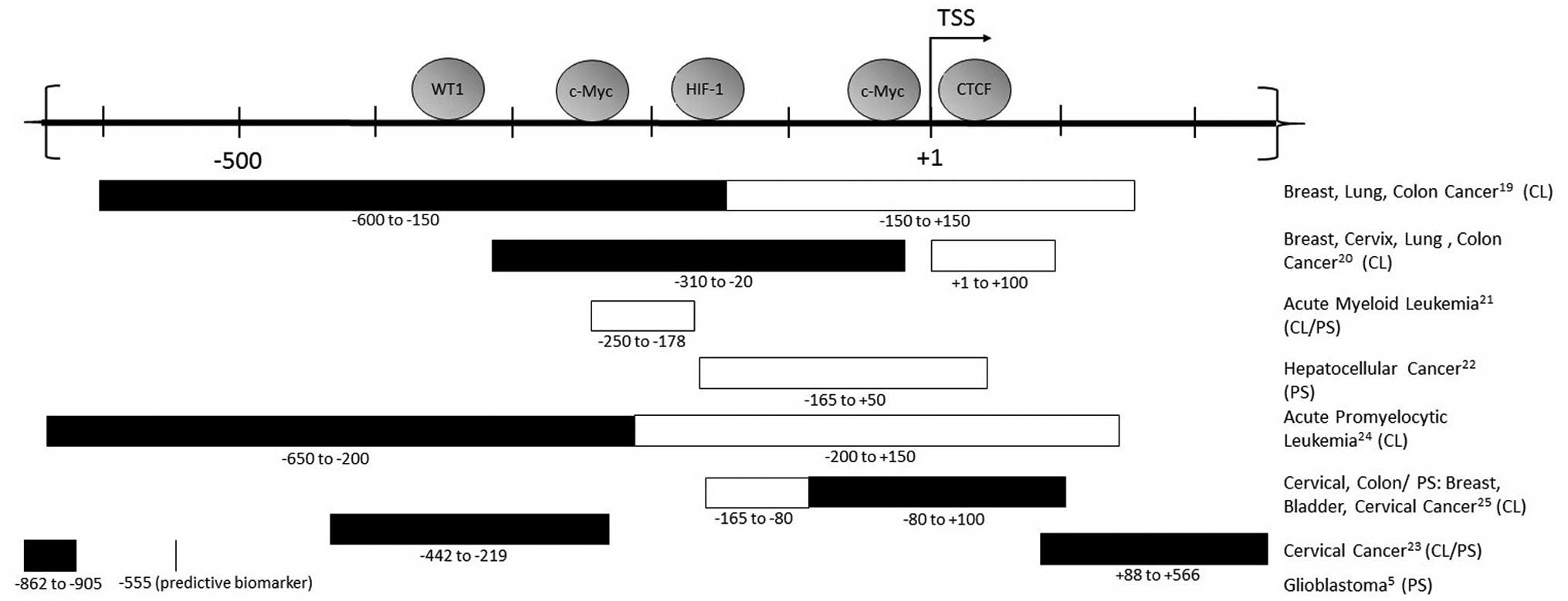
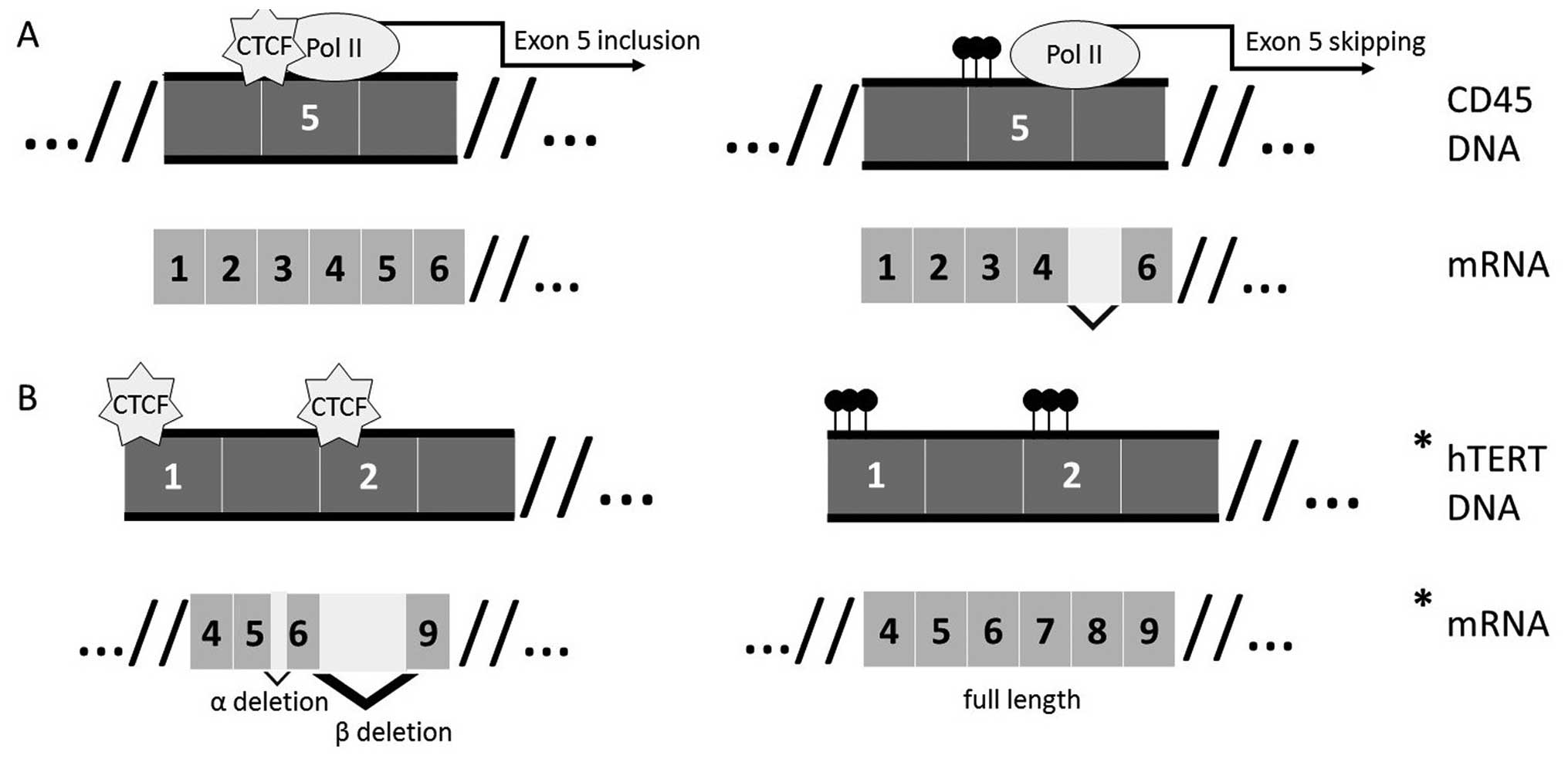
|
1
|
Lü MH, Liao ZL, Zhao XY, Fan YH, Lin XL,
Fang DC, Guo H and Yang SM: hTERT-based therapy: A universal
anticancer approach (Review). Oncol Rep. 28:1945–1952.
2012.PubMed/NCBI
|
|
2
|
Wong MS, Chen L, Foster C, Kainthla R,
Shay JW and Wright WE: Regulation of telomerase alternative
splicing: A target for chemotherapy. Cell Rep. 3:1028–1035. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Koziel JE, Fox MJ, Steding CE, Sprouse AA
and Herbert BS: Medical genetics and epigenetics of telomerase. J
Cell Mol Med. 15:457–467. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Umbricht CB, Sherman ME, Dome J, Carey LA,
Marks J, Kim N and Sukumar S: Telomerase activity in ductal
carcinoma in situ and invasive breast cancer. Oncogene.
18:3407–3414. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Castelo-Branco P, Choufani S, Mack S,
Gallagher D, Zhang C, Lipman T, Zhukova N, Walker EJ, Martin D,
Merino D, et al: Methylation of the TERT promoter and risk
stratification of childhood brain tumours: An integrative genomic
and molecular study. Lancet Oncol. 14:534–542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu X, Bishop J, Shan Y, Pai S, Liu D,
Murugan AK, Sun H, El-Naggar AK and Xing M: Highly prevalent TERT
promoter mutations in aggressive thyroid cancers. Endocr Relat
Cancer. 20:603–610. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kyo S, Takakura M, Fujiwara T and Inoue M:
Understanding and exploiting hTERT promoter regulation for
diagnosis and treatment of human cancers. Cancer Sci. 99:1528–1538.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adams RL: Eukaryotic DNA
methyltransferases - structure and function. BioEssays. 17:139–145.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jair KW, Bachman KE, Suzuki H, Ting AH,
Rhee I, Yen RW, Baylin SB and Schuebel KE: De novo CpG island
methylation in human cancer cells. Cancer Res. 66:682–692. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wan J, Oliver VF, Wang G, Zhu H, Zack DJ,
Merbs SL and Qian J: Characterization of tissue-specific
differential DNA methylation suggests distinct modes of positive
and negative gene expression regulation. BMC Genomics. 16:492015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deaton AM and Bird A: CpG islands and the
regulation of transcription. Genes Dev. 25:1010–1022. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Baylin SB and Jones PA: A decade of
exploring the cancer epigenome - biological and translational
implications. Nat Rev Cancer. 11:726–734. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stirzaker C, Millar DS, Paul CL, Warnecke
PM, Harrison J, Vincent PC, Frommer M and Clark SJ: Extensive DNA
methylation spanning the Rb promoter in retinoblastoma tumors.
