Introduction
Pineal parenchymal cells in the brain, which are
distinct from neuronal and glial cells, can give rise to tumors
such as pineocytomas and malignant pineoblastomas (PBs) (1). PB is a highly malignant, primitive
embryonal tumor of the pineal gland with a predilection for
children (2). PBs account for only
0.6% of pediatric brain tumors, but they display aggressive
behavior that is associated with low survival rates (3). Studies have shown that the 5-year
survival rate of PB patients is 58% (4), and the median survival following
surgical intervention is 25.7 months (5). We believe that the poor clinical
outcome for PB patients might be attributable to the effect of
refractory cells that show resistance to the current mode of
treatment (6,7).
Previous studies have suggested that a distinct
population of tumor-initiating cells known as cancer stem cells
(CSCs) exist in cancers such as leukemia (8), solid tumors of epithelial origin
(9), glioblastomas (GBMs)
(10,11) and medulloblastomas (10–14).
These cells are characterized by their ability to self-renew and
form secondary tumorspheres (TSs), ability to undergo limited
multipotent differentiation, and successful tumorigenesis upon
implantation (11,12,15).
It has been suggested that the incomplete elimination of these TSs
may give rise to cancer recurrence (16–18).
To the best of our knowledge, isolation of TSs from recurrent PB
(rPB) has not been reported. We confirmed our hypothesis that TSs
could be isolated from rPB and assessed their stemness, neuro-glial
differentiation and invasion characteristics in comparison with TSs
from GBM. A mouse orthotopic xenograft model was established to
compare histopathological properties and DNA fingerprint with the
original patient tumor. In the present study, we report the
isolation and the characterization of rPB TSs for cellular
immortalization in an rPB patient.
Materials and methods
Patient clinical information
A 6-year-old male patient was admitted to the
hospital for headache, nausea and vomiting, which had continued for
1 month. Magnetic resonance imaging (MRI) revealed a solid mass in
the pineal region with hydrocephalus (Fig. 1A). The patient underwent a series
of chemotherapy and radiation therapy following endoscopic third
ventriculostomy and biopsy, which were diagnosed as PB. The tumor
recurred on the left frontal lobe 10 months after the initial
therapy and was removed using an occipital transtentorial approach.
The final pathological diagnosis was also consistent with PB. A
year later, the tumor recurred in the fronto-parietal region and
the patient's condition deteriorated (Fig. 1B). The tumor was removed again and
a fresh tumor specimen was obtained in the operating room through
the cryostat laboratory. The patient relapsed with leptomeningeal
seeding 2 months after the surgery and subsequently passed away,
despite a series of additional surgery, chemotherapy and radiation
therapy. The patient diagnosed with GBM was a 61-year-old male, who
was presented with tingling sensation in the left hand and
twitching in the left arm and face. MRI showed a 3.5-cm-sized
enhanced mass in the right parietal lobe. The patient underwent
total removal of the tumor. Pathology showed GBM and the patient
underwent concurrent chemoradiation therapy as well as a series of
chemotherapy (temozolomide). Chemotherapy was reinitiated after
radiological evidence of tumor recurrence.
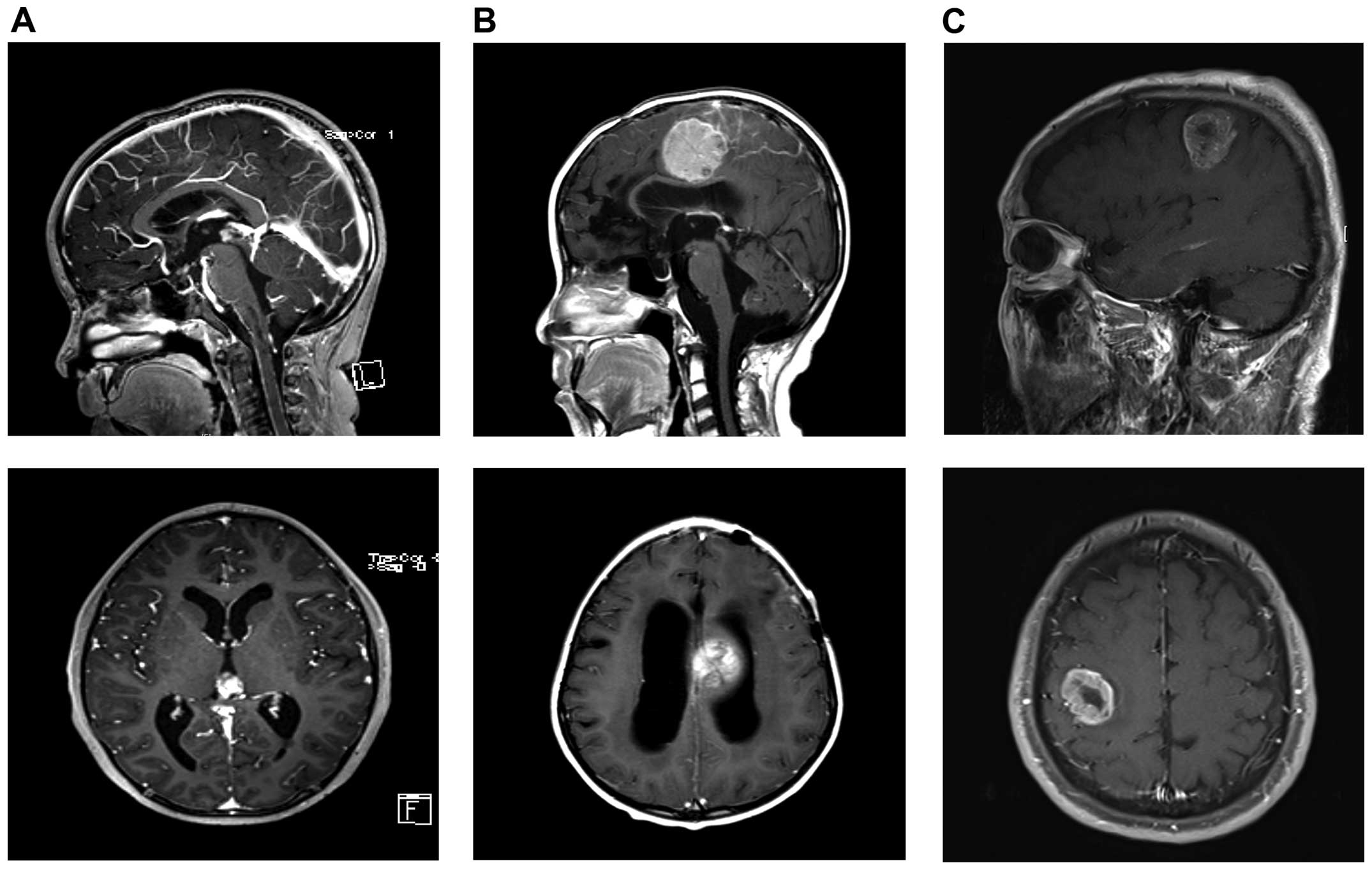 | Figure 1Patient images. (A) Images of the
initial PB. MRI showed enhanced pineal mass with hydrocephalus.
Top, T1 sagittal, enhanced; bottom, T1 axial, enhanced. (B) Images
of rPB. MRI showed enhanced left fronto-parietal mass that was
compressing the corpus callosum with hydrocephalus. Top, T1
sagittal, enhanced; bottom, T1 axial, enhanced. (C) Images of GBM.
