Introduction
Hepatocellular carcinoma (HCC) is the seventh most
common cancer in the world and the third leading cause of
cancer-related deaths (1).
Hepatitis B virus (HBV) and hepatitis C virus (HCV) are the main
causes of HCC, with the latter being the major cause of HCC in
Western countries and in Japan (1,2).
Surgical resection is one of the first treatments for HCC, though
the long-term survival rate after hepatic resection is poor due to
frequent recurrence (1). HCC
recurrence arises from multicentric HCC or intrahepatic metastasis
(3–5), but Wang et al indicated that
compared with intrahepatic metastatic HCC, multicentric recurrent
HCC is associated with a better prognosis (5). Although some cases of early-stage HCC
do not recur for a long period after therapeutic resection,
intrahepatic metastasis can in some cases occur early, within 12
months after resection. Thus, a comparative study of these two
types of HCC to identify factors that influence early intrahepatic
metastasis would be helpful for predicting HCC recurrence.
Several studies have demonstrated that
clinicopathological factors, such as tumor size, stage, grade and
viral load, are significantly related to intrahepatic recurrence
(6–8). Molecular prediction of intrahepatic
recurrence has been investigated by gene expression profiling
analysis in human HCC tissues, whereby transcriptomics between
cases of early and late recurrence was performed; some of these
molecules identified, however, were not validated by protein
expression in HCC tissues (9–12).
In addition, there have been a few proteomic studies focused on the
recurrence-related characteristics of HCC (13–15).
However, consensus results were not obtained because the
backgrounds of the subjects studied were complex, with the
complexity varying by study. For example, both HBV-positive and
HCV-positive HCCs were included, and various stages of HCC were
included in comparisons between early and late recurrence.
Therefore, multicentric HCC and intrahepatic metastatic HCC, which
are generated by distinct mechanisms and should be analyzed
separately, were not differentiated in previous studies.
In this study, we focused on cases of early-stage
HCV-positive HCC and compared the protein profiles of HCC samples
obtained at the first therapeutic resection between two groups:
early and late recurrence after the first resection. Early
recurrence in our study was considered to be intrahepatic
metastasis, for which characterization by HCC protein profiles may
have been possible. We then performed two-dimensional (2D) gel
electrophoretic proteomic analysis between the two groups of HCC to
clarify the molecular characteristics of early metastatic HCC.
Materials and methods
Liver samples for proteomics
Cancerous liver tissues were obtained by surgical
resection of 41 cases of hepatocellular carcinoma positive for HCV.
All cases had solitary tumors less than 6 cm in diameter and no
metastasis to lymph nodes or other organs (some vascular
invasion-positive samples were included). According to the period
from the first resection to recurrence, 11 cases were designated as
early recurrence (within 12 months), and 14 cases were designated
as late recurrence (no recurrence within 48 months). We performed
two-dimensional differential gel electrophoresis (2D-DIGE) analysis
using 11 and 10 cases from the early and late recurrence groups,
respectively. Our study protocol was approved by the Ethics
Committee of our School, and informed consent was obtained from the
patients. The HCC samples were stored at −80°C until use.
Protein extraction from HCC samples and
2D-DIGE
Protein was directly extracted from the cancerous
liver tissues by homogenization in urea-buffer (7 M urea, 2 M
thiourea, 4% w/v 3-(3-cholamidopropyl)
dimethylammonio-1-propanesulfonate, 1 mM phenylmethanesulfonyl
fluoride, and 30 mM Tris-HCl, pH 9.0) using a microtube
homogenizer, followed by incubation at room temperature for 30 min.
After sonication and centrifugation, the pH of the supernatant was
adjusted to 8.0–9.0 with urea-buffer, pH 9.5. The protein
concentration was determined using the Bradford method (Bio-Rad
Laboratories, Inc., Hercules, CA, USA) (16). Bovine serum albumin was used as a
standard. The extracted protein samples were stored at −80°C.
A 50-μg sample of protein was labeled with
300 pmol Cy5 (Minimal Labelling Dye, GE Healthcare, Little
Chalfont, UK) and an internal standard (a pool of an equal amount
of all samples) with 300 pmol Cy3 (Minimal Labelling Dye, GE
Healthcare) according to the manufacturer's instructions. The
labeled samples were stored at −80°C. 2D gel electrophoresis was
carried out as previously described (17). Briefly, a mixture of 50 μg
each of the Cy3-labeled internal standard and Cy5-labeled sample
was loaded onto a ReadyStrip IPG (pH 4–7, 18 cm length, Bio-Rad
Laboratories, Inc.) for isoelectric focusing with a CoolPhoreStar
IPG-IEF Type-P (Anatech, Tokyo, Japan).
For separation in the second dimension, sodium
dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was
carried out using a 12.5% polyacrylamide gel (190 × 173 × 1 mm)
with a CoolPhoreStar SDS-PAGE Dual-200 (Anatech). The FLA-5100
Fluorescent Image Analyzer (Fujifilm, Tokyo, Japan) was used for
acquisition of the gel images. Spot patterns and spot abundance
were analyzed by Progenesis SameSpots (Nonlinear Dynamics,
Newcastle upon Tyne, UK).
To determine the abundance of each spot, the
fluorescence intensity was calculated as a percent value per gel.
To normalize inter-gel differences, the Cy5 percent value was
normalized to Cy3 in each spot, and the normalized value was used
for the statistical analysis. Significantly different spots between
the early and late recurrence groups were selected using one-way
analysis of variance (P<0.05). Cluster analysis was performed
with GeneSpring ver. 13.0 (Agilent Technologies, Santa Clara, CA,
USA) using values normalized to the median of all samples.
Protein identification
To obtain gels for protein identification, 300–440
μg non-labeled mixed protein was loaded onto an IPG strip
and stained with Deep Purple Total Protein Stain (GE Healthcare)
according to the manufacturer's instructions. Differentially
expressed protein spots in sufficient abundance were automatically
excised, washed and destained using Xcise (Shimadzu Biotech, Kyoto,
Japan). The spots were then digested with trypsin, and the peptides
were desalinized using pipette-tip columns loaded with C18 resin
(ZipTip m-C18, Merck Millipore, Darmstadt, Germany). The peptides
were eluted onto a μFocus MALDI plate (Hudson Surface
Technology, Inc., Old Tappan, NJ, USA) for AXIMA-QIT (Shimadzu
Biotech) analysis. α-Cyano-4-hydroxycinnamic acid (CHCA) was used
as the MALDI-matrix. MALDI-TOF mass spectrometry was performed in
the positive reflectron mode at 20–23 kV voltage. Approximately
1000 laser shots of the MS spectra were summed. To validate the
protein identification, selected intense peptides were subjected to
MS/MS fragmentation.
LC-MS/MS or LC-MALDI analysis was performed for
low-abundance protein spots. For LC-MS/MS analysis, peptide samples
desalinized with a ZipTip m-C18 were collected in 1.5-ml tubes and
dried. The samples were then solubilized in 2% v/v
acetonitrile/0.1% v/v formic acid. The peptide samples were loaded
onto a 5-cm, 100-μm inner diameter reversed-phase C18 column
(HiQ sil C18W-3, KYA Technologies Corp., Tokyo, Japan) using the
DiNA nanoLC system (KYA Technologies Corp.) and separated on-line
with a QSTAR XL mass spectrometer (AB SCIEX Ltd., Framingham, MA,
USA) using a 40-min linear gradient from 2% v/v acetonitrile/0.1%
v/v formic acid to 41% v/v acetonitrile/0.1% v/v formic acid.
For LC-MALDI analysis, the trypsinized peptide
samples were separated using a Prominence nano (Shimadzu Biotech).
A 15-cm column with a 100-μm inner diameter,
MonoCap® for Fast-flow column (GL Sciences Inc., Tokyo,
Japan), was used for separation with a 30-min gradient from 5% v/v
acetonitrile/0.1% v/v trifluoroacetic acid to 43.25% v/v
acetonitrile/0.1% v/v trifluoroacetic acid. The separated peptide
samples were automatically spotted onto a μFocus MALDI plate
using AccuSpot (Shimadzu Biotech). One microliter of a CHCA matrix
solution (0.5 μg/μl) was applied and crystallized,
and then MALDI-TOF-MS was automatically performed (AXIMA
Performance, Shimadzu Biotech). All acquired peptide MS or MS/MS
data were analyzed with the database search engine MASCOT (Matrix
Science Inc., Boston, MA, USA). Protein and peptide identification
was based on the MASCOT definition.
