Introduction
Epithelial ovarian cancer (EOC) accounts for 4% of
all cancers in women and is the leading cause of death from
gynecologic malignancies (1).
Despite medical and surgical improvements, long-term survival rates
for EOC patients with advanced disease remain disappointing,
primarily due to its asymptomatic nature and the lack of reliable
methods for early diagnosis (2,3).
Indeed, most women are diagnosed with EOC when micro- and
macro-metastases are already present and currently, the 5-year EOC
survival rate is rather disappointing (<40%) (1). Thus, the identification of novel
pro-metastatic EOC molecules and associated pathways could provide
additional therapeutic targets for improved management of this
deadly disease.
There are many documented studies investigating
different post-translational modifications (PTMs) and their
association with cancer; among these, aberrant glycosylation has
displayed rather important roles in cancer progression (4,5),
including EOC dissemination (6,7).
Mucin-type O-glycosylation of proteins represents the most diverse
PTM form, and is considered to be a conserved type of glycosylation
found in many species and organ types (8). This type of PTM is rather complex and
involves the transfer of different monosaccharides to each of the
six O-linked glycans (9).
O-glycosylation is initiated by 20 GalNAc-transferases, a family of
enzymes known as the UDP N-acetylgalactosamine: polypeptide
N-acetyl galactosaminyl transferases (GalNAc-Ts) (9), which are responsible for the transfer
of the monosaccharide GalNAc from UDP-GalNAc to the hydroxyl group
of the serine, threonine or tyrosine residues found in the target
protein substrate (10).
Interestingly, the genes comprising this family of enzymes show
tissue-specific expression (11,12),
but also have overlapping substrate specificities, suggestive for
partial functional redundancy in their function (11). Several in vitro studies have
shown that GalNAc-Ts are differentially expressed in cells and
tissues during development (9).
Aberrant GalNAc-Ts expression patterns have been frequently
observed in cancer (13), which
warrants further studies on the possible implications of these
enzymes in tumor progression and their potential use as therapeutic
targets and/or prognostic markers (14).
We have previously identified the GalNAc-T3 (GALNT3)
gene as a potential EOC oncogene, highly expressed in advanced
disease, as GALNT3 expression was significantly associated with
poor outcome (15). We also
demonstrated that in EOC cells, GALNT3 might directly alter
biosynthesis and/or aberrant glycosylation of different
O-glycoproteins that could play an essential role in EOC
dissemination (15–17). However, to date, no other studies
have comprehensively assessed the implications of other members of
the GalNAc-Ts gene family in EOC dissemination. We thus decided to
examine the role of some of the GalNAc-Ts that have been reported
to play essential roles in the etiology of different cancer types.
Five members of this gene family (GALNT2, T4, T6, T9 and T14) have
previously displayed significant alterations in their expression in
different cancer types (reviewed in ref. 7). In silico analysis of the
GalNAc-Ts expression profiles from publicly available data were
also indicative for overexpression of some of these GalNAc-Ts
(including GALNT4, T6, and T14), as well as GALNT3, in ovarian HGSC
samples.
The above data prompted us to investigate the
expression levels of these five GalNAc-Ts (GALNT2, T4, T6, T9 and
T14) in EOC cells and EOC tumor tissues, and to correlate their
expression with the corresponding clinicopathological
characteristics. To our knowledge this is the first report that
comprehensively examines the expression profiles of several
GalNAc-Ts in EOC with the perspective of identifying novel
prognostic factors for this deadly disease. We show that most of
the GalNAc-Ts analyzed (including GALNT2, T6, T9 and T14) displayed
altered expression in EOC cells and tumors importantly, our data
are indicative for a significant association of two GalNAc-Ts
(GALNT6 and T14) with progression- free survival (PFS) values of
EOC patients, suggestive for oncogenic functions of these two
enzymes in EOC, and their potential use as novel EOC prognostic
biomarkers.
Materials and methods
Patient cohort
Patients included in this study were operated
between January 1998 to December 2015 for advanced ovarian cancer
at the CHUQ Hôtel-Dieu Hospital in Quebec City, Canada. Inclusion
criteria were: serous papillary carcinoma histology, FIGO stages
II, III or IV and chemotherapy received after the surgery (see
Table I for detailed
clinicopathological characteristics). All tumors were
histologically classified according to the criteria defined by the
World Health Organization (18).
Disease progression was evaluated following the guidelines of the
Gynecology Cancer Intergroup (18). PFS was defined as the time from
surgery to the first observation of disease progression, recurrence
or death. The follow-up was available until death or to the date
the study was closed (30 December 2016). Nineteen non-tumoral
(control) ovarian samples were derived from women subjected to
hysterectomy with oophorectomy due to non-ovarian pathologies.
 | Table IDetailed patients'
clinicopathological characteristics. |
Table I
Detailed patients'
clinicopathological characteristics.
| Variable | Range | N/Total | % |
|---|
| Age (years) | ≥65 | 54/162 | 33 |
| <65 | 108/162 | 67 |
| Median age | 60 | | |
| Tissue/tumor
type | Normal | 18/162 | 11 |
| LMP | 13/162 | 8 |
| High-grade | 131/162 | 81 |
| Grade | 3 | 126/131 | 96 |
| 2 | 5/131 | 4 |
| Stage | II | 5/131 | 4 |
| III | 92/131 | 70 |
| IV | 34/131 | 26 |
|
Chemotherapya | Platinum+Taxol | 120/131 | 92 |
| Other | 11/131 | 8 |
| CA125b (U/ml) | ≥800 | 59/118 | 50 |
| <800 | 59/118 | 50 |
| PFSc (months) | 0–6 | 53/124 | 43 |
| 7–24 | 44/124 | 35 |
| >25 | 27/124 | 22 |
Ethics statement approval and consent to
participate
The study was approved by the Clinical Research
Ethics Committee of the Hotel-Dieu de Québec Hospital, and all
patients signed an informed consent for voluntary participation and
agreed to report individual patient data.
Cell culture
The EOC cell lines SKOV3 and CaOV3 were purchased
from the American Tissue Type Collection; OV-90, OV2008, TOV-112
and TOV-21 cell lines were a kind gift from Dr Anne-Marie
Mes-Masson (Montreal University), A2780s and A2780cp cell lines
were a kind gift from Dr Benjamin Tsang (Ottawa University), the
OVCAR4 cell line was a kind gift from Dr Stephan Gobeil (Laval
University), and the two human ovarian surface epithelial (HOSE)
cell lines; HOSE 6.3 and 17.1 were a kind gift from Dr Francis
Jacob (University Hospital Basel). The cell lines were passaged in
different culture media supplemented with 10% fetal bovine serum,
and 1% penicillin streptomycin or 50 μg/ml of gentamicin as
described previously (15).
Western blotting
Western blot analysis was performed as previously
described (15). Briefly, protein
lysates of the EOC and HOSE cell lines, were prepared by suspending
cell pellets in Laemmli sample buffer containing 5%
β-mercaptoethanol. Ovarian tumor tissue and non-tumoral tissues
were homogenized and sonicated in RIPA buffer [50 mM Tris (pH 7.4),
150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X-100]
containing protease and phosphatase inhibitors, samples were then
incubated on ice for 15 min. Protein samples from cells and tissues
were measured using a BCA protein assay kit (Thermo Scientific
Pierce, Rockford, IL, USA). BSA standard or samples (20 μl)
were transferred to a 96 well plate to which 200 μl working
reagent was added (working reagent 50:1 ratio of assay reagents A
and B). The plate was incubated for 30 min at 37°C, the plates were
then analyzed with a spectrophotometer at 560 nm [iMARK microplate
reader (Bio-Rad Hercules, CA, USA)]. After centrifugation at 13,300
rpm for 15 min at 4°C, the supernatant was taken and 20–30
μg of the protein were used for sample preparation. Protein
lysates were separated by 6–12% Tris-glycine gel electrophoresis
and transferred onto a polyvinylidene difluoride membrane using a
semidry apparatus (Bio-Rad Laboratories, Mississauga, ON, Canada).
