Introduction
High-mobility group (HMG) proteins are heterogeneous
non-histone DNA-binding factors that organize active chromatin
(1). High-mobility group AT-hook
protein 2 (HMGA 2) belongs to this family and is located at
chromosome 12q14 and encodes an 109 amino acid protein. The
functional unit of all HMGA proteins are 3 copies of a conserved
DNA-binding peptide motif (‘AT-hooks’) that cause the binding to
adenine- thymine (AT)-rich regions of nuclear DNA (2,3).
HMGA 2 is expressed primarily in the early embryonic development,
and is suppressed in differentiated cells (4). Some reports suggest that HMG proteins
regulate the expression of one or more genes that control embryonic
cell growth and differentiation (1,4).
Furthermore, HMG may also affect the expression of oncogenes and
tumor suppressor genes (5). HMGA 2
is regulated by let-7, a tumor suppressor microRNA (miRNA), which
is downregulated in regards to cancer development (6). A directly regulatory relationship
between let-7a (member of let-7 family) and HMGA 2 regulation has
been confirmed in breast, lung and esophageal cancers (7–9).
HMGA 2 is not expressed in normal human adult tissue
but it can be detected in many human tumors including lipoma,
leiomyoma and pituitary tumors, which are benign tumors but they
can be very invasive (10,11). Furthermore, HMGA 2 is also
expressed in malignant neoplasms such as colorectal, lung, gastric,
ovarian and breast cancers, neuroblastoma and glioblastoma
(12–19).
Prior studies showed that miR-142-3p, a miRNA in
pluripotent stem cells, suppresses the expression of HMGA 2
targeting the 3′UTR, and thus, decreasing the expression of Sox2, a
transcription factor in stem cells. HMGA 2 increases the
transcription of Sox2 through directly binding to the Sox2 promoter
gene region. Patients with an upregulated HMGA 2 expression
demonstrated a poor overall survival (20). miRNA alterations are involved in
the initiation and progression of human cancer (6). Morishita et al (21) conducted a study demonstrating the
molecular mechanisms of HMGA 2 in tumor pathogenesis through
activation of the TGFβ signaling pathway, a major inducer of the
epithelial-mesenchymal-transition (EMT), in epithelial carcinomas.
Their results showed that HMGA 2 plays a critical role in inducing
tumor invasion, and metastasis in EMT by activating the TGFβ
signaling pathway.
A study by Liu et al (16) presented a significant correlation
of HMGA 2 expression, and glioblastoma cell proliferation, invasion
and survival. Glioblastomas are highly invasive, rapidly growing,
scatter along the white matter tracts and their structure is poorly
differentiated. Furthermore, glioblastoma represents the most
aggressive type of glial brain tumors. Treatment is rarely
effective despite gross total resection, chemotherapy, radiotherapy
or all of them (22). MGMT
(O-6-methylguanine-DNA methyltransferase) methylation status and
the analysis of IDH1 and IDH2 mutations has become diagnostic and
prognostic standards, and there are numerous studies investigating
new molecular markers for a better characterization of this tumor
entity (23–25).
Unfortunately, the recurrence of glioblastoma is
still inevitable after a median progression-free period of 6.8
months (26). Consequently, novel
therapies targeting cell proliferation and invasion e.g. through
gene therapy may become a more effective strategy (27).
Based on the available data, HMGA 2 could be a
biomarker and a potential target in glioblastoma therapy.
Therefore, in the present study, we investigated the expression of
HMGA 2 in glioblastoma patients, and we correlated the expression
data with clinical parameters, survival and MGMT methylation
status.
Materials and methods
Patient data
A retrospective analysis of medical records was
conducted and clinical data were extracted. Variables assessed
include: birth date, gender, date of diagnosis, date of
operation/reoperation, type/date of chemotherapy, type/date of
radiotherapy, MRI follow-up reports, relapse status, date of
relapse, date of last follow-up, and vital status at last
follow-up. All patients were periodically followed for survival.
The follow-up period was calculated from the date of surgery to the
date of last contact. The time of progression-free survival (PFS)
was defined as at the time of initial surgical therapy to tumor
recurrence.
Tissue specimens
A total of 44 diagnostically confirmed cases of
glioblastoma (WHO IV) were retrieved as formalin-fixed,
paraffin-embedded tissue blocks and kryopreserved tissue between
2006 and 2012 from the Department of Neuropathology, University of
Giessen, Giessen, Germany. Five normal brain tissue specimens were
used as reference for tumor-free brain tissue and a breast cancer
specimen was used as a positive control provided by the same
institution. The present study was approved by the local ethics
committee (application number: AZ 07/09).
RNA-isolation, quantitative real-time
PCR
RNA-isolation was performed from frozen specimens
stored in fluid nitrogen using the RNeasy Lipid Tissue Mini
kit® from Qiagen GmbH (Hilden, Germany). RNA
concentration was measured photometrically (peqLab
NanoDrop® 1000 Spectrophotometer; VWR International GmbH
Life Science Competence Center, Erlangen, Germany). Total RNA (1
μg) was used for cDNA synthesis with the QuantiTect®
reverse transcription kit (Qiagen). Quantitative real-time PCR
(qPCR) was performed along with the following human primers with
the Mastercycler Gradient Thermal Cycler® (Eppendorf,
Hamburg, Germany): actin-β, human (Hs99999903), IPO8, human
(Hs00914040), HMGA 2, human (Hs00171569_m1), TBP, human
(Hs00427620) (all from Life Technologies Inc., Carlsbad, CA, USA).
Reaction setup and cycling conditions adhered to the kit manual.
Reaction efficiency was determined using standard curves, gene
expression levels calculated by the ΔΔCt method and expressed as
efficiency corrected log2 values. Additionally, the linear n-fold
expression ratio of cases/controls was calculated by first
converting the log2 values to linear and calculating the ratio of
cases/controls to facilitate easy comprehension of the gene
expression changes.
Raw cycle threshold (Ct) data of qPCR experiments
were processed by subtracting the mean Ct of all endogenous control
genes (actin-β, IPO8 and TBP) from the Ct of the according gene of
interest (HMGA 2). Taking into account the exponential nature of
PCR methodology, relative expression was obtained from the
resulting ΔCt value using the formula of 2−ΔCt.
