Introduction
Testicular germ cell tumors (TGCTs) are the most
common solid tumors among adolescent and young adult male, of which
the incidence rates is actually increasing every year (1). Seminoma is the most common testicular
germ cell tumor which often develops in the cryptorchid testis and
occurs between 35 and 45 years (2). As we know, seminoma, like other
cancers, is considered as a histologically heterogeneous disease in
which gene aberrations have a prominent role in cancer occurrence,
progression, and metastasis (3,4).
Emerging evidence has revealed that multiple genes and molecular
pathways participate in the initiation and progression of human
cancers. Targeting of hub genes and key molecular pathways has been
recognized as a promising approach in the discovery and treatment
of cancer (5). However, only few
aspects of the mechanism and gene expression of this cancer are
well studied and remain largely unclear. Therefore, to get a better
understanding of the genetic etiology and molecular mechanism
involved in the occurrence, progression, and metastasis of seminoma
is extremely vital for acquiring more effective diagnostic
biomarkers and therapeutic strategies.
The conventional serum diagnostic markers, such as
α-fetoprotein (AFP), human chorionic gonadotrophin (hCG) and
lactate dehydrogenase (LDH), show some utility in the diagnosis and
follow-up purposes of TGCT (6).
However, AFP and hCG also exhibit certain limited sensitivity and
specificity, being indicative of yolk sac tumor (AFP) and
choriocarcinoma or syncytiotrophoblast (hCG) subtypes. Furthermore,
LDH is recognized as a very nonspecific biomarker. Therefore,
seminomas and non-seminomatous GCTs (NSGCTs) comprising a pure
embryonal carcinoma subtype are often negative for these
conventional markers (7–9).
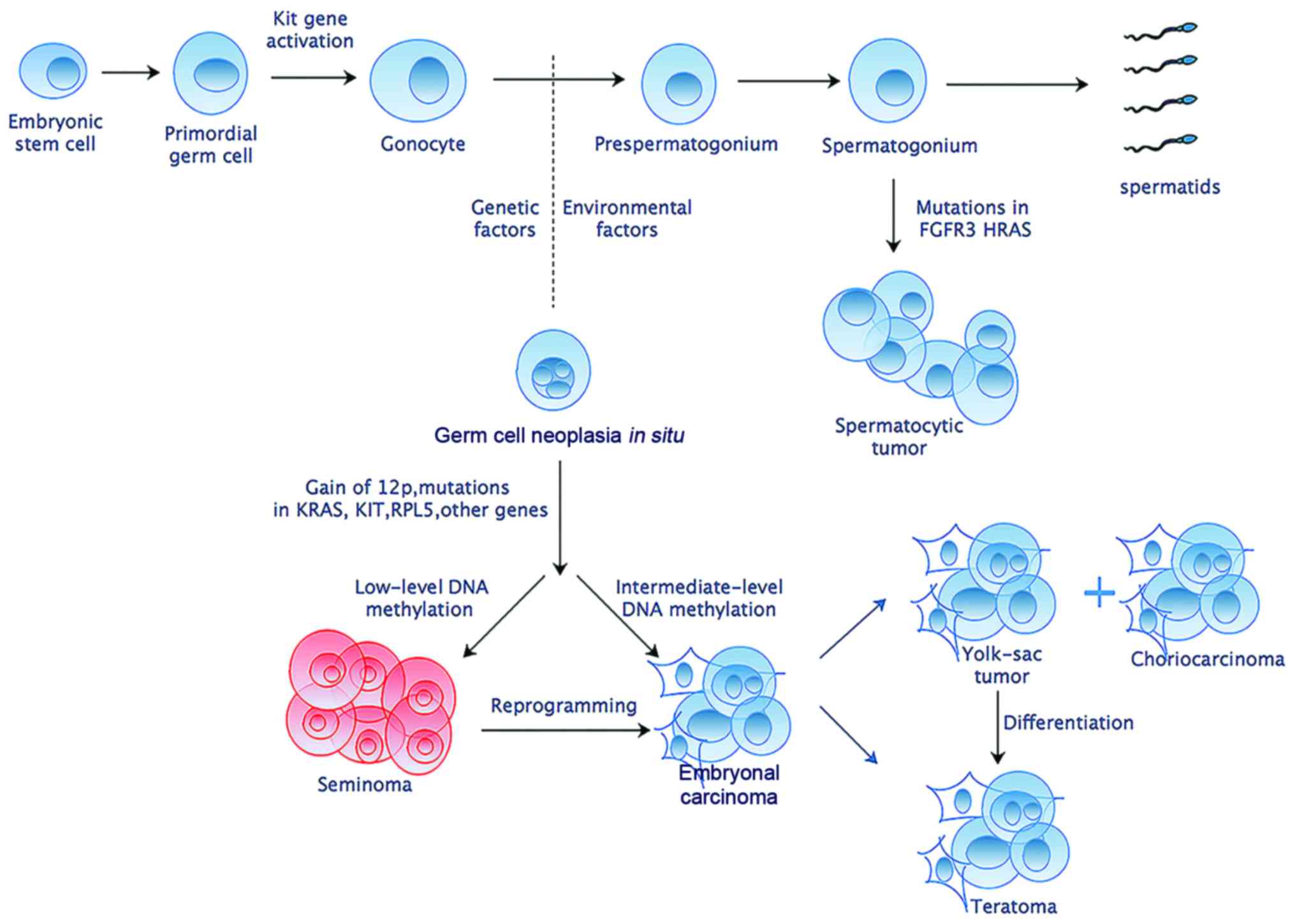
Accumulated studies have shed light on the
transformation of normal gonocytes into malignant germ cell tumors
(Fig. 1). Gain of chromosome arm
12p is considered to have a highest effect and be nearly universal
in TGCTs (10,11). Despite germline genome wide
association studies have confirmed several risk loci for TGCTs,
only Kit and Ras gene family have been implicated repeatedly in
different TGCTs (12). Besides,
genome-wide microarray screens have identified some elevated
expression of embryonic pluripotency-related genes such as
Pou5f1, Lin28, Nanog and Tfap2c in germ cell
neoplasia in situ, embryonal carcinoma and seminoma
(4). Despite these former efforts
to discover genetic foci of susceptibility, no validated molecular
biomarkers exist that can be used for precise screening, diagnostic
or therapeutic purposes.
In this study, we focused on the most common
testicular germ cell tumors - seminoma (roughly 56% of cases, peak
incidence at 35 years (13). We
used a systematic approach that can be used for acquiring novel
molecular biomarkers for seminoma. Based on data from GEO online
database, we explored the differently expressed genes (DEGs),
related molecular pathways and consequently constructed a
regulatory network. Then the top 10 hub genes were chosen and
determined by RT-qPCR and IHC assay, which could be used as
potential biomarkers for diagnosis and may also be related with
prognosis. Furthermore, these targets could possibly also give us a
novel insight into seminoma pathogenesis.
Materials and methods
Microarray data
The gene expression profiles of GSE18155, including
12 seminoma samples and 5 normal testis samples, were obtained from
National Center of Biotechnology Information (NCBI) GEO database
(GEO, http://www.ncbi.nlm.nih.gov/geo/). The GSE18155, which
was based on GPL96 [HG-U133A] Affymetrix Human Genome U133A array,
was submitted to the database by Matthew Jonathan Murray et
al.
Differentially expressed gene
analysis
The raw data were analyzed by using GCBI online
software (https://www.gcbi.com.cn/gclib/html/index). Upregulated
and downregulated genes were identified between seminoma and normal
controls. A classical criteria of t-test was used to identify DEGs
with a change ≥2-fold and defined a P-value cutoff <0.05 to be
statistically significant.
Functional and pathway enrichment
analysis of DEGs
It is well known that Database for Annotation
Visualization and Integrated Discovery (DAVID) is a common useful
method to perform Gene ontology analysis (GO) and Kyoto
Encyclopedia of Genes and Genomes (KEGG). We chose the human genome
as the background list and human was used as the species. P<0.05
was considered statistically significant.
PPI network construction and selection of
modules
We used online tool Search for the Retrieval of
Interacting Genes (STRING) database (http://www.string-db.org) to evaluate the
protein-protein interaction (PPI) information. To identify the
interactive relationship among DEGs, we imported the DEGs to STRING
and only experimentally validated interactions with a combined
score >0.7 was selected as significant. Then these significant
DEGs were mapped into Cytoscape plugin to create network
visualizations. Finally, we put the resulting PPI network to module
analysis with the Plugin MCODE with the default parameters (Degree
cutoff ≥2, Node score cutoff ≥2, K-core ≥2, and Max depth=100).
