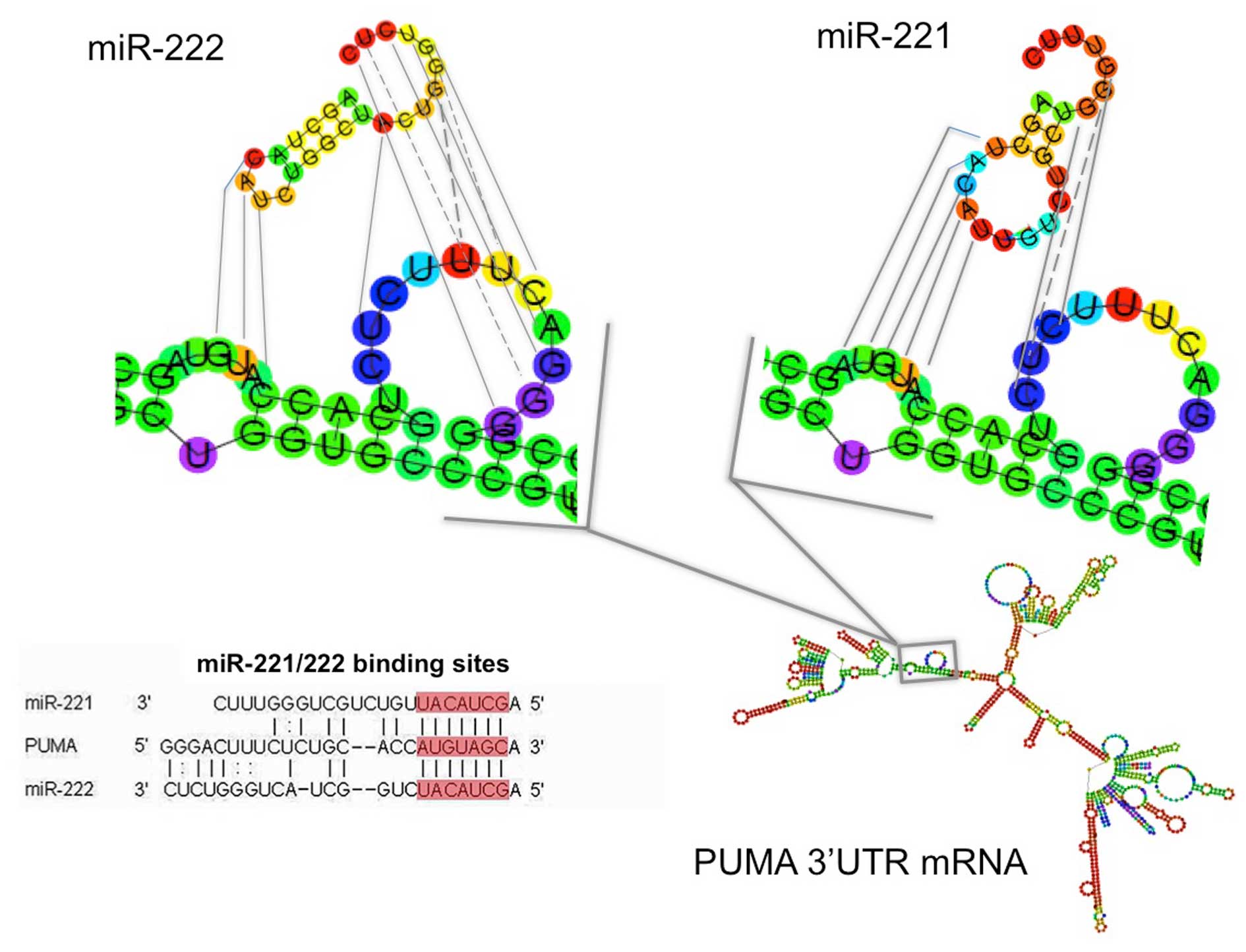
miR-222 (Fig. 1). The structure of
pre-miR-221 and pre-miR-222, together with their possible site of
interaction within the 3′-UTR sequences of PUMA, one of the major
mRNA targets of miR-221 and miR-222, is depicted in Fig. 2. The sequences of the PNAs utilized
in the present study are reported in Table I. In particular the design and
synthesis of the a221 PNAs were already reported (36), while the a222 PNAs were designed to
be complementary to the seed region of the mature miR-222 (full
complementarity to nt 1–18). Full-matched PNAs are expected to bind
to both pre-miR-221 and pre-miR-222 as well as to mature miR-221
and miR-222. An octaarginine peptide (R8) has been conjugated to
the PNAs, since it has been reported that this peptide displays
optimal uptake, while shorter oligoarginine sequences (R2, R4, R6)
give poorer results (11,12,36,42,43).
In order to verify the selectivity of the observed effects, PNAs
containing four mismatches were also synthesized, and their design
was carried out in order to avoid possible complex formation of the
off target sequences.
 | Table ISequences of PNA used in the present
study. |
Table I
Sequences of PNA used in the present
study.
| PNA | Sequence |
|---|
| R8-PNA-a221 | H-R8-AAA CCC AGC
AGA CAA TGT-Gly-NH2 |
|
R8-PNA-a221-MUT | H-R8-AAT CCC
ACC AGA GAA AGT-Gly-NH2 |
| R8-PNA-a222 | H-R8-CAG TAG CCA
GAT GTA GCT-Gly-NH2 |
|
R8-PNA-a222-MUT | H-R8-CAC TAG
CGA GAA GTT GCT-Gly-NH2 |
Validation of the PNA molecules targeting
miR-221 and miR-222: a Biacore study
The ability of the R8-PNA-a221 and R8-PNA-a222 to
specifically interact with pre-miR-221 and pre-miR-222 has been
validated by SPR-based analysis using the Biacore X100 biosensor.