In preliminary experiments, we analyzed samples from
the individual frontal, parietal, temporal, occipital and
cerebellar lobes for HMGA 2 expression using qPCR. No significant
differences in expression were found between these anatomical
regions (data not shown). Therefore, we did not further match the
control samples to the anatomical brain regions of the tumors.
Immunohistochemistry
Of all samples investigated using qPCR, a subset of
44 tumor samples and 5 non-tumorous brain tissue samples were
available as paraffin-embedded tissue and used for
immunohistochemistry. Samples from breast cancers were used as
positive controls. Immunohistochemical staining was performed using
the BenchMark XT IHC fully automated staining instrument (Ventana
Medical Systems, Tucson, AZ, USA) following the manufacturer's
instructions on 3-μm paraffin sections. A polyclonal rabbit
antibody against HMGA 2 (ab52039; Abcam, Cambridge, UK) was used
with a final dilution of 1:25.
Quantification of immunohistochemical
staining
The sections were microscopically (Leica DMLB
microscope; Leica Microsystems, Wetzlar, Germany) assessed in ×200
and x400 magnification by two investigators (C.W. and F.P.S.) who
were blinded to patient characteristics and outcome.
Immunoreactivity score (IRS) was determined using staining
intensity and number of positively stained cells (nuclear
expression). Staining intensity was determined on the following
scale: 0 (no staining), 1 (weak staining, light yellow), 2
(moderate staining, yellowish brown), and 3 (strong staining,
brown). In addition, the percentage of positive cells was
semi-quantitatively determined (0–100% in 10% steps). IRS was
calculated as the product of staining intensity and percentage of
positive tumor cells, resulting in a value ranging from 0 to 30.
The k-statistics of the analyzed immunohistochemical stained slides
revealed a kappa value of 0.534 for HMGA 2. Differences in
assessment were discussed until consensus was reached.
Calculation of HMGA 2 gene expression and
statistical analysis
qPCR and calculation of gene
expression
Gene expression calculations were performed using
the ΔΔCt method in GenEx 6 (Multid Analyses AB, Göteborg, Sweden).
Stable expression of housekeeping genes β-actin (ACTB),
TATA-binding protein (TBP) and importin (IPO) were assessed using
the NormFinder algorithm, hereby confirming suitability of all 3
genes to be used as references in our samples. Normalization of
HMGA 2 was performed against these three housekeeping genes and
expression levels were calculated for normal brain tissue and
glioblastoma specimens. To allow direct comparison, data are shown
as 2−ΔCt values unless otherwise indicated.
Statistical analysis
The following statistical assessments were performed
using the Mann-Whitney U test: gene expression levels in MGMT
methylated vs. unmethylated patients, gene expression levels
between patient groups, both for qPCR and immunohistochemical data.
The relationship between HMGA2 expression, regression-free survival