Immunohistochemistry (IHC)
Thirty-three cases were subjected to IHC: 13 and 20
cases of early and late recurrence, respectively, of which 2D-DIGE
analysis was performed for 10 and 6 cases, respectively. IHC was
performed using formalin-fixed paraffin-embedded tissues, as
previously described (18), with
some modifications. For antigen retrieval, the sections were
treated with 10 mM citrate buffer (pH 6.0) at 120°C for 15 min for
transglutaminase 2 (TGM2) or with 1X Target Retrieval Solution High
pH (pH 9.0) (Dako, Glostrup, Denmark) at 120°C for 15 min for
E-cadherin (CDH1). The first antibody reaction was performed with a
1/100 dilution of anti-human TGM2 (rabbit monoclonal, ab109200,
Abcam, Cambridge, UK) or a 1/50 dilution of anti-human CDH1 (mouse
monoclonal, M3612, Dako) at 37°C for 60 min; the second antibody
was reacted using Histofine Simple Stain MAX PO (MULTI; Nichirei,
Tokyo, Japan) for 30 min at room temperature. The TGM2-positivity
was determined using all fields of view (FOVs, 2.2-mm in diameter
per FOV) containing HCC (15–107 FOVs per case) as follows:
TGM2-positive HCC cells were observed in at least two different
FOVs per HCC. According to the staining intensity,
semi-quantitative scores were defined as follows: 0, negative; 1,
weakly positive; 2, moderately positive; 3, strongly positive.
Quantification of mRNA
In addition to the cases used for 2D-DIGE, another
15 cases were subjected to mRNA quantification: 9 and 6 cases from
the early and late recurrence groups, respectively. Total RNA was
isolated from frozen liver tissues and subjected to DNase I
treatment, followed by cDNA synthesis, as previously described
(19). Quantitative real-time
polymerase chain reaction (qPCR) was performed as previously
described (20) with minor
modifications; for an internal standard gene, 18S rRNA was
quantified with TaqMan gene expression assays using 1 ng cDNA. The
expression levels of the target gene mRNAs were determined by qPCR
using THUNDERBIRD® SYBR qPCR Mix (TOYOBO Co., Ltd.,
Osaka, Japan) following the manufacturer's protocol. SNAI1 and
TGFB1 were quantified using 50 ng cDNA, and other target genes were
quantified using 10 ng cDNA. The primer sequences and TaqMan assay
IDs used in this analysis are shown in Table I. The quantity of mRNA was
normalized against the quantity of 18S rRNA by the relative
quantitation (ΔCt) method, as previously described (20).
 | Table IPrimer sequences of the 7 genes
examined in this study. |
Table I
Primer sequences of the 7 genes
examined in this study.
| Accession no. | Symbol | Primer
sequence/TaqMan assay ID | Product size
(bp) |
|---|
| NM_004613.2 | TGM2 | F:
TCCCCACTCACCCCCTTTGC
R: TTCCTCTCTCACCCCAGCCC | 93 |
| NM_000660.5 | TGFB1 | F:
CATGGGGGCTGTATTTAAGG
R: GGGCAAAGGAATAGTGCAGA | 190 |
| NM_001530.3 | HIF1A | F:
ACCACTACCACTGCCACCAC
R: CCTTTTCCTGCTCTGTTTGG | 182 |
| NM_005985.3 | SNAI1 | F:
GGCCTAGCGAGTGGTTCTTC
R: TAGGGCTGCTGGAAGGTAAA | 169 |
| NM_003068.4 | SNAI2 | F:
ATGAGGAATCTGGCTGCTGT
R: GGCTTCGGAGTGAAGAAATG | 84 |
| NM_004360.3 | CDH1 | F:
TGGCTTCCCTCTTTCATCTC
R: TAGTTCCGCTCTGTCTTTGG | 171 |
| NR_003286.2 | 18S | Hs99999901_s1 | |
Western blot analysis
Western blotting was carried out as previously
described (17). Briefly, 20
μg of protein extracted in urea-buffer for 2D-DIGE was
separated on SDS-10% polyacrylamide gels and transferred to a
polyvinylidene difluoride membranes. The membranes were incubated
with antibodies and then with horseradish peroxidase-conjugated
secondary antibodies. The target protein was detected by ImmunoStar
Zeta or LD (Wako Pure Chemical Industries, Ltd., Osaka, Japan). The
chemiluminescence was acquired and quantified. The first antibodies
were as follows: 1/10,000 dilution of anti-human TGM2 (rabbit
monoclonal, ab109200, Abcam), 1/1,000 dilution of anti-human PTEN
(rabbit monoclonal, #9188, Cell Signaling Technology, Inc.,
Danvers, MA, USA), 1/1,000 dilution of anti-human phospho-Akt
(Ser473) (rabbit monoclonal, #4060, Cell Signaling Technology),
1/3,000 dilution of anti-human pan-Akt (rabbit monoclonal, #4691,
Cell Signaling Technology), 1/1,000 dilution of anti-human
phospho-FAK (Tyr397) (rabbit monoclonal, #8556, Cell Signaling
Technology), 1/2,000 dilution of anti-human FAK (rabbit monoclonal,
ab40794, Abcam) and 1/10,000 dilution of anti-human GAPDH (rabbit
polyclonal, ab37168, Abcam). The chemiluminescence of TGM2 and PTEN
was normalized against that of loading control GAPDH. The
phosphorylation rate of FAK and Akt was calculated as a fraction of
total FAK and total Akt, respectively.
Statistical analyses
Sex, cirrhosis and tumor grade were analyzed by the
χ2-test or Fisher's exact test, and other
clinicopathological features were analyzed by the Mann-Whitney
U-test. Comparisons of the mRNA and protein levels were analyzed by
Student's t-test, and the correlation of the mRNA and protein
levels between TGM2 and other genes was analyzed by Spearman's rank
correlation coefficient. The immunohistochemical scores for TGM2
were assessed by the Mann-Whitney U test and χ2-test. A
difference was considered significant if the P-value was <0.05.
All statistical analyses were performed using SPSS Statistics 19
(IBM, Armonk, NY, USA).
Results
Comparison of clinicopathological
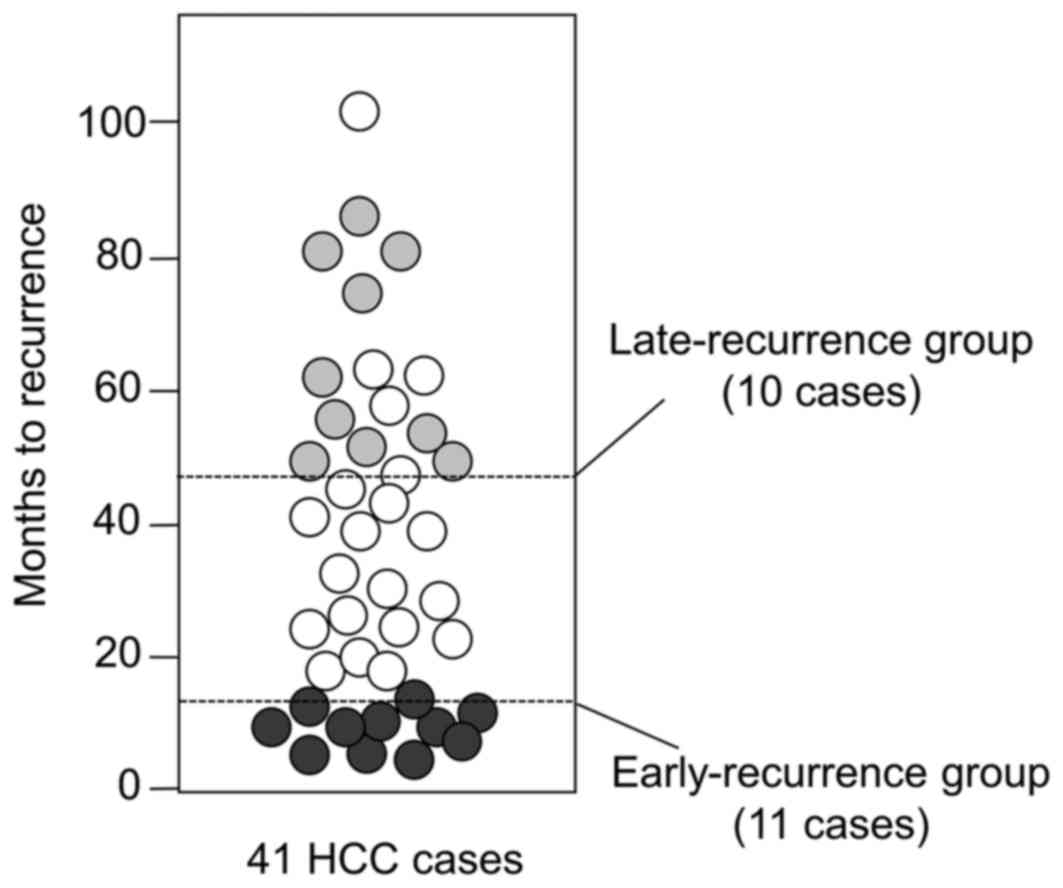
features of HCC cases between early and late recurrence groups
To characterize HCC prone to intrahepatic
metastasis, we isolated 41 rare cases at an early stage of primary
and solitary HCC; the average diameter was 32 mm. All cases were
positive for HCV because the HCC proteomics clearly differentiated
between HBV-positive and HCV-positive tumors (21). Among the 41 cases, 11 recurred
early, within 12 months, despite therapeutic resection; these cases
are considered to potentially have characteristics that predispose
to intrahepatic metastasis compared with the 10 cases that showed
no recurrence within 48 months after therapeutic resection
(Fig. 1). Comparison of the
clinicopathological features between the two groups (Table II) showed no significant
difference, except for AST (P=0.006).