Moreover, the function and pathway enrichment analysis were
performed for DEGs in the modules. P<0.05 was considered to be
significant.
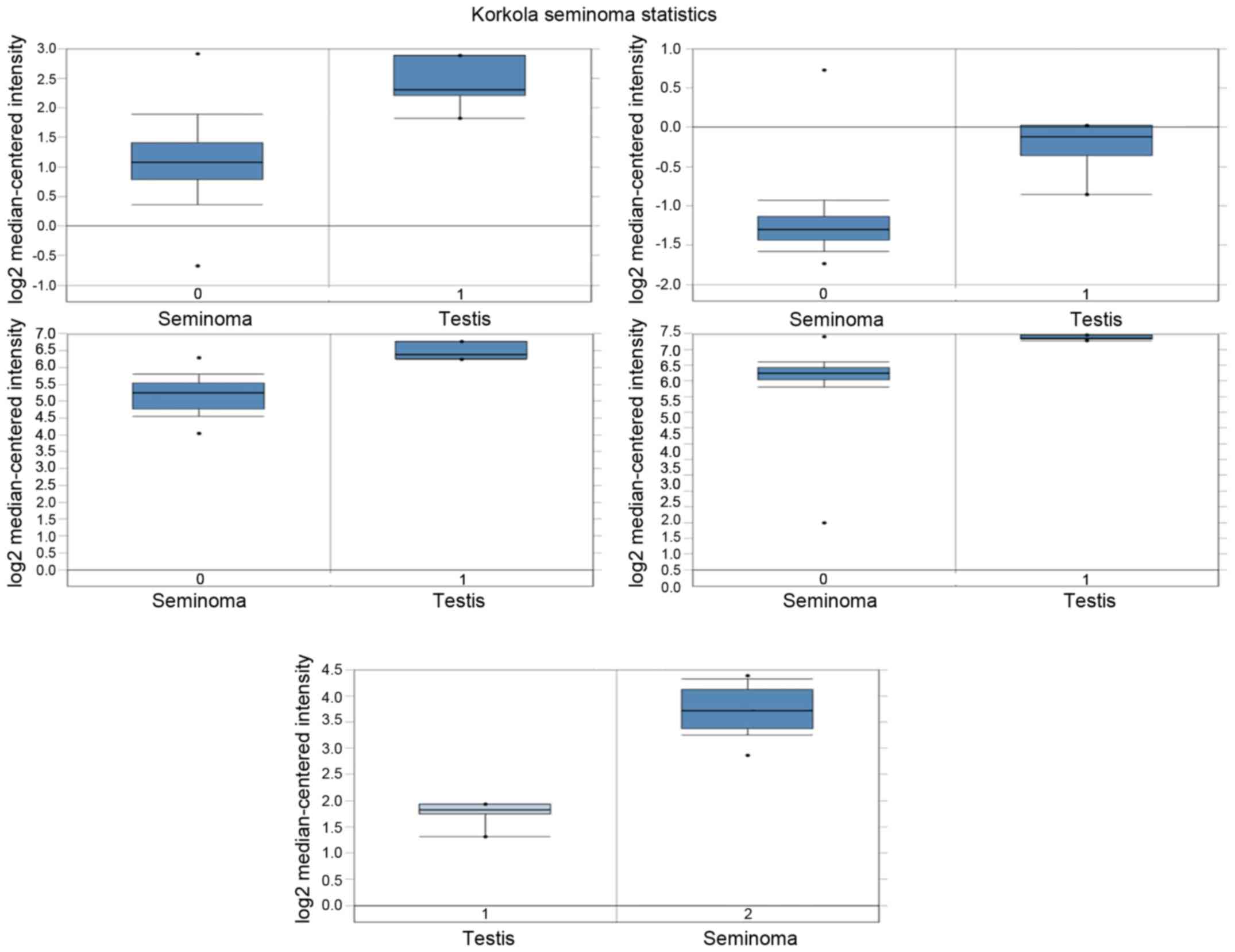
Analysis of hub gene mRNA and protein
expression in human seminoma
Top 5 hub gene protein expression in seminoma
tissues and normal tissues was determined from the human protein
atlas (www.proteinatlas.org). Meanwhile,
mRNA expression was determined via analysis of Korkola Seminoma
Statistics, which are available through Oncomine (Compendia
Biosciences, www.oncomine.org). High and low
groups were defined as above and below the mean, respectively.
Clinical specimens
Five tumor samples and paired adjacent non-cancerous
tissues were obtained from 5 seminoma patients subjected to
orchiectomy in the Sir Run Run Hospital of Zhejiang University who
were diagnosed with seminoma by more than two pathologists.
Quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from patient tissues using
TRIzol reagent (Invitrogen, USA) and was stored at −80°C until use.
Reverse Transcription system (Promega) was used for cDNA synthesis
according to the manufacturer's protocol. The mRNA expression
levels of hub genes were measured by quantitative real-time PCR
using the ABI PRISM 7500 Sequence Detector system (Applied
Biosystems, USA), and was normalized to an internal standard
(glyceraldehyde-3-phosphate dehydrogenase, GAPDH). PCR primer used
were as follows: AR: forward, 5′-TACCG CATGCACAAGTCCCG-3′; reverse,
5′-TCACTGGGTGTGGAAATAGA-3′. UBB: forward,
5′-GGTGAGCTTGTTTGTGTCCCTGT-3′; reverse,
5′-TCCACCTCAAGGGTGATGGTC-3′. UBC: forward, 5′-TGCACCTGGTACTCCGTC
TCA-3′; reverse, 5′-CAGTGAGTGTCTTCACGAAGATTTG-3′. CDK4: forward,
5′-ATGGCTACCTCTCGATATGAGC-3′; reverse, 5′-CATTGGGGACTCTCACACTCT-3′.
PTEN: forward, 5′-ACCCACACGACGGGAAGACA-3′; reverse,
5′-CTGTTTGTGGAAGAACTCTACTTTGATATCAC-3′. GAPDH: forward,
5′-AGACAGCCGCATCTTCTTGT-3′; reverse, 5′-TGATGGCAACAATGTCCACT-3′.
The reaction protocol involved heating for 3 min at 95°C, followed
by 45 cycles of amplification (15 sec at 95°C and 1 min at 60°C).
All reactions were done in triplicate.
Results
Identification of DEGs in seminoma
In order to reveal the different expression of genes
involved in seminoma, we selected the publicly available microarray
datasets (12 seminoma samples and 5 normal samples) from the GEO
database and used the GCBI analysis to identify DEGs between them.