and overall survival was analyzed using the Kaplan-Meier method and
the log-rank test. Survival analysis was performed by dividing
patients into low and high expression groups. Low expression was
defined as normalized gene expression levels equal or below the
mean expression of each respective tumor group, while all other
patients within this group were classified as high expression. To
analyze further the influence of HMGA 2 expression, we performed a
multivariate analysis of variance (MANOVA) stratified according to
age, HMGA 2 gene expression on mRNA and protein levels, MGMT
methylation status, progression-free survival (PFS) and overall
survival (OS). P-values of <0.05 were considered statistically
significant throughout the analyses.
Results
Patient collective
The patient collective included 44 gross total
resected glioblastoma (WHO grade IV) patients, 40 of whom were
diagnosed with primary, and 4 with secondary glioblastoma. Mean age
at diagnosis was 57.4±15.7 years. The study population consisted of
31 male and 13 female patients. The patients' median survival time
was 16 months (SE 2.8; 95% CI, 10.6–21.4). Two of the patients had
received chemotherapy or radiotherapy prior to first surgery.
Further details are listed in Table I. Five normal brain tissue
specimens were used as reference for tumor-free brain tissue.
 | Table ISummary of the baseline
characteristics. |
Table I
Summary of the baseline
characteristics.
| n (%) |
|---|
| Total cases | 44 (100) |
| Age at diagnosis
(years) | 57.4±15.7) |
| Gender |
| Male | 31 (70.5) |
| Female | 13 (29.5) |
| Survival |
| Unstratified
patients |
| 16 months (SE,
2.8, 95% CI, 10.6–21.4) | 100 |
| MGMT |
| Methylated: 22
months (SE, 3.7; 95% CI, 14.8–29.2) | 54.5 |
| Unmethylated: 11
months (SE, 2.2; 95% CI, 6.7–15.3) | 43.2 |
| Tumor entity |
| Glioblastoma | 44 (100) |
| Primary | 40 (90.9) |
| Secondary | 4 (9.1) |
| Methylation |
| MGMT |
|
Hypermethylated | 24 (54.5) |
| Not
hypermethylated | 19 (43.2) |
| Unavailable | 1 (2.3) |
| Neoadjuvant
treatment before 1st resection |
| Chemotherapy |
| Temozolomide | 2 (4.5) |
| Radiation |
| 60 Gy | 3 (6.8) |
| Initial
resection |
| Resection | 41 (93.2) |
| Missing data | 3 (6.8) |
| Adjuvant treatment
after initial resection |
| Chemotherapy |
| Total treated in
group | 37 (100) |
| Temozolomide | 37 (100) |
| Radiation |
| Total treated in
group | 38 (100) |
| 60-Gy
concomitant | 36 (94.7) |
| 60-Gy
stereotactic | 2 (5.3) |
| 1st recurrence |
| Total affected
patients | 24 (54.5) |
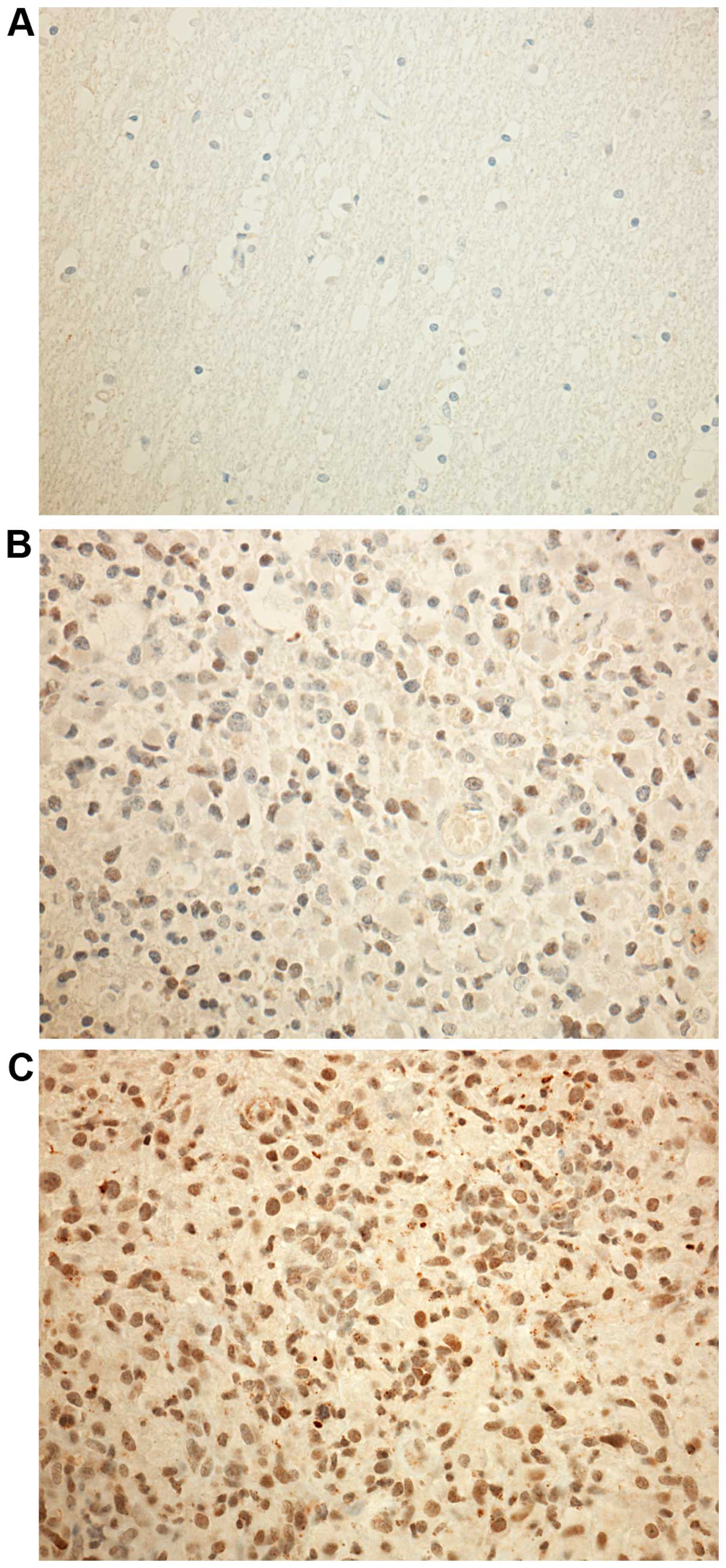
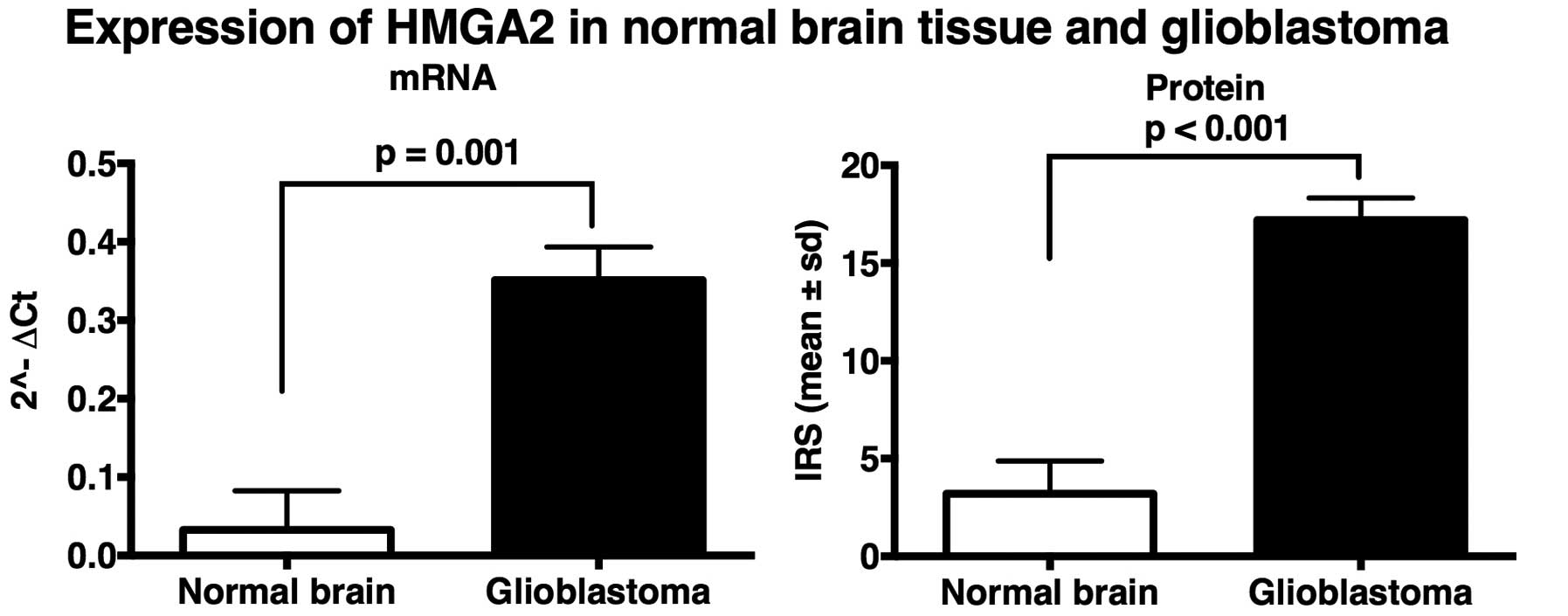
HMGA 2 mRNA expression in glioblastoma
and normal brain tissue
A subset of 40 glioblastoma specimens along with 5
normal brain tissues were analyzed performing qRT-PCR. The HMGA 2
expression on mRNA levels in glioblastomas were upregulated (mean,
0.35; SD, 0.27) compared to non-tumorous brain tissue (mean, 0.03;
SD, 0.05). HMGA 2 gene expression was significantly higher in
glioblastoma (P=0.001) (Figs. 1
and 2).