MRI showed enhanced mass with internal necrosis along the right
corona radiate in the right perirolandic area. Top, T1 sagittal,
enhanced; bottom, T1 axial, enhanced. |
Isolation and culture of rPB TSs
A surgical specimen from the rPB patient was freshly
obtained from the operating room. Informed consent was provided
according to the Declaration of Helsinki. Neuropathologists
diagnosed each surgical specimen according to the WHO
classification criteria (19). TSs
were isolated from the rPB specimen using a modification of
previous methods for TS isolation from human brain cancers
(11,13,15,20,21).
Briefly, the cell isolation procedure was performed within 60 min
of PB removal using a mechanical dissociation method. The surgical
specimen was minced and dissociated with a scalpel in Dulbecco's
modified Eagle's medium/Nutrient Mixture F-12 (DMEM/F-12;
Mediatech, Manassas, VA, USA) and then passed through a series of
100-μm nylon mesh cell strainers (BD Falcon; BD Biosciences
Franklin Lakes, NJ, USA). Cell suspensions were washed twice in
DMEM/F-12 and cultured in complete TS media composed of DMEM/F-12
containing B27 supplements (1X; Invitrogen, San Diego, CA, USA), 20
ng/ml of basic fibroblast growth factor (bFGF; Sigma, St. Louis,
MO, USA), 20 ng/ml of epidermal growth factor (EGF; Sigma), and 50
U/ml penicillin/50 mg/ml streptomycin (11,22–25).
In addition, glioblastoma (GBM) TSs (TS13-20), isolated from a
primary GBM patient, were used for comparison with rPB TSs.
Immunocytochemical staining
For investigation of surface and intracellular
antigen expression profiles, rPB TSs were transferred to cover
slides, fixed with 2% paraformaldehyde for 7 min, and then treated
with a 3:1 ratio of methanol and acetic acid for 3 min. The cells
were then washed and permeabilized by incubating with 0.1% Triton
X-100 for 10 min. After blocking with 1% bovine serum albumin (BSA;
Amresco, Solon, OH, USA) for 1 h, cells were incubated with primary
antibodies for 2 h at room temperature. The following antibodies
were used: rabbit anti-CD133 (1:250, ab19898; Abcam Dawinbio Inc.,
Hanam, Korea), rabbit anti-musashi (1:250, ab52865; Abcam) and
rabbit anti-podoplanin (1:250, ab10274; Abcam). Primary antibody
against CD133 was detected with goat anti-rabbit IgG conjugated
with Alexa Fluor 555 (1:2,000; Invitrogen), which is spectrally
similar to Cy3. Alexa Fluor 488-conjugated goat anti-rabbit IgG
(1:2,000; Invitrogen) was used to detect antibodies against musashi
and podoplanin. The cells were mounted with Vectashield H-1200
mounting media containing 4′6-diamidino-2-phenylindole (DAPI;
Vector Laboratories, Burlingame, CA, USA) to counterstain nuclei.
Phosphate-buffered saline (PBS; Dawinbio Inc., Hanam, Korea) was
used for all washing steps, and antibody diluent reagent solution
(Invitrogen) was used to dilute antibodies. As a negative control,
only the secondary antibody was used. A fluorescence microscope
(1X71; Olympus Korea, Co., Ltd., Seoul, Korea) and DP Controller
software (Olympus Korea) were used for observing and photographing
the cells.
Reverse-transcription polymerase chain
reaction (RT-PCR)
Total RNA was isolated from rPB TSs (TS13-19) and
GBM TSs (TS13-20) using an RNeasy kit (Qiagen, Hilden, Germany).
cDNA was synthesized from 2 μg of total RNA using the SuperScript
II First-Strand Synthesis system (Invitrogen). The following
oligonucleotide primer pairs were used for PCR: GAPDH
(glyceraldehyde-3-phosphate dehydrogenase), 5′-AGG GGT CTA CAT GGC
AAC TG-3′ and 5′-ACC CAG AAG ACT GTG GAT GG-3′; CD133, 5′-GCC AGC
CTC AGA CAG AAA AC-3′ and 5′-TAC CTG GTG ATT TGC CAC AA-3′;
podoplanin, 5′-CCA GCG AAG ACC GCT ATA AB-3′ and 5′-AGA GGA GCC AAG
TCT GGT GA-3′; musashi-1, 5′-ACC CCC ACA TTC TCT CAC TG-3′ and
5′-GAG ACA CCG GAG GAT GGT AA-3′; GFAP (glial fibrillary acidic
protein), 5′-AGA TCC ACG AGG AGG AGG TT-3′ and 5′-CGG CGT TCC ATT
TAC AAT CT-3′; Olig2 (oligodendrocyte transcription factor 2),
5′-CAG AAG CGC TGA TGG TCA TA-3′ and 5′-AAGGGTGTTACACGGCAGAC-3′;
TUBB3 (β-tubulin III), 5′-CAT CCA GAG CAA GAA CAG CA-3′ and 5′-GCC
TGG AGC TGC AAT AAG AC-3′; β-catenin, 5′-GCT TGG TTC ACC AGT GGA
TT-3′ and 5′-GAG TCC CAA GGA GAC CTT CC-3′; snail, 5′-GAG CAT ACA
GCC CCA TCA CT-3′ and 5′-TTG GAG CAG TTT TTG CAC TG-3′; Zeb1 (zinc
finger E-box binding homeobox 1), 5′-GAC AGG GCT GAA AGT AGT CAA
GC-3′ and 5′-GGT AGT TAG CAC GGG TTG GA-3′.
Quantitative real-time PCR
Total RNA from rPB TSs was prepared using an RNeasy
kit (Qiagen) and reverse transcribed using a First-Strand cDNA
Synthesis kit (Invitrogen) according to the manufacturer's
instructions. Changes in expression levels were quantitatively
analyzed using a real-time PCR machine (StepOne Plus; Applied
Biosystems, Foster City, CA, USA). Specific commercial TaqMan
probes (Applied Biosystems) were used to quantify levels of the
following mRNAs: CD133, podoplanin, nestin, musashi-1, Sox2, Oct4,
β-catenin, snail and Zeb1. For analysis of relative gene
expression, rPB TSs were tested in triplicate; transcript levels of
GBM TSs were monitored as an internal control.
Neuro-glial differentiation
The multipotency of rPB TSs was tested by examining
neural lineage expression by immunocytochemical staining, as
previously described (11,20,26).
Briefly, after being seeded onto chamber slides (Lab-Tek II; Nalge
Nunc International, Rochester, NY, USA), cells were grown in neural
differentiation media containing 10% fetal bovine serum (FBS;
Lonza) and 1X B27 supplement (Invitrogen) for up to 14 days. Cells
were then fixed with 2% paraformaldehyde for 7 min at 4°C, and
permeabilized by incubating with 0.1% Triton X-100 for 10 min.
After blocking with 1% BSA (Amresco) for 1 h, cells were
immunostained with the following antibodies: rabbit anti-GFAP
(1:200 dilution; Dako, Carpinteria, CA, USA), mouse anti-MBP
(myelin basic protein, 1:200 dilution; Chemicon International,
Inc., Temecula, CA, USA), mouse anti-NeuN (1:100 dilution;
Chemicon) and mouse anti-TUBB3 (Tuj1, 1:200 dilution; Chemicon).