Fig. 3A shows representative data
obtained by injecting 50 ng of pre-miR-221 on sensor chips to which
R8-PNA-a221, R8-PNA-a222, R8-PNA-a221-MUT and the R8-PNA-a222-MUT
have been immobilized. The hybridization between pre-miR-221 and
R8-PNA-a221 occurs within seconds (and the dissociation is very
slow, leading to persistence of high RUres values). On the
contrary, pre-miR-221 binds weakly to immobilized R8-PNA-a222
sensor chip. Importantly, the binding of pre-miR-221 to sensor
chips on which R8-PNA-a221-MUT and R8-PNA-a222-MUT were immobilized
is very low. This experiment suggests that the binding of
pre-miR-221 to the R8-PNA-a221 is selective. Similar conclusions
can be drawn from the experiment shown in Fig. 3B, which demonstrates that
pre-miR-222 binds to R8-PNA-a222 immobilized on the sensor chip
much more efficiently than to R8-PNA-a221. However, a certain
degree of cross-binging was observed for pre-miR-221 on R8-PNA-222
chip (higher than that of R8-PNA-a221-MUT), and for pre-miR-222 on
R8-PNA-221 sensor chips. As expected, no binding of pre-miR-210 was
found to either R8-PNA-a221 or R8-PNA-a222 (Fig. 3C). Table II summarizes the results of RUfin
and RUres obtained when pre-miR-221 and pre-miR-222 were injected
in sensor chips to which normal and mutated anti-miR-221 and
anti-miR-222 PNAs were immobilized.
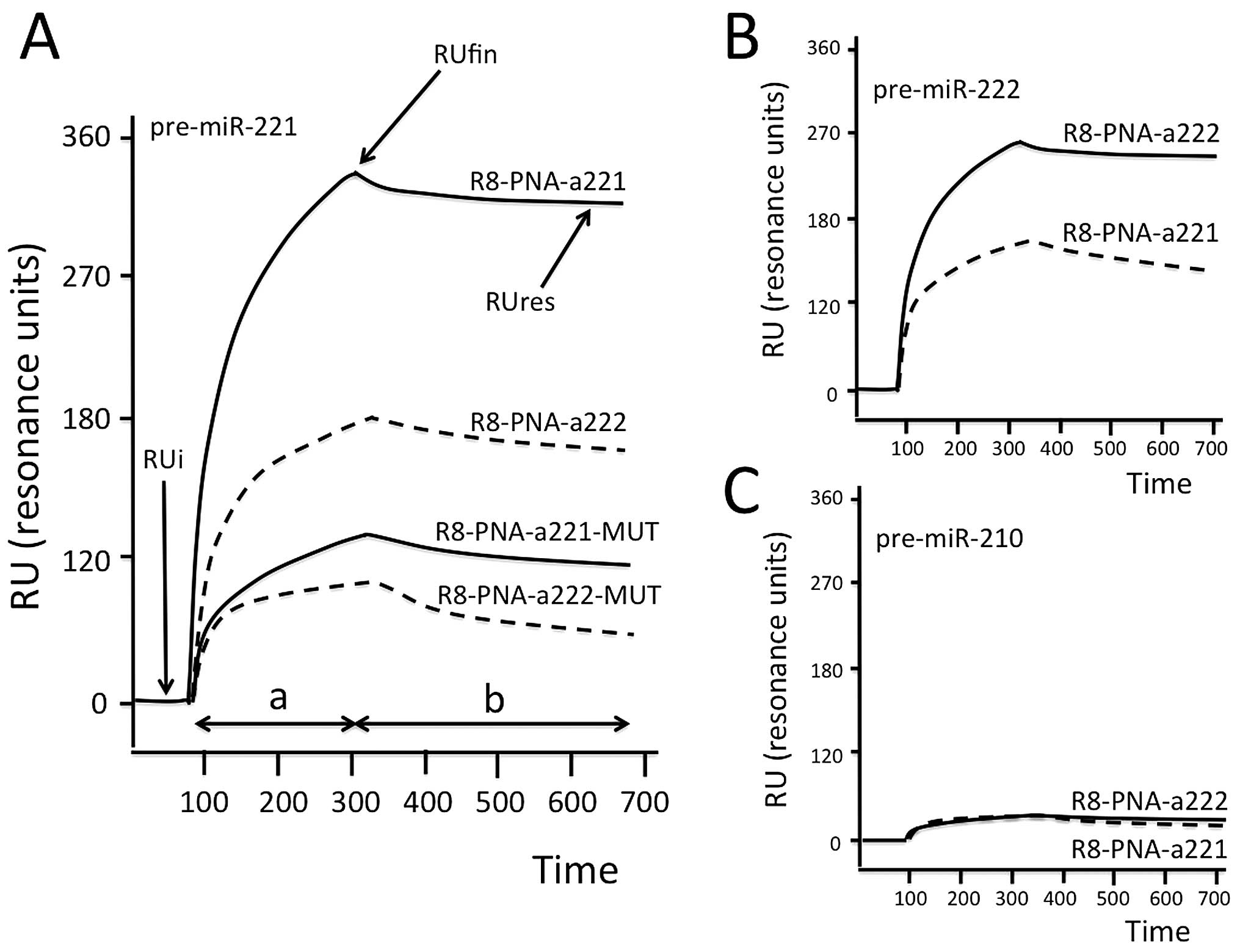 | Figure 3Biospecific interaction analysis. (A)
Hybridization between injected pre-miR-221 and sensor-chips to
which R8-PNA-a221, R8-PNA-a222, R8-PNA-a221-MUT and R8-PNA-a222-MUT
were, as indicated, immobilized. The SPR-based Biacore X100 was
used. (B and C) BIA showing the hybridization between injected
pre-miR-222 (B) and pre-miR-210 (C) and sensor-chips to which
R8-PNA-a221 or R8-PNA-a222 were, as indicated, immobilized. RUi,
initial RU, i.e., resonance unit (RU) values before the injection;
RUfin, final RU after injection; RUres, residual RU after
sensor-chip washing with HBS buffer; a, analyte injection step; b,
washing step. Time, seconds. |
 | Table IIBiacore analysis of the hybridization
between injected pre-miRNAs and PNAs immobilized on the
sensor-chips. |
Table II
Biacore analysis of the hybridization
between injected pre-miRNAs and PNAs immobilized on the
sensor-chips.