The membrane was blocked with 4% non-fat dry milk in TBST (20
mmol/l Tris-HCl, 0.5 M NaCl and 0.1% Tween-20), and the membranes
were incubated overnight with the primary antibody at 4°C or 1 h at
RT (for a list of all the antibodies used in this study refer to
Table II). After 3×15 min washes
with TBST (20 mmol/l Tris-HCl, 0.5 M NaCl and 0.1% Tween-20) at
room temperature, the membranes were incubated with horseradish
peroxidase-conjugated secondary antibody and detected with ECL
solution (Thermo Fisher Scientific, Waltham, MA, USA).
 | Table IIDilution and technique used for each
antibody in IHC and western blot analyses. |
Table II
Dilution and technique used for each
antibody in IHC and western blot analyses.
| Antibody | Species | Dilution TMA | Company | Retrieval | Incubation TMA | Dilution WB | Incubation WB |
|---|
| Anti-GALNT2 | Rabbit | 1:50 | Abcam | Microwave | 4°C overnight | 1:1,000 | 4°C overnight |
| Anti-GALNT4 | Rabbit | 1:100, 1:50,
1:25 | Proteintech |
Microwave/pronase | 4°C overnight | 1:500 | 4°C overnight |
| Anti-GALNT6 | Mouse | 1:75 | Abcam | Microwave | 4°C overnight | 1:1,000 | 4°C overnight |
| Anti-GALNT9 | Rabbit | 1:100 | LifeSpan
BioScience | Microwave | 4°C overnight | 1:1,000 | 4°C overnight |
| Anti-GALNT14 | Rabbit | 1:100 | Abcam | Microwave | 4°C overnight | 1:1,000 | 4°C overnight |
| Anti-β-actin | Mouse | N/A | Santa Cruz | N/A | N/A | 1:2,000 | 1 h/room
temperature |
Tissue micro arrays (TMAs) and
immunohistochemistry (IHC)
TMAs were constructed as previously described
(15). Briefly, one representative
block of each ovarian tumor and control ovarian tissue was selected
for the preparation of the tissue arrays. Three 0.6-mm cores of
tumor were taken from each tumor block and placed, 0.4 mm apart, on
a recipient paraffin block using a commercial tissue arrayer
(MTA-II arrayer) (Beecher Instruments, Silver Spring, MD, USA). The
cores were randomly placed on one of three recipient blocks to
avoid evaluation biases.
IHC analyses were performed, as previously described
(15). Briefly, 4-μm tissue
sections were deparaffinised and rehydrated in graded alcohols,
then incubated with blocking serum for 20 min. Following treatment
with 3% H2O2 for 10 min to quench the
endogenous peroxidise activity, sections were incubated with the
primary antibody overnight at 4°C (for a list of all the antibodies
used in this study refer to Table
II). Incubation and detection with SignalStain
3,3′-diaminoben-zidine (DAB) Substrate kit (IDetect Universal Mouse
Kit HRP-DAB; ID Labs, Buffalo, NY, USA) were done according to the
manufacturer's instructions. Sections were then counter-stained
with hematoxylin. Images were acquired using a Leica Confocal Scope
(TCS SP5 X; Leica Microsystems, Exton, PA, USA) and analyzed via
the Leica Application Suite Software (Leica Microsystems).
Scoring and statistical analysis
Protein expression was scored according to intensity
(value of 0 for absence, 1 for low, 2 for moderate, and 3 for high)
of staining based on manual visualization. A composite score was
defined as the product of staining intensity (nuclear, cytoplasmic,
or membranous depending on the expected staining). All slides were
independently scored in a blinded manner by 2 observers, and the
integration was >85%. In case of differences between the 2
scorings, the core was re-evaluated to reach a consensus. The
relationship between the protein expression of the listed genes in
HGSC and LMP tumors, and control ovarian tissues was evaluated by
the Wilcoxon two-sample test. A significant association was
considered when p-values were <0.05.
Multiple correspondence analysis (MCA) was used to
produce 2-dimensional displays of similarities of the relative
expression patterns amongst the five GalNAc-Ts staining intensity
in the different patient samples. MCA statistical analyses were
carried out using SPSS software, version 13.0. Survival analysis
results were visualized using Kaplan-Meier survival curve analysis
(SPSS software, version 13.0). A Kaplan-Meier curve and the
log-rank test were performed based on PFS values to test the effect
of the intensity of the gene (3, 2 vs. 0, 1) on disease
progression. The relationship between the GalNAc-Ts staining
intensity and PFS was determined by the non-parametric Mantel-Cox
log-rank test to compare survival distributions (SPSS software,
version 13.0). Bivariate and multivariate analyses, taking into
account standard or strongly associated prognostic variables, were
performed to identify independent prognostic factors and a
statistic test p-value of <0.05 was considered statistically
significant.
Results
Analyses of the expression profiles of
different members of the GalNAc-Ts family in both ovarian HGSC
tumors and EOC cell lines
Initially, we investigated GalNAc-Ts expression
profiles from publicly available data generated from samples found
in the independently derived Affymetrix GeneChip Human Genome U133
Plus 2.0 [HG-U133_Plus_2] (19).
The in silico data examined were based on global gene
expression analysis of differentially expressed candidate mRNAs in
21 moderately differentiated (MD) and poorly differentiated (PD)
serous ovarian carcinomas (SOC) (MD/PD SC), 13 serous ovarian
borderline (SOBT), and seven superficial scrapings from normal
ovary (SNO) samples (19). These
analyses showed significant correlation and high fold change in the
overexpression of several GalNAc-Ts in the SOC samples compared to
both the SBOT and SNO samples including: GALNT3, GALNT4, GALNT6,
and GALNT14. Based on the above in silico analyses, as well
as on literature data (as can be seen in Introduction), we decided
to further proceed with investigating the expression levels of five
GalNAc-Ts (GALNT2, T4, T6, T9 and T14) in EOC specimens by western
blotting. The protein expression levels of these GalNAc-Ts were
initially analyzed in nine EOC cell lines and in two HOSE cell
lines (Fig. 1A). As seen from
Fig. 1A, GALNT2 displayed lack of
expression in seven EOC cell lines studied and quite a weak
expression in SKOV3 and TOV112 EOC cells, compared to its rather
strong expression in the two control HOSE cell lines, indicative
for a suppression of this enzyme in EOC cells. GALNT4 showed very
weak, or lack of expression in all (both EOC and HOSE) cell lines
analyzed. GALNT6 expression was highly observed in three EOC cell
lines (TOV21, CaOV3 and OVCAR4) while showing no, or weak
expression in the two HOSE cell lines (Fig. 1A). Finally, both GALNT9 and GALNT14
displayed very high expression in all EOC cell lines and almost
lack of expression in the HOSE cell lines (Fig. 1A).
