Introduction
Epithelial ovarian cancer (EOC) has the highest
mortality rate among all gynaecological malignancies, metastasis is
the main cause of death in patients with EOC. Epithelial protein
lost in neoplasm (EPLIN), first discovered through its differential
expression between normal oral epithelial cells and human papilloma
virus (HPV)-immortalised oral epithelial cell lines (1), is a regulator of cytoskeletal
dynamics. It has been shown to influence actin stabilization, to
regulate actin turnover and to link the cadherin-catenin complex to
F-actin (2–4). EPLIN exists as two isoforms (EPLIN-α,
EPLIN-β) which arise due to transcription from two distinct
promoter regions (5,6). EPLIN-α appears to play key roles in
regulating actin dynamics and motility in normal cells. Cytoplasmic
expression of EPLIN-α has been detected in a fibrillar pattern,
similar to that of actin fibres (5). Research conducted in our laboratory
has previously revealed that EPLIN-α expression was dys-regulated
in clinical prostate and breast cancer samples and lower EPLIN-α
levels were associated with more aggressive cancer and poor
survival, and that EPLIN-α may impact on the angiogenic process
(7,8). It has also been suggested that the
loss of EPLIN-α expression in cancerous cells may contribute to
genomic instability and to the enhanced motility and invasiveness
of cancer cells (2–4,7–10).
All these findings indicate that EPLIN-α may act as a tumour
suppressor. Recent research has shown that the special
AT-rich-binding protein 2 (SATB2) plays a role as a novel regulator
of osteosarcoma invasion, in part via effects on EPLIN and the
cytoskeleton (11). Such findings
provide conclusive evidence that reduction of EPLIN has the
potential to disrupt cell-cell adhesion via disorganization of the
adherens junctions, which promotes IGF1R signalling. This is
followed by the attenuation of E-cadherin expression and the
formation of an EMT-like phenotype (12). Whilst the importance and role of
EPLIN-α in a number of cancers is beginning to become apparent, its
role in EOC is currently unknown.
In this study, we investigated the clinical and
biological role of EPLIN-α in human ovarian cancer. This study
provides evidence that EPLIN-α knockdown in ovarian cancer cells
can increase the aggressive nature of these cells.
Materials and methods
Clinical sample collection, processing
and IHC staining
All clinical samples examined in this study were
obtained from surgically removed ovarian tissues of inpatients in
Wuhan Tongji Hospital of Huazhong University of Science and
Technology (Wuhan, China) from 2013 to 2014; patients who had
received pre-operative radiotherapy or chemotherapy were excluded.
Immunohistochemistry was performed on 30 epithelial ovarian serous
carcinomas, 15 samples were non-metastatic and 15 had lymph node or
omentum metastases. All of the tumour samples were obtained from
the primary tumour site. Diagnosis was confirmed by histopathology
in all cases. All protocols were reviewed and approved by the
ethics committee of Wuhan Tongji Hospital, and all the patients
gave their written informed consent.
Tissue sections (4 μm) were prepared from
formalin-fixed paraffin embedded blocks. IHC was performed using
rabbit anti-EPLIN-α antibody (Calbiochem, Nottingham, UK) and the
Vectastain® Elite Universal ABC kit (Vector
Laboratories, Peterborough, UK). The de-paraffinized sections were
rehydrated in Tris-buffered saline (TBS). Antigen retrieval was
then performed by heating the samples for 20 min in a microwave in
1 mM EDTA buffer (pH 8.0). The sections were cooled and washed in
tap water (10 min). Non-specific binding was blocked with 5–10%
goat serum (90 min) and slides were then incubated with the primary
EPLIN-α antibody (1:100 in TBS) for 1 h. Following sequential
30-min incubations with mouse biotinylated secondary and ABC
complex, respectively, the target protein was visualised using
freshly prepared 3,3-diaminobenzidine (DAB) (Sigma-Aldrich, Poole,
UK). Slides were rinsed with water, counterstained with
haematoxylin, dehydrated, cleared in xylene and mounted in DPX.
Negative controls were prepared by substituting the primary
antibody with TBS. The sections were then viewed under the Leica
MC120 microscope, photographed and the intensity and localisation
of the staining was analysed.
Cell lines and culture conditions
Human ovarian epithelial-serous carcinoma cell lines
SKOV3 and COV504 (ECACC, European Collection of Animal Cell
Culture, Salisbury, UK) were routinely maintained in DMEM-F12
medium supplemented with 10% fetal bovine serum, penicillin (100
U/ml), streptomycin (100 μg/ml) and amphotericin B (0.25 μg/ml)
(Sigma). Cells were incubated at 37°C with 95% humidity in 5%
CO2.
Generation of EPLIN-α ribozyme
transgenes
Anti-Eplin-α hammerhead ribozyme transgenes were
synthesised and cloned into pEF6/V5-His-TOPO plasmid vector as
described in previous studies (13–15).
Purified EPLIN-α plasmids (8 μg) and control plasmid vectors were
then transfected into SKOV3 and COV504 cells (300 V, 1500 μF),
(1×106/ml) using a Gene Pulser X cell electroporator
(Bio-Rad Laboratories, Hemel Hempstead, UK). Transfected cells
underwent selection for ~2 weeks with blasticidin (5 μg/ml)
(Melford Laboratories, Ipswich, UK). Positively transfected cell
cultures were maintained in medium containing 0.5 μg/ml
blasticidin. The transfectants were verified for their expression
of Eplin-α mRNA and protein, and the successful clones were used in
subsequent studies.
RNA extraction and reverse transcription
PCR
Total cellular RNA was isolated from the EOC cells
using Tri-reagent according to the manufacturer's instructions
(Sigma-Aldrich). RNA concentration and quality were determined
through spectrophotometric measurement (NanoPhotometer, Implen,
Mϋnich, Germany). RNA (500 ng) was reverse transcribed into cDNA
using an Applied Biosystems high capacity reverse transcription kit
(Life Technologies, Paisley, UK). DNA quality was verified using
GAPDH PCR (sense, GGCTGCTTTTAACTCTGGTA; antisense,
GACTGTGGTCATGAGTCCTT) which was also used as a loading control.
