Introduction
Most eukaryotic cells are now recognized to shed
extracellular vesicles (EVs) into their environment (1,2).
Among these vesicles, some are known as membrane vesicles (MVs) or
ectosomes and originate from the plasma membrane, through a process
resembling viral budding, whereas a second class consists of
smaller vesicles (exosomes), which are released after plasma
membrane fusion of the so-called multivesicular bodies (MVBs),
which originate in the endosomal compartment (3).
Production of EVs, initially discovered in
transformed cells, has been demonstrated as a physiological
mechanism involved in the horizontal transfer of several kinds of
molecules in normal cells, among which different classes of cells
in the nervous system (4–7). Cancer cells, however, produce and
release much higher amounts of EVs (8), which can reach most biological
fluids, such as blood plasma (9),
breast milk (10), and saliva
(11,12), where they could be used as
biological markers of disease and even of the disease grade
(13–15).
Concerning protein content, EVs released from cancer
cells transport different classes of chaperones, tumor-specific
antigens, apoptosis-inducing proteins, such as FasL and TRAIL
(16,17), immune modulatory factors (18), and many other oncogenic molecules,
which, once transferred into surrounding cells, can facilitate
cancer development by suppressing immune responses, and stimulating
tumor growth, invasion and metastasis.
Recently, we reported that oligodendroglioma cells
release into EVs also the H1.0 linker histone variant (19). H1 linker histones constitute in
mammals the most heterogeneous family of histones. In humans, the
H1 family includes 11 members, among which 7 are expressed in
somatic cells (H1.1-H1.5; H1.0 and H1X), 3 are specifically
expressed in testis, and one is specifically expressed in the
oocyte (H1.oo) (20–22). Moreover, among the somatic
subtypes, H1.1-H1.5 are encoded by repeated genes which are
transcribed during the S phase of the cell cycle, in a
replication-dependent manner, to give mRNAs which are not
polyadenylated. On the other hand, H1.0 is transcribed in a
replication-independent way, into a polyadenylated mRNA (23), and is prevalent in differentiated
cells.
The presence of H1.0 in the EVs produced by
oligodendroglioma cells suggested, on one hand, that deregulation
of H1.0 histone expression can be linked to tumorigenesis, and, on
the other, that cancer cells can escape differentiation by
discarding this protein into EVs (19). To shed more light on this
hypothesis, in this study we analyzed a different kind of cancer
cells: A375 melanoma cells. Here we report that these cells
synthesize H1.0 histone and secrete a modified form of it into MVs.
In addition, we found that EVs released from melanoma cells also
contain H1.0 mRNA. Since post-transcriptional regulation of mRNA
trafficking, stability and translation depends on a number of
RNA-binding proteins (RBPs) (24),
and since a group of H1.0 mRNA-binding proteins have been already
found in the rat brain (25–28),
we also looked for RBPs with this ability in melanoma cells and
EVs. Here we show that H1.0-binding RBPs are indeed present in the
EVs released from A375 melanoma cells, and that one of these
proteins is MYEF2.
Materials and methods
Cell cultures
A375 melanoma cells (A375CL1006) were cultured in
Dulbecco's modified Eagle's medium - low glucose (DMEM 5546,
Sigma-Aldrich, MO, USA) supplemented with 10% heat-inactivated
fetal bovine serum (F7524 Sigma-Aldrich), 2 mM glutamine
(Euroclone, Milan, Italy), 0.1% MEM non essential amino acids
(Sigma-Aldrich), 1 mM sodium pyruvate (Euroclone), 100,000 U
penicillin, 100 mg streptomycin and 250 μg amphotericin B
(Sigma-Aldrich) per liter. Cells were maintained in humidified 5%
CO2/95% air, at 37°C. Some A375 melanoma cells were
cultured in the same medium without serum.
Immunofluorescence analyses
Cells were fixed in 96% ethanol and immune-stained
with rat anti-integrin β1 (1:100; in-house produced), and rabbit
anti-H1° (1:100; sc-67324 Santa Cruz, CA, USA).
The secondary antibodies used were fluorescein
isothiocyanate-conjugated anti-rat-(1:100; F1763), or
rhodamine-conjugated anti-rabbit-(1:200; T6778) immunoglobulins
(both from Sigma, MO, USA).
Nuclei were stained with
4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (H1200, Vector
Laboratories, Youngstown, OH, USA). Cells were observed in an
Olympus BX-50 microscope (Olympus Italia S.r.l., Segrate, Italy)
equipped with Vario Cam B/W camera (Nikon Instruments S.p.A.,
Calenzano, Italy).
Preparation of microvesicles from the
A375 medium
Vesicles were prepared from A375 FBS-free
conditioned medium, as follows: 24 h before collecting media, cells
were washed twice in phosphate-buffered saline (PBS), pH 7.5, and
then incubated with FCS-free DMEM. Conditioned media were
centrifuged at 2,000 × g for 10 min and then at 4,000 × g for 15
min. The supernatant was centrifuged at 105,000 × g (Ti70 Rotor,
Beckman) for 90 min at 4°C.
To separate exosomes from MVs, before
ultracentrifugation, medium was filtered with a 0.2-μm filter and
centrifuged at 10,000 × g for 30 min. The pellet containing MVs was
saved while the supernatant was finally centrifuged at 105,000 × g
for 90 min to obtain exosomes.
Pelleted vesicles were suspended in PBS and protein
concentration was determined using Qubit® Protein assay
kit (Q33211, Invitrogen, OR, USA).
Vesicles analyses
The NS300 (NanoSight, London, UK) instrument is
based on a conventional optical microscope and uses a laser light
source to illuminate nano-scale particles. This analysis allowed
measuring size and concentration of vesicles in a liquid medium
based on tracking of Brownian motion.
Purification of total cell extracts
Cells were collected and homogenized in nuclei
buffer (0.32 M sucrose; 50 mM sodium phosphate buffer, pH 6.5; 50
mM KCl; 0.5 mM spermine; 0.15 mM spermidine; 2 mM EDTA; 0.15 mM
EGTA), containing the protease inhibitors aprotinin (2 μg/ml),
antipain (2 μg/ml), leupeptin (2 μg/ml), pepstatin A (2 μg/ml),
benzamidine (1.0 mM), and phenylmethylsulfonyl fluoride (1.0 mM),
all purchased from (Sigma-Aldrich). Protein concentration was
determined according to Bradford (29).
Western blot analysis
Proteins (15 μg) were separated by electrophoresis
on denaturing 12.5% polyacrylamide slab gels (SDS-PAGE) and
transferred to PVDF membrane (IPVH00010, Immobilon P, Millipore,
MA, USA), as previously described (19). Samples on the membrane were
visualized by staining with Ponceau Red (Sigma-Aldrich) for 5 min.
Membranes were immune-stained with rabbit polyclonal anti-H1°
antibodies (1:500, sc-67324 Santa Cruz), mouse monoclonal
anti-Hsc70 antibodies (1:1,000, sc-7298, Santa Cruz), rabbit
polyclonal anti-SUMO1 (1:500, S373B, Santa Cruz). The secondary
antibodies were AP-conjugated anti-mouse (1:7,500, S372B) and
anti-rabbit (1:7,500, S373B) IgGs (Promega Corp., Madison, WI,
USA).
