Introduction
Osteosarcoma is the most common malignant bone tumor
with a 5-year survival rate <70% (1). Most patients present with lung
metastatic lesions (2,3) or bone metastatic lesions and are
incurable. Hence, a better understanding of the biological
processes underlying osteosarcoma cell motility, survival,
proliferation, invation and metastasis is needed to improve patient
survival. New gene targeting therapy is a strong hope for
osteosarcoma individual tumors.
The 90-kDa ribosomal S6 kinase (RSK) family, first
purified in 1985 (4), is activated
by the MAPK (mitogen-activated protein kinase) family members
ERK1/2 (extracellular signal-regulated kinase 1/2) in response to
growth factors, phorbolesters and other agonists (5–7). The
human RSK family contains four isoforms (RSK1-4) (8). RSKs are characterised by the
existence of two kinase domains that come into close proximity
following activating phosphorylation events and connected by a
regulatory linker region. Their downstream substrates include a
number of cytoplasmic and nuclear targets (CREB, c-Fos, c-Jun, TSC2
and filamin A) (8) that explain
their involvement in diverse cellular processes, such as cell
proliferation and survival. Increased expression of RSKs was shown
in breast (9) and prostate cancer
(10), and RSK2 activity has been
linked to cell transformation (11,12).
RSKs have been shown to phosphorylate filamin A (13), glycogen synthase kinase-3 (14–16)
and p27Kip (17), and also Bad
(18), c-Fos (19) and estrogen receptor (20). Evidence from human and mouse has
identified an important role for RSK2 in osteoblast differentiation
and function through phosphorylation of activating transcription
factor-4 (21) and in stimulation
of white adipose tissue mass via an unknown mechanism (22).
In this study, immunohistochemical staining revealed
that RSK2 was overexpressed in osteosarcoma samples compared with
normal matched tissues. Then we performed an shRNA in three human
osteosarcoma cell lines (MG-63, U2-OS, 143B) and demonstrated that
RSK2 silencing increased apoptosis and chemosensitivity, reduced
proliferation, migration and oncogenesis. This could potentially be
explained by activation of Bax and inhibition of Bcl2, c-Fos
phosphorylation and AKT, mTOR phosphorylation, a series of cell
factors that are associated with cell viability and apoptosis.
Thus, in osteosarcomas, our results suggest that knockdown of RSK2
increased cell apoptosis, enhanced cell chemosensitivity, inhibits
proliferation and migration, and weakened tumor formation, RSK2
might be a potential target of biotherapy to osteosarcomas.
Materials and methods
Reagents
Fetal bovine serum (FBS) and DMEM were purchased
from Gibco (San Francisco, CA, USA). Primary antibodies: rabbit
anti-human RSK2 was from Bioworld (USA). Rabbit anti-human total
c-Fos, rabbit anti-human total AKT, rabbit anti-human total mTOR,
and phosphorylated ERK, phosphorylated AKT, phosphorylated mTOR
were from Cell Signaling Technology (Boston, MA, USA). Rabbit
antihuman Bax, Bcl2, caspase-3, rabbit anti-human Ki67 nuclear
antigen, mouse anti-human PCNA, mouse anti-human β-actin and mouse
anti-human glyceraldehydes-3-phosphate dehydrogenase (GAPDH) were
from Santa Cruz Biotechnology (San Francisco, CA, USA). Horseradish
peroxidase-conjugated goat anti-rabbit and goat anti-mouse
secondary antibodies were from Zhong Shan Golden Bridge
Biotechnology (Beijing, China).
shRNA
The RSK2 shRNA were designed and synthesized by
Genechem Co. Ltd. (Shanghai, China) and the sequences targeting
RSK2 were 5′-TGCCACAATACCAACTAAA-3′. A non-specific scrambled shRNA
with a sequence of 5′-TTCTCCGAACGTGTCACGT-3′ was used as a negative
control. Transfection with shRNA was accomplished using cationic
liposome (Lipofectamine 2000; Invitrogen, Carlsbad, CA, USA),
according to the manufacturer's instructions.
Specimen collection
Specimens were collected from 30 patients with
osteosarcomas by excisional or needle biopsy at initial medical
examinations in Cancer Hospital of Guizhou Medical University
between October 2012 and September 2015. All tumor biopsies were
collected by excisional biopsy or needle core biopsy at the time of
initial diagnosis, before preoperative chemotherapy or
radiotherapy, with informed consent from patients/guardians and
approval from the relevant institutional Research Ethics
Committees. All specimens were confirmed by histopathological
examination. The patients were divided into IA, IB, IIA, IIB and
III grade according to the GTM staging (data not shown).
Immunohistochemistry (IHC)
Antigen retrieval on the deparaffinized sections was
performed by immersing the specimens in 0.1 M citrate buffer (pH
6.0), boiling the sections in the microwave for 10 min, and then
allowing the sections to cool to room temperature. Endogenous
peroxidase activity was blocked by immersing the sections in
methanol containing 3% hydrogen peroxide for 10 min. Then the
sections were blocked in FCS for 10 min at room temperature. The
sections were incubated overnight at 4°C with the RSK2 antibody
(1:50), washed in phosphate-buffered saline (PBS) three times for 5
min each. The sections were incubated with the secondary antibody
at 37°C for 30 min, washed in phosphate-buffered saline (PBS) three
times for 5 min each. Streptavidin conjugated peroxidase was added
for 10 min at room temperature. Diamino-benzidine substrate was
added for 5 min for visualizing. Immunohistochemical staining of
RSK2 was calculated as both percentage of positive cells and color
intensity. The percentage of the positivity of staining was graded
as 0 (negative), 1 (<10%), 2 (10–50%) and 3 (>50%). The
intensity of staining was scored as 0 (absent), 1 (light yellow), 2
(yellowish brown) and 3 (brown). The staining index (SI) was used
for assessing the expression of RSK2. SI, proportion score ×
intensity score: 0–2 was categorized as negative (1–2 as low
expression), 3–9 was positive (3–4 as moderate expression, 6 and 9
as high expression).
Cell lines and cell culture
Three human OS cell lines (MG63, 143B and U2OS) were
recently purchased from Shanghai Life Academy of Sciences cell
library (Shanghai, China). All cells were cultured and maintained
in Dulbecco's modified Eagle's medium (DMEM; Hyclone, Logan, UT,
USA) supplemented with 10% heat-inactivated fetal bovine serum
(FBS; Hyclone) and antibiotics (100 IU/ml penicillin and 100 mg/ml
streptomycin; Hyclone) in a humidified incubator with 5%
CO2 at 37°C.
Knockdown of RSK2 in OS cells
OS cells were seeded with DMEM supplemented with 10%
FBS in 96-well plates (Costar Corning Inc., NY, USA) at
1–2×105 cells/well or 6-well plates (Costar Corning
Inc.) at 1–2×106 cells/well and incubated overnight in
an incubator with 5% CO2 at 37°C. RSK2 shRNA and
scrambled shRNA were diluted in deionized distilled water (DDW)
according to the manufacturer's instructions. Diluted shRNAs were
complexed in 0.5 ml of cationic liposome dissolved in 1 ml DDW for
96-well plates (10 ml of cationic liposome for 6-well plates) and
were incubated at room temperature for 20 min. Then, 0.5 ml of
shRNA/liposome complexes were added to each well in 96-well plates
(10 ml for 6-well plates), and cells were incubated in an incubator
with 5% CO2 at 37°C.
Measurement of cell proliferation
The change of cell proliferation was observed with
Cell Counting Kit-8 (CCK-8) assay. All cells were seeded in 96-well
cell culture cluster plates at a density of 2×104
cells/well in 100 μl culture after transfecting RSK2-shRNA and
control shRNA. Then, 10 μl CCK-8 (Beyotime Institute of
Biotechnology, Beijing, China) reagents were added to each well for
2-h incubation at 37°C according to the manufacturer's instructions
after 24, 48, 72, 96 and 120 h. The absorbance value was read at
450 nm using an enzyme-labeled instrument. The experiments were
repeated three times.
Flow cytometry analysis (FCM) of cell
cycle distributions
OS cells were transfected with RSK2-shRNA and
scramble shRNA, then collected at 48 h after transfection in
suspension to each tube. For cell cycle analysis,
starvation-refeeding model was used. To begin with, OS cells were
incubated without fetal bovine serum for 48 h to synchronize cells,
then changed into complete medium and collected cells. Furthermore,
cells were fixed in 70% ethanol for ≥24 h at −20°C. Subsequently,
the cells incubated with 1 mg/ml RNase (Sigma, St. Louis, MO, USA)
for 30 min at 37°C in PBS, stained with 50 μg/ml propidium iodide
(Sigma) in PBS-Triton X-100 for an additional 20 min at 4°C, and
analyzed using a Becton-Dickinson flow cytometer BD FACScan (San
Jose, CA, USA) as well as CellQuest acquisition and analysis
programs. For cell apoptosis analysis, OS cells were transfected
with RSK2-shRNA and control shRNA, then the above cells were
collected in suspension to each tube and 60 μl Muse™ Annexin V and
Dead Cell Reagent (part no. 4700–1485, 100 tests/bottle) was added,
incubating for 20 min. The apoptosis assay was completed by flow
cytometer evaluation (BD Bioscience, Franklin Lakes, NJ, USA).
Cell migration assay
The Transwell chambers (Corning Inc.) were used to
analyse the migration of OS cells. The cells (2×105)
were seeded in the upper of the 8-μm pore size Transwell chambers
in 0.6 ml DMEM without serum and incubated in 6-well-plates with 2
ml 10% FBS supplemented DMEM. Non-migrated cells were removed by
cotton swab after incubated for 24 h. The cells were washed twice
with phosphate-buffered solution (PBS) and fixed with 4%
paraformaldehyde in PBS (pH 7.4) for 30 min at 4°C and stained with
100 ng/ml crystal violet for 10 min at room temperature. Cells were
washed twice with PBS and examined by a fluorescence
microscope.
