Introduction
Pancreatic ductal adenocarcinoma (PDAC), also known
as pancreatic cancer is one of the most fatal diseases, and the
overall 5-year survival rate among patients with pancreatic cancer
is <5%. In pancreatic cancer, 90% of tumors have activating
mutations in the KRAS2 oncogene (1). In addition, activation of hedgehog
signaling pathway is vital in tumor development. Activity of
hedgehog signaling pathway is gradually increased in the progress
from pancreatic intraepithelial neoplasia (PanIN) to PDAC (2–4).
Mutations of components lead to activation of hedgehog signaling
pathway in some types of tumors (5–8).
However, no mutations have been detected in components of this
pathway in pancreatic cancer. SUFU is an essential negative
regulator of mammalian hedgehog signaling pathway. Kasai et
al (9) provided evidence that
physical binding inhibition of GLI1 by SUFU is dismissed by
tethering of SUFU in the cytoplasm of PDAC cells by overexpressed
SCL/TAL1 interrupting locus (SIL) and KRAS mutation. However, Chen
et al (10) provided
convincing data indicating that mouse Sufu sequesters Gli2/3
protein in the cytoplasm and protects them from Spop-mediated
protein degradation, providing a Gli protein pool for the
production of Gli2/3 activators and repressors. Sufu is necessary
for maximal activation of hedgehog pathway. Thus Sufu has an
unexpected positive role in controlling mammalian hedgehog
signaling. Thus, we proceeded to clarify the function of SUFU in
human pancreatic cancer. There are two transcript variants of
SUFU published in NCBI website: transcript variant 1 of
SUFU (SUFUv1, NM_016169) and transcript variant 2 of SUFU
(SUFUv2, NM_001178133). We started by making cDNA clones of
alternative splicing transcript variants of SUFU gene in
human pancreatic cancer cells.
Materials and methods
Cell culture and RNA extraction
Seven pancreatic cancer cell lines SW1990, PATU8988,
AsPC-1, BxPC-3, CFPAC-1, Capan-2, PANC-1 were cultured in DMEM
medium supplemented with 10% FBS and incubated in a 5%
CO2 humidified incubator at 37°C. Total RNA was
extracted from cells using RNA iso Plus (Takara, Japan) according
to the manufacturer’s instructions. The yield of RNA was determined
using a spectrophotometer (NanoDrop 2000, Thermo Scientific, USA)
and the integrity was evaluated using agarose gel electrophoresis
stained with ethidium bromide.
3′ rapid amplification of cDNA ends
(3′RACE) and nested PCR
Two reference sequence transcript variants of
SUFU gene are published on NCBI. They are transcript variant
1 of SUFU (SUFUv1, NM_016169) and transcript variant 2 of
SUFU (SUFUv2, NM_001178133). 3′RACE was made to obtain the
3′ cDNA of SUFU transcripts.
3′-Full RACE Core Set Ver.2.0 (product code, D314,
Takara) kit contains: 3′RACE adaptor (a reverse transcription
primer adaptor which can addict to mature mRNA poly A tail by its
poly T sequences), M-MLV reverse transcriptase, 3′RACE outer primer
(reverse primer of the first nested PCR), 3′RACE inner primer
(reverse primer of the second nested PCR), etc. The steps are shown
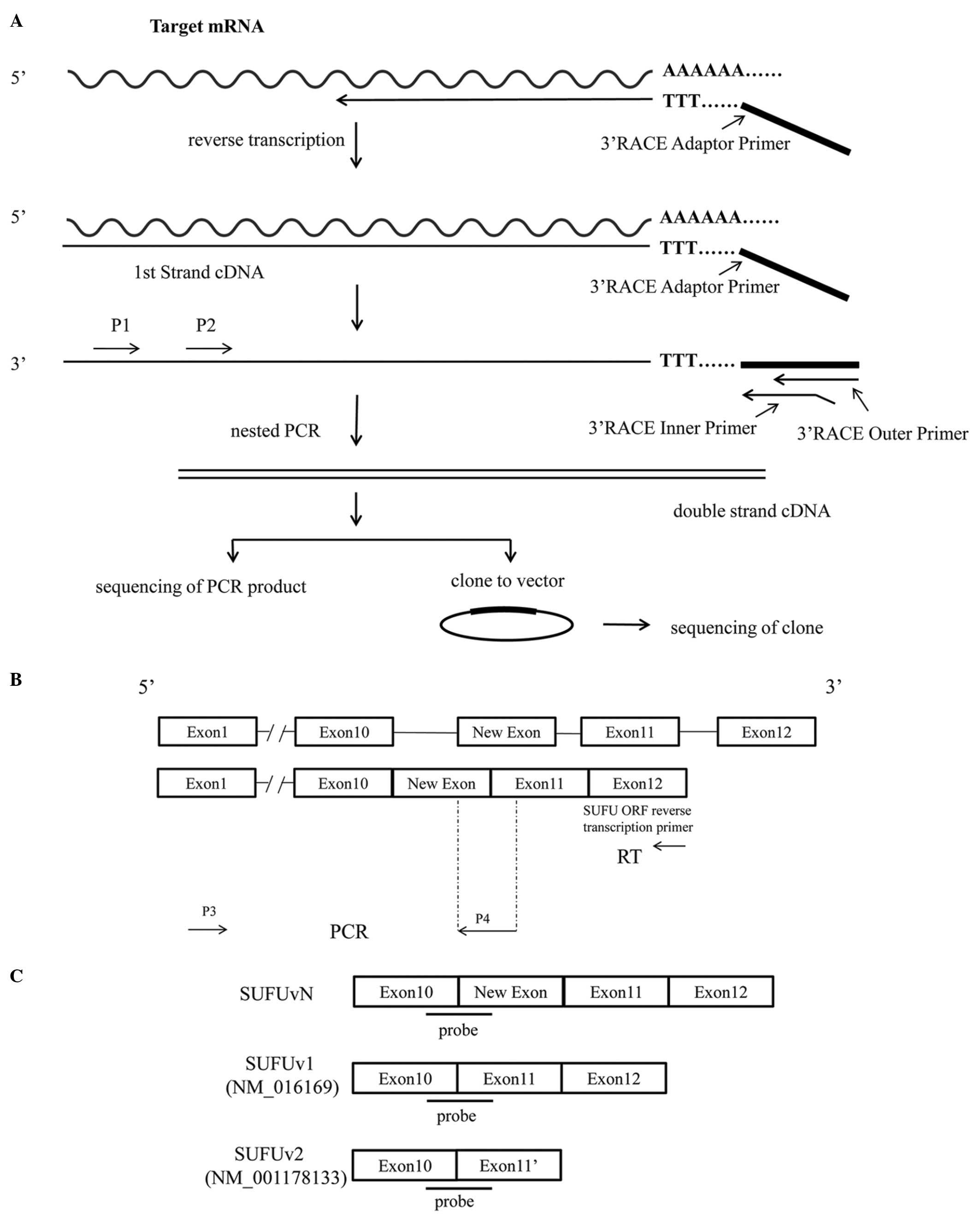
in Fig. 1A.