Reverse transcription (RT)-PCR
Total RNA was purified from cells according to
Chomczynski and Sacchi (30). RNA
from vesicles was prepared using either the Pure-Link RNA microKit
(12183016, Invitrogen) or the TRIzol® reagent
(15596-018, Invitrogen), according to the manufacturer's
instructions.
Synthesis of cDNA was performed using the
Superscript II Reverse Transcriptase kit (18064-022 Invitrogen),
following the manufacturer's protocol. The primer used for the
reverse transcription had the following sequence: (5′→3′) GGC TTT
CTT GGG CGT GGC AGC C.
PCR was performed using Taq DNA polymerase
(10342-020, Invitrogen), according to the manufacturer's
instructions, and using the following primers: forward, (5′→3′),
ATG ATC GTG GCT GCC ATC CAG GC; reverse, (5′→3′), GGC TTT CTT GGG
CGT GGC AGC C.
Amplification was performed by Mastercycler Thermal
Cycler 5345 (Eppendorf AG, Hamburg, Germany) using the following
program: 94°C for 30 sec, 35 cycles at 95°C for 30 sec, 52°C for 30
sec, 72°C for 30 sec, and 72°C for 5 min.
Preparation of in vitro transcripts and
T1 RNase protection assay
33P-radiolabeled H1.0 RNA was prepared as
previously described (25), using
as a template the plasmid pMH1.0 (31), which contains the H1.0 insert (EMBL
ID: X70685). In order to prepare H1.0 mRNA to be used for
chromatographic experiments, in some reactions transcription was
performed in the presence of biotin-21-UTP (AM8450, Ambion-Life
Technologies, Paisley, UK), a UTP analog which has biotin attached
to the pyrimidine ring by a 21-atom spacer arm (27).
For T1 RNase (EC 3.1.27.3; Roche, Switzerland)
protection assay, radiolabeled H1.0 RNA was mixed with either total
cell extracts or vesicles (15 μg), prepared as described above,
following the procedure previously described (32). RNA-protein complexes were analyzed
by denaturing electrophoresis on sodium dodecyl sulfate
(SDS)-polyacrylamide slab gel (PAGE). At the end of the run, the
gel was directly exposed to X-ray film for autoradiography
(Amersham Hyperfilm™, GE Healthcare, USA). The gels were stained
with Coomassie Brilliant Blue R-250 (Sigma-Aldrich), to confirm
loading of equal amounts of proteins per lane.
Chromatographic purification from A375
cell-released EVs of H1.0 RNA-binding factors
Streptavidin-conjugated paramagnetic beads (Z5481,
Magnesphere, Promega) were washed three times in PBS according to
the manufacturer's instructions, and then mixed with 450 μg of A375
EV proteins, in 500 μl (final volume) of binding buffer (BB: 75 mM
Tris-HCl, pH 7.5; 50 mM KCl; 5 mM dithiothreitol) (27), containing proteases and
phosphatases inhibitors (Sigma-Aldrich). Samples were incubated for
1 h, at 4°C, under shaking, to allow unspecific protein binding to
the particles (pre-clearing step). After centrifuging at 10,000 × g
for 5 min, the pre-cleared supernatants were used for the specific
binding reaction. The pre-cleared sample was divided into two
aliquots, one of which was mixed with H1.0 RNA (1.2 μg) in BB,
while the other one was used as an RNA-free control. Both samples
were incubated for 1 h at 4°C, after which fresh aliquots of
pre-washed beads were added, and incubation was continued for 1 h,
at 4°C, under shaking. Finally, the supernatants containing unbound
proteins were collected by a magnetic device (Magnesphere, Promega)
and frozen. Paramagnetic beads were washed four times in BB and
then resuspended in electrophoresis sample buffer, boiled and
centrifuged at 10,000 × g. The supernatants, which contain bound
proteins, were frozen and saved for analyses.
Silver staining
After SDS-PAGE, the gel was silver stained according
to Yan et al (33). The
region of interest was cut from the gel and analyzed by MALDI-TOF
mass spectrometry.
MALDI-TOF mass spectrometry
MALDI-TOF mass spectrometry analysis was performed
using the Voyager DE-PRO (Applied Biosystems, Foster City, CA, USA)
mass spectrometer as previously described (34). Briefly, silver stained band was in
gel-destained with K3[Fe(CN)6] and
Na2S2O3, reduced with
dithiothreitol, S-alkylated with iodoacetamide, and subsequently
digested with trypsin. The tryptic peptide extracts were desalted
by μZip-TipC18 (Millipore) and loaded on the MALDI target, using
the dried droplet technique and α-cyano-4-hydroxycinnamic acid as
matrix. The resulting mass spectrum, was elaborated using the
DataExplorer software (Applied Biosystems) and manually inspected
to obtain the corresponding peak lists. Internal mass calibration
was done using trypsin autolysis fragments at m/z 842.5100,
1045.5642, and 2211.1046 Da. Peptide mass fingerprinting was
compared to the theoretical masses from the Swiss-Prot.