| Injected
pre-miRNAs |
|---|
|
|
|---|
| pre-miR-221 | pre-miR-222 |
|---|
|
|
|
|---|
| Immobilized
PNAs | RUfin | RUres | RUfin | RUres |
|---|
| R8-PNA-a221 | 301.2±35.1 | 300.1±21.2 | 163.3±18.4 | 144.2±12.1 |
|
R8-PNA-a221-MUT | 87.4±7.1 | 57.2±6.5 | 129.8±9.7 | 100.1±9.1 |
| R8-PNA-a222 | 160.3±12.5 | 141.2±16.1 | 225.1±19.2 | 220.4±21.2 |
|
R8-PNA-a222-MUT | 54.2±4.1 | 42.3±3.1 | 140.7±11.5 | 126.1±11.2 |
R8-PNA-a221 and R8-PNA-a222: inhibitory
effects on miR-221 and miR-222 in glioma cell lines
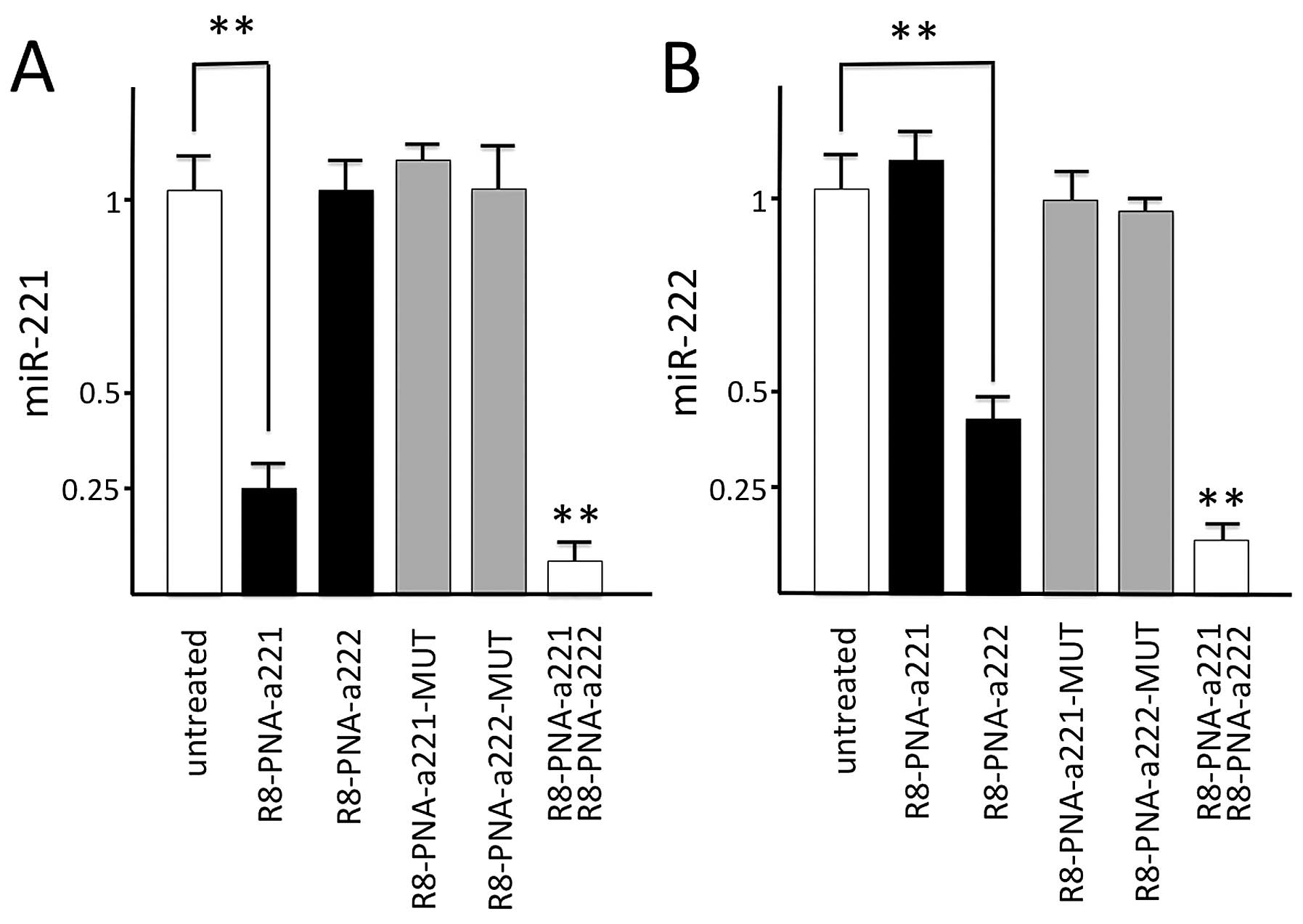
When the glioma cell line U251 was cultured in the
presence of the four PNAs, very different effects were obtained, as
shown in Fig. 4A. The results
demonstrate that the miR-221 signal was strongly reduced only when
RNA was isolated from U251 glioma cells cultured for 48 h in the
presence of R8-PNA-a221, while no major effects were observed with
R8-PNA-a222. Furthermore, no inhibitory effects were observed using
R8-PNA-a221-MUT and R8-PNA-a222-MUT. In agreement Fig. 4B shows that miR-222 specific
hybridization signal is reduced in samples isolated from U251 cells
treated in the presence of R8-PNA-a222, either used alone or in
combination with R8-PNA-a221, while no variation is observed after
treatment with R8-PNA-a221, R8-PNA-a221-MUT and R8-PNA-a222-MUT.
Altogether these experiments support the concept that the effects
of R8-PNA-a221 on miR-221 and of R8-PNA-a222 on miR-222 are
sequence-specific. The highest effect was obtained after
co-treatment with R8-PNA-a221 and R8-PNA-a222.
