Introduction
Oral cancer occurs at various parts of the oral
cavity, such as the tongue, gingiva, and buccal mucosa, and it
represents the sixth most common malignant neoplasm worldwide
(1). Annually, 28,030 new cases
and 5,850 deaths have been estimated in the United States (1). Oral squamous cell carcinoma
represents ~90% of oral cancer cases. As disease progression
depends on differences in oral regions, which vary in structure and
function, the choice of treatment depends on the anatomical primary
region of cancer. The current standard therapy is surgery followed
by radiotherapy for early and localized advanced oral cancer. The
survival rate has been reported to be up to 80% in the early stage
and only 20–30% in the late stage (2). Deficiency in oral function is
unavoidable after surgery for oral cancer, even with reconstructive
surgery, such as tissue transplantation (3). After surgery and radiotherapy, local
and regional recurrences have been reported in up to 90% of oral
cancer patients, and additional adjuvant chemotherapy has been
shown to improve survival (4–6).
Despite recent advances in imaging, surgery, radiation, and
systemic therapies, overall survival has improved by 15% in the
last 50 years and only 5% in the last 20 years (7). Therefore, further development of
non-surgical therapies, such as those targeting cancer-specific
molecules, with no or low risks of adverse effects are important
for oral cancer treatment.
To identify potential molecular targets for the
diagnosis and treatment of cancer, we performed genome-wide gene
expression analysis and subsequent tissue microarray analysis of
solid tumor tissues and various normal tissues and isolated several
oncoantigens involved in the development and/or progression of
various solid cancers (8–30). Since members of the kinesin
superfamily such as mitotic kinesins are involved in key functions
during intracellular transport and cell division of cancers, we
further focused on the genes encoding kinesin proteins that are
upregulated in the majority of oral cancers, but scarcely expressed
in normal organs. During this process, we identified kinesin family
member 11 (KIF11) as a candidate target molecule. KIF11 encodes a
motor protein that belongs to the kinesin-like protein family.
Members of this protein family are known to be involved in various
kinds of spindle dynamics. The function of this gene product
includes chromosome positioning, centrosome separation, and bipolar
spindle establishment during cell mitosis (31–33).
Additionally, KIF11 plays a role in the transport of secretory
proteins from the Golgi complex to the cell surface in non-mitotic
cells (34). Some reports stated
that KIF11 was expressed in some human cancers, including lung
cancer, glioblastoma, malignant mesothelioma, and gastric cancer
(35–38); however, no report revealed details
of the oncogenic function and prognostic value of this protein as a
therapeutic and diagnostic target for oral cancer. Therefore, the
present study aimed to assess the role of KIF11 in oral cancer and
evaluate its role as a prognostic biomarker and therapeutic target
for treating oral cancer.
Materials and methods
Cell lines and tissue samples
Five oral cancer cell lines (FaDu, SCC9, CAL27, HSC3
and HSC4) and human keratinocytes isolated from normal oral mucosa
(HOMK100) were used in this study. FaDu, SCC9, and CAL27 were
purchased from American Type Culture Collection (ATCC, Rockville,
MD, USA). HSC3 and HSC4 were provided by RIKEN BioResource Center
(Tsukuba, Japan), and HOMK100 was purchased from Cell Research
Corporation (Singapore) (Table I).
The five oral cancer cell lines were grown in monolayers in
appropriate medium supplemented with 10% fetal bovine serum (FBS)
and antibiotics (Thermo Fisher Scientific, Waltham, MA, USA) and
were maintained at 37°C in an atmosphere of humidified air. HOMK
cells were grown in medium supplemented with EpiLife defined growth
supplement (Gibco, Grand Island, NY, USA). Eight oral squamous cell
cancer tissue samples were obtained from ProteoGenex (Inglewood,
CA, USA), and a normal tongue tissue was obtained from Clontech
(Palo Alto, CA, USA). The population of the cancer cells in oral
cancer tissues used in this study ranged from 60 to 100% (mean
82%). In addition, we obtained 99 oral cancer and adjacent normal
oral tissue samples from oral cancer patients who had undergone
curative surgery at Kumamoto University for immunostaining on
tissue microarrays. Individual institutional ethics committees
approved this study and the use of all clinical materials.
 | Table IThe human oral cancer cell lines and
oral mucosa keratinocyte. |
Table I
The human oral cancer cell lines and
oral mucosa keratinocyte.
| Cell line | Histology | Resource
distributor |
|---|
| FaDu | Squamous cell
carcinoma of pharynx | ATCCa |
| SCC9 | Squamous cell
carcinoma of tongue | ATCCa |
| CAL 27 | Squamous cell
carcinoma of tongue | ATCCa |
| HSC3 | Squamous cell
carcinoma of tongue | RIKEN BRCb |
| HSC4 | Squamous cell
carcinoma of tongue | RIKEN BRCb |
| HOMK | Human oral mucosa
keratinocyte | Cell Research Corp.
Pte Ltd. |
Quantitative real-time PCR
Total RNA was extracted from cultured cells and
clinical tissues using the Maxwell 16 LEV simply RNA purification
kit (Promega, Madison, WI, USA) according to the manufacture's
protocol. The RNA from cultured cells and clinical tissues was
reverse transcribed to cDNA using Prime Script RT Master Mix
(Takara Bio, Otsu, Shiga, Japan). Real-time PCR experiments were
carried out with TaqMan Universal Master Mix II (Thermo Fisher
Scientific). Each experiment was performed in triplicate.
ACTB (Hs01060665_g1) as an internal control and KIF11
(Hs00189698_m1) primer were used (Applied Biosytems, Warrington,
UK). The reaction conditions were as follows: initial denaturation
for 2 min at 50°C and 10 min at 95°C followed by 40 cycles of
denaturation (15 sec at 95°C and 60 sec at 60°C). Each PCR product
was run in triplicate. The relative KIF11 mRNA expression
was calculated by 2−ΔΔCt.
Western blot analysis
Cells were lysed in Pierce RIPA buffer (Thermo
Scientific) that included a 1% protease inhibitor cocktail (Thermo
Scientific). After homogenization, the cell lysates were incubated
on ice for 30 min and centrifuged at 15,000 rpm for 15 min to
separate the supernatant from cellular debris. The amount of total
protein was estimated using the Qubit Protein assay kit (Thermo
Scientific), and the proteins were then mixed with SDS sample
buffer and incubated at room temperature for 5 min after boiling at
100°C for 5 min. After electrophoresis on 10%
Mini-Protean®TGX gels (Bio Rad, Hercules, CA, USA), the
proteins were transferred onto Trans-Blot®Turbo 0.2
μm PVDF membranes (Bio-Rad). The membranes were blocked
using the iBind solution kit (Thermo Scientific) and incubated with
rabbit anti-KIF11 antibody (catalog no. HPA010568; Sigma-Aldrich,
St. Louis, MO, USA) or rabbit anti-actin antibody (catalog no.
4970, 13E5; Cell Signaling Technology, Danvers, MA, USA). Membranes
were incubated with anti-rabbit horseradish peroxidase
(HRP)-conjugated secondary antibody (GE Healthcare,
Buckinghamshire, UK) for 60 min at room temperature. Protein bands
were visualized using Image Quant LAS 4000 mini (GE
Healthcare).
Immunocytochemical analysis
Cultured cells were plated onto Lab-Tek II chamber
slides (Nalge Nunc International, Rochester, NY, USA). The cells
were washed twice with phosphate-buffered saline (PBS) (-), fixed
with 4% paraformaldehyde for 10 min at room temperature, and
permeabilized with 0.1% Triton X-100 in PBS (-) for 3 min at room
temperature. Non-specific binding was blocked with CAS-Block
(Invitrogen, Carlsbad, CA, USA) for 7 min at room temperature
before the primary antibody reaction. Then, cells were incubated
with rabbit anti-KIF11 antibody (catalog no. HPA010568;
Sigma-Aldrich) in PBS (-) containing 1% bovine serum albumin (BSA)
for 60 min at room temperature in a wet box. After washing with PBS
(-), the cells were stained with Alexa 488-conjugated anti-rabbit
secondary antibody (Life Technologies, Grand Island, NY, USA) for
60 min at room temperature in a wet box to protect from light.
After washing again with PBS (-), the cells were mounted using
Vectashields mounting medium containing DAPI (Vector Laboratories,
Inc. Burlingame, CA, USA) and were visualized using BZ-X710
(Keyence, Osaka, Japan).