Results
A375 melanoma cells release both membrane
vesicles (MVs) and exosomes
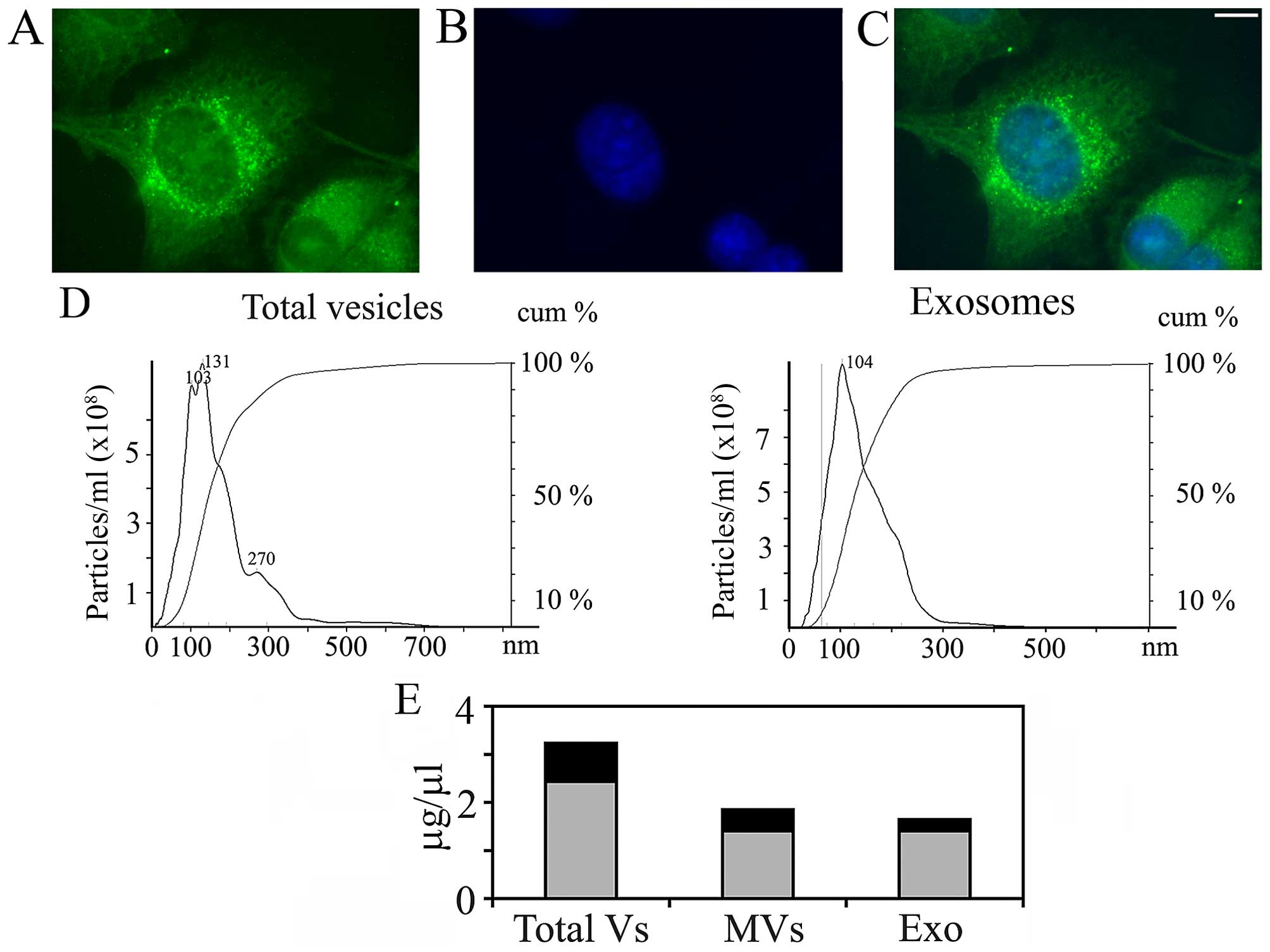
As shown in Fig. 1,
A375 melanoma cells produce and release into the culture medium
extracellular vesicles, at least in part from plasma membrane
regions enriched in integrin β1 (Fig.
1A–C). The vesicular population is actually a mixed one, as
demonstrated by NanoSight (Fig.
1D), which allowed measuring size and concentration of vesicles
in the culture medium, based on tracking of Brownian motion. In
addition, according to the NanoSight data (which are quantitative),
the EV population is composed mainly of exosomes (compare the
height of the peak at 103–131 nm, which corresponds to exosomes,
with the shoulder at 270 nm, which probably corresponds to MVs). In
some experiments, the medium in which melanoma cells had been
cultured was filtered and centrifuged at 10,000 × g for 30 min,
before ultracentrifugation, in order to pellet first only MVs. The
supernatant was then centrifuged at 105,000 × g for 90 min to
obtain a final pellet of exosomes. The NanoSight analysis of the
separated fractions gave only single peaks (Fig. 1D, right panel, where only the
analysis concerning purified exosomes is shown). The relative
concentrations (expressed as μg/μl of proteins) of the two
populations of vesicles obtained are reported in Fig. 1E.
EVs released from A375 melanoma cells
contain both H1.0 linker histone and the corresponding mRNA
H1.0 linker histone was first discovered in
non-dividing tissues (35,36), and, in general, accumulates in
differentiating cells at the end of the proliferative phase.
Recently, it was however found in total cell extracts and
extracellular vesicles from G26/24 dividing oligodendroglioma cells
(19). In this study we therefore
looked for the possibility that also melanoma cells synthesize and
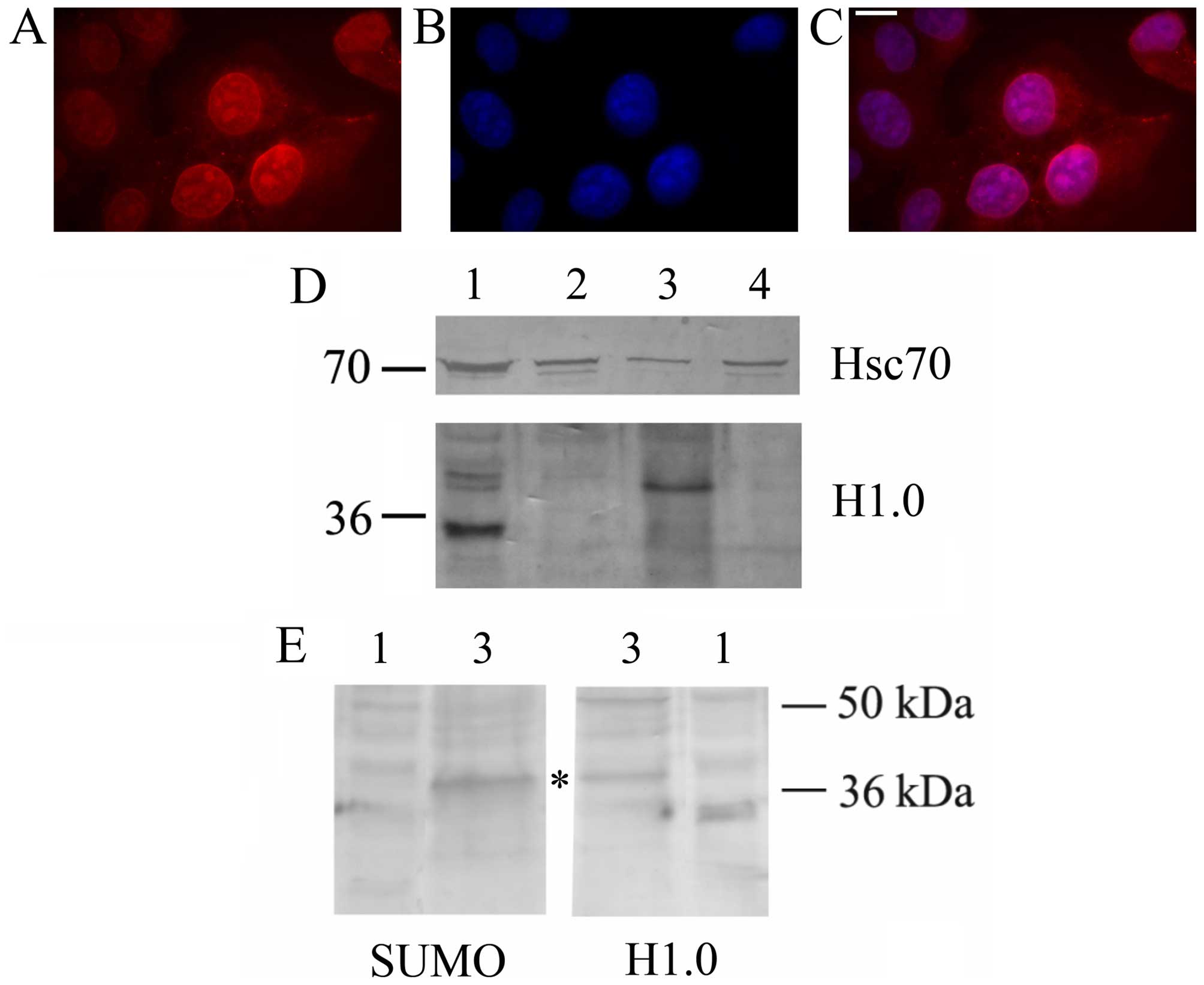
secrete this histone via EVs. As shown in Fig. 2, A375 cells indeed produce a
protein which is immune-stained by anti-H1.0 antibodies both in
immunofluorescence (Fig. 2A–C) and
western blot analyses (Fig. 2D).