 | Table IIClinicopathological features of the
two groups of early and late recurrence used in this study. |
Table II
Clinicopathological features of the
two groups of early and late recurrence used in this study.
| Features | Proteomics
| IHC
| mRNA quantification
|
|---|
Early
(n=11) | Late
(n=10) | P-value | Early
(n=13) | Late
(n=20) | P-value | Early
(n=9) | Late
(n=6) | P-value |
|---|
| Male/female | 7/4 | 6/4 | 1.000 | 10/3 | 15/5 | 1.000 | 8/1 | 5/1 | 1.000 |
| Age (range) | 68.6 (61–75) | 66.3 (59–72) | 0.242 | 68.1 (54–75) | 63.8 (49–71) | 0.031a | 63.8 (54–78) | 63.8 (51–68) | 0.800 |
| ICG-R15 (%)
(<10) | 21.1±2.5 | 16.2±1.5 | 0.314 | 17.9±2.3 | 14.5±1.1 | 0.518 | 19.9±5.5 | 11.8±2.2 | 0.491 |
| Alb (g/dl)
(6.7–8.3) | 3.60±0.12 | 3.69±0.10 | 0.640 | 3.74±0.13 | 3.92±0.12 | 0.356 | 3.89±0.17 | 4.37±0.21 | 0.061 |
| AST (IU/l)
(8–38) | 68.0±6.0 | 44.8±4.0 | 0.006a | 60.8±5.4 | 48.5±3.6 | 0.092 | 61.9±8.7 | 43.5±4.7 | 0.145 |
| ALT (IU/l)
(40–44) | 59.2±6.7 | 49.4±7.2 | 0.258 | 54.6±5.92 | 50.0±4.5 | 0.709 | 72.4±15.7 | 46.2±6.8 | 0.137 |
| T. bil (mg/dl)
(0.3-1.2) | 0.73±0.08 | 0.85±0.08 | 0.394 | 0.68±0.06 | 0.77±0.06 | 0.471 | 0.77±0.07 | 0.75±0.10 | 0.798 |
| AFP (ng/ml)
(<10) | 302±177 | 1497±1460 | 0.436 | 355±172 | 790±729 | 0.579 | 1495±1025 | 61±32 | 0.088 |
| Cirrhosis
(−/+) | 4/7 | 8/2 | 0.080 | 3/10 | 11/9 | 0.087 | 2/7 | 4/2 | 0.136 |
| Tumor size
(mm) | 30.8±2.7 | 37.1±4.2 | 0.229 | 33.0±2.8 | 29.1±2.8 | 0.223 | 33.7±3.3 | 22.8±3.9 | 0.028a |
| Tumor grade
(W/M/P) | 3/7/1 | 2/8/0 | 1.000 | 4/8/1 | 5/14/1 | 0.854 | 3/6/0 | 0/3/3 | 0.061 |
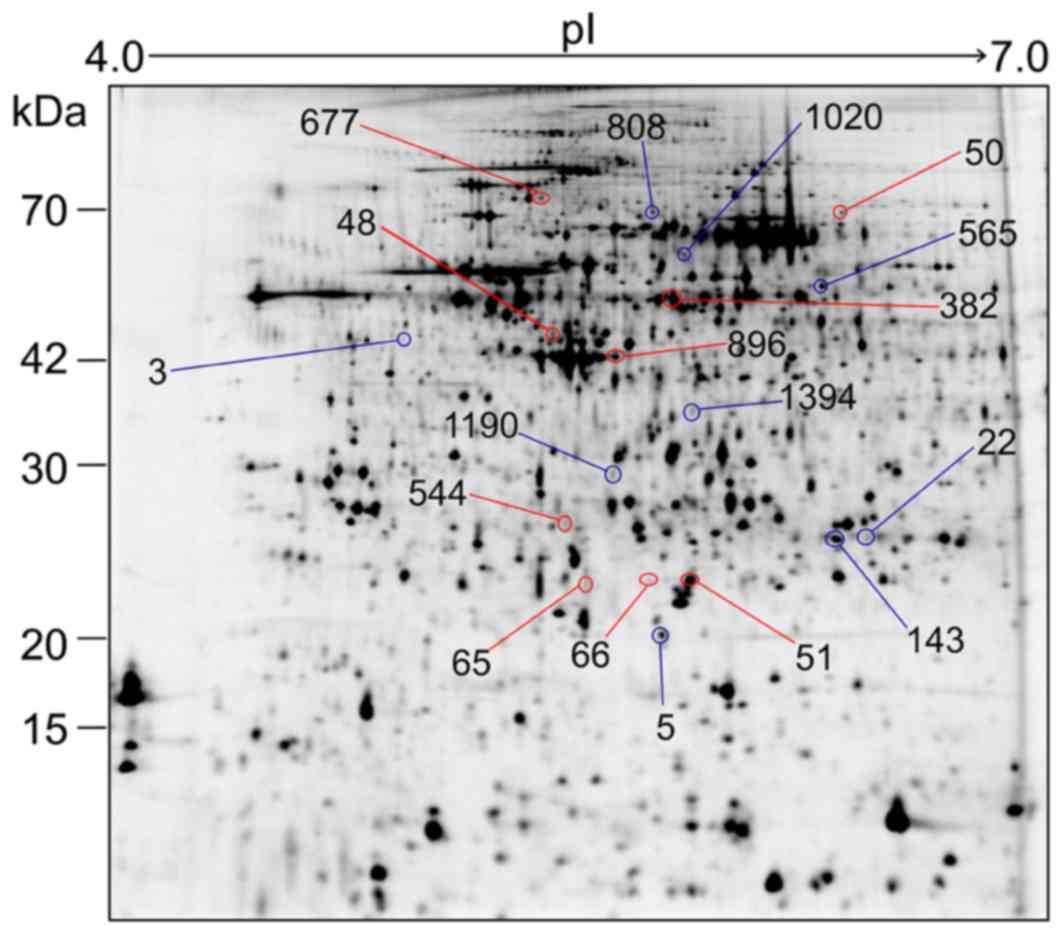
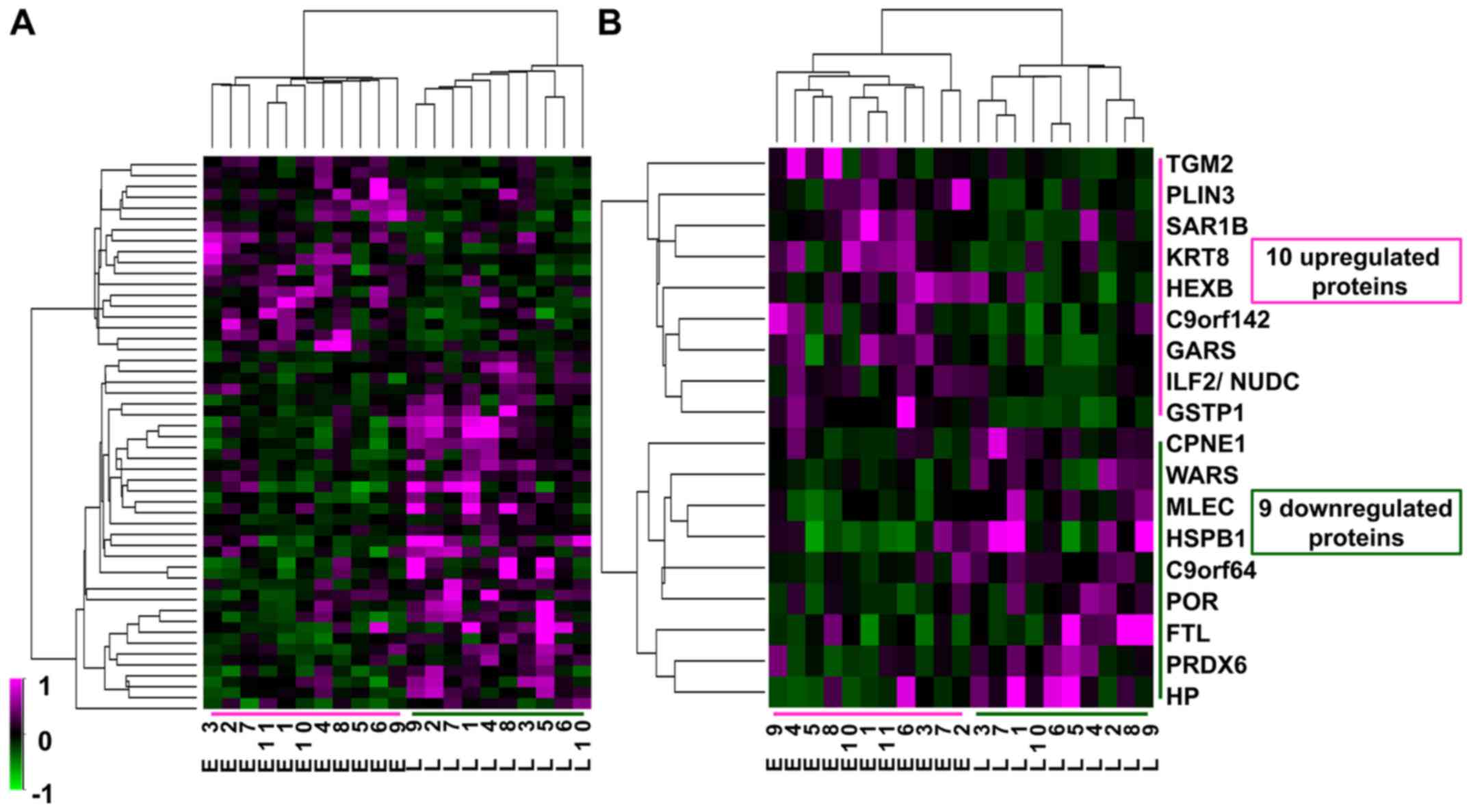
Differential proteins of HCC between
early and late recurrence groups
Twenty-one 2D-DIGE gel images were compared between
the early and late recurrence groups, and 1623 spots were detected
by Progenesis SameSpots (Fig. 2).
Among these 1623 spots, the normalized volumes of 51 were
significantly different between the 2 groups; 18 spots were
upregulated and 33 spots downregulated in the early group. Of
these, we identified 10 upregulated and 9 downregulated proteins
that were expressed in a sufficient quantity for identification
(Table III). Although the fold
change in differential expression for these proteins was not very
high, at less than 3-fold, the expression profiles of the 51
differential protein spots and of the 18 identified spots
classified the 21 HCC samples exactly into the two groups of
recurrence (Fig. 3). Therefore,
these proteins might be associated with intrahepatic recurrence of
HCV-positive HCC.
 | Table IIIUpregulated and downregulated
proteins in the early recurrence group. |
Table III
Upregulated and downregulated
proteins in the early recurrence group.
A, Upregulated
proteins
|
|---|
| Score
|
|---|
| MALDI-MS | MALDI-MS/MS
| LC-MS/MS | LC-MALDI |
|---|
| Spot no. | P-value change | Fold | Protein ID | Protein | Gene | Function | | Peak (m/z) | Score | | |