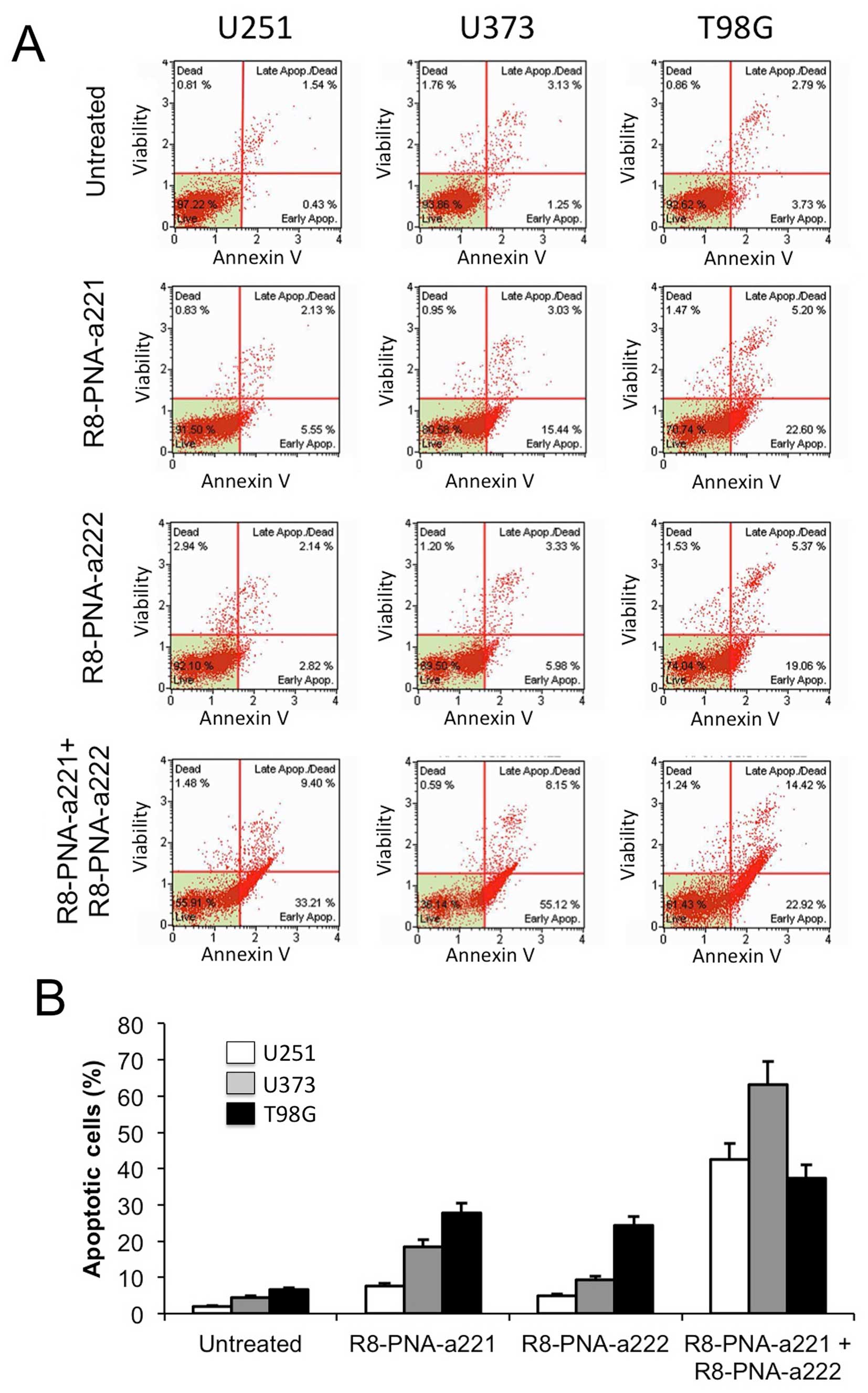
Co-treatment of U251, U373 and T98G
glioma cell lines with R8-PNA-a221 and R8-PNA-a222: effects on
apoptosis
When the glioma cell lines U251, U373 and T98G were
cultured in the presence of singularly administered R8-PNA-a221 or
R8-PNA-a222 a significant and dose-dependent increase of early and
late apoptotic cells was observed. Fig. 5A shows representative results
obtained after treatment of U251, U373 and T98G glioma cell lines
with 4 μM R8-PNA-a221, 4 μM R8-PNA-a222, or combined treatment with
2 μM R8-PNA-a221 plus 2 μM R8-PNA-a222, confirming the
pro-apoptotic effects of R8-PNA-a221 [as published by our group
(36)] and demonstrating the
pro-apoptotic effects of R8-PNA-a222 and the remarkable increased
effect obtained with the co-administration of the two PNAs. This
effect is superior, especially in the percentage of late apoptotic
cells, to that obtained using singularly administered 4 μM
R8-PNA-a221 and R8-PNA-a222. The full set of data obtained in three
independent experiments are comparatively presented in Fig. 5B.