Immunohistochemistry and tissue
microarray
Tumor tissue microarrays were constructed with 99
formalin-fixed, paraffin-embedded primary oral cancer samples, each
of which had been obtained as mentioned above. The tissue area for
sampling was selected on the basis of visual alignment with the
corresponding hematoxylin and eosin (H&E)-stained section on a
slide. For construction of the microarrays, 3, 4, or 5 tissue cores
(diameter, 0.6 mm; height, 3–4 mm) taken from a donor tumor block
were placed into a recipient paraffin block with a tissue
microarrayer (Beecher Instruments, Silver Spring, MD, USA). A core
of normal oral epithelial tissue was punched in each case, and 5
μm sections of the resulting microarray block were used for
immunohistochemical analysis.
To investigate the clinicopathological significance
of KIF11 expression in oral cancers, we stained tissue sections
using rabbit anti-KIF11 antibody (catalog no. HPA010568;
Sigma-Aldrich) and Envision + Kit/HRP (Dako Cytomation Carpenteria,
CA, USA). Tissue microarray slides were deparaffinized, and
heat-induced antigen retrieval was accomplished in Target Retrieval
Solution (pH 9.0) with a Pascal pressurized heating chamber (Dako
Cytomation). Anti-KIF11 antibody was added after endogenous
peroxidase blocking (Dako Cytomation) and protein blocking. Each
section was incubated with HRP-labeled anti-rabbit IgG as the
secondary antibody for 30 min. Substrate-chromogen was added, and
the specimens were counterstained with H&E. Three independent
investigators without prior knowledge of clinicopathological
information semiquantitatively assessed KIF11 positivity. The
intensity of KIF11 staining was evaluated using the following
criteria: strongly positive (scored as 2+), brown staining in
>70% of tumor cells completely obscuring cytoplasm; weakly
positive (scored as 1+), brown staining in 10–70% of tumor cells;
and absent (scored as 0), brown staining in <10% of tumor cells.
Cases were considered as strongly positive only if two or more
investigators independently identified them as strongly
positive.
RNA interference assay
To evaluate the biological functions of KIF11 in
oral cancer cells, we used small-interfering RNA (siRNA) duplexes
(Sigma-Aldrich). The sequences targeting each gene were as follows:
si-KIF11-#1, 5′-CAGAUUGAUGUUUACCGAATT-3′; si-KIF11-#2,
5′-GAAACUUACUGAUAAUGGUTT-3′; si-EGFP, 5′-GAAGCAGCACGACUUCUUCTT-3′;
si-LUC, 5′-CGUACGCGGAAUACUUCGATT-3′. The oral cancer cell lines
HSC4 and FaDu were transfected with either of the siRNAs using
Lipofectamine 2000 reagent (Invitrogen) according to the
manufacturer's instructions. Cell numbers and cell viability were
measured using the colony formation assay with Giemsa staining and
MTT assay with cell counting kit-8 solution (Dojindo Laboratories,
Kumamoto, Japan) at 7 days after transfection. These cells were
also used for a caspase-3/7 assay (Image-iT™ LIVE Green Caspase-3
and -7 Detection kit, Thermo Fisher Scientific) at 3 days after
transfection.
KIF11 inhibitor assay
To evaluate the essential role of the kinesin motor
ATPase activity of KIF11 in oral cancer cells, we treated HSC4
cells with 0, 0.1, 1.0 or 10 nM of a small compound inhibitor
against KIF11 (SB743921; Medkoo Biosciences, Chapel Hill, NC, USA),
which blocks the ATPase activity of KIF11. These cells were used
for MTT assay, flow cytometry, and live-cell imaging.
Flow cytometric analysis
Flow cytometric analysis was performed using the
Cycletest Plus DNA reagent kit and the FACS Verse system (BD
Bioscience, San Jose, CA, USA). In this analysis, 72 h after siRNA
transfection into HSC4 and FaDu cells or 48 h after treatment of
HSC4 cells with SB743921, 1.0×106 oral cancer cells were
collected for staining DNA ploidy. The sample was filtered through
a 50-μm nylon mesh, and it was stored in the dark on ice and
kept ready for analyzing the cell cycle within 3 h using a flow
cytometer (BD FACS Verse). Additionally, 20,000 ungated cells were
analyzed for DNA content. Evaluation of apoptosis was performed
using the Annexin V assay at 72 h after transfection of siRNAs into
HSC4 and FaDu cells.
Live-cell imaging
Live-cell imaging was performed using the Evos FL
Auto cell imaging system (Life Technologies) to monitor
cytokinetics after transfection of siRNAs for KIF11 or SB743921
treatment. HSC4 cells were seeded into 35 mm glass dishes in RPMI
containing 10% FBS. Forty-eight hours after transfection of siRNAs
for KIF11 or control siRNA into cells, or immediately after
treatment of cells with SB743921 or control PBS (-), live-cell
imaging was started and images were captured every 20 or 30 min for
24 h.
Statistical analysis
We examined the correlation of KIF11 protein
expression levels determined in tissue microarray analysis with
relevant variables, such as patient sex, patient age, primary tumor
region, pT factor (pathologic tumor classification), and pN factor
(pathologic lymph node classification). Tumor-specific survival
curves were constructed from the date of surgery to the time of
death related to oral cancer or to the last follow-up observation.
Kaplan-Meier curves were constructed for each relevant variable and
for KIF11 expression. Differences in survival times among patient
subgroups were analyzed using the log-rank test. Univariate and
multivariate analyses were performed with Cox proportional hazards
models to determine the prognostic factors in patients with oral
cancer. We first analyzed associations between death and possible
prognostic factors, including strong KIF11 expression, age, sex,
primary region, pT factor, and pN factor. Then, a multivariate Cox
analysis was applied in stepwise procedures that always forced
strong KIF11 expression into the model. All statistical analyses
were performed using the StatView statistical program (SAS, Cary,
NC, USA).
Results
KIF11 expression in oral cancer cell
lines and tissues
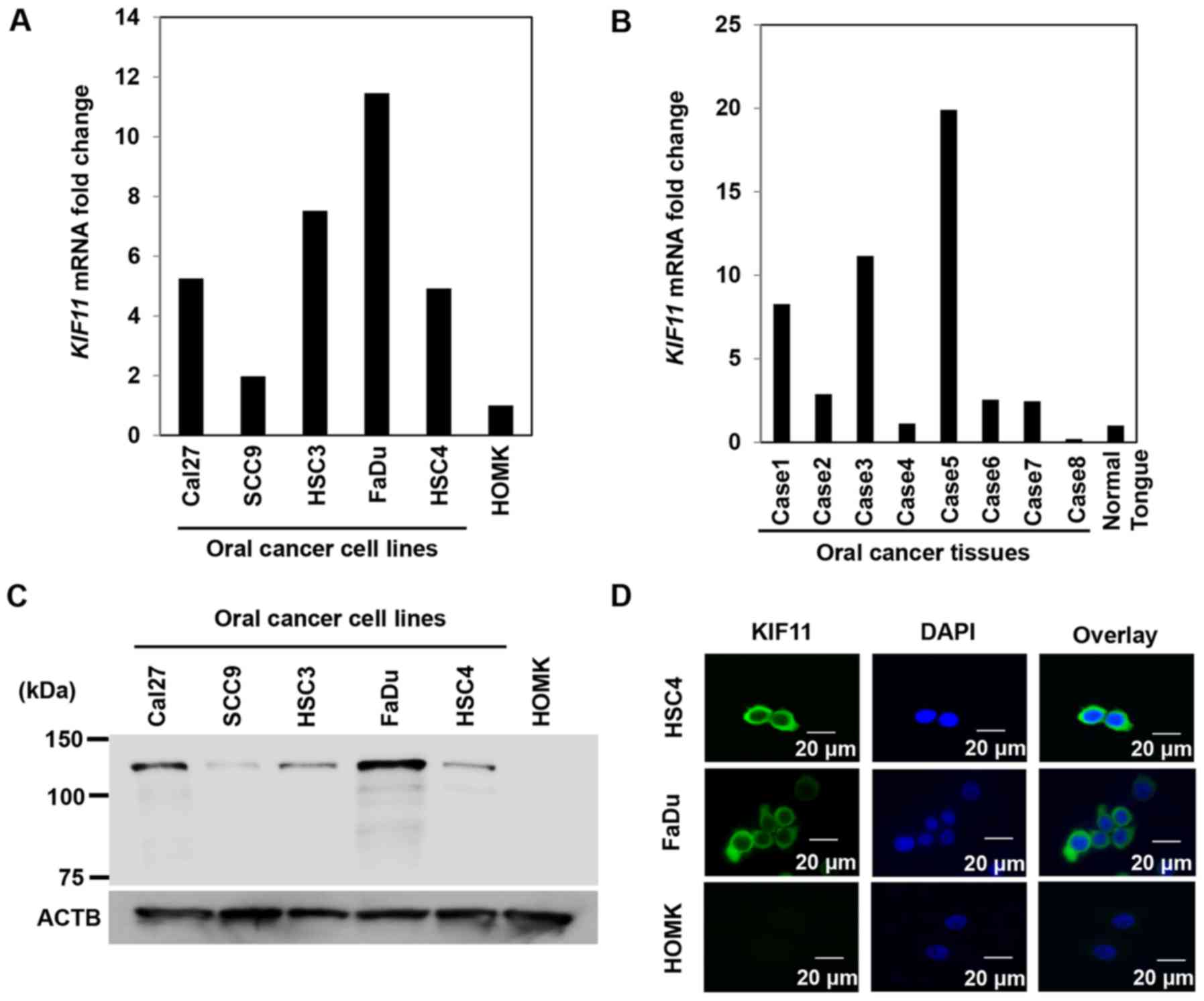
On real-time PCR, we noted high expression of KIF11
in all five oral cancer cell lines but scarce expression in normal
oral epithelial cells (HOMK) (Fig.