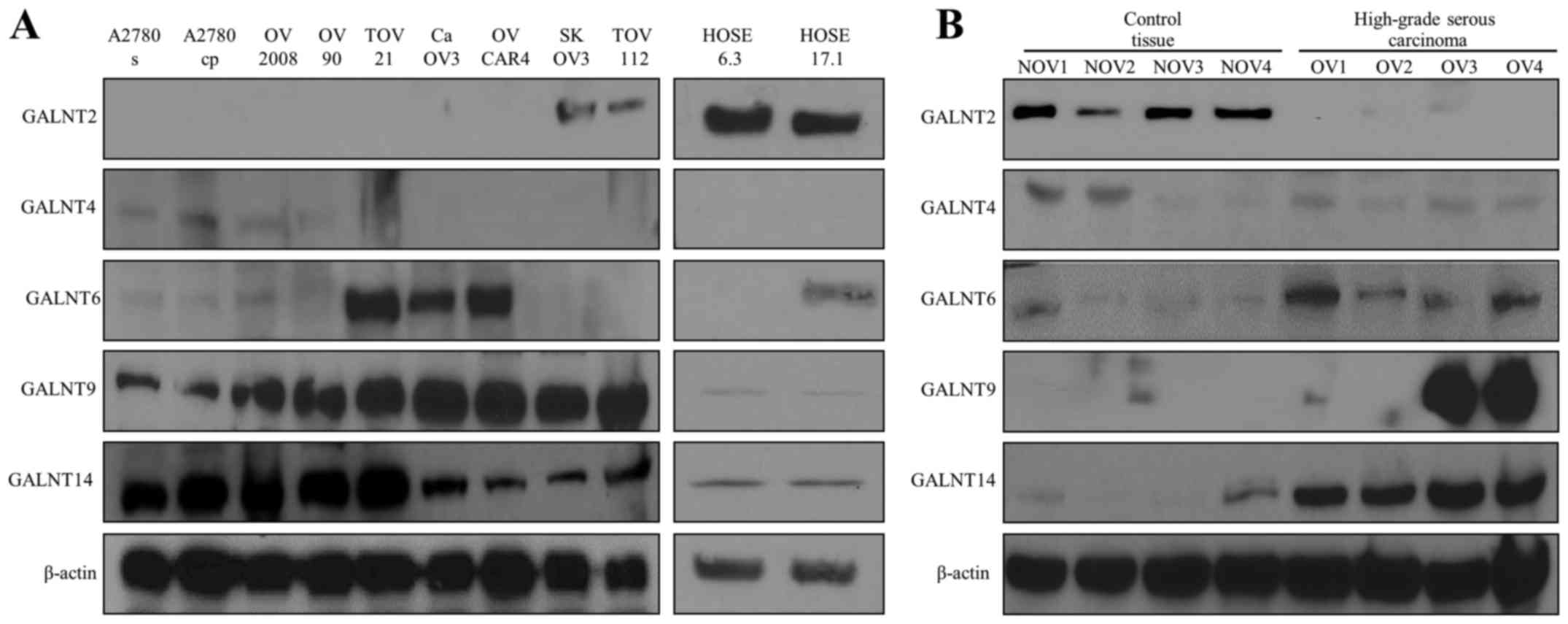 | Figure 1GalNAc-Ts expression in EOC cells and
EOC tumors. (A) Western blot analysis of endogenous GALNT2, T4, T6,
T9 and T14 protein expression in different EOC cell lines and the
HOSE cell lines. (B) Western blot analysis of endogenous GALNT2,
T4, T6, T9 and T14 protein expression in 4 HG ovarian samples
(OG1-OG4) and 4 non-tumoral ovarian tissue (control tissue) samples
(NOV1-NOV4). β-actin was used as a loading control. EOC, epithelial
ovarian cancer; HOSE, human ovarian surface epithelial; NOV, normal
ovarian; OV, ovarian tumor. |
Further analyses of the protein expression levels of
the selected GalNAc-Ts in four ovarian HGSC tumor samples and four
control ovarian tissue samples were quite confirmatory to the data
obtained with the EOC and HOSE (control) cell lines (Fig. 1B). Indeed, GALNT2 showed a
relatively high expression in all control samples, while no
expression was observed in the HGSCs, while GALNT4 showed an
analogous pattern of rather weak or no expression between control
tissues and HGSC tumors. Correspondingly, GALNT6 showed a
relatively high expression in the HGSCs tumor samples compared to
control tissues, and again, GALNT9 and GALNT14 displayed the
strongest overexpression levels in HGSCs, relative to the controls
(Fig. 1B).
IHC analysis of the expression patterns
of five GalNAc-Ts in numerous EOC tumors using TMAs
We next proceeded with evaluating the GalNAc-Ts
expression levels by IHC in TMAs comprising triplicate cores of 131
HGSCs and 13 LMP tumors, and including 18 control ovarian tissue
specimens. Table I shows the major
clinical characteristics of these patients for whom extensive
follow-up clinical data were available. The age ranged from 38 to
91 years (median: 60 years). HGSC tumors were grade 3 [126 patients
(96%) and grade 2 (5 patients (4%)]. Grade 3 tumors included stage
III (70%) and stage IV (26%) tumors, while grade 2 tumors included
stage II (4%). Most patients (92%) received standard treatment of
cytoreductive surgery followed by platinum/taxane chemotherapy
regimens. The median baseline CA125 was ~800. Forty-three percent
of the patients had a progression or a recurrence within the first
6 months of follow-up; for 35% of the patients the PFS interval was
in the range of 7 to 24 months, and 22% of the patients displayed
PFS values >25 months. The mean score for expression was defined
by the extent and intensity of immunohistochemistry staining for
GALNT2, T4, T6, T9 and T14 (Fig.
2). As expected, GALNT2 displayed highly significant expression
(strong cytoplasmic granular staining) in control tissues compared
to negative staining in both LMP (p<0.0001) and HGSC tumors
(p<0.0001) (Fig. 2A). As
previously, GALNT4 showed negative expression in all cases (tumor
samples and control tissues), despite repeated attempts with
various retrieval systems, antibody concentration, incubating time,
or signal enhancements systems (Fig.
2B). GALNT6 showed a strong diffuse cytoplasmic staining which
was exclusively present in HGSCs compared to control tissues
(p=0.0001) and LMP tumors (p<0.0001) (Fig. 2C). A similar expression pattern was
also observed for GALNT9 and GALNT14, as both these GalNAc-Ts were
significantly overexpressed in HGSC tumors, when compared to
control tissues (p<0.0001), and (p<0.0001) respectively
(Fig. 2D and E); however, GALNT9
also displayed significant overexpression in LMP tumors compared to
control samples (p=0.0146) (Fig.
2D), while no significant difference in GALNT14 protein
expression was observed between LMP tumors and control tissues
(p=0.0859) (Fig. 2E).
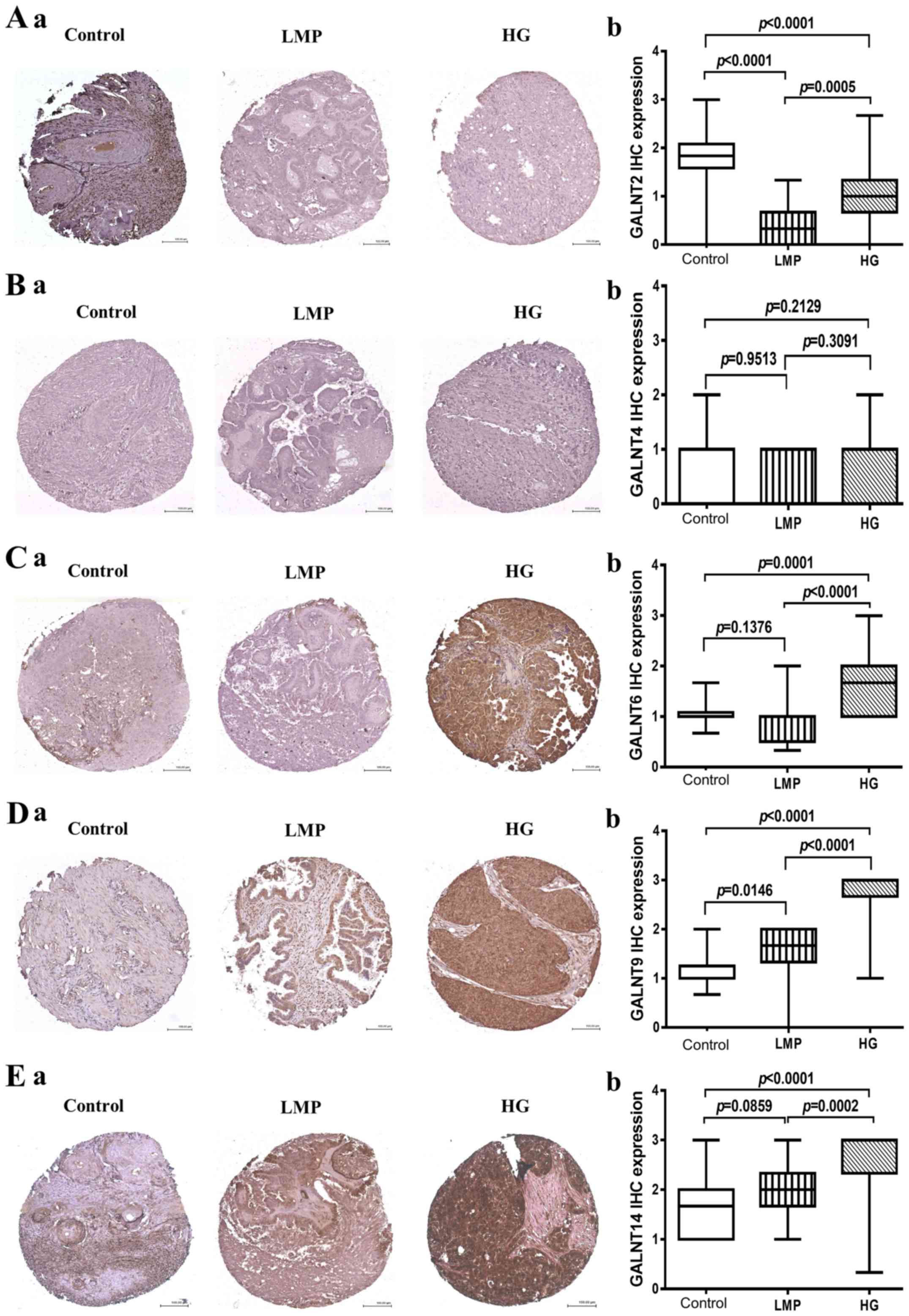 | Figure 2GalNAc-Ts protein expression in HG
samples compared to LMP and non-tumoral ovarian samples. (A) a,
GALNT2 staining patterns in representative cores in control ovarian
tissues, LMP tumors and HG tumors. b, Box-plot presentation of
GALNT2 protein expression levels in control ovarian tissues, LMP
tumors and HG tumors. (B) a, GALNT4 staining patterns in