We performed reverse transcription. The reaction
consisted of 1 μl total RNA (500 ng/μl), 1 μl 3′RACE adaptor, 0.25
μl M-MLV reverse transcriptase, 0.25 μl RNase inhibitor, 2 μl 5×
M-MLV buffer, 1 μl dNTP mixture and 4.5 μl RNase-free
dH2O, in a total volume of 10 μl. Reaction was performed
in a GeneAmp® PCR System 9700 (Applied Biosystems, USA)
for 60 min at 42°C, followed by heat inactivation of RT for 15 min
at 70°C.
First cycle of nested PCR
The forward primer of the first nested PCR was
designed by us according to the kit instructions (Table I). We call this forward primer P1.
It is situated in the 9th exon of SUFUv1. The first base pair of P1
is situated in the 1,218 bp of the whole RNA sequences. The reverse
primer of the first nested PCR is in the kit called 3′RACE outer
primer. The reaction consisted of 3 μl reverse transcription
product, 7 μl 1× cDNA dilution buffer, 2 μl P1 (10 μmol/l), 2 μl
3′RACE outer primer, 5 μl 10× Ex Taq buffer (including
Mg2), 0.25 μl Ex Taq enzyme and 30.75 μl
dH2O. Reactions were performed in a GeneAmp PCR System
9700 for 3 min at 94°C, followed by 25 cycles of 94°C for 30 sec,
55°C for 30 sec, 72°C for 5 min, and a last step of 72°C for 10
min.
 | Table IPrimers used in 3′RACE and RT-PCR of
SUFUvN 5′cDNA. |
Table I
Primers used in 3′RACE and RT-PCR of
SUFUvN 5′cDNA.
| Primers | Sequences |
|---|
| First cycle of nested
PCR | |
| P1 | 5′-CGC AAA GAC AGC
CTG GAA AGT GAC-3′ |
| 3′RACE outer
primer |
5′-TACCGTCGTTCCACTAGTGATTT-3′ |
| Second cycle of
nested PCR | |
| P2 | 5′-TCC TGC ATG GAC
GGC ACT TTA C-3′ |
| 3′RACE inner
primer |
5′CGCGGATCCTCCACTAGTGATTTCAC
TATAGG-3′ |
| RT | |
| SUFU ORF reverse
transcription primer |
5′-CTAGTGTAGCGGACTGTCGAACA-3′ |
| PCR | |
| P3 |
5′-CTCCAGGTTACCGCTATCG TC-3′ |
| P4 |
5′-TTCGGTCAACAGAATCAGGTTTC-3′ |
Second cycle of nested PCR
The forward primer of the second nested PCR was also
designed by us according to the kit instruction (Table I). We call this forward primer P2.
It is situated in the 10th exon of SUFUv1. The first base pair of
P2 is situated in the 1,357 bp of the whole RNA sequences. The
reverse primer of the second nested PCR is in the kit called 3′RACE
inner primer. The reaction consisted of 1 μl the first nested PCR
product, 8 μl dNTP mixture, 5 μl 10× Ex Taq buffer (including
Mg2), 0.5 μl Ex Taq enzyme, 2 μl P2 (10 μmol/l), 2 μl
3′RACE inner primer and 31.5 μl dH2O. Reactions were
performed in a GeneAmp PCR System 9700 for 3 min at 94°C, followed
by 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 5 min,
and a last step of 72°C for 10 min.
The product of second nested PCR was ligated with
pMD-18T vector, and infected DH5α. Then, we selected 10 positive
clones, and made sequence detection.
RT-PCR to identify the 5′ cDNA sequence
of novel transcript variant of SUFU (SUFUvN)
We performed reverse transcription by SUFU ORF
reverse transcription primer (Table
I), then, by PCR. The forward primer of PCR was designed by us
called P3 (Table I). It is
situated in the first exon of SUFUv1. Reverse primer of PCR was
also designed by us called P4 (Table
I). It contains partial 11th exon of SUFUv1 and partial new
exon. Product of PCR was used for sequence detection. The steps are
shown in Fig. 1B.
Construction of expression clones of
SUFUvN and SUFUv1
The forward primer terminal is added with protection
base pairs, BamHI enzyme sites and Kozak sequences. The
reverse primer terminal is added with protection base pairs,
AgeI enzyme sites. The forwad primer and reverse primer were
designed to clone open reading frame (ORF) of SUFU. We
performed reverse transcription by SUFU ORF reverse transcription
primer (Table I), then by PCR. The
sequence of forward primer is: 5′-cgggatccGGGATGGC
GGAGCTGCGGCCTA-3′. The sequence of reverse primer is:
5′-gcgcaccggtCTAGTGTAG CGGACTGTCGAACA-3′. BamHI and
AgeI double enzyme digestion was made in both PCR product
and pcDNA3.1mychisA (+) expression vector. After ligation of PCR
product with pcDNA3.1mychisA (+) expression vector and DH5α
infection we selected positive clones and sequence detection was
performed.
Cell transfection
SW1990 cells were transiently transfected with empty
vector, SUFUv1 and SUFUvN expression vector, respectively, by use
of Lipofectamine 2000 kit (Takara) according to the manufacturer’s
instructions. The cells were cultured for 24 h after transfection.