Eplin-α mRNA levels were assessed using primers (sense,
AAGCAAAAATGAAAACTAAG; antisense, GACACCCACCTTAGCAATAG). PCR was
carried out in an Applied Biosystems thermocycler using a Go Taq
green PCR reaction mix (Promega UK, Southampton, UK). Cycling
conditions were 94°C for 5 min, followed by 28 cycles of 94°C for
30 sec, 55°C for 30 sec and 72°C for 30 sec. This was followed by a
final 7-min extension period at 72°C. The products were visualized
on 2% agarose gel stained with SYBRSafe (Life Technologies).
Immunofluorescence staining
Cells were seeded at a density of 20,000 cells per
well in an 8-well chamber slide (Merck-Millipore, UK). Following an
overnight incubation, the medium was aspirated and the cells were
fixed in 4% formalin (4°C, 20 min). Following fixation, the cells
were rehydrated in phosphate-buffered saline (PBS) for 20 min at
room temperature before being permeabilised for 5 min in a 0.1%
Triton in PBS. Non-specific binding was blocked by 1-h incubation
in phosphate-buffered saline (PBS) containing 5–10% goat serum.
Cells were incubated for 1 h with Eplin-α antibody (1:100) in PBS
blocking solution (Calbiochem). Slides were washed 3×5 min in PBS
then incubated on a shaker platform in the dark for 1 h with FITC
conjugated anti-mouse secondary antibody (Insight Biotechnology
Ltd., Middlesex, UK) and 1:1,000 DAPI (Roche, Hertfordshire, UK).
Slides were finally washed 3×5 min PBS, mounted with Fluorsave
(Merk-Millipore, UK) and visualised using an EVOS fluorescence auto
imaging system (Life Technologies).
Western blot analysis
Cell lines were grown to 70% confluence, monolayers
were washed with PBS and lysed in ice cold lysis buffer (50 mm
Tris, 150 mM NaCl, 5 mM EGTA, 1% Triton X-100 pH 7.5) supplemented
with protease inhibitor cocktail (Roche). Lysates were clarified by
centrifugation (12,000 rpm, 15 min, 4°C) and the protein
concentrations in the supernatants were determined using the DC
Protein Assay kit (Bio-Rad, Hemel Hempstead, UK). Protein was
reduced and denatured by boiling (5 min) in Laemmli buffer
(Sigma-Aldrich) and 20 μg protein samples were resolved by SDS-PAGE
and transferred onto nitrocellulose membrane (GE Healthcare Life
Sciences, Buckinghamshire, UK). After blocking for 1 h in 5%
skimmed milk (TBS/Tween: 140 mM NaCl; 50 mM Tris, 0.05% Tween pH
7.4), blots were incubated overnight at 4°C with primary antibodies
Eplin-α (1:500 prepared in TBS/Tween/1% milk) and GAPDH (1:1,000 in
TBS/Tween/1% milk) (Santa Cruz Biotechnology, Heidelberg, Germany)
was used as a loading control. Blots were washed with TBS/Tween and
bound antibodies were detected after 1-h incubation (room
temperature) with appropriate horseradish peroxidase-conjugated
secondary antibody (1:1,000, Sigma-Aldrich). Following 3×5 min
TBS/Tween washes, protein bands were visualized using enhanced
chemiluminescence (Luminata Forte, Millipore, Herefordshire, UK),
and photographed using a UVITech imager (UVITech, Inc., Cambridge,
UK).
Cell proliferation assay
Cells were seeded into 96-well plates at a seeding
density of 3,000 cells per well with 12 replicates/experiment.
Cells were fixed with 4% formalin after 1, 3 and 5 days growth.
Fixed cells were stained with 0.5% crystal violet, washed and
dried. Dye was re-solubilised in 200 μl acetic acid/well and
absorbance was determined at 540 nm using an ELx800 multi-plate
reader (BioTek UK, Bedfordshire, UK). Each experiment was repeated
at least 3 times. For each cell line, analysis compared cell number
(absorbance) on days 3 and 5 relative to day 1.
Cell adhesion assay
Cell-matrix adhesion was examined using an in
vitro Matrigel adhesion assay adapted from a previously
described method (16–18). Cells were seeded into 96-well
plates pre-coated with 5 μg/well Matrigel basement membrane matrix
(BD Biosciences, Oxford, UK). After 40 min of incubation (37°C) the
cells were washed with PBS to remove unbound cells. The remaining
adherent cells were fixed with 4% formalin, stained with 0.5%
crystal violet, visualized under a microscope (x20) and cell number
counted per field of view. Four counts were made from each 6
replicate wells and results were expressed as mean cell
number/well. Each experiment was repeated 3 times.
Cell invasion assay
Cell invasive capability was examined using an in
vitro Matrigel invasion assay. Transwell inserts (Greiner
Bio-One, Stonehouse, UK) with 8.0-μm pore size were coated with 50
μg Matrigel (BD Biosciences), dried at 55°C and rehydrated with 100
μl serum-free medium before seeding 4,000 cells per insert. After
48 h of incubation at 37°C, non-invasive cells and Matrigel were
removed from the inside of the inserts with a cotton swab. Cells
that had invaded to the underside of the insert were fixed (4%
formalin), stained with 0.5% crystal violet and washed. Cell
invasion was quantitated by counting the cell number in 4 fields of
view (x20 magnification). Data were analysed as mean cell number
per field of view for 3 independent experiments with 3 replicates
per experiment. Results were confirmed by incubating the stained
inserts in 10% acetic acid. Absorbance of solubilized crystal
violet was determined at 540 nm.
Migration assay
A cellular wounding assay was used to study
directional cell migration in vitro as previously described
(19). In brief, cells were
cultured to confluence in a 24-well plate before scratching the
cell monolayer with a 10-μl pipette tip. The closure of the induced
wound, through the migration of cells, was tracked and recorded
over a 36-h period using an automated cell imaging system EVOS
(Life Technologies). Using ImageJ software, the relative distance
cells migrated was calculated using multiple measurements of the
width of wound gap after 12, 24 and 36 h compared to 0 h.
Electric cell-substrate impedance sensing
(ECIS)-based attachment and migration assay
Cell attachment and migration were further studied
using an ECIS Z-Theta instrument and 96W1E arrays (Applied
Biophysics, Inc., NY, USA) as previously described (7). Briefly, 40,000 cells per well were
added to the ECIS arrays. Impedance and resistance of the cell
layer was immediately recorded for a period of ≤5 h. When
confluence was reached, the monolayer in each well was electrically
wounded at 2,600 μA and 6,0000 Hz for 20 sec to create a 250-μm
wound per well. Impedance and resistance of the wounded cells as
they migrated in the wound was then recorded for a period of up to
10 h. Data were analysed using the ECIS software, supplied by the
manufacturer.
