Introduction
Gastric cancer rates as the fourth most common
malignancy worldwide and is the third most common cause of death
from a malignant disease, after lung and liver cancer (1). Eastern Asia accounts for more than
50% of cases registered globally. A gradual decrease in the number
of cases has been observed in regions such as the United States,
Western and Central Europe (2,3).
In 2012, the incidence of gastric cancer in the
Czech Republic was 14.6 cases per 100,000 inhabitants. Most cases
are diagnosed in the late stages of the disease and only palliative
treatment remains possible. Chemotherapy is often embarked upon as
part of palliative treatment and most patients receive the same
medications, overall survival varies greatly from patient to
patient, however. One possible cause of this variability could be
the gene expression changes occurring in cancer tissue, which may
alter the effect of cytostatics. Evaluating the expression of
specific genes including genes for microRNA (miRNA) could help
single out chemotherapy efficacy predictors in gastric cancer
patients undergoing palliative treatment (4,5). By
identifying patients with chemoresistant tumors, we hope to spare
them the strain of inefficient chemotherapy.
The tissue samples were the same as those used by
the pathologists when making the diagnoses, to measure the levels
of chemotherapy response predictors. The use of efficacy predictors
makes up an important part of the development of new targeted
therapy drugs and their introduction into clinical practice; this
is no less the case for conventional chemotherapeutics.
The ability of cancer cells to overcome the effects
of chemotherapy by changing the expression of repair genes, enzymes
partaking in nucleic acid metabolism and genes involved in
apoptosis limits the success of chemotherapy. Eukaryotic cells
react to DNA damage by activating repair mechanisms with these key
functions: DNA damage detection, stopping replication of damaged
DNA, repair of damaged area if possible or else the induction of
apoptosis. Nucleotide excision repair (NER), base excision repair
(BER), mismatch repair (MMR) and homologous recombination repair
(HRR) belong to the most important repair mechanisms of eukaryotic
cells (6,7). Damage to these mechanisms means that
mutations are passed on to the next cell generation. Impaired
function of DNA repair mechanisms is thus linked to both ageing and
cancerogenesis. However, increased activity of these mechanisms can
hinder chemotherapy by forestalling further effective damage to DNA
and thereby preventing the activation of apoptosis in proliferating
tumor cells (8).
Cisplatin acts cytotoxically by creating adducts,
which participate in crosslinking DNA, and in so doing activates
programmed cell death. Decrease in the scale of damage to DNA,
whether as a result of fewer adducts being created (in lower drug
dosage, for instance) or because of their repair, can lead to a
decrease of efficacy of this chemotherapeutic substance (9).
5-Fluorouracil (5-FU) is one of the most commonly
used chemotherapeutics in gastric cancer treatment regimens
(10). The primary site of action
for 5-FU is thymidylate synthase (TS), an essential enzyme in de
novo biosynthetic pathway of deoxythymidylate (dTMP).
Thymidylate synthase catalyzes the reductive methylation of dUMP
(deoxyuridine-5-prime monophosphate or deoxyuridylate) to dTMP
(deoxythymidine-5-prime monophosphate or deoxythymidylate) using
5,10-methylenetetrahydrofolate as a cofactor. Maintaining a dTMP
pool is crucial for DNA replication and repair.
We measured the levels of mRNA of the excision
repair cross-complementary group 1 gene (ERCC1), ribonucleotide
reductase subunit M1 gene (RRM1), breast cancer 1 gene (BRCA1) and
TS, all of which participate in repair to damaged DNA. We
determined their relationship to time to progression (TTP), using
the definition of progression according to RECIST (response
evaluation criteria in solid tumours) criteria (11), and overall survival (OS). Table I lists the basic characteristics of
the genes of interest.
 | Table IPrimer sequences for quantitative
reverse transcription polymerase chain reaction (qRT-PCR) with the
probes of Universal Probe Library. |
Table I
Primer sequences for quantitative
reverse transcription polymerase chain reaction (qRT-PCR) with the
probes of Universal Probe Library.
| Symbol | Gene name | Function of the
product of this gene | Primer sequence
5′-3′ | UPL probe |
|---|
| Reference
genes |
| GAPDH |
Glyceraldehyde-3-phosphate
dehydrogenase | Enzyme of
glycolysis. |
AGCCACATCGCTCAGACAC
GCCCAATACGACCAAATCC | 60 |
| HPRT | Hypoxanthine
guanine phosphoribosyltransferase | Purine salvage
pathway. |
TGACCTTGATTTATTTTGCATACC
CGAGCAAGACGTTCAGTCCT | 73 |
| Predictors of
treatment response |
| ERCC1 | Excision repair
cross-complementary group 1 | Nucleotide excision
repair pathway, catalyzes the 5′ incision in the process of
excising the DNA lesion. Studied as a predictor of platinum
chemotherapy drugs. |
GAAATTTGTGATACCCCTCGAC
GATCGGAATAAGGGCTTGG | 79 |
| RRM1 | Ribonucleotide
reductase subunit M1 | Subunit of an
enzyme essential for the production of deoxyribonucleotides needed
for DNA synthesis. Studied as a predictor of vinorelbine. |
AAGCACCCTGACTATGCTATCC
GTTATAGAGGTCTTCCATCACATCAC | 71 |
| BRCA1 | Breast cancer
1 | Protein involved in
DNA repair of double-stranded breaks, and recombination. Studied as
a predictor of platinum drugs. |
TTGTTGATGTGGAGGAGCAA
CAGATTCCAGGTAAGGGGTTC | 11 |
| TS | Thymidylate
synthase | Methylation of
deoxyuridylate to deoxythymidylate needed for DNA synthesis.
Studied as a predictor of 5-fluorouracil and folate
antimetabolites. |
CCCAGTTTATGGCTTCCAGT
GCAGTTGGTCAACTCCCTGT | 43 |
In addition to genes whose products are directly
involved in DNA repair and nucleic acid metabolism, we also focused
on gene products that seem to affect the expression of genes
involved in the above mentioned processes. MicroRNAs (also known as
miRNAs or miRs), small non-coding RNA molecules 18–23 nucleotides
in length, make up an immense group of regulatory molecules
involved in carcinogenesis. The human genome may encode over 2,500
miRNAs, which may target ~60% of mammalian genes and are abundant
in many human cell types (see miRNA database available online at
www.mirbase.org). MicroRNAs participate in the
post-transcriptional regulation of gene expression controlling
development and maintain diverse cellular processes including
proliferation, apoptosis, differentiation, motility and
morphogenesis.
The effect of microRNA regulatory networks in cancer
tissue can be oncogenic (by targeting tumor suppressor genes) or
tumor-suppressive (by post-transcriptional repression of oncogenes)
(12). However, the final effect
of any particular miRNA is time and tissue dependent (13).
Many studies have described changes in expression of
miRNAs and their involvement in carcinogenesis, tumor progression,
invasion, metastasis and the effects of treatment in gastric cancer
tissue. MicroRNAs may become valuable diagnostic markers and
therapeutic targets for gastric cancer (14). Based on our research of published
literature, we chose 29 miRNAs, which have a potential role in
carcinogenesis or drug metabolism and therefore could be expected
to influence the efficacy of treatment. A list of the main
characteristics of miRNAs of interest is shown in Table II.
 | Table IIThe analysed microRNAs and their
involvement in the cancer process. |
Table II
The analysed microRNAs and their
involvement in the cancer process.
