Introduction
Breast cancer is one of the most frequently
diagnosed malignant cancers in women worldwide (1). Breast cancer is the second leading
cause of cancer-related deaths among females, with a yearly toll of
more than 40,000 deaths in the United States alone (2). Furthermore, the number of cases is
increasing, which represents a serious threat to women's health and
their quality of life. Therefore, research in the diagnosis and
treatment of breast cancer is very important (3).
MicroRNAs (miRNAs) play important roles in cancers
(4). miRNAs form a class of small
non-coding RNAs (5) which function
by binding to the 3′ terminal untranslated region of targeted mRNAs
to inhibit their translation (6).
Furthermore, miRNAs are important for regulating various biological
processes including proliferation, migration and inhibition of
apoptosis (7). miR-145 is a
microRNA which has been widely studied in cancer and has been shown
to be expressed at low levels in a wide variety of tumors,
including bladder cancer (8),
colorectal cancer (9), and lung
cancer (10).
In this study, we examined the role of miR-145 in
human breast cancer. Expression levels of miR-145 in breast cancer
tissue and adjacent normal tissue were first analyzed using
quantitative real-time polymerase chain reaction (qRT-PCR)
(3). We then transfected breast
cancer cells with hsa-miR-145 mimics, hsa-miR-145 inhibitor, mimics
negative control (mimics NC) and inhibitor negative control
(inhibitor NC). Cell proliferation was analyzed by colony formation
assay and MTT cell proliferation assay. Cell migration was assessed
using Transwell and wound-healing assays. Previous studies
indicated that transforming growth factor-β1 (TGF-β1) played an
important role in growth and metastasis of breast cancer cells
(11). To detect the relationship
between TGF-β1 and miR-145, we used western blot analysis to
analyze protein expression of TGF-β1.
Materials and methods
Breast cancer and normal tissues
The ethics committee of Jiangsu University
(Zhenjiang, Jiangsu, China) approved all aspects of this study.
Documented informed consent was obtained from all subjects. Breast
cancer tissues and corresponding adjacent normal tissues were
obtained from the Department of Surgery, the Second People's
Hospital of Kunshan, China. Cancer status was histologically
confirmed, and all tissues were immediately stored at -80°C.
Cell culture
Breast cancer cells (MCF-7 and MDA-MB 231) were
provided from Nanjing University and grown in DMEM (Gibco,
Carlsbad, CA, USA) containing 10% fetal bovine serum (ExCell
Biology, Shanghai, China) at 37°C in a humidified chamber.
Cell transfection
Breast cancer cells were seeded in 6-well plates.
After 24 h, the cells were transfected with 100 nM of miR-145
mimics, miR-145 inhibitor, mimics NC or inhibitor NC (provided by
GenePharma, Shanghai, China), using Lipofectamine-2000 (Invitrogen,
Carlsbad, CA, USA) according to the manufacturer's protocol. The
sequences are listed in Table
I.
 | Table ISequences of miR-145 mimics, miR-145
inhibitor and negative controls. |
Table I
Sequences of miR-145 mimics, miR-145
inhibitor and negative controls.
| Name | Sequence |
|---|
| Hsa-miR-145
mimic | |
| Sense |
5′-GUCCAGUUUUCCCAGGAAUCCCU-3′ |
| Antisense |
5′-GGAUUCCUGGGAAAACUGGACUU-3′ |
| Mimic-negative
control | |
| Sense |
5′-UUCUUCGAACGUGUCACGUTT-3′ |
| Antisense |
5′-ACGUGACACGUUCGGAGAATT-3′ |
| Hsa-miR-145
inhibitor |
5′-AGGGAUUCCUGGGAAAACUGGAC-3′ |
| Inhibitor negative
control |
5′-CAGUACUUUUGUGUAGUACAA-3′ |
Quantitative real-time PCR
Total cellular RNA was isolated from tissue samples
and cells using TRIzol reagent (Invitrogen) according to the
manufacturer's protocol. RNA (1 μg) was reverse transcribed
into cDNA using reverse transcriptase (GenePharma). The expression
levels of miR-145 were evaluated by quantitative real-time PCR
performed using a SYBR green-containing PCR kit (GenePharma). A
CFX-96 real-time fluorescence thermal cycler (Bio-Rad, Hercules,
CA, USA) was used for quantitative miRNA and mRNA detection. U6
snRNA was used for normalization of miR-145 expression levels and
GAPDH was used for normalization of TGF-β1 mRNA expression levels.
Primer sequences are listed in Table
II. miR-145 and TGF-β1 mRNA expression levels were calculated
using the 2−ΔCt method relative to U6 snRNA and
GAPDH.
 | Table IISpecific primers for target and
control genes. |
Table II
Specific primers for target and
control genes.
| Name | Sequence |
|---|
| miR-145 | F:
5′-CAGTCTTGTCCAGTTTTCCCAG-3′ |
| R:
5′-TATGCTTGTTCTCGTCTCTGTGTC-3′ |
| U6snRNA | F:
5′-ATTGGAACGATACAGAGAAGATT-3′ |
| R:
5′-GGAACGCTTCACGAATTTG-3′ |
| TGFB1 | F:
5′-GTACCTGAACCCGTGTTGCT-3′ |
| R:
5′-GTATCGCCAGGAATTGTTGC-3′ |
| GAPDH | F:
5′-GAAGGTGAAGGTCGGAGTC-3′ |
| R:
5′-GAAGATGGTGATGGGATTTC-3 |
Cell proliferation assay
The effect of miR-145 on proliferation was measured
using Thiazolyl Blue Tetrazolium Bromide-MTT (Amresco, Solon, OH,
USA). Briefly, 4×103 cells transfected with miR-145
mimics, miR-145 inhibitor, mimics NC or inhibitor NC were plated in
96-well plates. MTT reagent was added to each well 24, 48 and 72 h
after seeding. After 4 h incubation, the reagent was removed and
dimethyl sulfoxide was added to each well. The absorbance value in
each well was measured with an FLx800 Fluorescence Microplate
Reader (BioTek, Winooski, VT, USA) at 490 nm.
Colony formation assay
Breast cancer cells transfected with miR-145 mimics,
miR-145 inhibitor, mimics NC or inhibitor NC were seeded into
six-well plates and incubated for 12 days. Colonies were fixed with
formaldehyde, stained with crystal violet and counted. Assays were
performed in triplicate.