Semi-quantitative RT-PCR
Total RNA in cells was extracted using RNAiso Plus
(Invitrogen). The concentration of these RNA samples was then
measured using spectrophotometer at 260 and 280 nm (A260/280) and
the RNA samples were reverse-transcribed into cDNA using the
Primescript RT reagent kit (Takara Biotechnology, Dalian, China).
The primer sequence for RSK2 was 5′-GGGACCAACTGCCACAATAC-3′
(forward) and 5′-TGACTGATTACGGTTCAAAGCA-3′ (reverse), and for
GAPDH, was 5′-CTTTGGTATCGTGGAAGGACTC-3′ (forward) and reverse
5′-GTAGAGGCAGGGATGATGTTCT-3. Amplification conditions were as
follows: 95°C for 30 sec, followed by 40 cycles at 95°C for 15 sec,
and 60°C for 45 sec in a final volume of 25 μl containing 2× PCR Mi
× 12.5 μl, 20 μmol specific forward and reverse primers 1.0 μl,
ddH2O 9.5 and 1 μl cDNA as a template. Products were
electrophoresed on a 2% agarose gel, and densitometric analysis of
DNA bands was tested by Quantity One 4.6 computer software
(Bio-Rad, Hercules, CA, USA).
Western blotting
Cells were treated with shRNA in 100-mm cell culture
dishes (Costar Corning Inc.). Cells were harvested at 24, 48, and
72 h after transfection with shRNA. Cells were washed twice
ice-cold phosphate buffer saline (PBS, pH 7.4; Sigma-Aldrich) and
lysed with 100 μl lysis buffer (Beyotime Institute of
Biotechnology) containing phosphatase inhibitors, proteinase
inhibitor and PMSF. The protein concentration of each lysate was
determined by Bradford protein assay using bovine serum albumin
(Beyotime Institute of Biotechnology) and were denatured with
SDS-PAGE loading buffer. Proteins (30 mg for each sample) were
subjected to SDS-PAGE using 10% polyacrylamide gels (Beyotime
Institute of Biotechnology) and electro-transferred into
polyvinylidene difluoride membrane (PVDF, 0.45). Membranes were
blotted with 5% BSA (Beyotime Institute of Biotechnology) or 5%
skimmed milk powder in PBST for 2 h at room temperature. Membranes
were incubated overnight at 4°C with primary antibodies against
β-actin (ABM-0001, Zoonbio, Nanjing, China) and RSK2 (Bioworld,
USA) at a dilution of 1:1,000 in antibody dilution solution.
Membranes were washed 3 times for 10 min each with Tween-PBS and
incubated with goat anti-rabbit or mouse horseradish peroxidase
conjugate IgG Abgent (San Diego, CA, USA) at a dilution of 1:3,000
in Tween-PBS. The Fusion computer software (Vilber Lourmat, French)
was used for visualization.
Xenograft tumor model
In this study, the male nude mice (4 weeks) were
purchased from the experimental animal center of Chongqing Medical
University. All the mouse experiments were approved by the Ethics
Committee of Chongqing Medical University. The OS cells were
injected subcutaneously into the nude mice at a density of
5×106 cells per 100 μl PBS. Tumor volume was measure at
7, 14, 21, 28 days after injection. Mice were sacrificed on day 28,
the xenograft tumors were dissected and embedded in paraffin for HE
staining and IHC as described above.
Proliferation index in xenograft
tumors
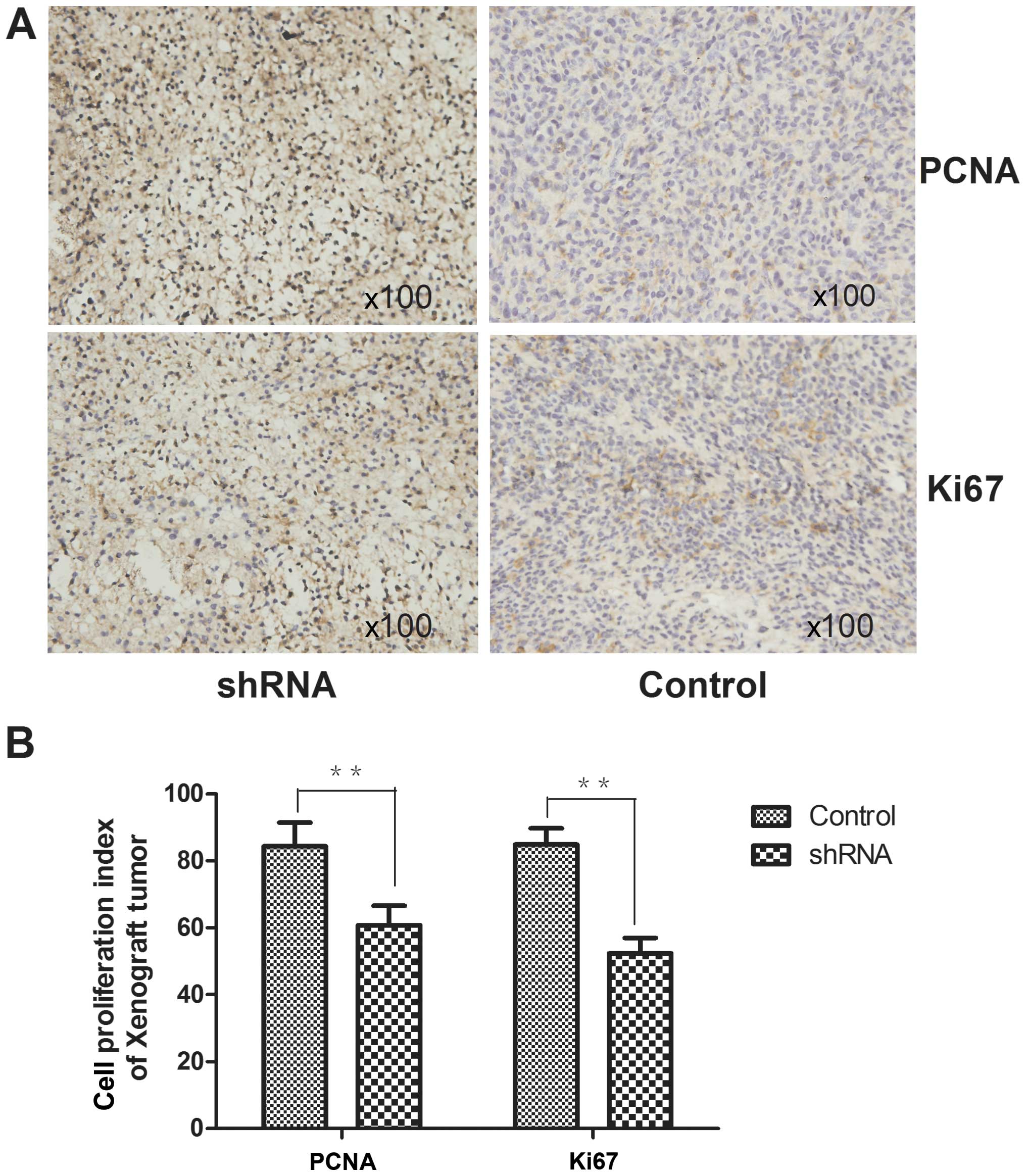
IHC staining for the expression of Ki67 and PCNA
(proliferating cell nuclear antigen) xenograft tumor tissues were
carried out. The proliferation index (Ki-67 and PCNA index) was
measured (the percentage of positive cells from five randomly
fields under a light microscopy at ×400 magnification).
Chemosensitivity
The change of cell chemosensitivity was observed
with a CCK-8 colorimeter. Cells were divided into two groups
(pre-transfection and post-transfection). All cells were seeded
onto 96-well plates at a density of 2,000 cells/well, cisplatin and
doxorubicin with different concentrations and 10 μl CCK-8 were
added to each well. The absorbance value was read at 450 nm using
an enzyme-labeled instrument.
Statistical analysis
Data were expressed as the mean ± standard
deviation. Student's t-test, one-way ANOVA or Chi-square test
followed by the Tukey post-hoc test for data with multiple
comparisons was performed using commercial statistical software
(SPSS Inc., Chicago, IL, USA) to determine significant differences
between groups, and P-values at <0.05 were considered
statistically significant.