Co-treatment of T98G glioma cells with
R8-PNA-a221 and R8-PNA-a222 in the presence of temozolomide (TMZ):
effects on apoptosis
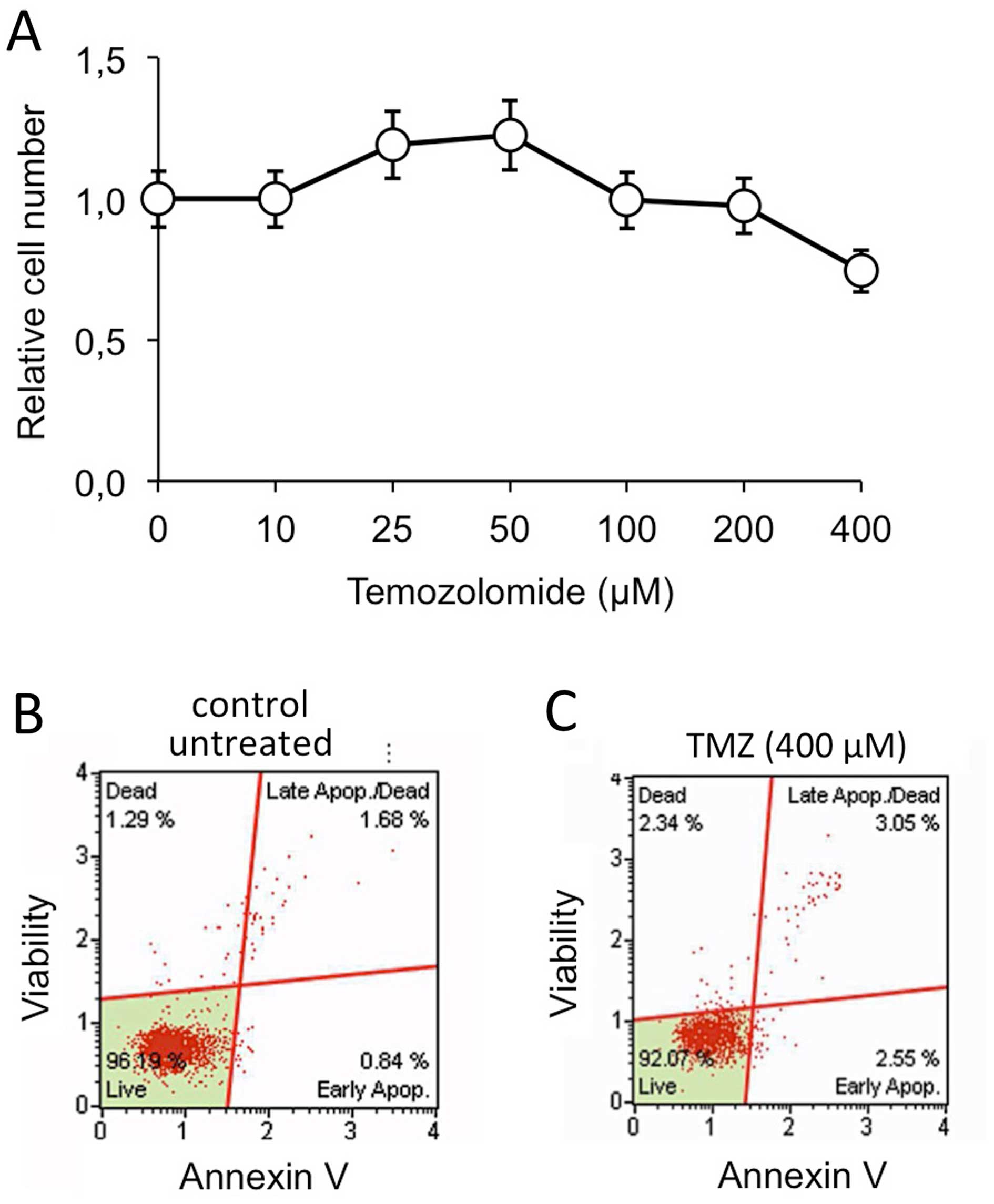
As shown in Fig. 6,
the T98G cell line is resistant to temozolomide (TMZ) (Fig. 6A) and no increase of apoptosis was
observed when these cells were treated with TMZ (Fig. 6B and C). We have previously
reported that when TMZ-treatment was conducted in the presence of
R8-PNA-a221 a pro-apototic effect was observed much higher than the
effect of singularly administered R8-PNA-a221, supporting a
possible synergistic effect of TMZ and R8-PNA-a221 on apoptosis of
T98G cells (36). Therefore, in
our study T98G cells were treated with 2 and 4 μM R8-PNA-a221 or
R8-PNA-a222 in the presence of 400 μM TMZ. The data were compared
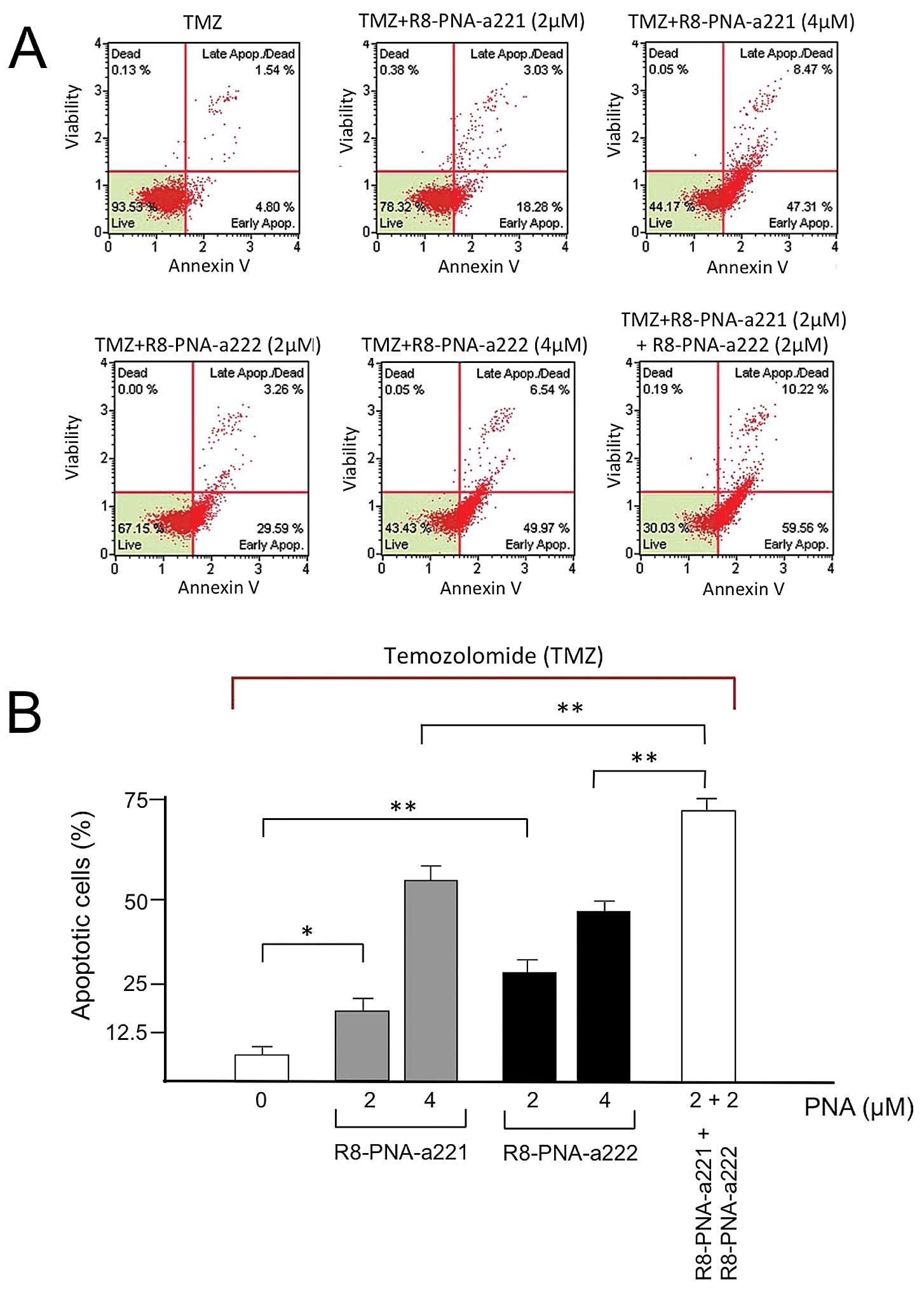
with the results obtained after 2 μM R8-PNA-a221 plus 2 μM
R8-PNA-a222 treatment in the presence of TMZ. The remarkable
results obtained show that co-administration of R8-PNA-a221 and
R8-PNA-a222 induced apoptosis of TMZ-treated T98G cells at a level
higher than that obtained following singular administration of
R8-PNA-a221 or R8-PNA-a222. Representative data are shown in
Fig. 7A, the full set of data
obtained in three independent experiments are comparatively
presented in Fig. 7B.