Then the cells were harvested for further studies.
Western blotting
The empty vector, SUFUv1 and SUFUvN expression
vector transfected SW1990 cells were collected. Cells were
dissolved in cell lysis buffer. Protein of pancreatic cancer tissue
was extracted by RIPA lysis buffer. The lysates were cooled with
ice for 30 min, and then centrifuged at 13,000 × g for 30 min.
Proteins (10 μg) in the collected supernatant were separated by
SDS-PAGE on 12% gels and then transferred to PVDF membranes. The
membrane, after a block with 5% skim milk, was incubated with
primary antibodies to SUFU (rabbit anti-human monoclonal antibody,
catalog no. NBP1-40515, Novus, USA) and GAPDH (ab97626, Abcam,
Cambridge, UK). This SUFU antibody is made by N terminal of SUFU
antigen, so it can bind the N terminal of protein encoded by
SUFUv1, SUFUv2 and SUFUvN (SUFU isoform 1, isoform 2 and isoform
N). Primary antibody is 1:200 diluted to detect SUFU isoforms. The
secondary antibody is HRP labeled goat anti-rabbit polyclonal IgG
antibody. GAPDH is used as inner control. ECL kit (product code,
34096, Thermo Fisher Scientific, USA) and the camera are used to
capture the results.
Patients, tissue samples and total RNA
extraction of tissue samples
Tumors were obtained from 40 adult patients
diagnosed with pancreatic ductal adenocarcinoma who undergwent
surgery at The Second Military Medical University affiliated
Changhai Hospital between 2013 and 2014. This study was approved by
the Ethics Committee of Changhai Hospital. The total RNA of the
pancreatic cancer tissue was extracted by mirVana™ miRNA isolation
kit (am1561, Takara). DNA contamination was removed by DNase in
this kit. Total RNA was stored at −70°C.
Real-time quantitative RT-PCR using
TaqMan MGB probe method to detect transcription levels of different
SUFU variants in PDAC tissues of 40 patients
The 18s control (Hs99999901_s1) MGB probe and
primers kit were purchased from Invitrogen Co., USA. The primer
pairs and the TaqMan MGB probe sequences of SUFU different variants
were designed by Primer Express 3.0 software and were synthesized
by Invitrogen Co. The sequences are shown in Table II, and the probes are shown in
Fig. 1C.
 | Table IIProbes detecting different
transcription variants of SUFU. |
Table II
Probes detecting different
transcription variants of SUFU.
| Transcript
variants | Primer pairs and MGB
probe |
|---|
| NM_016169 |
| Forward primer |
CACTGAGGAGCATCCTTACGC |
| Reverse primer |
AGCTGTACTCTTTGGGAAGTTTGAA |
| Probe |
FAM-CCTGGTTACAAATTCTGTT-MGB |
| NM_001178133 |
| Forward primer |
CCACTGAGGAGCATCCTTACG |
| Reverse primer |
GCTGAAAATTGGAGATGCTGACT |
| Probe |
FAM-CTGGTTACAACTCTGAACC-MGB |
| Sufu-vN |
| Forward primer |
AGGCGCCTTTGCCACTG |
| Reverse primer |
ACACAGGAAGGTGAGCACACAG |
| Probe |
FAM-TGGTTACAAAGACCTCCGT-MGB |
Quantification was performed with a two-step
reaction process: reverse transcription (RT) and PCR. Each RT
reaction consisted of 1 μg RNA, 2 μl of PrimerScript buffer, 0.5 μl
of oligodT, 2 μl of random 6 mers and 0.5 μl of PrimerScript RT
Enzyme Mix I (Takara), in a total volume of 10 μl. Reactions were
performed in a GeneAmp PCR System 9700 (Applied Biosystems) for 15
min at 37°C, followed by heat inactivation of RT for 5 sec at 85°C.
Real-time PCR was performed using LightCycler® 480 II
Real-time PCR Instrument (Roche, Swiss) with 20 μl PCR reaction
mixture that included 10 μl of 2× TaqMan mix (Takara), 1 μl of 20×
probe and primers mixture (Invitrogen), 2 μl of cDNA, and 7 μl of
nuclease-free water. Reactions were incubated in a 384-well optical
plate (Roche) at 95°C for 10 min, followed by 40 cycles of 95°C for
10 sec, 60°C for 30 sec. Each sample was run in triplicate for
analysis. At the end of the PCR cycles, melting curve analysis was
performed to validate the specific generation of the expected PCR
product. The expression levels of mRNAs were normalized to 18s rRNA
and were calculated using the 2−ΔΔCt method.
Statistical analyses
The relation of SUFU variant mRNA RQ values (−ΔΔCt)
with clinical characteristics were assessed by Mann-Whitney U test.
Statistical analysis was processed by the SPSS 18.0 software
package. P-values <0.05 were considered statistically
significant.