| Symbol | miRBase accession
no. | Role in gastric
cancer/Prediction potential | Ref. |
|---|
| miR-15b | MIMAT0000417 | Regulates cisplatin
resistance and metastasis by targeting PEBP4 in lung adenocarcinoma
cells | (40) |
| | Modulates multidrug
resistance by targeting BCL2 in human gastric cancer cells | (41) |
| miR-16 | MIMAT0000069 | Associated with
chemosensitivity in gastric cancer | (42) |
| | Modulates multidrug
resistance by targeting BCL2 in human gastric cancer cells | (41) |
| miR-21 | MIMAT0000076 | Stimulates gastric
cancer growth and invasion by inhibiting many tumor suppressors
(PTEN, PDCD4) | (43) |
| | Confers cisplatin
resistance in gastric cancer cells by regulating PTEN | (44) |
| miR-27a | MIMAT0000084 | Functions as an
oncogene in gastric adenocarcinoma by targeting prohibitin | (45) |
| | Potential biomarker
for predicting resistance to fluoropyrimidine-based
chemotherapy | (46) |
| miR-34a | MIMAT0000255 | Inhibits the
growth, invasion and metastasis of gastric cancer by targeting
PDGFR and MET expression | (47) |
| | Regulates
cisplatin-induced gastric cancer cell death by modulating
PI3K/AKT/survivin pathway | (48) |
| miR-99a-3p | MIMAT0004511 | Predicts
fluoropyrimidine-based chemotherapy response in patients with
advanced colorectal cancer | (49) |
| miR-101 | MIMAT0000099 | Downregulated in
gastric cancer and involved in cell migration and invasion | (50) |
| | Enhances cisplatin
sensitivity in bladder cancer cells | (51) |
| miR-106a | MIMAT0000103 | Confers cisplatin
resistance by regulating PTEN/Akt pathway in gastric cancer
cells | (52) |
| | Induces multidrug
resistance in gastric cancer by targeting RUNX3 | (53) |
| miR-107 | MIMAT0000104 | Significantly
dysregulated in gastric adenocarcinoma tissues | (54) |
| | Regulates cisplatin
chemosensitivity of A549 non-small cell lung cancer cell line by
targeting cyclin dependent kinase 8 | (55) |
| miR-141 | MIMAT0000432 | Inhibits tumor
growth and metastasis in gastric cancer | (56) |
| | Overexpression of
miR-141 results in enhanced resistance to cisplatin in gastric
cancer cells | (57) |
| miR-143 | MIMAT0000435 | Suppresses gastric
cancer cell growth and induces apoptosis | (58) |
| | Involved in
cisplatin resistance of gastric cancer cells via targeting IGF1R
and BCL2 | (59) |
| miR-145 | MIMAT0000437 | Suppress
invasion-metastasis cascade in gastric cancer | (60) |
| | Reverses 5-FU
resistance in tumor xenograft models | (61) |
| miR-150 | MIMAT0000451 | Promotes gastric
cancer proliferation by negatively regulating the pro-apoptotic
gene EGR2 | (22) |
| | Reduces cisplatin
chemosensitivity and promotes invasiveness of muscle-invasive
bladder cancer cells | (62) |
| miR-181b | MIMAT0000257 | Aberrantly
overexpressed in gastric cancer cells and primary gastric cancer
tissues | (26) |
| | Prognostic
significance in gastric cancer patients treated with
S-1/oxaliplatin or doxifluridine/oxaliplatin | (25) |
| miR-192 | MIMAT0000222 | miR-215/192
significantly upregulated in gastric cancer tissues from
gastrectomy | (28) |
| | miR-192/miR-215
influence 5-FU resistance in colorectal cancer cell lines | (63) |
| miR-193a-3p | MIMAT0000459 | Associated with
precancerous lesions of gastric cancer | (64) |
| | Regulates the
multi-drug resistance of bladder cancer | (65) |
| miR-202 | MIMAT0002811 | Inhibits the
expression of γ-catenin and BCL-2; miR-202 has decreased expression
in gastric cancer | (66) |
| miR-206 | MIMAT0000462 | Suppresses gastric
cancer cell growth and metastasis | (67) |
| | Inhibition of
gastric cancer progression through the c-Met pathway | (68) |
| miR-211 | MIMAT0000268 | Associated with
gastric cancers as potential biomarkers for gastric cancer
diagnosis and treatment | (69) |
| | Downregulation of
ribonucleotide reductase | (70) |
| miR-218 | MIMAT0000275 | Inhibits invasion
and metastasis of gastric cancer | (71) |
| | Regulates cisplatin
chemosensitivity in breast cancer by targeting BRCA1 | (72) |
| miR-221 | MIMAT0000278 | miR-221/222 are
encoded in tandem and they have the same seed sequence; miR-221 and
miR-222 regulate gastric carcinoma cell proliferation and
radioresistance by targeting PTEN | (33) |
| miR-222 | MIMAT0000279 |
| miR-224 | MIMAT0000281 | Promotes
chemoresistance of lung adenocarcinoma cells to cisplatin | (73) |
| | 5-FU
chemosensitivity is significantly increased in miR-224 knockdown
cells | (74) |
| miR-342-3p | MIMAT0000753 | Upregulation is
associated with chemosensitivity in gastric cancer | (42) |
| miR-372-3p | MIMAT0000724 | Maintains oncogene
characteristics by targeting TNFAIP1 and affects NF-κB signaling in
human gastric carcinoma cells | (75) |
| miR-375 | MIMAT0000728 | Downregulated in
gastric cancer, inhibits cell migration and invasion by targeting
JAK2 | (76) |
| | Predictive for
response for non-small cell lung cancer treated with
cisplatin-vinorelbine A | (77) |
| miR-509-3p | MIMAT0002881 | Inhibits cell
proliferation and migration by targeting CDK2, Rac1, and
PIK3C2A | (78) |
| miR-575 | MIMAT0003240 | Significantly
upregulated in gastric cancer | (79) |
| miR-520h | MIMAT0002867 | Downregulates
histone deacetylase 1 and so contributes to the chemotherapeutic
effect of doxorubicin | (39) |
| | Controls ABCG2
level and thereby anticancer drug response | (80) |
Materials and methods
Ethics statement
The present study was approved by the ethics
committee of the University Hospital in Pilsen (decision from
11.7.2012 to the grant NT14227). Anonymised data were used to
conduct this study.
Patients
This was a retrospective study. The patients were
treated at the Complex Oncology Center of the University Hospital
in Pilsen between 1st January 2000 and 30th June 2013. The
inclusion criteria of this study were: patients with gastric
cancer, with no gastric resection treatment, who underwent
palliative chemotherapy only. We evaluated nearly 1,300 cases, all
of which were treated at the Complex Oncology Center, but our
inclusion criteria meant only 54 cases could be used in the present
study. Stage of disease was determined using the TNM
(tumor-node-metastasis) system of the International Union Against
Cancer (IUCC, 7th edition) (15).
All patients were in the fourth stage of the disease. Each
diagnosis of gastric cancer was verified by a pathologist.
Tissue samples
Biopsy tissue samples, gathered, using endoscopy,
from gastric cancer patients for diagnostic purposes prior to
chemotherapy, were processed by standard laboratory techniques at
the Institute of Pathology of the University Hospital in Pilsen,
Czech Republic. FFPE tissue samples were stored at room temperature
until use. Paraffin sections (4-μm thick) were stained with
hematoxylin and eosin (H&E), microscopically verified by
pathologists and examined in order to identify sites with cancer
cells and sites of adjacent non-cancerous epithelial tissue
suitable for macrodissection. Areas selected for expression
analysis were highlighted manually.
RNA isolation
Total RNA (including microRNA) was extracted from
10-μm FFPE sections following macrodissection of tumor tissue and
adjacent non-cancerous tissue using the miRNeasy FFPE kit (Qiagen,
Hilden, Germany) as we previously described (16). The paired samples (tumor and
adjacent non-cancerous tissue) were only available from 18
patients. The 10-μm sections corresponded to H&E
representatives, on the areas highlighted for macrodissection.
Quantitative estimation of protein coding
gene expression
Quantitative estimation of mRNA of selected genes
(BRCA 1, ERCC1, RRM1 and TS) was performed by a real-time RT-PCR
method with Universal Probe Library (UPL) probes (Roche, Mannheim,
Germany). Reverse transcription was performed on 50 ng of total RNA
with Superscript III reverse transcriptase (Life Technologies,
Carlsbad, CA, USA) and random hexamers as primers. The sequences of
primers and corresponding UPL probes generated by ProbeFinder
software (Roche) are shown in Table
I. The primers were synthesized by East Port Praha (Prague,
Czech Republic). The quantitative estimation was performed in
technical duplicates on Stratagene Mx3005P apparatus (Agilent
Technologies, Santa Clara, CA, USA). The 20-μl PCR reactions
included 1.0 μl of RT product, FastStart TaqMan Probe Master
(Roche), 2.5 μl of each primer and 2.5 μl of UPL probe. The
reactions were incubated in 96-well plates at 95°C for 10 min and
then followed by 48 cycles of 95°C for 10 sec and 60°C for 30
sec.