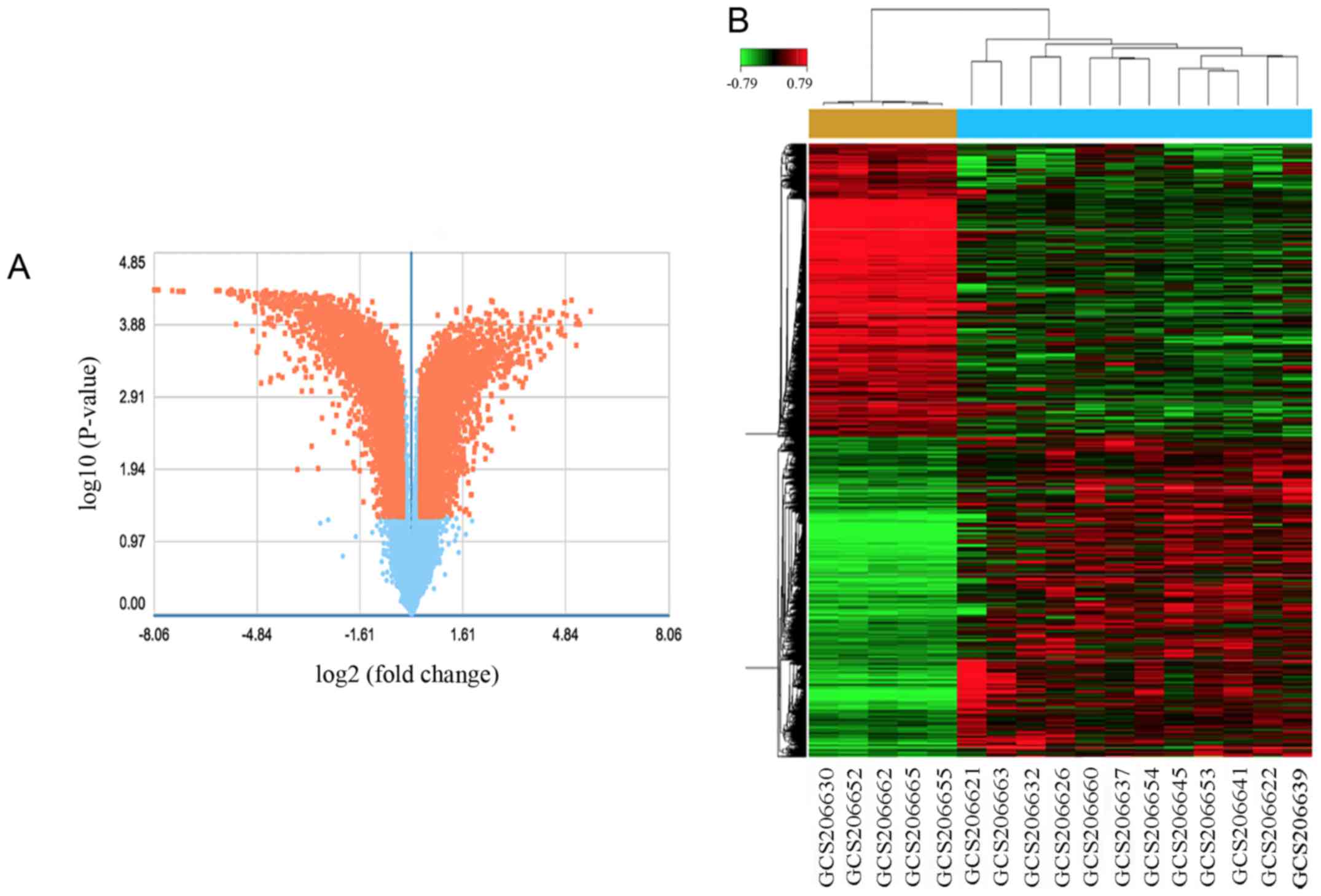
With the criteria P<0.05 and fold control (FC) ≥2, a total of
3,502 genes were identified after the analysis of GSE18155, among
which, 1,563 were upregulated and 1,939 were downregulated. DEGs
expression volcano plot and heat map are presented in Fig. 2.
GO term enrichment analysis
DEGs (3,502) were then used for functional analysis
via online biological classification tool DAVID (14). GO analysis was carried out which
covered three aspects, named molecular function (MF), cellular
component (CC) and biological process (BP) (15). The results revealed that DEGs were
enriched in 873 BP terms, 250 CC terms and 208 MF terms under the
condition of FDR<0.05, P<0.05. GO analysis results showed
that upregulated DEGs were significantly enriched in biological
processes (BP), including immune response, cell adhesion, cell
migration and cell proliferation (Table I). The downregulated DEGs were
significantly enriched in biological processes, including cell
circle, spermatogenesis, and reproduction (Table I). For molecular function (MF), the
upregulated DEGs were enriched in protein binding and poly(A) RNA
binding, and the downregulated DEGs were enriched in protein
binding and ATP binding (Table I).
Besides, GO cell component (CC) analysis showed that the
upregulated DEGs were significantly enriched in the cytoplasm,
extracellular exosome, while downregulated DEGs were enriched in
cytoplasm and nucleus (Table
I).
 | Table IGene ontology analysis of
differentially expressed genes associated with seminoma. |
Table I
Gene ontology analysis of
differentially expressed genes associated with seminoma.
| Expression | Category | Go term | Gene count % | P-value |
|---|
| Upregulated | GOTERM_BP_FAT | Immune
response | 23.3 | 2.0E-57 |
| GOTERM_BP_FAT | Cell adhesion | 19.1 | 3.3E-26 |
| GOTERM_BP_FAT | Cell
proliferation | 18.2 | 1.1E-17 |
| GOTERM_BP_FAT | Cell migration | 13.0 | 5.3E-17 |
| GOTERM_CC_FAT | Cytoplasm | 36.5 | 1.1E-12 |
| GOTERM_CC_FAT | Plasma
membrane | 27.4 | 7.5E-7 |
| GOTERM_CC_FAT | Extracellular
exosome | 27.3 | 1.9E-31 |
| GOTERM_MF_FAT | Protein
binding | 61.8 | 1.2E-25 |
| GOTERM_MF_FAT | PolyA RNA
binding | 12.2 | 1.4E-15 |
| GOTERM_MF_FAT | Cadherin binding in
cell adhesion | 4.30 | 1.4E-10 |
| Downregulated | GOTERM_BP_FAT | Reproduction | 14.7 | 3.7E-26 |
| GOTERM_BP_FAT | Cell cycle | 13.9 | 9.8E-12 |
| GOTERM_BP_FAT |
Spermatogenesis | 9.10 | 6.0E-40 |
| GOTERM_BP_FAT | Male gamete
generation | 9.10 | 7.6E-40 |
| GOTERM_CC_FAT | Cytoplasm | 36.5 | 2.0E-19 |
| GOTERM_CC_FAT | Nucleus | 35.2 | 8.4E-12 |
| GOTERM_CC_FAT | Cytosol | 23.9 | 3.4E-13 |
| GOTERM_MF_FAT | Protein
binding | 56.5 | 4.8E-20 |
| GOTERM_MF_FAT | ATP binding | 10.7 | 2.4E-5 |
| GOTERM_MF_FAT | Ubiquitin-protein
transferase activity | 3.4 | 3.3E-6 |
KEGG pathway analysis
To further reveal the functions of DEGs, the DEGs
with FDR <0.05 were entered into DAVID for KEGG pathway
enrichment analysis (16). The
most significantly enriched molecular pathways of the upregulated
DEGs and downregulated DEGs analyzed by KEGG are showed in Table II, from which we can draw the
conclusion that the upregulated DEGs were enriched in cell adhesion
molecules, PI3K-AKT signaling pathway, pathways in cancer, p53
signaling pathway, and NF-κB signaling pathway, while the
downregulated DEGs were enriched in metabolic pathway, cGMP-PKG
signaling pathways, FoxO signaling pathways and Wnt signaling
pathway (Table II).
 | Table IIKEGG pathway analysis of
differentially expressed genes associated with seminoma. |
Table II
KEGG pathway analysis of
differentially expressed genes associated with seminoma.