Transwell invasion assay
Transwell invasion assays were performed using
Transwell plates (Corning Inc., Corning, NY, USA) containing
membranes with 8 μm pores. Breast cancer cells were
transfected with miR-145 mimics, miR-145 inhibitor, mimics NC or
inhibitor NC. After 48 h, 4×104 cells were resuspended
in 200 μl serum-free medium and seeded into the upper
chamber. Culture medium containing 10% fetal bovine serum was added
to the lower chamber as the chemoattractant. The cells were
incubated in a humidified incubator at 37°C for 14 h, and cells
which had migrated to the lower surface of the insert were stained
and counted. Assays were performed in triplicate.
Wound scratch assay
Wound scratch assays were performed in 6-well
Transwell chambers (Corning). Cells transfected with miR-145
mimics, miR-145 inhibitor, mimics NC or inhibitor NC were plated
and grown to 100% confluence Scratches were created with sterile
pipette tips. The wounds were photographed at 0 and 48 h and
monitored following standard protocols.
Western blot analysis
We use a modified radio immunoprecipitation assay
lysis buffer (Beyotime, Shanghai, China) and phenylmethanesulfonyl
fluoride (Beyotime) to extract total cellular protein from cells 48
h after they were transfected with miR-145 mimics, miR-145
inhibitor, mimics NC or inhibitor NC. Equal amounts of protein
lysates (100–150 μg) were separated by 10% sodium dodecyl
sulfate polyacrylamide gel electrophoresis (Beyotime) and then
transferred to polyvinylidene fluoride membranes (Beyotime). The
membranes were blocked with 5% nonfat milk for 2 h and then
incubated with primary anti-TGF-β1 (Bioworld Technology, Inc, St.
Louis, MN, USA, BS1361, dilution of 1:500) and anti-GAPDH (Ruiying,
RLM3029, dilution of 1:1000) at 4°C overnight. Membranes were
washed three times in TBST (Tris Buffered Saline, with Tween-20),