All the samples were also assessed for the
expression of reference genes GAPDH
(glyceraldehyde-3-phosphate dehydrogenase) and HPRT
(hypoxanthine-guanine phosphoribosyltransferase). Due to generally
low HPRT expression and small yield of RNA isolated from FFPE
tissue (small tissue samples), we were unable to measure the
expression of HPRT in all of our samples. Therefore, we did not use
HPRT data for normalization along with GAPDH and total RNA.
Quantitative estimation of microRNA
expression
A quantitative estimation of selected microRNAs
(Table II) was performed by a RT
real-time PCR method using TaqMan® MicroRNA assays
(Applied Biosystems, Foster City, CA, USA) in technical duplicates
on Stratagene Mx3005P apparatus (Agilent Technologies). A two-step
protocol requires reverse transcription with a miRNA-specific
primer, followed by a real-time PCR with TaqMan® probes.
The assays target only mature miRNAs, not their precursors. We used
RNU6B (U6snRNA) as a normalizer.
Processing of real-time PCR data
Samples were assessed in technical duplicates. The
Ct values were corrected using calibrators to eliminate differences
between the individual runs of the real-time PCR apparatus. In
cases where there was a discrepancy between the results obtained
from both technical duplicates, the sample assessment was repeated.
The results are presented as normalized values as a ratio of the
number of copies of the given gene to that of the reference gene.
To obtain gene expression data we used the ΔΔCt approach
(2−ΔΔCt algorithm).
Statistical analysis
The statistical analysis was performed using SAS 9.3
software (SAS, Institute Inc., Cary, NC, USA). The statistical
results were calculated by a Wilcoxon non-parametric two sample
test. For the maximum hazard ratio (OS, TTP) the Cox regression
hazard model was used. After finding an ‘optimal cut off’ given by
the lowest P-value of log-rank test for the examined markers, the
Kaplan-Meier survival distribution functions determined by the
‘optimal cut off’ in given groups and subgroups were generated.
Results
Prior to our analysis of the relationship between
gene expression and TTP and OS, we compared gene expression in
cancer and adjacent non-cancer epithelial gastric tissue. We found
the expression of miR-221 in cancer tissue to be significantly
higher, while the expression of miR-202 and miR-509 proved to be
lower compared to healthy tissue (P-value 0.013, 0.011 and 0.018,
respectively). However, when drawing conclusions from this
analysis, we have to take into account the low number of paired
samples, caused by the lack of non-cancerous tissue in some of the
FFPE samples.
The Cox regression hazard model was used to
determine the relation of marker level to OS or TTP. Results for
all markers are summarized in Table
III. From the set of the 29 microRNAs of interest, we found
high expression levels of miR-150, miR 342-3p, miR-181b, miR-221,
miR-224 and low levels of miR-520h related to shorter TTP. High
levels of miR-150, miR-192, miR-224, miR-375 and miR-342-3p related
to shorter OS. For markers with statistically significant results,
optimal cut off values were chosen and Kaplan-Meier survival
distribution functions for OS and TPP were generated.
 | Table IIIRelation between level of given
marker and TTP or OS (Cox regresion hazard model). |
Table III
Relation between level of given
marker and TTP or OS (Cox regresion hazard model).
| All patients | 5-FU alone | 5-FU/cisplatin |
|---|
|
|
|
|
|---|
| OS | TTP | OS | TTP | OS | TTP |
|---|
|
|
|
|
|
|
|
|---|
| Marker | P-value | HR | P-value | HR | P-value | HR | P-value | HR | P-value | HR | P-value | HR |
|---|
| ERCC1 | 0.9896 | 1.000 | 0.6452 | 0.547 | 0.2203 | 1.083 | 0.1464 | 1.103 | 0.2740 | * | 0.0679 | * |
| RRM1 | 0.8615 | * | 0.8486 | * | 0.7395 | * | 0.7137 | * | 0.4886 | * | 0.1703 | * |
| BRCA1 | 0.1658 | 17.340 | 0.4511 | 4.261 | 0.1881 | * | 0.2095 | * | 0.9996 | * | 0.4076 | * |
| TS | 0.3422 | * | 0.4118 | * | 0.0524 | 0.985 | 0.4693 | * | 0.5148 | * | 0.3000 | * |
| miR-15b | 0.3772 | 1.247 | 0.5953 | 1.199 | 0.2288 | 2.749 | 0.7050 | 1.380 | 0.2717 | 1.468 | 0.4768 | 1.297 |
| miR-16 | 0.4178 | 1.003 | 0.7514 | 1.001 | 0.4816 | 1.003 | 0.6070 | 1.002 | 0.1216 | 1.053 | 0.1344 | 1.050 |
| miR-21 | 0.5122 | 1.001 | 0.7040 | 1.001 | 0.5362 | 1.001 | 0.5916 | 1.001 | 0.4965 | 1.007 | 0.3586 | 1.011 |
| miR-27a | 0.1592 | 1.105 | 0.3201 | 1.072 | 0.1169 | 1.315 | 0.4536 | 1.141 | 0.1402 | 1.387 | 0.0716 | 1.568 |
| miR-34a | 0.5279 | 1.014 | 0.8185 | 1.005 | 0.5976 | 1.019 | 0.8735 | 1.006 | 0.0567 | 1.536 | 0.0806 | 1.626 |
| miR-99a-3p | 0.1951 | * | 0.7863 | 2.705 | 0.4441 | * | 0.7846 | 5.008 | 0.4712 | * | 0.7964 | 5.091 |
| miR-101 | 0.3734 | 4.040 | 0.6222 | 2.142 | 0.3617 | 4.703 | 0.6637 | 2.110 | 0.9744 | 1.299 | 0.0966 | * |
| miR-106a | 0.4283 | 1.034 | 0.5075 | 1.028 | 0.4467 | 1.038 | 0.6843 | 1.022 | 0.4066 | 1.106 | 0.7731 | 1.039 |
| miR-107 | 0.4980 | 1.829 | 0.6703 | 1.438 | 0.6056 | 1.713 | 0.9425 | 0.928 | 0.1013 | * | 0.1739 | * |
| miR-141 | 0.2236 | 1.241 | 0.1042 | 1.295 | 0.0646 | 1.946 | 0.1968 | 1.609 | 0.7076 | 1.134 | 0.2205 | 1.460 |
| miR-143 | 0.5144 | 1.002 | 0.8042 | 1.001 | 0.5847 | 1.002 | 0.6593 | 1.002 | 0.0619 | 1.395 | 0.0537 | 2.322 |
| miR-145 | 0.4063 | 1.001 | 0.8176 | 1.000 | 0.5373 | 1.001 | 0.6376 | 1.001 | 0.0727 | 1.057 | 0.1059 | 1.121 |
| miR-150 | 0.0494 | 1.004 | 0.0056 | 1.006 | 0.0438 | 1.039 | 0.0743 | 1.034 | 0.1311 | 1.004 | 0.2351 | 1.046 |
|
miR-181b | 0.1406 | 1.061 | 0.0882 | 1.063 | 0.0564 | 1.130 | 0.0333 | 1.777 | 0.3327 | 1.185 | 0.2270 | 1.082 |
| miR-192 | 0.7227 | 1.011 | 0.8569 | 1.006 | 0.0233 | 1.200 | 0.3684 | 1.080 | 0.8814 | 0.992 | 0.5269 | 0.959 |
| miR-193a-3p | 0.2323 | 4.798 | 0.7265 | 1.577 | 0.2401 | 7.129 | 0.5706 | 2.464 | 0.3158 | * | 0.0606 | * |
| miR-202 | 0.3803 | 7.022 | 0.6739 | 0.320 | 0.4932 | 6.787 | 0.6010 | 4.384 | 0.1694 | * | 0.0946 | * |
| miR-206 | 0.0594 | * | 0.4452 | * | 0.1810 | * | 0.2963 | * | 0.2441 | * | 0.1440 | * |
| miR-211 | 0.1548 | * | 0.2151 | * | 0.1155 | * | 0.3240 | * | 0.5506 | * | 0.4309 | * |
| miR-218 | 0.0639 | * | 0.6789 | 2.981 | 0.4176 | * | 0.0687 | * | 0.0961 | * | 0.0847 | * |
| miR-221 | 0.1934 | 1.051 | 0.3627 | 1.034 | 0.6144 | 1.032 | 0.7147 | 1.023 | 0.0160 | 2.438 | 0.0371 | 2.099 |
| miR-222 | 0.5086 | 1.001 | 0.6742 | 1.000 | 0.5679 | 1.001 | 0.6445 | 1.001 | 0.2627 | 1.012 | 0.2698 | 1.012 |
| miR-224 | 0.0175 | 7.609 | 0.2724 | 2.620 | 0.1441 | 4.532 | 0.3603 | 2.602 | 0.0283 | 322.120 | 0.0367 | 436.694 |
|
miR-342-3p | 0.0286 | 1.261 | 0.0144 | 1.383 | 0.0443 | 2.516 | 0.1531 | 2.077 | 0.1383 | 1.272 | 0.1641 | 1.692 |
| miR-372-3p | 0.6113 | 1.000 | 0.1651 | 7.159 | 0.5731 | * | 0.5128 | * | 0.2737 | 6.344 | 0.2863 | 6.080 |
| miR-375 | 0.1968 | * | 0.9422 | 1.000 | 0.4974 | 1.001 | 0.9324 | 1.011 | 0.0427 | 1.362 | 0.8672 | 1.000 |
| miR-509-3p | 0.2446 | 3.507 | 0.6196 | * | 0.6909 | * | 0.5540 | * | 0.1246 | * | 0.1607 | * |
| miR-575 | 0.1999 | * | 0.4849 | * | 0.4778 | * | 0.5664 | * | 0.6211 | * | 0.5531 | * |
|
miR-520h | 0.1712 | * | 0.1190 | * | 0.3799 | * | 0.2106 | * | 0.0977 | * | 0.0483 | 0.584 |
The treatment regimen of all patients included 5-FU.