| Expression | Pathway | Genes | Gene count % | P-value |
|---|
| Upregulated | Cell adhesion
molecules | CD2CD4, CD86, CD8A,
CDH1, CDH3, CDH5, CLDN6, CLDN7, ITGA4, SDC2, SDC3, VCAM, ITGAL,
ITGAM, ITGB2, ICAM1, ICAM2, ICAM3, HLA-A, HLA-B, HLA-C, SELPLG,
SIGLEC1, HLA-E, HLA-F, HLA-G, HLA-DMA, HLA-DMB, HLA-DPA1, HLDPB1,
PTPRF, SELL, HLA-DQB1, HLA-DRA, HLA-DRB1, NLGN4X, NRCAM, | 3.5 | 1.9E-11 |
| PI3K-AKT signaling
pathway | MCL1, KIT, KRAS,
MyB, TCL1A, TCL1B, CREB3L2, COL3A1, COL4A1, TP53, COL4A2, COL5A1,
COL6A2, CCND2, CCND3, CDK2, CDK4, EFNA4, EIF4B EIF4EBP1, FGF13,
FGFR1, FGFR2, FN1, GyS1, HSP90AB1, IGF1, ITGA4 | 3.7 | 1.5E-2 |
| Pathways in
cancer | ITGA5, IFNAR2,
IL2RB, IL2RG, IL6R, LPAR6, PIK3CD, RAC1, yWHAZ, BAX, CXCL12, CXCR4,
CEBPA, E2F3, KIT, KRAS, NFKBIA, SKP2, SMAD3, WNT2B, ADCy1, ADCy7,
CDH1, COL4A1, COL4A2, CSF1R, CDK2, CDK4, FGF13, FGFR1, FGFR2, FN1,
FZD5, HSP90AB1, GF1, LPAR6, MMP2, MMP9 | 3.7 | 9.3E-2 |
| Cell cycle | E2F3, SKP2, SMAD3,
CDC25B, CCNA2, CCND2, CCND3, CDK2, CDK4, MCM2, yWHAZ, MCM3, MCM4,
MCM5, MCM6, MCM7, PRKDC, SFN, STAG2, SMC1A, TGFB1, TP53 | 2.1 | 2.1E-4 |
| NF-κB signaling
pathway | BCL2A1, CCL19,
CCL4, CXCL12, CFLAR, CD14, ERC1, LCK, LyN, NFKBIA, ICAM1 LAT, Ly96,
LTB, PLCG2, PLAU, VCAM1 | 1.4 | 4.0E-3 |
| Downregulated | Metabolic
pathways | ADO, HMGCS1,
HMGCS2, HIBCH, ABAT, DHCR7, ALG8, ALG9, CDS1, NME5, NME7, OCRL,
UAP1, ACAT1, ACO1, ACSBG2, ACSL6, AK1, ADSS, ALDH1A1, ALDH1A2,
ALDH3A2, ALDH6A1, ALDH9A1, AGPS, ALLC, ASS1, B3GALT4, BCAT1, CERS1,
CHKA, CHPT1, CHPF, CKB, CKM, CyP11A1 | 8 | 6.0E-4 |
| cGMP-PKG signaling
pathway | ATP1A3, ATP2B4,
GNA11, GNAI1, ATF2, ADORA1, ADCy2, CREB1, CALM1, IRS2, MyL9,
MyLK3MyLK, PIK3R3, PLN, PPP1R12A, PPP3CA, PPP3CB, PPP3CC, SLC25A31,
SLC25A4 | 1.5 | 1.5E-1 |
| Oocyte meiosis | SKP1, ADCy2, ADCy9,
AR, CALM1, CDC25C, CDC27, CUL1, PTTG1, PGR, PRKACG, PPP1CC,
PPP2R5C, PPP2R1B, PPP3CA, PPP3CC, STAG3, SMC3, yWHAZ, | 1.4 | 1.5E-4 |
| FoxO signaling
pathway | BRAF, BCL6,
BCL2L11, GABARAP, RBL2, SMAD4, CDKN1A, CDKN2D, FOXG1, IRS2, MAPK13,
MAPK8, PTEN, PLK2, PLK4, PRKAA1, SOS2 | 1.2 | 1.1E-2 |
| Wnt signaling
pathway | SKP1, SMAD4,
CSNK2A1, CSNK2A2, CSNK2B, CUL1, DAAM1, DAAM2, LEF1, MAPK8, PRKCB,
PRKACG, PPP3CA, PPP3CB | 1.1 | 5.5E-2 |
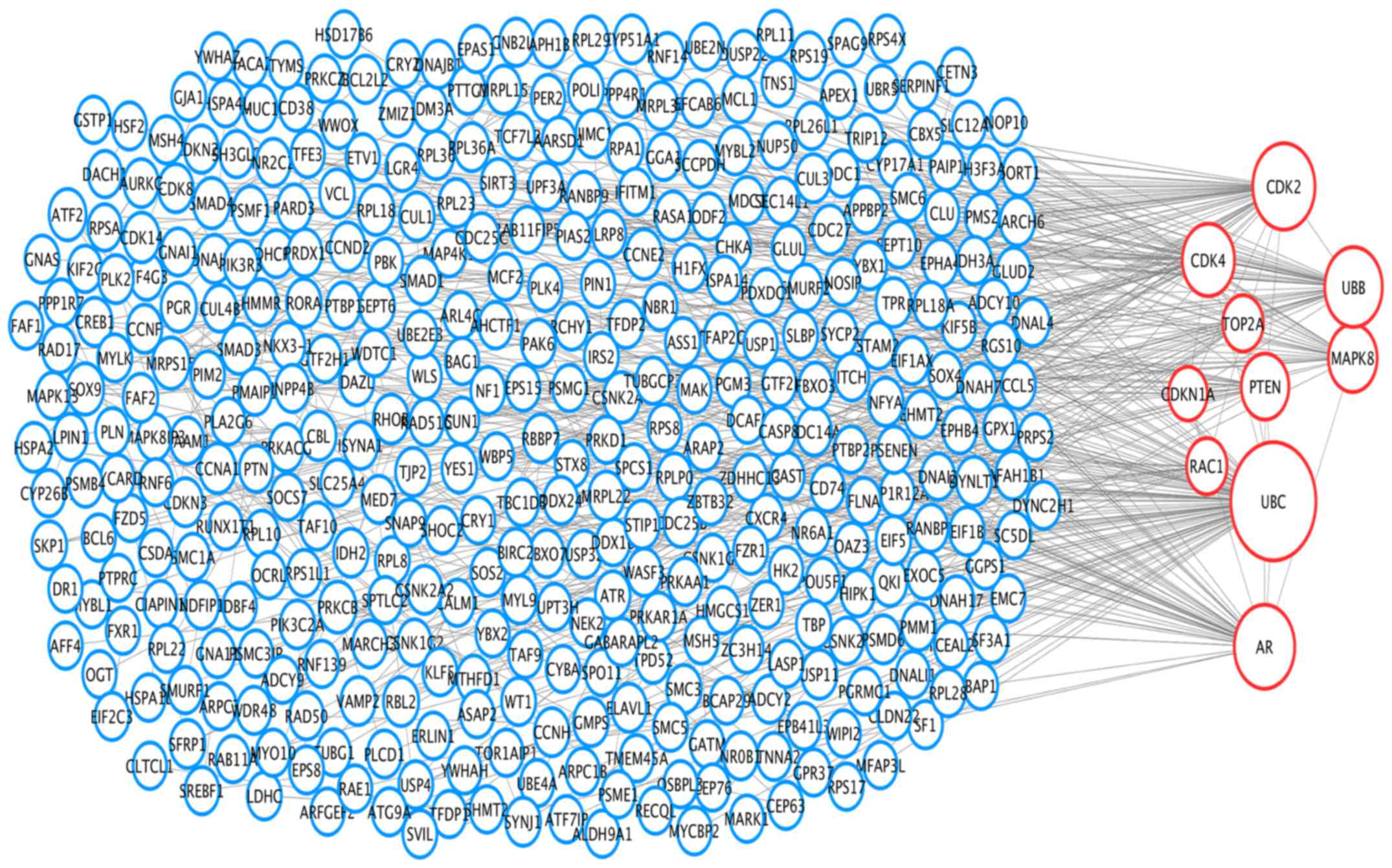
PPI networks and module analysis
To analyze the interaction and acquire hub genes of
potentially diagnosis-related DEGs, protein-protein interactome was
constructed using STRING (17).
