Introduction
Osteosarcoma (OS), a primary malignant bone tumor,
which is characterized by a highly malignant tendency to rapidly
damage the surrounding tissues and to metastasize (1). The global incidence rate is estimated
at 4 million per year, with a peak incidence in children and
adolescents (2). Despite current
therapeutic strategies including multi-agent chemotherapy, surgery
and radiotherapy, the survival of OS patients remains poor, since
~80% of patients with surgical treatment eventually develop
recurrent metastatic OS (3), with
5-year survival rate of only 50–60% (4). Therefore, it is essential to clarify
the molecular mechanism underlying OS tumorigenesis and investigate
the new molecular targets inhibiting tumor growth and metastasis in
OS clinical diagnosis and treatment.
Increasing evidence have demonstrated that
non-coding RNAs (ncRNAs) including microRNA (miRNA) and long
non-coding RNA (lncRNA) have been well reported as a novel class of
potential therapeutic targets and clinical biomarkers for cancers
(5–7). miRNAs are small ncRNAs averaging
20–24 nucleotides, which modulate translational suppression or mRNA
degradation by binding to their 3′-untranslated region (3′-UTR)
(8). Additionally, lncRNAs, are
defined as ncRNAs more than 200 nucleotides in length, have been
identified to have key regulatory roles via regulating gene
expression, such as intergenic regions, overlapping, antisense and
intronic to protein-coding genes (9–11).
The growing number of evidence demonstrated that lncRNAs have an
important role in tumor biology, including tumor initiation,
progression and metastasis (12,13).
lncRNAs aberrant expressions have been confirmed in different
cancers, such as hepatocellular carcinoma, gastric, lung and
cervical cancer (10,11,14,15).
In OS, some lncRNAs were confirmed to act as tumor suppressors
(MEG3, HIF2PUT and TUSC7) or exhibit oncogenic properties (H19,
MALAT1, ANCR, HOTAIR and TUG1) to control OS progression by
modulating cell cycle or apoptosis to regulate cell proliferation
or migration 16). Tumor suppressor candidate 7 (TUSC7), a potential
tumor suppressor, which was remarkably downregulated in OS tissues,
and low TUSC7 expression is associated with poor survival in OS
patients (17). Li et al
(18) confirmed that lncRNA
HOPTTIP was overexpressed in OS tissues, and was correlated with
advanced clinical stage and distant metastasis. Inhibition of
HOPTTIP in OS cells dramatically repressed cell proliferation,
migration and invasion. Therefore, the biological role of lncRNAs
were further investigated in OS to develop a novel prognostic
marker and potential therapeutic target in OS patients.
It is well documented that many miRNAs have been
identified to act as tumor suppressor genes or oncogenes in cancers
and could block cell signaling pathways to suppress different
biological processes, such as cell proliferation, apoptosis,
differentiation and migration (19,20).
Recently, lncRNA was identified to be a sponge for mediating the
miRNA expression and activity as competing endogenous RNA (ceRNA)
(21–23). A recent study demonstrated that
lncRNA small nucleolar RNA host gene 1 (SNHG1) was upregulated in
hepatocellular carcinoma (HCC) tissues and cells, and promoted HCC
cells proliferation and cycle progression (24). Growing evidence uncovered that
ectopic expression of SNHG1 could function as oncogenes in various
cancers including lung and breast cancer (24–26).
However, the roles of SNHG1 in OS have not yet been elucidated.
Moreover, increasing evidence confirmed that SNHG1 acts as crucial
regulator to modulate various cancer progressions through
competitively binding miRNAs including miR-199a-3p and miR-195
(27,28). These inspired us to explore the
roles of SNHG1 in OS and investigate the relationship between SNHG1
and miRNAs in the carcinogenesis of OS.
In the present study, our results showed that SNHG1
was upregulated in OS tissues and cell lines, knockdown of SNHG1
suppressed cell growth and metastasis in vitro and in
vivo. Subsequently, we explored the molecular mechanism of
SNHG1 in OS progression and the reciprocal regulation between SNHG1
and miR-326. Our findings uncovered that the SNHG1/miR-326/human
nin one binding protein (NOB1) signaling pathway play an important
role in OS tumorigenesis and suggested the potential value of SNHG1
in OS clinical diagnosis and treatment.
Materials and methods
Patient tissue samples and OS cell
lines
Forty-five paired tissue samples of OS and adjacent
normal tissues were obtained from patients in the Department of
Trauma and Orthopedics, Shanghai General Hospital, Shanghai Jiao
Tong University. These patients did not receive preoperative radio
therapy or chemotherapy before surgical resection. Histological
diagnosis and differentiation were assessed independently by three
pathologists according to the WHO classification system (29). Fresh specimens were stored at −70°C
until use. All patients agreed to provide their tumor tissues for
clinical research through signing written informed consent. The
Institute Research Medical Ethics Committee of China Medical
University approved the study protocol. The human OS cell lines
MG-63, U2OS, Saos-2 and SOSP-9607 were purchased from the the
American Type Culture Collection (ATCC; Rockville, MD, USA) and
grown in RPMI-1640 medium supplemented with 10% fetal bovine serum
(FBS; Sigma-Aldrich, St. Louis, MO, USA), 100 IU/ml penicillin and
100 mg/ml streptomycin, at 37°C in a humidified atmosphere
containing 5% CO2. The osteoblastic cell line hFOB 1.19
was maintained in Dulbecco's modified Eagle's minimal essential
medium (DMEM)/F12; Gibco, Carlsbad, CA, USA) supplemented with 10%
FBS, 100 IU/ml penicillin and 100 mg/ml streptomycin.