10 min per wash, and incubated with HRP (horseradish
peroxidase)-conjugated secondary antibodies (anti-mouse or
anti-rabbit IgG, Ruiying, RS0001 and RS0002, dilution of 1:2000)
for 2 h at room temperature. Finally, immunoblots were visualized
using chemiluminescence reagents (ECL, Beyotime).
Statistical analyses
Results were expressed as means ± standard deviation
(SD) using the GraphPad Prism V5.0 software program (GraphPad, San
Diego, CA, USA). P-values were obtained from t-tests for either
paired or independent samples. P-value <0.05 was considered to
indicate a statistically significant difference.
Results
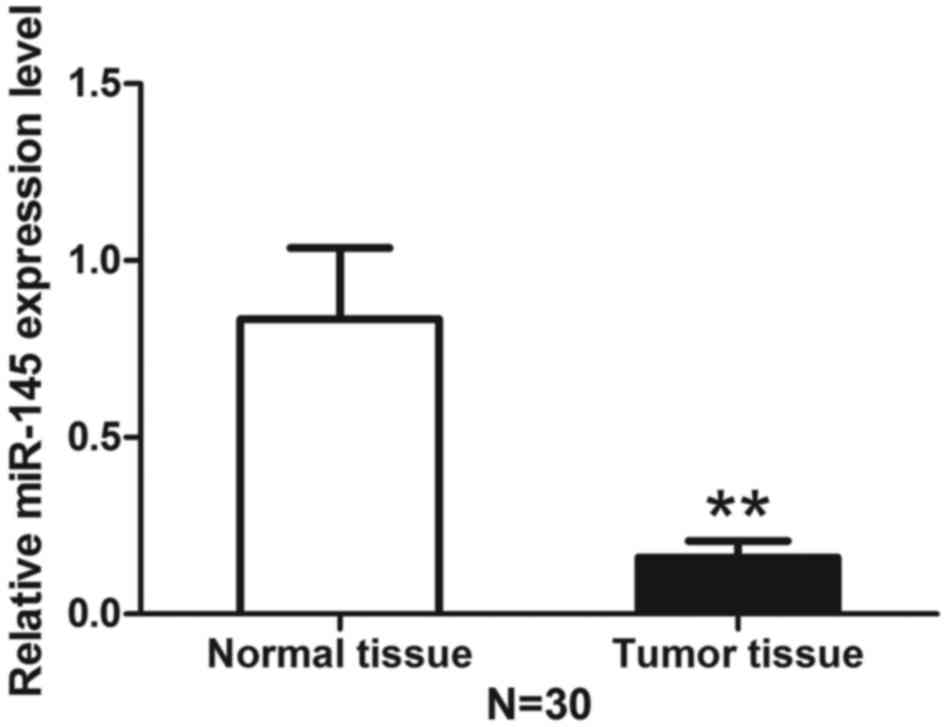
Expression level of miR-145 is decreased
in breast cancer tissues compared with adjacent normal tissue
Total RNA extracted from breast cancer tissue and
corresponding adjacent normal tissue was analyzed using qRT-PCR to
determine the level of miR-145. The expression level of miR-145 was
decreased in breast cancer tissues compared with normal adjacent
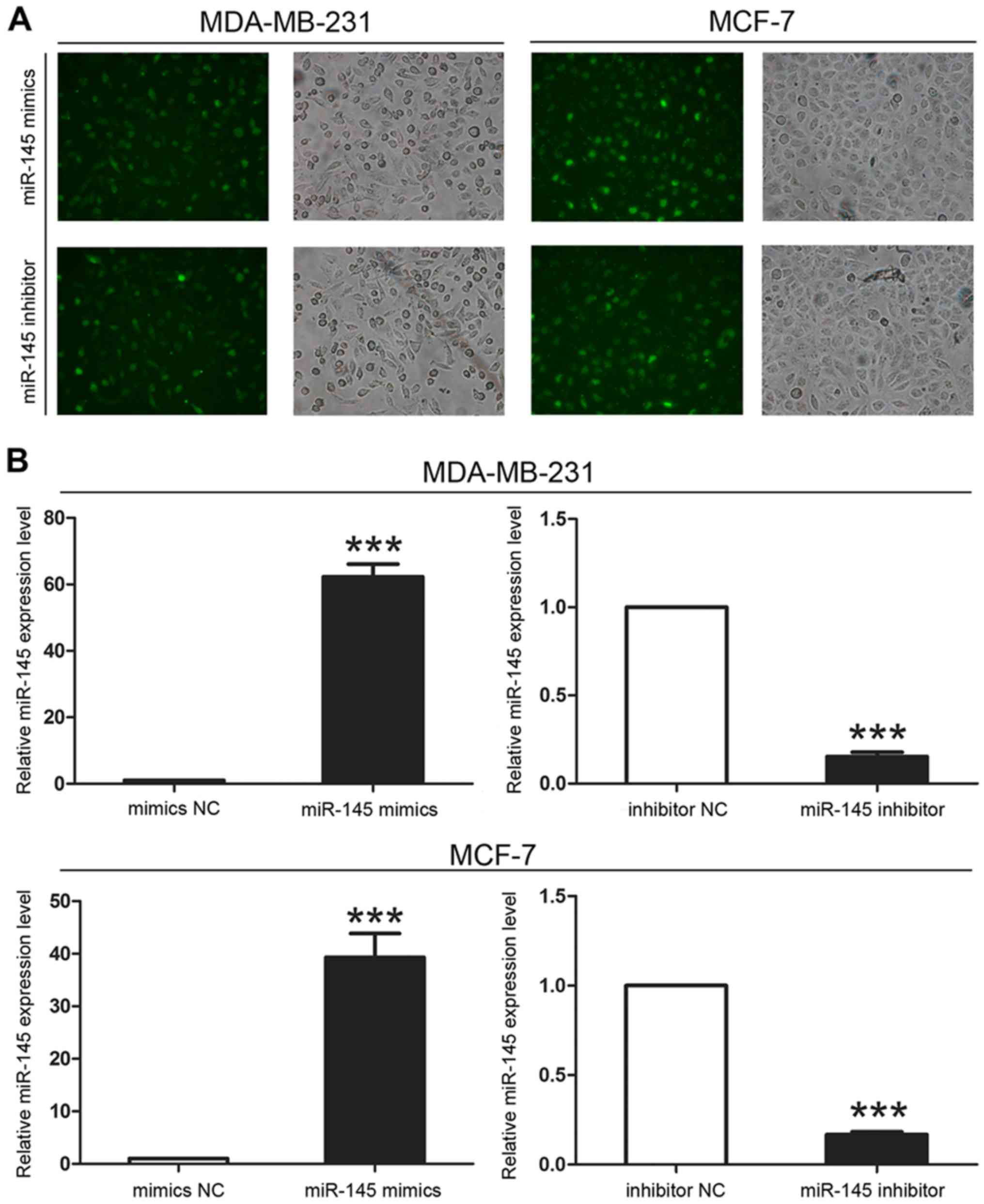
tissues (Fig. 1). We then
transfected miR-145 mimics or inhibitor into breast cancer cell
lines (MDA-MB-231 and MCF-7) to confirm their effect on miR-145
expression. After 6 h, we evaluated transfection efficiency by
fluorescence microscopy and verified miR-145 expression levels by
qRT-PCR and found that the transfection was successful (Fig. 2).
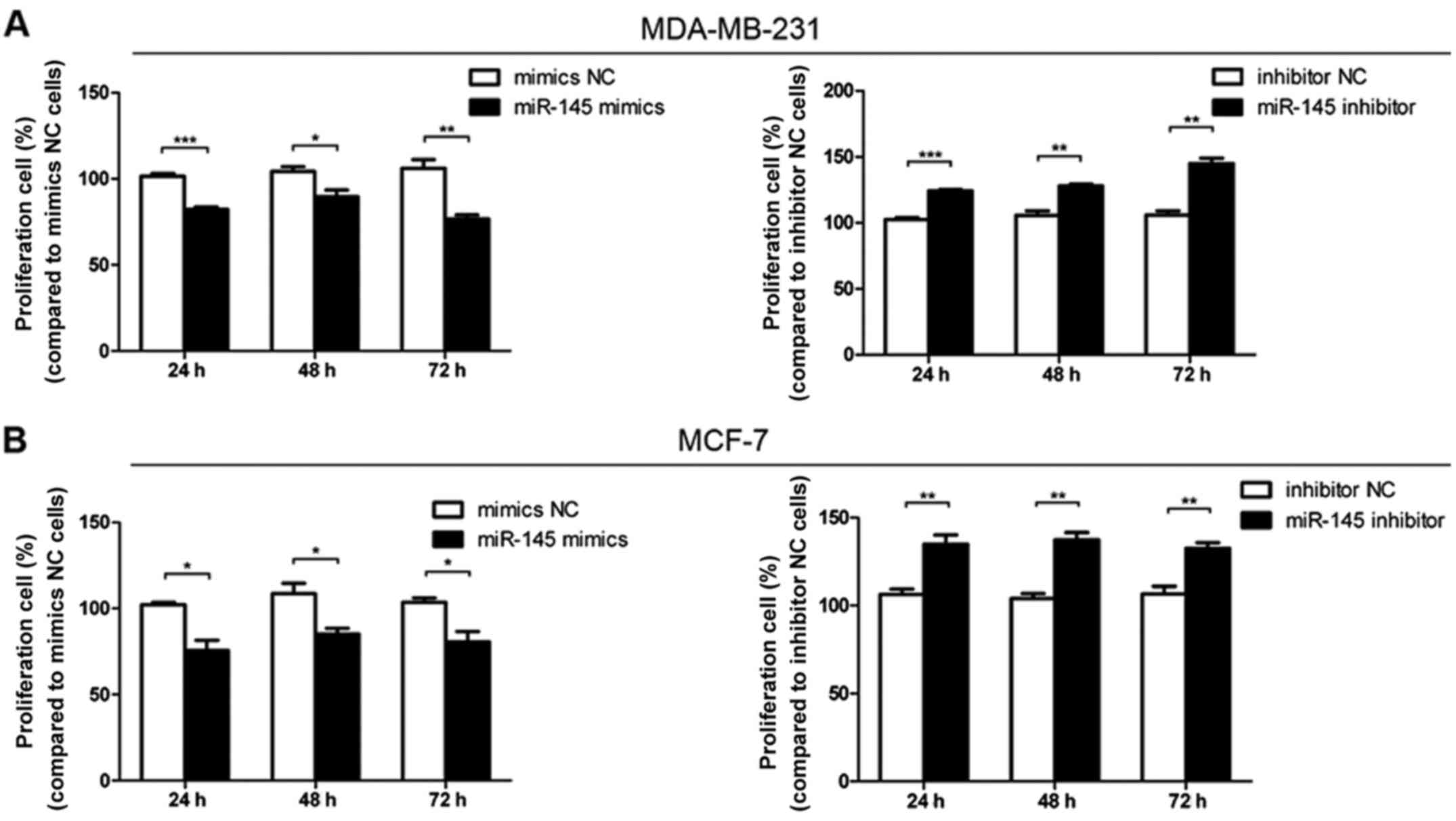
miR-145 inhibits the proliferation of
breast cancer cells
Cell proliferation assays indicated that
proliferation was inhibited when cells were transfected with
miR-145 mimics compared with mimics NC transfection (Fig. 3). Furthermore, cell proliferation
increased following transfection with miR-145 inhibitor compared
with inhibitor NC transfection (Fig.