We noted a definite correlation between high levels of miR-150 and
miR-342-p in cancerous tissue and shorter TTP as well as OS. High
expression of miR-224, however, only proved to have a relation to
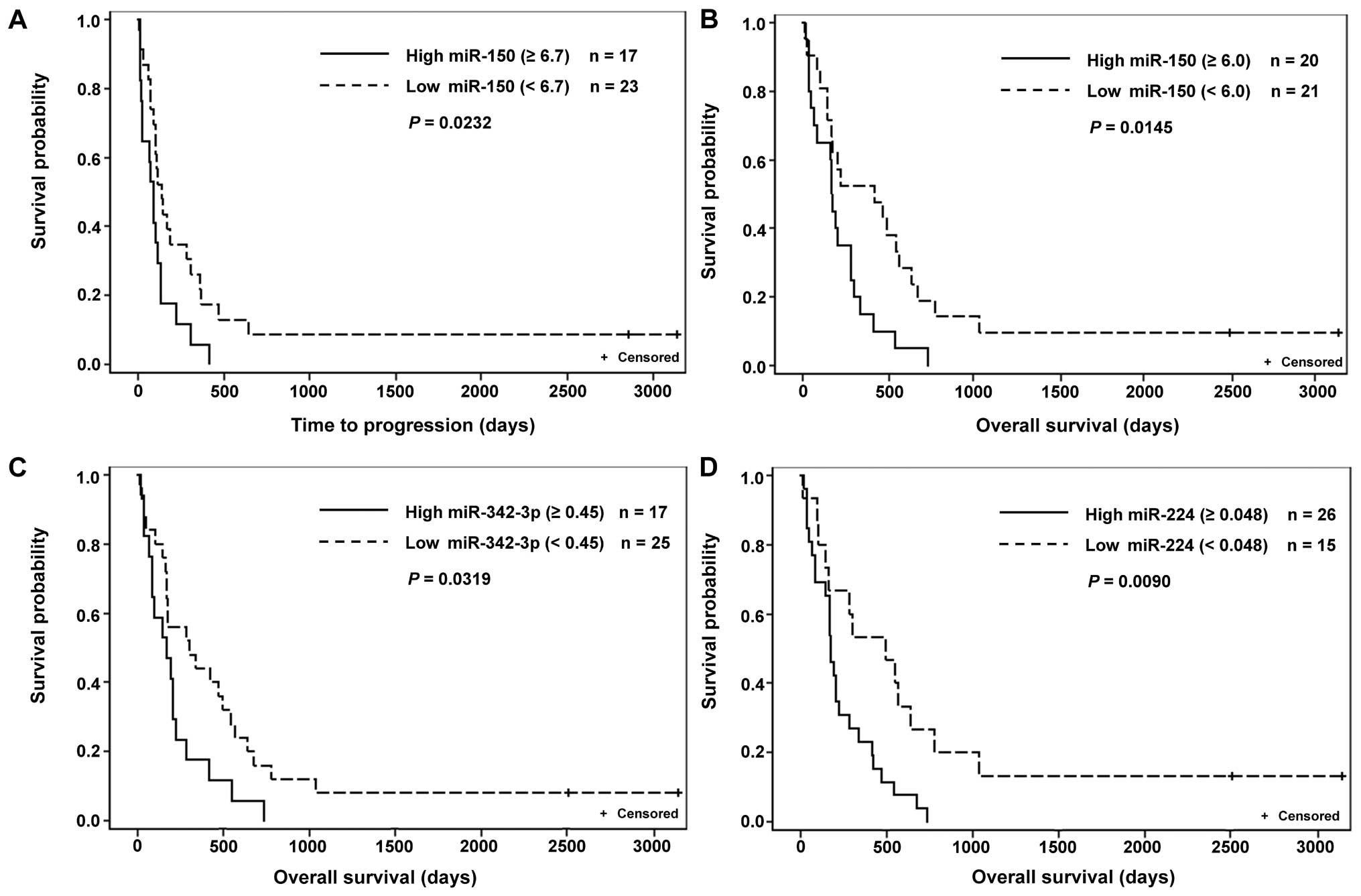
shorter OS (Table IV and Fig. 1).
 | Table IVRelation between level of given
microRNA and TTP or OS (Kaplan-Meier estimation). |
Table IV
Relation between level of given
microRNA and TTP or OS (Kaplan-Meier estimation).