Then we put the genes with combined score ≥0.7 into Cytoscape for
further analysis. The PPI network included 499 nodes and 577
interactions (Fig. 3). The top 10
hub nodes with higher degrees were screened, including ubiquitin C
(Ubc), ubiquitin B (Ubb), androgen receptor (Ar), phosphatase and
tensin homolog (Pten), cyclin-dependent kinase 2 (Cdk2),
cyclin-dependent kinase 4 (Cdk4), mitogen-activated protein kinase
8 (Mapk8), topoisomerase (DNA) IIα (Top2a), cyclin-dependent kinase
inhibitor 1a (Cdkn1a), ras-related C3 botulinum toxin substrate 1
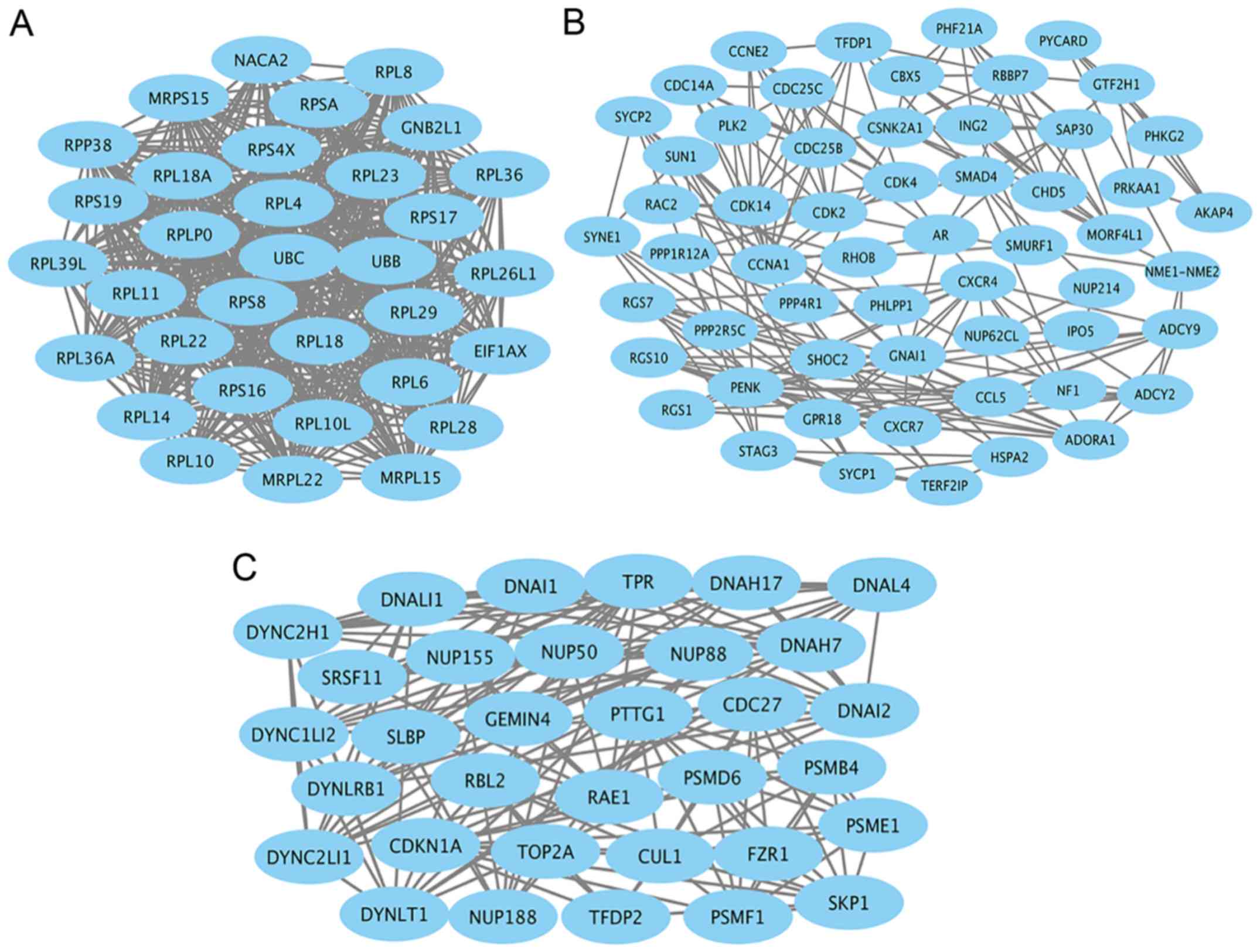
(Rac1) (Fig. 3). Module genes of
PPI analysis were identified by MCODE. The top 3 significant
modules (Fig. 4) were selected and
the functional annotation of the genes involved in the top 3
modules were analyzed by DAVID (Tables IIITable IV–V), showing that module genes were related
mainly to the cell cycle, cell proliferation, pathways in cancer,
and PI3K-AKT signaling pathway, all of which have a close relation
with tumor biology and could probably affect survival and prognosis
in patients with seminoma.
 | Table IIIFunctional annotation of the
significant module 1. |
Table III
Functional annotation of the
significant module 1.
| Category | Term | Gene | P-value |
|---|
| GOTERM_BP_FAT | Translation | EIF1AX, MRPL15,
MRPL22, MRPS15, RPL10L, RPL11, RPL14, RPL18A, RPL22, RPL23,
RPL26L1, RPL28, RPL29, RPL36, RPL36A | 2.4E-33 |
| GOTERM_BP_FATE | RBB2 signaling
pathway | UBC, UBB | 6. 4E-2 |
| KEEN PATHWAy | Ribosome | MRPL15, MRPL22,
MRPS15, RPL10L, RPL11, RPL14, RPL18, RPL23, RPL26, L1, RPL28,
RPL29, RPL36, RPL36A, RPL4, RPL6, RPL8 | 2.6E-38 |
 | Table IVFunctional annotation of the
significant module 2. |
Table IV
Functional annotation of the
significant module 2.
| Category | Term | Gene | P-value |
|---|
| GOTERM_BP_FAT | Cell cycle | SUN1, CSNK2A1,
CDC14A, CDC25B, CDC25C, CCNA1, CCNE2, CDK14, CDK2, CDK4, HSPA2,
ING2, NUP214, PLK2, PRKAA1, RHOB, STAG3 | 6.6E-10 |
| GOTERM_BP_FAT | Protein
phosphorylation | CCL5, CXCR4,
NME1-NME2, PHLPP1, PyCARD, SMAD4, RAC2, AR, ADORA1, ADCy2, ADCy9,
CDC25B, PLK2, PRKAA, 1PPP4R1 | 1.3E-10 |
| GOTERM_BP_FAT | Cell
proliferation | CCL5, NME1-NME2,
PyCARD, RBBP7, SMAD4, ADORA1, CSNK2A1, AR, CDC14A, CDC25B, CDC25C,
CHD5, CDK2, CDK4, ING2, NF1, PENK | 1.8E-7 |
| KEEN PATHWAy | Cell cycle | SMAD4, CDC14A,
CDC25B, CDC25C, CCNA1, CCNE2, CDK2, CDK4, TFDP1 | 6.3E-8 |
| KEEN PATHWAy | Pathways in
cancer | AR, CXCR4, SMAD4,
ADCy2, ADCy9, CCNE2, CDK2, CDK4, RAC2, GNAI1 | 4.9E-5 |
| KEEN PATHWAy | PI3K-AKT signaling
pathway | PHLPP1, CCNE2,
CDK2, CDK4, PRKAA1, PPP2R5C | 2.0E-2 |
 | Table VFunctional annotation of the
significant module 3. |
Table V
Functional annotation of the
significant module 3.