representative cores in control ovarian tissues, LMP tumors and HG
tumors. b, Box-plot presentation of GALNT4 protein expression
levels in control ovarian tissues, LMP tumors and HG tumors. (C) a,
GALNT6 staining patterns in representative cores in control ovarian
tissues, LMP tumors and HG tumors. b, Box-plot presentation of
GALNT6 protein expression levels in control ovarian tissues, LMP
tumors and HG tumors. (D) a, GALNT9 staining patterns in
representative cores in control ovarian tissues, LMP tumors and HG
tumors. b, Box-plot presentation of GALNT9 protein expression
levels in control ovarian tissues, LMP tumors and HG tumors. (E) a,
GALNT14 staining patterns in representative cores in control
ovarian tissues, LMP tumors and HG tumors. b, Box-plot presentation
of GALNT14 protein expression levels in control ovarian tissues,
LMP tumors and HG tumors. All p-values were derived from log-rank
tests. Error bars denote standard deviation of each mean
calculation. HG, high-grade; LMP, low-malignant potential. |
The above results, as well as data from our previous
study (15) are indicative for the
simultaneous overexpression of several GalNAc-Ts in EOC cell lines
and tumor tissues (including GALNT3, T6, T9 and T14). Since a
considerable degree of redundancy between the different members of
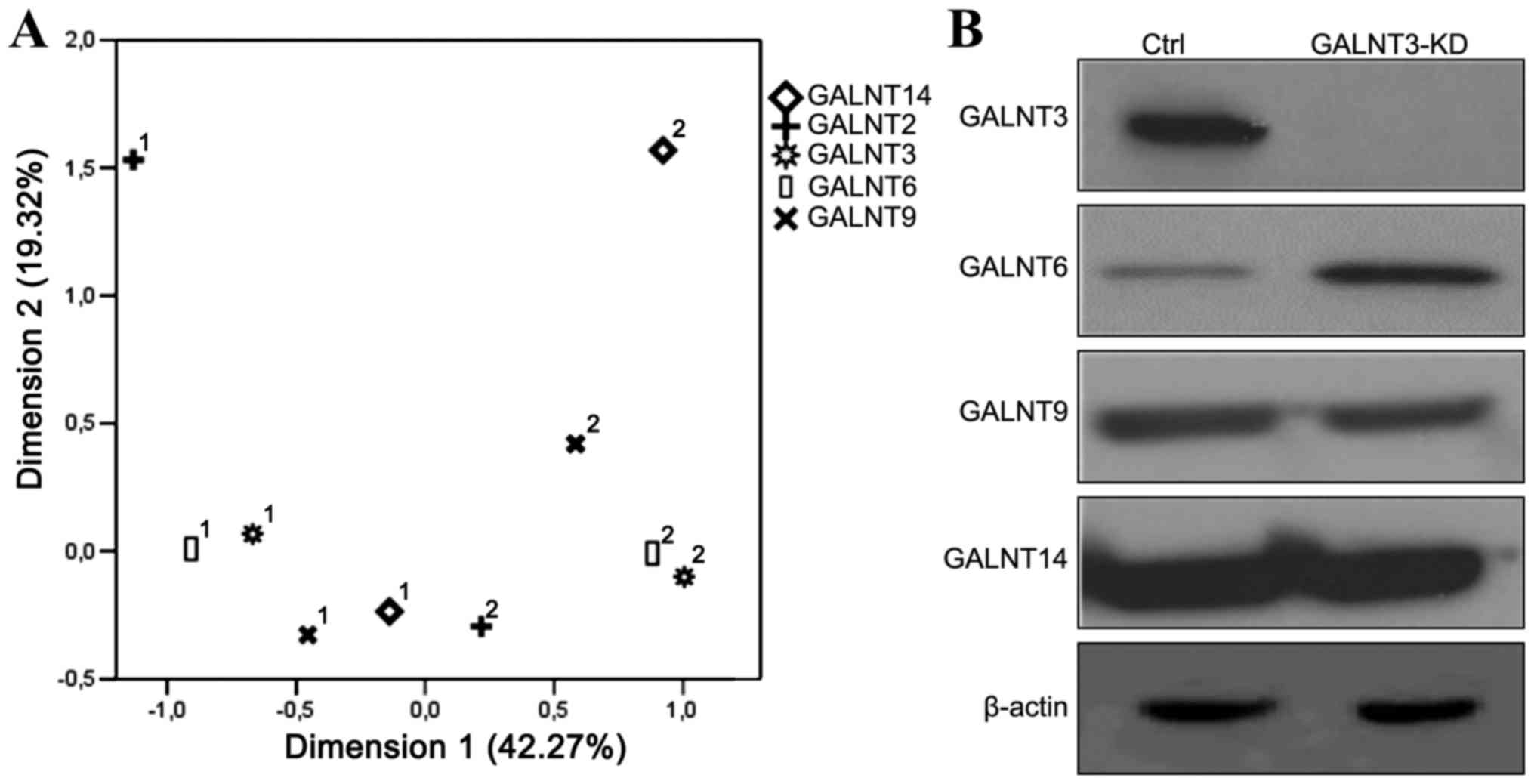
the GalNAc-Ts gene family has been frequently observed (20), we decided to apply the MCA approach
in order to more deeply investigate the extent of overlapping
expression of these GalNAc-Ts among the EOC tumor samples included
in this study. Two quantitative variables are included in the
analysis based on staining intensity: i) for positive staining in
patient samples, and ii) for negative staining in patient samples.
The two dimensions of the MCA explained 42.27% (dimension 1) and
19.32% (dimension 2) of the total data variability, respectively
(Fig. 3A). These analyses were
indicative for a strong and a highly overlapping relationship
between GALNT3 and GALNT6, as additionally, some overlapping
relationship between GALNT6 and GALNT9 cannot be excluded (Fig. 3A). GALNT14 showed the highest
diversity between staining 1 and 2, with no observed overlap with
the other 4 genes (Fig. 3A).
GALNT2 confirmed our previous data showing a complete inverse
relationship to the other 4 genes, as GALNT2 staining expression 1
and 2 showed a high inverse correlation to expression pattern
observed for GALNT14, and overall differential expression to those
of GALNT3, T6 and T9 (Fig. 3A).
The MCA-suggested GALNT3/T6 redundancy was further confirmed by
examining protein expression profiles of GALNT6, T9 and T14 in the
GALNT3 knockdown (KD) clone, previously generated in the EOC cell
line A2780s (15). As shown in
Fig. 3B, we observed a clear
upregulation in GALNT6 protein expression in the GALNT3 KD clone,
while no expression alterations were observed for both GALNT6 and
T14.
Association of GalNAc-Ts expression with
clinicopathological data
We further proceeded to investigate the prognostic
values of the four differentially expressed GalNAc-Ts (T2, T6, T9
and T14) in EOC patients, as we evaluated the relationship between
the TMA immunostaining of the 4 GalNAc-Ts and the patient PFS data
using Kaplan-Meier survival curve analyses. PFS follow-up data were
available for 124 patients (Table
I). The analyses were based on the staining intensities: low
(includes staining levels of <2) and high (includes staining
levels of >2) (Fig. 4). No
significant differences were observed between GALNT2 and T9
expression levels and the patient PFS values (log-rank=2.715,
p=0.099) and (log-rank=0.995, p=0.319) respectively (Fig. 4A and C), which suggests that
staining intensities for both GALNT2 and T9 in pre-treatment
surgical EOC specimens are not predictive of PFS. However, there
was a significant association between PFS and the expression of the
two GalNAc-Ts T6 and T14 (log-rank=5.119, p=0.024) and
(log-rank=5.770, p=0.016) respectively (Fig. 4B and D).