Results
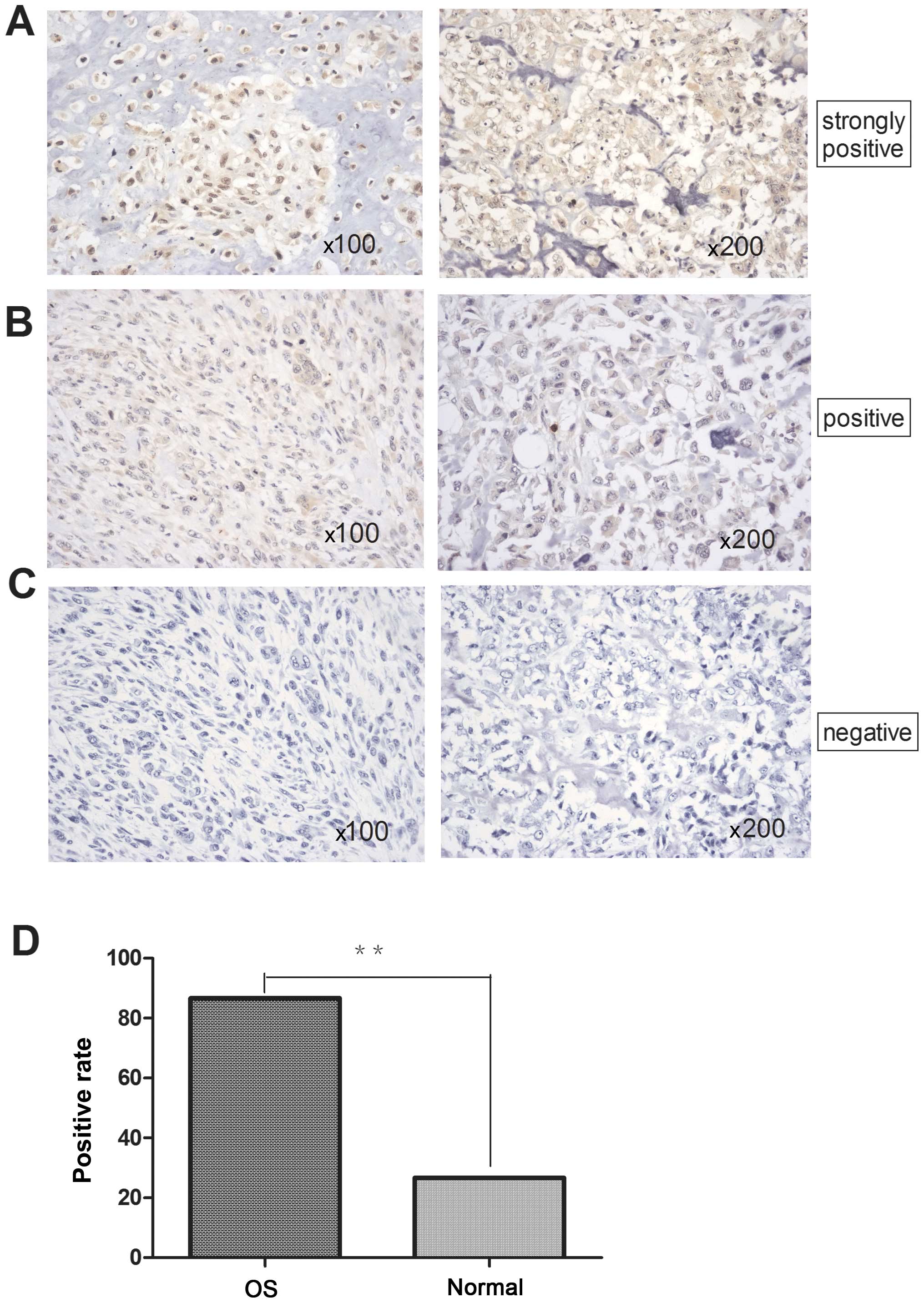
Overexpression of RSK2 gene in human
specimens
The expression levels of RSK2 gene in osteosarcomas
and normal bone tissues were comparatively analyzed using IHC. The
expression of RSK2 (not shown) in osteosarcomas was significantly
higher than normal bone tissues. The positive rate was 86.67 and
26.67% respectively, the difference was statistically significant
(P<0.01) (Table I and Fig. 1).
 | Table IExpression of RSK2 protein in
osteosarcomas and normal tissues. |
Table I
Expression of RSK2 protein in
osteosarcomas and normal tissues.
| Positive | Negative | Total |
|---|
| OS | 26 | 4 | 30 |
| Normal | 4 | 11 | 15 |
| Total | 30 | 15 | 45 |
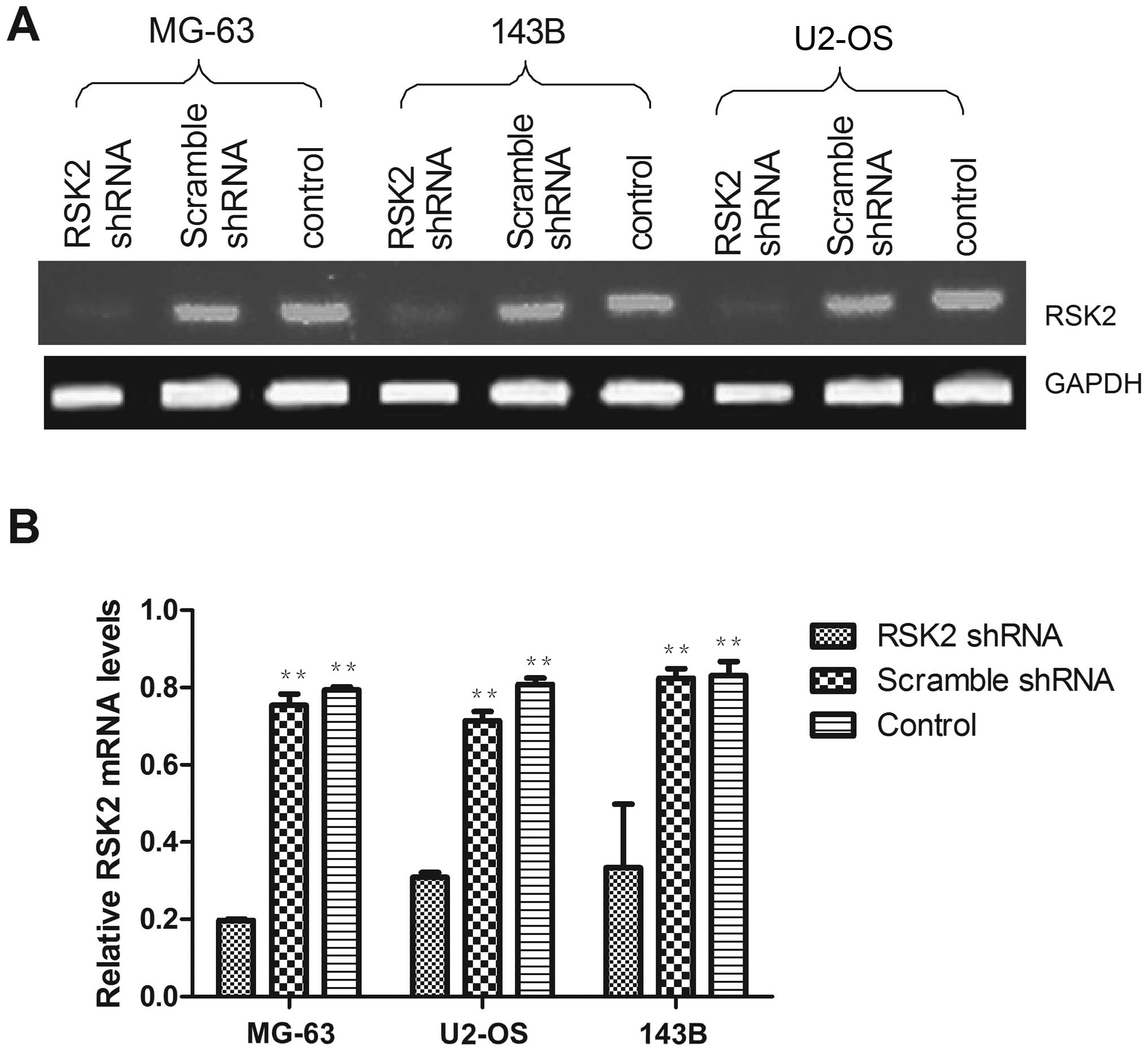
Knockdown of RSK2 in OS cell lines
To verify the transfection efficiency of the shRNA,
RSK2 mRNA expression was measured at 48 h after transfection
(Fig. 2). RSK2 mRNA expression was
significantly decreased after transfection with shRNA, while
scramble shRNA had only negligible effects on RSK2 expression.
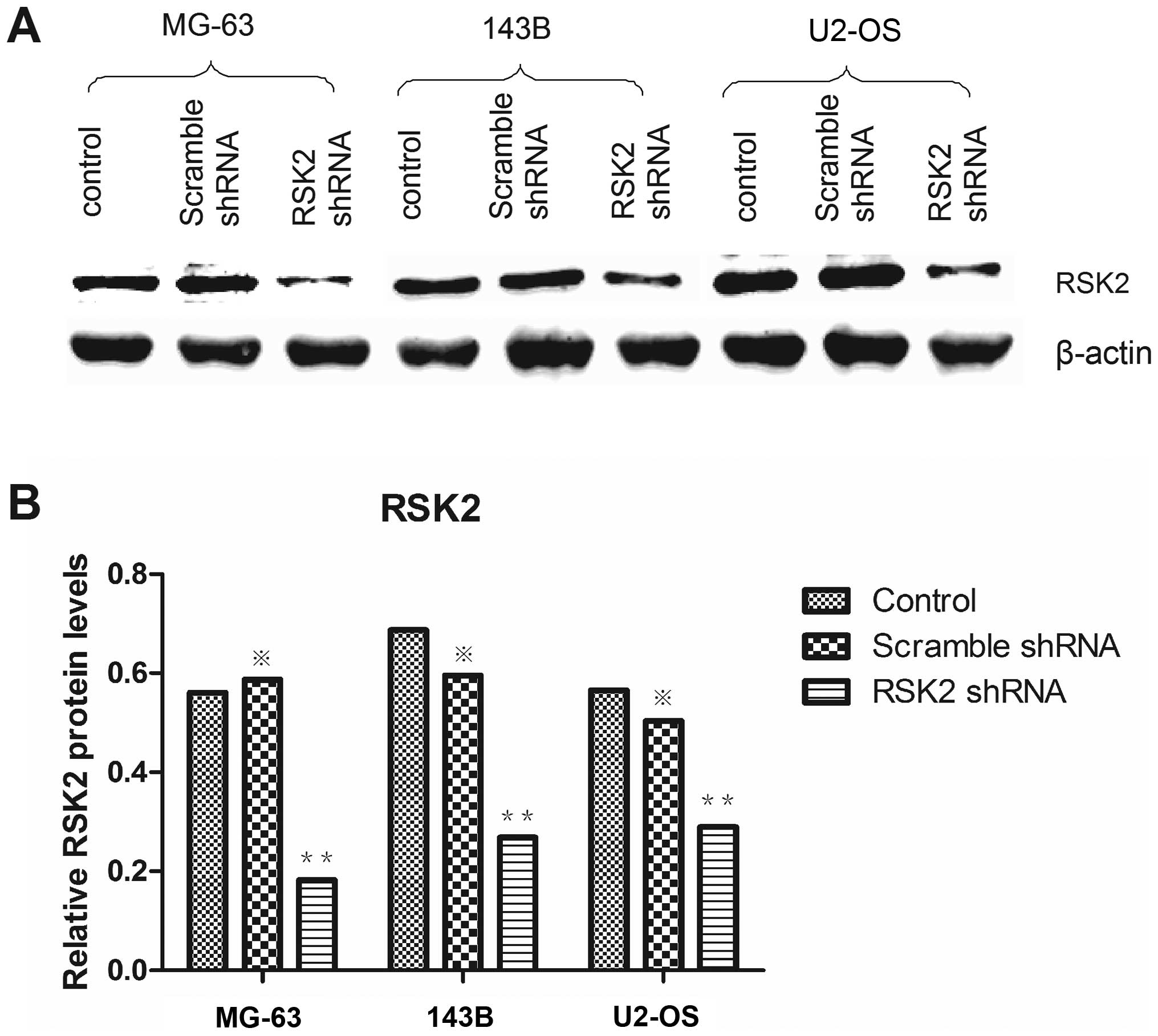
Western blot analysis confirmed that RSK2 protein was significantly
down regulated in all OS cells at 48 h after transfection with RSK2
shRNA (Fig. 3); scrambled shRNA
did not affect RSK2 protein expression in any cell lines. According
to the results above, RSK2 shRNA could effectively knock down the
RSK2 gene in mRNA level and the protein level.