| Category | Term | Gene | P-value |
|---|
| GOTERM_BP_FAT | Cell cycle
process | RBL2, SKP1, CDC27,
CUL1, CDKN1A, DyNLT1, PSME1, SMB4, TOP2A, TFDP2, SLBP, TPR | 7.6E-13 |
| KEEN PATHWAy | Cell cycle | RBL2, SKP1, CDC27,
CUL1, CDKN1A, PTTG1, TFDP2 | 4.0E-6 |
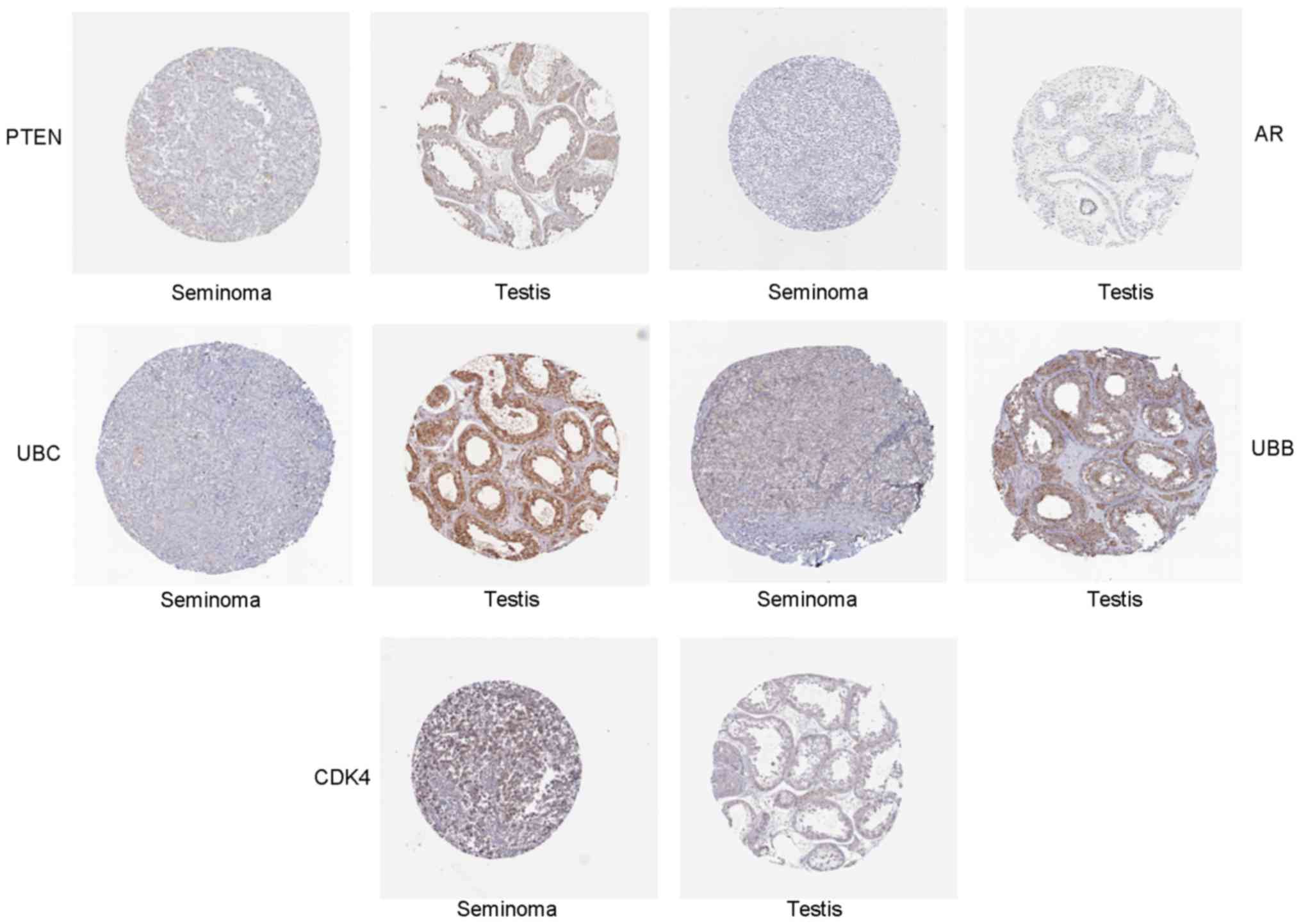
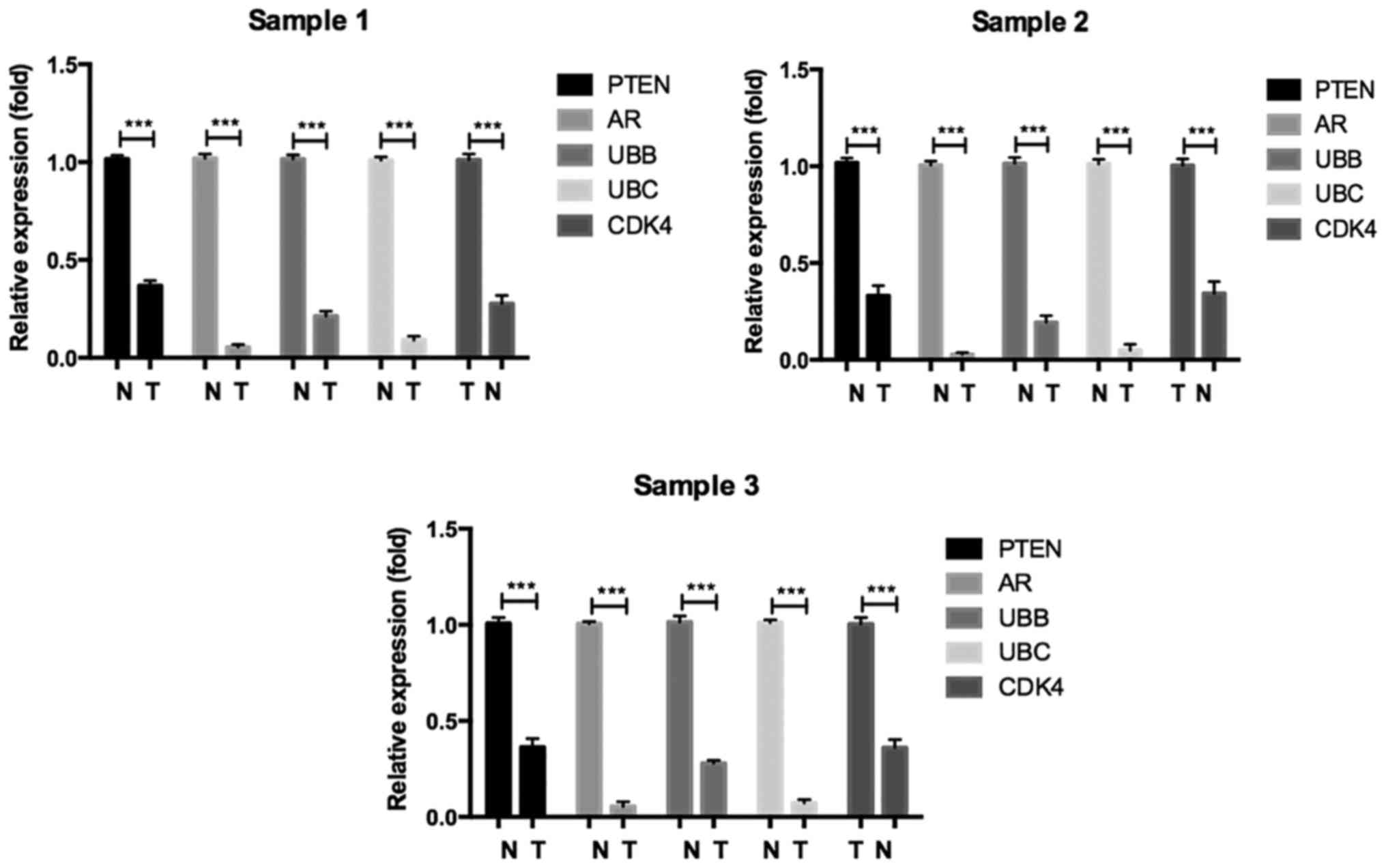
Validation of hub genes via RT-PCR and
IHC
To confirm the key genes identified by above
analyses, RT-PCR assay and IHC of 5 hub genes (Ubb, Ubc, Ar, Pten
and Cdk4) were conducted by using online database Oncomine and the
human protein atlas. Ubb, Ubc, Ar and Pten were downregulated DEGs,
while Cdk4 was an upregulated DEG. The RT-PCR results are shown in
Fig. 5. The relative expression
levels of Cdk4 was increased, but that of Ubb, Ubc, Ar, Pten were
decreased in seminoma compared with normal controls, which was
consistent with the DEG analysis. Furthermore, the differences in
hub gene expression levels were further conformed by IHC (Fig. 6). Moreover, we evaluated identified
hub genes RNA expression by RT-qPCR in paired seminoma tissues. The
result was consistent with above analysis (Fig. 7). In conclusion, our results
demonstrated that these hub genes were significantly differentially
expressed in seminoma.
Discussion
Testicular germ cell cancer is the most common tumor
among young men (aged 15–40 years) around the world (18). It is evident that there is a strong
relationship between congenital anomaly (cryptorchidism) and
testicular cancer (19). However,
the risk factors and genetic etiology for testicular cancer are
largely unknown. Consequently, understanding the molecular
mechanism of TGCG is of critical importance for diagnosis and
treatment. In this study, we first aimed to explore the mechanism
of seminoma, which comprises 56% of total TGCGs. Nowadays,
microarray and high-throughput sequencing technology give us a deep
insight into the expression levels of thousands of genes in human
genome simultaneously, as a result, it has been widely used to
predict the potential diagnostic biomarkers and therapeutic targets
for different cancers (20).
In this study, we compared gene expression profiles
of seminoma tumor with normal tissues from the GEO database and
identified 1,563 upregulated and 1,939 downregulated DEGs. The GO
term analysis showed that upregulated DEGs were mainly composed of
molecules participating in cell adhesion, PI3K-AKT signaling
pathway and cell cycle. PI3K-Akt pathway is known to be a vital
signaling pathway, which can promote EMT transition and have an
effect on the occurrence and progression of many types of cancers
(21,22). The downregulated DEGs were involved
in metabolic molecular pathways, cGMP-PKG signaling pathway and
Oocyte meiosis. A recent study suggested cGMP-dependent protein
kinase G was involved in maintaining stemness of cancer stem cells
and targeting this molecular pathway could effectively prevent
initiation, metastasis, and relapse of the cancer (23). Hence, GO analysis revealed several
possible biological processes, molecular functions and cellular
components which might be involved in the initiation and
development of seminoma. Function annotation and KEGG suggested
that DEGs between tumor and normal controls were greater in cell
cycle, pathways in cancer, cell proliferation and ERBB2 signaling
pathway. It is consistent with the knowledge that defective
function of cell cycle and increased cell proliferation are the
main cause for cancer initiation and progression (24–26).