Results
Identification of 3′- and 5′ cDNA
sequences of novel SUFU transcript variant (SUFUvN)
Following the strategy described in Fig. 1A, the 3′RACE amplification for 3′
cDNA of SUFU in pancreatic cancer cells was performed. The
products of first cycle (using P1 and 3′RACE outer primer) and
second cycle of nested PCR (using P2 and 3′RACE inner primer) are
shown in Fig. 2A. A cDNA fragment
of ~1,000 bp products from SW1990 cells was cloned into pMD18-T and
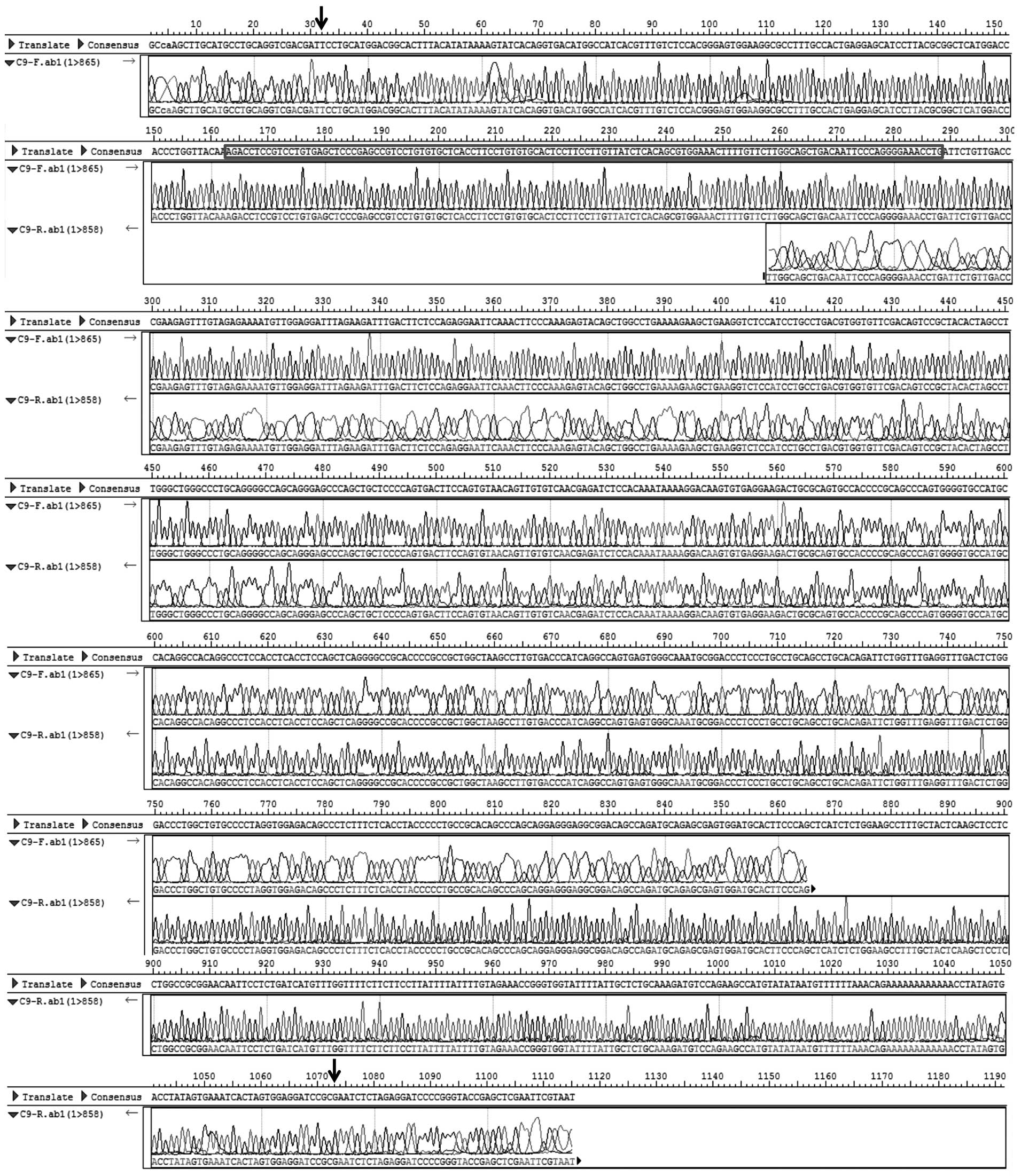
sequenced. As shown in Figs. 2B
and 3, a novel cDNA fragment
sequence was found at 3′ termination of SUFU gene compared
with cDNA of transcript variant 1 of SUFU (SUFUv1,
NM_016169) and transcript variant 2 of SUFU (SUFUv2,
NM_001178133) published in NCBI database. This fragment sequence
contains a new protein-coding exon (126 bp) the same as a fragment
of the intron sequence (102,625,811–102,625,936 of the human
chromosome 10, hg38 version) between exon 10 and 11 of SUFUv1. We
hypothesized that there exists a novel alternative splicing
transcript variant of SUFU (SUFUvN) which contains an
additional new exon compared with SUFUv1.
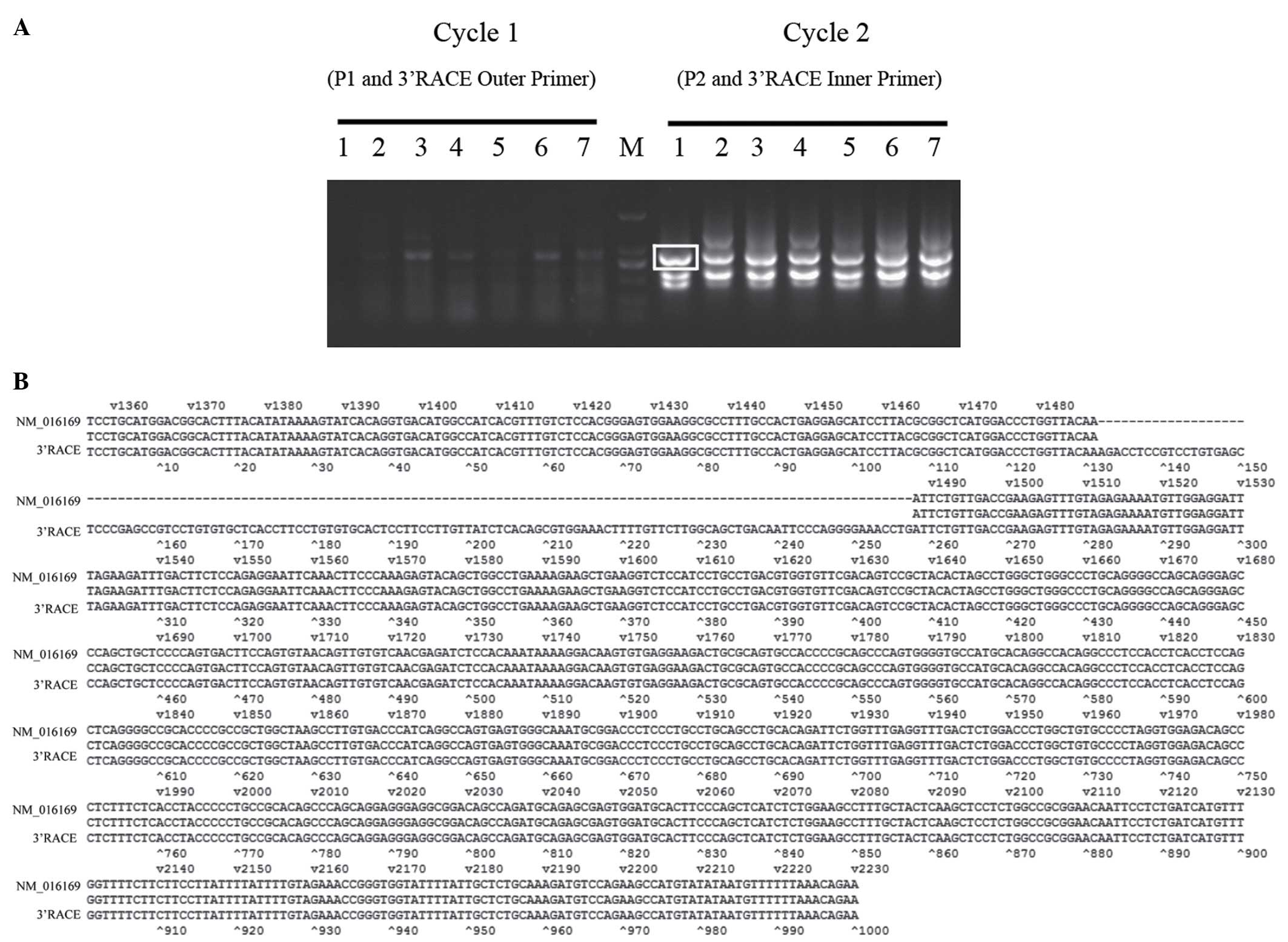 | Figure 23′RACE of SUFU. (A) The
products of the first cycle and second cycle of 3′RACE nested PCR
amplification for SUFU cDNA from pancreatic cancer cells.