| | | Patients below
cut-off | Patients above
cut-off | |
|---|
| | |
|
| |
|---|
| Marker | No. of
patients | Cut-off | N | Median (days) | N | Median (days) | P-value |
|---|
| Time to progression
(TTP) |
| miR-150 | 40 | 45 | 37 | 113 | 3 | 26 | 0.0016 |
| | 6.7 | 23 | 138 | 17 | 90 | 0.0232 |
| miR-342-3p | 42 | 2.7 | 40 | 106.5 | 1 | 12 | 0.0006 |
| | 0.6 | 31 | 113 | 10 | 66 | 0.0997 |
| Overall survival
(OS) |
| miR-150 | 41 | 45 | 38 | 215 | 3 | 69 | 0.0020 |
| | 6 | 21 | 424 | 20 | 172.5 | 0.0145 |
| miR-342-3p | 42 | 0.45 | 25 | 304 | 17 | 170 | 0.0319 |
| miR-224 | 41 | 0.048 | 15 | 494 | 26 | 175 | 0.0090 |
In the subgroup of patients receiving 5-FU as the
only treatment, we recorded a relation between high levels of
miR-181b and shorter TTP and between high levels of miR-150,
miR-192 and miR-342-p and shorter OS. In the subgroup treated with
both 5-FU and cisplatin, we noted that high levels of miR-221,
miR-224 and low levels of miR-520 related to shorter TTP and high
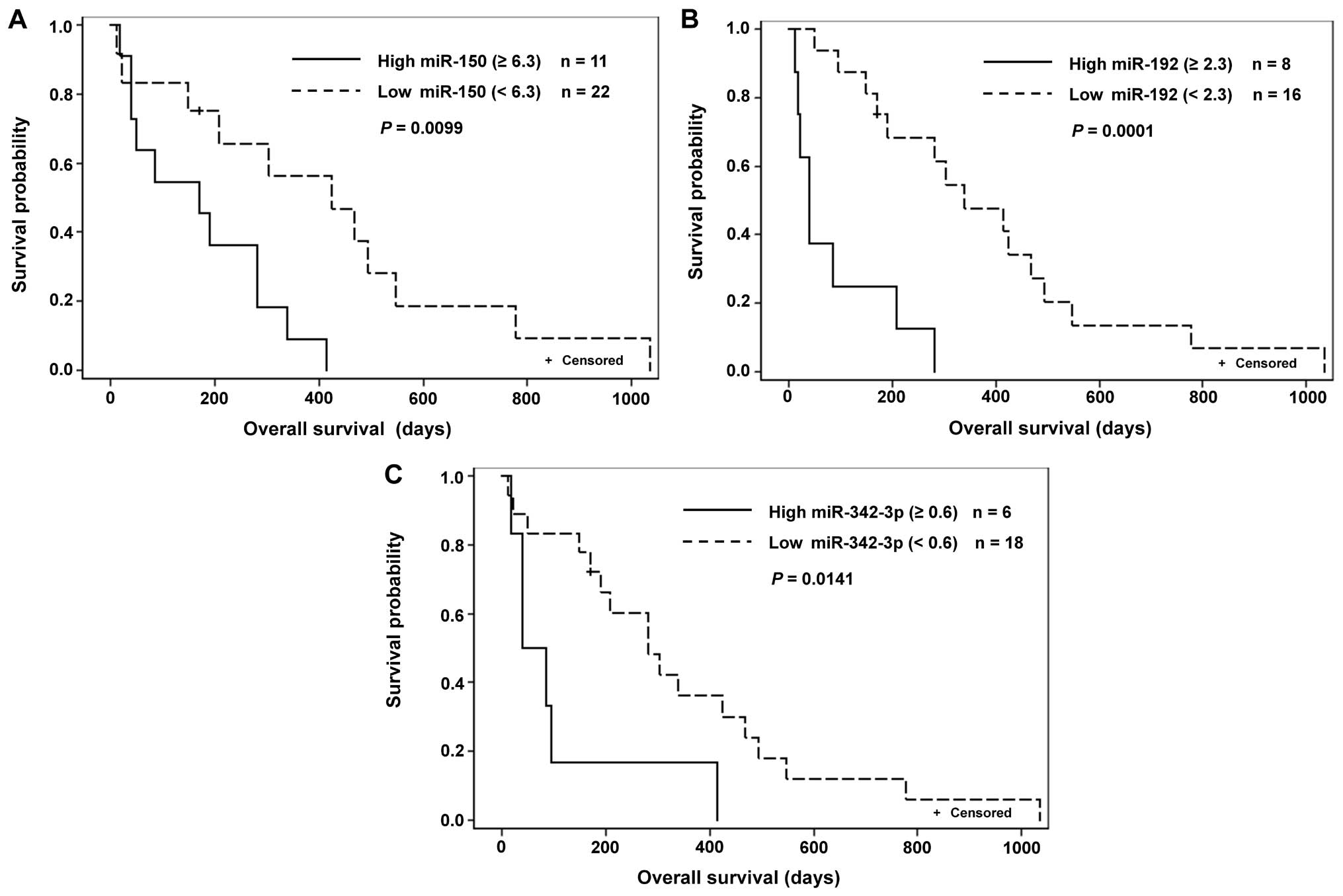
levels of miR-221, miR-224 and miR-375 to shorter OS (Table V and Fig. 2).
 | Table VRelation between level of given
marker and TTP or OS based on the treatment (Kaplan-Meier
estimation). |
Table V
Relation between level of given
marker and TTP or OS based on the treatment (Kaplan-Meier
estimation).
| Marker | No. of
patients | Cut-off | Patients below
cut-off | Patients above
cut-off | P-value | Relation to
OS/TTP |
|---|
|
|
|---|
| N | Median (days) | N | Median (days) |
|---|
| Treatment |
|
5-fluorouracil |
| TS | 14 | 0.008 | 11 | 282 | 3 | 547 | 0.0226 | OS |
| miR-150 | 23 | 6.300 | 12 | 424 | 11 | 170 | 0.0099 | OS |
| miR-181b | 21 | 0.260 | 2 | 13.5 | 19 | 147 | 0.0038 | TTP |
| miR-192 | 24 | 2.300 | 16 | 339 | 8 | 39 | 0.0001 | OS |
| miR-342-3p | 24 | 0.600 | 18 | 282 | 6 | 62.5 | 0.0141 | OS |
|
5-fluorouracil/cisplatin |
| miR-221 | 10 | 0.600 | 4 | 732 | 6 | 129.5 | 0.0038 | OS |
| 10 | 1.500 | 4 | 262.5 | 6 | 67 | 0.0356 | TTP |
| miR-224 | 9 | 0.150 | 5 | 684.5 | 4 | 84 | 0.0049 | OS |
| 9 | 0.150 | 5 | 108 | 4 | 43 | 0.0027 | TTP |
| miR-375 | 9 | 26.000 | 5 | 637 | 4 | 76.5 | 0.0027 | OS |
| miR-520h | 10 | 40.000 | 8 | 88 | 2 | 563.5 | 0.0265 | TTP |
Discussion
The present study focused on patients in advanced
stages of gastric cancer. Therefore, these patients could not have
the tumors surgically removed and underwent palliative treatment
only. The main clinical concern in such cases is deciding which
chemotherapeutic regimen is indicated. The question was whether we
could predict the effect of chemotherapy and thereby determine, if
the aggressive chemotherapeutic treatment, which inevitably
decreases quality of life, would prolong survival. If a low effect
of treatment is predicted, it would be appropriate to offer to
those patients inclusion in new ongoing studies.
In our laboratory assessment we used tissue samples
taken gastroscopically for routine diagnostic purposes and
macrodissected a sample of cancerous tissue, verified by a
pathologist from FFPE sections, to analyse the expression of genes
influencing the effects of therapy. This approach can be easily
translated into clinical practice. We conducted expression analysis
from a large number of tumor cells. Cancerous tissue is
heterogeneous by nature, and originates in the process of clonal
evolution, and therefore examining the collective changes in
expression of a variety of cancer cell types (gathered by
macrodissection) provides a more complex insight regarding
prognosis. We set out to ascertain the prognostic potential of
chosen miRNAs, which influence apoptosis and cell proliferation and
in so doing interact with the mechanism of chemotherapy indicated
in the cases we examined. However, we could not leave out
monitoring protein coding genes frequently investigated as possible
treatment outcome predictors.
Many studies have noted the prognostic value of low
ERCC1 expression in gastric cancer patients undergoing
chemotherapy; a meta-analysis published in 2015 concluded ERCC1 may
be a useful prognostic factor for gastric cancer and furthermore
that low mRNA levels of ERCC1 appear to be associated with a
significant OS benefit to patients treated with platinum-based
chemotherapy (17). However, the
predictive value of the ERCC1 gene for survival and response to
platinum-based chemotherapy in gastric cancer remains controversial
(18). We found no statistically
significant relation between the levels of ERCC1 and survival. It
is possible our results reflect our test group, as in all cases we
examined 5-FU was part of the treatment regimen but platinum-based
chemotherapy was only used in a subgroup, whose analysis was
affected by the small number of patients.
The very nature of the TS enzyme makes its
expression the most commonly examined in relation to 5-FU
chemotherapy. We determined high levels of TS predict longer OS
(Table V). Similar results were
published by Wei et al (19), who conducted their analysis on a
group of patients treated in the same way and also used RT-PCR to
assess TS expression. On the other hand, some studies have shown
high levels of TS to have the opposite effect (20).
From the set of the 29 microRNAs of interest, we
found that high expression levels of miR-150, miR-181b, miR-192,
miR-221, miR-224, miR 342-3p, miR-375 and low expression of
miR-520h relate to unfavourable outcomes for patients (shorter TTP
or OS) Table III.
MicroRNAs have the potential to become accurate,
easily measurable biomarkers, with features fortuitous for
diagnostic testing methods, such as stability in FFPE tissue, blood
and perhaps other bodily fluids (21).
Wu et al (22) found miR-150 was overexpressed in
gastric cancer cell lines and tissue samples and demonstrated
overexpression of miR-150 could promote proliferation and growth of
cancer cells by targeting the tumor-suppressor EGR2. In
undifferentiated gastric cancer, higher miR-150 levels appeared to
be associated with shorter postoperative patient survival, however,
miR-150 was deemed to be an insufficiently independent prognostic
factor in these cases (23). In
the study of Chen et al (24), miR-150 showed decreased expression
in gastric cancer patients compared to healthy test subjects. In
the present study, higher levels of miR-150 showed a relation to
shorter TTP and OS (Table IV;
Figs. 1A and B and 2A).