HMGA 2 protein expression in glioblastoma
and normal brain tissue
Forty-four glioblastoma specimens along with 5
normal brain tissues were analyzed by immunohistochemistry
confirming the expression difference seen in the qRT-PCR analysis:
IRS of HMGA 2 was significantly higher in glio-blastoma tissue
(mean, 17.21; SD, 7.43) than in normal brain tissue (mean, 3.20;
SD, 1.68) (P<0.001). Thus, HMGA 2 gene expression was
significantly higher in glioblastoma than in non-tumorous brain
tissue (Figs. 1 and 2).
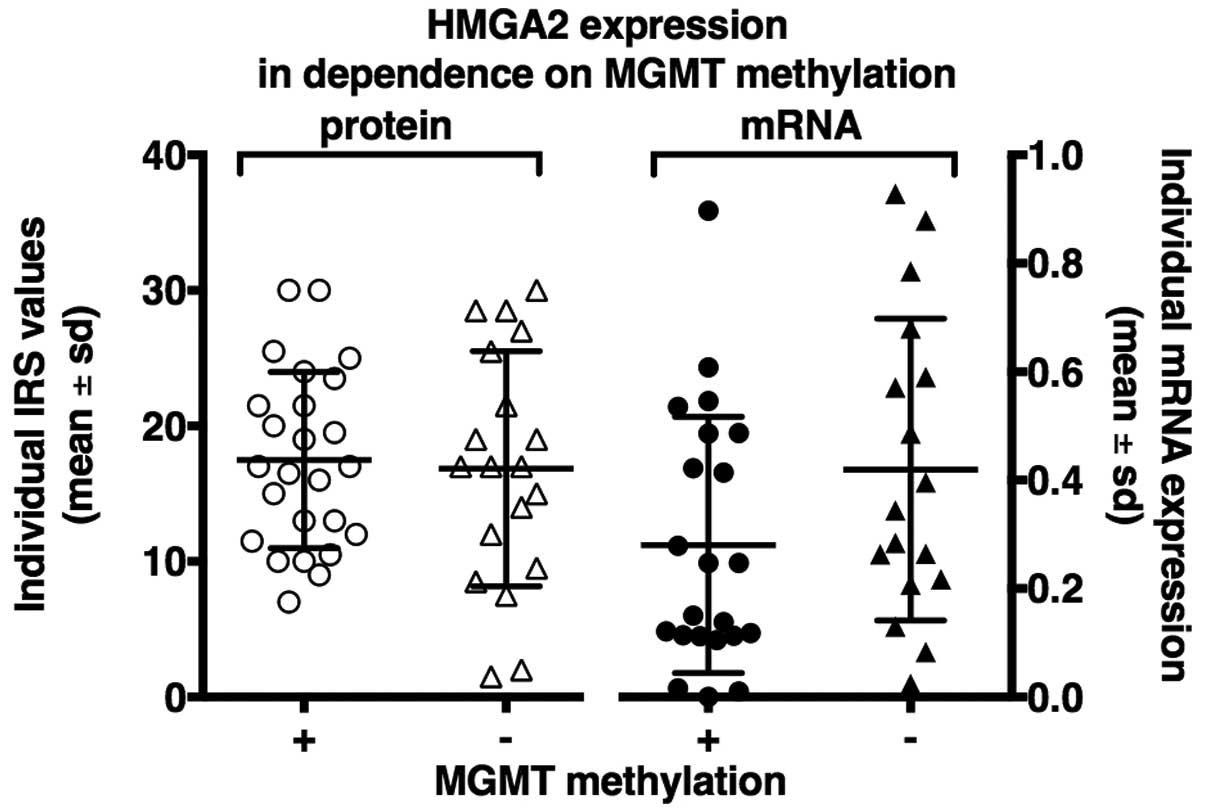
HMGA 2 expression as a parameter of MGMT
methylation status
The analysis of HMGA 2 expression as a parameter of
MGMT methylation status showed no significant differences in qPCR
and immunohistochemistry analysis. On protein level there were no
expression differences of HMGA 2 between methylated and
unmethylated patients (P=0.87). On mRNA levels a slightly higher
HMGA 2 expression was seen for unmethylated patients (P=0.09)
(Fig. 3).