Cancer Res. 57:2229–2237. 1997.PubMed/NCBI
|
|
14
|
Smith IM, Glazer CA, Mithani SK, Ochs MF,
Sun W, Bhan S, Vostrov A, Abdullaev Z, Lobanenkov V, Gray A, et al:
Coordinated activation of candidate proto-oncogenes and cancer
testes antigens via promoter demethylation in head and neck cancer
and lung cancer. PLoS One. 4:e49612009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
16
|
Tahira AC, Kubrusly MS, Faria MF, Dazzani
B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC
and Reis EM: Long noncoding intronic RNAs are differentially
expressed in primary and metastatic pancreatic cancer. Mol Cancer.
10:1412011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hangauer MJ, Vaughn IW and McManus MT:
Pervasive transcription of the human genome produces thousands of
previously unidentified long intergenic noncoding RNAs. PLoS Genet.
9:e10035692013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Choi JH, Park SH, Park J, Park BG, Cha SJ,
Kong KH, Lee KH and Park AJ: Site-specific methylation of CpG
nucleotides in the hTERT promoter region can control the expression
of hTERT during malignant progression of colorectal carcinoma.
Biochem Biophys Res Commun. 361:615–620. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zinn RL, Pruitt K, Eguchi S, Baylin SB and
Herman JG: hTERT is expressed in cancer cell lines despite promoter
DNA methylation by preservation of unmethylated DNA and active
chromatin around the transcription start site. Cancer Res.
67:194–201. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guilleret I and Benhattar J: Unusual
distribution of DNA methylation within the hTERT CpG island in
tissues and cell lines. Biochem Biophys Res Commun. 325:1037–1043.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pettigrew KA, Armstrong RN, Colyer HA,
Zhang SD, Rea IM, Jones RE, Baird DM and Mills KI: Differential
TERT promoter methylation and response to 5-aza-2′-deoxycytidine in
acute myeloid leukemia cell lines: TERT expression, telomerase
activity, telomere length, and cell death. Genes Chromosomes
Cancer. 51:768–780. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Iliopoulos D, Satra M, Drakaki A,
Poultsides GA and Tsezou A: Epigenetic regulation of hTERT promoter
in hepatocellular carcinomas. Int J Oncol. 34:391–399.
2009.PubMed/NCBI
|
|
23
|
de Wilde J, Kooter JM, Overmeer RM,
Claassen-Kramer D, Meijer CJ, Snijders PJ and Steenbergen RD: hTERT
promoter activity and CpG methylation in HPV-induced
carcinogenesis. BMC Cancer. 10:2712010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Azouz A, Wu YL, Hillion J, Tarkanyi I,
Karniguian A, Aradi J, Lanotte M, Chen GQ, Chehna M and
Ségal-Bendirdjian E: Epigenetic plasticity of hTERT gene promoter
determines retinoid capacity to repress telomerase in
maturation-resistant acute promyelocytic leukemia cells. Leukemia.
24:613–622. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Renaud S, Loukinov D, Abdullaev Z,
Guilleret I, Bosman FT, Lobanenkov V and Benhattar J: Dual role of
DNA methylation inside and outside of CTCF-binding regions in the
transcriptional regulation of the telomerase hTERT gene. Nucleic
Acids Res. 35:1245–1256. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kumari A, Srinivasan R, Vasishta RK and
Wig JD: Positive regulation of human telomerase reverse
transcriptase gene expression and telomerase activity by DNA
methylation in pancreatic cancer. Ann Surg Oncol. 16:1051–1059.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lopatina NG, Poole JC, Saldanha SN, Hansen
NJ, Key JS, Pita MA, Andrews LG and Tollefsbol TO: Control
mechanisms in the regulation of telomerase reverse transcriptase
expression in differentiating human teratocarcinoma cells. Biochem
Biophys Res Commun. 306:650–659. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu J, Zhao Y and Wang S: Chromatin and
epigenetic regulation of the telomerase reverse transcriptase gene.