Module analysis of the PPI network revealed that the development of
seminoma was associated with cell cycle, cell proliferation and
pathways in cancer, which was consistent with our pathway analysis.
Some hub genes, including Ubc, Ubb, Mapk8, Ar, Pten, Cdk2, Cdk4,
Rac1, Top2a and Cdkn1a have been identified based on the
degree, which can provide a new sight for the therapeutic strategy
in seminoma by constructing the PPI network.
When genes such as the cell cycle regulators
cyclin-dependent kinases (Cdk) mutate, they may cause cells
to multiply uncontrollably growth and thus result in cancer
formation. Recent studies have established cell cycle kinases as
anticancer drug targets by using Cdk4/6 inhibitors (27,28).
Among the DEGs of seminoma in the present study, there were several
genes deeply associated with the cell cycle, such as Cdk2, Cdk4,
Top2a, Cdc14a, Cdc25c, Ccna1 and Ccne2, indicating
Cdk2, Cdk4 and Top2a may serve as critical genetic
aberrations in seminoma.
Ar, a downregulated DE gene, was identified
as a hub gene of seminoma in this study. Androgens exert many
biological effects on many tissues through the androgen receptor
(Ar) (29,30). Previous studies indicated that
activation of androgen/Ar signaling had a positive role on
prostate cancer cell growth (31,32).
In the case of testes, androgen/Ar signaling was known to be
indispensable for normal development and function (33). Recently, a study revealed that
activation of androgen/Ar signal suppressed cell growth of
testicular cancer in vitro and in vivo (34). In our study, Ar expression
was downregulated as confirmed by RT-PCR assays and IHC, indicating
Ar is significantly negatively associated with seminoma.
Pten is a tumor suppressor and known to
negatively regulate PI3K signaling to inhibit cell growth.
Consequently, its suppression leads to increased proliferation and
invasion ability and promotes tumorigenesis. The decreased
expression of Pten is also related with the transformation
of carcinoma in situ (CIS) cells into cancerous tumors
(35,36). Furthermore, loss of PIK3IP1,
an additional negative regulator of PIK kinase, contributed to
increased relapse rate in TGCTs (37). This observation suggests the
negative regulators of PI3K signaling may also be related to TGCT
progression/prognosis. However, literature focused on Pten
expression in seminoma is still lacking. In our study, we
discovered that Pten was a downregulated DEG and
significantly differently expressed based on RT-PCR assays and IHC
in seminoma. These results revealed a negative role of Pten
on seminoma pathogenesis, which was in accordance with previous
studies.
Ubb and Ubc are members of the
ubiquitin gene family. Ubiquitin is a small, highly conserved
protein expressed in all eukaryotic cells, which can be covalently
linked to certain target proteins to mark them for degradation by
the ubiquitin-proteasome system (UPS) (38). The abundance of cellular ubiquitin
is in a dynamic balance, which is ultimately maintained by de
novo synthesis of ubiquitin from ubiquitin gene transcripts.
Human ubiquitin is encoded by a family of multiple genes, composed
of Uba52, Uba80, Ubb and Ubc, of which Ubb and
Ubc are inducible by various cell stresses (39–41).
Former studies found that there are some alterations in the UB
system in many types of human cancers and the deregulation of its
components have been found to play key roles in cellular processes
relevant to tumorigenesis (42,43).
Besides, reduced Ubb expression inhibited TSA-induced
apoptotic cascade in tumor cells (38). In this study, we found the
expression of Ubb and Ubc were significantly
decreased in seminoma tissue via RT-PCR and IHC assay, which
indicated the UB system is possibly involved in the tumorigenesis
of seminoma.
In conclusion, with the microarray gene expression
profiling, this study provides a comprehensive bioinformatic
analysis of DEGs, which may be related to initiation and
progression of seminoma. Results from our study provide a cluster
of potential diagnostic-related genes and molecular pathways for
future investigation and may be helpful for revealing the molecular
mechanisms of seminoma. While, further molecular biological
experiments are essential to demonstrate certain function of these
hub genes and key molecular pathways in seminoma.
Abbreviations:
|
GEO
|
Gene Expression Omnibus
|
|
DEGs
|
different expression genes
|
|
KEEG
|
Kyoto Encyclopedia of Genes and
Genomes
|
|
STRING
|
Search Tool the Retrieval of
Interacting Genes
|
|
GO
|
Gene ontology
|
|
PPI
|
protein-protein interaction
|
|
IHC
|
immunohistochemistry
|
|
RT-qPCR
|
reverse transcription-polymerase chain
reaction
|
|
TGCTs
|
testicular germ cell tumors
|
|
NSGCTs
|
non-seminomatous germ cell tumors
|
|
AFP
|
α-fetoprotein
|
|
HCG
|
human chorionic gonadotrophin
|
|
LDH
|
lactate dehydrogenase
|
|
MF
|
molecular function
|
|
CC
|
cellular component
|
|
BP
|
biological process
|
|
POU5F1
|
POU class 5 homeobox 1
|
|
NANGO
|
Nanog homeobox
|
|
LIN28
|
Lin-28 homolog
|
|
TFAP2C
|
transcription factor AP-2γ
|
|
UBC
|
ubiquitin C
|
|
UBB
|
ubiquitin B
|
|
AR
|
androgen receptor
|
|
PTEN
|
phosphatase and tensin homolog
|
|
CDK
|
cyclin-dependent kinases
|
|
AKT
|
RAC-α serine/threonine-protein
kinase
|
|
MAPK8
|
mitogen-activated protein kinase 8
|
|
TOP2A
|
topoisomerase (DNA) IIα
|
|
CDKN1A
|
cyclin-dependent kinase inhibitor
1a
|
|
RAC1
|
Ras-related C3 botulinum toxin
substrate 1
|
|
UPS
|
ubiquitin-proteasome system
|
|
WNT
|
wingless-type
|
|
PI3K
|
phosphatidylinositol-4,5-bisphosphate
3-kinase
|
References
|
1
|
Manku G, Hueso A, Brimo F, Chan P,
Gonzalez-Peramato P, Jabado N, Gayden T, Bourgey M, Riazalhosseini
Y and Culty M: Changes in the expression profiles of claudins
during gonocyte differentiation and in seminomas. Andrology.
4:95–110. 2016. View Article : Google Scholar
|
|
2
|
Litchfield K, Summersgill B, Yost S,
Sultana R, Labreche K, Dudakia D, Renwick A, Seal S, Al-Saadi R,
Broderick P, et al: Whole-exome sequencing reveals the mutational
spectrum of testicular germ cell tumours. Nat Commun. 6:59732015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aran V, Victorino AP, Thuler LC and
Ferreira CG: Colorectal cancer: Epidemiology, disease mechanisms
and interventions to reduce onset and mortality. Clin Colorectal
Cancer. 15:195–203. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Taylor-Weiner A, Zack T, O'Donnell E,
Guerriero JL, Bernard B, Reddy A, Han GC, AlDubayan S, Amin-Mansour
A, Schumacher SE, et al: Genomic evolution and chemoresistance in
germ-cell tumours. Nature. 540:114–118. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu T, Li R, Zhao H, Deng J, Long Y, Shuai
MT, Li Q, Gu H, Chen YQ and Leng AM: eIF4E promotes tumorigenesis
and modulates chemosensitivity to cisplatin in esophageal squamous
cell carcinoma. Oncotarget. 7:66851–66864. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chieffi P and Chieffi S: Molecular
biomarkers as potential targets for therapeutic strategies in human
testicular germ cell tumors: An overview. J Cell Physiol.
228:1641–1646. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Trabert B, Chen J, Devesa SS, Bray F and
McGlynn KA: International patterns and trends in testicular cancer
incidence, overall and by histologic subtype, 1973–2007. Andrology.