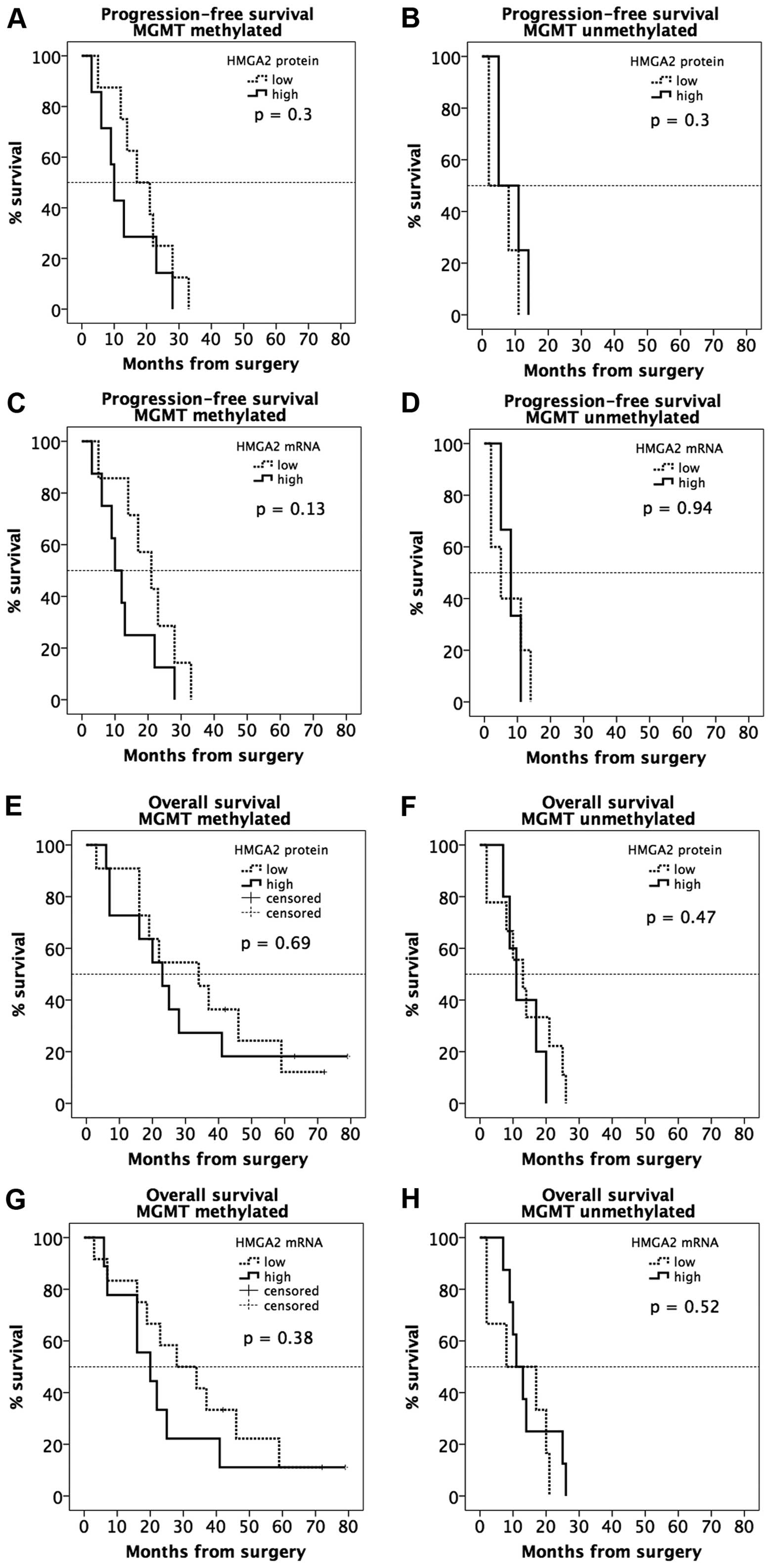
Progression-free survival (PFS) time as a
parameter of MGMT-methylation status and HMGA 2 mRNA and protein
expression
Progression-free survival (PFS) time in primary
glioblastoma patients with surgical resection and adjuvant combined
radio-chemotherapy with temozolomide and 60-Gy concomitant
irradiation, stratified by MGMT promoter methylation was shorter
with HMGA 2 upregulation (Fig. 4.)
On mRNA levels 8 patients with positive MGMT promoter methylation
status who had primary glioblastoma with high HMGA 2 levels had a
median PFS time of 10.0 months (SE, 2.1; 95% CI, 5.8–14.2), whereas
7 patients with primary glioblastoma and low HMGA 2 expression and
methylated MGMT had a median PFS time of 21.0 months (SE, 5.2; 95%
CI 10.7–31.3) (P=0.14).
Three negative MGMT promoter methylation status
patients with high HMGA 2 levels had a median PFS time of 8.0
months (SE, 2.4; 95% CI, 3.2–12.8), whereas 5 patients with primary
glioblastoma and low HMGA 2 expression and negative MGMT promoter
methylation status had a median PFS of 5.0 months (SE, 3.3, 95% CI
0.0–11.4) (P=0.90).
Overexpression of HMGA 2 showed a tendency to a
shorter PFS time. These results on mRNA level were confirmed by IRS
and showed no statistical significance (Fig. 4).
Overall survival (OS) time as a parameter
of MGMT-methylation status and HMGA 2 mRNA and protein
expression
The effect of HMGA 2 expression on overall survival
(OS) time in primary glioblastoma patients with surgical resection
and adjuvant combined radio-chemotherapy with temozolomide and
60-Gy concomitant irradiation, stratified by MGMT promoter
methylation, showed a shorter OS time when HMGA 2 expression was
upregulated (Fig. 4).
On mRNA levels 9 patients with positive MGMT
promoter methylation status who had primary glioblastoma with high
HMGA 2 levels had a median OS time of 20 months (SE, 6.0; 95% CI,
8.3–31.7), whereas 12 patients with primary glioblastoma and low
HMGA 2 expression and positive MGMT promoter methylation status had
a median OS time of 28.0 months (SE, 9.5; 95% CI, 9.3–46.7)
(P=0.38). Eight negative MGMT promoter methylation status patients
with high HMGA 2 levels had a median OS of 11 months (SE, 2.1; 95%
CI, 6.8–15.2), whereas, 6 patients with primary glioblastoma and
low HMGA 2 expression and negative MGMT promoter methylation status
had a median OS time of 8 months (SE, 5.5; 95% CI, 0.0–20.8)
(P=0.52).
Overexpression of HMGA 2 was associated with a
tendency to a shorter OS time. These results on mRNA level were
confirmed by IRS and showed no statistical significance (Fig. 4).
Multivariate analysis
The multivariate analysis of variance (MANOVA)
showed no statistical significance except for HMGA 2 expression in
tumor vs. normal brain tissue (P<0.05).
Discussion
In the past decades advances in neuroimaging,
treatment paradigms and molecular approaches enabled neurosurgeons
to better understand glioblastoma and its treatment (22,23,28–30).
Numerous studies have been done on the invasive behavior of
malignant gliomas, and there are many studies investigating new
molecular markers to better characterize this tumor entity and
improve clinical decision (23,31–38).
The current standard of care consists of microsurgical gross total
resection, if possible, concurrent radiotherapy, and temozolomide
chemotherapy followed by adjuvant temozolomide (39,40).
Despite ongoing research efforts and aggressive
treatment the median survival of glioblastoma patients still yields
poor outcomes. The complete resection of the tumor is almost
impossible because of the invasive growth of tumor cells and the
lack of a clear border (41,42).
New chemotherapy approaches with temozolomide showed an improvement
of survival from 12 to 14.6 months (43).