|---|
| 677 | 0.047 | 1.6 | P21980 | Protein-glutamine
γ-glutamyltransferase 2 | TGM2 | Protein
cross-linking | 105 | | | | |
| 51 | 0.001 | 1.4 | P09211 | Glutathione
S-transferase P | GSTP1 | The electrophiles
detoxification | 74 | 1337.7
1550.9
1883.9 | 41
38
61 | | |
| 382 | 0.022 | 1.4 | P05787 | Keratin, type II
cytoskeletal 8 | KRT8 | Intermediate
filament | 198 | | | | |
| 544 | 0.026 | 1.4 | P07686 | β-hexosaminidase
subunit β | HEXB | Ganglioside GM2
degradation | | | | 165 | |
| 48 | 0.028 | 1.4 | O60664 | Perilipin-3 | PLIN3 | Protein
transport | | | | 285 | |
| 66 | 0.030 | 1.4 | Q9Y6B6 | GTP-binding protein
SAR1b | SAR1B | Protein
transport | | | | 90 | |
| 50 | 0.031 | 1.4 | P41250 | Glycine-tRNA
ligase | GARS | tRNA
aminoacylation | | | | | 311 |
| 65 | 0.039 | 1.4 | Q9BUH6 | Protein PAXX | C9orf142 | DNA double-strand
break repair | | | | 124 | |
| 896 (mixed) | 0.044 | 1.2 | Q12905 | Interleukin
enhancer-binding factor 2 | ILF2 | Interleukin 2 gene
transcription regulator | | | | 95 | |
| | | Q9Y266 | Nuclear migration
protein nudC | NUDC | Mitotic spindles
formation | | | | 224 | |
B, Downregulated
proteins
|
|---|
| Score
|
|---|
| MALDI-MS | MALDI-MS/MS
| LC-MS/MS | LC-MALDI |
|---|
| Spot no. | P-value change | Fold | Protein ID | Protein | Gene | Function | | Peak (m/z) | Score | | |
|---|
| 3 | 0.027 | 2.7 | P00738 | Haptoglobin | HP | Iron
homeostasis | | | | 134 | |
| 5 | 0.028 | 1.9 | P02792 | Ferritin light
chain | FTL | Iron
homeostasis | 140 | 1607.8 | 81 | | |
| 143 | 0.040 | 1.6 | P04792 | Heat shock protein
β-1 | HSPB1 | Anti-oxidative
stress | | 1906.0 | 36 | | 218 |
| 1190 | 0.012 | 1.4 | Q14165 | Malectin | MLEC | Protein
glycosylation | 68 | 1592.8 | 58 | | |
| 22 | 0.020 | 1.4 | P30041 |
Peroxiredoxin-6 | PRDX6 | Anti-oxidative
stress Phospholipid turnover regulation | | | | 81 | |
| 565 | 0.048 | 1.3 | P23381 | Tryptophan-tRNA
ligase, cytoplasmic | WARS | tRNA aminoacylation
Angiogenesis regulation | 83 | | | | |
| 1020 | 0.020 | 1.3 | Q99829 | Copine-1 | CPNE1 | Membrane
trafficking NF-κB transcription repressor | | | | 197 | |
| 808 | 0.043 | 1.2 | P16435 | NADPH-cytochrome
P450 reductase | POR | Electron transfer
to cytochrome P450 | | | | 276 | |
| 1394 | 0.048 | 1.2 | Q5T6V5 | UPF0553 protein
C9orf64 | C9orf64 | Unknown | 99 | | | | |
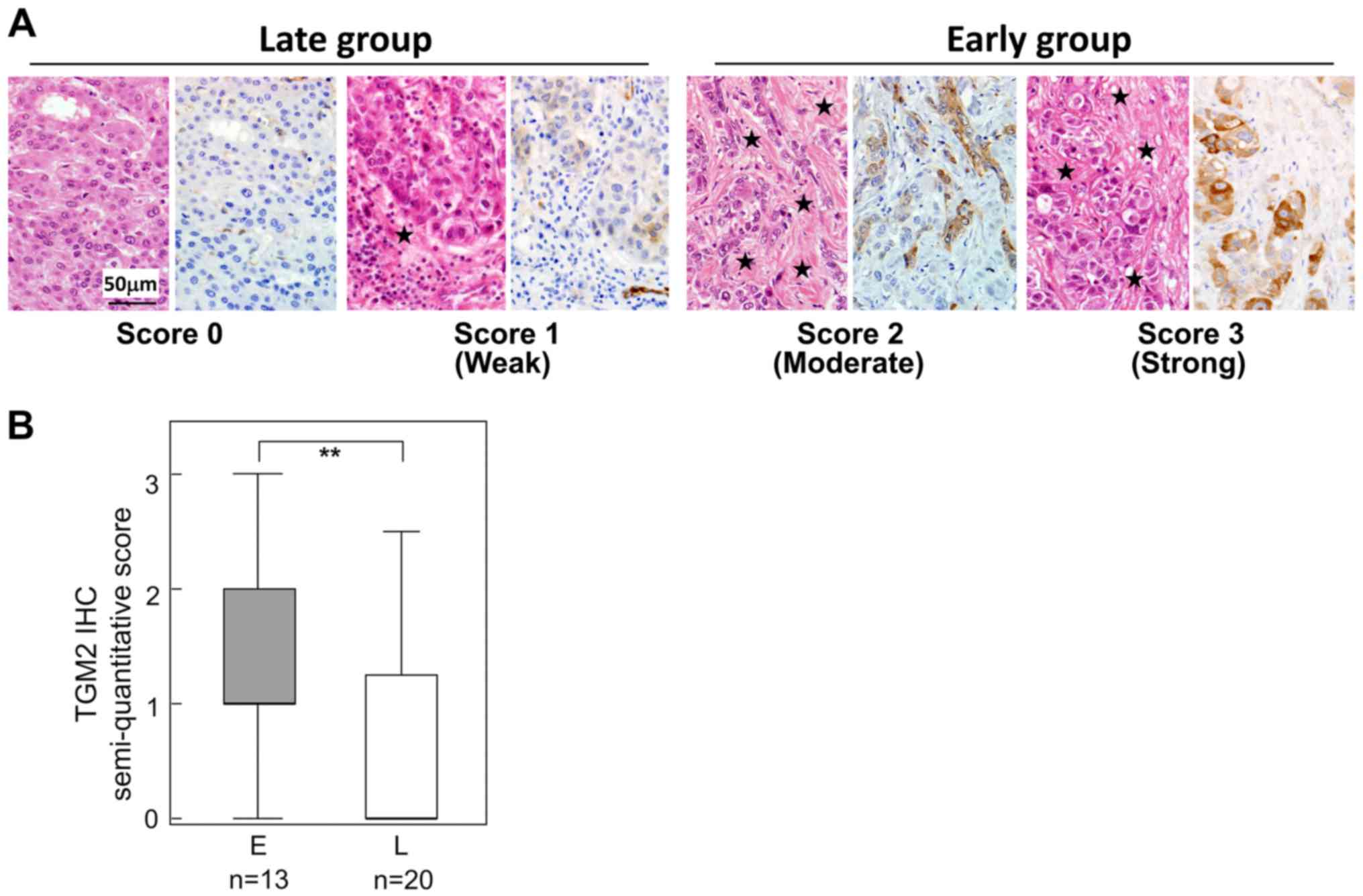
Immunohistochemistry of TGM2
TGM2 was found to be a significantly upregulated
protein in the early group. To determine the localization of TGM2
and to validate this differential expression, we examined TGM2 IHC
33 HCC tissues; the clinicopathological features between 13 early
and 20 late recurrence cases are compared in Table II. In most HCC cases, intra-tumor
vascular endothelial cells were positive for TGM2 (data not shown).
HCC cells were also positive for TGM2, though the frequency of
positivity differed by case (Fig.