Protein Cell. 1:22–32. 2010. View Article : Google Scholar
|
|
29
|
Guilleret I and Benhattar J: Demethylation
of the human telomerase catalytic subunit (hTERT) gene promoter
reduced hTERT expression and telomerase activity and shortened
telomeres. Exp Cell Res. 289:326–334. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsujioka T, Yokoi A, Itano Y, Takahashi K,
Ouchida M, Okamoto S, Kondo T, Suemori S, Tohyama Y and Tohyama K:
Five-aza-2′-deoxycytidine-induced hypomethylation of cholesterol
25-hydroxylase gene is responsible for cell death of
myelodysplasia/leukemia cells. Sci Rep. 5:167092015. View Article : Google Scholar
|
|
31
|
Prendergast G and Ziff E:
Methylation-sensitive sequence-specific DNA binding by the c-Myc
basic region. Science. 251:186–189. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sitaram RT, Degerman S, Ljungberg B,
Andersson E, Oji Y, Sugiyama H, Roos G and Li A: Wilms’ tumour 1
can suppress hTERT gene expression and telomerase activity in clear
cell renal cell carcinoma via multiple pathways. Br J Cancer.
103:1255–1262. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Drummond IA, Rupprecht HD, Rohwer-Nutter
P, Lopez-Guisa JM, Madden SL, Rauscher FJ III and Sukhatme VP: DNA
recognition by splicing variants of the Wilms’ tumor suppressor,
WT1. Mol Cell Biol. 14:3800–3809. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shin KH, Kang MK, Dicterow E and Park NH:
Hypermethylation of the hTERT promoter inhibits the expression of
telomerase activity in normal oral fibroblasts and senescent normal
oral keratinocytes. Br J Cancer. 89:1473–1478. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Renaud S, Loukinov D, Bosman FT,
Lobanenkov V and Benhattar J: CTCF binds the proximal exonic region
of hTERT and inhibits its transcription. Nucleic Acids Res.
33:6850–6860. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Feldmann A, Ivanek R, Murr R, Gaidatzis D,
Burger L and Schübeler D: Transcription factor occupancy can
mediate active turnover of DNA methylation at regulatory regions.
PLoS Genet. 9:e10039942013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Choi JH, Min NY, Park J, Kim JH, Park SH,
Ko YJ, Kang Y, Moon YJ, Rhee S, Ham SW, et al: TSA-induced DNMT1
downregulation represses hTERT expression via recruiting CTCF into
demethylated core promoter region of hTERT in HCT116. Biochem
Biophys Res Commun. 391:449–454. 2010. View Article : Google Scholar
|
|
38
|
Meeran SM, Patel SN and Tollefsbol TO:
Sulforaphane causes epigenetic repression of hTERT expression in
human breast cancer cell lines. PLoS One. 5:e114572010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kornblihtt AR: Promoter usage and
alternative splicing. Curr Opin Cell Biol. 17:262–268. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Nieto Moreno N, Giono LE, Cambindo Botto
AE, Muñoz MJ and Kornblihtt AR: Chromatin, DNA structure and
alternative splicing. FEBS Lett. 589:3370–3378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cramer P, Cáceres JF, Cazalla D, Kadener
S, Muro AF, Baralle FE and Kornblihtt AR: Coupling of transcription
with alternative splicing: RNA pol II promoters modulate SF2/ASF
and 9G8 effects on an exonic splicing enhancer. Mol Cell.
4:251–258. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Schor IE, Gómez Acuña LI and Kornblihtt
AR: Coupling between transcription and alternative splicing. Cancer
Treat Res. 158:1–24. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Naftelberg S, Schor IE, Ast G and
Kornblihtt AR: Regulation of alternative splicing through coupling
with transcription and chromatin structure. Annu Rev Biochem.
84:165–198. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Davies RC, Calvio C, Bratt E, Larsson SH,
Lamond AI and Hastie ND: WT1 interacts with the splicing factor
U2AF65 in an isoform-dependent manner and can be incorporated into
spliceosomes. Genes Dev. 12:3217–3225. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Markus MA, Heinrich B, Raitskin O, Adams
DJ, Mangs H, Goy C, Ladomery M, Sperling R, Stamm S and Morris BJ:
WT1 interacts with the splicing protein RBM4 and regulates its
ability to modulate alternative splicing in vivo. Exp Cell Res.