As already reported for other tumor cells, melanoma cells release
EVs (both MVs and exosomes) which contain the Hsc70 chaperone
(19). Interestingly, they also
secrete an anti-H1.0 antibody-positive protein which, however, is
larger than expected, and is specifically sorted to MVs. Since
other proteins sorted to vesicles bear specific post-translational
modifications, such as sumoylation (37), we looked for the presence of a SUMO
moiety on this larger H1.0. As shown in Fig. 2E, anti-SUMO1 antibodies not only
recognized a protein of about 38 kDa, but this band exactly
co-migrates with the slow migrating protein recognized by the
anti-H1.0 antibodies (Fig. 2E,
asterisk).
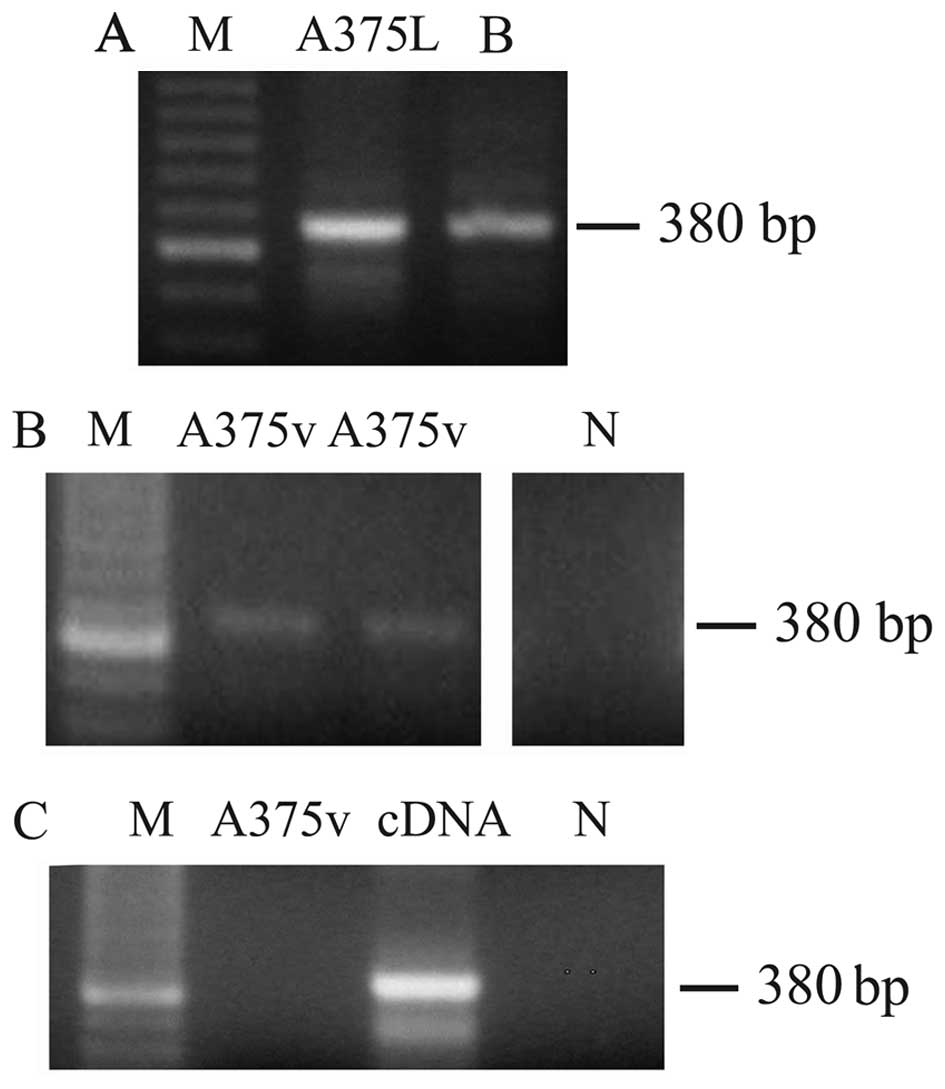
We then analyzed both total cell lysates and the
vesicular fraction for the presence of H1.0 mRNA. We used as a
template for reverse transcription (RT)-PCR total RNA prepared from
either whole melanoma cells or EVs (MVs plus exosomes). As shown in
Fig. 3, a band of the expected
size [380 base pairs (bp)] was found both in cells (Fig. 3A, A375L) and vesicles (Fig. 3B, A375v), thus indicating that,
indeed, not only H1.0 protein but also the corresponding mRNA was
sorted to EVs. As internal references for these RT-PCR experiments
we used two positive controls: total RNA from adult rat brain
(Fig. 3A, lane B), and the cDNA
insert encoding H1.0 (Fig. 3C,
lane cDNA); one sample which did not contain RNA was included in
all the experiments as a negative control (Fig. 3B and C, lane N). Furthermore, to be
sure that we were amplifying RNA and not contaminating DNA, we also
performed a PCR reaction not preceded by the RT step; in these PCR
control experiments no band was seen (Fig. 3C, A375v).