4). In addition, the TGM2-positive HCC cells tended to be
adjacent to fibrous stroma (Fig.
4A). When the TGM2 positivity was semi-quantitatively scored
(Fig. 4A), TGM2-positive HCC cells
were significantly increased in the early recurrence group compared
with those of the late recurrence group (Fig. 4B) (Table IV).
 | Table IVTGM2 expression in HCC in the early
and late recurrence groups. |
Table IV
TGM2 expression in HCC in the early
and late recurrence groups.
| TGM2 expression in
HCC
| Total | P-value |
|---|
| Negative | Positive |
|---|
| Early | 2 | 11 | 13 | 0.004 |
| Late | 14 | 6 | 20 | |
| Total | 16 | 17 | 33 | |
TGM2-associated pathways in HCC
TGM2 is a multifunctional protein involved in many
biological processes, such as angiogenesis, cell adhesion,
migration, survival and the epithelial-mesenchymal transition (EMT)
(22). Thus, we investigated two
major TGM2-mediated pathways that might influence tumor recurrence
(Fig. 5A). EMT plays an important
role in tumor metastasis (23). To
examine whether upregulation of TGM2 is associated with EMT, we
measured mRNA expression of five genes, TGFB1, HIF1A, SNAI1,
SNAI2 and CDH1, related to EMT, and the mRNA levels were
compared between the two groups; Table II shows the clinicopathological
comparison of 9 early and 6 late recurrence cases. TGFB1 signaling
and hypoxia induce EMT-inducer genes, such as SNAI1 and
SNAI2, and then these transcription factors downregulate
E-cadherin (24,25). TGM2 mRNA is also upregulated
by TGFB and HIF1 (26), and the
TGM2 protein negatively regulates CDH1 at the
transcriptional level (27).
Correlation analyses revealed that the TGM2 mRNA level was
strongly correlated with TGFB1 and moderately with
HIF1A and SNAI1 (Fig.
5B). CDH1 was also strongly correlated with TGM2
(Fig. 5B). In addition, the
expression levels of these EMT-related genes were not significantly
different between the early and late groups (Fig. 5C). IHC showed that the
CDH1-staining pattern did not depend on negative staining of TGM2;
TGM2-positive and CDH1-negative HCC cells were not consistently
observed (Fig. 5D).
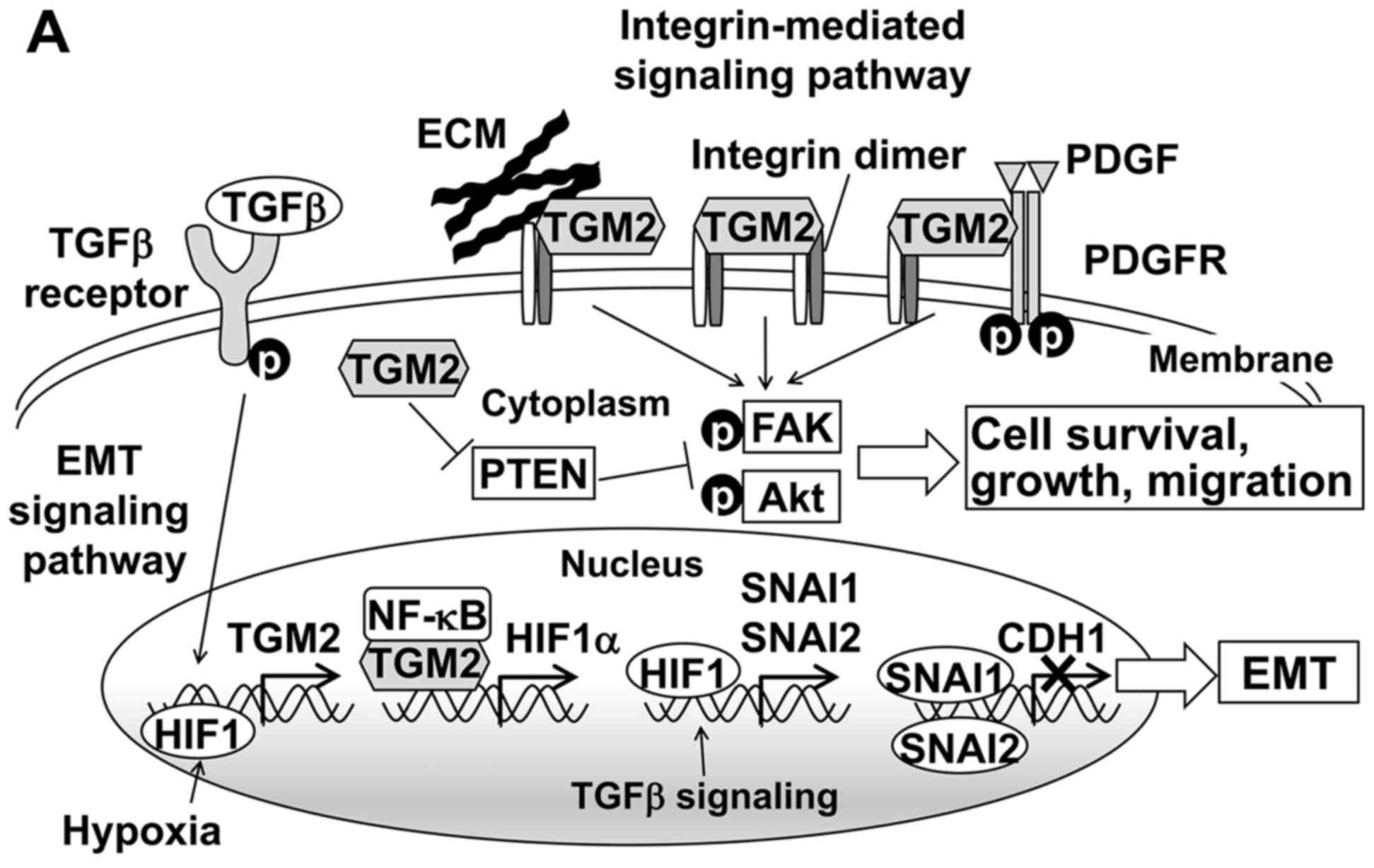 | Figure 5TGM2-associated pathways and
downstream gene expression related to EMT. (A) TGM2 mediates the
EMT and integrin pathways. TGFβ signaling and HIF1 induce the
transcriptional activation of the TGM2. Upregulated TGM2 protein
forms a complex with NF-κB and then translocates to the nucleus to
promote HIF-1α transcription. Increased HIF1α induces the
expression of transcription factors such as SNAI1 and SNAI2, which
transcriptionally repress the E-cadherin. On the cell surface, TGM2
interacts with both integrins and extracellular matrix (ECM)
proteins such as fibronectin to mediate stable complex formation
and induce integrin-integrin clustering. TGM2 also enhances the
PDGFR-integrin association. These interactions activate downstream
target proteins such as FAK and Akt by phosphorylation.