The primary antibodies were detected with Cy3-conjugated anti-mouse
or anti-rabbit secondary antibodies (1:200 dilution; Jackson
ImmunoResearch Laboratories, West Grove, PA, USA), as appropriate.
Nuclei were counterstained with DAPI (Vector Laboratories). Slides
were examined and photographed using a fluorescence microscope.
Three-dimensional (3D) invasion
assay
For invasion assays, collagen I/Matrigel matrices
were prepared from 2.4 mg/ml of high-concentration rat tail
collagen type I (Corning Life Sciences, Tewksbury, MA, USA), 2.1
mg/ml of Matrigel (Corning Life Sciences), 10% NaHCO3
and 2X TS culture medium. The solution was well mixed and kept at
4°C before use. Single rPB TSs and GBM TSs were each placed
individually onto 100 μl collagen I/Matrigel matrices in a 96-well
plate and plates were incubated at 37.4°C in a 5% CO2
environment for 30 min. After full gelation of the matrix, 100 μl
of TS culture medium was added on top. The dynamic morphology of
rPB TSs and GBM TSs was observed by collecting images using an
inverted microscope (Optinity KI 400; Intron Biotechnology, Inc.,
Seongnam, Korea). For quantification, the maximal area covered by
migrating edges of cells was used as the parameter for defining
invasiveness, calculated as the invaded area at a certain
time/spheroid area at initial time x 100. Data were analyzed using
ToupView image analysis software (x64 v3.7.1460; AmScope, Irvine,
CA, USA).
Orthotopic rPB TS xenograft
Four-to 8-week-old male athymic nude mice (Central
Lab. Animal Inc., Seoul, Korea) were used for experiments. Mice
were housed in micro-isolator cages under sterile conditions and
observed for at least 1 week before study initiation to ensure
proper health. Lighting, temperature and humidity were controlled
centrally. All experimental procedures were approved by the Yonsei
University College of Medicine Institutional Animal Care and Use
Committee. Mice were anesthetized with a solution of Zoletil (30
mg/kg; Virbac Korea, Co., Ltd., Seoul, Korea) and xylazine (10
mg/kg; Bayer Korea, Seoul, Korea), delivered intraperitoneally. rPB
TSs were implanted into the right frontal lobe of nude mice using a
guide-screw system within the skull, as previously described
(22,24,25,27–29).
Mice received 5×105 rPB TSs via a Hamilton syringe
(Dongwoo Science, Co., Seoul, Korea), inserted to a depth of 4.5
mm. rPB TSs were injected into three mice simultaneously using a
multiple micro-infusion syringe pump (Harvard Apparatus, Holliston,
MA, USA) at a speed of 0.5 μl/min. Body weights of mice were
checked every other day. If body weight decreased by >15%
compared to the original weight, mice were euthanized according to
the study protocol. Formalin-fixed, paraffin-embedded tissue blocks
were used for the generation of slides for histologic examination.
Hematoxylin and eosin (H&E)-stained sections were reviewed by a
pathologist.
DNA extraction and forensic short tandem
repeat (STR) typing
DNA was extracted from paraffin embedded tissues and
their stem cells using a QIAamp DNA Mini kit (Qiagen) according to
the manufacturer's instructions. DNA extracted from reference
tissues and stem cells was measured using a NanoDrop 1000
Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and
diluted to 1 ng/μl. For specific detection of human DNA in
xenografts extracted from mouse tissue, DNA was quantified using a
Quantifiler Duo DNA Quantification kit (Applied Biosystems),
according to the manufacturer's instructions. The DNA quantity
ranged from 42 to 126 pg/μl. Extracted DNA was tested for the
genotypes of amelogenin and 23 forensic STRs (D3S1358, D1S1656,
D2S441, D10S1248, D13S317, D16S539, D18S51, D2S1338, CSF1PO, TH01,
vWA, D21S11, D7S820, D5S818, TPOX, D8S1179, D12S391, D19S433, FGA,
D22S1045, Penta E, Penta D and DYS391). PCR amplification using
PowerPlex Fusion (Promega Corp., Madison, MI, USA), Kplex-16
(BioQuest, Inc., Seoul, Korea) and Euplex-13 (BioQuest), performed
according to the manufacturer's instruction using 1 ng of DNA
extracted from human tissue and stem cells, were used to maximize
STR genotypes obtained from degraded DNA in paraffin-embedded
tissue. DNA extracted from mouse tissue was amplified by PCR
essentially as described by the manufacturer, except that 5 μl of
the extracted DNA was used and two additional PCR cycles were
performed to increase the PCR yield of each multiplex system. All
PCR amplifications were performed in duplicate in two additional
independent experiments. PCR products were separated by capillary
electrophoresis using an ABI PRISM 3130xl Genetic Analyzer (Applied
Biosystems), and the results were analyzed using GeneMapper ID
Software version 3.2 (Applied Biosystems). The genotypes of
amelogenin and STRs were determined based on observation of each
allele at least two additional times in independent tests.
Gene expression microarray analysis and
gene set enrichment analysis
Total RNA was extracted from rPB-TSs and 4 GBM-TSs
that were established using a Qiagen miRNA kit according to the
manufacturer's protocol. Expression profiles of TSs from rPB and
GBM (control) were obtained using an Illumina HumanHT-12 v4
Expression BeadChip (Illumina, Inc., San Diego, CA, USA). Data were
variance stabilizing transformed and normalized with the quantile
normalization method using R/Bioconductor lumi package. Complete
linkage hierarchical clustering with distance metric by taking
distance = (1-corelation)/2, was performed and depicted as heatmap.
Comparative Marker Viewer version 7.13 of GenePattern module was
used for differential expression of two groups. Gene set enrichment
analysis was performed using gene sets of hallmark signatures and
those composed of genes upregulated in metastasis and epithelial
mesenchymal transition, all of them provided by the Molecular
Signatures Database (v5.0 MSigDB) (30).
Statistical analysis
Data are expressed as means ± standard deviations.
Comparisons between two groups were done using the Student's
t-test. P-values <0.05 or false discovery rate <25% were
considered statistically significant.