EVs released from A375 melanoma cells
also contain H1.0 mRNA-binding proteins
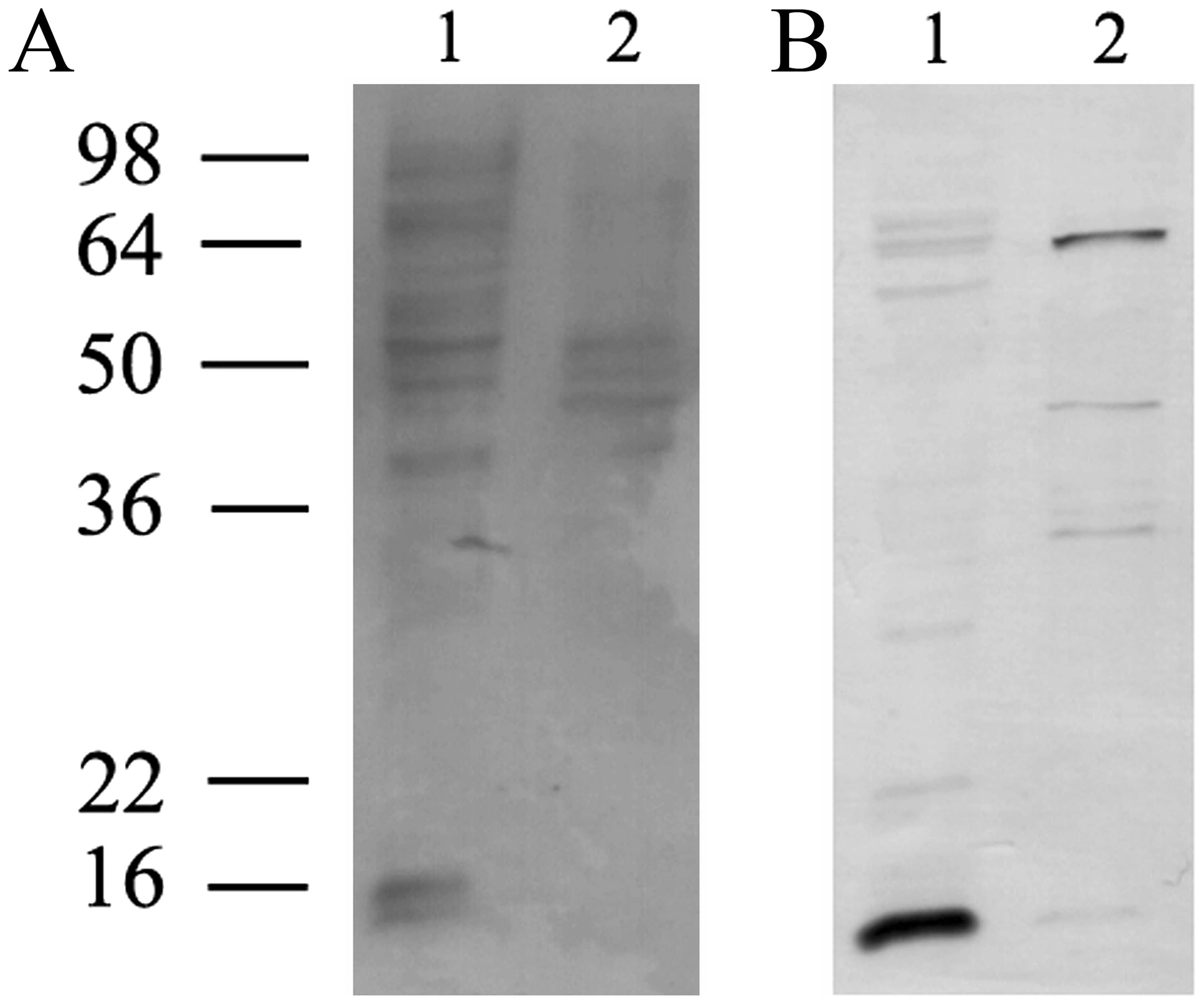
The presence of H1.0 RNA in EVs prompted us to also
look for RNA-binding proteins (RBPs) in the vesicles. We applied a
T1 RNase protection assay used by us for many years to study
protein-RNA interactions (25,32).
Briefly, proteins from freeze fractured vesicles were incubated
with ~5.0×106 cpm of 33P-labeled, in
vitro transcribed H1.0 RNA, in order to allow formation of
non-covalent protein-RNA complexes. The putative complexes were
then treated with T1 RNase to digest all the RNA but the sequences
protected by proteins. Finally, the complexes were cross-linked by
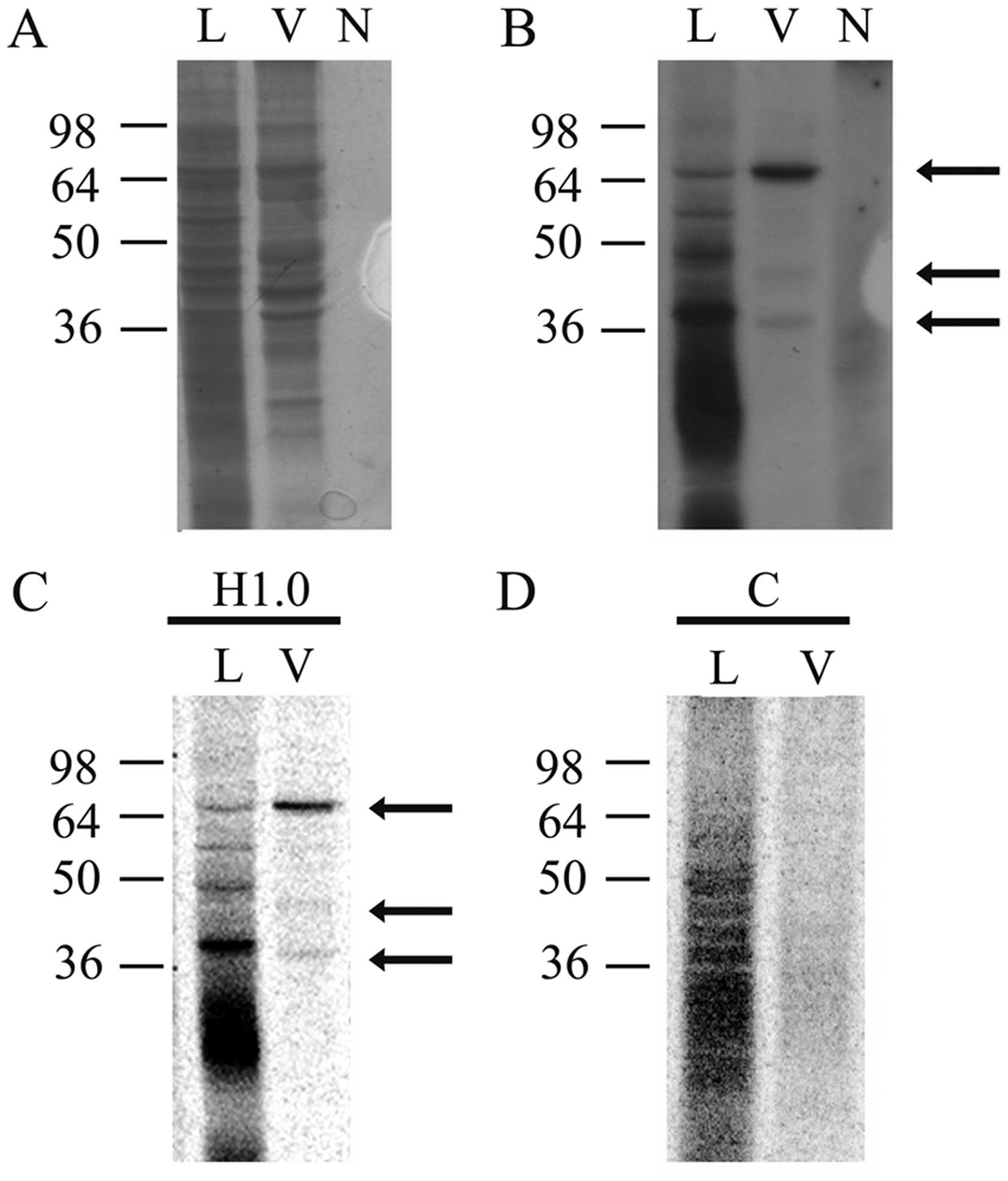
UV treatment and analyzed by denaturing PAGE. As shown in Fig. 4, many H1.0 RNA-binding proteins are
present in melanoma cells (Fig.