Lane M, DNA marker (2,000, 1,000, 750, 500, 250 and 100 bp). Lanes
1–7, SW1990, PATU8988, AsPC-1, BxPC-3, CFPAC-1, Capan-2, PANC-1.
The indicated band ~1,000 bp in SW1990 was cloned and sequenced.
(B) Sequence alignment of NM_016169 and 3′RACE clone. The mismatch
sequence is the new exon. |
In order to identify the full length of SUFUvN cDNA
squence, the 5′ cDNA fragment of SUFUvN was amplified by RT-PCR
following the strategy described in Fig. 1B, in which a specific primer 4 (P4)
covering the new exon and exon 11 of SUFUv1 pairing with a specific
primer 3 (P3) siting in exon 1 of SUFUv1 were used. As shown in
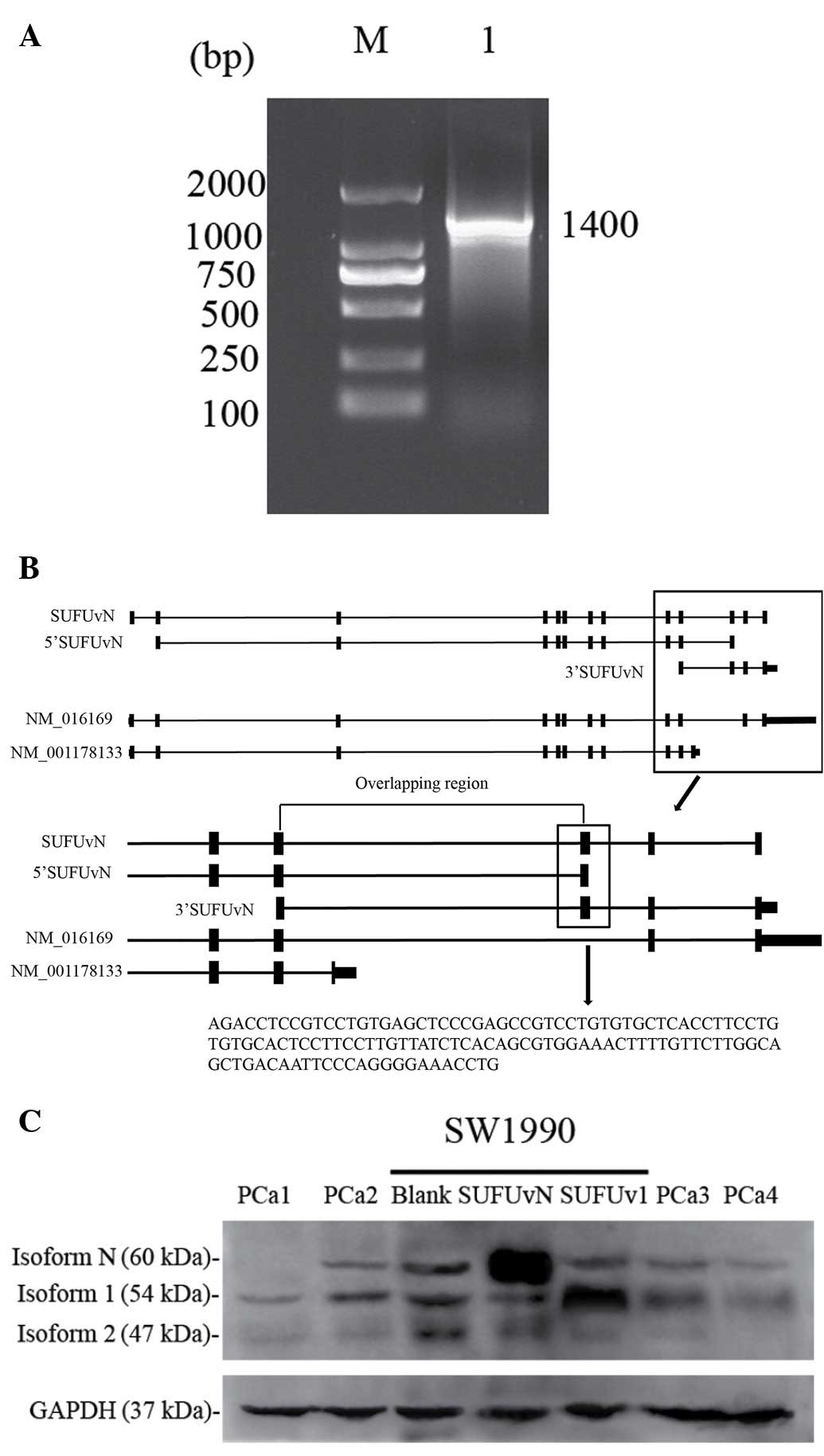
Fig. 4A, a cDNA fragment of 1,400
bp products from SW1990 cells was obtained by RT-PCR and its
sequence was the same as the exon 2-exon 10 of SUFUv1 cDNA. The
combination of exon 10 and new exon was also been detectable in
1,400 bp product (data not shown). Thus, the new exon is combined
with exon 1–10 in 5′-terminal and with exon 11–12 in 3′
terminal.