312:3379–3388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Mohamed AM, Balsat M, Thenoz M, Koering C,
Payen-Gay L, Cheok M, Mortada H, Auboeuf D, Pinatel C, El-Hamri M,
et al: Oncogene- and drug resistance-associated alternative exon
usage in acute myeloid leukemia (AML). Oncotarget. 7:2889–2909.
2016.
|
|
47
|
Katuri V, Gerber S, Qiu X, McCarty G,
Goldstein SD, Hammers H, Montgomery E, Chen AR and Loeb DM: WT1
regulates angiogenesis in Ewing Sarcoma. Oncotarget. 5:2436–2449.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Cunningham TJ, Palumbo I, Grosso M, Slater
N and Miles CG: WT1 regulates murine hematopoiesis via maintenance
of VEGF isoform ratio. Blood. 122:188–192. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Amin EM, Oltean S, Hua J, Gammons MV,
Hamdollah-Zadeh M, Welsh GI, Cheung MK, Ni L, Kase S, Rennel ES, et
al: WT1 mutants reveal SRPK1 to be a downstream angiogenesis target
by altering VEGF splicing. Cancer Cell. 20:768–780. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Radan L, Hughes CS, Teichroeb JH, Vieira
Zamora FM, Jewer M, Postovit LM and Betts DH: Microenvironmental
regulation of telomerase isoforms in human embryonic stem cells.
Stem Cells Dev. 23:2046–2066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Yatabe N, Kyo S, Maida Y, Nishi H,
Nakamura M, Kanaya T, Tanaka M, Isaka K, Ogawa S and Inoue M:
HIF-1-mediated activation of telomerase in cervical cancer cells.
Oncogene. 23:3708–3715. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Nishi H, Nakada T, Kyo S, Inoue M, Shay JW
and Isaka K: Hypoxia-inducible factor 1 mediates upregulation of
telomerase (hTERT). Mol Cell Biol. 24:6076–6083. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Anderson CJ, Hoare SF, Ashcroft M,
Bilsland AE and Keith WN: Hypoxic regulation of telomerase gene
expression by transcriptional and post-transcriptional mechanisms.
Oncogene. 25:61–69. 2006.
|
|
54
|
Kechris K, Yang YH and Yeh RF: Prediction
of alternatively skipped exons and splicing enhancers from exon
junction arrays. BMC Genomics. 9:5512008. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kornblihtt AR: CTCF: From insulators to
alternative splicing regulation. Cell Res. 22:450–452. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shukla S, Kavak E, Gregory M, Imashimizu
M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R and
Oberdoerffer S: CTCF-promoted RNA polymerase II pausing links DNA
methylation to splicing. Nature. 479:74–79. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Paredes SH, Melgar MF and Sethupathy P:
Promoter-proximal CCCTC-factor binding is associated with an
increase in the transcriptional pausing index. Bioinformatics.
29:1485–1487. 2013. View Article : Google Scholar :
|
|
58
|
Adelman K and Lis JT: Promoter-proximal
pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev
Genet. 13:720–731. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Ong CT and Corces VG: CTCF: An
architectural protein bridging genome topology and function. Nat
Rev Genet. 15:234–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Stadhouders R, Thongjuea S, Andrieu-Soler
C, Palstra RJ, Bryne JC, van den Heuvel A, Stevens M, de Boer E,
Kockx C, van der Sloot A, et al: Dynamic long-range chromatin
interactions control Myb proto-oncogene transcription during
erythroid development. EMBO J. 31:986–999. 2012. View Article : Google Scholar :
|
|
61
|
Sebestyén E, Zawisza M and Eyras E:
Detection of recurrent alternative splicing switches in tumor
samples reveals novel signatures of cancer. Nucleic Acids Res.
43:1345–1356. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ulaner GA, Hu JF, Vu TH, Giudice LC and
Hoffman AR: Tissue-specific alternate splicing of human telomerase
reverse transcriptase (hTERT) influences telomere lengths during
human development. Int J Cancer. 91:644–649. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Lincz LF, Mudge LM, Scorgie FE, Sakoff JA,
Hamilton CS and Seldon M: Quantification of hTERT splice variants
in melanoma by SYBR green real-time polymerase chain reaction
indicates a negative regulatory role for the β deletion variant.