Furthermore, cytosolic TGM2 downregulates the tumor suppressor PTEN
at the protein level and consequently activates FAK and Akt. As a
result, TGM2 promotes cell survival, growth and migration. Based on
Nurminskaya et al (22),
Verma et al (29) and
Agnihotri et al (30). (B)
mRNA expression of TGM2 and EMT-related genes. The quantity of
target mRNA was normalized against that of 18S rRNA, and Spearman's
rank correlation coefficient were performed between TGM2 and the
other genes. Gray circles indicate 9 cases of early recurrence;
white circles indicate 6 cases of late recurrence. (C) mRNA
expression of TGM2 and EMT-related genes in the early and late
recurrence groups. Two-group comparisons were performed using the
Student's t-test. The bar graph represents the mean ± SD. E, early
recurrence group; L, late recurrence group. Clinicopathological
features were compared between the two groups (Table II). (D) Representative images of
H&E staining (upper panels) and corresponding CDH1 (middle
panels) and TGM2 (lower panels) staining in HCC samples. Three
representative cases are shown: TGM2-negative, TGM2 moderately
positive and TGM2 strongly positive HCC cells. Magnification for
all the panels is shown by a scale bar in the upper left panel. |
On the other hand, TGM2 also activates integrin
signaling pathways (Fig. 5A). Such
TGM2-mediated integrin signaling enhances not only tumor cell
adhesion and migration but also survival and growth (28). Focal adhesion kinase (FAK) and
protein kinase B (Akt) are the downstream targets of TGM2-mediated
integrin signaling. In addition, TGM2 downregulates the tumor
suppressor phosphatase PTEN at a protein level and subsequently
leads to the phosphorylation of FAK and Akt (29). Therefore, we examined whether the
FAK and Akt proteins were phosphorylated and whether PTEN was
downregulated in the HCC tissues of the early recurrence group.
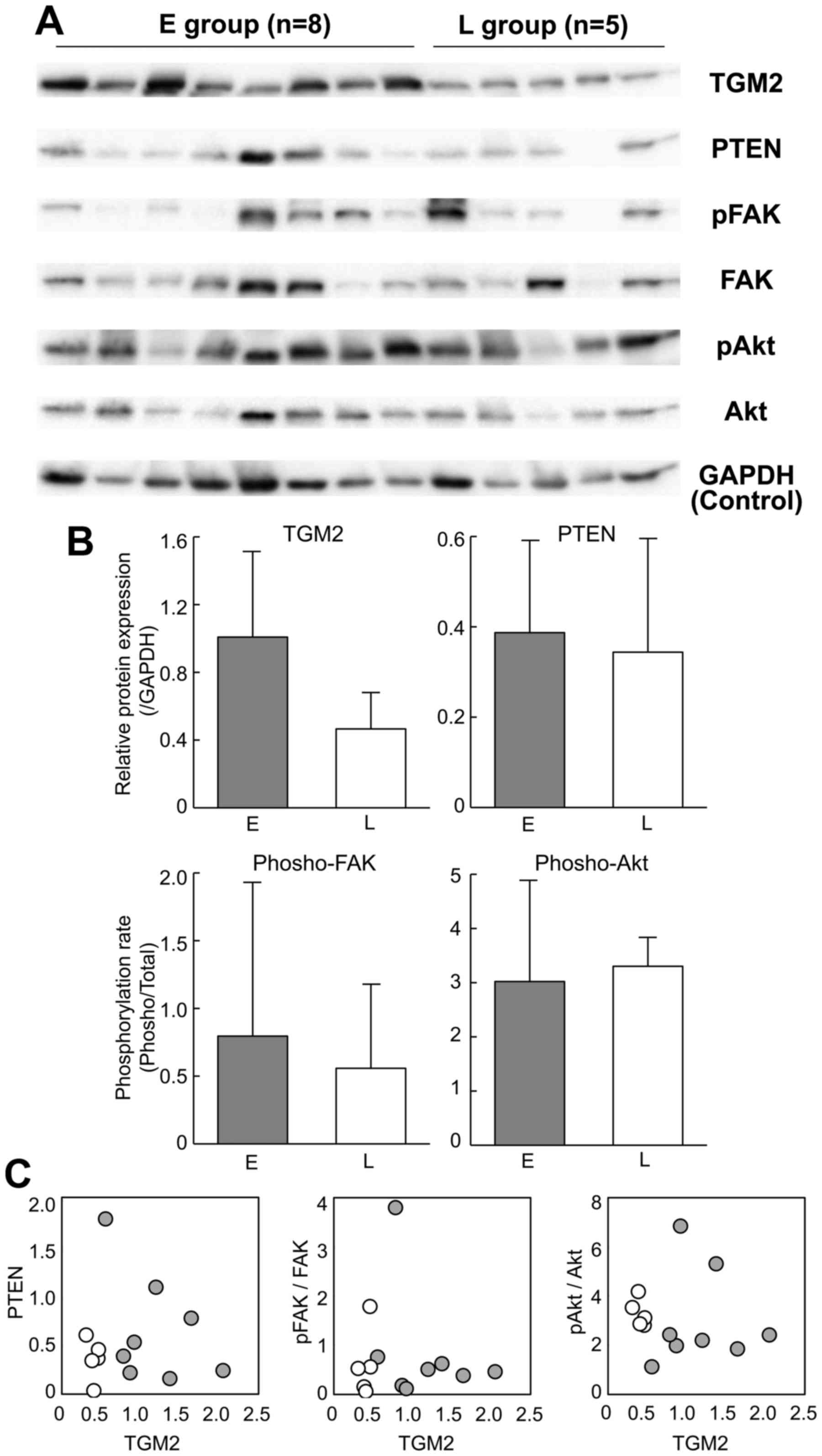
Western blot analysis confirmed the upregulation of TGM2 in the
early recurrence group of HCC (Fig. 6A
and B, P=0.06 by Student's t-test). However, the
phosphorylation rates of FAK and Akt were not significantly
different between the early and late recurrence groups (Fig. 6B) and were not correlated with the
TGM2 abundance (Fig. 6C). The PTEN
expression was also similar between the recurrence groups (Fig. 6B) and was not downregulated in
correlation with the TGM2 expression (Fig. 6C).
Discussion
To strictly characterize intrahepatic metastasis of
HCC, this study focused on primary, solitary and early-stage HCC
cases that were positive for HCV. We then compared the proteomes of
two groups of HCC cases: those recurring early and those recurring
late. We identified 10 upregulated and 9 downregulated proteins in
the early recurrence group (Table
III). As the expression profiles of these proteins
discriminated the two groups, i.e., early and late recurrence
(Fig. 3), it is clear that HCCs
recurring early and late have individually characteristic proteins.
These proteins are related to a variety of functions, such as iron
storage, antioxidant activity, protein transport and lipid
metabolism (Table III), and
might be candidates for recurrence markers.