Statistical analysis
All statistical analysis was performed using the
paired t-test for normally distributed data (data were tested for
normal distribution before further statistical analyses were
carried out). Differences were considered to be statistically
significant at p<0.05.
Results
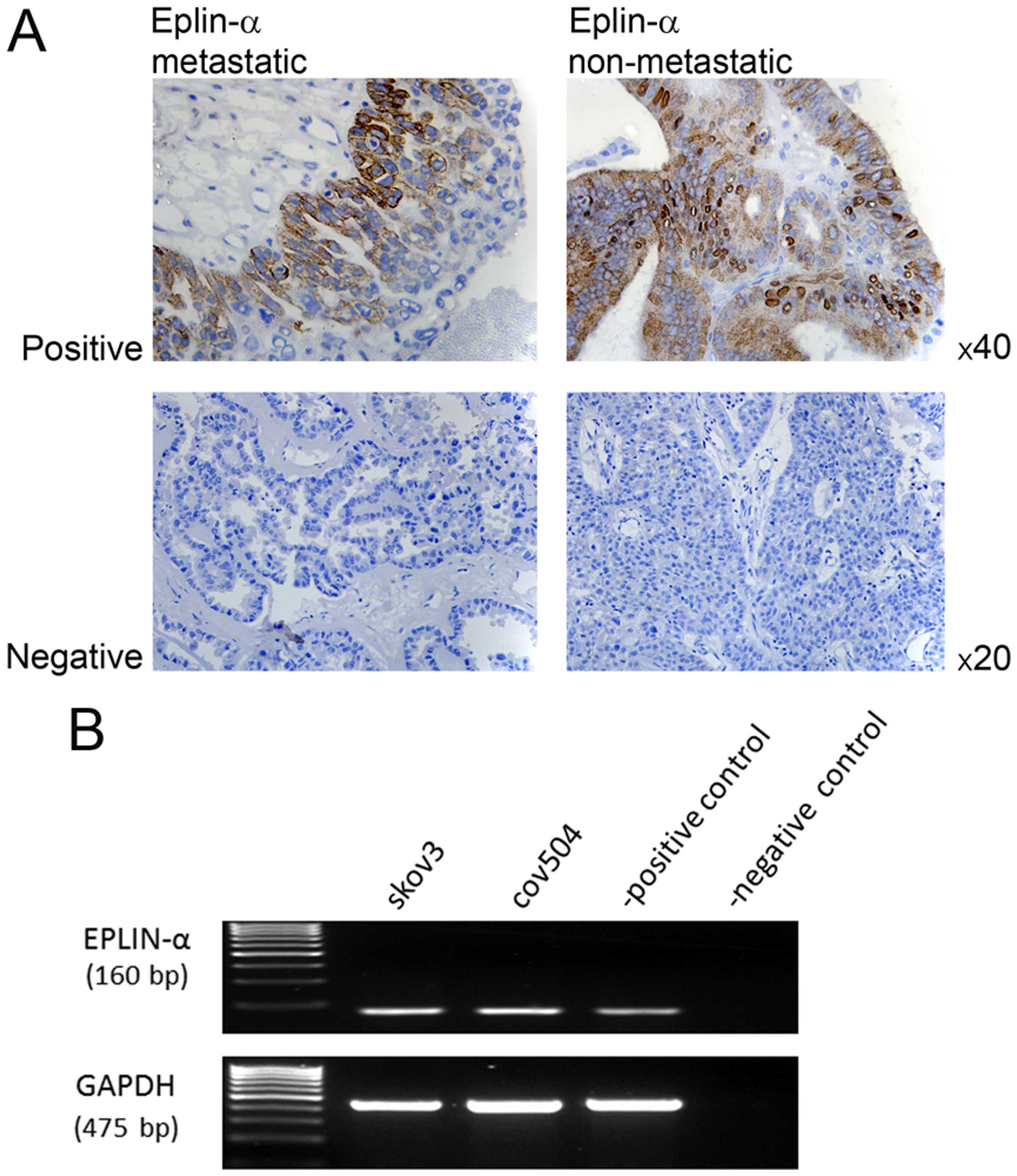
Expression of Eplin-α in human ovarian
tissues and EOC cells
IHC staining of 15 sections of non-metastastic and
15 sections of metastatic epithelial cancerous ovarian growths was
used to assess Eplin-α expression pattern in the clinical setting.
Eplin-α expression was detected in 6/30 tissue samples examined;
2/15 non-metastatic tumours and 4/15 metastatic tumours were
localised in epithelial cell cytoplasm. Where staining was observed
there was no difference in the intensity of the stain between
non-metastatic and metastatic samples. Images are shown of both
metastatic and non-metastatic samples, representing the range of
staining detected (Fig. 1A).
The mRNA expression of Eplin-α was also examined in
two EOC cell lines using RT-PCR. Eplin-α mRNA was expressed at
relatively high levels in all cell lines (Fig. 1B).