3). These data showed that miR-145 could decrease the
proliferation of breast cancer cells.
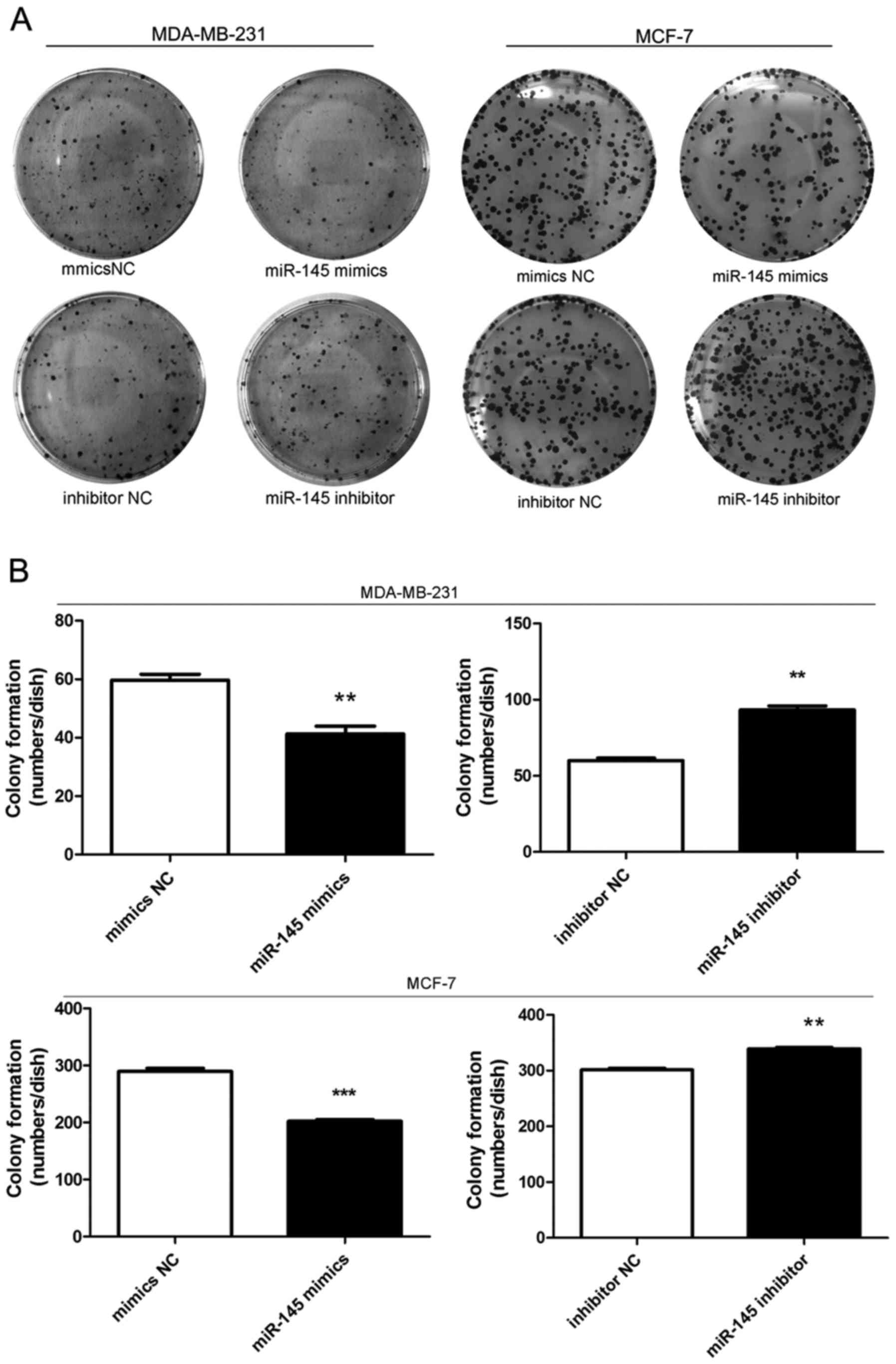
miR-145 decreases colony formation of
breast cancer cells
Colony formation assays showed that cell colony
formation was inhibited when cells were transfected with miR-145
mimics compared with mimics NC transfection (Fig. 4). In contrast, cells transfected
with miR-145 inhibitor formed more colonies than inhibitor
NC-transfected cells (Fig. 4).
These results showed that miR-145 could inhibit colony formation in
breast cancer cells.