Influence of RSK2 inhibition on cell
apoptosis and cell cycle progression
At 48 h after transfection with shRNA, Annexin V-PI
staining was performed to detect apoptotic cells and cell cycle
progression. After transfection with RSK2 shRNA, apoptosis was
significantly increased in all OS cells compared to cells without
transfection (Fig. 4). The cell
cycle analysis showed that cells were arrested in the G1/S phase,
while blockage was not observed in the untransfected OS cells.
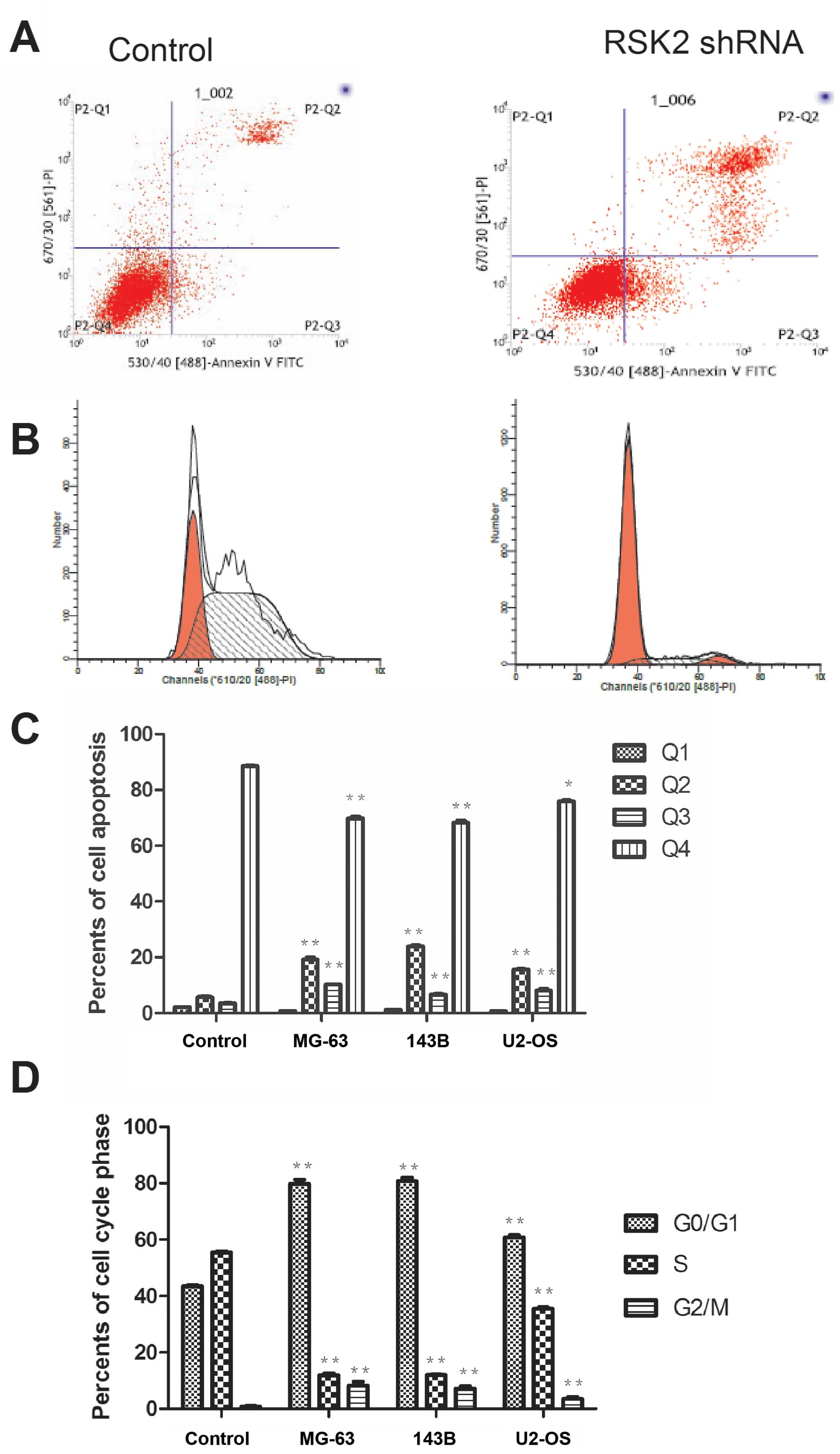 | Figure 4OS cell apoptosis and cell cycle were
analysised by Annexin V-PI staining. (A) Representative images
showing gross increased counts of apoptotic cell in three OS cell
lines. (B) Representative images showing cell cycle arrest induced
by RSK2 shRNA at 48 h in three OS cell lines. (C) The percentage of
OS cells in 4 quadrants respectively. Q1, dead cells, Q2, late
apoptotic cells, Q3, early apoptotic cells, and Q4, live cells. The
values represent the means ± SD of three experiments performed in
triplicate. *P≤0.05, **P≤0.01, versus control
as determined by the Student's t-test. (D) The percentage of cell
cycle phase. The cells were fixed by 70% ethylalcohol before
detection. The values represent the means ± SD of three experiments
performed in triplicate. *P≤0.05, **P≤0.01
versus control, as determined by the Student's t-test. |
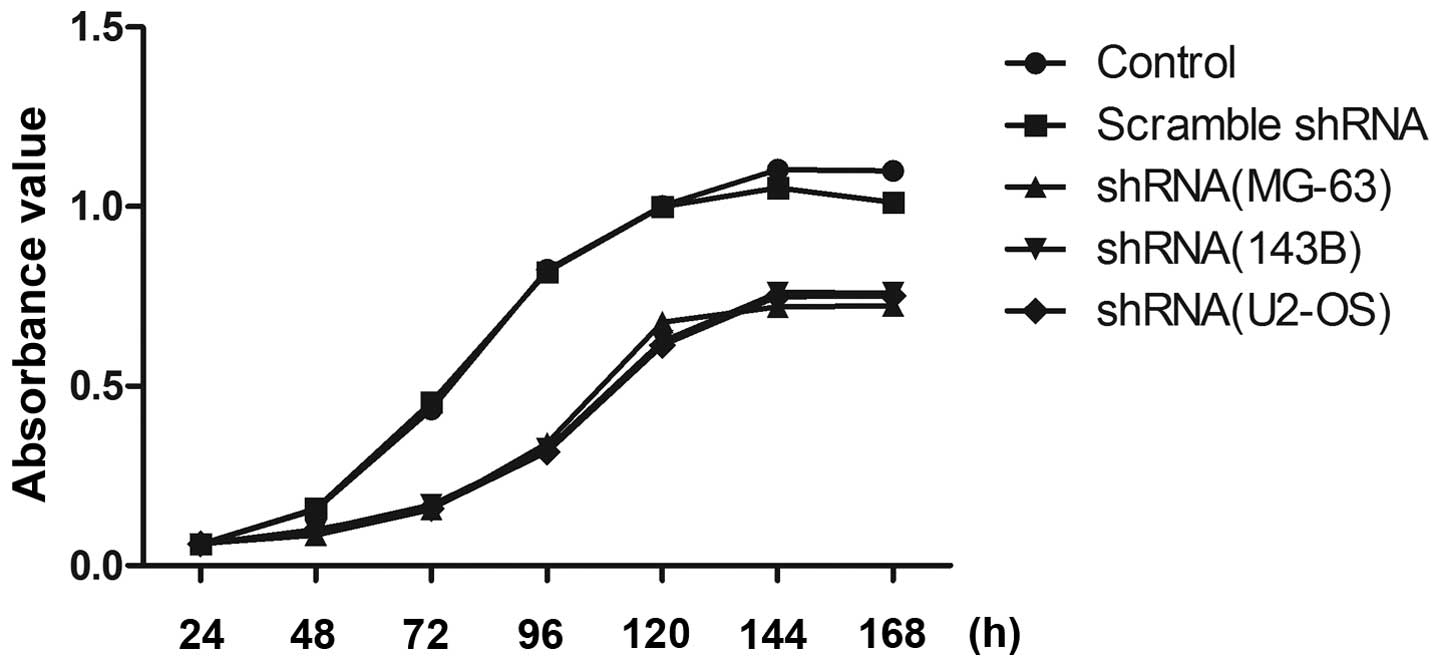
Influence of RSK2 inhibition on cell
proliferation
At 48 h after transfection with shRNA, cell
viability was evaluated (Fig. 5).
After transfection with RSK2 shRNA, the viability of all OS cells
was significantly decreased as compared to that of the control. In
contrast, cells transfected with scrambled shRNA exhibited no
changes in cell viability as compared to the control. Of the OS
cell lines, three exhibited no difference in cell viability
compared to each other.