Discussion
Gliomas express miR-221 at high levels, promoting
malignant progression through activation of the Akt pathway and
inhibition of p27Kip1 (44–46);
in addition, miR-221 mediated downregulation of other genes such as
PUMA (35), ICAM-1 (47), TIMP-3 (48) and PTEN (49) might be associated with glioma onset
and progression. Therefore, miR-221 appears to be a specific target
for treatments against gliomas (29–35).
We have previously reported that a PNA targeting the miR-221 can be
internalized by glioma cells when it is linked to an octaarginine
tail (R8), leading to inhibition of miR-221 (36). No inhibitory effects were observed
on miR-222; moreover, the mutant R8-PNA-a221-MUT was inactive in
inhibiting miR-221, using RT-qPCR as the validation assay. This
effect of anti-miR-221 PNA was associated with the activation of
the apoptotic pathway.
However, the same site recognized by miR-221 in the
3′-UTR of target mRNAs can be also identified by miR-222 (see the
example of molecular interactions we have reported in Fig. 2 using PUMA 3′-UTR mRNA as model
system). Results very similar to those reported in Fig. 2 can be obtained using 3′-UTR
sequences of other miR-221 target mRNAs, such as
p27Kip1, PTEN and TIMP-3 (data not shown). Therefore,
targeting of miR-221 with anti-miRs, including PNA-based anti-miR,
might not be sufficient to obtain full suppression of miR-221
biological activity due to the presence of miR-222 in target cells.
Since miR-221 and miR-222 belong to a same transcriptional unit and
are, as expected, co-expressed in glioma cell lines, including
those used in the present report, we were interested to determine
whether co-administration of anti-miR peptide nucleic acids
recognizing miR-221 and miR-222 leads to more efficient inhibitory
activity on miR-221/222 dependent functions, promoting
pro-apoptotic effects on brain tumor cells.
The major results of the present investigation are
the following: a) R8-conjugated PNAs show biological activity
against miR-221 (R8-PNA-a221) and miR-222 (R8-PNA-a222); b) SPR-BIA
analysis shows that the recognition of R8-PNA-a221 is selective for
miR-221 sequences and that of R8-PNA-a222 is selective for miR-222,
with only partial cross-binding; c) no interactions of miR-221 and
miR-222 occurs with mutated R8-PNA-a221-MUT and R8-PNA-a222-MUT,
respectively; d) when R8-PNA-a221 and R8-PNA-a222 are singularly
administered to glioma cells, specific inhibition of hybridization
to miR-221 and miR-222, respectively, are obtained following
RT-qPCR analysis; e) both R8-PNA-a221 and R8-PNA-a222 induce
apoptosis of U251, U373 and T98G glioma cell lines; f) the
treatment of T98G cells with R8-PNA-a221 and R8-PNA-a222 reversed
the resistance of the cells to apoptosis induced by temozolomide
(TMZ); g) when the R8-PNA-a221 and R8-PNA-a222 are co-administered
the pro-apoptotic effects on U251, U373 and T98G cells and the
reversion of TMZ resistance in T98G cells are much more
evident.
In conclusion, our results support the concept that
anti-miR strategy led to therapeutic relevant inhibition of miRNA
dependent effects (18,20,21,25–28)
and that PNA-based anti-miR molecules are very promising reagents
to regulate tumor cell growth; further research on PNA analogues to
increase efficiency of delivery, stability and control of
intracellular distribution for specific targets, i.e., mature
miRNA, pre-miRNA or pri-miRNA, are further steps for the selection
of best candidate drugs. Finally, our study strongly indicates that
the combined treatment of tumor cells with PNAs targeting both
miR-221 and miR-222, or, more generally, multiple miR targets,
might lead to significant improvement in the efficacy of the
treatment. This last conclusion supports also the concept of
designing multifunctional PNA-containing systems or nanocarriers
(50), enabling to perform
targeting of different miRNA sequences.
Acknowledgements
This study was supported by CIB, by COFIN-2009 and
by AIRC (IG 13575: peptide nucleic acids targeting oncomiR and
tumor-suppressor miRNAs: cancer diagnosis and therapy). G.C. was
funded by Verona Brain Research Foundation.