1A). In addition, real-time PCR identified higher KIF11
expression levels in 6 of 8 oral cancer tissues than in normal oral
epithelia (Fig. 1B). Western blot
analysis detected high levels of KIF11 protein expression in all
five oral cancer cell lines but hardly detected KIF11 protein
expression in normal epithelial cells (Fig. 1C). Furthermore, immunocytochemical
analysis identified the KIF11 protein in the cytoplasm of cancer
cells that expressed KIF11 (Fig.
1D).
Association of KIF11 expression with poor
prognosis in oral cancer patients
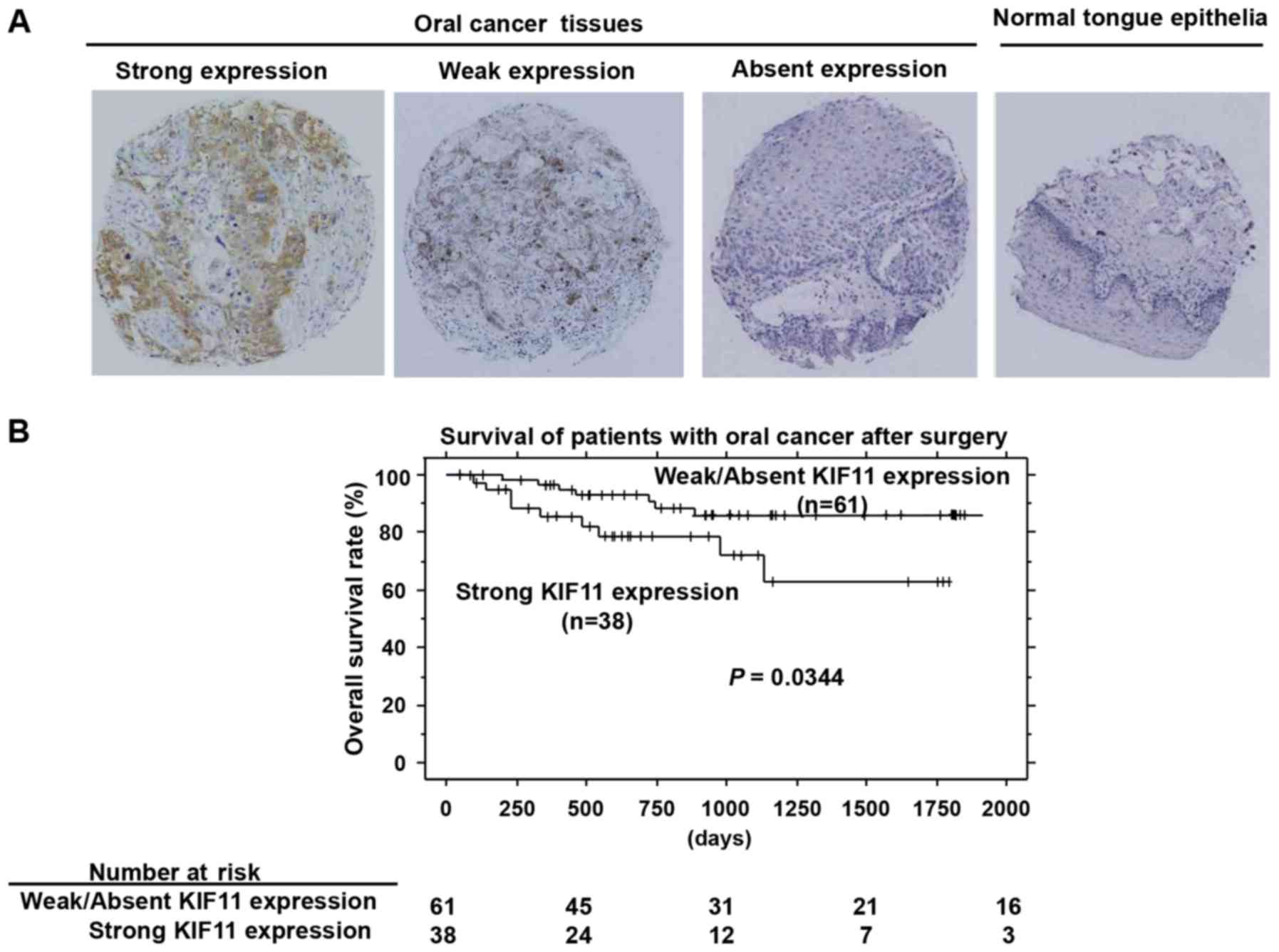
Immunohistochemical analysis using tissue
microarrays of 99 cases of oral cancers that underwent radical
operation demonstrated that KIF11 staining was mainly observed in
the cytoplasm of cancer cells. KIF11 was expressed in 64 of the 99
(64.6%) oral cancer cases (Fig.
2A). Strong, weak, and absent expressions were observed in 38
(38.4%), 26 (26.3%), and 35 (35.3%) of the 99 cases, respectively.
In contrast, positive staining was not observed in adjacent normal
tongue epithelial tissues. In the assessment of the association
between KIF11 expression and clinical parameters, a significant
correlation was noted between strong KIF11 positivity and pN factor
(higher in N1-2, P=0.0371 by Fisher's test; Table II). Furthermore, strong KIF11
expression was significantly correlated with shorter patient
survival compared to survival with weak or absent KIF11 expression
(P=0.0344, by log-rank test; Fig.
2B). We performed univariate analysis to investigate the
correlation of patient prognosis with other clinicopathological
factors, including age (<65 vs. ≥65 years), sex (female vs.
male), tumor location (tongue vs. other locations), pT
classification (T1-2 vs. T3-4), pN classification (N0 vs. N1-2),
and KIF11 expression status (weak/absent vs. strong). Among those
parameters, strong KIF11 expression, advanced pT stage, and
advanced pN stage were significantly associated with poorer
prognosis in oral cancer patients (P=0.0425, 0.0161 and 0.0021,
respectively, Table III).
Multivariate analysis showed that strong KIF11 expression was an
independent prognostic factor (P=0.0444, Table III).
 | Table IIAssociation of KIF11 protein
expression in oral cancer tissues with patients'
characteristics. |
Table II
Association of KIF11 protein
expression in oral cancer tissues with patients'
characteristics.
| Parameters | Total n=99 | Strong KIF11
expression, n=38 | Weak KIF11
expression, n=26 | Absent KIF11
expression, n=35 | P-value strong vs.
weak/absent |
|---|
| Sex |
| Male | 56 | 16 | 9 | 18 | >0.9999 |
| Female | 43 | 22 | 17 | 17 | |
| Age (years) |
| <65 | 42 | 14 | 12 | 16 | 0.2975 |
| ≥65 | 57 | 24 | 14 | 19 | |
| Region |
| Tongue | 52 | 22 | 18 | 12 | 0.5397b |
| Gingiva | 17 | 8 | 2 | 7 | |
| Buccal mucosa | 7 | 3 | 2 | 2 | |
| Others | 23 | 5 | 4 | 14 | |
| pT factor |
| T1-2 | 78 | 27 | 20 | 31 | 0.6155 |
| T3-4 | 21 | 9 | 4 | 8 | |
| pN factor |
| N0 | 89 | 31 | 24 | 34 | 0.0371a |
| N1-2 | 10 | 7 | 2 | 1 | |
 | Table IIICox's proportional hazards model
analysis of prognostic factors in patients with oral cancer. |
Table III
Cox's proportional hazards model
analysis of prognostic factors in patients with oral cancer.