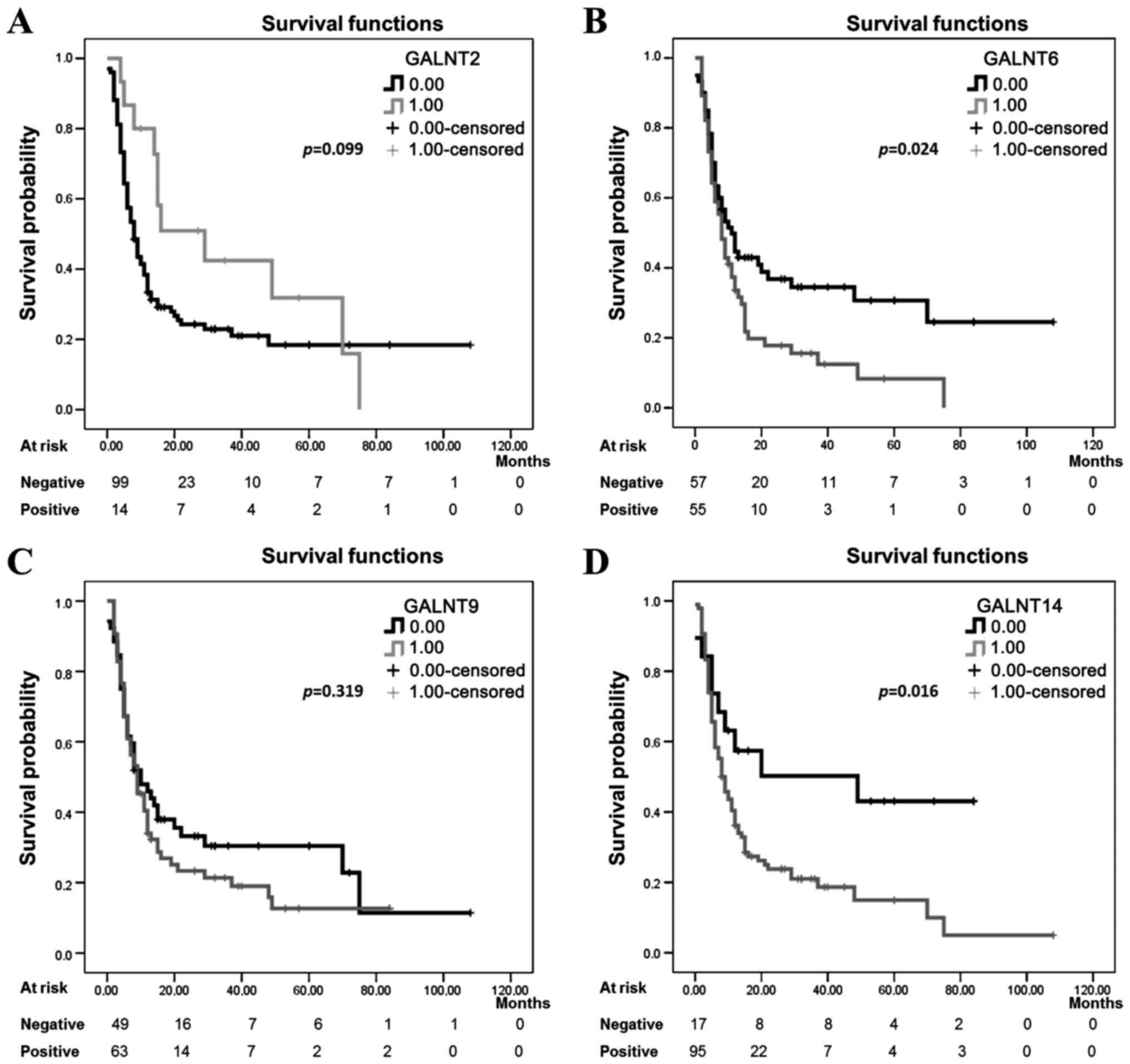 | Figure 4Kaplan-Meier progression-free
survival curves showing the association between GalNAc-Ts
expression patterns and prognosis in HG ovarian cancer.
Kaplan-Meier survival curve analysis of HG cases for
progression-free survival (PFS), presented as survival probability,
of patients whose tumors show a defined staining pattern. For
evaluation of GalNAc-Ts staining, HG ovarian cancer subjects were
divided into two groups, those with positive staining (3 or 2 gene
staining intensity) defined here as 1,00 on the Kaplan-Meier curves
and those with negative staining (1 or 0 gene staining intensity)
defined here as 0.00 on the Kaplan-Meier curves. The staining
pattern for (A) GALNT2 protein, negative (n=101), and positive
(n=15). (B) GALNT6 protein, negative (n=60), and positive (n=56).
(C) GALNT9 protein, negative (n=52), and positive (n=64). (D)
GALNT14 protein, negative (n=19), and positive (n=97). All p-values
were derived from log-rank tests. HG, high-grade. |
Bivariate and multivariate analyses to predict PFS
were also performed on all the 4 genes. Multivariate analyses
taking into account standard or strongly associated prognostic
variables (age, grade, stage and initial CA125) were performed to
identify independent prognostic factors. Multivariate analyses
showed a significant association for both GALNT6 and GALNT14 and
the higher risk of progression (HR, 1.672; CI, 1.043–2.682;
p=0.033) and (HR, 2.163; CI, 1.032–4.534; p= 0.041) respectively
(Table III), but not for GALNT2
and GALNT9 (HR, 0.339; CI, 0.103–1.119; p=0.076) and (HR, 1.470;
CI, 0.939–2.300; p=0.092) respectively (Table III). Taken together, our findings
indicate that GALNT6 and T14 may represent useful markers to
predict PFS of women with high grade serous EOC.
 | Table IIICox regression analysis to predict
progression-free survival (PFS). |
Table III
Cox regression analysis to predict
progression-free survival (PFS).
| Marker | Value | Frequency N
(%) | Event n (%) | Crude
| Adjusted
|
|---|
| HR | 95% CI | P-value | HRa | 95% CIb | P-value |
|---|
| GALNT2 | Negative | 101 (87.1%) | 78 (77.2%) | 1.0 | | | 1.0 | | |
| Positive | 15 (12.9%) | 11 (73.3%) | 0.599 | 0.317–1.130 | 0.113 | 0.339 | 0.103–1.119 | 0.076 |
| GALNT6 | Negative | 60 (51.7%) | 40 (66.7%) | 1.0 | | | 1.0 | | |
| Positive | 56 (48.3%) | 49 (87.5%) | 1.596 | 1.047–2.434 | 0.030 | 1.672 | 1.043–2.682 | 0.033 |
| GALNT9 | Negative | 52 (44.8%) | 37 (71.2%) | 1.0 | | | 1.0 | | |
| Positive | 64 (55.2%) | 52 (81.3%) | 1.232 | 0.807–1.880 | 0.334 | 1.470 | 0.939–2.300 | 0.092 |
| GALNT14 | Negative | 20 (17.2%) | 11 (55.0%) | 1.0 | | | 1.0 | | |
| Positive | 96 (82.8%) | 78 (81.3%) | 2.185 | 1.117–4.275 | 0.022 | 2.163 | 1.032–4.534 | 0.041 |
Discussion
Accumulating data are indicative for the important
role of glycosylation perturbations in carcinogenesis, including
also the abnormal expression of glycans, exclusively involved in
embryonic development under normal conditions (21). Glycan alterations can be associated
with cancer cell signaling and communication, tumor cell
dissociation, migration and invasion, cell-matrix interactions,
tumor angiogenesis, immune modulation and metastasis formation
(22,23), and can serve as important
biomarkers and/or therapeutic targets (24).
Altered expression of glycans in cancer is
frequently attributed to aberrant expression of different members
of the GalNAc-Ts family in malignant tumors compared to non-tumoral
tissue (7,14). The deregulation in the expression
of the different GalNAc-Ts allows them to play diverse roles in
carcinogenesis (14). We have
previously reported that one member of these GalNAc-Ts (GALNT3)
represents a potential EOC oncogene, as its expression
significantly correlated with shorter PFS intervals in EOC patients
with advanced disease (15);
however, data concerning the implication of other members of the
GalNAc-Ts gene family in EOC dissemination are rather scarce.
Indeed, only one study was suggestive for a possible role of
GALNT14 in mediating the malignant behavior of EOC cells (25). Otherwise, some of the most
extensively studied GalNAc-Ts in cancer include GALNT2, T4, T6, T9
and T14 (7), which represent the
transferases included in the present study. Thus, concerning
GALNT2, only one study is suggestive for a possible oncogenic role
of this transferase in oral cancer (26), while other studies in different
cancer types, including neuroblastoma, liver and gastric cancer,
are strongly supportive for a role of GALNT2 in suppressing
tumorigenesis (27–29). GALNT4 has also displayed discrepant
roles in different cancers, as an implication of GALNT4 in breast
carcinogenesis has been repeatedly demonstrated (30–32);
however low GALNT4 expression was associated with poor PFS and
overall survival of clear cell renal cell carcinoma patients
(33). GALNT6 has been extensively
studied for its implication in the malignant transformation and
metastasis of epithelial cancers (34–37),
and especially in breast cancer (38–40),
where GALNT6 has been suggested as a novel marker for breast cancer
detection and potential therapeutic target (41–44).