To the best of our knowledge, this is the first
study analyzing on HMGA 2 expression in glioblastoma with respect
to the MGMT methylation status. Our results showed that HMGA 2
overexpression has an influence on progression-free and overall
survival time of glioblastoma patients.
Previous research showed that HMGA 2 is not
expressed in normal brain tissue (16). Also in the present series no HMGA 2
expression was found in five normal brain tissue controls.
Furthermore, HMGA 2 is associated with poor
prognosis, malignancy and invasiveness in tumors such as gastric
and lung cancer, retinoblastoma and pituitary adenoma (11,14,15,44).
The expression of HMGA 2 correlates with the degree of malignancy
of astrocytic brain tumors and shows an increase in higher grade
gliomas. The highest HMGA 2 overexpression was detected in
glioblastoma (16).
Inhibition of HMGA 2 leads to tumor growth
inhibition and an increase of apoptosis in ovarian cancer (13). Halle et al (45) showed a possible mechanism to
regulate the mRNA level of HMGA 2 due to repression of let-7a
through local drug delivery with the convection-enhanced delivery
(CED) technique. They showed an in vivo and in vitro
de-repression of HMGA 2 after intratumoral therapy with CED. Thus,
miRNAs and especially oncogenic miRNAs have the potential to impact
on future cancer therapy (45).
In line with the study of Liu et al (16) in glioblastoma samples and on other
human malignancies, the present study study showed that HMGA 2 was
expressed in glioblastoma and not in normal brain tissue.
Furthermore, overexpression of HMGA 2 in glioblastoma is reported
to be closely correlated with poor survival prognosis (16,44).
In the present study, the overall and tumor progression-free
survival time tended to be shorter in the group of patients with
overexpression of HMGA 2, on IRS and qPCR levels.
Previous studies showed that MGMT promoter
methylation is used to identify patients who benefit from
alkylating chemotherapy. Additionally, MGMT methylation leads to a
longer PFS and OS in glioblastoma patients (40,46,47).
Liu et al (44) reported that aberrant HMGA 2 was
associated with long-term survival of glioblastoma patients.
However, in the present study patients with HMGA 2 showed a
tendency to a shorter survival time (PFS and OS). Moreover, our
results showed that the MGMT promoter methylation did not lead to a
longer survival time in the group of patients with HMGA 2
overexpression.
Furthermore, Lee et al (48) presented in HMGA2 knocked down tumor
cells a reduction of cell invasion and migration. They showed the
downregulation of multiple EMT-factors such as N-cadherine
(mesenchymal marker), β-catenin, transcriptional factors like Snail
and Zeb 1 and upregulation of E-cadherine (epithelial marker).
HMGA2 overexpression through its relationship to EMT-pathway seems
to intensify invasion of cancer cells.
Morishita et al (21) showed in their in vitro
experiments that HMGA 2 converts non-invasive cell types into their
invasive counterparts through the induction EMT. Cells at the
‘invasive front’ of human tumors preferentially express HMGA 2
where the tumor cells exhibit the EMT. According to their results
HMGA 2 is localized to the ‘invasive front’ of tumors and enables
tumor cells to migrate. These findings could be a further
explanation for infiltration of glioblastomas into normal brain
tissue and, thus, the impossibility of a complete tumor resection
and curative treatment so far. This hypothesis is speculative
because it is based on other tumor entities and not
glioblastomas.
As Halle et al (45) showed, the de-repression of miRNA
levels of HMGA 2 through anti-let-7a application via
convection-enhanced delivery (CED), HMGA 2 could be a potential
target in future glioblastoma therapies with strategies for
manipulating the expression of HMGA 2-regulating miRNAs.
There are several limitations of the present study.
First of all there is the retrospective character of the study with
the well-known shortcomings of this study design. Furthermore, the
study population is small and heterogeneous, so that a selection
bias cannot be excluded.
In conclusion, the present study indicated that HMGA
2 overexpression had a tendency towards poor prognosis of
glioblastoma patients independent of their MGMT methylation status.
The high expression of HMGA 2 could lead to shorter survival time
and poor prognosis, whereas, glioblastoma patients with low HMGA 2
expression have longer survival times (OS and PFS). HMGA 2 is an
informative biomarker, which is associated with poor prognosis of
patients with glioblastoma. This hypothesis may have potential
implications for glioblastoma survival prediction, the choice of
treatment regimens and may be helpful to create novel strategies
for glioma therapy and prevention.
Acknowledgements
The authors thank sincerely Nga Rötering for
excellent technical assistance, Dr Karl Quint for assistance in
statistical analysis, Sabine Gräf and Boyan Garvalov for the
analysis of MGMT promoter methylation.
Abbreviations:
|
EMT
|
epithelial-mesenchymal-transition
|
|
GBM
|
glioblastoma multiforme
|
|
HMG
|
high-mobility group
|
|
IHC
|
immunohistochemistry
|
|
IRS
|
immunoreactive score
|
|
MGMT
|
O-6-methylguanine-DNA
methyltransferase
|
|
mRNA
|
messenger RNA
|
|
OS
|
overall survival
|
|
PFS
|
progression-free survival
|
|
qPCR
|
real-time quantitative PCR
|
|
RCT
|
radiochemotherapy
|
|
RNA
|
ribonucleic acid
|
References
|
1
|
Grosschedl R, Giese K and Pagel J: HMG
domain proteins: Architectural elements in the assembly of
nucleoprotein structures. Trends Genet. 10:94–100. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monzen K, Ito Y, Naito AT, Kasai H, Hiroi
Y, Hayashi D, Shiojima I, Yamazaki T, Miyazono K, Asashima M, et
al: A crucial role of a high mobility group protein HMGA2 in
cardiogenesis. Nat Cell Biol. 10:567–574. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang X, Liu X, Li AYJ, Chen L, Lai L, Lin
HH, Hu S, Yao L, Peng J, Loera S, et al: Overexpression of HMGA2
promotes metastasis and impacts survival of colorectal cancers.
Clin Cancer Res. 17:2570–2580. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fusco A and Fedele M: Roles of HMGA
proteins in cancer. Nat Rev Cancer. 7:899–910. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Akai T, Ueda Y, Sasagawa Y, Hamada T, Date
T, Katsuda S, Iizuka H, Okada Y and Chada K: High mobility group
I-C protein in astrocytoma and glioblastoma. Pathol Res Pract.