Influence of RSK2 inhibition on cell
chemosensitivity
The sensitivity of OS cells to cisplatin and
doxorubicin after transfection with shRNA were evaluated. After
transfection with RSK2 shRNA, the IC50s of cisplatin and
doxorubicin decreased in all three cell lines. In contrast, the
IC50s of these drugs were not significantly affected by
transfection with scrambled shRNA in any cell line (data not shown)
(Table II).
 | Table IIThe 50% inhibitory concentrations of
cisplatin and doxobubicin in OS cell lines. |
Table II
The 50% inhibitory concentrations of
cisplatin and doxobubicin in OS cell lines.
| Cisplatin
(ng/μl) | Doxorubicin
(ng/μl) |
|---|
|
|
|
|---|
| Cell lines | Prea | Postb | Prea | Postb |
|---|
| MG-63 | 4.66 | 3.4 | 2.08 | 1.11 |
| 143B | 4.71 | 3.91 | 2.13 | 1.04 |
| U2-OS | 4.83 | 3.79 | 2.05 | 1.10 |
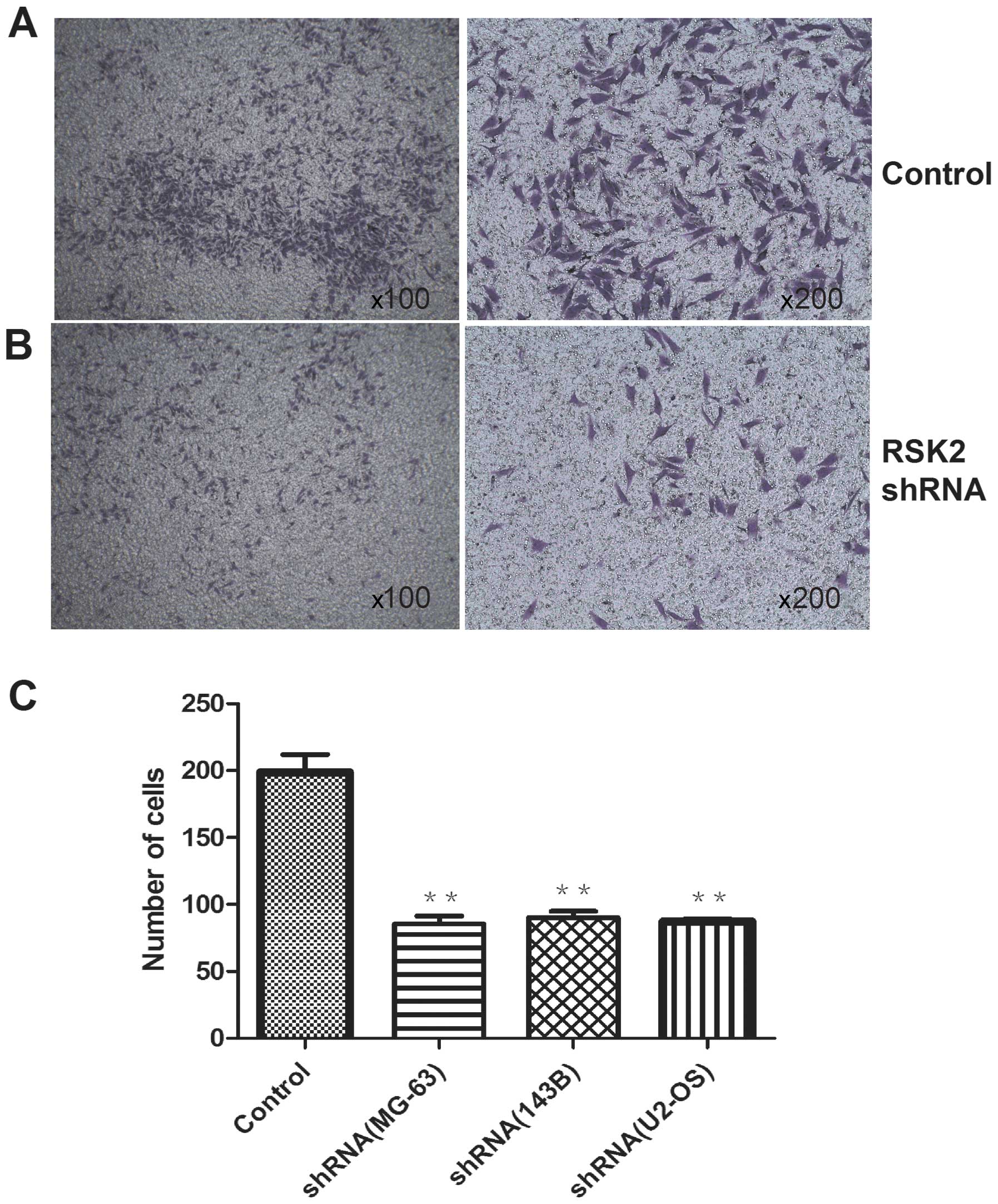
Influence of RSK2 knockdown on migration
activity
The migration activity of OS cells after
transfection with RSK2 shRNA at 48 h was weaker than that of
control. In addition, the three cell lines showed no difference by
naked eye and the differences were not significant (Fig. 6).
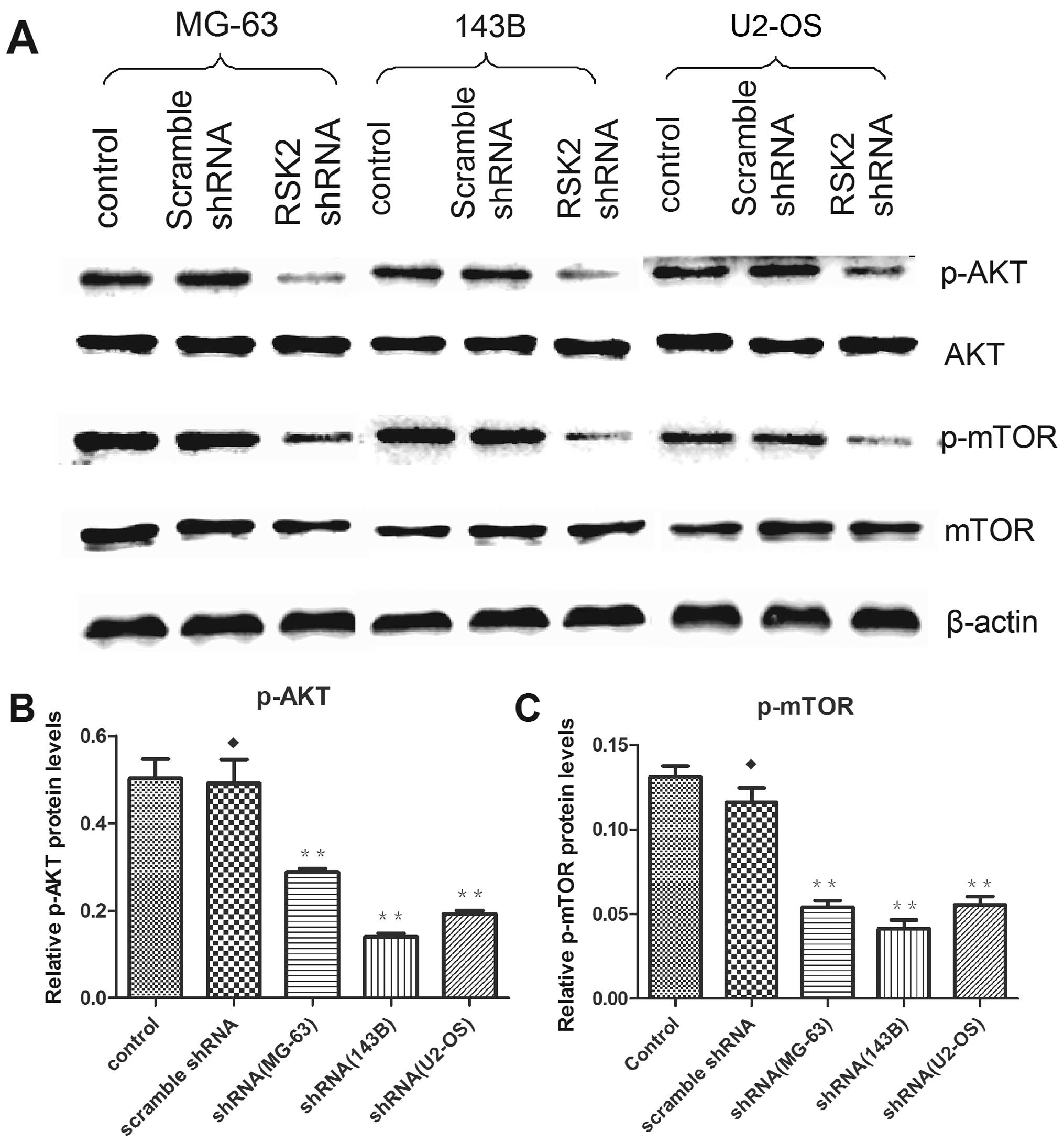
Influence of RSK2 inhibition on protein
expression
At 48 h after transfection with RSK2 shRNA,
expression of related proteins were altered in OS cell lines as
compared to control and scrambled shRNA-transfected cells. In
addition, RSK2 knockdown weakened the expression of p-AKT, and
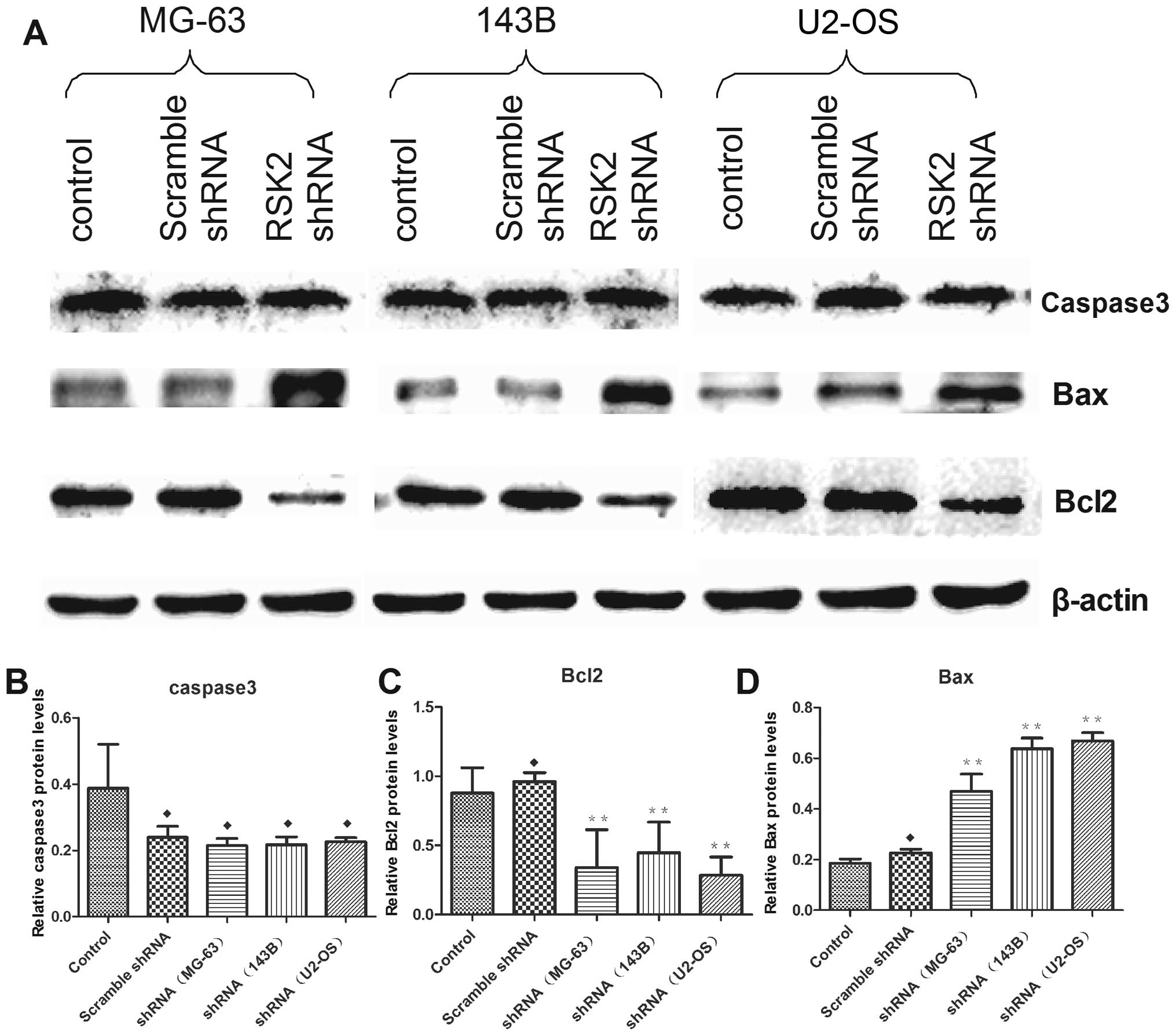
p-mTOR (Fig. 7). Knockdown of RSK2
also weakened the expression of Bcl2, enhanced the expression of
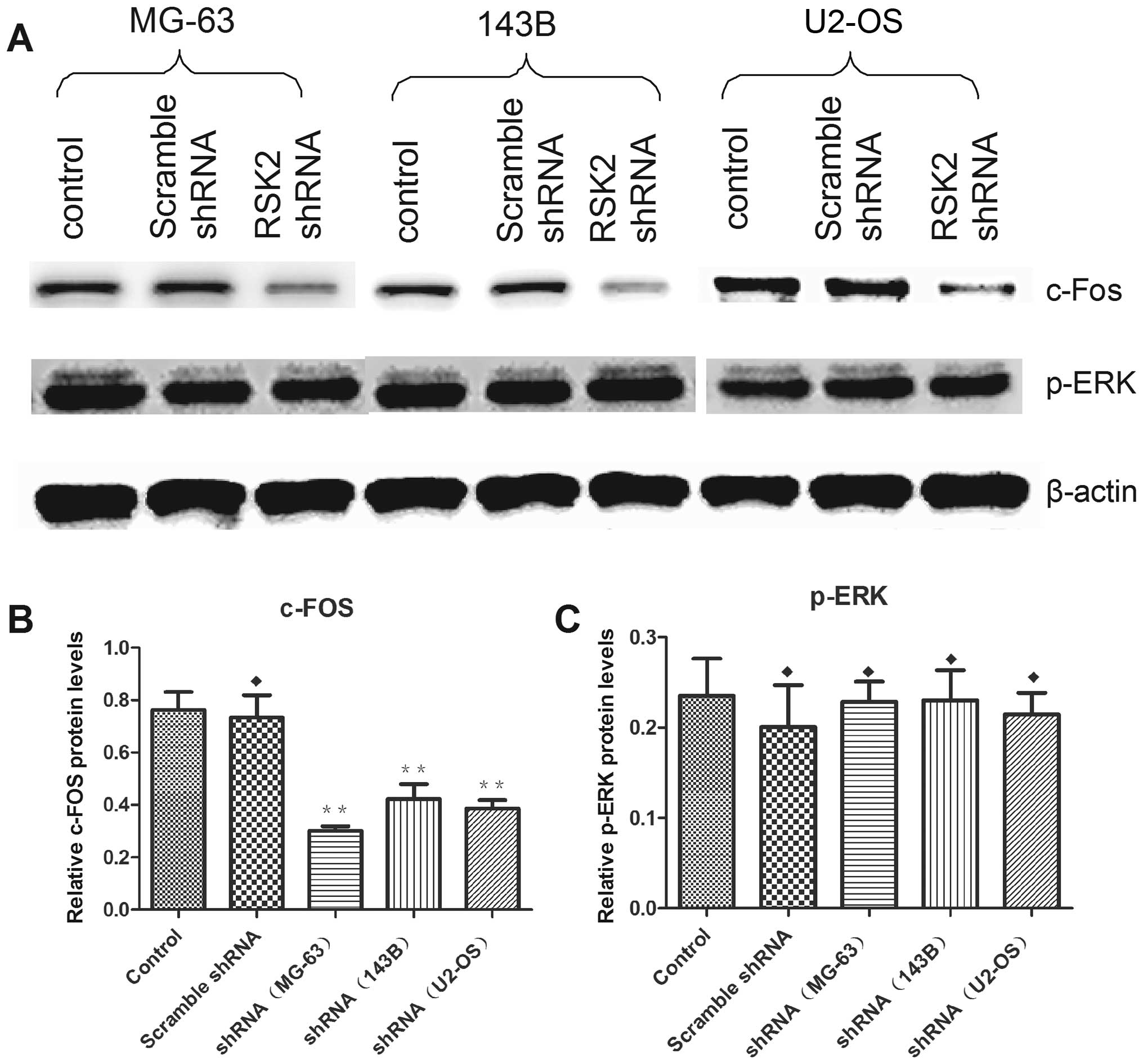
Bax, but did not influence the expression of caspase-3 (Fig. 8). In addition, inhibition of RSK2
influenced the expression of c-Fos, but could not reversely
stimulate the expression of p-ERK (Fig. 9).
Influence of RSK2 knockdown on xenograft
tumors
The activity of OS cells to produce xenograft tumor
was tested in nude mice. The cells after transfection with RSK2
shRNA were considered to produce less xenografted tumors than that
of the control group. In addition, the tumors of the control group
were smaller than the experimental group. The expression of
proliferating cell nuclear antigen and pi67 in the xenograft tumor
induced by OS cell transfection with RSK2 shRNA was significantly
lower than that of the untransfected cells (Fig. 10).
Discussion