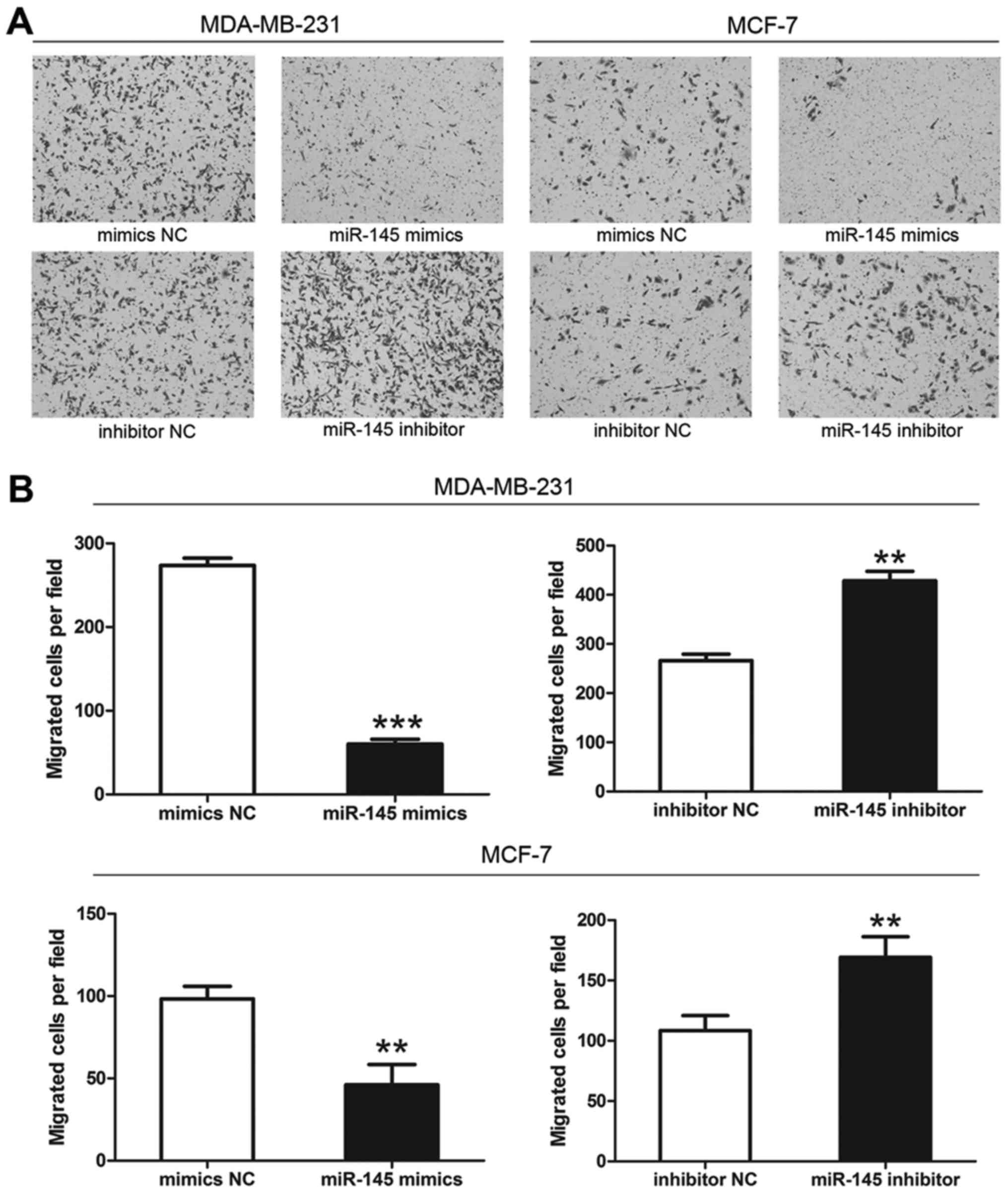
miR-145 influences the invasive ability
of breast cancer cells
Transwell invasion assays showed that the number of
membrane-penetrating cells transfected with miR-145 mimics was
significantly less than the number transfected with mimics NC
(Fig. 5). Moreover, the number of
membrane-penetrating cells transfected with miR-145 inhibitor was
significantly greater than the number of cells transfected with
inhibitor NC (Fig. 5). We
therefore concluded that miR-145 could inhibit breast cancer cell
invasion (Fig. 5).
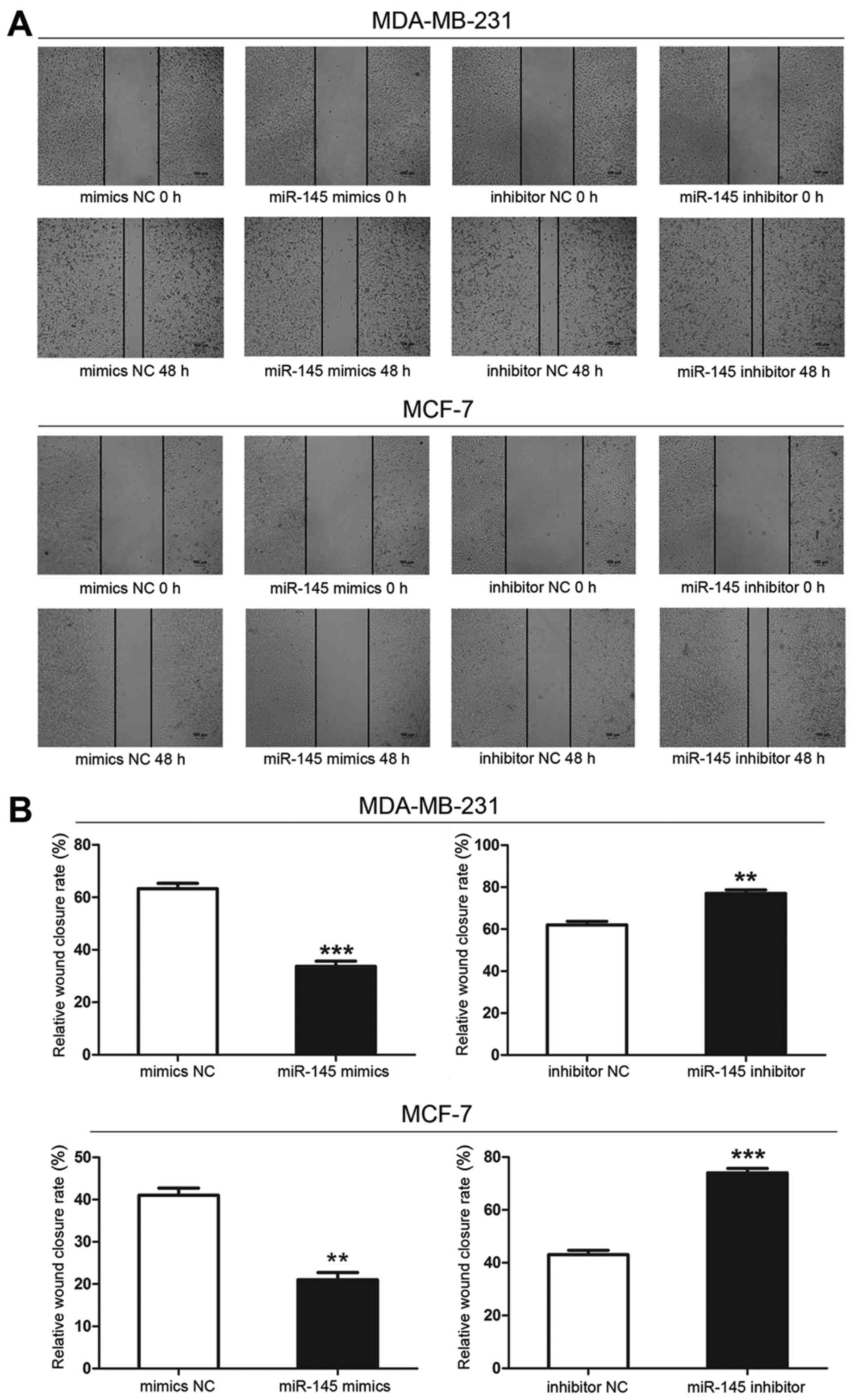
miR-145 decreases the migration of breast
cancer cells
Wound scratch assays showed that the wound size in
the miR-145-transfected group was greater at 48 h than that in the
mimics negative control groups, indicating that cells transfected
with miR-145 mimics migrated more slowly than cells transfected
with mimics NC. Cells transfected with miR-145 inhibitor migrated
the most rapidly of all transfected cells. Therefore, miR-145
decreased the migratory ability of breast cancer cells (Fig. 6).