Results
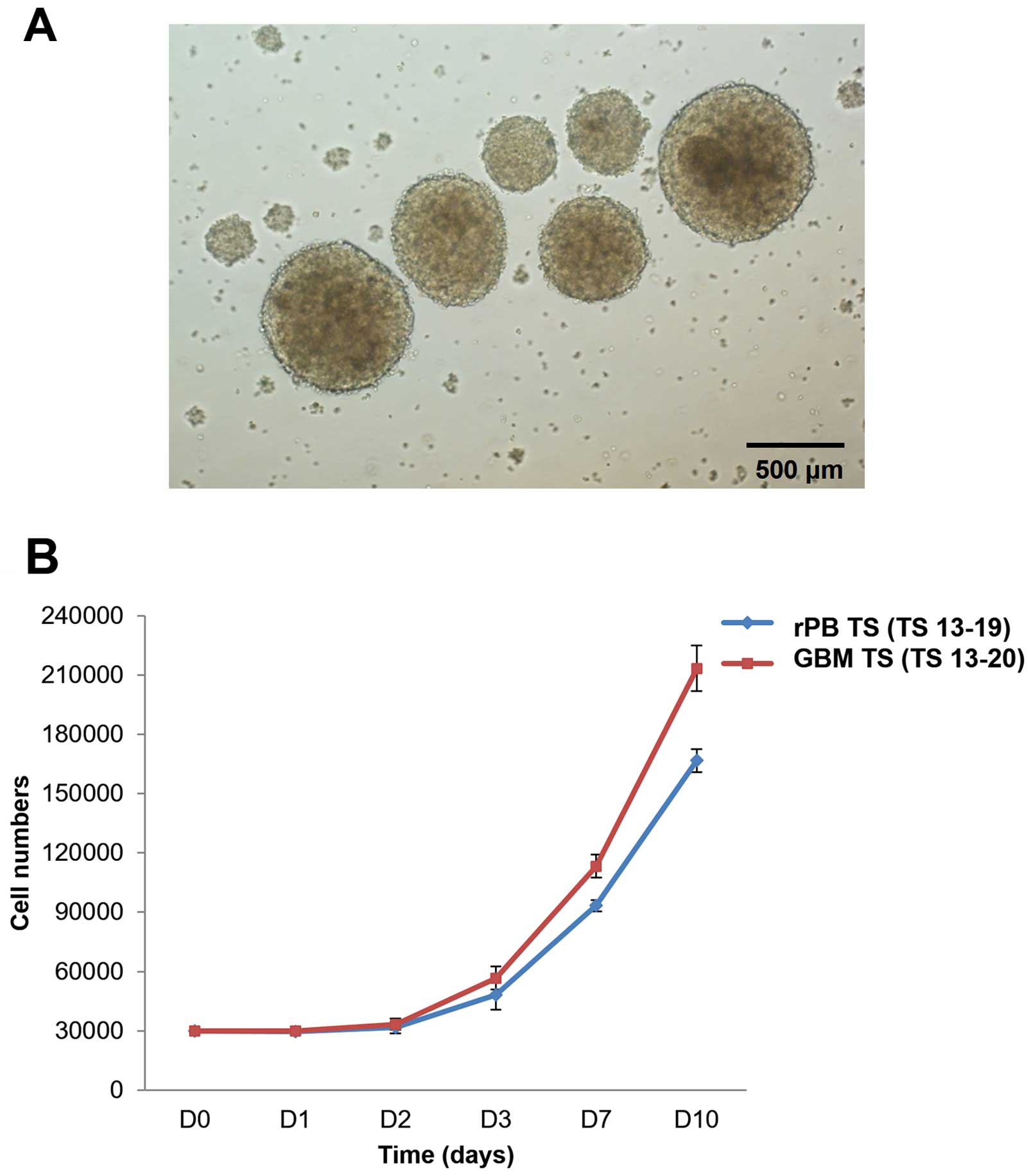
Morphology and growth characteristics of
rPB TSs
Cells isolated from the rPB specimen yielded
spheroids when cultured in TS complete media (Fig. 2A). These spheroids (TS 13-19),
termed rPB TSs, proliferated ~5- to 6-fold in 10 days. This
proliferation pattern was similar to that of GBM TSs (TS 13-20),
which grew ~7-fold in 10 days (Fig.
2B).
Stemness of rPB TSs
To examine the stemness of rPB TSs, we used
neurosphere formation assays, immunocytochemical staining and
RT-PCR and qPCR analyses. Results from these studies were compared
with those for GBM TSs. Neurosphere assays confirmed the presence
of cells with extensive self-renewal ability in the rPB specimen.
The size of neurospheres increased over time for both rPB TSs and
GBM TSs (Fig. 3A), as could be
seen grossly by light microscopy and as quantified by sphere radius
measurements on days 3, 7 and 10. Immunocytochemical staining of
rPB TSs identified cells expressing markers associated with stem
cells and brain tumor stem cells (31), including CD133, podoplanin and
musashi (Fig. 3B). To further
assess the stemness of rPB TSs at the gene level, we used RT-PCR
and qPCR analyses to measure CD133, podoplanin, nestin, musashi,
Sox2 and Oct4 expression levels. Whereas both rPB TSs and GBM TSs
expressed all stem cell surface markers, expression of CD133,
podoplanin, musashi and Sox2 was significantly higher in rPB TSs
(Fig. 3C and D), whereas
expression of nestin and Oct4 was higher in GBM TSs.
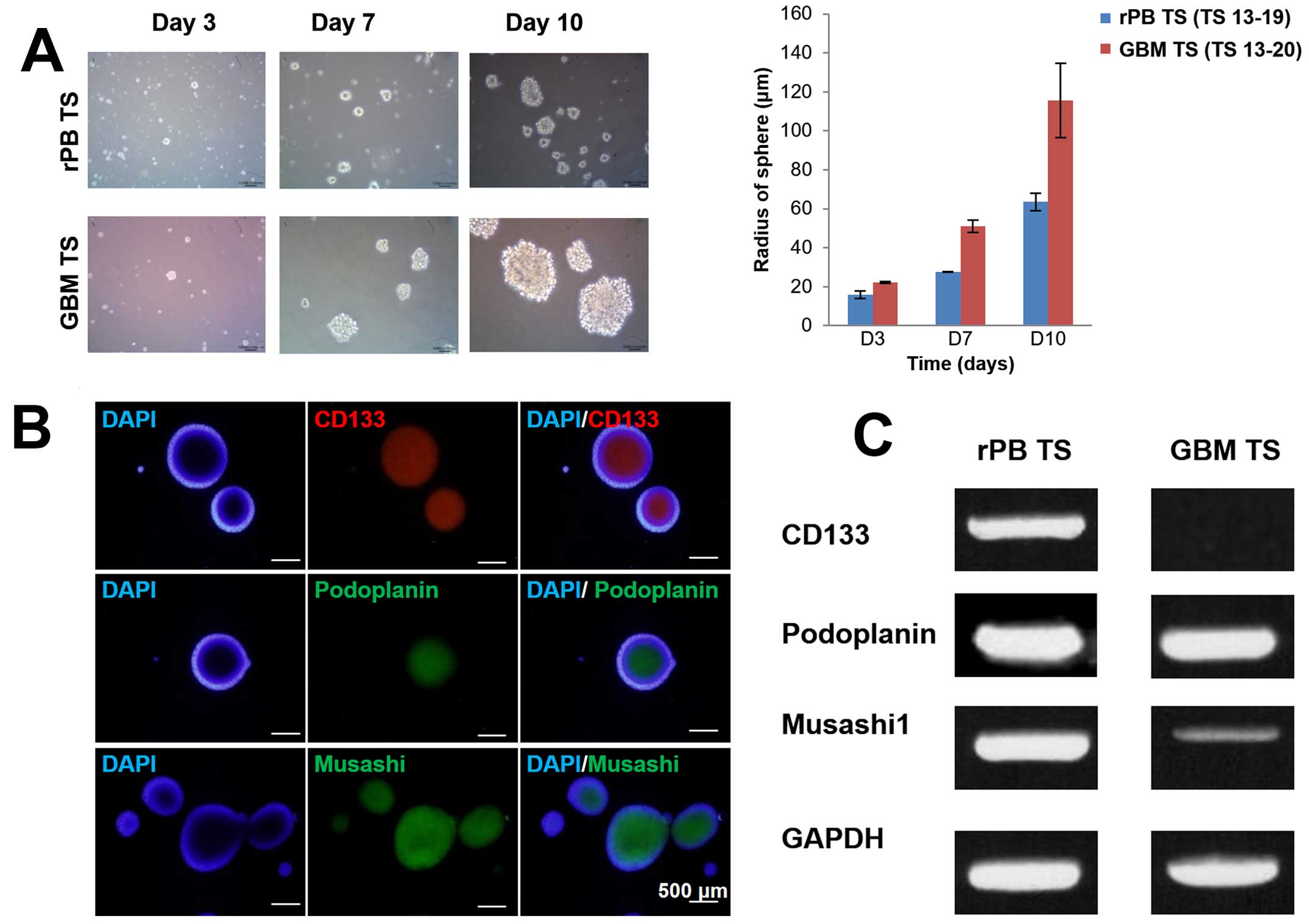 | Figure 3Stemness and differentiation
potential of rPB TSs characterized by neurosphere formation,
immunocytochemical staining, RT-PCR and qPCR. (A) In vitro
formation of TSs from single rPB TS cells (original magnification,
x100) and GBM TS cells on days 3, 7 and 10. The bar graph compares
the radii of rPB TSs with those of GBM TSs on days 3, 7 and 10. (B)
Immunocytochemical staining of rPB TS cells for CD133, podoplanin
and musashi; nuclei were counterstained with DAPI (original
magnification, x100). (C) RT-PCR analysis of rPB TSs and GBM TSs
for CD133, podoplanin and musashi. GAPDH was used as a control. (D)
qPCR analysis of rPB TSs and GBM TSs for CD133, podoplanin, nestin,
musashi, Sox2 and Oct4. Each sample was analyzed in triplicate.