In this study, we concentrated on TGM2 upregulation
in HCCs prone to recurrence. This multifunctional protein is
related to cancer progression, chemoresistance, invasiveness and
metastasis and is upregulated in various cancers (26); therefore, we investigated two major
TGM2-mediated pathways that might influence tumor recurrence
(Fig. 5A). First, in the EMT
pathway, TGM2 signaling negatively regulates CDH1 at the
transcriptional level (27,30);
thus, because CDH1 downregulation is a characteristic of EMT
(24), increases in TGM2 may
result in EMT and cancer metastasis promotion (23). However, contrary to the
expectation, TGM2 mRNA was positively correlated with CDH1 mRNA
rather than negatively correlated, yet co-expression of both
proteins was not always observed. Recently, Fang et al
suggested a new metastatic model of HCC independent of EMT
(31). Therefore, upregulation of
TGM2 might not be associated with EMT but might contribute to early
HCC recurrence via a process not related to EMT. Second, in the
integrin signaling pathway, FAK and Akt are downstream target
proteins activated by phosphorylation through TGM2-integrin
signaling, resulting in the activation of tumor cell growth,
survival, motility and invasiveness (22). However, the linkage was not
observed in the early recurrence group of HCC. Thus, although
multiple TGM2-mediated pathways have been clarified in experimental
models in vitro and in vivo, these are not simply
observed in human pathological tissues.
Recently, Tatsukawa et al demonstrated the
complexity of TGM2-mediated signaling (32); multifunctional TGM2 is involved in
many cellular processes and has different functions, which vary
among organs, cell types and subcellular protein localization
(22,32). Even in the liver, TGM2 has opposing
roles, such as anti-apoptotic and pro-apoptotic or fibrogenic and
anti-fibrogenic roles, depending on the experimental animal model
used (32). Thus, it is yet
unknown by what molecular mechanism TGM2 is involved in the early
recurrence of HCC. Considering our observation of TGM2-positive HCC
cells adjacent to fibrous stroma (Fig.
4A), it is possible that TGM2 might be associated with early
recurrence via ECM-integrin signaling and 'locally' activates
downstream targets such as FAK and Akt. Further analyses are needed
to clarify the contribution to early HCC recurrence by TGM2.
Another upregulated protein in the early group, PAXX
(the C9orf142 gene), is involved in repairing DNA
double-strand breaks via the non-homologous end joining pathway
(33–35). Upregulation of PAXX may indirectly
indicate that severe DNA damage is a characteristic of HCC early
recurrence.
In contrast, copine-1 (CPNE1), tryptophanyl-tRNA
synthetase (WARS) and heat shock protein beta-1 (HSPB1, also known
as HSP27) were downregulated in the early group. CPNE1 acts as a
transcriptional repressor of NF-κB (36). NF-κB has dual role in HCC, as a
tumor promoter and a tumor suppressor (37), but NF-κB overexpression is
associated with poor overall survival in most solid tumor cases
(38). Thus, downregulation of
CPNE1 in early recurrence HCC may result in constitutive NF-κB
expression and contribute to a highly malignant phenotype.
Ghanipour et al reported that low expression of WARS is
associated with a metastatic phenotype of colorectal cancer
(39). In contrast, WARS was found
to be upregulated in malignant ovarian cancer (40) and oral squamous cell carcinoma
(41). Thus, Lee et al
suggested that WARS has paradoxical effect on tumor invasiveness in
different tumor types (41).
In the present study, WARS was downregulated in
early recurrence HCC; conversely, Taoka et al reported WARS
upregulation in early recurrent HCC (15). Such an inconsistency was also
observed for HSPB1. Upregulation of heat shock proteins such as
HSPB1, HSP70 and HSP90 is often observed in many types of cancer,
including HCC, due to cancer cell adaptation to stress conditions
(42). HSPB1 upregulation has been
associated with metastatic or invasive HCC (43), and Taoka et al found HSPB1
upregulation in early recurrence HCC (15). However, HSPB1 protein spots in our
study indicated downregulation in early recurrence HCC. There are
two possibilities for such discrepancies. One possibility is a
difference in the clinicopathological factors used for defining
early and late recurrence groups, such as tumor size, grade, time
to recurrence, and viral infection status. We strictly selected our
HCC cases and categorized them into two groups; therefore, such
differences in background factors may result in discrepancies. The
other possibility is a difference in the proteomic methodology,
i.e., protein separation and protein quantification. Taoka et
al separated protein samples by SDS-PAGE, and all identified
peptides corresponding to a single protein, including modified or
degraded forms, were used to determine protein abundance (15) by emPAI (44). In contrast, we used the 2D-DIGE
method and focused on a single protein spot to i) identify a
protein and ii) determine abundance according to the fluorescence
intensity. Therefore, it is critical for clinical proteomics to
precisely estimate the background clinicopathological features and
appropriately incorporate these features into the study.
Overall, this study suggests that TGM2 upregulation
is a characteristic of early HCC recurrence and that it might be a
helpful marker for the early detection of HCC recurrence.
Acknowledgments
We are indebted to Dr H. Nakayama (Nihon University
School of Medicine) for providing the clinical data of the
patients. The human liver samples were provided by Dr T. Mamiya
(Nihon University School of Medicine) and Professor N. Kokudo
(Graduate School of Medicine, University of Tokyo). We also thank
Dr F. Fuchinoue (Nihon University School of Medicine) for helpful
suggestions on the pathological data and Ms. Y. Hirotani (Nihon
University School of Medicine) for technical assistance. This work
was supported by a grant for 'Open Research Center' Project for
Private Universities: matching fund subsidy from MEXT (2005), Nihon
University Multidisciplinary Research Grants for 2009, and
Grant-in-Aid for Scientific Research (C) 25430142 from MEXT (2013).
English editorial assistance was provided by American Journal
Experts.
References
|
1
|
Yang JD and Roberts LR: Hepatocellular
carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 7:448–458.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ikai I, Arii S, Okazaki M, Okita K, Omata
M, Kojiro M, Takayasu K, Nakanuma Y, Makuuchi M, Matsuyama Y, et
al: Report of the 17th Nationwide Follow-up Survey of Primary Liver
Cancer in Japan. Hepatol Res. 37:676–691. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chen YJ, Yeh SH, Chen JT, Wu CC, Hsu MT,
Tsai SF, Chen PJ and Lin CH: Chromosomal changes and clonality
relationship between primary and recurrent hepatocellular
carcinoma. Gastroenterology. 119:431–440. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Nomoto S, Kinoshita T, Kato K, Otani S,
Kasuya H, Takeda S, Kanazumi N, Sugimoto H and Nakao A:
Hypermethylation of multiple genes as clonal markers in
multicentric hepatocellular carcinoma. Br J Cancer. 97:1260–1265.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang B, Xia CY, Lau WY, Lu XY, Dong H, Yu
WL, Jin GZ, Cong WM and Wu MC: Determination of clonal origin of
recurrent hepatocellular carcinoma for personalized therapy and
outcomes evaluation: A new strategy for hepatic surgery. J Am Coll
Surg. 217:1054–1062. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Portolani N, Coniglio A, Ghidoni S,
Giovanelli M, Benetti A, Tiberio GA and Giulini SM: Early and late
recurrence after liver resection for hepatocellular carcinoma:
Prognostic and therapeutic implications. Ann Surg. 243:229–235.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shindoh J, Hasegawa K, Matsuyama Y, Inoue
Y, Ishizawa T, Aoki T, Sakamoto Y, Sugawara Y, Makuuchi M and
Kokudo N: Low hepatitis C viral load predicts better long-term
outcomes in patients undergoing resection of hepatocellular
carcinoma irrespective of serologic eradication of hepatitis C
virus. J Clin Oncol. 31:766–773. 2013. View Article : Google Scholar
|
|
8
|
Cheng Z, Yang P, Qu S, Zhou J, Yang J,
Yang X, Xia Y, Li J, Wang K, Yan Z, et al: Risk factors and
management for early and late intrahepatic recurrence of solitary
hepatocellular carcinoma after curative resection. HPB Oxf.
17:422–427. 2015. View Article : Google Scholar
|
|
9
|
Iizuka N, Oka M, Yamada-Okabe H, Nishida
M, Maeda Y, Mori N, Takao T, Tamesa T, Tangoku A, Tabuchi H, et al:
Oligonucleotide microarray for prediction of early intrahepatic
recurrence of hepatocellular carcinoma after curative resection.
Lancet. 361:923–929. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurokawa Y, Matoba R, Takemasa I, Nagano
H, Dono K, Nakamori S, Umeshita K, Sakon M, Ueno N, Oba S, et al:
Molecular-based prediction of early recurrence in hepatocellular
carcinoma. J Hepatol. 41:284–291. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang SM, Ooi LL and Hui KM: Identification
and validation of a novel gene signature associated with the
recurrence of human hepatocellular carcinoma. Clin Cancer Res.