Similarly to GALNT2 and T4, literature data for the role of GALNT9
in carcinogenesis is rather contradictory, since a protective role
of GALNT9 was suggested in neuroblastoma and breast cancer
dissemination (45,46), while a recent report was indicative
for an oncogenic function of this enzyme in colorectal cancer
(47). Interestingly, GALNT9 was
among the genes identified as potentially hypomethylated and
overexpressed in advanced EOC as reported in our previous study
(48). Finally, GALNT14 has been
characterized as an oncogene in breast and lung cancer (49–51);
moreover, recent data are strongly indicative for a role of this
transferase in mediating chemotherapy resistance in different
cancer types (52–58).
These reports are indicative for either protective
or tumorigenic roles of the different GalNAc-Ts in cancer,
suggesting a highly complex and rather specific O-glycosylation
pattern of glycoproteins in the different cancer types. In this
study we initially analyzed the expression levels of the selected
five members of the polypeptide GalNAc-Ts family in EOC cell lines
and EOC tumor samples (HGSCs) by western blotting. Similar, if not
identical patterns of expression were observed for these GalNAc-Ts
in both EOC cell lines and EOC tumors, as GALNT2 displayed very low
or lack of expression in all EOC specimens studied, compared to
control samples (including HOSE cells and non-tumoral ovarian
tissues), while the expression levels of GALNT6, T9 and T14 were
significantly higher in EOC cells and EOC tumors, as compared to
very low/lack of expression in the corresponding controls (Fig. 1). Only GALNT4 showed very subtle,
or absence of any expression in all EOC and control specimens
analyzed, indicative for no implications of this enzyme in EOC
progression (Fig. 1). These
observations were further confirmed by performing IHC analysis of
the protein expression levels in numerous EOC tumors and control
tissues, using TMAs. Indeed, GALNT6, T9 and T14 exhibited very
strong staining in HGSC tissues and very weak/no staining in the
LMP tumors and control tissues, while GALNT2 displayed a rather
inverse staining pattern indicative for a subtle or no expression
in both HGSC and LMP tumors, compared to relatively strong
expression in control tissues (Fig.
2). Again, lack of any staining was observed for GALNT4 in all
(EOC tumor and control) tissue samples analyzed (Fig. 2). With the exception of GALNT4, our
data are rather in accordance with previous studies for the roles
of these GalNAc-Ts in different carcinoma types as summarized
above, and suggest for a possible correlation of GALNT6, T9 and T14
expression with EOC progression, while a putative protective role
of GALNT2 in EOC dissemination cannot be excluded.
Moreover, Kaplan-Meier survival curves and
consecutive Cox-regression analyses revealed that the expression
levels of two of the GalNAc-Ts analyzed (GALNT6 and GALNT14) were
significantly related with poor PFS in the studied cohort of HG
serous EOC patients (Fig. 4B and D
and Table III), which suggests
the putative use of these two transferases as novel prognostic
serous EOC biomarkers. These results, as well as data from our
previous study (15), are
indicative for the significant inverse association of three members
of the GalNAc-Ts gene family (GALNT3, T6 and T14) with disease
progression in HGSC patients.
Furthermore, the simultaneous overexpression of four
GalNAc-Ts (GALNT3, T6, T9 and T14) in advanced EOC raises the
question about the specific and/or redundant functions of the
members of this gene family in normal and pathological conditions,
including EOC. Notably, it is known that the different GalNAc-Ts do
have partially overlapping but distinct substrate specificities,
which may result in these GalNAc-Ts having partial functional
redundancy (59). So far, twenty
members of the human GalNAc-Ts gene family have been identified
(20), as such an abundance of
GalNAc-Ts could provide substantial biosynthetic back-up. Although
different GalNAc-Ts are differentially expressed within tissues,
between cells, within a single tissue, and in different patterns at
different stages in the development and differentiation, it is now
becoming clear that a subset of the GalNAc-Ts display both distinct
and overlapping substrate specificities (11,13).
Some of the mammalian GalNAc-transferase isoforms have been grouped
into subfamilies based on their high homology (11,20).
One such example is the human subfamily comprised of GALNT3 and
GALNT6, displaying 65% homology in their coding sequence, although
this homology does not provide complete functional redundancy
(59). Interestingly, GALNT6
displayed quite similar oncogenic functions in breast cancer
(modulating aberrant O-glycosylation and MUC1 stabilization)
(39), as those found by us for
GALNT3 in EOC (15). Increased
GALNT3 and T6 co-expression has been detected in pancreatic
(60) and renal (61) carcinomas, suggestive for GALNT3/T6
complementary correlations. As shown (60,61),
the upregulation of GALNT3 within the malignant transformation and
progression of these cancers could partly depend on that of GALNT6
in both synergistic and compensatory ways. Moreover, not a separate
GALNT3 or GALNT6 KD, but only double GALNT3/GALNT6 KD was shown to
inhibit TGFβ-induced epithelial-to-mesenchymal transition (EMT) in
prostate cancer cells (35).
Similarly, our western blotting and consecutive MCA analyses
described herein (Fig. 3) are
strongly suggestive for a possible functional overlap and
redundancy of GALNT3 and T6 in EOC. Based on the above
considerations we can suggest that different GalNAc-Ts perform
redundant and/or overlapping functions in disease progression of
women with HGSC. We believe that these relationships need to be
examined further, both in vitro and in vivo to better
understand how these transferases function together in initiating
the biosynthesis of specific target glycoproteins, and if they
actually demonstrate compensatory methods that may enhance their
implication in disease progression.
In conclusion, we have shown that four members of
the GalNAc-Ts gene family are differentially expressed in EOC
specimens, as GALNT6, T9 and T14 were significantly over-expressed
in both EOC cell lines and HGSCs, while GALNT2 displayed an
inversed expression pattern indicative for very weak or no
expression in both EOC cells and EOC tumors, compared to relatively
strong expression in HOSE cells and control tissues. Importantly,
GALNT6 and GALNT14 expression significantly correlated with poor
prognosis of EOC patients with advanced disease. These data and our
previously published data are indicative for a possible implication
of different members of the GalNAc-Ts gene family (including
GALNT2, T3, T6, T9 and T14) in modulating EOC progression, as
GALNT6 and GALNT14, together with the previously characterized
GALNT3, could represent novel prognostic EOC biomarkers. Moreover,
our results are suggestive for overlapping functions of some
GalNAc-Ts, and especially GALNT3 and GALNT6, in EOC, in conformity
with GALNT3/T6 functional redundancy described in other cancer
types. Further functional studies are warranted to more completely
elucidate in vitro and in vivo the individual and/or
synergistic implications of the members of the GalNAc-Ts gene
family in ovarian tumorigenesis.
Acknowledgments
This study was supported by a grant to D.B. from the
Cancer Research Society of Canada. Clinical specimens were provided
by the 'Banque de tissus et de données of the Réseau de recherche
sur le cancer' of the 'Fonds de recherche du Québec - Santé
(FRQ-S)', associated with the Canadian Tumor Repository Network
(CTRNet). We would also like to thank Ms. Anne-Sophie Julien from
the 'Plateforme de Recherche Clinique du CHU de Québec-Université
Laval' for helping us with the statistical analyses and
interpretation of the data generated. In addition, we would like to
thank Mr. Khaly Mbodji from the Department of Radi-oncologie and
the 'Centre de recherché du Chu de Québec' for his help with the
statistical analyses.
Glossary
Abbreviations
Abbreviations:
|
EOC
|
epithelial ovarian cancer
|
|
GalNAc-Ts
|
GalNAc transferases
|
|
HGSC
|
high-grade serous carcinoma
|
|
HOSE
|
human ovarian surface epithelial
|
|
IHC
|
immunohistochemistry
|
|
KD
|
knockdown
|
|
LMP
|
low-malignant potential
|
|
MD
|
moderately differentiated
|
|
MCA
|
multiple correspondence analyses
|
|
PD
|
poorly differentiated
|
|
PTM
|
post-translational modifications
|
|
PFS
|
progression-free survival
|
|
SOC
|
serous ovarian carcinomas
|
|
SNO
|
superficial scrapings from normal
ovaries
|
|
TMAs
|
tissue microarrays
|
References
|
1
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fruscio R, Corso S, Ceppi L, Garavaglia D,
Garbi A, Floriani I, Franchi D, Cantù MG, Bonazzi CM, Milani R, et
al: Conservative management of early-stage epithelial ovarian
cancer: Results of a large retrospective series. Ann Oncol.