*P<0.05, ***P<0.001, n=3 for Student's
t-test. (E) Immunocytochemical staining against GFAP, MBP, NeuN and
TUBB3 from rPB TSs grown in neural differential media (original
magnification, x200). (F) RT-PCR analysis for selected
differentiation markers including GFAP, Olig2 and TUBB3 from rPB
TSs and GBM TSs grown in neural differential media. GAPDH was used
as a control. Each sample was analyzed in triplicate.
*P<0.05. |
Neuro-glial differentiation of rPB
TSs
To assess the multilineage differentiation capacity
of rPB TSs, we cultured them in neuro-glial differentiation media
as described in Materials and methods. Immunocytochemical staining
with Tuj1 demonstrated that rPB TSs expressed TUBB3, a marker for
immature neurons; however, they did not express other
differentiation markers such as GFAP, MBP, or NeuN, which are
specific for astrocytes, oligodendrocytes and mature neurons,
respectively (Fig. 3E). RT-PCR
analyses showed that rPB TSs expressed higher level of TUBB3, but
lower levels of GFAP and Olig2 compared with GBM TSs (Fig. 3F). These results make sense given
that pineocytes are specialized neurons (32), and give rise to tumors that are
distinct from tumors of glial origin, such as GBMs, which express
surface markers for oligodendrocytes, astrocytes and mature and
immature neurons.
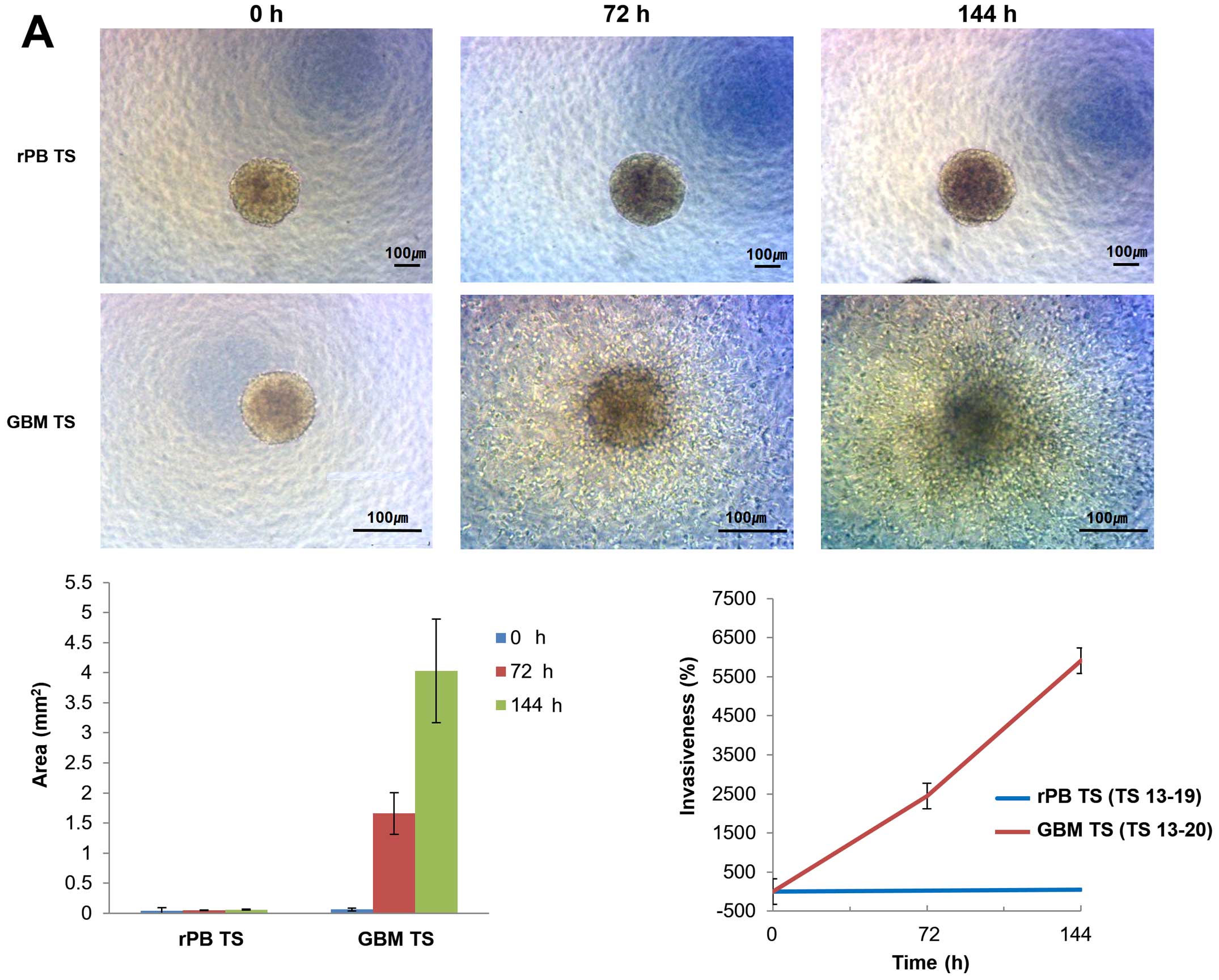
Invasiveness of rPB TSs
rPB TSs invasion patterns were investigated using 3D
invasion assays. Light microscopy showed no overt evidence for
invasion by rPB TSs at 0, 72 or 144 h, whereas GBM TSs were clearly
invasive at 72 and 144 h (Fig.