| Variables | Hazards ratio | 95% CI |
Unfavorable/favorable | P-value |
|---|
| Univariate
analysis |
| Strong KIF11
expression | 2.798 | 1.035–7.560 | Strong/weak and
absent | 0.0425a |
| Age (years) | 2.803 | 0.901–8.715 | ≥65/<65 | 0.0749 |
| Sex | 2.048 | 0.761–5.510 | Male/female | 0.1557 |
| Region | 1.403 | 0.521–3.777 |
Tongue/othersb | 0.5026 |
| T-factor | 3.369 | 1.253–9.058 | T3-4/T1-2 | 0.0161a |
| N-factor | 6.393 | 1.957–20.887 | N1-2/N0 | 0.0021a |
| Multivariate
analysis |
| Strong KIF11
expression | 2.801 | 1.026–7.648 | Strong/weak and
absent | 0.0444a |
| T-factor | 2.212 | 0.712–6.872 | T3-4/T1-2 | 0.1699 |
| N-factor | 3.857 | 0.983–15.125 | N1-2/N0 | 0.0528 |
Growth inhibition in oral cancer cells by
knockdown of KIF11 expression
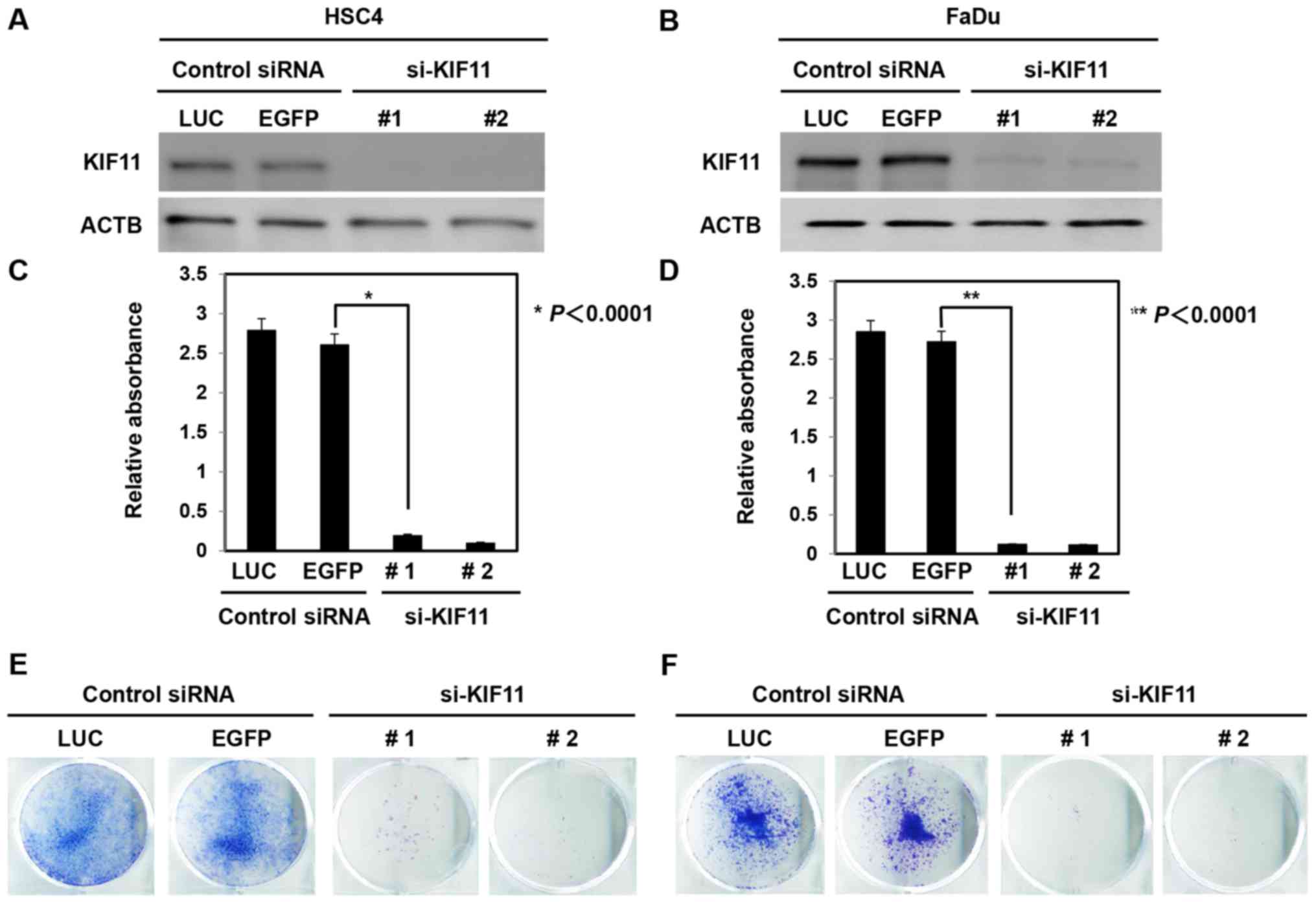
To confirm whether KIF11 expression could affect the
growth of oral cancer cells, siRNAs against KIF11 (siKIF11-#1 and
siKIF11-#2) and control siRNAs (si-LUC and si-EGFP) were
transfected into oral cancer cells (HSC4 and FaDu). Western
blotting revealed that si-KIF11 decreased KIF11 protein levels in
cancer cells compared to the levels in si-controls (Fig. 3A and B). In addition, suppression
of KIF11 expression significantly inhibited the cell viability of
HSC4 and FaDu cells (Fig. 3C and
D). The colony formation assay also showed that knockdown of
KIF11 expression decreased the numbers of HSC4 and FaDu cells
(Fig. 3E and F).
Induction of apoptosis in oral cancer
cells by knockdown of KIF11 expression
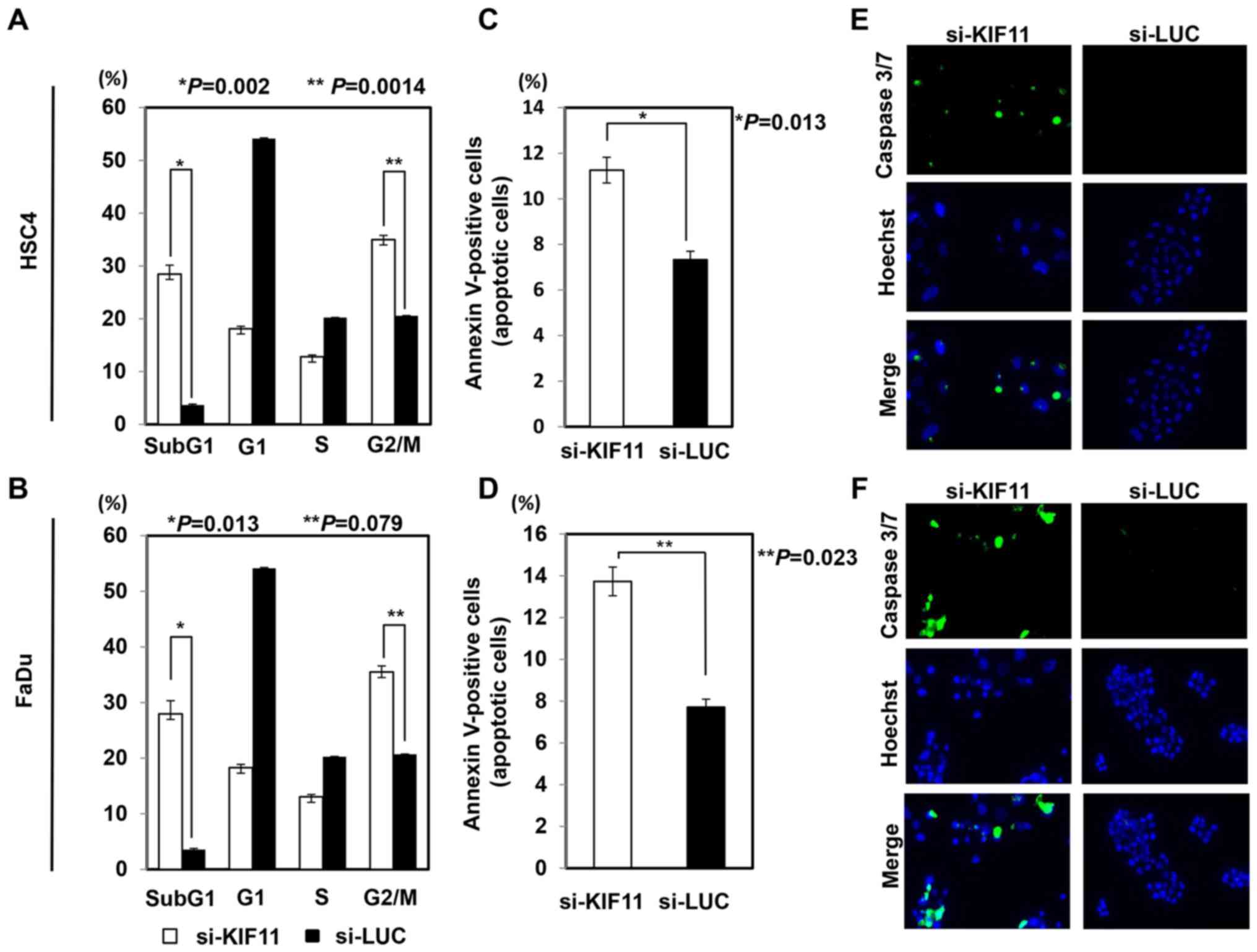
To elucidate the effect of KIF11 on cell cycle and
cell death, we performed flow cytometric analysis after
transfection of si-KIF11 or control siRNAs into HSC4 and FaDu
cells. The proportion of cells at subG1 and G2/M phases was
significantly higher for cells transfected with si-KIF11 than for
those with control siRNAs (Fig. 4A and
B). The Annexin V assay showed that inhibition of KIF11
expression with si-KIF11 significantly increased the occurrence of
apoptosis in HSC4 and FaDu cells compared to that in cells with
control siRNAs (Fig. 4C and D).