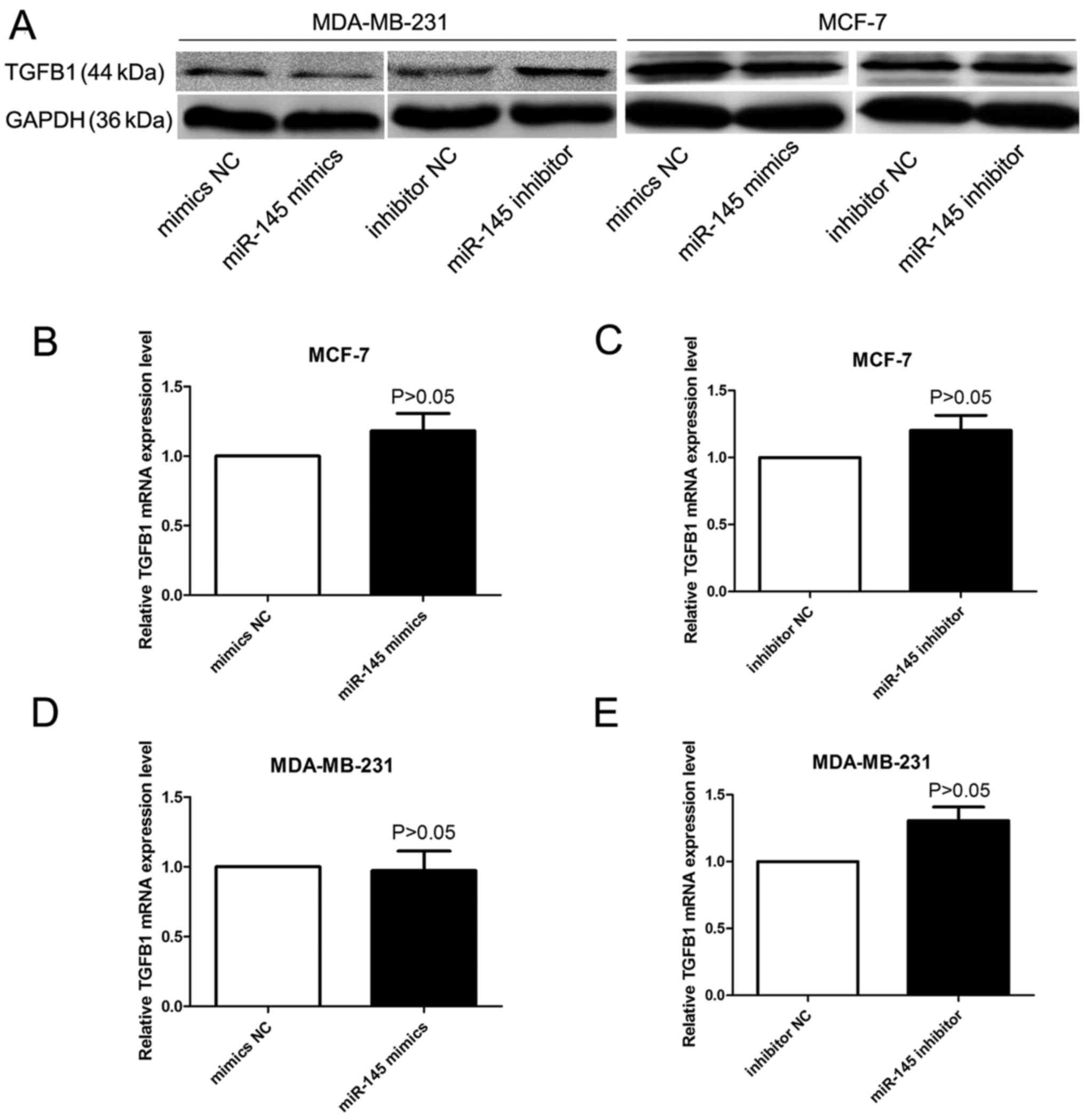
TGF-β1 mRNA and protein expression
The impact of miR-145 on TGF-β1 expression was
detected by qRT-PCR assays and western blot analysis. Although we
did not find apparent effects on TGF-β1 mRNA expression in miR-145
mimics or miR-145 inhibitor transfected cells (Fig. 7B–E), western blot analysis showed
that TGF-β1 protein expression was significantly decreased or
increased in the breast cancer cells after treatment of miR-145
mimics or miR-145 inhibitor (Fig.
7A).
Discussion
Breast cancer remains the second leading cause of
cancer-related deaths among females (1), and evidence suggests that 20–50% of
patients first diagnosed with primary breast cancer will eventually
develop metastatic disease (2).
Breast cancer is a heterogeneous disease with varied morphological
appearances, molecular features, behavior and response to therapy
(12). Unfortunately, current
treatment strategies for breast cancer metastasis still largely
rely on the use of systemic cytotoxic agents, which usually cause
severe side effects, and in many cases have limited long-term
success (2). Consequently,
developing targeted treatment for breast cancer is of great
importance (13). miRNAs are
critical regulators of gene expression. Since overexpression of
oncogenes or lower expression of tumor suppressor genes will
eventually lead to the occurrence of cancer (14), miRNA expression and cancer may be
closely linked.
Since miRNAs were first discovered in the early
1990s, they have been determined to play an increasingly important
role in the development and progression of cancer (15). In addition, abnormal miRNA
expression can affect cancer-related processes such as cell
proliferation (16), apoptosis,
invasion (16,17), and metastasis (17). Regulation of miRNA-mediated gene
expression is vitally important in many biological processes such
as inflammation and neurodegeneration (18). Recent studies have shown that some
miRNAs can function as oncogenes or tumor suppressors and regulate
cellular proliferation and apoptosis (15), but the role of miRNAs in mediating
cancer metastasis remains unexplored (17,19).