Neoplasia. 10:1131–1137. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mavrogiannou E, Strati A, Stathopoulou A,
Tsaroucha EG, Kaklamanis L and Lianidou ES: Real-time RT-PCR
quantification of human telomerase reverse transcriptase splice
variants in tumor cell lines and non-small cell lung cancer. Clin
Chem. 53:53–61. 2007. View Article : Google Scholar
|
|
65
|
Listerman I, Sun J, Gazzaniga FS, Lukas JL
and Blackburn EH: The major reverse transcriptase-incompetent
splice variant of the human telomerase protein inhibits telomerase
activity but protects from apoptosis. Cancer Res. 73:2817–2828.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Kolquist KA, Ellisen LW, Counter CM,
Meyerson M, Tan LK, Weinberg RA, Haber DA and Gerald WL: Expression
of TERT in early premalignant lesions and a subset of cells in
normal tissues. Nat Genet. 19:182–186. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Meyerson M, Counter CM, Eaton EN, Ellisen
LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ,
Liu Q, et al: hEST2, the putative human telomerase catalytic
subunit gene, is up-regulated in tumor cells and during
immortalization. Cell. 90:785–795. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Yi X, Shay JW and Wright WE: Quantitation
of telomerase components and hTERT mRNA splicing patterns in
immortal human cells. Nucleic Acids Res. 29:4818–4825. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kotoula V, Hytiroglou P, Pyrpasopoulou A,
Saxena R, Thung SN and Papadimitriou CS: Expression of human
telomerase reverse transcriptase in regenerative and precancerous
lesions of cirrhotic livers. Liver. 22:57–69. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ohyashiki JH, Hisatomi H, Nagao K, Honda
S, Takaku T, Zhang Y, Sashida G and Ohyashiki K: Quantitative
relationship between functionally active telomerase and major
telomerase components (hTERT and hTR) in acute leukaemia cells. Br
J Cancer. 92:1942–1947. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Barclay JY, Morris AG and Nwokolo CU:
HTERT mRNA partially regulates telomerase activity in gastric
adenocarcinoma and adjacent normal gastric mucosa. Dig Dis Sci.
50:1299–1303. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Rha SY, Jeung HC, Park KH, Kim JJ and
Chung HC: Changes of telomerase activity by alternative splicing of
full-length and β variants of hTERT in breast cancer patients.
Oncol Res. 18:213–220. 2009. View Article : Google Scholar
|
|
73
|
Liu Y, Wu BQ, Zhong HH, Tian XX and Fang
WG: Quantification of alternative splicing variants of human
telomerase reverse transcriptase and correlations with telomerase
activity in lung cancer. PLoS One. 7:e388682012. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wang Y, Meeker AK, Kowalski J, Tsai HL,
Somervell H, Heaphy C, Sangenario LE, Prasad N, Westra WH, Zeiger
MA, et al: Telomere length is related to alternative splice
patterns of telomerase in thyroid tumors. Am J Pathol.
179:1415–1424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Wong MS, Wright WE and Shay JW:
Alternative splicing regulation of telomerase: A new paradigm?
Trends Genet. 30:430–438. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Kilian A, Bowtell DD, Abud HE, Hime GR,
Venter DJ, Keese PK, Duncan EL, Reddel RR and Jefferson RA:
Isolation of a candidate human telomerase catalytic subunit gene,
which reveals complex splicing patterns in different cell types.
Hum Mol Genet. 6:2011–2019. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Saebøe-Larssen S, Fossberg E and
Gaudernack G: Characterization of novel alternative splicing sites
in human telomerase reverse transcriptase (hTERT): Analysis of
expression and mutual correlation in mRNA isoforms from normal and
tumour tissues. BMC Mol Biol. 7:262006. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Withers JB, Ashvetiya T and Beemon KL:
Exclusion of exon 2 is a common mRNA splice variant of primate
telomerase reverse transcriptases. PLoS One. 7:e480162012.
View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Khosravi-Maharlooei M, Jaberipour M,
Hosseini Tashnizi A, Attar A, Amirmoezi F and Habibagahi M:
Expression pattern of alternative splicing variants of human
telomerase reverse Transcriptase (hTERT) in cancer cell lines was
not associated with the origin of the cells. Int J Mol Cell Med.
4:109–119. 2015.PubMed/NCBI
|
|
80
|
Jafri MA, Ansari SA, Alqahtani MH and Shay
JW: Roles of telomeres and telomerase in cancer, and advances in
telomerase-targeted therapies. Genome Med. 8:692016. View Article : Google Scholar : PubMed/NCBI
|