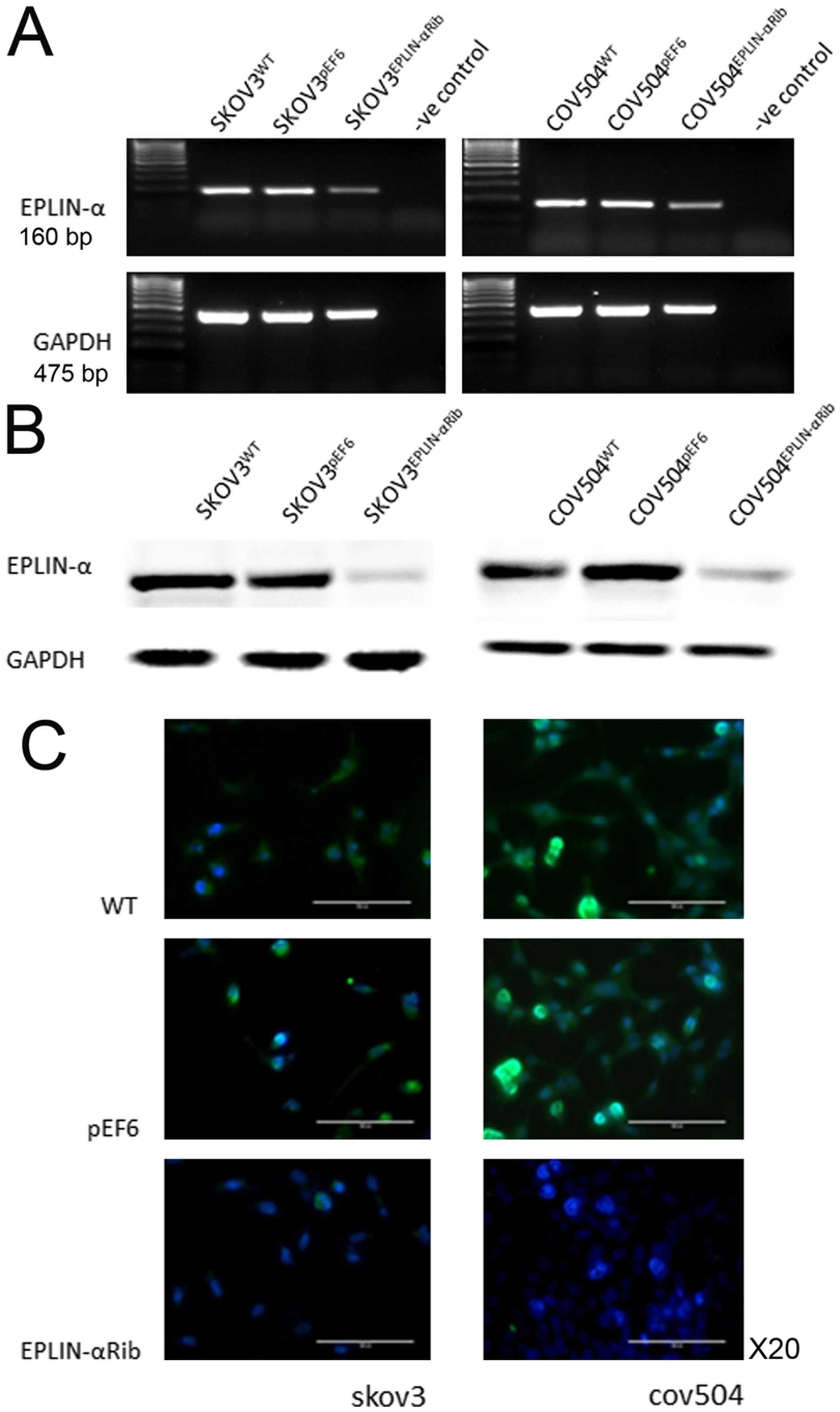
Knock-down of Eplin-α in EOC cells
To investigate the impact of Eplin-α on a range of
behavioural functions of ovarian cancer cells, Eplin-α hammerhead
ribozyme transgenes were utilised to knock down Eplin-α. After
selection using blasticidin, the reduced expression of Eplin-α in
the transfected cells was verified using RT-PCR, immunofluorescent
staining and western blotting (Fig.
2). Decreased expression of both Eplin-α mRNA (~50%) (Fig. 2A) and protein (~80%) (Fig. 2B) was seen in
SKOV3Eplin-αRib, in comparison with the controls
(wild-type SKOV3WT and empty plasmid
SKOV3pEF6). Knock-down of Eplin-α mRNA (~40%) and
protein (~80%) was also confirmed in COV504Eplin-αRib
cells, in comparison with the COV504WT and
COV504pEF6 control cells. Immunofluorescent staining was
carried out to examine the expression and localisation of the
Eplin-α in the transfected cells (Fig.
2C). Eplin-α staining (green), was predominantly associated
with the cytoplasm. Control cells, both wild-type and empty vector
transfectants, had strong staining intensity in two EOC cell lines,
with the majority of the cells showing more intense and frequently
observed staining. However, in the transfected knocked down cells
Eplin-α had minimal staining levels.
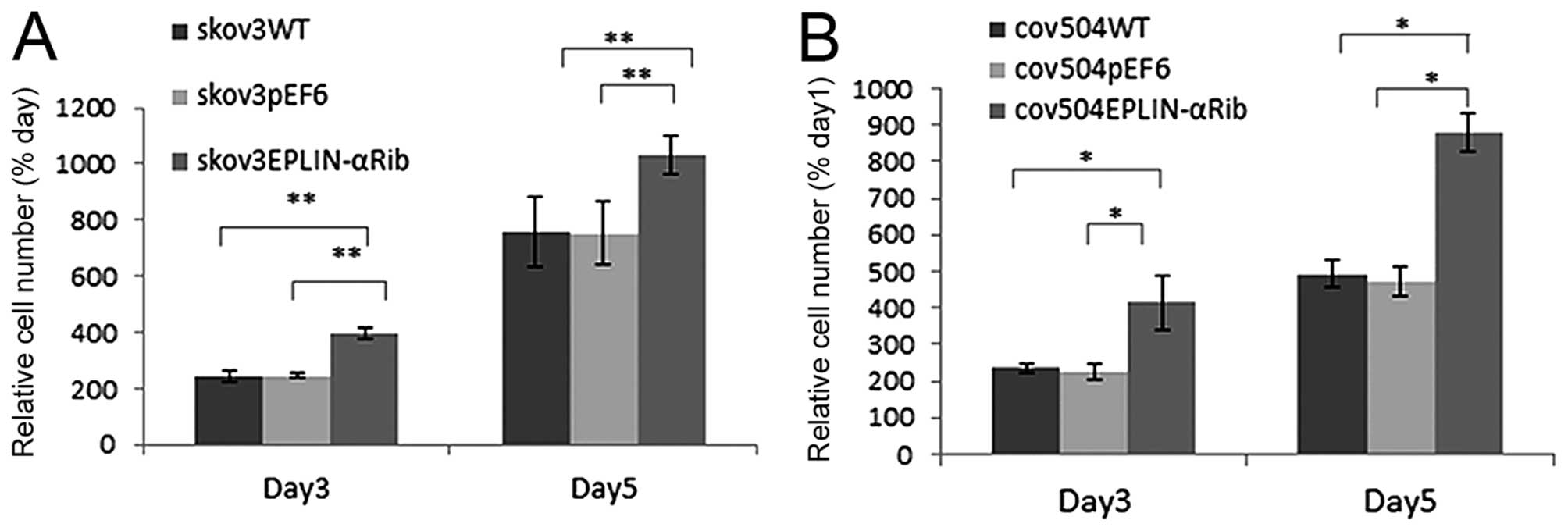
Regulation of Eplin-α expression affects
the rate of cell growth of EOC cells
The growth capacity of the EOC cells following
Eplin-α knock-down was examined and compared to the wild-type and
empty vector control cells using an in vitro cell growth
assay (Fig. 3). In all transfected
cells, knock-down of Eplin-α protein increased growth rate by both
day 3 and day 5. In SKOV3Eplin-αRib cells, the mean cell
number at day 5 was increased by 36% (p<0.01) compared to pEF6
control, in COV504Eplin-αRib cells, growth rate
increased by 46% (p<0.05) (Fig.