3:4–12. 2015. View
Article : Google Scholar
|
|
8
|
Gilligan TD, Seidenfeld J, Basch EM,
Einhorn LH, Fancher T, Smith DC, Stephenson AJ, Vaughn DJ, Cosby R
and Hayes DF; American Society of Clinical Oncology: American
Society of Clinical Oncology Clinical Practice Guideline on uses of
serum tumor markers in adult males with germ cell tumors. J Clin
Oncol. 28:3388–3404. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Murray MJ, Huddart RA and Coleman N: The
present and future of serum diagnostic tests for testicular germ
cell tumours. Nat Rev Urol. 13:715–725. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hanna NH and Einhorn LH: Testicular cancer
- discoveries and updates. N Engl J Med. 371:2005–2016. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sheikine Y, Genega E, Melamed J, Lee P,
Reuter VE and Ye H: Molecular genetics of testicular germ cell
tumors. Am J Cancer Res. 2:153–167. 2012.
|
|
12
|
Kanetsky PA, Mitra N, Vardhanabhuti S, Li
M, Vaughn DJ, Letrero R, Ciosek SL, Doody DR, Smith LM, Weaver J,
et al: Common variation in KITLG and at 5q31.3 predisposes to
testicular germ cell cancer. Nat Genet. 41:811–815. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rijlaarsdam MA and Looijenga LH: An
oncofetal and developmental perspective on testicular germ cell
cancer. Semin Cancer Biol. 29:59–74. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
15
|
Consortium TGO: Gene ontology: tool for
the unification of biology. The Gene Ontology Consortium Nat Genet.
25:25–29. 2000.
|
|
16
|
Ogata H, Goto S, Sato K, Fujibuchi W, Bono
H and Kanehisa M: KEGG: Kyoto Encyclopedia of Genes and Genomes.
Nucleic Acids Res. 27:29–34. 1999. View Article : Google Scholar
|
|
17
|
Köhler S, Bauer S, Horn D and Robinson PN:
Walking the interactome for prioritization of candidate disease
genes. Am J Hum Genet. 82:949–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chia VM, Quraishi SM, Devesa SS, Purdue
MP, Cook MB and McGlynn KA: International trends in the incidence
of testicular cancer, 1973–2002. Cancer Epidemiol Biomarkers Prev.
19:1151–1159. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cook MB, Akre O, Forman D, Madigan MP,
Richiardi L and McGlynn KA: A systematic review and meta-analysis
of perinatal variables in relation to the risk of testicular cancer
- experiences of the son. Int J Epidemiol. 39:1605–1618. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peng J, Wu Y, Tian X, Pang J, Kuai L, Cao
F, Qin X, Zhong J, Li X, Li Y, et al: High-throughput sequencing
and co-expression network analysis of lncRNAs and mRNAs in early
brain injury following experimental subarachnoid haemorrhage. Sci
Rep. 7:465772017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jain A, Tripathi R, Turpin CP, Wang C and
Plattner R: Abl kinase regulation by BRAF/ERK and cooperation with
Akt in melanoma. Oncogene. Apr 3–2017.Epub ahead of print.
View Article : Google Scholar : 2017.
|
|
22
|
Chen L, Fu H, Luo Y, Chen L, Cheng R,
Zhang N and Guo H: cPLA2α mediates TGF-β-induced
epithelial-mesenchymal transition in breast cancer through PI3k/Akt
signaling. Cell Death Dis. 8:e27282017. View Article : Google Scholar
|
|
23
|
Liu N, Mei L, Fan X, Tang C, Ji X, Hu X,
Shi W, Qian Y, Hussain M, Wu J, et al: Phosphodiesterase 5/protein
kinase G signal governs stemness of prostate cancer stem cells
through Hippo pathway. Cancer Lett. 378:38–50. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Perez R, Wu N, Klipfel AA and Beart RW Jr:
A better cell cycle target for gene therapy of colorectal cancer:
Cyclin G. J Gastrointest Surg. 7:884–889. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tominaga O1, Nita ME, Nagawa H, Fujii S,
Tsuruo T and Muto T: Expressions of cell cycle regulators in human
colorectal cancer cell lines. Jpn J Cancer Res. 88:855–860. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen HQ, Xiao YX, She ZY, Tan FQ and Yang
WX: A novel role of KIF3b in the seminoma cell cycle. Exp Cell Res.
352:95–103. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sherr CJ, Beach D and Shapiro GI:
Targeting CDK4 and CDK6: From discovery to therapy. Cancer Discov.
6:353–367. 2016. View Article : Google Scholar :
|
|
28
|
Asghar U, Witkiewicz AK, Turner NC and
Knudsen ES: The history and future of targeting cyclin-dependent
kinases in cancer therapy. Nat Rev Drug Discov. 14:130–146. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oury F, Sumara G, Sumara O, Ferron M,
Chang H, Smith CE, Hermo L, Suarez S, Roth BL, Ducy P, et al:
Endocrine regulation of male fertility by the skeleton. Cell.
144:796–809. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Rao PM, Kelly DM and Jones TH:
Testosterone and insulin resistance in the metabolic syndrome and
T2DM in men. Nat Rev Endocrinol. 9:479–493. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Qi J, Tripathi M, Mishra R, Sahgal N,
Fazli L, Ettinger S, Placzek WJ, Claps G, Chung LW, Bowtell D, et
al: The E3 ubiquitin ligase Siah2 contributes to
castration-resistant prostate cancer by regulation of androgen
receptor transcriptional activity. Cancer Cell. 23:332–346. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ito S, Ueda T, Ueno A, Nakagawa H,
Taniguchi H, Hongo F, Kamoi K, Okihara K, Kawauchi A and Miki T:
Paired box 2 upregulates androgen receptor gene expression in
androgen-independent prostate cancer. FEBS J. 281:4506–4518. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang RS, Yeh S, Tzeng CR and Chang C:
Androgen receptor roles in spermatogenesis and fertility: Lessons
from testicular cell-specific androgen receptor knockout mice.
Endocr Rev. 30:119–132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Nakagawa H, Ueda T, Ito S, Shiraishi T,
Taniguchi H, Kayukawa N, Nakanishi H, Ushijima S, Kanazawa M,
Nakamura T, et al: Androgen suppresses testicular cancer cell
growth in vitro and in vivo. Oncotarget. 7:35224–35232. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Di Vizio D, Cito L, Boccia A, Chieffi P,
Insabato L, Pettinato G, Motti ML, Schepis F, D'Amico W, Fabiani F,
et al: Loss of the tumor suppressor gene PTEN marks the transition
from intra-tubular germ cell neoplasias (ITGCN) to invasive germ
cell tumors. Oncogene. 24:1882–1894. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
McIver SC, Stanger SJ, Santarelli DM,
Roman SD, Nixon B and McLaughlin EA: A unique combination of male
germ cell miRNAs coordinates gonocyte differentiation. PLoS One.
7:e355532012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gilbert DC, McIntyre A, Summersgill B,
Missiaglia E, Goddard NC, Chandler I, Huddart RA and Shipley J:
Minimum regions of genomic imbalance in stage I testicular
embryonal carcinoma and association of 22q loss with relapse. Genes
Chromosomes Cancer. 50:186–195. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu P, Tian Y, Chen G, Wang B, Gui L, Xi L,
Ma X, Fang Y, Zhu T, Wang D, et al: Ubiquitin B: An essential
mediator of trichostatin A-induced tumor-selective killing in human
cancer cells. Cell Death Differ. 17:109–118. 2010. View Article : Google Scholar
|
|
39
|
Nenoi M: Induced accumulation of
polyubiquitin gene transcripts in HeLa cells after UV-irradiation
and TPA-treatment. Int J Radiat Biol. 61:205–211. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Finch JS, St John T, Krieg P, Bonham K,
Smith HT, Fried VA and Bowden GT: Overexpression of three ubiquitin
genes in mouse epidermal tumors is associated with enhanced
cellular proliferation and stress. Cell Growth Differ. 3:269–278.
1992.PubMed/NCBI
|
|
41
|
Ryu KY, Sinnar SA, Reinholdt LG, Vaccari
S, Hall S, Garcia MA, Zaitseva TS, Bouley DM, Boekelheide K, Handel
MA, et al: The mouse polyubiquitin gene Ubb is essential for
meiotic progression. Mol Cell Biol. 28:1136–1146. 2008. View Article : Google Scholar :
|
|
42
|
Ciechanover A and Schwartz AL: The
ubiquitin system: Pathogenesis of human diseases and drug
targeting. Biochim Biophys Acta. 1695:3–17. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Hoeller D and Dikic I: Targeting the
ubiquitin system in cancer therapy. Nature. 458:438–444. 2009.
View Article : Google Scholar : PubMed/NCBI
|