4A). From 0 to 144 h, the area occupied by rPB TSs increased
~1.2-fold, whereas that of GBM TSs increased by 63.7-fold. At 144
h, the areas were significantly different between the groups
(P<0.05). The relative invasiveness, calculated by comparing
maximal areas, was 43% for rPB TSs and 5911% for GBM TSs
(P<0.005). An RT-PCR analysis of epithelial-mesenchymal
transition (EMT) markers detected expression of β-catenin and Zeb1
in rPB TSs and GBM TSs, and additionally detected snail in GBM TSs
(Fig. 4B). A qPCR analysis found
that all three EMT genes were expressed in both rPB TSs and GBM
TSs, but were expressed at significantly lower levels in rPB TSs
(Fig. 4C).
Gene expression profile of rPB TSs
compared with GBM TSs and gene set enrichment analysis
To assess the difference in the expression of genes
between rPB TSs and GBM TSs, we performed whole-genome expression
profiling. rPB TSs showed a gene expression profile distinct from
that of GBM TSs of different origin, as depicted in the heatmap
shown in Fig. 4D. Differential
expression analysis revealed 4809 genes with false discovery rate
<25%. Highly ranked genes in the comparative marker selection
analysis included ARID3B, NOP56, RPOS4Y1 and JARID2, all of which
were known to be expressed in embryonal tumors or cell lines
derived from them. Gene set enrichment analysis revealed the
activation of myc-targeted genes in rPB TSs with statistical
significance whereas gene sets upregulated in metastasis and
epithelial mesenchymal transition were activated in GBM TS.
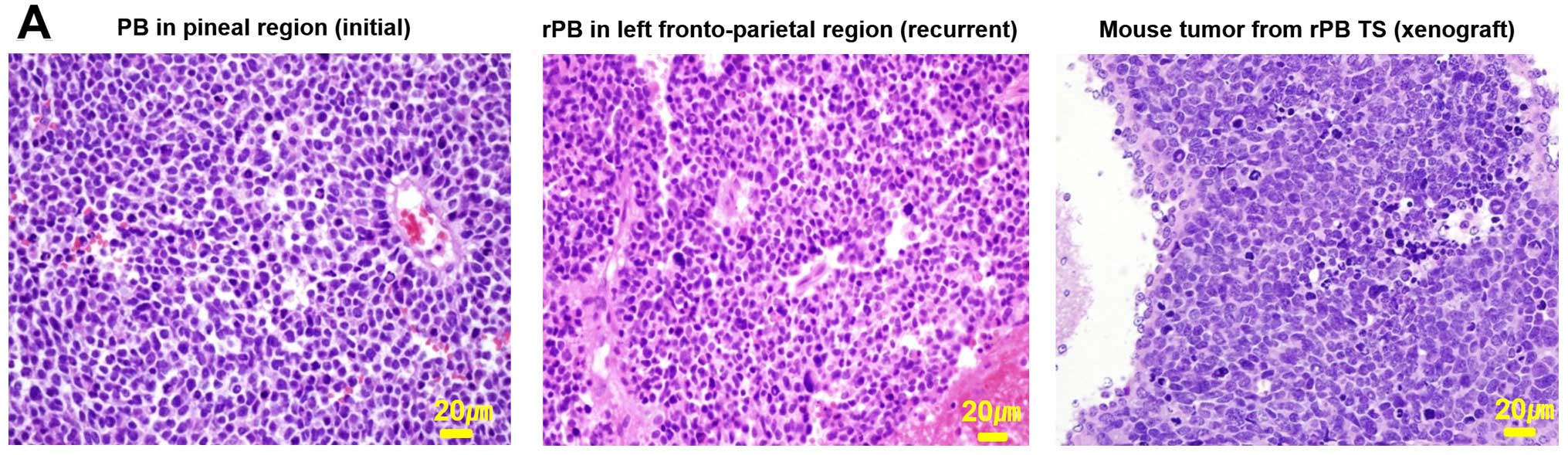
Establishment of orthotopic mouse model
from patient-derived rPB TSs
To determine the tumorigenic potential of rPB TSs,
we injected them into the right cerebrum of male athymic nude mice.
Mice were euthanized when their weight decreased by >15%
relative to their original body weight. A cross-sectional slice
showed the presence of an intracerebral xenograft tumor (Fig. 5A, right image). To determine
whether the xenograft tumor replicated phenotypes of the parent
tumor, we compared histological features of xenograft tumors with
those of the parent tumor tissue obtained in the initial surgery
(Fig. 5A, right image,
corresponding to the tumor in Fig.
1A) and from surgery after recurrence (Fig. 5A, middle image, corresponding to
the tumor in Fig. 1B). A
pathologist confirmed that histopathological findings of all three
tissues showed common characteristics of embryonal tumors,
including high cellularity, increased mitotic index and high
nucleus-cytoplasm ratio. A test of the genotypes of amelogenin and
23 forensic STRs (D3S1358, D1S1656, D2S441, D10S1248, D13S317,
D16S539, D18S51, D2S1338, CSF1PO, TH01, vWA, D21S11, D7S820,
D5S818, TPOX, D8S1179, D12S391, D19S433, FGA, D22S1045, Penta E,
Penta D and DYS391) showed that all autosomal STR profiles were
identical, indicating that the genomic alterations found in the
parental rPB were replicated in the corresponding xenograft tumors
(Fig. 5B).
Discussion
In the present study, we describe the first report
of the isolation of TSs from an rPB specimen. We confirmed that the
rPB TS cells have strong self-renewal and tumor-initiating
capacity, while lacking the ability for multi-lineage
differentiation typical of GBM TSs. In addition, 3D invasion and
gene expression studies showed that rPB TSs are not as invasive as
GBM TSs. An orthotopic xenograft model created using rPB TSs
replicated the parent tumor histopathologically as well as
genetically, supporting the potential use of rPB TSs for
patient-derived xenograft (PDX) models of PB.
The invasive behavior of rPB TSs and GBM TSs was
different, with rPB TSs showing no overt evidence of invasion in 3D
invasion assays and GBM TSs, demonstrating a clear invasive pattern
(Fig. 4A). These features
correlate with the characteristic invasive patterns of the two
tumors as observed in MRI. PBs feature a well-circumscribed
(33,34), lobulated, and solid lesion with
avid contrast enhancement (35),
as evident in MRIs of both the patient's original and recurred PB
(Fig. 1B). In contrast, GBMs are
highly infiltrative and have irregular borders; their mass
infiltrates the brain parenchyma via white matter tracks (Fig. 1C) (36), thereby increasing the isotropic
component in diffusion tensor imaging (DTI) (37). Some studies have shown that PBs can
invade the surrounding brain parenchyma (38,39)
and may disseminate through the cerebrospinal fluid (40,41).
However, we were unable to find many studies showing radiologically
infiltrative PBs, suggesting that PBs with invasiveness comparable
to that of GBMs are rare. Such a low degree of invasiveness of rPB
TSs compared with GBM TSs might result from the lower expression
levels of EMT-associated genes in rPB TSs (Fig. 4B and C). In many different cancers,
a number of transcription factors, including snail, slug, Zeb1 and
twist, have been identified as important EMT regulators (42). Studies have also shown that
β-catenin, a downstream target of AKT, is capable of modulating the
aggressive phenotype of cancer cells (42,43).
Compared with GBM TSs, the levels of EMT genes, including
β-catenin, snail and Zeb1, were all significantly lower in rPB TSs
(Fig. 4B). In particular, the
greater invasiveness of GBM TSs compared with rPB TSs demonstrated
by our data might be attributable to the high expression level of
snail, which is known to be at least ten times more potent than
Zeb1 in repressing standard epithelial markers, such as E-cadherin
and Mucin-1 (44).