Compared to normal gastric tissue samples, there is
an overexpression of miR-181b in gastric tumors. Lower levels of
miR-181b relate to longer OS of patients on regimens based on 5-FU
and platinum derivatives (25).
Furthermore, overexpression of miR-181b was found to downregulate
the tissue inhibitor of metalloproteinases 3 (TIMP3) (26). Our results hint at the association
of higher miR-181b levels to shorter TTP of patients treated with
5-FU (Table V). The results of
another study show the ambiguous nature of the effects of certain
miRNA; Chen et al (27)
observed miR-181b was downregulated in human gastric adenocarcinoma
tissue samples compared to adjacent normal gastric tissue and also
described how miR-181b could suppress tumor cell proliferation by
downregulating the expression of cAMP responsive element binding
protein 1 (CREB1).
Our analysis of miR-192 levels showed its high
expression related to shorter OS in the group of patients treated
with 5-FU (Table V and Fig. 2B). To the best of our knowledge, no
other study dealing with the predictive value of miR-192 in gastric
cancer patients treated with 5-FU has been published. Xu et
al (28) found miR-192 to be
upregulated in gastric cancer tissue samples obtained by
gastrectomy. The upregulation of both miR-192 and miR-215 was
related to clinical characteristic such as lymph node metastases,
while the inhibition of miR-215 or miR-192 significantly decreased
gastric cancer cell invasion. The results reported by Chen et
al (29) demonstrate that
elevated circulating miR-192 has the potential to improve the early
detection of distant metastases of GC.
Recently published studies show that miR-221 is an
oncogenic microRNA involved in several malignancies (30,31).
We found higher miR-221 expression in tumor samples in comparison
to adjacent noncancerous tissue. This is in accordance with Liu
et al (32) who found
miR-221 was upregulated in 88% of gastric cancer tissue samples.
Moreover, we observed a relation of high miR-221 expression to
shorter TTP and OS in 5-FU monotherapy treated patients (Table V). The influence of miR-221 on the
effect of chemotherapy is corroborated by published experiments
conducted on the human gastric cancer cell line SGC7901 showing the
knockdown of miR-221 inhibited cell growth and invasion and
increased the radiosensitivity of the cells (33).
We found higher levels of miR-224 indicate shorter
OS (Table IV and Fig. 1D). Mao et al (34) concluded that miR-224 is
overexpressed in human gastric cancer cells. Reducing the
expression of miR-224 can effectively inhibit growth and promote
apoptosis of gastric cancer cells. These results are also supported
by the study of Liu et al (35) who investigated the expression of
miR-224 in the human gastric cancer cell line SGC-7901. In
examining the effects of miR-224 mimics, they observed miR-224
could negatively regulate the expression of Raf-1 kinase inhibitor
protein (RKIP). RKIP contributes to the suppression of
proliferation and invasion of gastric cells.
We determined higher levels of miR-342-3p correlated
to shorter TTP and OS in 5-FU monotherapy treated patients
(Table V and Fig. 2C); similar results were described
in colorectal cancer. High levels of miR-342-3p were associated
with shorter survival time (36).
Kim et al (37) screened
miRNAs associated with response to chemotherapy using microarrays
and found miR-342-p belongs to the miRNAs, whose upregulation is
associated with chemosensitivity in gastric cancer.
Our observations of the relation of high miR-375
expression to shorter OS in 5-FU monotherapy treated patients
(Table V) could be explained by
the findings of Liu et al (38), who showed that miR-375
downregulated p53 expression through an interaction with the 3′ UTR
region of p53. In addition, they observed the expression of miR-375
desensitized cells to ionizing radiation and etoposide.
We demonstrated that higher levels of miR-520h
correlated to longer TTP in 5-FU and cisplatin therapy treated
patients (Table V). Shen et
al (39) found that miR-520h
downregulates histone deacetylase 1 and, thus, contributes to the
chemotherapeutic effect of doxorubicin.
Summarizing the aforelisted results, amongst the
miRNAs we examined, we found six miRNAs (miR-150, miR-181, miR-221,
miR-224, miR-342-p and miR-520h) with a relation to TTP, which
could serve as predictors of the effectiveness of treatment. These
results merit multifactorial analysis, which we were, however,
unable to perform due to the limited number of samples.
In our experience, microRNAs can generally be
assessed with more precision and ease than mRNA of coding genes.
This is essentially due to the fact that miRNA analysis is less
demanding in terms of both quality and quantity of isolated RNA,
features problematic in samples of RNA extracted from FFPE tissue.
FFPE tissue samples are routinely taken and analysed during
standard gastric cancer management and that is why we believe
microRNAs could become clinically applicable predictors of the
effectiveness of palliative treatment in gastric cancer
patients.
Acknowledgements
The study was supported by the grant of Ministry of
Health of the Czech republic NT14227 - Determining predictive
factors for a therapeutic effect of chemotherapy in patients with
stomach cancer (2013–2015, MZ0/NT).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rahman R, Asombang AW and Ibdah JA:
Characteristics of gastric cancer in Asia. World J Gastroenterol.
20:4483–4490. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Coupland VH, Allum W, Blazeby JM, Mendall
MA, Hardwick RH, Linklater KM, Møller H and Davies EA: Incidence
and survival of oesophageal and gastric cancer in England between
1998 and 2007, a population-based study. BMC Cancer. 12:112012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cidon EU, Ellis SG, Inam Y, Adeleke S,
Zarif S and Geldart T: Molecular targeted agents for gastric
cancer: A step forward towards personalized therapy. Cancers
(Basel). 5:64–91. 2013. View Article : Google Scholar
|
|
5
|
Wang JL, Hu Y, Kong X, Wang ZH, Chen HY,
Xu J and Fang JY: Candidate microRNA biomarkers in human gastric
cancer: A systematic review and validation study. PLoS One.
8:e736832013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Menon V and Povirk L: Involvement of p53
in the repair of DNA double strand breaks: Multifaceted roles of
p53 in homologous recombination repair (HRR) and non-homologous end
joining (NHEJ). Subcell Biochem. 85:321–336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Melis JPM, van Steeg H and Luijten M:
Oxidative DNA damage and nucleotide excision repair. Antioxid Redox
Signal. 18:2409–2419. 2013. View Article : Google Scholar :
|
|
8
|
Tian H, Gao Z, Li H, Zhang B, Wang G,
Zhang Q, Pei D and Zheng J: DNA damage response - a double-edged
sword in cancer prevention and cancer therapy. Cancer Lett.
358:8–16. 2015. View Article : Google Scholar
|
|
9
|
Vasquez KM: Targeting and processing of
site-specific DNA interstrand crosslinks. Environ Mol Mutagen.
51:527–539. 2010.PubMed/NCBI
|
|
10
|
Wagner AD, Unverzagt S, Grothe W, Kleber
G, Grothey A, Haerting J and Fleig WE: Chemotherapy for advanced
gastric cancer. Cochrane Database Syst Rev. (3):
CD0040642010.PubMed/NCBI
|
|
11
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar
|
|
12
|
Hagan JP and Croce CM: MicroRNAs in
carcinogenesis. Cytogenet Genome Res. 118:252–259. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Suzuki HI, Katsura A, Matsuyama H and
Miyazono K: MicroRNA regulons in tumor microenvironment. Oncogene.
34:3085–3094. 2015. View Article : Google Scholar
|
|
14
|
Jiang C, Chen X, Alattar M, Wei J and Liu
H: MicroRNAs in tumorigenesis, metastasis, diagnosis and prognosis
of gastric cancer. Cancer Gene Ther. 22:291–301. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sobin LH, Gospodarowicz MK and Wittekind
Ch: TNM classification of malignant tumours. 7th edition.
Wiley-Blackwell; Hoboken, NJ: 2009
|
|
16
|
Kalfert D, Pesta M, Kulda V, Topolcan O,
Ryska A, Celakovsky P, Laco J and Ludvikova M: MicroRNA profile in
site-specific head and neck squamous cell cancer. Anticancer Res.