Clone of full length of SUFUvN
RT-PCR product of SUFU transcript was double enzyme
digested by BamHI and AgeI and was ligated with
expression vector pcDNA3.1mychisA (+). After infection to DH5α,
positive clones were selected and sequenced. From the sequencing
and alignment result (data not shown), we identified two kinds of
SUFU cDNA clones. They are SUFUv1 and full length of SUFUvN as
described above. Figure of BLAT in UCSC web site show us SUFUvN,
5′cDNA of SUFUvN, 3′cDNA of SUFUvN, and other two SUFU variants
(Fig. 4B).
Identification of protein expression
encoded by SUFUvN (isoform N)
There were three bands present in the western
blotting figure of PDAC tissue (Fig.
4C). Compared with lanes with SUFUv1 and SUFUvN expression
vector transfected SW1990 serum, two bands were identified to be
protein encoded by SUFUv1 (isoform 1) and isoform N in PDAC
tissues. Isoform 1 is ~54 kDa, while isoform N is ~60 kDa. The
additional band ~47 kDa may be protein encoded by SUFUv2 (isoform
2).
Detection and clinical relevance of
SUFUvN in PDAC tissue
We designed TaqMan MGB probes to identify different
transcript variants of SUFU. Each probe is described in the figure,
so each probe can uniquely detect the corresponding SUFU variant
(Fig. 1C). After detecting
transcription levels of SUFU variants in PDAC tissues of 40
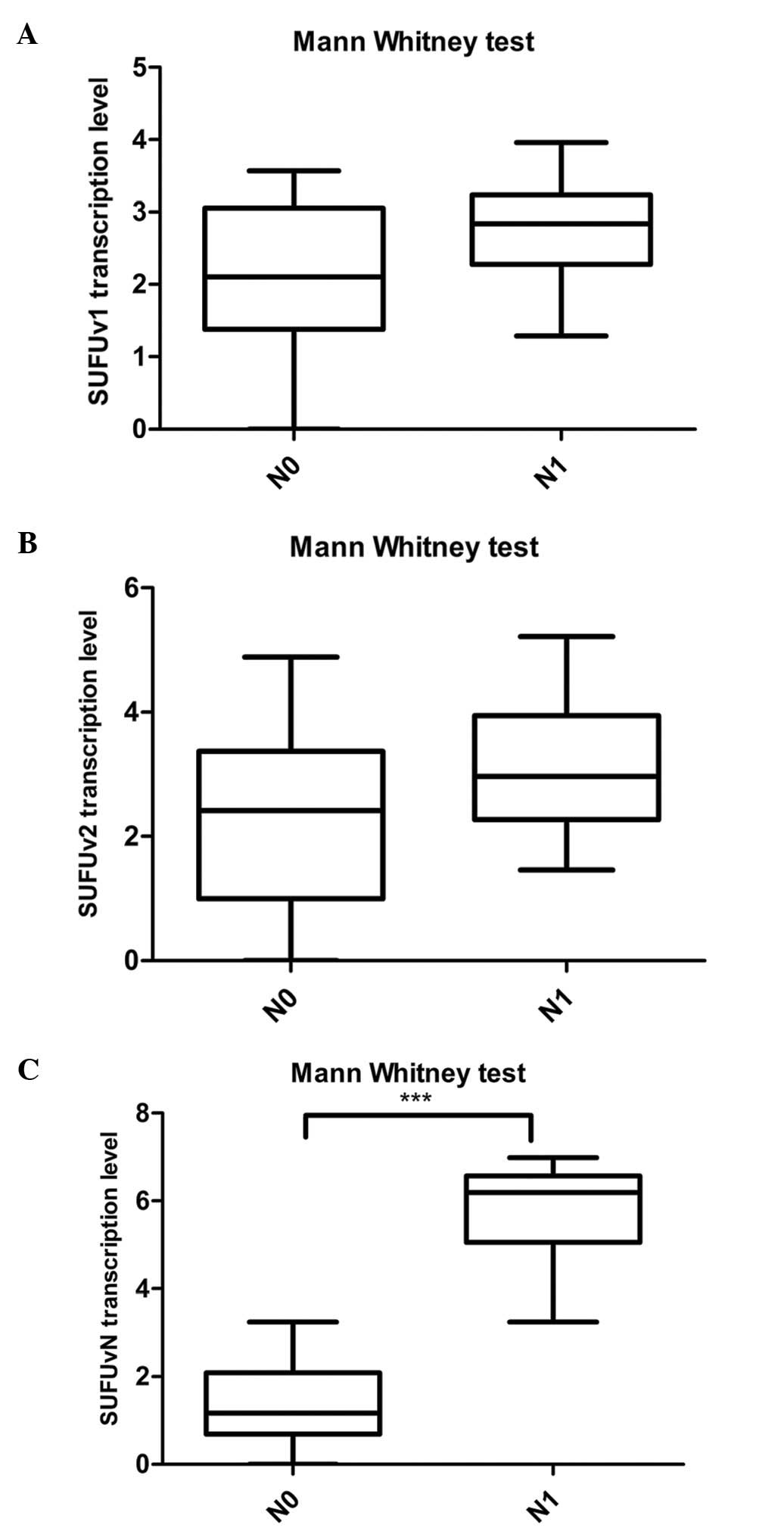
patients, we found that SUFUvN transcription levels are
significantly higher in PDAC tissues of N1 stage patients than in
N0 stage patients, while there is no relationship between other
variants transcription levels and lymph node metastasis (Fig. 5). As shown in Table III, we found that the elevated
transcription of SUFUvN was related with high N stage, while no
correlation was found with other clinical features, including T
stage, age of onset, gender, tumor size and tumor
differentiation.