The prognosis of OS patients remains unsatisfactory
despite the development and advances in the diagnosis, and
treatment technology. The patients with OS often have high
metastasis rate postoperatively, and chemoresistance. To explore an
effective therapy method is necessary, and also a huge challenge.
Tumorigenesis of OS is associated with biological events involved
in the process of OS. Several studies have shown that RSK2 is
overexpressed in prostate cancer tissues and stimulate
proliferation in prostate cancer cells (10,23),
multiple myeloma (12,24), non-small cell lung cancer (NSCLC)
(25), skin cancer (26), mammary cancer (27), and in head and neck squamous cell
carcinoma (28). Mutations in the
human X chromosomal gene, RSK2 (RPS6KA3), cause the Coffin-Lowry
syndrome (29–32).
Our study showed that median expression levels of
RSK2 protein in clinical specimens collected from 30 patients with
osteosarcoma were significantly elevated compared to the expression
levels of para-tumor tissue and normal bone tissues. The expression
was mostly located in the cell nucleus and cytoplasm, effectly
supporting that RSK1/2/3 are present in the cytoplasm of quiescent
cells, but translocate to the nucleus after stimulation (33–37).
However, the result are based on a small number of samples, and a
limited geographical area. More specimens need to be collected for
further study.
We investigated the effects of RSK2 knockdown in OS
cell lines in order to determine the potential efficacy of
RSK2-targeted therapy for the treatment of OS. As the expression of
RSK2 was most significantly decreased at 48 h after transfected
with shRNA (data not shown), the difference in the viability of the
three OS cell lines was detected at 48 h after transfection with
RSK2 shRNA, as well as the other phenomena, including
chemosensitivity, apoptosis and migration.
Cell viability was decreased in all OS cell lines
following transfection with RSK2 shRNA. The percentage of apoptotic
cells, assayed by flow cytometry, was obviously higher than the
control group and the empty vector transfected group. RSK2 protein
is synthesized and expressed at high levels during the G1/S-phase
of the cancer cell division cycle, effectively supporting that the
RSK2 could regulate G1 phase progression by controlling the
activity of the CDK2 (cyclin-dependent kinase 2) inhibitor p27kip1
and negatively regulating GSK3, which targets c-Myc and cyclin D1
for degradation (15,38).