13:6275–6283. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoshioka S, Takemasa I, Nagano H, Kittaka
N, Noda T, Wada H, Kobayashi S, Marubashi S, Takeda Y, Umeshita K,
et al: Molecular prediction of early recurrence after resection of
hepatocellular carcinoma. Eur J Cancer. 45:881–889. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yokoo H, Kondo T, Okano T, Nakanishi K,
Sakamoto M, Kosuge T, Todo S and Hirohashi S: Protein expression
associated with early intrahepatic recurrence of hepatocellular
carcinoma after curative surgery. Cancer Sci. 98:665–673. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan GS, Lim KH, Tan HT, Khoo ML, Tan SH,
Toh HC and Ching Ming Chung M: Novel proteomic biomarker panel for
prediction of aggressive metastatic hepatocellular carcinoma
relapse in surgically resectable patients. J Proteome Res.
13:4833–4846. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taoka M, Morofuji N, Yamauchi Y, Ojima H,
Kubota D, Terukina G, Nobe Y, Nakayama H, Takahashi N, Kosuge T, et
al: Global PROTOMAP profiling to search for biomarkers of
early-recurrent hepatocellular carcinoma. J Proteome Res.
13:4847–4858. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bradford MM: A rapid and sensitive method
for the quantitation of microgram quantities of protein utilizing
the principle of protein-dye binding. Anal Biochem. 72:248–254.
1976. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yamaguchi H, Hasegawa K and Esumi M:
Protein from the fraction remaining after RNA extraction is useful
for proteomics but care must be exercised in its application. Exp
Mol Pathol. 95:46–50. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Esumi M, Ishibashi M, Yamaguchi H,
Nakajima S, Tai Y, Kikuta S, Sugitani M, Takayama T, Tahara M,
Takeda M, et al: Transmembrane serine protease TMPRSS2 activates
hepatitis C virus infection. Hepatology. 61:437–446. 2015.
View Article : Google Scholar
|
|
19
|
Yamaguchi H, Matsumoto S, Ishibashi M,
Hasegawa K, Sugitani M, Takayama T and Esumi M: β-Glucuronidase is
a suitable internal control gene for mRNA quantitation in
pathophysiological and non-pathological livers. Exp Mol Pathol.
95:131–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Numaguchi S, Esumi M, Sakamoto M, Endo M,
Ebihara T, Soma H, Yoshida A and Tokuhashi Y: Passive cigarette
smoking changes the circadian rhythm of clock genes in rat
intervertebral discs. J Orthop Res. 34:39–47. 2016. View Article : Google Scholar
|
|
21
|
Kim W, Oe Lim S, Kim JS, Ryu YH, Byeon JY,
Kim HJ, Kim YI, Heo JS, Park YM and Jung G: Comparison of proteome
between hepatitis B virus- and hepatitis C virus-associated
hepatocellular carcinoma. Clin Cancer Res. 9:5493–5500.
2003.PubMed/NCBI
|
|
22
|
Nurminskaya MV and Belkin AM: Cellular
functions of tissue transglutaminase. Int Rev Cell Mol Biol.
294:1–97. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Naber HP, Drabsch Y, Snaar-Jagalska BE,
ten Dijke P and van Laar T: Snail and Slug, key regulators of
TGF-β-induced EMT, are sufficient for the induction of single-cell
invasion. Biochem Biophys Res Commun. 435:58–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Catalano V, Turdo A, Di Franco S, Dieli F,
Todaro M and Stassi G: Tumor and its microenvironment: A
synergistic interplay. Semin Cancer Biol. 23:522–532. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang L, Xu AM and Liu W: Transglutaminase
2 in cancer. Am J Cancer Res. 5:2756–2776. 2015.PubMed/NCBI
|
|
27
|
Shao M, Cao L, Shen C, Satpathy M,
Chelladurai B, Bigsby RM, Nakshatri H and Matei D:
Epithelial-to-mesenchymal transition and ovarian tumor progression
induced by tissue transglutaminase. Cancer Res. 69:9192–9201. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mangala LS, Fok JY, Zorrilla-Calancha IR,
Verma A and Mehta K: Tissue transglutaminase expression promotes
cell attachment, invasion and survival in breast cancer cells.
Oncogene. 26:2459–2470. 2007. View Article : Google Scholar
|
|
29
|
Verma A, Guha S, Wang H, Fok JY, Koul D,
Abbruzzese J and Mehta K: Tissue transglutaminase regulates focal
adhesion kinase/AKT activation by modulating PTEN expression in
pancreatic cancer cells. Clin Cancer Res. 14:1997–2005. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agnihotri N, Kumar S and Mehta K: Tissue
transglutaminase as a central mediator in inflammation-induced
progression of breast cancer. Breast Cancer Res. 15:2022013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang JH, Zhou HC, Zhang C, Shang LR, Zhang
L, Xu J, Zheng L, Yuan Y, Guo RP, Jia WH, et al: A novel vascular
pattern promotes metastasis of hepatocellular carcinoma in an
epithelial-mesenchymal transition-independent manner. Hepatology.
62:452–465. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tatsukawa H, Furutani Y, Hitomi K and
Kojima S: Transglutaminase 2 has opposing roles in the regulation
of cellular functions as well as cell growth and death. Cell Death
Dis. 7:e22442016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xing M, Yang M, Huo W, Feng F, Wei L,
Jiang W, Ning S, Yan Z, Li W, Wang Q, et al: Interactome analysis
identifies a new paralogue of XRCC4 in non-homologous end joining
DNA repair pathway. Nat Commun. 6:62332015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Craxton A, Somers J, Munnur D, Jukes-Jones
R, Cain K and Malewicz M: XLS (c9orf142) is a new component of
mammalian DNA double-stranded break repair. Cell Death Differ.
22:890–897. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ochi T, Blackford AN, Coates J, Jhujh S,
Mehmood S, Tamura N, Travers J, Wu Q, Draviam VM, Robinson CV, et
al: DNA repair. PAXX, a paralog of XRCC4 and XLF, interacts with Ku
to promote DNA double-strand break repair. Science. 347:185–188.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ramsey CS, Yeung F, Stoddard PB, Li D,
Creutz CE and Mayo MW: Copine-I represses NF-kappaB transcription
by endoproteolysis of p65. Oncogene. 27:3516–3526. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xia Y, Shen S and Verma IM: NF-κB, an
active player in human cancers. Cancer Immunol Res. 2:823–830.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu D, Wu P, Zhao L, Huang L, Zhang Z, Zhao
S and Huang J: NF-κB expression and outcomes in solid tumors: A
systematic review and meta-analysis. Medicine (Baltimore).
94:e16872015. View Article : Google Scholar
|
|
39
|
Ghanipour A, Jirström K, Pontén F,
Glimelius B, Påhlman L and Birgisson H: The prognostic significance
of tryptophanyl-tRNA synthetase in colorectal cancer. Cancer
Epidemiol Biomarkers Prev. 18:2949–2956. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Morita A, Miyagi E, Yasumitsu H, Kawasaki
H, Hirano H and Hirahara F: Proteomic search for potential
diagnostic markers and therapeutic targets for ovarian clear cell
adenocarcinoma. Proteomics. 6:5880–5890. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lee CW, Chang KP, Chen YY, Liang Y, Hsueh
C, Yu JS, Chang YS and Yu CJ: Overexpressed tryptophanyl-tRNA
synthetase, an angiostatic protein, enhances oral cancer cell
invasiveness. Oncotarget. 6:21979–21992. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang C, Zhang Y, Guo K, Wang N, Jin H, Liu
Y and Qin W: Heat shock proteins in hepatocellular carcinoma:
Molecular mechanism and therapeutic potential. Int J Cancer.
138:1824–1834. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang RC, Huang CY, Pan TL, Chen WY, Ho CT,
Liu TZ and Chang YJ: Proteomic characterization of Annexin l (ANX1)
and heat shock protein 27 (HSP27) as biomarkers for invasive
hepatocellular carcinoma cells. PLoS One. 10:e01392322015.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ishihama Y, Oda Y, Tabata T, Sato T,
Nagasu T, Rappsilber J and Mann M: Exponentially modified protein
abundance index (emPAI) for estimation of absolute protein amount
in proteomics by the number of sequenced peptides per protein. Mol
Cell Proteomics. 4:1265–1272. 2005. View Article : Google Scholar : PubMed/NCBI
|