 | Table IIIRelation of SUFU variant transcription
level in the pancreatic cancer tissues with clinical
characteristics of pancreatic cancer patients. |
Table III
Relation of SUFU variant transcription
level in the pancreatic cancer tissues with clinical
characteristics of pancreatic cancer patients.
| | SUFUvN | SUFUv1 | SUFUv2 |
|---|
| |
|
|
|
|---|
| Characteristics | N | −ΔΔCt | P-value | −ΔΔCt | P-value | −ΔΔCt | P-value |
|---|
| N stage |
| N0 | | 1.39±0.91 | <0.0001 | 2.13±1.04 | 0.08 | 2.38±1.49 | 0.05 |
| N1 | | 5.84±1.07 | | 2.80±0.68 | | 3.22±1.11 | |
| T stage |
| T0–2 | | 2.85±2.44 | 0.23 | 2.44±1.20 | 0.95 | 2.38±1.47 | 0.29 |
| T3–4 | | 3.98±2.43 | | 2.48±0.79 | | 3.00±1.29 | |
| Age (years) |
| <65 | | 3.56±2.56 | 0.86 | 2.40±1.05 | 0.64 | 2.97±1.37 | 0.16 |
| ≥65 | | 3.77±2.25 | | 2.66±0.37 | | 2.31±1.31 | |
| Gender |
| Male | | 3.68±2.40 | 0.83 | 2.47±0.99 | 1.00 | 2.76±1.39 | 0.84 |
| Female | | 3.47±2.67 | | 2.47±0.81 | | 2.89±1.38 | |
| Tumor size
(cm) |
| <3 | | 3.18±2.58 | 0.80 | 2.81±0.73 | 0.22 | 2.51±1.37 | 0.55 |
| ≥3 | | 3.76±2.45 | | 2.35±0.97 | | 2.90±1.37 | |
| Tumor
differentiation |
| Poor | | 3.30±2.55 | 0.98 | 2.11±0.99 | 0.33 | 2.67±1.51 | 0.76 |
| Well | | 3.67±2.48 | | 2.53±0.92 | | 2.82±1.36 | |
Discussion
Hedgehog (Hh) signaling pathway plays an important
role in pathogenesis and development of pancreatic cancer, so
investigating this pathway is very important. Mammalian Hh pathway
is composed of Hh ligand, target cell receptor patched (Ptch),
smoothened (Smo), and transcription factor family Gli. Gli family
contains Gli1, Gli2 and Gli3. Gli2 and Gli3 are the targets of
primary Hh signaling. When Hh ligand is absent, Gli2/3 undergo
proteolysis to generate a transcriptional repressor form. They are
processed into active form when Hh ligand is present. Gli1 cannot
undergo the same kind of regulated proteolysis and is used
exclusively as gene activator. Gli1 is not only a transcription
activator but also a transcription target gene of Hh signaling
pathway. Activation of Gli2 and/or Gli3 by Hh often leads to
expression of Gli1, which then becomes important in a second wave
of activation (11). The formation
of either Gli repressor or activator are dependent on primary cilia
structure. As a physically binding protein of Gli transcription
factor, Sufu is recognized as a negative regulator of hedgehog
pathway. However, Chen et al (10) provided convincing data indicating
that mouse Sufu regulates Gli2/3 stability by antagonizing the Spop
protein of the BTB domain containing protein family. Spop binds to
Gli2 and Gli3 and promotes their ubiquitination and degradation in
a proteasome-dependent manner. The overexpression of Sufu appears
to block Spop-dependent Gli protein reduction. They speculated that
Sufu sequesters Gli2/3 protein in the cytoplasm and protects them
from Spop-mediated protein degradation, providing a Gli protein
pool for the production of Gli2/3 activators and repressors. Sufu
is necessary for maximal activation of Hh pathway. Thus Sufu is a
positive regulator of Hh signaling pathway in this aspect.
Clarification is required to establish whether Sufu is a negative
or positive regulator of the Hh pathway. We found a novel
alternative splicing transcript of SUFU named SUFUvN by
3′RACE of SUFU gene and cDNA clone in human pancreatic
cancer SW1990 cells. In SUFUvN, a new exon is inserted, which is
from intron between 10th and 11th exon of SUFUv1 (NM_016169). The
inserted exon makes the carboxyl terminal of protein encoded from
SUFUvN (isoform N) different from protein encoded from SUFUv1
(isoform 1) and may alter the function of the expressed protein. A
further statistical analysis of clinical data revealed that
transcription of SUFUvN is related to metastasis of lymph nodes. A
study made by another group gives us some tips on SUFU and GLI
interaction (12). The study found
the ability of GLI binding of different SUFU isoforms will differ
because of their different carboxyl terminal structure. To our
knowledge, direct binding of Sufu to all three Gli transcription
factors has been well documented (13–17).
Sufu binds Gli: the amino and carboxyl terminal of Sufu bind the
carboxyl and amino terminal of Gli, respectively. Thus we
hypothesize that SUFU functions mentioned in our experiment are
loaded by different variants of it according to their affinity to
GLI. SUFUvN is possibly a positive regulator of Hh pathway because
it has more affinitive to GLI2 and GLI3, while SUFUv1 is more
likely to be a negative regulator of Hh pathway because of its
affinity to GLI1. So, in pancreatic cancer tissue, endogenous
SUFUvN binds with GLI2/3 to antagonize their degradation. High
level of endogenous SUFUvN leads to more activation of Hh pathway,
and accelerates tumor metastasis.
Research reports describing gene alternative
splicing variants and different function of their proteins are
limited. It is partially due to the complicated and limited methods
in studying splicing variant. In our study, we designed TaqMan MGB
probe and primers according to the unique exon of different
variants. This can distinguish transcription of different variants.