24:138–144. 2013. View Article : Google Scholar
|
|
3
|
Alouini S: Management of ovarian cancer
has changed. Gynecol Oncol. 126:313author reply 314. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pinho SS and Reis CA: Glycosylation in
cancer: Mechanisms and clinical implications. Nat Rev Cancer.
15:540–555. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Stowell SR, Ju T and Cummings RD: Protein
glycosylation in cancer. Annu Rev Pathol. 10:473–510. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Abbott KL: Glycomic analysis of ovarian
cancer: Past, present, and future. Cancer Biomark. 8:273–280. 2011.
View Article : Google Scholar
|
|
7
|
Sheta R and Bachvarov D: Role of aberrant
glycosylation in ovarian cancer dissemination. Biomedecial Rev.
25:83–92. 2014. View Article : Google Scholar
|
|
8
|
Tabak LA: The role of mucin-type O-glycans
in eukaryotic development. Semin Cell Dev Biol. 21:616–621. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ten Hagen KG, Fritz TA and Tabak LA: All
in the family: the UDP-GalNAc:polypeptide
N-acetylgalactosaminyltransferases. Glycobiology. 13:1R–16R. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hang HC and Bertozzi CR: The chemistry and
biology of mucin-type O-linked glycosylation. Bioorg Med Chem.
13:5021–5034. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bennett EP, Mandel U, Clausen H, Gerken
TA, Fritz TA and Tabak LA: Control of mucin-type O-glycosylation: A
classification of the polypeptide GalNAc-transferase gene family.
Glycobiology. 22:736–756. 2012. View Article : Google Scholar :
|
|
12
|
Bennett EP, Hassan H, Mandel U,
Mirgorodskaya E, Roepstorff P, Burchell J, Taylor-Papadimitriou J,
Hollingsworth MA, Merkx G, van Kessel AG, et al: Cloning of a human
UDP-N-acetyl-alpha-D-Galactosamine:polypeptide
N-acetylgalactosaminyltransferase that complements other
GalNAc-transferases in complete O-glycosylation of the MUC1 tandem
repeat. J Biol Chem. 273:30472–30481. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Beaman EM and Brooks SA: The extended
ppGalNAc-T family and their functional involvement in the
metastatic cascade. Histol Histopathol. 29:293–304. 2014.
|
|
14
|
Brooks SA, Carter TM, Bennett EP, Clausen
H and Mandel U: Immunolocalisation of members of the polypeptide
N-acetylgalactosaminyl transferase (ppGalNAc-T) family is
consistent with biologically relevant altered cell surface
glycosylation in breast cancer. Acta Histochem. 109:273–284. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang ZQ, Bachvarova M, Morin C, Plante M,
Gregoire J, Renaud MC, Sebastianelli A and Bachvarov D: Role of the
polypeptide N-acetylgalactosaminyltransferase 3 in ovarian cancer
progression: Possible implications in abnormal mucin
O-glycosylation. Oncotarget. 5:544–560. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheta R, Woo CM, Roux-Dalvai F, Fournier
F, Bourassa S, Droit A, Bertozzi CR and Bachvarov D: A metabolic
labeling approach for glycoproteomic analysis reveals altered
glycoprotein expression upon GALNT3 knockdown in ovarian cancer
cells. J Proteomics. 145:91–102. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sheta R, Roux-Dalvai F, Woo CM, Fournier
F, Bourassa S, Bertozzi CR, Droit A and Bachvarov D: Proteomic
dataset for altered glycoprotein expression upon GALNT3 knockdown
in ovarian cancer cells. Data Brief. 8:342–349. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taylor PT and Haverstick D: Re: New
guidelines to evaluate the response to treatment in solid tumors
(ovarian cancer). J Natl Cancer Inst. 97:151author reply 152. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Elgaaen BV, Olstad OK, Sandvik L, Odegaard
E, Sauer T, Staff AC and Gautvik KM: ZNF385B and VEGFA are strongly
differentially expressed in serous ovarian carcinomas and correlate
with survival. PLoS One. 7:e463172012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Marth JD: Complexity in O-linked
oligosaccharide biosynthesis engendered by multiple polypeptide
N-acetylgalactosaminyltransferases. Glycobiology. 6:701–705. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dube DH and Bertozzi CR: Glycans in cancer
and inflammation-potential for therapeutics and diagnostics. Nat
Rev Drug Discov. 4:477–488. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hakomori S: Glycosylation defining cancer
malignancy: New wine in an old bottle. Proc Natl Acad Sci USA.
99:10231–10233. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Christiansen MN, Chik J, Lee L, Anugraham
M, Abrahams JL and Packer NH: Cell surface protein glycosylation in
cancer. Proteomics. 14:525–546. 2014. View Article : Google Scholar
|
|
24
|
Fuster MM and Esko JD: The sweet and sour
of cancer: Glycans as novel therapeutic targets. Nat Rev Cancer.
5:526–542. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang R, Yu C, Zhao D, Wu M and Yang Z: The
mucin-type glycosylating enzyme polypeptide
N-acetylgalactosaminyltransferase 14 promotes the migration of
ovarian cancer by modifying mucin 13. Oncol Rep. 30:667–676. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lin MC, Huang MJ, Liu CH, Yang TL and
Huang MC: GALNT2 enhances migration and invasion of oral squamous
cell carcinoma by regulating EGFR glycosylation and activity. Oral
Oncol. 50:478–484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ho WL, Chou CH, Jeng YM, Lu MY, Yang YL,
Jou ST, Lin DT, Chang HH, Lin KH, Hsu WM, et al: GALNT2 suppresses
malignant phenotypes through IGF-1 receptor and predicts favorable
prognosis in neuroblastoma. Oncotarget. 5:12247–12259. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu YM, Liu CH, Hu RH, Huang MJ, Lee JJ,
Chen CH, Huang J, Lai HS, Lee PH, Hsu WM, et al: Mucin
glycosylating enzyme GALNT2 regulates the malignant character of
hepatocellular carcinoma by modifying the EGF receptor. Cancer Res.
71:7270–7279. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu SY, Shun CT, Hung KY, Juan HF, Hsu CL,
Huang MC and Lai IR: Mucin glycosylating enzyme GALNT2 suppresses
malignancy in gastric adenocarcinoma by reducing MET
phos-phorylation. Oncotarget. 7:11251–11262. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wright PK, May FE, Darby S, Saif R,
Lennard TW and Westley BR: Estrogen regulates vesicle trafficking
gene expression in EFF-3, EFM-19 and MCF-7 breast cancer cells. Int
J Clin Exp Pathol. 2:463–475. 2009.PubMed/NCBI
|
|
31
|
Niang B, Jin L, Chen X, Guo X, Zhang H, Wu
Q, Padhiar AA, Xiao M, Fang D and Zhang J: GalNAc-T4 putatively
modulates the estrogen regulatory network through FOXA1
glycosylation in human breast cancer cells. Mol Cell Biochem.
411:393–402. 2016. View Article : Google Scholar
|
|
32
|
Zhang J, Zhang Z, Wang Q, Xing XJ and Zhao
Y: Overexpression of microRNA-365 inhibits breast cancer cell
growth and chemoresistance through GALNT4. Eur Rev Med Pharmacol
Sci. 20:4710–4718. 2016.PubMed/NCBI
|
|
33
|
Liu Y, Liu W, Xu L, Liu H, Zhang W, Zhu Y,
Xu J and Gu J: GALNT4 predicts clinical outcome in patients with
clear cell renal cell carcinoma. J Urol. 192:1534–1541. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Z, Yamada S, Wu Y, Wang KY, Liu YP,
Uramoto H, Kohno K and Sasaguri Y: Polypeptide
N-acetylgalactosaminyltransferase-6 expression independently
predicts poor overall survival in patients with lung adenocarcinoma
after curative resection. Oncotarget. 7:54463–54473. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Freire-de-Lima L, Gelfenbeyn K, Ding Y,
Mandel U, Clausen H, Handa K and Hakomori SI: Involvement of
O-glycosylation defining oncofetal fibronectin in
epithelial-mesenchymal transition process. Proc Natl Acad Sci USA.
108:17690–17695. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tarhan YE, Kato T, Jang M, Haga Y, Ueda K,
Nakamura Y and Park JH: Morphological changes, cadherin switching,
and growth suppression in pancreatic cancer by GALNT6 knockdown.