The caspase-3/7 assay also detected activation of caspase-3/7 more
frequently in HSC4 and FaDu cells transfected with si-KIF11 than in
cells with control siRNAs (Fig. 4E and
F). We further monitored morphological changes through
live-cell imaging of HSC4 cells transfected with siRNAs against
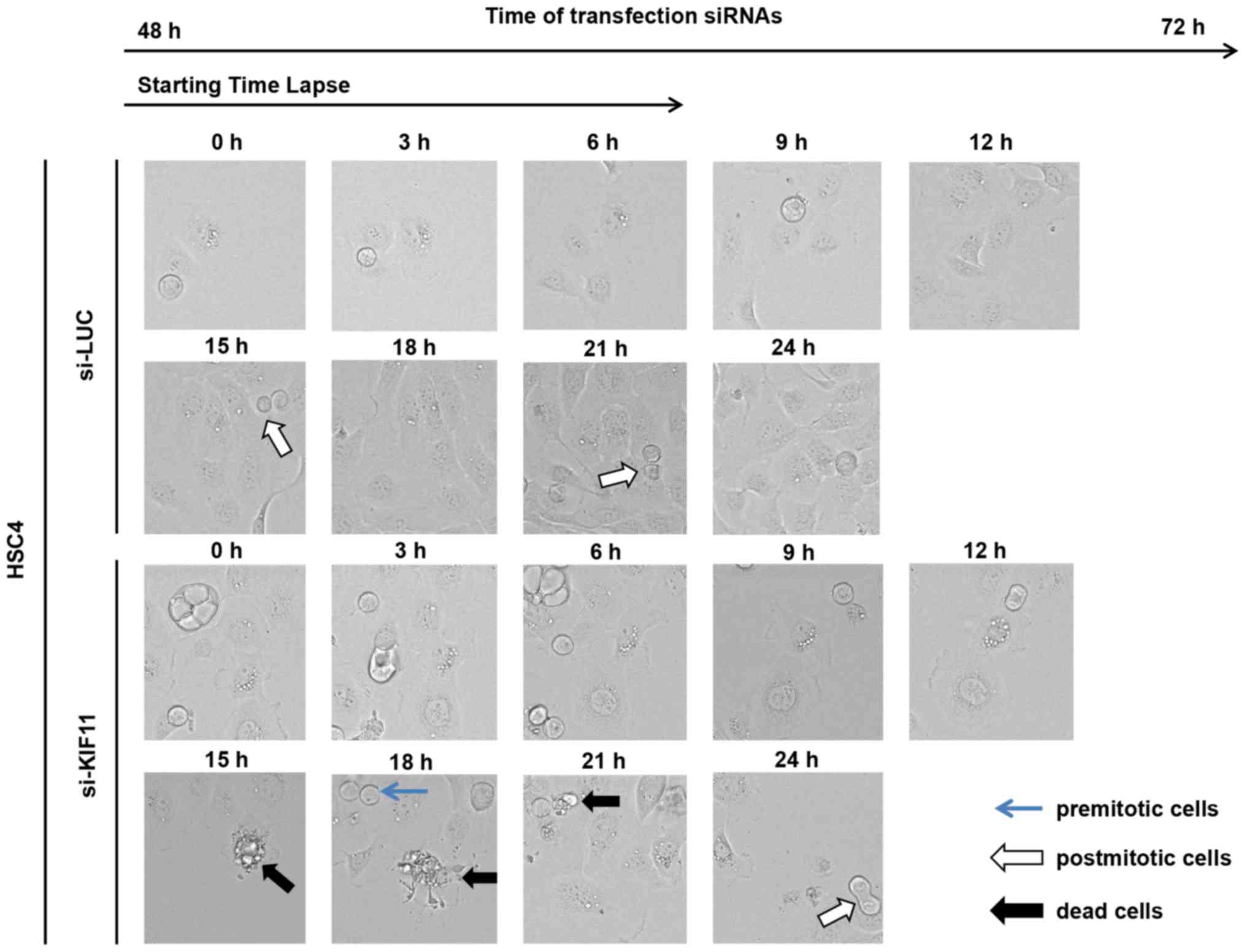
KIF11 (Fig. 5). During 24 h of
monitoring with time-lapse imaging, we detected regular cell
divisions of HSC4 cells transfected with si-control, while we
observed very few cell divisions and subsequent death of HSC4 cells
transfected with si-KIF11.
Growth inhibition in oral cancer cells by
blocking the kinesin motor ATPase activity of KIF11
To clearly evaluate the functional role of the
kinesin motor ATPase activity of KIF11 in oral cancer cells, we
incubated HSC4 cells in medium with or without SB743921, which is a
small compound that selectively blocks the ATPase activity of
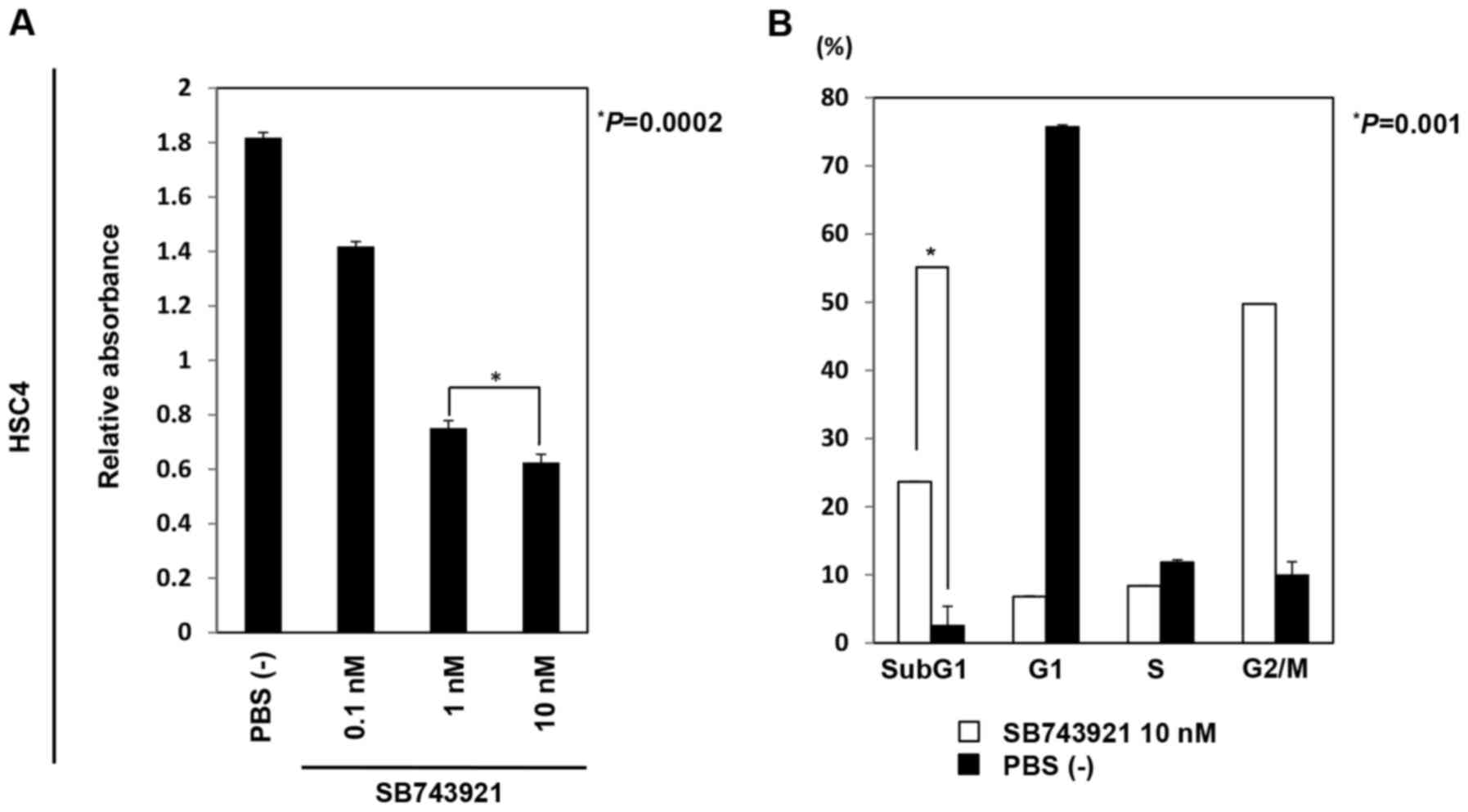
KIF11. After 72 h of incubation with SB743921, MTT assay showed
that the viability of HSC4 cells significantly decreased in a
dose-dependent manner (Fig. 6A).