Although the corresponding protein isoforms can be recognized by
western blotting because of the different molecular weights, the
subcellular situation of different protein isoforms can not be
distinguished by immunohistochemistry stain in tissue section
unless unique antibodies are produced. Recognition of subcellular
situation of different isoforms is very helpful for studing their
functions. Additionally, if in situ hybridization of
different variant mRNA is required, new technology and method
should be applied, for example the LNA probe (18,19).
In addition, the crystal and small-angle X-ray scattering
structures of full-length human SUFU and its complex with the key
SYGHL motif conserved in all GLIs have been reported by Cherry
et al (20). It is
demonstrated that GLI binding is associated with major
conformational changes in SUFU. If alternative splicing
transcription variants of SUFU including SUFUvN were to be
added to their study in the future, we could improve the
understanding of the affinity between SUFU variants and GLIs.
Acknowledgements
This study was supported by grants from National
Natural Science Foundation of China (nos. 81272663 and 81472279 to
Jun Gao, no. 30910103911 to Zhaoshen Li), Shanghai Science and
Technology Committee Major Project on Basic Research (no.
11441901800 to Jun Gao), and Shanghai Municipal Bureau of Health
Research Topic (no. 2012-2015 to Shude Li). The authors also would
like to thank Oebiotech Company for gene expression detection
systems.
References
|
1
|
Hidalgo M: Pancreatic cancer. N Engl J
Med. 362:1605–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berman DM, Karhadkar SS, Maitra A, Montes
De Oca R, Gerstenblith MR, Briggs K, Parker AR, Shimada Y, Eshleman
JR, Watkins DN, et al: Widespread requirement for Hedgehog ligand
stimulation in growth of digestive tract tumours. Nature.
425:846–851. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thayer SP, di Magliano MP, Heiser PW,
Nielsen CM, Roberts DJ, Lauwers GY, Qi YP, Gysin S, Fernández-del
Castillo C, Yajnik V, et al: Hedgehog is an early and late mediator
of pancreatic cancer tumorigenesis. Nature. 425:851–856. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hidalgo M and Maitra A: The hedgehog
pathway and pancreatic cancer. N Engl J Med. 361:2094–2096. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ingham PW: The patched gene in development
and cancer. Curr Opin Genet Dev. 8:88–94. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Reifenberger J, Wolter M, Knobbe CB,
Köhler B, Schönicke A, Scharwächter C, Kumar K, Blaschke B, Ruzicka
T and Reifenberger G: Somatic mutations in the PTCH, SMOH, SUFUH
and TP53 genes in sporadic basal cell carcinomas. Br J Dermatol.
152:43–51. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Taylor MD, Liu L, Raffel C, Hui CC,
Mainprize TG, Zhang X, Agatep R, Chiappa S, Gao L, Lowrance A, et
al: Mutations in SUFU predispose to medulloblastoma. Nat Genet.
31:306–310. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie J, Murone M, Luoh SM, Ryan A, Gu Q,
Zhang C, Bonifas JM, Lam CW, Hynes M, Goddard A, et al: Activating
smoothened mutations in sporadic basal-cell carcinoma. Nature.
391:90–92. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kasai K, Inaguma S, Yoneyama A, Yoshikawa
K and Ikeda H: SCL/TAL1 interrupting locus derepresses GLI1 from
the negative control of suppressor-of-fused in pancreatic cancer
cell. Cancer Res. 68:7723–7729. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen MH, Wilson CW, Li YJ, Law KK, Lu CS,
Gacayan R, Zhang X, Hui CC and Chuang PT: Cilium-independent
regulation of Gli protein function by Sufu in Hedgehog signaling is
evolutionarily conserved. Genes Dev. 23:1910–1928. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Østerlund T and Kogerman P: Hedgehog
signalling: How to get from Smo to Ci and Gli. Trends Cell Biol.
16:176–180. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tostar U, Finta C, Rahman MF, Shimokawa T
and Zaphiropoulos PG: Novel mechanism of action on Hedgehog
signaling by a suppressor of fused carboxy terminal variant. PLoS
One. 7:e377612012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kogerman P, Grimm T, Kogerman L, Krause D,
Undén AB, Sandstedt B, Toftgård R and Zaphiropoulos PG: Mammalian
suppressor-of-fused modulates nuclear-cytoplasmic shuttling of
Gli-1. Nat Cell Biol. 1:312–319. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pearse RV II, Collier LS, Scott MP and
Tabin CJ: Vertebrate homologs of Drosophila suppressor of fused
interact with the gli family of transcriptional regulators. Dev
Biol. 212:323–336. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Stone DM, Murone M, Luoh S, Ye W, Armanini
MP, Gurney A, Phillips H, Brush J, Goddard A, de Sauvage FJ, et al:
Characterization of the human suppressor of fused, a negative
regulator of the zinc-finger transcription factor Gli. J Cell Sci.
112:4437–4448. 1999.PubMed/NCBI
|
|
16
|
Dunaeva M, Michelson P, Kogerman P and
Toftgard R: Characterization of the physical interaction of Gli
proteins with SUFU proteins. J Biol Chem. 278:5116–5122. 2003.
View Article : Google Scholar
|
|
17
|
Merchant M, Vajdos FF, Ultsch M, Maun HR,
Wendt U, Cannon J, Desmarais W, Lazarus RA, de Vos AM and de
Sauvage FJ: Suppressor of fused regulates Gli activity through a
dual binding mechanism. Mol Cell Biol. 24:8627–8641. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nuovo GJ, Elton TS, Nana-Sinkam P, Volinia
S, Croce CM and Schmittgen TD: A methodology for the combined in
situ analyses of the precursor and mature forms of microRNAs and
correlation with their putative targets. Nat Protoc. 4:107–115.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Urbanek MO, Nawrocka AU and Krzyzosiak WJ:
Small RNA detection by in situ hybridization methods. Int J Mol
Sci. 16:13259–13286. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cherry AL, Finta C, Karlström M, Jin Q,
Schwend T, Astorga-Wells J, Zubarev RA, Del Campo M, Criswell AR,
de Sanctis D, et al: Structural basis of SUFU-GLI interaction in
human Hedgehog signalling regulation. Acta Crystallogr D Biol
Crystallogr. 69:2563–2579. 2013. View Article : Google Scholar : PubMed/NCBI
|