Further study demonstrated that the expression
levels of apoptosis related genes, including Bax, Bcl2, and
caspase-3, were affected by knockdown of RSK2 by shRNA in three OS
cells lines, increasing expression of Bax, reducing expression of
Bcl2, explaining the increased apoptosis. Several groups have shown
that RSK activation or overexpression inhibited cell death via
inactivation of the Bcl-2 homology 3-only pro-apoptotic protein,
Bad (18,39,40).
RSK2 could interact with PEA-15/PED, increased its expression and
reduced apoptosis (37,41,42).
In our study, we suggested that inhibition of RSK2 might induce a
preferential decrease in cell viability by apoptosis and delay the
progressive pathological process of different types of OS
cells.
After transfection with RSK2 shRNA, the sensitivity
of OS cells to cisplatin and doxorubicin was increased as compared
to that of control. Other groups have found that RSK2 activity was
regulated by its interaction with PEA-15 and ERK, increased
expression of PEA-15/PED could reduce the sensitivity of tumor
cells (37,42,43).
RSK2 stimulated the phosphorylation of IκB and activated the NF-κB
signaling pathway (43,44). Moreover, activated NF-κB induced
the production of chemoresistance genes and proteins, including
P-glycoprotein and ABC transporters (45–47).
In this study, Bcl2, downstream of NF-κB and one of the indicators
of apoptosis, was downregulated after transfected with RSK2 shRNA,
while Bax was upregulated. So we considered that the potential
mechanism of increased chemosensitivity was related with the
activation of NF-κB/Bcl2/Bax pathway. Further studies, including
related gene expression, related protein expression and functional
assessment based on RSK2 inhibition, are required to determine the
molecular mechanisms through which RSK2 affects chemosensitivity in
OS cells.
Some research groups have reported that regulation
of c-Fos by RSK2 was shown to play important roles in bone
homoeostasis and tumorigenesis. In the absence of RSK2,
c-Fos-dependent osteosarcoma formation is impaired (48). The lack of c-Fos phosphorylation
leads to reduced c-Fos protein levels, which are thought to be
responsible for decreased proliferation and increased apoptosis of
transformed osteoblasts (49).
c-Fos transgenic mice crossed with RSK2 null mice produce offspring
whose tumors have increased levels of apoptosis and decreased
proliferation compared to c-Fos transgenic animals expressing
wild-type RSK2 (50).
Interestingly, our data supported the hypothesis that RSK2
inhibition induced downregulation of c-Fos protein in OS cell
lines, resulting in decreased proliferation and increased
apoptosis, effectively supporting that RSK2 is essential for c-Fos
transactivation because it stabilizes c-fos protein (19,48,51).
It has been verified that RSK2 could promote the
expression of AKT through the increased combination with
keratinocyte growth factor receptor (KGFP) on epithelial cells
(52). mTOR (a downstream factor
of AKT) is an essential regulator of ribosome biogenesis, mRNA
translation and cell growth, and its activity is controlled by
several growth-regulating pathways. Activated RSKs promotes mTOR
signaling through the phosphorylation of TSC2 on Ser1798, which
prevents its GAP (guanine nucleotide-activating protein) activity
towards the small GTPase Rheb (53–55).
More recently, RSK was shown to phosphorylate Raptor, an important
mTORC1 (mTOR complex 1) scaffolding protein, providing another link
between the Ras/MAPK and mTOR signaling pathways (56). In this study, inhibition with RSK2
shRNA induced downregulated expression of phosphorylated AKT and
mTOR. RSK2 shRNA acted on the OS cells through AKT/mTOR signaling
pathway.
PCNA and Ki67 are key controllers of multiple
processes in DNA and chromatin metabolism, regulating replication,
repair and chromatin assembly through interaction with a huge
number of partner proteins. They exist only in proliferative cells
and tumor cells and are used to detect the cell's proliferative
activity. In our study, the rate of positive cells of PCNA and Ki67
in xenografted tumors induced by untransfected cells are obviously
higher than in the transfected groups. This verified that knockdown
of RSK2 inhibited the proliferation of OS cells in nude mice.
There are now three different potent and highly
specific inhibitors of RSK2 (BI-D1870, SL0101, FMK) (57). However, we did not study their
effects on OS cells. Further studies are required to verify whether
RSK2 inhibitors may be effective for all types of OS and whether
there are side effects of RSK2 inhibitors in vivo.
In conclusion, this study demonstrated that
inhibition of RSK2 decreased cell viability through the induction
of apoptosis, blocked the cell cycle progress, enhanced
chemosensitivity, and weakened migration in OS cell lines. These
findings suggested that RSK2 may partially support cell survival
and maintain the aggressive biological behavior in OS. Therefore,
RSK2 may be an effective target for novel therapeutics or
combination therapies with conventional anticancer drugs for OS.
Further studies are required to fully elucidate the mechanism of
RSK2 in OS, and to verify whether RSK2 plays a role in
vivo.
Acknowledgements
We would like to acknowledge the service provided by
Chongqing Key Laboratory of Molecular Oncology and Epigenetics. We
thank Prof. Zhen-Ming Hu for guidance in the whole progress of
experiments.
References
|
1
|
Ottaviani G and Jaffe N: The epidemiology
of osteosarcoma. Cancer Treat Res. 152:3–13. 2009. View Article : Google Scholar
|
|
2
|
Sztán M, Pápai Z, Szendrôi M, Looij M and
Oláh E: Allelic losses from chromosome 17 in human osteosarcomas.
Pathol Oncol Res. 3:115–120. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dahlin DC: Pathology of osteosarcoma. Clin
Orthop Relat Res. 111:23–32. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Erikson E and Maller JL: A protein kinase
from Xenopus eggs specific for ribosomal protein S6. Proc Natl Acad
Sci USA. 82:742–746. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dalby KN, Morrice N, Caudwell FB, Avruch J
and Cohen P: Identification of regulatory phosphorylation sites in
mitogen-activated protein kinase (MAPK)-activated protein
kinase-1a/p90rsk that are inducible by MAPK. J Biol Chem.
273:1496–1505. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Frödin M, Jensen CJ, Merienne K and
Gammeltoft S: A phosphoserine regulated docking site in the protein
kinase RSK2 that recruits and activates PDK1. EMBO J. 19:2924–2934.
2000. View Article : Google Scholar
|
|
7
|
Blenis J, Chung J, Erikson E, Alcorta DA
and Erikson RL: Distinct mechanisms for the activation of the RSK
kinases/MAP2 kinase/pp90rsk and pp70-S6 kinase signaling systems
are indicated by inhibition of protein synthesis. Cell Growth
Differ. 2:279–285. 1991.PubMed/NCBI
|
|
8
|
Anjum R and Blenis J: The RSK family of
kinases: Emerging roles in cellular signalling. Nat Rev Mol Cell
Biol. 9:747–758. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smith JA, Poteet-Smith CE, Xu Y, Errington
TM, Hecht SM and Lannigan DA: Identification of the first specific
inhibitor of p90 ribosomal S6 kinase (RSK) reveals an unexpected
role for RSK in cancer cell proliferation. Cancer Res.
65:1027–1034. 2005.PubMed/NCBI
|
|
10
|
Clark DE, Errington TM, Smith JA, Frierson
HF Jr, Weber MJ and Lannigan DA: The serine/threonine protein
kinase, p90 ribosomal S6 kinase, is an important regulator of
prostate cancer cell proliferation. Cancer Res. 65:3108–3116.
2005.PubMed/NCBI
|
|
11
|
Cho YY, Yao K, Kim HG, Kang BS, Zheng D,
Bode AM and Dong Z: Ribosomal S6 kinase 2 is a key regulator in
tumor promoter induced cell transformation. Cancer Res.
67:8104–8112. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kang S, Dong S, Gu TL, Guo A, Cohen MS,
Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, et al:
FGFR3 activates RSK2 to mediate hematopoietic transformation
through tyrosine phosphorylation of RSK2 and activation of the
MEK/ERK pathway. Cancer Cell. 12:201–214. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Woo MS, Ohta Y, Rabinovitz I, Stossel TP
and Blenis J: Ribosomal S6 kinase (RSK) regulates phosphorylation
of filamin A on an important regulatory site. Mol Cell Biol.
24:3025–3035. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cohen P and Frame S: The renaissance of
GSK3. Nat Rev Mol Cell Biol. 2:769–776. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sutherland C, Leighton IA and Cohen P:
Inactivation of glycogen synthase kinase-3 beta by phosphorylation:
New kinase connections in insulin and growth-factor signalling.
Biochem J. 296:15–19. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stambolic V and Woodgett JR: Mitogen
inactivation of glycogen synthase kinase-3 beta in intact cells via
serine 9 phosphorylation. Biochem J. 303:701–704. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Larrea MD, Hong F, Wander SA, da Silva TG,
Helfman D, Lannigan D, Smith JA and Slingerland JM: RSK1 drives
p27Kip1 phosphorylation at T198 to promote RhoA inhibition and
increase cell motility. Proc Natl Acad Sci USA. 106:9268–9273.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bonni A, Brunet A, West AE, Datta SR,
Takasu MA and Greenberg ME: Cell survival promoted by the Ras-MAPK
signaling pathway by transcription-dependent and -independent
mechanisms. Science. 286:1358–1362. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Murphy LO, Smith S, Chen RH, Fingar DC and
Blenis J: Molecular interpretation of ERK signal duration by
immediate early gene products. Nat Cell Biol. 4:556–564.
2002.PubMed/NCBI
|
|
20
|
Joel PB, Smith J, Sturgill TW, Fisher TL,
Blenis J and Lannigan DA: pp90rsk1 regulates estrogen
receptor-mediated transcription through phosphorylation of Ser-167.