Abbreviations:
|
PNA
|
peptide nucleic acid
|
|
Fl
|
fluorescein
|
|
RT-qPCR
|
reverse transcription quantitative
polymerase-chain reaction
|
|
SDS
|
sodium dodecylsulphate
|
|
SDS-PAGE
|
SDS-polyacrylamide-gel
electrophoresis
|
|
TMZ
|
temozolomide
|
|
PUMA
|
p53-upregulated modulator of
apoptosis
|
|
PTEN
|
phosphatase and tensin homolog
|
|
TIMP3
|
metalloproteinase inhibitor 3
|
|
ICAM-1
|
intercellular adhesion molecule 1
|
|
SPR
|
surface plasmon resonance
|
|
HBS
|
HEPES-buffered saline
|
References
|
1
|
Nielsen PE, Egholm M, Berg RH and Buchardt
O: Sequence-selective recognition of DNA by strand displacement
with a thymine-substituted polyamide. Science. 254:1497–1500. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nielsen PE: Targeting double stranded DNA
with peptide nucleic acid (PNA). Curr Med Chem. 8:545–550. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Borgatti M, Lampronti I, Romanelli A,
Pedone C, Saviano M, Bianchi N, Mischiati C and Gambari R:
Transcription factor decoy molecules based on a peptide nucleic
acid (PNA)-DNA chimera mimicking Sp1 binding sites. J Biol Chem.
278:7500–7509. 2003. View Article : Google Scholar
|
|
4
|
Gambari R: Peptide-nucleic acids (PNAs): A
tool for the development of gene expression modifiers. Curr Pharm
Des. 7:1839–1862. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Gambari R: Biological activity and
delivery of peptide nucleic acids (PNA)-DNA chimeras for
transcription factor decoy (TFD) pharmacotherapy. Curr Med Chem.
11:1253–1263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nielsen PE: Peptide nucleic acids (PNA) in
chemical biology and drug discovery. Chem Biodivers. 7:786–804.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hatamoto M, Ohashi A and Imachi H: Peptide
nucleic acids (PNAs) antisense effect to bacterial growth and their
application potentiality in biotechnology. Appl Microbiol
Biotechnol. 86:397–402. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gambari R, Borgatti M, Bezzerri V, Nicolis
E, Lampronti I, Dechecchi MC, Mancini I, Tamanini A and Cabrini G:
Decoy oligodeoxyribonucleotides and peptide nucleic acids-DNA
chimeras targeting nuclear factor kappa-B: Inhibition of IL-8 gene
expression in cystic fibrosis cells infected with Pseudomonas
aeruginosa. Biochem Pharmacol. 80:1887–1894. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pandey VN, Upadhyay A and Chaubey B:
Prospects for antisense peptide nucleic acid (PNA) therapies for
HIV. Expert Opin Biol Ther. 9:975–989. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nielsen PE: Gene targeting and expression
modulation by peptide nucleic acids (PNA). Curr Pharm Des.
16:3118–3123. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Manicardi A, Fabbri E, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Gambari R, Marchelli R and Corradini R:
Cellular uptakes, biostabilities and anti-miR-210 activities of
chiral arginine-PNAs in leukaemic K562 cells. ChemBioChem.
13:1327–1337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fabbri E, Manicardi A, Tedeschi T, Sforza
S, Bianchi N, Brognara E, Finotti A, Breveglieri G, Borgatti M,
Corradini R, et al: Modulation of the biological activity of
microRNA-210 with peptide nucleic acids (PNAs). ChemMedChem.
6:2192–2202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gambari R, Fabbri E, Borgatti M, Lampronti
I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R and
Corradini R: Targeting microRNAs involved in human diseases: A
novel approach for modification of gene expression and drug
development. Biochem Pharmacol. 82:1416–1429. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fabani MM and Gait MJ: miR-122 targeting
with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids
(PNA), and PNA-peptide conjugates. RNA. 14:336–346. 2008.
View Article : Google Scholar :
|
|
15
|
Fabani MM, Abreu-Goodger C, Williams D,
Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ and Vigorito E:
Efficient inhibition of miR-155 function in vivo by peptide nucleic
acids. Nucleic Acids Res. 38:4466–4475. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Brown PN and Yin H: PNA-based microRNA
inhibitors elicit anti-inflammatory effects in microglia cells.
Chem Commun (Camb). 49:4415–4417. 2013. View Article : Google Scholar
|
|
17
|
Brognara E, Fabbri E, Aimi F, Manicardi A,
Bianchi N, Finotti A, Breveglieri G, Borgatti M, Corradini R,
Marchelli R, et al: Peptide nucleic acids targeting miR-221
modulate p27Kip1 expression in breast cancer MDA-MB-231
cells. Int J Oncol. 41:2119–2127. 2012.PubMed/NCBI
|
|
18
|
Cheng CJ, Bahal R, Babar IA, Pincus Z,
Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM,
et al: MicroRNA silencing for cancer therapy targeted to the tumour
microenvironment. Nature. 518:107–110. 2015. View Article : Google Scholar :
|
|
19
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Piva R, Spandidos DA and Gambari R: From
microRNA functions to microRNA therapeutics: Novel targets and
novel drugs in breast cancer research and treatment (Review). Int J
Oncol. 43:985–994. 2013.PubMed/NCBI
|
|
21
|
Taylor MA and Schiemann WP: Therapeutic
opportunities for targeting microRNAs in cancer. Mol Cell Ther.
2:1–13. 2014. View Article : Google Scholar
|
|
22
|
Song MS and Rossi JJ: The anti-miR21
antagomir, a therapeutic tool for colorectal cancer, has a
potential synergistic effect by perturbing an
angiogenesis-associated miR30. Front Genet. 4:3012014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nana-Sinkam SP and Croce CM: Clinical
applications for microRNAs in cancer. Clin Pharmacol Ther.
93:98–104. 2013. View Article : Google Scholar
|
|
24
|
Hermansen SK and Kristensen BW: MicroRNA
biomarkers in glioblastoma. J Neurooncol. 114:13–23. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shu M, Zheng X, Wu S, Lu H, Leng T, Zhu W,
Zhou Y, Ou Y, Lin X, Lin Y, et al: Targeting oncogenic miR-335
inhibits growth and invasion of malignant astrocytoma cells. Mol
Cancer. 10:592011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chan XH, Nama S, Gopal F, Rizk P, Ramasamy
S, Sundaram G, Ow GS, Ivshina AV, Tanavde V, Haybaeck J, et al:
Targeting glioma stem cells by functional inhibition of a
prosurvival oncomiR-138 in malignant gliomas. Cell Reports.