35:2455–2463. 2015.PubMed/NCBI
|
|
17
|
Song P, Yin Q, Lu M, Fu BO, Wang B and
Zhao Q: Prognostic value of excision repair cross-complementation
group 1 expression in gastric cancer: A meta-analysis. Exp Ther
Med. 9:1393–1400. 2015.PubMed/NCBI
|
|
18
|
Yao A, Wang Y, Peng X, Ye R, Wang Q, Qi Y
and Zhou F: Predictive value of excision repair
cross-complementation group 1 expression for platinum-based
chemotherapy and survival in gastric cancer: A meta-analysis. J
Cancer Res Clin Oncol. 140:2107–2117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wei J, Zou Z, Qian X, Ding Y, Xie L,
Sanchez JJ, Zhao Y, Feng J, Ling Y, Liu Y, et al: ERCC1 mRNA levels
and survival of advanced gastric cancer patients treated with a
modified FOLFOX regimen. Br J Cancer. 98:1398–1402. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hu HB, Kuang L, Zeng XM, Li B, Liu EY and
Zhong MZ: Predictive value of thymidylate synthase expression in
gastric cancer: A systematic review with meta-analysis. Asian Pac J
Cancer Prev. 13:261–267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Faruq O and Vecchione A: microRNA:
Diagnostic Perspective. Front Med (Lausanne). 2:512015.
|
|
22
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: MiR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Katada T, Ishiguro H, Kuwabara Y, Kimura
M, Mitui A, Mori Y, Ogawa R, Harata K and Fujii Y: microRNA
expression profile in undifferentiated gastric cancer. Int J Oncol.
34:537–542. 2009.PubMed/NCBI
|
|
24
|
Chen G, Tang Y, Wu JH and Liu FH: Role of
microRNAs in diagnosis and treatment of the pathogenesis of gastric
cancer. Int J Clin Exp Med. 7:5947–5957. 2014.
|
|
25
|
Jiang J, Zheng X, Xu X, Zhou Q, Yan H,
Zhang X, Lu B, Wu C and Ju J: Prognostic significance of miR-181b
and miR-21 in gastric cancer patients treated with S-1/Oxaliplatin
or Doxifluridine/oxaliplatin. PLoS One. 6:e232712011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Guo JX, Tao QS, Lou PR, Chen XC, Chen J
and Yuan GB: miR-181b as a potential molecular target for
anticancer therapy of gastric neoplasms. Asian Pac J Cancer Prev.
13:2263–2267. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen L, Yang Q, Kong W-Q, Liu T, Liu M, Li
X and Tang H: MicroRNA-181b targets cAMP responsive element binding
protein 1 in gastric adenocarcinomas. IUBMB Life. 64:628–635. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu YJ and Fan Y: MiR-215/192 participates
in gastric cancer progression. Clin Transl Oncol. 17:34–40. 2015.
View Article : Google Scholar
|
|
29
|
Chen Q, Ge X, Zhang Y, Xia H, Yuan D, Tang
Q, Chen L, Pang X, Leng W and Bi F: Plasma miR-122 and miR-192 as
potential novel biomarkers for the early detection of distant
metastasis of gastric cancer. Oncol Rep. 31:1863–1870.
2014.PubMed/NCBI
|
|
30
|
Sun T, Yang M, Kantoff P and Lee GS: Role
of microRNA-221/-222 in cancer development and progression. Cell
Cycle. 8:2315–2316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang J, Liu S, Sun GP, Wang F, Zou YF,
Jiao Y, Ning J and Xu J: Prognostic significance of
microRNA-221/222 expression in cancers: Evidence from 1,204
subjects. Int J Biol Markers. 29:e129–e141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu K, Li G, Fan C, Diao Y, Wu B and Li J:
Increased expression of MicroRNA-221 in gastric cancer and its
clinical significance. J Int Med Res. 40:467–474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F,
Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z and Chun-Sheng
K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Mao S, He N, Xin L, Zeng F and Cao J:
Effect of antisense miR-224 on gastric cancer cell proliferation
and apoptosis. Zhonghua Zhong Liu Za Zhi. 36:92–96. 2014.(In
Chinese). PubMed/NCBI
|
|
35
|
Liu H, Li P, Li B, Sun P, Zhang J, Wang B
and Jia B: RKIP suppresses gastric cancer cell proliferation and
invasion and enhances apoptosis regulated by microRNA-224. Tumour
Biol. 35:10095–10103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H
and Yu H: Prognostic value of miR-221-3p, miR-342-3p and miR-491-5p
expression in colon cancer. Am J Transl Res. 6:391–401.
2014.PubMed/NCBI
|
|
37
|
Kim CH, Kim HK, Rettig RL, Kim J, Lee ET,
Aprelikova O, Choi IJ, Munroe DJ and Green JE: miRNA signature
associated with outcome of gastric cancer patients following
chemotherapy. BMC Med Genomics. 4:792011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu Y, Xing R, Zhang X, Dong W, Zhang J,
Yan Z, Li W, Cui J and Lu Y: miR-375 targets the p53 gene to
regulate cellular response to ionizing radiation and etoposide in
gastric cancer cells. DNA Repair (Amst). 12:741–750. 2013.
View Article : Google Scholar
|
|
39
|
Shen Q, Yao Q, Sun J, Feng L, Lu H, Ma Y,
Liu L, Wang F, Li J, Yue Y, et al: Downregulation of histone
deacetylase 1 by microRNA-520h contributes to the chemotherapeutic
effect of doxorubicin. FEBS Lett. 588:184–191. 2014. View Article : Google Scholar
|
|
40
|
Zhao Z, Zhang L, Yao Q and Tao Z: miR-15b
regulates cisplatin resistance and metastasis by targeting PEBP4 in
human lung adenocarcinoma cells. Cancer Gene Ther. 22:108–114.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xia L, Zhang D, Du R, Pan Y, Zhao L, Sun
S, Hong L, Liu J and Fan D: miR-15b and miR-16 modulate multidrug
resistance by targeting BCL2 in human gastric cancer cells. Int J
Cancer. 123:372–379. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kim CH, Kim HK, Rettig RL, Kim J, Lee ET,
Aprelikova O, Choi IJ, Munroe DJ and Green JE: miRNA signature
associated with outcome of gastric cancer patients following
chemotherapy. BMC Med Genomics. 4:792011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Li L, Zhou L, Li Y, Lin S and Tomuleasa C:
MicroRNA-21 stimulates gastric cancer growth and invasion by
inhibiting the tumor suppressor effects of programmed cell death
protein 4 and phosphatase and tensin homolog. J BUON. 19:228–236.
2014.PubMed/NCBI
|
|
44
|
Yang SM, Huang C, Li XF, Yu MZ, He Y and
Li J: miR-21 confers cisplatin resistance in gastric cancer cells
by regulating PTEN. Toxicology. 306:162–168. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar
|
|
46
|
Huang D, Wang H, Liu R, Li H, Ge S, Bai M,
Deng T, Yao G and Ba Y: miRNA27a is a biomarker for predicting
chemosensitivity and prognosis in metastatic or recurrent gastric
cancer. J Cell Biochem. 115:549–556. 2014. View Article : Google Scholar
|
|
47
|
Peng Y, Guo JJ, Liu YM and Wu XL:
MicroRNA-34A inhibits the growth, invasion and metastasis of
gastric cancer by targeting PDGFR and MET expression. Biosci Rep.
34:342014. View Article : Google Scholar
|
|
48
|
Cao W, Yang W, Fan R, Li H, Jiang J, Geng
M, Jin Y and Wu Y: miR-34a regulates cisplatin-induce gastric
cancer cell death by modulating PI3K/AKT/survivin pathway. Tumour
Biol. 35:1287–1295. 2014. View Article : Google Scholar
|
|
49
|
Molina-Pinelo S, Carnero A, Rivera F,
Estevez-Garcia P, Bozada JM, Limon ML, Benavent M, Gomez J, Pastor
MD, Chaves M, et al: MiR-107 and miR-99a-3p predict chemotherapy
response in patients with advanced colorectal cancer. BMC Cancer.
14:6562014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang HJ, Ruan HJ, He XJ, Ma YY, Jiang XT,
Xia YJ, Ye ZY and Tao HQ: MicroRNA-101 is down-regulated in gastric
cancer and involved in cell migration and invasion. Eur J Cancer.