4B, lane L); in the vesicles, however, only three main bands
are clearly visible, at ~65, 45 and 38 kDa, respectively (Fig. 4B, lane V). In Fig. 4A we show the stained proteins
present in the same gel that was then dried and used for
fluorography (Fig. 4B). In this
gel a negative control was also included, which was obtained by
treating H1.0 RNA with T1 nuclease and UV, but in the absence of
proteins (Fig. 4B, lane N).
Finally, we also probed specificity of these bands by repeating the
analysis with H1.0 RNA (Fig. 4C,
H1.0) and, in parallel, with a second RNA (Fig. 4C and D), encoding CSD-C2/PIPPin
protein (26,38). As clearly shown in Fig. 4D, although control RNA (lane C) can
be bound by several proteins in melanoma cells (Fig. 4D, lane L), no band is visible in
the vesicle fraction (Fig. 4D,
lane V). On the other hand, H1.0 RNA (Fig. 4C) again formed the complexes seen
in Fig. 4B.
Enrichment of the H1.0 RNA-binding
proteins present in EVs by affinity chromatography and analysis by
MALDI-TOF mass spectrometry
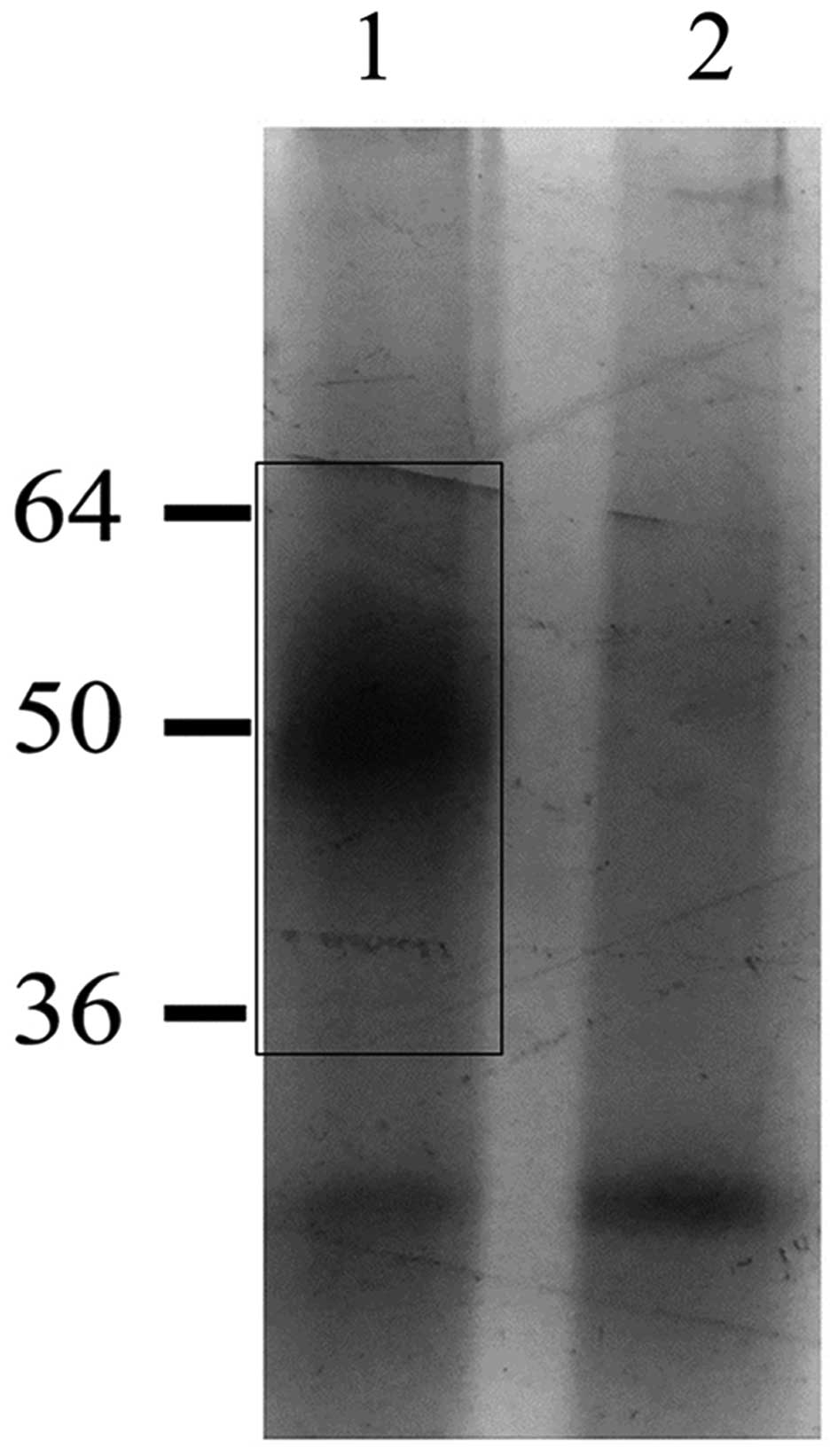
Then we tried to enrich the H1.0 RNA-binding
proteins evidenced in EVs by using a protocol based on affinity
chromatography on biotinylated H1.0 RNA, as already described
(27). The protein fractions
obtained from chromatography in the presence (Fig. 5, lane 1) or in the absence
(Fig. 5, lane 2) of H1.0 RNA were
analyzed by SDS-PAGE and the gel was silver stained. The region
indicated by the rectangle in Fig.
5 (lane 1), which clearly contains proteins not present in the
negative control sample (Fig. 5,
lane 2), was cut from the gel and analyzed by MALDI-TOF mass
spectrometry. The highest score was found for myelin expression
factor 2 (MYEF2) (Table I), a
protein mainly known as a DNA-binding repressor of the gene
encoding the myelin basic protein (39,40).
 | Table ISynopsis of information on the
protein identified as MYEF 2.a |
Table I
Synopsis of information on the
protein identified as MYEF 2.a
| Protein name | Gene name | AC | Mowse score | Sequence coverage
(%) | Theoretical MW
(Da)-pI |
|---|
| Myelin expression
factor 2 | MYEF2 | Q9P2K5 | 136 | 19 | 64121-8.86 |
Finally, we analyzed the proteins present in the
freeze fractured vesicles from melanoma cells by western blot with
anti-MYEF2 antibodies. As shown in Fig. 6, indeed, we observed three bands
immune-stained by these antibodies (of ~65, 45 and 34 kDa,
respectively).