3).
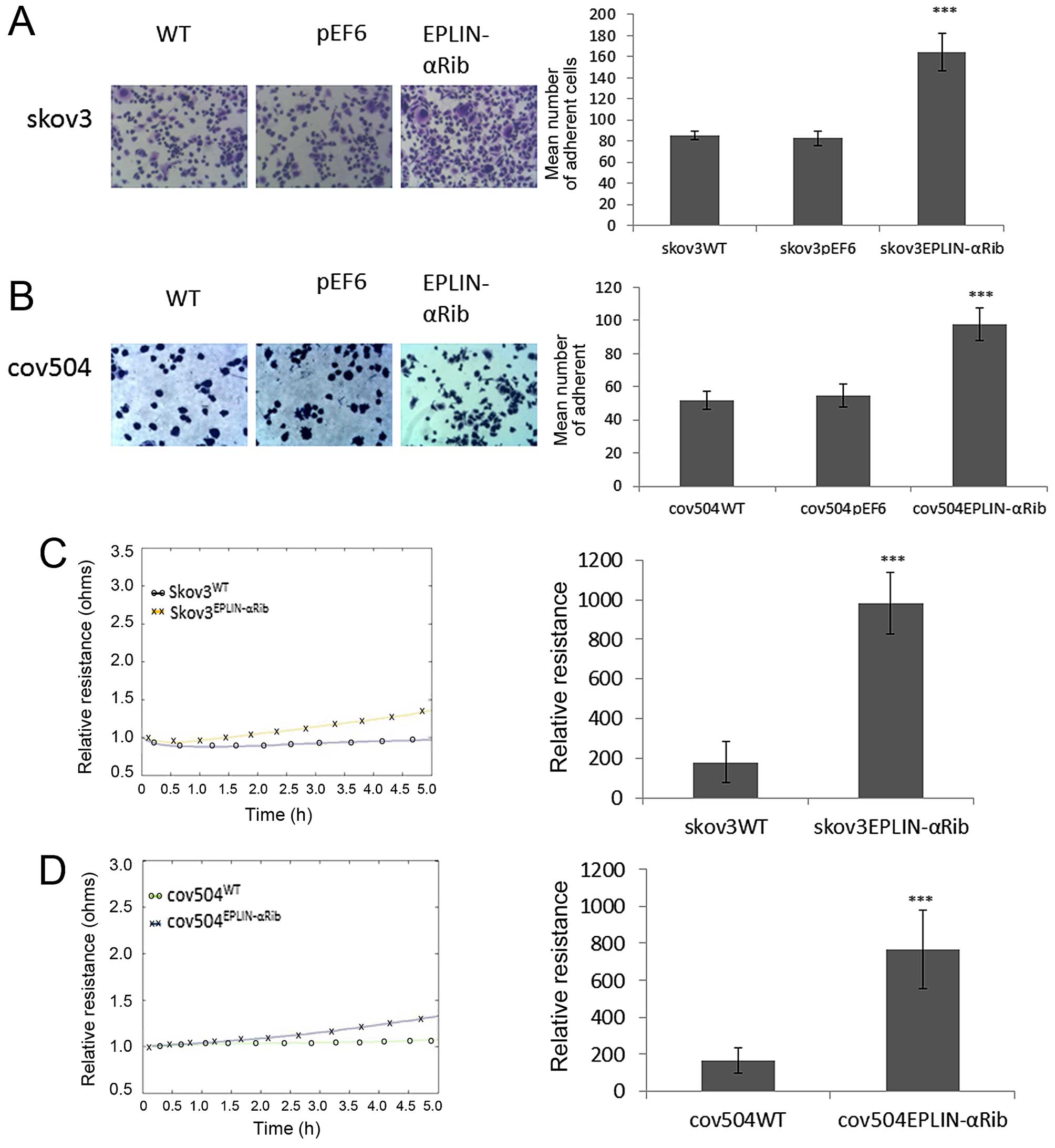
Effect of knocked down Eplin-α on
cell-matrix adhesion in EOC cells
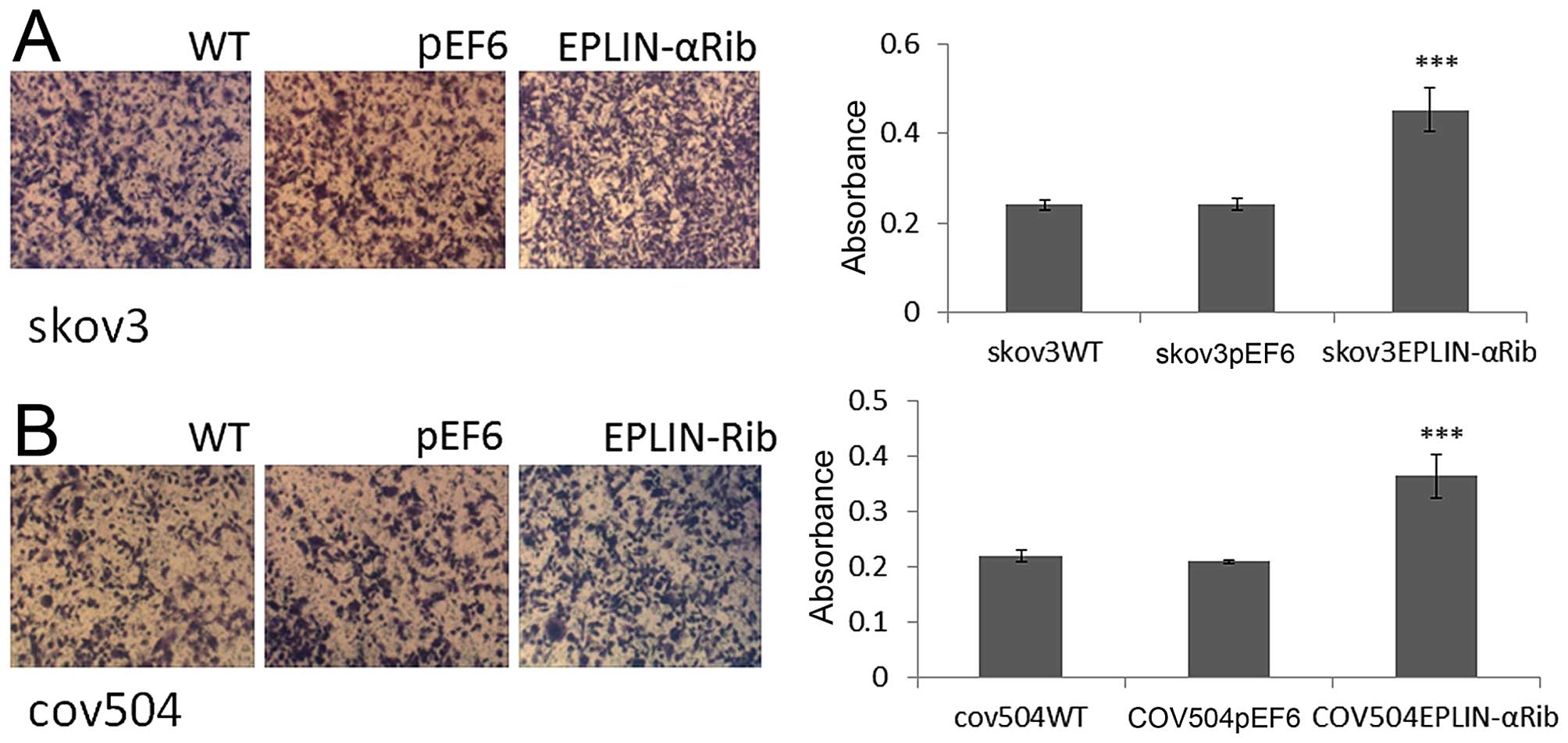
The effect of Eplin-α on the ability of EOC cells to
adhere to Matrigel matrix was examined (Fig. 4A and B). Knock-down of Eplin-α
protein caused a significant (p<0.001) increase of ~40% on
cell-matrix adhesion in the SKOV3 cells compared to both WT and
pEF6 controls (Fig. 4A). Compared
with COV504WT and COV504pEF6, the number of
COV504Eplin-αRib cells that adhered was also
significantly increased (p<0.001) by ~40% (Fig. 4B). The ECIS system was also used to
confirm the accelerative effect of reduced expression of Eplin-α on
SKOV3, COV504 cell adhesion. This was measured by change in
resistance formed over the growth surface as cells attached from 0
to 5 h (Fig. 4C and D). Compared