Examples of xenograft models that have been created
to recapitulate human glioma in animals include: i) xenografts
based on human glioma-derived cell lines passaged in neuro-basal
medium; ii) biopsy spheroid xenograft models or PDX; and iii) human
glioma monolayer cell lines established in serum-containing media
(45). Our xenograft model is
conceptually in line with the first approach. One advantage of this
approach lies in the ease of propagating TSs to yield sufficient
material for potential biomarker and therapeutics discovery, while
recapitulating the histology of the patient tumor (45). While it is true that this approach
might allow clonal selection to take place during passage, thereby
changing the genetic and epigenetic information, the results of our
genetic studies show that such selection did not take place in the
xenograft model. By establishing an orthotopic mouse model from
patient-derived rPB TSs, we have provided the first demonstration
of the feasibility of creating a PDX model using TSs from rPB.
Although we did not use mutational status, DNA copy number
variation, or gene expression to evaluate the genetic correlation
between our xenograft model and the original tumor as have some
previous reports on PDX (46), we
believe that our data (Fig. 5A and
B) provide sufficient histological and genetic evidence to
demonstrate the identity of the orthotopic xenograft model with the
parent tumor. In this regard, we suggest that our mouse orthotopic
xenograft model of rPB TSs represents an alternative method for
establishing a PDX.
Studies have shown that biopsy spheroid xenograft
models, or PDXs, can well replicate the invasive characteristics
and other histological features of the original patient tumor,
including human-derived microvasculature, host extracellular matrix
and resident macrophages (45,47).
However, one major disadvantage of this model is time; it can take
2–11 months for the initial engraftment and an additional 8–18
months for the minimum of three passages required to develop
xenografts that resemble the parent tumor (45,48).
Another potential difficulty presented by the genetic heterogeneity
of PDXs might be standardization of the model. Notwithstanding
these potential limitations, we believe that our xenograft model
using rPB TSs is not only efficient, but also more effective in
establishing a standardized in vivo model of PBs that is
genetically indistinguishable from the patient tumor. In the
future, incorporating genome sequencing, proteomics and
metabolomics would improve this platform for use in studying cancer
stem-cell biology and developing novel cancer therapeutics
(46).
Acknowledgements
We thank Dr Hyun-Jung Kee, Department of Surgery,
Yonsei University College of Medicine, Seoul, Korea, for providing
help with array experiments. The present study was supported by a
faculty research grant from the Yonsei University College of
Medicine for 2013 (6-2013-0034), and by grants from the Basic
Science Research Program through the National Research Foundation
of Korea (NRF) funded by the Ministry of Education, Science and
Technology (NRF-2013R1A1A2006427), and the Korean Health Technology
R&D Project, Ministry of Health & Welfare, Republic of
Korea (HI13C1509).
Abbreviations:
|
CSC
|
cancer stem cell
|
|
PDX
|
patient-derived xenograft
|
|
rPB
|
recurrent pineoblastoma
|
|
TS
|
tumorsphere
|
References
|
1
|
Lutterbach J, Fauchon F, Schild SE, Chang
SM, Pagenstecher A, Volk B, Ostertag C, Momm F and Jouvet A:
Malignant pineal parenchymal tumors in adult patients: patterns of
care and prognostic factors. Neurosurgery. 51:44–55; discussion
55–46. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kobayashi T and Lunsford L: Pineal Region
Tumors, Diagnosis and Treatment Options. Karger; Pittsburgh, PA:
2009, View Article : Google Scholar
|
|
3
|
Friedrich C, von Bueren AO, von Hoff K,
Gerber NU, Ottensmeier H, Deinlein F, Benesch M, Kwiecien R,
Pietsch T, Warmuth-Metz M, et al: Treatment of young children with
CNS-primitive neuroectodermal tumors/pineoblastomas in the
prospective multicenter trial HIT 2000 using different chemotherapy
regimens and radiotherapy. Neuro Oncol. 15:224–234. 2013.
View Article : Google Scholar :
|
|
4
|
Schild SE, Scheithauer BW, Schomberg PJ,
Hook CC, Kelly PJ, Frick L, Robinow JS and Buskirk SJ: Pineal
parenchymal tumors. Clinical, pathologic, and therapeutic aspects.
Cancer. 72:870–880. 1993. View Article : Google Scholar
|
|
5
|
Lee JY, Wakabayashi T and Yoshida J:
Management and survival of pineoblastoma: an analysis of 34 adults
from the brain tumor registry of Japan. Neurol Med Chir (Tokyo).
45:132–141; discussion 141–132. 2005. View Article : Google Scholar
|
|
6
|
Miller S, Rogers HA, Lyon P, Rand V,
Adamowicz-Brice M, Clifford SC, Hayden JT, Dyer S, Pfister S,
Korshunov A, et al: Genome-wide molecular characterization of
central nervous system primitive neuroectodermal tumor and
pineoblastoma. Neuro Oncol. 13:866–879. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huntly BJ and Gilliland DG: Leukaemia stem
cells and the evolution of cancer-stem-cell research. Nat Rev
Cancer. 5:311–321. 2005. View
Article : Google Scholar
|
|
9
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: Accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Singh SK, Hawkins C, Clarke ID, Squire JA,
Bayani J, Hide T, Henkelman RM, Cusimano MD and Dirks PB:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kong BH, Park NR, Shim JK, Kim BK, Shin
HJ, Lee JH, Huh YM, Lee SJ, Kim SH, Kim EH, et al: Isolation of
glioma cancer stem cells in relation to histological grades in
glioma specimens. Childs Nerv Syst. 29:217–229. 2013. View Article : Google Scholar
|
|
12
|
Hussein D, Punjaruk W, Storer LC, Shaw L,
Othman RT, Peet A, Miller S, Bandopadhyay G, Heath R, Kumari R, et
al: Pediatric brain tumor cancer stem cells: Cell cycle dynamics,
DNA repair, and etoposide extrusion. Neuro Oncol. 13:70–83. 2011.
View Article : Google Scholar :
|
|
13
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828. 2003.
|
|
14
|
Sutter R, Shakhova O, Bhagat H, Behesti H,
Sutter C, Penkar S, Santuccione A, Bernays R, Heppner FL, Schüller
U, et al: Cerebellar stem cells act as medulloblastoma-initiating
cells in a mouse model and a neural stem cell signature
characterizes a subset of human medulloblastomas. Oncogene.
29:1845–1856. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sulman E, Aldape K and Colman H: Brain
tumor stem cells. Curr Probl Cancer. 32:124–142. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gupta PB, Onder TT, Jiang G, Tao K,
Kuperwasser C, Weinberg RA and Lander ES: Identification of
selective inhibitors of cancer stem cells by high-throughput
screening. Cell. 138:645–659. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nautiyal J, Kanwar SS, Yu Y and Majumdar
AP: Combination of dasatinib and curcumin eliminates
chemo-resistant colon cancer cells. J Mol Signal. 6:72011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar
|
|
19
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwak J, Shin HJ, Kim SH, Shim JK, Lee JH,
Huh YM, Kim EH, Park EK, Chang JH, Kim SH, et al: Isolation of
tumor spheres and mesenchymal stem-like cells from a single
primitive neuroectodermal tumor specimen. Childs Nerv Syst.