200:619–624. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shell S, Park SM, Radjabi AR, Schickel R,
Kistner EO, Jewell DA, Feig C, Lengyel E and Peter ME: Let-7
expression defines two differentiation stages of cancer. Proc Natl
Acad Sci USA. 104:11400–11405. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guo L, Chen C, Shi M, Wang F, Chen X, Diao
D, Hu M, Yu M, Qian L and Guo N: Stat3-coordinated
Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain
oncostatin M-driven epithelial-mesenchymal transition. Oncogene.
32:5272–5282. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee YS and Dutta A: The tumor suppressor
microRNA let-7 represses the HMGA2 oncogene. Genes Dev.
21:1025–1030. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu Q, Liu T, Zheng S, Gao X, Lu M,
Sheyhidin I and Lu X: HMGA2 is down-regulated by microRNA let-7 and
associated with epithelial-mesenchymal transition in oesophageal
squamous cell carcinomas of Kazakhs. Histopathology. 65:408–417.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fedele M, Battista S, Kenyon L,
Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R,
Pierantoni GM, Outwater E, et al: Overexpression of the HMGA2 gene
in transgenic mice leads to the onset of pituitary adenomas.
Oncogene. 21:3190–3198. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Qian ZR, Asa SL, Siomi H, Siomi MC,
Yoshimoto K, Yamada S, Wang EL, Rahman MM, Inoue H, Itakura M, et
al: Overexpression of HMGA2 relates to reduction of the let-7 and
its relationship to clinicopathological features in pituitary
adenomas. Mod Pathol. 22:431–441. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rizzi C, Cataldi P, Iop A, Isola M, Sgarra
R, Manfioletti G and Giancotti V: The expression of the
high-mobility group A2 protein in colorectal cancer and surrounding
fibroblasts is linked to tumor invasiveness. Hum Pathol.
44:122–132. 2013. View Article : Google Scholar
|
|
13
|
Malek A, Bakhidze E, Noske A, Sers C,
Aigner A, Schäfer R and Tchernitsa O: HMGA2 gene is a promising
target for ovarian cancer silencing therapy. Int J Cancer.
123:348–356. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Motoyama K, Inoue H, Nakamura Y, Uetake H,
Sugihara K and Mori M: Clinical significance of high mobility group
A2 in human gastric cancer and its relationship to let-7 microRNA
family. Clin Cancer Res. 14:2334–2340. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Di Cello F, Hillion J, Hristov A, Wood LJ,
Mukherjee M, Schuldenfrei A, Kowalski J, Bhattacharya R, Ashfaq R
and Resar LM: HMGA2 participates in transformation in human lung
cancer. Mol Cancer Res. 6:743–750. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu B, Pang B, Hou X, Fan H, Liang N,
Zheng S, Feng B, Liu W, Guo H, Xu S, et al: Expression of
high-mobility group AT-hook protein 2 and its prognostic
significance in malignant gliomas. Hum Pathol. 45:1752–1758. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashar HR, Fejzo MS, Tkachenko A, Zhou X,
Fletcher JA, Weremowicz S, Morton CC and Chada K: Disruption of the
architectural factor HMGI-C: DNA-binding AT hook motifs fused in
lipomas to distinct transcriptional regulatory domains. Cell.
82:57–65. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tallini G, Vanni R, Manfioletti G,
Kazmierczak B, Faa G, Pauwels P, Bullerdiek J, Giancotti V, Van Den
Berghe H and Dal Cin P: HMGI-C and HMGI(Y) immunoreactivity
correlates with cytogenetic abnormalities in lipomas, pulmonary
chondroid hamartomas, endometrial polyps, and uterine leiomyomas
and is compatible with rearrangement of the HMGI-C and HMGI(Y)
genes. Lab Invest. 80:359–369. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Giannini G, Di Marcotullio L, Ristori E,
Zani M, Crescenzi M, Scarpa S, Piaggio G, Vacca A, Peverali FA,
Diana F, et al: HMGI(Y) and HMGI-C genes are expressed in
neuroblastoma cell lines and tumors and affect retinoic acid
responsiveness. Cancer Res. 59:2484–2492. 1999.PubMed/NCBI
|
|
20
|
Chiou GY, Chien CS, Wang ML, Chen MT, Yang
YP, Yu YL, Chien Y, Chang YC, Shen CC, Chio CC, et al: Epigenetic
regulation of the miR142-3p/interleukin-6 circuit in glioblastoma.
Mol Cell. 52:693–706. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Morishita A, Zaidi MR, Mitoro A,
Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J and Chada K:
HMGA2 is a driver of tumor metastasis. Cancer Res. 73:4289–4299.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, Taphoorn MJ, Janzer RC, Ludwin SK, Allgeier A, Fisher B,
Belanger K, et al: European Organisation for Research and Treatment
of Cancer Brain Tumour and Radiation Oncology Groups; National
Cancer Institute of Canada Clinical Trials Group: Effects of
radiotherapy with concomitant and adjuvant temozolomide versus
radiotherapy alone on survival in glioblastoma in a randomised
phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet
Oncol. 10:459–466. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weller M, Pfister SM, Wick W, Hegi ME,
Reifenberger G and Stupp R: Molecular neuro-oncology in clinical
practice: A new horizon. Lancet Oncol. 14:e370–e379. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
von Neubeck C, Seidlitz A, Kitzler HH,
Beuthien-Baumann B and Krause M: Glioblastoma multiforme: Emerging
treatments and stratification markers beyond new drugs. Br J
Radiol. 88:201503542015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Okonogi N, Shirai K, Oike T, Murata K,
Noda SE, Suzuki Y and Nakano T: Topics in chemotherapy,
molecular-targeted therapy, and immunotherapy for newly-diagnosed
glioblastoma multiforme. Anticancer Res. 35:1229–1235.