Previous studies have shown that some miRNAs can
influence the development of breast cancer, including miR-720
(20), miR-362 (21), miR-125b-1 (22) and miR-493 (23), among others. The role of miR-145
has been widely studied in cancer, and a number of studies have
shown abnormal miR-145 expression in a wide variety of tumors,
including colon cancer (24),
bladder cancer (8), retinoblastoma
(25) and colorectal cancer
(9). In this study, we used
qRT-PCR to detect the expression of miR-145 and found that miR-145
expression was significantly lower in breast cancer tissues
compared with corresponding adjacent normal tissues. We thus
hypothesized that miR-145 may act as a tumor suppressor. Cell
proliferation, invasion and migration were significantly decreased
by overexpression of miR-145 in breast cancer cells as shown by MTT
assay, colony formation assay, wound healing assay and Transwell
assay, whereas low expression of miR-145 significantly enhanced
cell growth, invasion and migration. It has been shown that miR-145
inhibits cell proliferation, invasion and metastasis by acting as a
tumor suppressor in other types of cancer, such as bladder cancer
(8), gastric cancer (26), lung cancer (27) and ovarian cancer (28). Previous studies have established
that microRNA-145 targets TRIM2 and exerts tumor-suppressing
functions in ovarian cancer (29).
Thus, these previous studies are consistent with our current data
suggesting that miR-145 plays a role as a tumor suppressor in
regulating the growth of breast cancer.
From previous studies we know that TGF-β1 may play
an important role in growth and metastasis of breast cancer cells
(11). During tumor development,
tumor cells secrete large amounts of active TGF-β1 which may
promote tumor invasion and metastasis (30). In this study, we demonstrated
TGF-β1 as a factor mediating the effect of miR-145. The human TGF-β
family comprises three isoforms: TGF-β1, TGF-β2 and TGF-β3
(31). As the major isoform,
TGF-β1 has been reported to be overexpressed in many tumors
including bladder cancer (32),
colorectal cancer (33), cervical
neoplasia (34) and pancreatic
cancer (35), and associated with
tumor transformation and progression (36). Furthermore, as a member of the
transforming growth factor family, TGF-β1 is involved in enhancing
tumor growth, progression and transformation, as well as the
invasive and metastatic potential of tumors (36–38).
Finally, we explored the relationship between
miR-145 and TGF-β1 expression. We used qRT-PCR assays and western
blot analysis to assess the impact of miR-145 on TGF-β1 expression.
We did not find apparent effects on TGF-β1 mRNA expression in
miR-145 mimics or miR-145 inhibitor transfected cells, but western
blot analysis showed that TGF-β1 protein expression was
significantly decreased or increased in the breast cancer cells
after treatment of miR-145 mimics or miR-145 inhibitor. When a
miRNA is perfectly complementary to its target, it can specifically
degrade the target mRNA (39). We
did not find hsa-miR-145 binding site of TGF-β1 mRNA, thus we did
not find apparent effects on TGF-β1 mRNA expression. The mechanism
of miR-145 inhibiting TGF-β1 protein expression requires further
study.
In conclusion, in this study, we explore the role of
miR-145 in breast cancer development and progression. Our data
suggests that miR-145 may act as a tumor-suppressor miRNA in breast
cancer through inhibiting proliferation and migration of breast
cancer cells. In addition, miR-145 inhibits the protein expression
of TGF-β1 which may in turn contribute to tumor formation.
Acknowledgments
This work was supported by the Open Projects of the
State Key Laboratory of Immunology of China (grant no. 20150511)
and the Science Foundation of Kunshan (grant no.
KS1331).
References
|
1
|
Gündüz UR, Gunaldi M, Isiksacan N, Gündüz
S, Okuturlar Y and Kocoglu H: A new marker for breast cancer
diagnosis, human epididymis protein 4: A preliminary study. Mol
Clin Oncol. 5:355–360. 2016.PubMed/NCBI
|
|
2
|
Lu J, Steeg PS, Price JE, Krishnamurthy S,
Mani SA, Reuben J, Cristofanilli M, Dontu G, Bidaut L, Valero V, et
al: Breast cancer metastasis: Challenges and opportunities. Cancer
Res. 69:4951–4953. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang C, Liu K, Li T, Fang J, Ding Y, Sun
L, Tu T, Jiang X, Du S, Hu J, et al: miR-21: A gene of dual
regulation in breast cancer. Int J Oncol. 48:161–172. 2016.
|
|
4
|
Nogales-Cadenas R, Cai Y, Lin JR, Zhang Q,
Zhang W, Montagna C and Zhang ZD: MicroRNA expression and gene
regulation drive breast cancer progression and metastasis in PyMT
mice. Breast Cancer Res. 18:752016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Carthew RW and Sontheimer EJ: Origins and
mechanisms of miRNAs and siRNAs. Cell. 136:642–655. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iorio MV, Ferracin M, Liu CG, Veronese A,
Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M,
et al: MicroRNA gene expression deregulation in human breast
cancer. Cancer Res. 65:7065–7070. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kou B, Gao Y, Du C, Shi Q, Xu S, Wang CQ,
Wang X, He D and Guo P: miR-145 inhibits invasion of bladder cancer
cells by targeting PAK1. Urol Oncol. 32:846–854. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qin J, Wang F, Jiang H, Xu J, Jiang Y and
Wang Z: MicroRNA-145 suppresses cell migration and invasion by
targeting paxillin in human colorectal cancer cells. Int J Clin Exp
Pathol. 8:1328–1340. 2015.PubMed/NCBI
|
|
10
|
Zhang Y and Lin Q: MicroRNA-145 inhibits
migration and invasion by down-regulating FSCN1 in lung cancer. Int
J Clin Exp Med. 8:8794–8802. 2015.PubMed/NCBI
|
|
11
|
Park SJ, Kim JG, Kim ND, Yang K, Shim JW
and Heo K: Estradiol, TGF-β1 and hypoxia promote breast cancer
stemness and EMT-mediated breast cancer migration. Oncol Lett.