with the appropriate WT and pEF6 controls, the resistance was
significantly increased in SKOV3Eplin-αRib and
COV504Eplin-αRib cells, confirming that low expression
of Eplin-α in ovarian cells increased the adhesive capability.
Effect of knocked down Eplin-α on the
invasion of EOC cells
The potential biological relevance of reduced
Eplin-α expression was further investigated using in vitro
invasion assays over the artificial basement membrane, Matrigel.
Reduced expression of Eplin-α in all of these cell lines caused a
50% increase (p<0.001) in basal invasion compared to both WT and
pEF6 controls (Fig. 5).
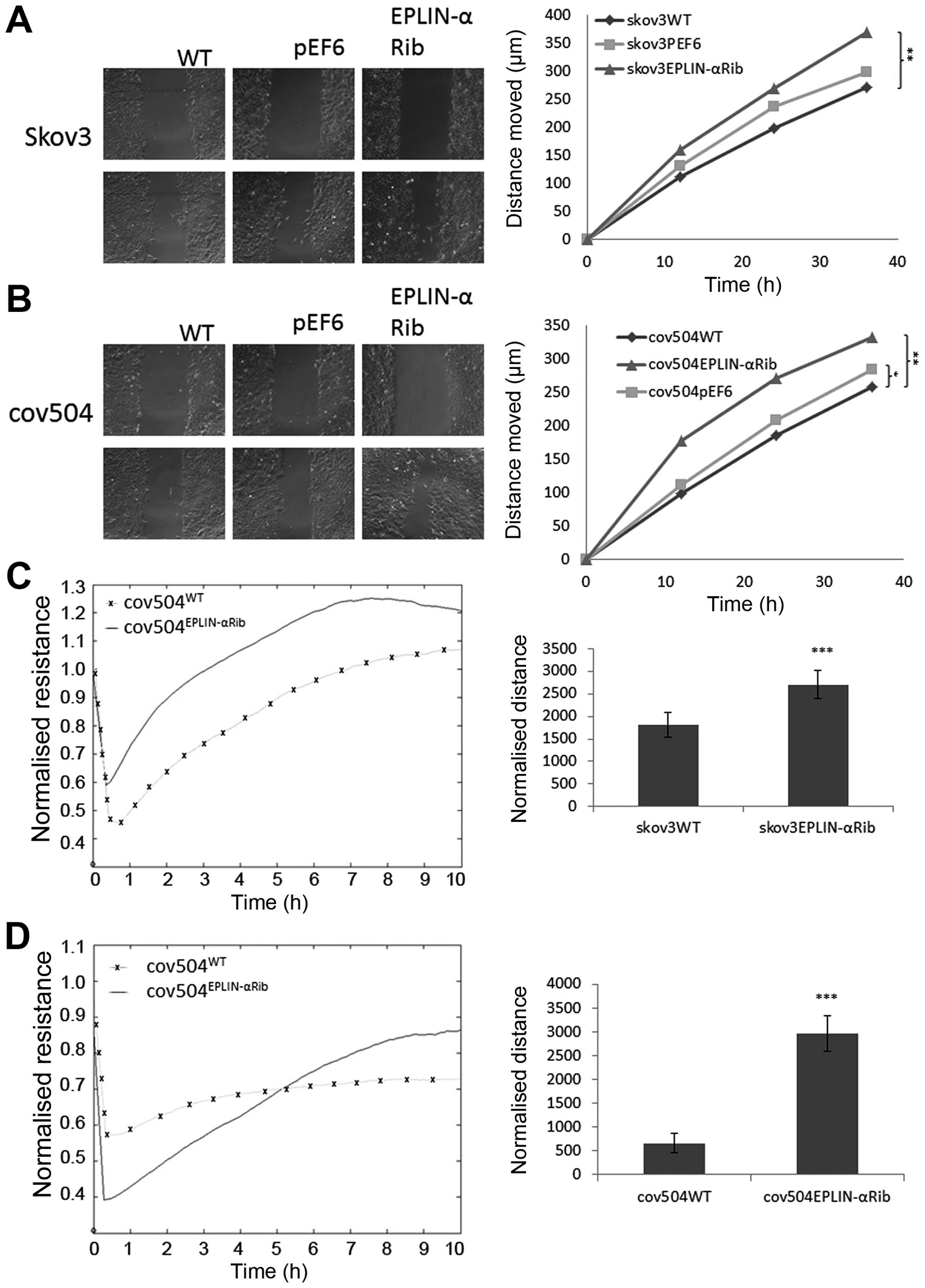
Effect of Eplin-α on wounding/migration
of EOC cells
A cellular wounding assay (Fig. 6A and B) was used to compare the
ability of wild-type and knock-down Eplin-α cells to migrate.