Furthermore, flow cytometric analysis performed 48 h after SB743921
treatment revealed that the population of cells at sub-G1 and G2/M
phases was significantly higher than that for cells without
SB743921 treatment (Fig. 6B). In
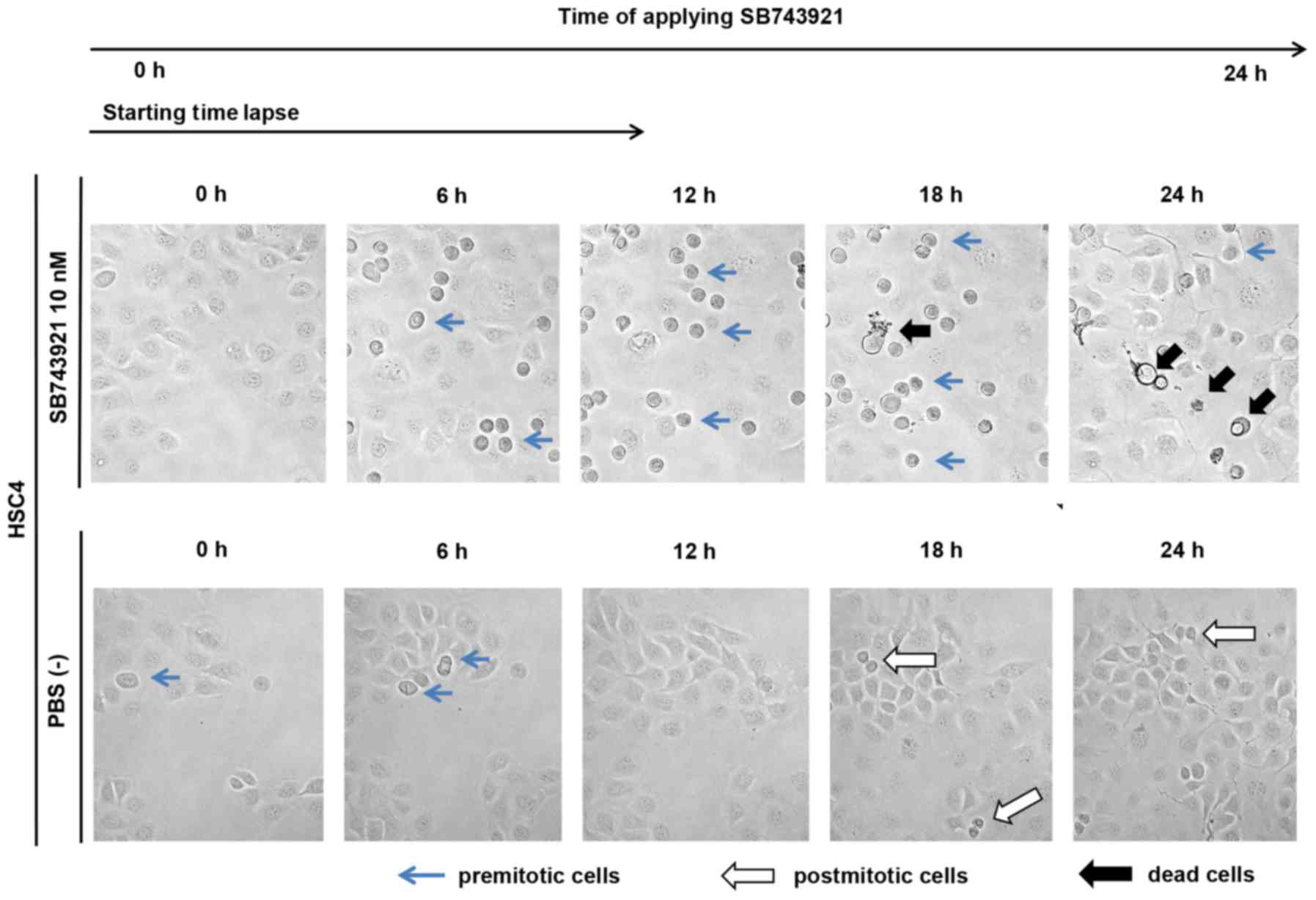
live-cell imaging, the control cells without SB743921 treatment
showed normal cell division, whereas the cells with SB743921
treatment showed cell cycle arrest at the M phase and subsequent
cell death (Fig. 7).
Discussion
Management of advanced oral cancer involves
multiple-modality therapy with surgery, radiation, and
chemotherapy. Current understanding of molecular mechanisms and
basic signaling pathways in the pathogenesis of oral cancer has led
to the development of some new molecular targeted therapies, such
as monoclonal antibodies and other molecular targeted agents.
Molecular targeted agents are expected to have a high clinical
effect against cancer cells, because of their specific anticancer
mechanisms of action. To date, the FDA has approved cetuximab,
nivolumab, and pembrolizumab as drugs for the treatment of patients
with relapsed or advanced head and neck cancers (39,40).
Although these drugs have been shown to improve the prognoses of
patients with oral cancer, their efficacies and benefits remain
limited. Therefore, the identification of new molecular targeted
drugs for oral cancer is desired.
In this study, KIF11 was expressed in the majority
of oral cancer cells and tissues but was scarcely detected in
healthy tongue tissue. In addition, strong KIF11 expression was
significantly correlated with poor prognosis in oral cancer
patients. Furthermore, the suppression of KIF11 reduced oral cancer
cell growth and promoted apoptosis. According to bioGPS (http://biogps.org/#goto=welcome), KIF11 is
not expressed in normal tissues or organs, except early blood
cells, such as lymphoblasts and early erythroids. To assess the
mechanism of KIF11 activation in oral cancers, we screened for
information on genetic aberrations of KIF11 using a public
database involving comparative genome hybridization and genome
sequencing. According to cBioportal for Cancer Genomics (http://www.cbioportal.org/), among 915 cases of head
and neck squamous cell carcinoma, missense mutations and deletions
as well as genetic amplification of KIF11 were detected in
only 9 cases (0.9%). Thus, we suggest that the overexpression of
KIF11 could be caused by epigenetic mechanisms. To our knowledge,
our study is the first to report on the functional and clinical
relevance of KIF11 in oral cancer.
KIF11 has been reported to play an essential role in
centrosome separation by cross-linking microtubules in the mitotic
spindle (41). Suppression of
KIF11 increased the proportion of cells in the G2/M phase and
sub-G1 phase, which suggests that cell death eventually occurred
through G2/M arrest and subsequent apoptosis of cancer cells,
indicating the important role of KIF11 during G2/M phase transition
and cell cycle checkpoints in some cancer cells, such as non-small
cell lung cancer (NSCLC) and head and neck squamous cell carcinoma
(HNSCC) cells (42,43). In controlling spindle formation
during mitosis, KIF11 has been suggested to be phosphorylated
through the signal pathways involving various mitotic kinases such
as cyclin-dependent kinase 1 (CDK1), NIMA-related kinase 6 (NEK6)
and aurora kinase A (AURKA) in some cell lines (31,32,44,45).