Discussion
Construction of a complete multicellular organism
requires a precise and complex array of events which involves cell
proliferation as well as cell death, cell migration and
differentiation, formation of a network of cell-to-cell and
cell-to-ECM contacts, and exchange of chemical signals of many
kinds. Among the ways used by cells to exchange information,
production of extracellular vesicles has been recognized as a
physiological process, which is highly enhanced in transformed
cells. Cancer cells rely on EVs production for modifying to their
own advantage activities and properties of surrounding cells. In
addition, they probably use EVs to discard proteins otherwise able
to counteract tumorigenesis. In both normal and cancer cells,
indeed, the transcriptional potential of the cell nucleus is
controlled by availability of specific transcription factors, and
by the structural organization of chromatin. The structural
organization of chromatin is, in turn, regulated by a series of
dynamic events, involving chromatin remodelling factors (41) as well as the synthesis and
incorporation of replacement histone variants (42,43),
such as the linker H1.0 histone, which has been reported, long ago,
to be specifically accumulated during terminal differentiation of
several cell types (35,36,44).
Surprisingly, we recently found that in glial tumor
cells concentration of both H1.0 mRNA and protein is high and not
linked to a decrease of proliferation rate (19). Moreover, these cells secrete H1.0
by sorting it to EVs (19). In
this study we therefore analysed tumor cells of different origin,
A375 melanoma cells, and found that also these cells produce H1.0.
In addition, like oligodendroglioma cells, they secrete it into
EVs. In this case, however, the histone shows an apparent molecular
mass higher than expected. Since it has been reported that some
proteins, specifically sorted to EVs, are modified by sumoylation
(45), the presence of a SUMO
moiety in the putative H1.0 sorted to EVs was investigated. The
obtained results suggest that this is indeed the case. We conclude
that the still unknown mechanism responsible for the direct
correlation between H1.0 expression and differentiation does not
work in cancer cells. Moreover, we can hypothesize that secretion
of EVs from these cells could be also involved in eliminating
proteins (such as the H1.0 histone) that could be able to
counteract proliferation.
Interestingly, as demonstrated by RT-PCR, EVs
released from melanoma cells also contain H1.0 mRNA. This
observation prompted us to look for H1.0 mRNA-binding proteins. We
used an affinity chromatography approach that allowed to search,
only in the presence of a biotinylated RNA, a group of proteins,
which were further analysed by MALDI-TOF mass spectrometry. Among
these proteins, the most prevalent was myelin expression factor 2
(MYEF2). This protein has been identified in undifferentiated
cells, as a repressor able to bind directly to and to repress the
promoter of the gene encoding the mouse myelin basic protein
(39); it also contains two RNA
recognition motifs (RRM) which were shown to bind DNA (40); finally, it has also been reported
to form a complex with Runt-related transcription factor 1 (RUNX1),
an essential transcription factor involved in generating
hematopoietic stem cells (46). On
the basis of these previous observations, MYEF2 expression in
cancer cells is not surprising. In addition, since this protein
contains RNA-recognition motifs, its binding to an mRNA is not
surprising either. However, the fact that this repressor protein,
mostly expressed in undifferentiated cells, binds to the mRNA
encoding the differentiation-linked H1.0 histone, probably
participating in its elimination from the cells via EVs, is in our
opinion of great importance and sheds new light on the biochemical
mechanisms involved in tumorigenesis. Moreover, H1.0 mRNA could in
turn function as a MYEF2-carrier: once entered a new cell, MYEF2
could indeed also function as a transcriptional repressor, thus
conditioning the expression properties of the receiving cell.
Acknowledgements
This study was supported by University of Palermo
(Università degli Studi di Palermo, Palermo, Italy; ex 60%). We
thank Dr Giulio Ghersi for the kind gift of anti-integrin β1
antibodies. We also thank Dr Roberto Ghiandoni for helpful advice
with the NanoSight.
References
|
1
|
Iraci N, Leonardi T, Gessler F, Vega B and
Pluchino S: Focus on extracellular vesicles: Physiological role and
signalling properties of extracellular membrane vesicles. Int J Mol
Sci. 17:E1712016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tkach M and Théry C: Communication by
extracellular vesicles: Where we are and where we need to go. Cell.
164:1226–1232. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cocucci E and Meldolesi J: Ectosomes and
exosomes: Shedding the confusion between extracellular vesicles.
Trends Cell Biol. 25:364–372. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schiera G, Proia P, Alberti C, Mineo M,
Savettieri G and Di Liegro I: Neurons produce FGF2 and VEGF and
secrete them at least in part by shedding extracellular vesicles. J
Cell Mol Med. 11:1384–1394. 2007. View Article : Google Scholar
|
|
5
|
Proia P, Schiera G, Mineo M, Ingrassia
AMR, Santoro G, Savettieri G and Di Liegro I: Astrocytes shed
extracellular vesicles that contain fibroblast growth factor-2 and
vascular endothelial growth factor. Int J Mol Med. 21:63–67.
2008.
|
|
6
|
Prada I, Furlan R, Matteoli M and Verderio
C: Classical and unconventional pathways of vesicular release in
microglia. Glia. 61:1003–1017. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schiera G, Di Liegro CM and Di Liegro I:
Extracellular membrane vesicles as vehicles for brain cell-to-cell
interactions in physiological as well as pathological conditions.
BioMed Res Int. 2015:1529262015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lorico A, Corbeil D, Pawelek JM and
Alessandro R: Transmission of Information in Neoplasia by
Extracellular Vesicles. BioMed Res Int. 2015:2895672015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Arraud N, Linares R, Tan S, Gounou C,
Pasquet JM, Mornet S and Brisson AR: Extracellular vesicles from
blood plasma: Determination of their morphology, size, phenotype
and concentration. J Thromb Haemost. 12:614–627. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen T, Xi QY, Ye RS, Cheng X, Qi QE, Wang
SB, Shu G, Wang LN, Zhu XT, Jiang QY, et al: Exploration of
microRNAs in porcine milk exosomes. BMC Genomics. 15:1002014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gallo A, Tandon M, Alevizos I and Illei
GG: The majority of microRNAs detectable in serum and saliva is
concentrated in exosomes. PLoS One. 7:e306792012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ogawa Y, Taketomi Y, Murakami M, Tsujimoto
M and Yanoshita R: Small RNA transcriptomes of two types of
exosomes in human whole saliva determined by next generation
sequencing. Biol Pharm Bull. 36:66–75. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pap E, Pállinger E and Falus A: The role
of membrane vesicles in tumorigenesis. Crit Rev Oncol Hematol.
79:213–223. 2011. View Article : Google Scholar
|
|
14
|
Giusti I, D'Ascenzo S and Dolo V:
Microvesicles as potential ovarian cancer biomarkers. BioMed Res
Int. 2013:7030482013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ban LA, Shackel NA and McLennan SV:
Extracellular vesicles: A new frontier in biomarker discovery for
non-alcoholic fatty Liver disease. Int J Mol Sci. 17:172016.
View Article : Google Scholar
|
|
16
|
D'Agostino S, Salamone M, Di Liegro I and
Vittorelli ML: Membrane vesicles shed by oligodendroglioma cells
induce neuronal apoptosis. Int J Oncol. 29:1075–1085.