Neoplasia. 18:265–272. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Gomes J, Marcos NT, Berois N, Osinaga E,
Magalhães A, Pinto-de-Sousa J, Almeida R, Gärtner F and Reis CA:
Expression of UDP-N-acetyl-D-galactosamine: Polypeptide
N-acetylgalactosaminyltransferase-6 in gastric mucosa, intestinal
metaplasia, and gastric carcinoma. J Histochem Cytochem. 57:79–86.
2009. View Article : Google Scholar :
|
|
38
|
Liesche F, Kölbl AC, Ilmer M, Hutter S,
Jeschke U and Andergassen U: Role of
N-acetylgalactosaminyltransferase 6 in early tumorigenesis and
formation of metastasis. Mol Med Rep. 13:4309–4314. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Park JH, Nishidate T, Kijima K, Ohashi T,
Takegawa K, Fujikane T, Hirata K, Nakamura Y and Katagiri T:
Critical roles of mucin 1 glycosylation by transactivated
polypeptide N-acetylgalactosaminyltransferase 6 in mammary
carcinogenesis. Cancer Res. 70:2759–2769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Park JH, Katagiri T, Chung S, Kijima K and
Nakamura Y: Polypeptide N-acetylgalactosaminyltransferase 6
disrupts mammary acinar morphogenesis through O-glycosylation of
fibronectin. Neoplasia. 13:320–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Berois N, Mazal D, Ubillos L, Trajtenberg
F, Nicolas A, Sastre-Garau X, Magdelenat H and Osinaga E:
UDP-N-acetyl-D-galactosamine: Polypeptide
N-acetylgalactosaminyltransferase-6 as a new immunohistochemical
breast cancer marker. J Histochem Cytochem. 54:317–328. 2006.
View Article : Google Scholar
|
|
42
|
Freire T, Berois N, Sóñora C, Varangot M,
Barrios E and Osinaga E: UDP-N-acetyl-D-galactosamine:polypeptide
N-acetylgalactosaminyltransferase 6 (ppGalNAc-T6) mRNA as a
potential new marker for detection of bone marrow-disseminated
breast cancer cells. Int J Cancer. 119:1383–1388. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Patani N, Jiang W and Mokbel K: Prognostic
utility of glycosyltransferase expression in breast cancer. Cancer
Genomics Proteomics. 5:333–340. 2008.
|
|
44
|
Kölbl AC, Hiller RA, Ilmer M, Liesche F,
Heublein S, Schröder L, Hutter S, Friese K, Jeschke U and
Andergassen U: Glycosyltransferases as marker genes for the
quantitative polymerase chain reaction-based detection of
circulating tumour cells from blood samples of patients with breast
cancer undergoing adjuvant therapy. Mol Med Rep. 12:2933–2938.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pangeni RP, Channathodiyil P, Huen DS,
Eagles LW, Johal BK, Pasha D, Hadjistephanou N, Nevell O, Davies
CL, Adewumi AI, et al: The GALNT9, BNC1 and CCDC8 genes are
frequently epigenetically dysregulated in breast tumours that
metastasise to the brain. Clin Epigenetics. 7:572015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Berois N, Gattolliat CH, Barrios E,
Capandeguy L, Douc-Rasy S, Valteau-Couanet D, Bénard J and Osinaga
E: GALNT9 gene expression is a prognostic marker in neuroblastoma
patients. Clin Chem. 59:225–233. 2013. View Article : Google Scholar
|
|
47
|
Tuupanen S, Hänninen UA, Kondelin J, von
Nandelstadh P, Cajuso T, Gylfe AE, Katainen R, Tanskanen T,
Ristolainen H, Böhm J, et al: Identification of 33 candidate
oncogenes by screening for base-specific mutations. Br J Cancer.
111:1657–1662. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Keita M, Wang ZQ, Pelletier JF, Bachvarova
M, Plante M, Gregoire J, Renaud MC, Mes-Masson AM, Paquet ÉR and
Bachvarov D: Global methylation profiling in serous ovarian cancer
is indicative for distinct aberrant DNA methylation signatures
associated with tumor aggressiveness and disease progression.
Gynecol Oncol. 128:356–363. 2013. View Article : Google Scholar
|
|
49
|
Huanna T, Tao Z, Xiangfei W, Longfei A,
Yuanyuan X, Jianhua W, Cuifang Z, Manjing J, Wenjing C, Shaochuan
Q, et al: GALNT14 mediates tumor invasion and migration in breast
cancer cell MCF-7. Mol Carcinog. 54:1159–1171. 2015. View Article : Google Scholar
|
|
50
|
Wu C, Guo X, Wang W, Wang Y, Shan Y, Zhang
B, Song W, Ma S, Ge J, Deng H, et al:
N-Acetylgalactosaminyltransferase-14 as a potential biomarker for
breast cancer by immunohistochemistry. BMC Cancer. 10:1232010.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Kwon OS, Oh E, Park JR, Lee JS, Bae GY,
Koo JH, Kim H, Choi YL, Choi YS, Kim J, et al: GalNAc-T14 promotes
metastasis through Wnt dependent HOXB9 expression in lung
adenocarcinoma. Oncotarget. 6:41916–41928. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Wagner KW, Punnoose EA, Januario T,
Lawrence DA, Pitti RM, Lancaster K, Lee D, von Goetz M, Yee SF,
Totpal K, et al: Death-receptor O-glycosylation controls tumor-cell
sensitivity to the proapoptotic ligand Apo2L/TRAIL. Nat Med.
13:1070–1077. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wu C, Shan Y, Liu X, Song W, Wang J, Zou
M, Wang M and Xu D: GalNAc-T14 may be involved in regulating the
apoptotic action of IGFBP-3. J Biosci. 34:389–395. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Liang KH, Lin CL, Chen SF, Chiu CW, Yang
PC, Chang ML, Lin CC, Sung KF, Yeh C, Hung CF, et al: GALNT14
genotype effectively predicts the therapeutic response in
unresectable hepatocellular carcinoma treated with transcatheter
arterial chemoembolization. Pharmacogenomics. 17:353–366. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lin WR, Chiang JM, Liang KH, Lim SN, Lai
MW, Tsou YK, Hsieh TY, Hsu CK and Yeh CT: GALNT14 genotype predicts
postoperative outcome of stage III colorectal cancer with
oxaliplatin as adjuvant chemotherapy. Medicine (Baltimore).
95:e34872016. View Article : Google Scholar
|
|
56
|
Liang KH, Lin CC and Yeh CT: GALNT14 SNP
as a potential predictor of response to combination chemotherapy
using 5-FU, mitoxantrone and cisplatin in advanced HCC.
Pharmacogenomics. 12:1061–1073. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Soria JC, Márk Z, Zatloukal P, Szima B,
Albert I, Juhász E, Pujol JL, Kozielski J, Baker N, Smethurst D, et
al: Randomized phase II study of dulanermin in combination with
paclitaxel, carboplatin, and bevacizumab in advanced non-small-cell
lung cancer. J Clin Oncol. 29:4442–4451. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Thorburn A, Behbakht K and Ford H: TRAIL
receptor-targeted therapeutics: Resistance mechanisms and
strategies to avoid them. Drug Resist Updat. 11:17–24. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bennett EP, Hassan H, Mandel U,
Hollingsworth MA, Akisawa N, Ikematsu Y, Merkx G, van Kessel AG,
Olofsson S and Clausen H: Cloning and characterization of a close
homologue of human UDP-N-acetyl-alpha-D-galactosamine:Polypeptide
N-acetylgalactosaminyltransferase-T3, designated GalNAc-T6.
Evidence for genetic but not functional redundancy. J Biol Chem.
274:25362–25370. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li Z, Yamada S, Inenaga S, Imamura T, Wu
Y, Wang KY, Shimajiri S, Nakano R, Izumi H, Kohno K, et al:
Polypeptide N-acetylgalactosaminyltransferase 6 expression in
pancreatic cancer is an independent prognostic factor indicating
better overall survival. Br J Cancer. 104:1882–1889. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Kitada S, Yamada S, Kuma A, Ouchi S,
Tasaki T, Nabeshima A, Noguchi H, Wang KY, Shimajiri S, Nakano R,
et al: Polypeptide N-acetylgalactosaminyl transferase 3
independently predicts high-grade tumours and poor prognosis in
patients with renal cell carcinomas. Br J Cancer. 109:472–481.
2013. View Article : Google Scholar : PubMed/NCBI
|