According to Oncomine database (www.oncomine.org), CDK1, NEK6 and AURKA
as well as KIF11 were frequently overexpressed in oral
cancer tissues. Therefore functional association between KIF11
protein and these kinases might control cell cycle progression of
oral cancer cells. Further detailed studies including novel pathway
analyses are warranted to clarify the involvement of KIF11 in oral
cancer cell proliferation and survival. On the other hand, histone
deacetylase 1 (HDAC1) is colocalized with KIF11 during mitosis and
influences the ATPase activity of KIF11 by deacetylating KIF11
(46). KIF11 deacetylation by
HDAC1 or other proteins may also promote centromere separation and
bipolar spindle formation, leading to metaphase progression and
complete cell division in oral cancer cells.
Kinesin inhibitors have been shown to exhibit strong
antitumor activity, and many clinical trials on kinesin inhibitors
are currently on going (39,40).
Recently, small molecules targeting the ATPase activity of KIF11
have been develo ped (31,32,39,40).
Accordingly, we focused on SB743921 as a selective KIF11 inhibitor.
Our study showed that SB743921 decreased the viability of HSC4
cells through G2/M arrest and subsequent apoptosis. On live-cell
imaging, cell waggling and filopodia formation required in the
process of polarization in cell mitosis were not observed under
SB743921 treatment when compared with the findings in the PBS (-)
group. Migrating cells typically form filopodia that extend from
the cell surface (47). Previous
basic research also showed that SB743921 promotes mitotic arrest
and subsequent cell death in certain cancer cell lines (48). In addition, SB743921 caused
regression of various types of tumors in human tumor xenograft
models in vivo, including colon (Colo205), lung (H69), and
breast (MCF7) cancer cell xenografts (49,50).
On the other hand, KIF11 inhibitors exhibited powerful anticancer
activity in gemcitabine-resistant bladder cancer cells, and it was
suggested that SB743921 helps overcome chemotherapy resistance in
cancer cells (51). In a previous
clinical trial, KIF11 inhibitors helped overcome imatinib
resistance in chronic myeloid leukemia cells (52). Targeting KIF11 with small molecule
inhibitors, such as SB743921, may be a powerful approach for oral
cancer treatment, including metastatic or chemoresistant
cancer.
In conclusion, our findings suggest that KIF11
should be considered as a typical oncoprotein in oral cancer.
Importantly, KIF11 has a pivotal role in oral cancer growth and
survival, and it can be a prognostic factor for oral cancer.
Therefore, targeting KIF11 is expected to involve novel treatment
strategies, such as immunotherapies and molecular targeted
therapies, which could have many powerful biological effects
against cancer with minimal side effects in healthy cells.
Acknowledgments
This study was supported in part by Grant-in-Aid for
Scientific Research (B) and Grant-in-Aid for Scientific Research on
Innovative Areas from The Japan Society for the Promotion of
Science (JSPS KAKENHI grant number JP: 15H04761 and 16H06277). Y.D.
is a member of Shiga Cancer Treatment Project supported by Shiga
Prefecture (Japan).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dumache R, Rogobete AF, Andreescu N and
Puiu M: Genetic and epigenetic biomarkers of molecular alterations
in oral carcinogenesis. Clin Lab. 61:1373–1381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Manrique OJ, Leland HA, Langevin CJ, Wong
A, Carey JN, Ciudad P, Chen HC and Patel KM: Optimizing outcomes
following total and subtotal tongue reconstruction: A systematic
review of the contemporary literature. J Reconstr Microsurg.
33:103–111. 2017. View Article : Google Scholar
|
|
4
|
Bernier J, Domenge C, Ozsahin M,
Matuszewska K, Lefèbvre JL, Greiner RH, Giralt J, Maingon P,
Rolland F, Bolla M, et al: European Organization for Research and
Treatment of Cancer Trial 22931: Postoperative irradiation with or
without concomitant chemotherapy for locally advanced head and neck
cancer. N Engl J Med. 350:1945–1952. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cooper JS, Pajak TF, Forastiere AA, Jacobs
J, Campbell BH, Saxman SB, Kish JA, Kim HE, Cmelak AJ, Rotman M, et
al: Radiation Therapy Oncology Group 9501/Intergroup: Postoperative
concurrent radiotherapy and chemotherapy for high-risk
squamous-cell carcinoma of the head and neck. N Engl J Med.
350:1937–1944. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Winquist E, Oliver T and Gilbert R:
Postoperative chemoradiotherapy for advanced squamous cell
carcinoma of the head and neck: A systematic review with
meta-analysis. Head Neck. 29:38–46. 2007. View Article : Google Scholar
|
|
7
|
Chinn SB and Myers JN: Oral cavity
carcinoma: Current management, controversies, and future
directions. J Clin Oncol. 33:3269–3276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Daigo Y and Nakamura Y: From cancer
genomics to thoracic oncology: Discovery of new biomarkers and
therapeutic targets for lung and esophageal carcinoma. Gen Thorac
Cardiovasc Surg. 56:43–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Hayama S, Murakami H, Takeshima Y, Inai K, Nishimura H, et
al: Increases of amphiregulin and transforming growth factor-alpha
in serum as predictors of poor response to gefitinib among patients
with advanced non-small cell lung cancers. Cancer Res.
65:9176–9184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ishikawa N, Daigo Y, Yasui W, Inai K,
Nishimura H, Tsuchiya E, Kohno N and Nakamura Y: ADAM8 as a novel
serological and histochemical marker for lung cancer. Clin Cancer
Res. 10:8363–8370. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kakiuchi S, Daigo Y, Ishikawa N, Furukawa
C, Tsunoda T, Yano S, Nakagawa K, Tsuruo T, Kohno N, Fukuoka M, et
al: Prediction of sensitivity of advanced non-small cell lung
cancers to gefitinib (Iressa, ZD1839). Hum Mol Genet. 13:3029–3043.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kato T, Daigo Y, Hayama S, Ishikawa N,
Yamabuki T, Ito T, Miyamoto M, Kondo S and Nakamura Y: A novel
human tRNA-dihydrouridine synthase involved in pulmonary
carcinogenesis. Cancer Res. 65:5638–5646. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kikuchi T, Daigo Y, Katagiri T, Tsunoda T,
Okada K, Kakiuchi S, Zembutsu H, Furukawa Y, Kawamura M, Kobayashi
K, et al: Expression profiles of non-small cell lung cancers on
cDNA microarrays: Identification of genes for prediction of
lymph-node metastasis and sensitivity to anti-cancer drugs.
Oncogene. 22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Suzuki C, Daigo Y, Ishikawa N, Kato T,
Hayama S, Ito T, Tsuchiya E and Nakamura Y: ANLN plays a critical
role in human lung carcinogenesis through the activation of RHOA
and by involvement in the phosphoinositide 3-kinase/AKT pathway.
Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kakiuchi S, Daigo Y, Tsunoda T, Yano S,
Sone S and Nakamura Y: Genome-wide analysis of organ-preferential
metastasis of human small cell lung cancer in mice. Mol Cancer Res.
1:485–499. 2003.PubMed/NCBI
|
|
17
|
Taniwaki M, Daigo Y, Ishikawa N, Takano A,
Tsunoda T, Yasui W, Inai K, Kohno N and Nakamura Y: Gene expression
profiles of small-cell lung cancers: Molecular signatures of lung
cancer. Int J Oncol. 29:567–575. 2006.PubMed/NCBI
|
|
18
|
Oshita H, Nishino R, Takano A, Fujitomo T,
Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y, et
al: RASEF is a novel diagnostic biomarker and a therapeutic target
for lung cancer. Mol Cancer Res. 11:937–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hayama S, Daigo Y, Yamabuki T, Hirata D,
Kato T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Phosphorylation and activation of cell division cycle associated 8
by aurora kinase B plays a significant role in human lung
carcinogenesis. Cancer Res. 67:4113–4122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ishikawa N, Daigo Y, Takano A, Taniwaki M,
Kato T, Tanaka S, Yasui W, Takeshima Y, Inai K, Nishimura H, et al:
Characterization of SEZ6L2 cell-surface protein as a novel
prognostic marker for lung cancer. Cancer Sci. 97:737–745. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kato T, Sato N, Hayama S, Yamabuki T, Ito
T, Miyamoto M, Kondo S, Nakamura Y and Daigo Y: Activation of
Holliday junction recognizing protein involved in the chromosomal
stability and immortality of cancer cells. Cancer Res.