2006.PubMed/NCBI
|
|
17
|
Lo Cicero A, Schiera G, Proia P, Saladino
P, Savettieri G, Di Liegro CM and Di Liegro I: Oligodendroglioma
cells shed microvesicles which contain TRAIL as well as molecular
chaperones and induce cell death in astrocytes. Int J Oncol.
39:1353–1357. 2011.PubMed/NCBI
|
|
18
|
Liu Y, Gu Y and Cao X: The exosomes in
tumor immunity. OncoImmunology. 4:e10274722015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schiera G, Di Liegro CM, Saladino P, Pitti
R, Savettieri G, Proia P and Di Liegro I: Oligodendroglioma cells
synthesize the differentiation-specific linker histone H1° and
release it into the extracellular environment through shed
vesicles. Int J Oncol. 43:1771–1776. 2013.PubMed/NCBI
|
|
20
|
Crane-Robinson C: Linker histones: History
and current perspective. Biochim Biophys Acta. 1859.431–435.
2016.
|
|
21
|
Izzo A and Schneider R: The role of linker
histone H1 modifications in the regulation of gene expression and
chromatin dynamics. Biochim Biophys Acta. 1859.486–495. 2016.
|
|
22
|
Roque A, Ponte I and Suau P: Interplay
between histone H1 structure and function. Biochim Biophys Acta.
1859.444–454. 2016.
|
|
23
|
Millán-Ariño L, Izquierdo-Bouldstridge A
and Jordan A: Specificities and genomic distribution of somatic
mammalian histone H1 subtypes. Biochim Biophys Acta. 1859.510–519.
2016.
|
|
24
|
Di Liegro CM, Schiera G and Di Liegro I:
Regulation of mRNA transport, localization and translation in the
nervous system of mammals (Review). Int J Mol Med. 33:747–762.
2014.PubMed/NCBI
|
|
25
|
Scaturro M, Nastasi T, Raimondi L,
Bellafiore M, Cestelli A and Di Liegro I: H1(0) RNA-binding
proteins specifically expressed in the rat brain. J Biol Chem.
273:22788–22791. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nastasi T, Scaturro M, Bellafiore M,
Raimondi L, Beccari S, Cestelli A and Di Liegro I: PIPPin is a
brain-specific protein that contains a cold-shock domain and binds
specifically to H1 degrees and H3.3 mRNAs. J Biol Chem.
274:24087–24093. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Scaturrok M, Sala A, Cutrona G, Raimondi
L, Cannino G, Fontana S, Pucci-Minafra I and Di Liegro I:
Purification by affinity chromatography of H1o RNA-binding proteins
from rat brain. Int J Mol Med. 11:509–513. 2003.PubMed/NCBI
|
|
28
|
Saladino P, Di Liegro CM, Proia P, Sala A,
Schiera G, Lo Cicero A and Di Liegro I: RNA-binding activity of the
rat calmodulin-binding PEP-19 protein and of the long PEP-19
isoform. Int J Mol Med. 29:141–145. 2012.
|
|
29
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chomczynski P and Sacchi N: Single-step
method of RNA isolation by acid guanidinium
thiocyanate-phenol-chloroform extraction. Anal Biochem.
162:156–159. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Castiglia D, Gristina R, Scaturro M and Di
Liegro I: Cloning and analysis of cDNA for rat histone H1(0).
Nucleic Acids Res. 21:16741993. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Di Liegro CM, Schiera G, Proia P, Saladino
P and Di Liegro I: Identification in the rat brain of a set of
nuclear proteins interacting with H1° mRNA. Neuroscience.
229:71–76. 2013. View Article : Google Scholar
|
|
33
|
Yan JX, Wait R, Berkelman T, Harry RA,
Westbrook JA, Wheeler CH and Dunn MJ: A modified silver staining
protocol for visualization of proteins compatible with
matrix-assisted laser desorption/ionization and electrospray
ionization-mass spectrometry. Electrophoresis. 21:3666–3672. 2000.
View Article : Google Scholar
|
|
34
|
Cancemi P, Di Cara G, Albanese NN,
Costantini F, Marabeti MR, Musso R, Riili I, Lupo C, Roz E and
Pucci-Minafra I: Differential occurrence of S100A7 in breast cancer
tissues: A proteomic-based investigation. Proteomics Clin Appl.
6:364–373. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Panyim S and Chalkley R: A new histone
found only in mammalian tissues with little cell division. Biochem
Biophys Res Commun. 37:1042–1049. 1969. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zlatanova J and Doenecke D: Histone H1
zero: A major player in cell differentiation? FASEB J. 8:1260–1268.
1994.PubMed/NCBI
|
|
37
|
Villarroya-Beltri C, Gutiérrez-Vázquez C,
Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N,
Martinez-Herrera DJ, Pascual-Montano A, Mittelbrunn M and
Sánchez-Madrid F: Sumoylated hnRNPA2B1 controls the sorting of
miRNAs into exosomes through binding to specific motifs. Nat
Commun. 4:29802013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Castiglia D, Scaturro M, Nastasi T,
Cestelli A and Di Liegro I: PIPPin, a putative RNA-binding protein
specifically expressed in the rat brain. Biochem Biophys Res
Commun. 218:390–394. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Haas S, Steplewski A, Siracusa LD, Amini S
and Khalili K: Identification of a sequence-specific
single-stranded DNA binding protein that suppresses transcription
of the mouse myelin basic protein gene. J Biol Chem.
270:12503–12510. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Muralidharan V, Tretiakova A, Steplewski
A, Haas S, Amini S, Johnson E and Khalili K: Evidence for
inhibition of MyEF-2 binding to MBP promoter by MEF-1/Pur alpha. J
Cell Biochem. 66:524–531. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zaret KS and Mango SE: Pioneer
transcription factors, chromatin dynamics, and cell fate control.
Curr Opin Genet Dev. 37:76–81. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gaume X and Torres-Padilla ME: Regulation
of reprogramming and cellular plasticity through histone exchange
and histone variant incorporation. Cold Spring Harb Symp Quant
Biol. Nov 18–2015.(Epub ahead of print). pii: 027458. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zink L-M and Hake SB: Histone variants:
Nuclear function and disease. Curr Opin Genet Dev. 37:82–89. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Scaffidi P: Histone H1 alterations in
cancer. Biochim Biophys Acta. 1859.533–539. 2016.
|
|
45
|
Kunadt M, Eckermann K, Stuendl A, Gong J,
Russo B, Strauss K, Rai S, Kügler S, Falomir Lockhart L, Schwalbe
M, et al: Extracellular vesicle sorting of α-Synuclein is regulated
by sumoylation. Acta Neuropathol. 129:695–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
van Riel B, Pakozdi T, Brouwer R, Monteiro
R, Tuladhar K, Franke V, Bryne JC, Jorna R, Rijkers EJ, van Ijcken
W, et al: A novel complex, RUNX1-MYEF2, represses hematopoietic
genes in erythroid cells. Mol Cell Biol. 32:3814–3822. 2012.
View Article : Google Scholar : PubMed/NCBI
|