2:591–602. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wagenaar TR, Zabludoff S, Ahn SM, Allerson
C, Arlt H, Baffa R, Cao H, Davis S, Garcia-Echeverria C, Gaur R, et
al: Anti-miR-21 suppresses hepatocellular carcinoma growth via
broad transcriptional network de-regulation. Mol Cancer Res.
13:1009–1021. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ma L, Reinhardt F, Pan E, Soutschek J,
Bhat B, Marcusson EG, Teruya-Feldstein J, Bell GW and Weinberg RA:
Therapeutic silencing of miR-10b inhibits metastasis in a mouse
mammary tumor model. Nat Biotechnol. 28:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shah MY and Calin GA: MicroRNAs miR-221
and miR-222: A new level of regulation in aggressive breast cancer.
Genome Med. 3:562011. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lambertini E, Lolli A, Vezzali F,
Penolazzi L, Gambari R and Piva R: Correlation between Slug
transcription factor and miR-221 in MDA-MB-231 breast cancer cells.
BMC Cancer. 12:4452012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J
Biol Chem. 282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yang J, Zhang JY, Chen J, Xu Y, Song NH
and Yin CJ: Prognostic role of microRNA-221 in various human
malignant neoplasms: A meta-analysis of 20 related studies. PLoS
One. 9:e876062014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li W, Guo F, Wang P, Hong S and Zhang C:
miR-221/222 confers radioresistance in glioblastoma cells through
activating Akt independent of PTEN status. Curr Mol Med.
14:185–195. 2014. View Article : Google Scholar
|
|
34
|
Chen L, Zhang J, Han L, Zhang A, Zhang C,
Zheng Y, Jiang T, Pu P, Jiang C and Kang C: Downregulation of
miR-221/222 sensitizes glioma cells to temozolomide by regulating
apoptosis independently of p53 status. Oncol Rep. 27:854–860.
2012.
|
|
35
|
Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han
L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al: MiR-221 and
miR-222 target PUMA to induce cell survival in glioblastoma. Mol
Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Brognara E, Fabbri E, Bazzoli E, Montagner
G, Ghimenton C, Eccher A, Cantù C, Manicardi A, Bianchi N, Finotti
A, et al: Uptake by human glioma cell lines and biological effects
of a peptide-nucleic acids targeting miR-221. J Neurooncol.
118:19–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jensen KK, Orum H, Nielsen PE and Nordén
B: Kinetics for hybridization of peptide nucleic acids (PNA) with
DNA and RNA studied with the BIAcore technique. Biochemistry.
36:5072–5077. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Corradini R, Feriotto G, Sforza S,
Marchelli R and Gambari R: Enhanced recognition of cystic fibrosis
W1282X DNA point mutation by chiral peptide nucleic acid probes by
a surface plasmon resonance biosensor. J Mol Recognit. 17:76–84.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao X, Gu Y, Jiang L, Wang Y, Liu F, Xu Y,
Deng J, Nan Y, Zhang L, Ye J, et al: A new approach to screening
cancer stem cells from the U251 human glioma cell line based on
cell growth state. Oncol Rep. 29:1013–1018. 2013.
|
|
40
|
Abdullah Thani NA, Sallis B, Nuttall R,
Schubert FR, Ahsan M, Davies D, Purewal S, Cooper A and Rooprai HK:
Induction of apoptosis and reduction of MMP gene expression in the
U373 cell line by polyphenolics in Aronia melanocarpa and by
curcumin. Oncol Rep. 28:1435–1442. 2012.PubMed/NCBI
|
|
41
|
Pen A, Durocher Y, Slinn J, Rukhlova M,
Charlebois C, Stanimirovic DB and Moreno MJ: Insulin-like growth
factor binding protein 7 exhibits tumor suppressive and vessel
stabilization properties in U87MG and T98G glioblastoma cell lines.
Cancer Biol Ther. 12:634–646. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Rothbard JB, Kreider E, VanDeusen CL,
Wright L, Wylie BL and Wender PA: Arginine-rich molecular
transporters for drug delivery: Role of backbone spacing in
cellular uptake. J Med Chem. 45:3612–3618. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Abes R, Arzumanov A, Moulton H, Abes S,
Ivanova G, Gait MJ, Iversen P and Lebleu B: Arginine-rich cell
penetrating peptides: Design, structure-activity, and applications
to alter pre-mRNA splicing by steric-block oligonucleotides. J Pept
Sci. 14:455–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bhatia B: On the move: p27Kip1
drives cell motility in glioma cells. Cell Cycle. 9:1231–1240.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lu X, Zhao P, Zhang C, Fu Z, Chen Y, Lu A,
Liu N, You Y, Pu P and Kang C: Analysis of miR-221 and p27
expression in human gliomas. Mol Med Rep. 2:651–656.
2009.PubMed/NCBI
|
|
46
|
Gillies JK and Lorimer IA: Regulation of
p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle.
6:2005–2009. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ueda R, Kohanbash G, Sasaki K, Fujita M,
Zhu X, Kastenhuber ER, McDonald HA, Potter DM, Hamilton RL, Lotze
MT, et al: Dicer-regulated microRNAs 222 and 339 promote resistance
of cancer cells to cytotoxic T-lymphocytes by down-regulation of
ICAM-1. Proc Natl Acad Sci USA. 106:10746–10751. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zerrouqi A, Pyrzynska B, Febbraio M, Brat
DJ and Van Meir EG: P14ARF inhibits human glioblastoma-induced
angiogenesis by upregulating the expression of TIMP3. J Clin
Invest. 122:1283–1295. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang Y, Wang X, Zhang J, Sun G, Luo H,
Kang C, Pu P, Jiang T, Liu N and You Y: MicroRNAs involved in the
EGFR/PTEN/AKT pathway in gliomas. J Neurooncol. 106:217–224. 2012.
View Article : Google Scholar
|
|
50
|
Bertucci A, Lülf H, Septiadi D, Manicardi
A, Corradini R and De Cola L: Intracellular delivery of peptide
nucleic acid and organic molecules using zeolite-L nanocrystals.
Adv Healthcare Mater. 3:1812–1817. 2014. View Article : Google Scholar
|