29:2229–2239. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kim SM, Kang SG, Park NR, Mok HS, Huh YM,
Lee SJ, Jeun SS, Hong YK, Park CK and Lang FF: Presence of glioma
stroma mesenchymal stem cells in a murine orthotopic glioma model.
Childs Nerv Syst. 27:911–922. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kang SG, Cheong JH, Huh YM, Kim EH, Kim SH
and Chang JH: Potential use of glioblastoma tumorsphere: Clinical
credentialing. Arch Pharm Res. 38:402–407. 2015. View Article : Google Scholar
|
|
23
|
Kong BH, Moon JH, Huh YM, Shim JK, Lee JH,
Kim EH, Chang JH, Kim DS, Hong YK, Kim SH, et al: Prognostic value
of glioma cancer stem cell isolation in survival of primary
glioblastoma patients. Stem Cells Int. 838950:20142014.
|
|
24
|
Kong BH, Shin HD, Kim SH, Mok HS, Shim JK,
Lee JH, Shin HJ, Huh YM, Kim EH, Park EK, et al: Increased in vivo
angiogenic effect of glioma stromal mesenchymal stem-like cells on
glioma cancer stem cells from patients with glioblastoma. Int J
Oncol. 42:1754–1762. 2013.PubMed/NCBI
|
|
25
|
Shin GY, Shim JK, Lee JH, Shin HJ, Lee SJ,
Huh YM, Kim EH, Park EK, Kim SH, Chang JH, et al: Changes in the
biological characteristics of glioma cancer stem cells after serial
in vivo subtransplantation. Childs Nerv Syst. 29:55–64. 2013.
View Article : Google Scholar
|
|
26
|
Kim YG, Jeon S, Sin GY, Shim JK, Kim BK,
Shin HJ, Lee JH, Huh YM, Lee SJ, Kim EH, et al: Existence of glioma
stroma mesenchymal stemlike cells in Korean glioma specimens.
Childs Nerv Syst. 29:549–563. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lal S, Lacroix M, Tofilon P, Fuller GN,
Sawaya R and Lang FF: An implantable guide-screw system for brain
tumor studies in small animals. J Neurosurg. 92:326–333. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lim HY, Kim KM, Kim BK, Shim JK, Lee JH,
Huh YM, Kim SH, Kim EH, Park EK, Shim KW, et al: Isolation of
mesenchymal stem-like cells in meningioma specimens. Int J Oncol.
43:1260–1268. 2013.PubMed/NCBI
|
|
29
|
Kang SG, Shinojima N, Hossain A, Gumin J,
Yong RL, Colman H, Marini F, Andreeff M and Lang FF: Isolation and
perivascular localization of mesenchymal stem cells from mouse
brain. Neurosurgery. 67:711–720. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hemmati HD, Nakano I, Lazareff JA,
Masterman-Smith M, Geschwind DH, Bronner-Fraser M and Kornblum HI:
Cancerous stem cells can arise from pediatric brain tumors. Proc
Natl Acad Sci USA. 100:15178–15183. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Smith AB, Rushing EJ and Smirniotopoulos
JG: From the archives of the AFIP: lesions of the pineal region:
radiologic-pathologic correlation. Radiographics. 30:2001–2020.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fang AS and Meyers SP: Magnetic resonance
imaging of pineal region tumours. Insights Imaging. 4:369–382.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tong T, Zhenwei Y and Xiaoyuan F: MRI and
1H-MRS on diagnosis of pineal region tumors. Clin Imaging.
36:702–709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Papaioannou G, Sebire NJ and McHugh K:
Imaging of the unusual pediatric ‘blastomas’. Cancer Imaging.
9:1–11. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Daumas-Duport C, Meder JF, Monsaingeon V,
Missir O, Aubin ML and Szikla G: Cerebral gliomas: Malignancy,
limits and spatial configuration. Comparative data from serial
stereo-taxic biopsies and computed tomography (a preliminary study
based on 50 cases). J Neuroradiol. 10:51–80. 1983.
|
|
37
|
Price SJ, Jena R, Burnet NG, Hutchinson
PJ, Dean AF, Peña A, Pickard JD, Carpenter TA and Gillard JH:
Improved delineation of glioma margins and regions of infiltration
with the use of diffusion tensor imaging: An image-guided biopsy
study. AJNR Am J Neuroradiol. 27:1969–1974. 2006.PubMed/NCBI
|
|
38
|
Korogi Y, Takahashi M and Ushio Y: MRI of
pineal region tumors. J Neurooncol. 54:251–261. 2001. View Article : Google Scholar
|
|
39
|
Palled S, Kalavagunta S, Beerappa Gowda J,
Umesh K, Aal M, Abdul Razack TP, Gowda V and Viswanath L: Tackling
a recurrent pinealoblastoma. Case Rep Oncol Med.
135435:20142014.
|
|
40
|
Gasparetto EL, Cruz LC Jr, Doring TM,
Araújo B, Dantas MA, Chimelli L and Domingues RC:
Diffusion-weighted MR images and pineoblastoma: Diagnosis and
follow-up. Arq Neuropsiquiatr. 66:64–68. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tate M, Sughrue ME, Rutkowski MJ, Kane AJ,
Aranda D, McClinton LS, McClinton LS, Barani IJ and Parsa AT: The
long-term postsurgical prognosis of patients with pineoblastoma.
Cancer. 118:173–179. 2012. View Article : Google Scholar
|
|
42
|
Wang H, Zhang G, Zhang H, Zhang F, Zhou B,
Ning F, Wang HS, Cai SH and Du J: Acquisition of
epithelial-mesenchymal transition phenotype and cancer stem
cell-like properties in cisplatin-resistant lung cancer cells
through AKT/beta-catenin/ Snail signaling pathway. Eur J Pharmacol.
723:156–166. 2014. View Article : Google Scholar
|
|
43
|
Heuberger J and Birchmeier W: Interplay of
cadherin-mediated cell adhesion and canonical Wnt signaling. Cold
Spring Harb Perspect Biol. 2:a0029152010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaufhold S and Bonavida B: Central role of
Snail1 in the regulation of EMT and resistance in cancer: A target
for therapeutic intervention. J Exp Clin Cancer Res. 33:622014.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Huszthy PC, Daphu I, Niclou SP, Stieber D,
Nigro JM, Sakariassen PO, Miletic H, Thorsen F and Bjerkvig R: In
vivo models of primary brain tumors: Pitfalls and perspectives.
Neuro Oncol. 14:979–993. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tentler JJ, Tan AC, Weekes CD, Jimeno A,
Leong S, Pitts TM, Arcaroli JJ, Messersmith WA and Eckhardt SG:
Patient-derived tumour xenografts as models for oncology drug
development. Nat Rev Clin Oncol. 9:338–350. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bjerkvig R, Tonnesen A, Laerum OD and
Backlund EO: Multicellular tumor spheroids from human gliomas
maintained in organ culture. J Neurosurg. 72:463–475. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang J, Miletic H, Sakariassen PO, Huszthy
PC, Jacobsen H, Brekkå N, Li X, Zhao P, Mørk S, Chekenya M, et al:
A reproducible brain tumour model established from human
glioblastoma biopsies. BMC Cancer. 9:4652009. View Article : Google Scholar : PubMed/NCBI
|