Mol Cell Biol. 18:1978–1984. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yang X, Matsuda K, Bialek P, Jacquot S,
Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes
TM, et al: ATF4 is a substrate of RSK2 and an essential regulator
of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell.
117:387–398. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
El-Haschimi K, Dufresne SD, Hirshman MF,
Flier JS, Goodyear LJ and Bjørbaek C: Insulin resistance and
lipodystrophy in mice lacking ribosomal S6 kinase 2. Diabetes.
52:1340–1346. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Burd CJ, Petre CE, Morey LM, Wang Y,
Revelo MP, Haiman CA, Lu S, Fenoglio-Preiser CM, Li J, Knudsen ES,
et al: Cyclin D1b variant influences prostate cancer growth through
aberrant androgen receptor regulation. Proc Natl Acad Sci USA.
103:2190–2195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kang S, Elf S, Dong S, Hitosugi T, Lythgoe
K, Guo A, Ruan H, Lonial S, Khoury HJ, Williams IR, et al:
Fibroblast growth factor receptor 3 associates with and tyrosine
phosphorylates p90 RSK2, leading to RSK2 activation that mediates
hematopoietic transformation. Mol Cell Biol. 29:2105–2117. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dehan E, Bassermann F, Guardavaccaro D,
Vasiliver-Shamis G, Cohen M, Lowes KN, Dustin M, Huang DC, Taunton
J and Pagano M: betaTrCP- and Rsk1/2-mediated degradation of BimEL
inhibits apoptosis. Mol Cell. 33:109–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cho YY, Lee MH, Lee CJ, Yao K, Lee HS,
Bode AM and Dong Z: RSK2 as a key regulator in human skin cancer.
Carcinogenesis. 33:2529–2537. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Czaplinska D, Turczyk L, Grudowska A,
Mieszkowska M, Lipinska AD, Skladanowski AC, Zaczek AJ, Romanska HM
and Sadej R: Phosphorylation of RSK2 at Tyr529 by FGFR2-p38
enhances human mammary epithelial cells migration. Biochim Biophys
Acta. 1843:2461–2470. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kang S, Elf S, Lythgoe K, Hitosugi T,
Taunton J, Zhou W, Xiong L, Wang D, Muller S, Fan S, et al: p90
ribosomal S6 kinase 2 promotes invasion and metastasis of human
head and neck squamous cell carcinoma cells. J Clin Invest.
120:1165–1177. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zeniou-Meyer M, Gambino F, Ammar MR,
Humeau Y and Vitale N: The Coffin-Lowry syndrome-associated protein
RSK2 and neurosecretion. Cell Mol Neurobiol. 30:1401–1406. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jurkiewicz D, Jezela-Stanek A, Ciara E,
Piekutowska-Abramczuk D, Kugaudo M, Gajdulewicz M, Chrzanowska K,
Popowska E and Krajewska-Walasek M: Four novel RSK2 mutations in
females with Coffin-Lowry syndrome. Eur J Med Genet. 53:268–273.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Abidi F, Jacquot S, Lassiter C, Trivier E,
Hanauer A and Schwartz CE: Novel mutations in Rsk-2, the gene for
Coffin-Lowry syndrome (CLS). Eur J Hum Genet. 7:20–26. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Trivier E, De Cesare D, Jacquot S,
Pannetier S, Zackai E, Young I, Mandel JL, Sassone-Corsi P and
Hanauer A: Mutations in the kinase Rsk-2 associated with
Coffin-Lowry syndrome. Nature. 384:567–570. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen RH, Sarnecki C and Blenis J: Nuclear
localization and regulation of erk- and rsk-encoded protein
kinases. Mol Cell Biol. 12:915–927. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lenormand P, Sardet C, Pagès G, L'Allemain
G, Brunet A and Pouysségur J: Growth factors induce nuclear
translocation of MAP kinases (p42mapk and p44mapk) but not of their
activator MAP kinase kinase (p45mapkk) in fibroblasts. J Cell Biol.
122:1079–1088. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao Y, Bjørbaek C, Weremowicz S, Morton
CC and Moller DE: RSK3 encodes a novel pp90rsk isoform with a
unique N-terminal sequence: Growth factor-stimulated kinase
function and nuclear translocation. Mol Cell Biol. 15:4353–4363.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Richards SA, Dreisbach VC, Murphy LO and
Blenis J: Characterization of regulatory events associated with
membrane targeting of p90 ribosomal S6 kinase 1. Mol Cell Biol.
21:7470–7480. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vaidyanathan H and Ramos JW: RSK2 activity
is regulated by its interaction with PEA-15. J Biol Chem.
278:32367–32372. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Fujita N, Sato S and Tsuruo T:
Phosphorylation of p27Kip1 at threonine 198 by p90 ribosomal
protein S6 kinases promotes its binding to 14–3-3 and cytoplasmic
localization. J Biol Chem. 278:49254–49260. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tan Y, Ruan H, Demeter MR and Comb MJ: p90
(RSK) blocks bad-mediated cell death via a protein kinase
C-dependent pathway. J Biol Chem. 274:34859–34867. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bertolotto C, Maulon L, Filippa N, Baier G
and Auberger P: Protein kinase C theta and epsilon promote T-cell
survival by a rsk-dependent phosphorylation and inactivation of
BAD. J Biol Chem. 275:37246–37250. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Vaidyanathan H, Opoku-Ansah J, Pastorino
S, Renganathan H, Matter ML and Ramos JW: ERK MAP kinase is
targeted to RSK2 by the phosphoprotein PEA-15. Proc Natl Acad Sci
USA. 104:19837–19842. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Formisano P, Ragno P, Pesapane A, Alfano
D, Alberobello AT, Rea VE, et al: PED/PEA-15 interacts with the 67
kDa laminin receptor and regulates cell adhesion, migration,
proliferation and apoptosis. J Cell Mol Med. 16:1435–1446. 2012.
View Article : Google Scholar
|
|
43
|
Cude K, Wang Y, Choi HJ, Hsuan SL, Zhang
H, Wang CY and Xia Z: Regulation of the G2-M cell cycle progression
by the ERK5-NFkappaB signaling pathway. J Cell Biol. 177:253–264.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Peng C, Cho YY, Zhu F, Xu YM, Wen W, Ma
WY, Bode AM and Dong Z: RSK2 mediates NF-{kappa}B activity through
the phosphorylation of IkappaBalpha in the TNF-R1 pathway. FASEB J.
24:3490–3499. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ke SZ, Ni XY, Zhang YH, Wang YN, Wu B and
Gao FG: Camptothecin and cisplatin upregulate ABCG2 and MRP2
expression by activating the ATM/NF-κB pathway in lung cancer
cells. Int J Oncol. 42:1289–1296. 2013.PubMed/NCBI
|
|
46
|
Luo L, Sun YJ, Yang L, Huang S and Wu YJ:
Avermectin induces P-glycoprotein expression in S2 cells via the
calcium/calmodulin/NF-κB pathway. Chem Biol Interact. 203:430–439.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bongiovanni L, Mazzocchetti F, Malatesta
D, Romanucci M, Ciccarelli A, Buracco P, De Maria R, Palmieri C,
Martano M, Morello E, et al: Immunohistochemical investigation of
cell cycle and apoptosis regulators (survivin, β-catenin, p53,
caspase 3) in canine appendicular osteosarcoma. BMC Vet Res.
8:782012. View Article : Google Scholar
|
|
48
|
Bakiri L, Reschke MO, Gefroh HA, Idarraga
MH, Polzer K, Zenz R, Schett G and Wagner EF: Functions of Fos
phosphorylation in bone homeostasis, cytokine response and
tumourigenesis. Oncogene. 30:1506–1517. 2011. View Article : Google Scholar
|
|
49
|
David JP, Mehic D, Bakiri L, Schilling AF,
Mandic V, Priemel M, Idarraga MH, Reschke MO, Hoffmann O, Amling M,
et al: Essential role of RSK2 in c-Fos-dependent osteosarcoma
development. J Clin Invest. 115:664–672. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen RH, Abate C and Blenis J:
Phosphorylation of the c-Fos transrepression domain by
mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase.
Proc Natl Acad Sci USA. 90:10952–10956. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Chen RH, Juo PC, Curran T and Blenis J:
Phosphorylation of c-Fos at the C-terminus enhances its
transforming activity. Oncogene. 12:1493–1502. 1996.PubMed/NCBI
|
|
52
|
Pan ZZ, Devaux Y and Ray P: Ribosomal S6
kinase as a mediator of keratinocyte growth factor-induced
activation of Akt in epithelial cells. Mol Biol Cell. 15:3106–3113.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma L, Chen Z, Erdjument-Bromage H, Tempst
P and Pandolfi PP: Phosphorylation and functional inactivation of
TSC2 by Erk implications for tuberous sclerosis and cancer
pathogenesis. Cell. 121:179–193. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Roux PP, Ballif BA, Anjum R, Gygi SP and
Blenis J: Tumor-promoting phorbol esters and activated Ras
inactivate the tuberous sclerosis tumor suppressor complex via p90
ribosomal S6 kinase. Proc Natl Acad Sci USA. 101:13489–13494. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Rolfe M, McLeod LE, Pratt PF and Proud CG:
Activation of protein synthesis in cardiomyocytes by the
hypertrophic agent phenylephrine requires the activation of ERK and
involves phosphorylation of tuberous sclerosis complex 2 (TSC2).
Biochem J. 388:973–984. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Carrière A, Cargnello M, Julien LA, Gao H,
Bonneil E, Thibault P and Roux PP: Oncogenic MAPK signaling
stimulates mTORC1 activity by promoting RSK-mediated raptor
phosphorylation. Curr Biol. 18:1269–1277. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Nguyen TL: Targeting RSK: An overview of
small molecule inhibitors. Anticancer Agents Med Chem. 8:710–716.
2008. View Article : Google Scholar : PubMed/NCBI
|