Reduced expression of Eplin-α in the two EOC cell lines caused a
marked (≤30%) increase in migration capability compared to the WT
control (p<0.05). The reduced ability of Eplin-α expression to
increase ovarian cell motility was also confirmed by measuring the
ability of cell lines to recover from an electrical wound generated
using the ECIS system (Fig. 6C and
D). Measurements taken 10 h post-wound also showed that the
migration capacity of SKOV3Eplin-αRib and
COV504Eplin-αRib were markedly increased (p<0.001) in
comparison with wild-type control cells.
Discussion
Uncontrolled tumour cell proliferation and robust
neovascularization are prominent features of aggressive ovarian
cancers, distant metastasis being a key factor in the poor
prognosis associated with this cancer. Although great efforts in
antiovarian cancer therapy have been made in the past decades, the
5-year survival rates for ovarian cancer patients are still poor,
and effective drugs to cure ovarian cancer patients are absent. To
improve the prognosis, assessment and treatment of EOC patients, it
is crucial that we identify the key molecular regulators of
tumourigenesis and understand the key molecular pathways involved.
However, many of these tumourigenesis mechanisms remain largely
unknown.
Eplin-α (epithelial protein lost in neoplasm) has
previously been found to localise with actin stress fibres and
plays an important role in regulating actin dynamics and linking
the catenin-cadherin complex to F-actin (2–4),
suggesting a key role for this molecule in regulating cellular
motility. It has been previously reported that Eplin-α has been
found to be downregulated in a number of oral, breast, esophageal
and prostate (5,7,8,20)
cancer cell lines compared to their normal counterparts. Previous
studies from our laboratories have provided data supporting a
tumour/metastasis suppressive role for EPLIN-α, where enhanced
levels of EPLIN-α can negatively impact on key metastatic and
angiogenic traits in vitro and in vivo (7,8,21).
Our study aimed to determine whether there was a relationship
between Eplin-α protein expression and the aggressiveness of
clinical ovarian cancer. Immunohistochemical analysis demonstrated
that only a small number of ovarian cancer tissues expressed
Eplin-α protein, but conclusions were limited by the relatively
small sample size used. The majority of tumours, both metastatic
and non-metastatic, were negative for Eplin-α (80%), and in the
tissues where positive staining was seen there appeared to be no
distinct difference in the localisation and intensity of stain. Due
to the relatively small number of clinical samples included in our
study, the results did not enable statistical analysis. In some
tumour types, including breast and esophageal cancer, EPLIN-α may
be considered a suitable biomarker for tumour progression, with
high EPLIN-α being associated with favourable prognosis and reduced
EPLIN-α a poorer outcome (7,20).
EPLIN-α expression is variable between individuals which may
reflect a range of differences in ovarian cancer aetiology or
disease stages when samples were taken. Conclusive results can only
be obtained if a larger cohort is studied.
A relatively small number of in vitro studies
have reported in which the function of EPLIN-α is characterized.
These studies with a panel of human cell lines have shown EPLIN-α
is differentially expressed with an inverse correlation between
cell differentiation, invasive capability and EPLIN-α expression
(5,7,8,20),
for example, MDA-MB-231 cells, considered highly invasive, were
negative for Eplin-α expression (7). Our present investigation showed that
the ovarian tumour cell lines, SKOV3 and COV504, which both
demonstrated reasonable (but not high) invasive capability, also
expressed a relatively high level of EPLIN-α mRNA and protein
suggesting other factors in addition to EPLIN-α expression may also
be involved in regulating invasion. However, cellular function
tests did demonstrate that the presence of EPLIN-α was related to
the inhibition of the ovarian cancer cell aggressiveness.
Knock-down of EPLIN-α expression resulted in an increase in the
in vitro growth rate of SKOV3 and COV504 cells during the 3
and 5-day incubation periods (p<0.01, p<0.05). Knock-down of
EPLIN-α expression also impacted cell-matrix adhesion significantly
decreasing it compared to that of pEF6 control cells.
We have demonstrated here that knock-down of EPLIN-α
expression resulted in a strong increase in ovarian cancer cell
line growth, adhesion, invasion and migration, in comparison with
control cells. The inhibitory effect of EPLIN-α on ovarian cancer
cell growth is in agreement with the findings in breast, prostate,
esophageal and endothelial cell lines (7,8,20,21).
Although the precise molecular mechanisms by which EPLIN-α inhibits
tumour growth remains unknown, research studies provide conclusive
evidence that reduction of EPLIN has the potential to disrupt
cell-cell adhesion via disorganization of the adherens junctions,
which promotes IGF1R signalling. This is followed by the
attenuation of E-cadherin expression and an EMT-like phenotype
(12). Further studies are
required to reveal the exact molecular mechanisms and signalling
pathways through which EPLIN-α modulates cancer cell migration and
invasion.