46:2295–2303. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bu Q, Fang Y, Cao Y, Chen Q and Liu Y:
Enforced expression of miR-101 enhances cisplatin sensitivity in
human bladder cancer cells by modulating the cyclooxygenase-2
pathway. Mol Med Rep. 10:2203–2209. 2014.PubMed/NCBI
|
|
52
|
Fang Y, Shen H, Li H, Cao Y, Qin R, Long
L, Zhu X, Xie C and Xu W: miR-106a confers cisplatin resistance by
regulating PTEN/Akt pathway in gastric cancer cells. Acta Biochim
Biophys Sin (Shanghai). 45:963–972. 2013. View Article : Google Scholar
|
|
53
|
Zhang Y, Lu Q and Cai X: MicroRNA-106a
induces multidrug resistance in gastric cancer by targeting RUNX3.
FEBS Lett. 587:3069–3075. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang S, Lv C, Jin H, Xu M, Kang M, Chu H,
Tong N, Wu D, Zhu H, Gong W, et al: A common genetic variation in
the promoter of miR-107 is associated with gastric adenocarcinoma
susceptibility and survival. Mutat Res. 769:35–41. 2014. View Article : Google Scholar
|
|
55
|
Zhang Z, Zhang L, Yin ZY, Fan XL, Hu B,
Wang LQ and Zhang D: miR-107 regulates cisplatin chemosensitivity
of A549 non-small cell lung cancer cell line by targeting cyclin
dependent kinase 8. Int J Clin Exp Pathol. 7:7236–7241. 2014.
|
|
56
|
Zuo QF, Zhang R, Li BS, Zhao YL, Zhuang Y,
Yu T, Gong L, Li S, Xiao B and Zou QM: MicroRNA-141 inhibits tumor
growth and metastasis in gastric cancer by directly targeting
transcriptional co-activator with PDZ-binding motif, TAZ. Cell
Death Dis. 6:e16232015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhou X, Su J, Zhu L and Zhang G:
Helicobacter pylori modulates cisplatin sensitivity in gastric
cancer by down-regulating miR-141 expression. Helicobacter.
19:174–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q,
Dan ZL, Tian DA and Zhang P: MicroRNA-143 suppresses gastric cancer
cell growth and induces apoptosis by targeting COX-2. World J
Gastroenterol. 19:7758–7765. 2013. View Article : Google Scholar
|
|
59
|
Zhuang M, Shi Q, Zhang X, Ding Y, Shan L,
Shan X, Qian J, Zhou X, Huang Z, Zhu W, et al: Involvement of
miR-143 in cisplatin resistance of gastric cancer cells via
targeting IGF1R and BCL2. Tumour Biol. 36:2737–2745. 2015.
View Article : Google Scholar
|
|
60
|
Gao P, Xing AY, Zhou GY, Zhang TG, Zhang
JP, Gao C, Li H and Shi DB: The molecular mechanism of microRNA-145
to suppress invasion-metastasis cascade in gastric cancer.
Oncogene. 32:491–501. 2013. View Article : Google Scholar
|
|
61
|
Liu RL, Dong Y, Deng YZ, Wang WJ and Li
WD: Tumor suppressor miR-145 reverses drug resistance by directly
targeting DNA damage-related gene RAD18 in colorectal cancer.
Tumour Biol. 36:5011–5019. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lei Y, Hu X, Li B, Peng M, Tong S, Zu X,
Wang Z, Qi L and Chen M: miR-150 modulates cisplatin
chemosensitivity and invasiveness of muscle-invasive bladder cancer
cells via targeting PDCD4 in vitro. Med Sci Monit. 20:1850–1857.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Boni V, Bitarte N, Cristobal I, Zarate R,
Rodriguez J, Maiello E, Garcia-Foncillas J and Bandres E:
miR-192/miR-215 influence 5-fluorouracil resistance through cell
cycle-mediated mechanisms complementary to its post-transcriptional
thymidilate synthase regulation. Mol Cancer Ther. 9:2265–2275.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Wang XW, Wu Y, Wang D and Qin ZF: MicroRNA
network analysis identifies key microRNAs and genes associated with
precancerous lesions of gastric cancer. Genet Mol Res.
13:8695–8703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Deng H, Lv L, Li Y, Zhang C, Meng F, Pu Y,
Xiao J, Qian L, Zhao W, Liu Q, et al: miR-193a-3p regulates the
multi-drug resistance of bladder cancer by targeting the LOXL4 gene
and the oxidative stress pathway. Mol Cancer. 13:2342014.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhao Y, Li C, Wang M, Su L, Qu Y, Li J, Yu
B, Yan M, Yu Y, Liu B, et al: Decrease of miR-202-3p expression, a
novel tumor suppressor, in gastric cancer. PLoS One. 8:e697562013.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ren J, Huang HJ, Gong Y, Yue S, Tang LM
and Cheng SY: MicroRNA-206 suppresses gastric cancer cell growth
and metastasis. Cell Biosci. 4:262014. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zheng Z, Yan D, Chen X, Huang H, Chen K,
Li G, Zhou L, Zheng D, Tu L and Dong XD: MicroRNA-206: Effective
inhibition of gastric cancer progression through the c-Met pathway.
PLoS One. 10:e01287512015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Yan W, Wang S, Sun Z, Lin Y, Sun S, Chen J
and Chen W: Identification of microRNAs as potential biomarker for
gastric cancer by system biological analysis. BioMed Res Int.
2014:9014282014. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Maftouh M, Avan A, Funel N, Frampton AE,
Fiuji H, Pelliccioni S, Castellano L, Galla V, Peters GJ and
Giovannetti E: miR-211 modulates gemcitabine activity through
downregulation of ribonucleotide reductase and inhibits the
invasive behavior of pancreatic cancer cells. Nucleosides
Nucleotides Nucleic Acids. 33:384–393. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Tie J, Pan Y, Zhao L, Wu K, Liu J, Sun S,
Guo X, Wang B, Gang Y, Zhang Y, et al: MiR-218 inhibits invasion
and metastasis of gastric cancer by targeting the Robo1 receptor.
PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
He X, Xiao X, Dong L, Wan N, Zhou Z, Deng
H and Zhang X: MiR-218 regulates cisplatin chemosensitivity in
breast cancer by targeting BRCA1. Tumour Biol. 36:2065–2075. 2015.
View Article : Google Scholar
|
|
73
|
Wang H, Zhu LJ, Yang YC, Wang ZX and Wang
R: MiR-224 promotes the chemoresistance of human lung
adenocarcinoma cells to cisplatin via regulating G1/S
transition and apoptosis by targeting p21WAF1/CIP1. Br J
Cancer. 111:339–354. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Amankwatia EB, Chakravarty P, Carey FA,
Weidlich S, Steele RJ, Munro AJ, Wolf CR and Smith G: MicroRNA-224
is associated with colorectal cancer progression and response to
5-fluorouracil-based chemotherapy by KRAS-dependent and
-independent mechanisms. Br J Cancer. 112:1480–1490. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhou C, Li X, Zhang X, Liu X, Tan Z, Yang
C and Zhang J: microRNA-372 maintains oncogene characteristics by
targeting TNFAIP1 and affects NF-κB signaling in human gastric
carcinoma cells. Int J Oncol. 42:635–642. 2013.
|
|
76
|
Xu Y, Jin J, Liu Y, Huang Z, Deng Y, You
T, Zhou T, Si J and Zhuo W: Snail-regulated MiR-375 inhibits
migration and invasion of gastric cancer cells by targeting JAK2.
PLoS One. 9:e995162014. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Berghmans T, Ameye L, Willems L, Paesmans
M, Mascaux C, Lafitte JJ, Meert AP, Scherpereel A, Cortot AB,
Cstoth I, et al; European Lung Cancer Working Party. Identification
of microRNA-based signatures for response and survival for
non-small cell lung cancer treated with cisplatin-vinorelbine A
ELCWP prospective study. Lung Cancer. 82:340–345. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yoon S, Han E, Choi YC, Kee H, Jeong Y,
Yoon J and Baek K: Inhibition of cell proliferation and migration
by miR-509-3p that targets CDK2, Rac1, and PIK3C2A. Mol Cells.
37:314–321. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
80
|
To KKW, Robey RW, Knutsen T, Zhan Z, Ried
T and Bates SE: Escape from hsa-miR-519c enables drug-resistant
cells to maintain high expression of ABCG2. Mol Cancer Ther.
8:2959–2968. 2009. View Article : Google Scholar : PubMed/NCBI
|