2015.PubMed/NCBI
|
|
26
|
Weller M, Felsberg J, Hartmann C, Berger
H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn
JC, et al: Molecular predictors of progression-free and overall
survival in patients with newly diagnosed glioblastoma: A
prospective translational study of the German Glioma Network. J
Clin Oncol. 27:5743–5750. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kim SM, Woo JS, Jeong CH, Ryu CH, Lim JY
and Jeun SS: Effective combination therapy for malignant glioma
with TRAIL-secreting mesenchymal stem cells and lipoxygenase
inhibitor MK886. Cancer Res. 72:4807–4817. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Macdonald DR, Kiebert G, Prados M, Yung A
and Olson J: Benefit of temozolomide compared to procarbazine in
treatment of glioblastoma multiforme at first relapse: Effect on
neurological functioning, performance status, and health related
quality of life. Cancer Invest. 23:138–144. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Venur VA, Peereboom DM and Ahluwalia MS:
Current medical treatment of glioblastoma. Cancer Treat Res.
163:103–115. 2015. View Article : Google Scholar
|
|
30
|
Mabray MC, Barajas RF Jr and Cha S: Modern
brain tumor imaging. Brain Tumor Res Treat. 3:8–23. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hoelzinger DB, Mariani L, Weis J, Woyke T,
Berens TJ, McDonough WS, Sloan A, Coons SW and Berens ME: Gene
expression profile of glioblastoma multiforme invasive phenotype
points to new therapeutic targets. Neoplasia. 7:7–16. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Perego C, Vanoni C, Massari S, Raimondi A,
Pola S, Cattaneo MG, Francolini M, Vicentini LM and Pietrini G:
Invasive behaviour of glioblastoma cell lines is associated with
altered organisation of the cadherin-catenin adhesion system. J
Cell Sci. 115:3331–3340. 2002.PubMed/NCBI
|
|
33
|
Sturm D, Witt H, Hovestadt V, Khuong-Quang
DA, Jones DT, Konermann C, Pfaff E, Tönjes M, Sill M, Bender S, et
al: Hotspot mutations in H3F3A and IDH1 define distinct epigenetic
and biological subgroups of glioblastoma. Cancer Cell. 22:425–437.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Badie B and Schartner J: Role of microglia
in glioma biology. Microsc Res Tech. 54:106–113. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Giese A, Bjerkvig R, Berens ME and
Westphal M: Cost of migration: Invasion of malignant gliomas and
implications for treatment. J Clin Oncol. 21:1624–1636. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Beije N, Kraan J, Taal W, van der Holt B,
Oosterkamp HM, Walenkamp AM, Beerepoot L, Hanse M, van Linde ME,
Otten A, et al: Prognostic value and kinetics of circulating
endothelial cells in patients with recurrent glioblastoma
randomised to bevacizumab plus lomustine, bevacizumab single agent
or lomustine single agent. A report from the Dutch Neuro-Oncology
Group BELOB trial. Br J Cancer. 113:226–231. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deviers A, Ken S, Filleron T, Rowland B,
Laruelo A, Catalaa I, Lubrano V, Celsis P, Berry I, Mogicato G, et
al: Evaluation of the lactate-to-N-acetyl-aspartate ratio defined
with magnetic resonance spectroscopic imaging before radiation
therapy as a new predictive marker of the site of relapse in
patients with glioblastoma multiforme. Int J Radiat Oncol Biol
Phys. 90:385–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hutterer M, Nowosielski M, Haybaeck J,
Embacher S, Stockhammer F, Gotwald T, Holzner B, Capper D, Preusser
M, Marosi C, et al: A single-arm phase II Austrian/German
multi-center trial on continuous daily sunitinib in primary
glioblastoma at first recurrence (SURGE 01-07). Neuro Oncol.
16:92–102. 2014. View Article : Google Scholar
|
|
39
|
Lacroix M, Abi-Said D, Fourney DR,
Gokaslan ZL, Shi W, DeMonte F, Lang FF, McCutcheon IE, Hassenbusch
SJ, Holland E, et al: A multivariate analysis of 416 patients with
glioblastoma multiforme: Prognosis, extent of resection, and
survival. J Neurosurg. 95:190–198. 2001. View Article : Google Scholar
|
|
40
|
Hegi ME, Diserens AC, Gorlia T, Hamou MF,
de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani
L, et al: MGMT gene silencing and benefit from temozolomide in
glioblastoma. N Engl J Med. 352:997–1003. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nestler U, Lutz K, Pichlmeier U, Stummer
W, Franz K, Reulen HJ and Bink A: 5-ALA Glioma Study Group:
Anatomic features of glioblastoma and their potential impact on
survival. Acta Neurochir (Wien). 157:179–186. 2015. View Article : Google Scholar
|
|
42
|
Matsukado Y, MacCarty CS and Kernohan JW:
The growth of glioblastoma multiforme (astrocytomas, grades 3 and
4) in neurosurgical practice. J Neurosurg. 18:636–644. 1961.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Becker KP and Yu J: Status quo -
standard-of-care medical and radiation therapy for glioblastoma.
Cancer J. 18:12–19. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu Y, Shete S, Etzel CJ, Scheurer M,
Alexiou G, Armstrong G, Tsavachidis S, Liang FW, Gilbert M, Aldape
K, et al: Polymorphisms of LIG4, BTBD2, HMGA2, and RTEL1 genes
involved in the double-strand break repair pathway predict
glioblastoma survival. J Clin Oncol. 28:2467–2474. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Halle B, Marcusson EG, Aaberg-Jessen C,
Jensen SS, Meyer M, Schulz MK, Andersen C and Kristensen BW:
Convection-enhanced delivery of an anti-miR is well-tolerated,
preserves anti-miR stability and causes efficient target
de-repression: A proof of concept. J Neurooncol. 126:47–45. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Stupp R, van den Bent MJ and Hegi ME:
Optimal role of temozolomide in the treatment of malignant gliomas.
Curr Neurol Neurosci Rep. 5:198–206. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gilbert MR, Wang M, Aldape KD, Stupp R,
Hegi ME, Jaeckle KA, Armstrong TS, Wefel JS, Won M, Blumenthal DT,
et al: Dose-dense temozolomide for newly diagnosed glioblastoma: A
randomized phase III clinical trial. J Clin Oncol. 31:4085–4091.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lee J, Ha S, Jung CK and Lee HH:
High-mobility-group A2 overexpression provokes a poor prognosis of
gastric cancer through the epithelial-mesenchymal transition. Int J
Oncol. 46:2431–2438. 2015.PubMed/NCBI
|