Our studies suggest that decreased expression of
EPLIN-α is seen in cancerous tissue where it may be associated with
a poorer prognosis making EPLIN-α loss a potential prognostic
marker. As far as the authors are aware, this is the first study
examining the expression of EPLIN-α in human EOC tissue and
reporting the effect of reduced EPLIN-α expression on EOC cell line
behaviour.
In conclusion, this study shows that the knock-down
of EPLIN-α protein can increase the aggressiveness of human ovarian
cancer, furthermore, it suggests that preventing EPLIN-α
degradation, or partially restoring EPLIN-α expression, could be a
possible novel strategy to treat aggressive ovarian cancer growth
and metastasis. These data clearly indicate that EPLIN-α may
potentially have use as a prognostic indicator and that the
molecule may act as a protective factor in patients with EOC.
Acknowledgements
The authors are grateful to Cancer Research Wales
and the Albert Hung Foundation for their support and funding of
this study. Dr Rong Liu is a recipient of Cardiff University China
Medical Scholarship.
References
|
1
|
Chang DD, Park NH, Denny CT, Nelson SF and
Pe M: Characterization of transformation related genes in oral
cancer cells. Oncogene. 16:1921–1930. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song Y, Maul RS, Gerbin CS and Chang DD:
Inhibition of anchorage-independent growth of transformed NIH3T3
cells by epithelial protein lost in neoplasm (EPLIN) requires
localization of EPLIN to actin cytoskeleton. Mol Biol Cell.
13:1408–1416. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Maul RS, Song Y, Amann KJ, Gerbin SC,
Pollard TD and Chang DD: EPLIN regulates actin dynamics by
cross-linking and stabilizing filaments. J Cell Biol. 160:399–407.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Abe K and Takeichi M: EPLIN mediates
linkage of the cadherin catenin complex to F-actin and stabilizes
the circumferential actin belt. Proc Natl Acad Sci USA. 105:13–19.
2008. View Article : Google Scholar
|
|
5
|
Maul RS and Chang DD: EPLIN, epithelial
protein lost in neoplasm. Oncogene. 18:7838–7841. 1999. View Article : Google Scholar
|
|
6
|
Chen S, Maul RS, Kim HR and Chang DD:
Characterization of the human EPLIN (epithelial protein lost in
neoplasm) gene reveals distinct promoters for the two EPLIN
isoforms. Gene. 248:69–76. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jiang WG, Martin TA, Lewis-Russell JM,
Douglas-Jones A, Ye L and Mansel RE: Eplin-alpha expression in
human breast cancer, the impact on cellular migration and clinical
outcome. Mol Cancer. 7:71–80. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sanders AJ, Martin TA, Ye L, Mason MD and
Jiang WG: EPLIN is a negative regulator of prostate cancer growth
and invasion. J Urol. 186:295–301. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang S, Wang X, Osunkoya AO, Iqbal S,
Wang Y, Chen Z, Müller S, Chen Z, Josson S, Coleman IM, et al:
EPLIN downregulation promotes epithelial-mesenchymal transition in
prostate cancer cells and correlates with clinical lymph node
metastasis. Oncogene. 30:4941–4952. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Han MY, Kosako H, Watanabe T and Hattori
S: Extracellular signal-regulated kinase/mitogen-activated protein
kinase regulates actin organization and cell motility by
phosphorylating the actin cross-linking protein EPLIN. Mol Cell
Biol. 27:8190–8204. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Seong BKA, Lau J, Adderley T, Kee L,
Chaukos D, Pienkowska M, Malkin D, Thorner P and Irwin MS: SATB2
enhances migration and invasion in osteosarcoma by regulating genes
involved in cytoskeletal organization. Oncogene. 34:3582–3592.
2015. View Article : Google Scholar
|
|
12
|
Steder M, Alla V, Meier C, Spitschak A,
Pahnke J, Fürst K, Kowtharapu BS, Engelmann D, Petigk J, Egberts F,
et al: DNp73 exerts function in metastasis initiation by
disconnecting the inhibitory role of EPLIN on IGF1R-AKT/STAT3
signaling. Cancer Cell. 24:512–527. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jiang WG, Watkins G, Douglas-Jones A,
Mokbel K, Mansel RE and Fodstad O: Expression of Com-1/P8 in human
breast cancer and its relevance to clinical outcome and ER status.
Int J Cancer. 117:730–737. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jiang WG, Davies G and Fodstad O: Com-1/P8
in oestrogen regulated growth of breast cancer cells, the ER-beta
connection. Biochem Biophys Res Commun. 330:253–262. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang WG, Davies G, Martin TA, Kynaston H,
Mason MD and Fodstad O: Com-1/p8 acts as a putative tumour
suppressor in prostate cancer. Int J Mol Med. 18:981–986.
2006.PubMed/NCBI
|
|
16
|
Jiang WG, Davies G, Martin TA, Parr C,
Watkins G, Mason MD and Mansel RE: Expression of membrane type-1
matrix metalloproteinase, MT1-MMP in human breast cancer and its
impact on invasiveness of breast cancer cells. Int J Mol Med.
17:583–590. 2006.PubMed/NCBI
|
|
17
|
Jiang WG, Davies G, Martin TA, Parr C,
Watkins G, Mason MD, Mokbel K and Mansel RE: Targeting matrilysin
and its impact on tumor growth in vivo: The potential implications
in breast cancer therapy. Clin Cancer Res. 11:6012–6019. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang WG, Hiscox S, Singhrao SK, Nakamura
T, Puntis MC and Hallett MB: Inhibition of HGF/SF-induced membrane
ruffling and cell motility by transient elevation of cytosolic free
Ca2+. Exp Cell Res. 220:424–433. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiang WG, Hiscox S, Hallett MB, Horrobin
DF, Scott C and Puntis MCA: Inhibition of invasion and motility of
human colon cancer cells by gamma linolenic acid. Br J Cancer.
71:744–752. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liu Y, Sanders AJ, Zhang L and Jiang WG:
EPLIN-Alpha expression in human oesophageal cancer and its impact
on cellular aggressiveness and clinical outcome. Anticancer Res.
32:1283–1289. 2012.PubMed/NCBI
|
|
21
|
Sanders AJ, Ye L, Mason MD and Jiang WG:
The impact of EPLINα (Epithelial protein lost in neoplasm) on
endothelial cells, angiogenesis and tumorigenesis. Angiogenesis.
13:317–326. 2010. View Article : Google Scholar : PubMed/NCBI
|