67:8544–8553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Suzuki C, Takahashi K, Hayama S, Ishikawa
N, Kato T, Ito T, Tsuchiya E, Nakamura Y and Daigo Y:
Identification of Myc-associated protein with JmjC domain as a
novel therapeutic target oncogene for lung cancer. Mol Cancer Ther.
6:542–551. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takahashi K, Furukawa C, Takano A,
Ishikawa N, Kato T, Hayama S, Suzuki C, Yasui W, Inai K, Sone S, et
al: The neuromedin U-growth hormone secretagogue receptor
1b/neurotensin receptor 1 oncogenic signaling pathway as a
therapeutic target for lung cancer. Cancer Res. 66:9408–9419. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Taniwaki M, Takano A, Ishikawa N, Yasui W,
Inai K, Nishimura H, Tsuchiya E, Kohno N, Nakamura Y and Daigo Y:
Activation of KIF4A as a prognostic biomarker and therapeutic
target for lung cancer. Clin Cancer Res. 13:6624–6631. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yamabuki T, Takano A, Hayama S, Ishikawa
N, Kato T, Miyamoto M, Ito T, Ito H, Miyagi Y, Nakayama H, et al:
Dikkopf-1 as a novel serologic and prognostic biomarker for lung
and esophageal carcinomas. Cancer Res. 67:2517–2525. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fujitomo T, Daigo Y, Matsuda K, Ueda K and
Nakamura Y: Identification of a nuclear protein, LRRC42, involved
in lung carcinogenesis. Int J Oncol. 45:147–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nguyen MH, Koinuma J, Ueda K, Ito T,
Tsuchiya E, Nakamura Y and Daigo Y: Phosphorylation and activation
of cell division cycle associated 5 by mitogen-activated protein
kinase play a crucial role in human lung carcinogenesis. Cancer
Res. 70:5337–5347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hayama S, Daigo Y, Kato T, Ishikawa N,
Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S and Nakamura Y:
Activation of CDCA1-KNTC2 members of centromere protein complex,
involved in pulmonary carcinogenesis. Cancer Res. 66:10339–10348.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kobayashi Y, Takano A, Miyagi Y, Tsuchiya
E, Sonoda H, Shimizu T, Okabe H, Tani T, Fujiyama Y and Daigo Y:
Cell division cycle-associated protein 1 overexpression is
essential for the malignant potential of colorectal cancers. Int J
Oncol. 44:69–77. 2014. View Article : Google Scholar
|
|
30
|
Thang PM, Takano A, Yoshitake Y, Shinohara
M, Murakami Y and Daigo Y: Cell division cycle associated 1 as a
novel prognostic biomarker and therapeutic target for oral cancer.
Int J Oncol. 49:1385–1393. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Blangy A, Lane HA, d'Hérin P, Harper M,
Kress M and Nigg EA: Phosphorylation by P34c dc2 regulates spindle
association of human Eg5, a kinesin-related motor essential for
bipolar spindle formation in vivo. Cell. 83:1159–1169. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Rapley J, Nicolàs M, Groen A, Regué L,
Bertran MT, Caelles C, Avruch J and Roig J: The NIMA-family kinase
Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for
mitotic spindle formation. J Cell Sci. 121:3912–3921. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ferenz NP, Gable A and Wadsworth P:
Mitotic functions of kinesin-5. Semin Cell Dev Biol. 21:255–259.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wakana Y, Villeneuve J, van Galen J,
Cruz-Garcia D, Tagaya M and Malhotra V: Kinesin-5/Eg5 is important
for transport of CARTS from the trans-Golgi network to the cell
surface. J Cell Biol. 202:241–250. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schneider MA, Christopoulos P, Muley T,
Warth A, Klingmueller U, Thomas M, Herth FJ, Dienemann H, Mueller
NS, Theis F, et al: AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five
specific mitosis-associated genes correlate with poor prognosis for
non-small cell lung cancer patients. Int J Oncol. 50:365–372. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Venere M, Horbinski C, Crish JF, Jin X,
Vasanji A, Major J, Burrows AC, Chang C, Prokop J, Wu Q, et al: The
mitotic kinesin KIF11 is a driver of invasion, proliferation, and
self-renewal in glioblastoma. Sci Transl Med. 7:304ra1432015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kato T, Lee D, Wu L, Patel P, Young AJ,
Wada H, Hu HP, Ujiie H, Kaji M, Kano S, et al: Kinesin family
members KIF11 and KIF23 as potential therapeutic targets in
malignant pleural mesothelioma. Int J Oncol. 49:448–456. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Imai T, Oue N, Nishioka M, Mukai S, Oshima
T, Sakamoto N, Sentani K, Matsusaki K, Yoshida K and Yasui W:
Overexpression of KIF11 in gastric cancer with intestinal mucin
phenotype. Pathobiology. 84:16–24. 2017. View Article : Google Scholar
|
|
39
|
Vermorken JB, Mesia R, Rivera F, Remenar
E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol
D, et al: Platinum-based chemotherapy plus cetuximab in head and
neck cancer. N Engl J Med. 359:1116–1127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ferris RL, Blumenschein G Jr, Fayette J,
Guigay J, Colevas AD, Licitra L, Harrington K, Kasper S, Vokes EE,
Even C, et al: Nivolumab for recurrent squamous-cell carcinoma of
the head and neck. N Engl J Med. 375:1856–1867. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wojcik EJ, Buckley RS, Richard J, Liu L,
Huckaba TM and Kim S: Kinesin-5: Cross-bridging mechanism to
targeted clinical therapy. Gene. 531:133–149. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sarli V and Giannis A: Targeting the
kinesin spindle protein: Basic principles and clinical
implications. Clin Cancer Res. 14:7583–7587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Martens-de Kemp SR, Nagel R, Stigter-van
Walsum M, van der Meulen IH, van Beusechem VW, Braakhuis BJ and
Brakenhoff RH: Functional genetic screens identify genes essential
for tumor cell survival in head and neck and lung cancer. Clin
Cancer Res. 19:1994–2003. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bertran MT, Sdelci S, Regué L, Avruch J,
Caelles C and Roig J: Nek9 is a Plk1-activated kinase that controls
early centrosome separation through Nek6/7 and Eg5. EMBO J.
30:2634–2647. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ma HT, Erdal S, Huang S and Poon RY:
Synergism between inhibitors of Aurora A and KIF11 overcomes
KIF15-dependent drug resistance. Mol Oncol. 8:1404–1418. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nalawansha DA, Gomes ID, Wambua MK and
Pflum MKH: HDAC inhibitor-induced mitotic arrest is mediated by
eg5/kif11 acetylation. Cell Chem Biol. 24:481–492.e5. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Meyen D, Tarbashevich K, Banisch TU,
Wittwer C, Reichman-Fried M, Maugis B, Grimaldi C, Messerschmidt EM
and Raz E: Dynamic filopodia are required for chemokine-dependent
intracellular polarization during guided cell migration in vivo.
eLife. 4:42015. View Article : Google Scholar
|
|
48
|
LoRusso PM, Goncalves PH, Casetta L,
Carter JA, Litwiler K, Roseberry D, Rush S, Schreiber J, Simmons
HM, Ptaszynski M, et al: First-in-human phase 1 study of filanesib
(ARRY-520), a kinesin spindle protein inhibitor, in patients with
advanced solid tumors. Invest New Drugs. 33:440–449. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Good JA, Wang F, Rath O, Kaan HY,
Talapatra SK, Podgórski D, MacKay SP and Kozielski F: Optimized
S-trityl-L-cysteine-based inhibitors of kinesin spindle protein
with potent in vivo antitumor activity in lung cancer xenograft
models. J Med Chem. 56:1878–1893. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Talapatra SK, Anthony NG, Mackay SP and
Kozielski F: Mitotic kinesin Eg5 overcomes inhibition to the phase
I/II clinical candidate SB743921 by an allosteric resistance
mechanism. J Med Chem. 56:6317–6329. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sun L, Lu J, Niu Z, Ding K, Bi D, Liu S,
Li J, Wu F, Zhang H, Zhao Z, et al: A potent chemotherapeutic
strategy with eg5 inhibitor against gemcitabine resistant bladder
cancer. PLoS One. 10:e01444842015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yin Y, Sun H, Xu J, Xiao F, Wang H, Yang
Y, Ren H, Wu CT, Gao C and Wang L: Kinesin spindle protein
inhibitor SB743921 induces mitotic arrest and apoptosis and
overcomes imatinib resistance of chronic myeloid leukemia cells.
Leuk Lymphoma. 56:1813–1820. 2015. View Article : Google Scholar
|