11:1895–1902. 2016.PubMed/NCBI
|
|
12
|
Rakha EA, Reis-Filho JS, Baehner F, Dabbs
DJ, Decker T, Eusebi V, Fox SB, Ichihara S, Jacquemier J, Lakhani
SR, et al: Breast cancer prognostic classification in the molecular
era: The role of histological grade. Breast Cancer Res. 12:2072010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu K, Zhang C, Li T, Ding Y, Tu T, Zhou
F, Qi W, Chen H and Sun X: Let-7a inhibits growth and migration of
breast cancer cells by targeting HMGA1. Int J Oncol. 46:2526–2534.
2015.PubMed/NCBI
|
|
14
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer.
94:776–780. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hiyoshi Y, Kamohara H, Karashima R, Sato
N, Imamura Y, Nagai Y, Yoshida N, Toyama E, Hayashi N, Watanabe M,
et al: MicroRNA-21 regulates the proliferation and invasion in
esophageal squamous cell carcinoma. Clin Cancer Res. 15:1915–1922.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Vaknin-Dembinsky A, Charbit H, Brill L,
Abramsky O, Gur-Wahnon D, Ben-Dov IZ and Lavon I: Circulating
microRNAs as biomarkers for rituximab therapy, in neuromyelitis
optica (NMO). J Neuroinflammation. 13:1792016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Y, Deng X, Zeng X and Peng X: The role
of miR-148a in cancer. J Cancer. 7:1233–1241. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li LZ, Zhang CZ, Liu LL, Yi C, Lu SX, Zhou
X, Zhang ZJ, Peng YH, Yang YZ and Yun JP: miR-720 inhibits tumor
invasion and migration in breast cancer by targeting TWIST1.
Carcinogenesis. 35:469–478. 2014. View Article : Google Scholar
|
|
21
|
Ni F, Gui Z, Guo Q, Hu Z, Wang X, Chen D
and Wang S: Downregulation of miR-362-5p inhibits proliferation,
migration and invasion of human breast cancer MCF7 cells. Oncol
Lett. 11:1155–1160. 2016.PubMed/NCBI
|
|
22
|
Cisneros-Soberanis F, Andonegui MA and
Herrera LA: miR-125b-1 is repressed by histone modifications in
breast cancer cell lines. Springerplus. 5:9592016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao L, Feng X, Song X, Zhou H, Zhao Y,
Cheng L and Jia L: miR-493-5p attenuates the invasiveness and
tumorigenicity in human breast cancer by targeting FUT4. Oncol Rep.
36:1007–1015. 2016.PubMed/NCBI
|
|
24
|
Li C, Xu N, Li YQ, Wang Y and Zhu ZT:
Inhibition of SW620 human colon cancer cells by upregulating
miRNA-145. World J Gastroenterol. 22:2771–2778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Sun Z, Zhang A, Jiang T, Du Z, Che C and
Wang F: MiR-145 suppressed human retinoblastoma cell proliferation
and invasion by targeting ADAM19. Int J Clin Exp Pathol.
8:14521–14527. 2015.
|
|
26
|
Jiang SB, He XJ, Xia YJ, Hu WJ, Luo JG,
Zhang J and Tao HQ: MicroRNA-145-5p inhibits gastric cancer
invasiveness through targeting N-cadherin and ZEB2 to suppress
epithelial-mesenchymal transition. Onco Targets Ther. 9:2305–2315.
2016.PubMed/NCBI
|
|
27
|
Li Y, Li Y, Liu J, Fan Y, Li X, Dong M,
Liu H and Chen J: Expression levels of microRNA-145 and
microRNA-10b are associated with metastasis in non-small cell lung
cancer. Cancer Biol Ther. 17:272–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong R, Liu X, Zhang Q, Jiang Z, Li Y, Wei
Y, Li Y, Yang Q, Liu J, Wei JJ, et al: miR-145 inhibits tumor
growth and metastasis by targeting metadherin in high-grade serous
ovarian carcinoma. Oncotarget. 5:10816–10829. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chen X, Dong C, Law PT, Chan MT, Su Z,
Wang S, Wu WK and Xu H: MicroRNA-145 targets TRIM2 and exerts
tumor-suppressing functions in epithelial ovarian cancer. Gynecol
Oncol. 139:513–519. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ding MJ, Su KE, Cui GZ, Yang WH, Chen L,
Yang M, Liu YQ and Dai DL: Association between transforming growth
factor-β1 expression and the clinical features of triple negative
breast cancer. Oncol Lett. 11:4040–4044. 2016.PubMed/NCBI
|
|
31
|
Zheng W: Genetic polymorphisms in the
transforming growth factor-beta signaling pathways and breast
cancer risk and survival. Methods Mol Biol. 472:265–277. 2009.
View Article : Google Scholar
|
|
32
|
Eder IE, Stenzl A, Hobisch A, Cronauer MV,
Bartsch G and Klocker H: Transforming growth factors-beta 1 and
beta 2 in serum and urine from patients with bladder carcinoma. J
Urol. 156:953–957. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsushima H, Kawata S, Tamura S, Ito N,
Shirai Y, Kiso S, Imai Y, Shimomukai H, Nomura Y, Matsuda Y, et al:
High levels of transforming growth factor beta 1 in patients with
colorectal cancer: Association with disease progression.
Gastroenterology. 110:375–382. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Comerci JT Jr, Runowicz CD, Flanders KC,
De Victoria C, Fields AL, Kadish AS and Goldberg GL: Altered
expression of transforming growth factor-beta 1 in cervical
neoplasia as an early biomarker in carcinogenesis of the uterine
cervix. Cancer. 77:1107–1114. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Friess H, Yamanaka Y, Büchler M, Ebert M,
Beger HG, Gold LI and Korc M: Enhanced expression of transforming
growth factor beta isoforms in pancreatic cancer correlates with
decreased survival. Gastroenterology. 105:1846–1856. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sheen-Chen SM, Chen HS, Sheen CW, Eng HL
and Chen WJ: Serum levels of transforming growth factor beta1 in
patients with breast cancer. Arch Surg. 136:937–940. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen JY, Liu JH, Wu HD, Lin KH, Chang KC
and Liou YM: Transforming growth factor-β1 T869C gene polymorphism
is associated with acquired sick sinus syndrome via linking a
higher serum protein level. PLoS One. 11:e01586762016. View Article : Google Scholar
|
|
38
|
Kelly RJ and Morris JC: Transforming
growth factor-beta: A target for cancer therapy. J Immunotoxicol.
7:15–26. 2010. View Article : Google Scholar
|
|
39
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA miR-196 acts upstream of Hoxb8 and Shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|