Quantitative real-time PCR
Total RNA was extracted from frozen tissues and cell
samples with TRIzol reagent (Invitrogen, Carlsbad, CA, USA)
according to the manufacturer's instructions. High Capacity cDNA
Reverse Transcription kit and TaqMan MicroRNA Reverse Transcription
kit were applied to lncRNAs and miRNA reverse transcription,
respectively (Applied Biosystems, Foster City, CA, USA).
Quantitative RT-PCR was performed by TaqMan Universal Master Mix II
with TaqMan non-coding RNA assays of SNHG1 and GAPDH or with TaqMan
microRNA assays of miR-326 and U6 (Applied Biosystems). GAPDH and
U6 were used as internal controls. The relative expression levels
were determined using the 2−ΔΔCt method.
Oligonucleotide and plasmid
transfection
Small interfering RNA si-SNHG1 (5′-GAG AGC TCT GTT
GTT GCA ATG TTCA-3′) and scrambled negative control si-Scramble
(5′-GAG TCT CGT TGC GTT GTA ATG ATCA-3′) oligonucleotides were
obtained from Shanghai GenePharma Co., Ltd. (Shanghai, China). The
miR-326 mimics or inhibitor and corresponding negative control (NC)
were obtained from Guangzhou RiboBio Co., Ltd. (Guangzhou, China).
To overexpress SNHG1, pcDNA-SNHG1 was amplified by PCR, and the
final product was inserted into the pcDNA3.1 plasmid (Invitrogen).
SNHG1 mutants were performed by the QuikChange Site-Directed
Mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the
manufacturer's instructions. The OS cells were seeded into each
well of a 6-well plate, cultured in the appropriate culture media
containing 10% FBS. Then, the cells were transfected with si-SNHG1,
si-Scramble, miR-326 mimics, miR-326 inhibitor, pcDNA-SNHG1 and
pcDNA-mut-SNHG1. Cells were collected 48 h after transfection.
Cell Counting kit-8 assay
The proliferation of cells was determined by the
Cell Counting kit-8 (CCK-8) assay according to the manufacturer's
instructions. After transfection, the OS cells (5×104
cells/well) were seeded in 96-well plate with 100 μl DMEM
medium supplemented with 10% FBS. After 48-h incubation, CCK-8
reagent (10 μl) was added to each well and continuously
cultured for 1 h in 5% CO2 (Thermo Fisher Scientific,
Waltham, MA, USA). The absorbance rate at 450 nm was measured by
microplate reader (Bio-Rad Laboratories, Hercules, CA, USA). All
experiments were performed in quintuplicate on three separate
occasions.
Analysis of apoptosis
Approximately 48 h after transfection,
1×106 cells were collected and washed twice with
HEPES-buffered saline. After treatment with trypsin, cells were
fixed with 70% ice-cold methanol at 4°C for 30 min. Cells were then
resuspended in binding buffer and stained with 5 μl of
Annexin V-FITC (BD Biosciences, Mountain View, CA, USA) and 1
μl of propidium iodide (PI, 50 μg/ml) (BD
Biosciences). Flow cytometric evaluation was performed within 5
min. Stained cells were measured by flow cytometry (FACSCalibur; BD
Biosciences). The measurements were performed independently for at
least three times with similar results.
Western blot analysis
OS cells were treated with ice-cold whole cell
extraction buffer (pH 7.6) combing the protease inhibitors [0.5 mM
DTT, 20 mM HEPES, 0.2 mM EDTA, 75 mM NaCl, 2.5 mM MgCl2,
0.1% Triton X-100, 50 mM NaF, 0.1 mM Na3VO4,
and the protease inhibitors including 1 mg/ml aprotinin, 0.5 mg/ml
leupeptin and 100 mg/ml 4-(2-aminoethyl) benzenesulfonyl fluoride].
The BCA protein assay kit (Pierce, Rockford, IL, USA) was used to
measure protein concentration. Equal amounts of protein samples (60
mg) were separated in 10% SDS-polyacrylamide gels (Sigma-Aldrich)
and then transferred onto polyvinylidene difluoride (PVDF)
membranes (BD Biosciences, San Diego, CA, USA). The membranes were
blocked with 5% skim milk at room temperature for 1 h and incubated
with primary antibodies at 4°C overnight against cle-caspase-3,
ZEB1, vimentin, N-Cadherin and NOB1, which were purchased from
Santa Cruz Biotechnology (Santa Cruz, CA, USA). The goat anti-mouse
IgG horseradish peoxidase antibodies were obtained from Santa Cruz
Biotechnology. The bands were scanned using the ChemiDoc XRS +
Imaging System (Bio-Rad Laboratories) and quantified using Quantity
One v4.6.2 software (Bio-Rad Laboratories).
Xenograft tumor model
All animal procedures were carried out with the
protocols approved by the Animal Care Committee of the Shanghai
General Hospital, Shanghai Jiao Tong University. The BALB/C athymic
nude mice (4 weeks of age) were obtained from the Cancer Institute
of the Chinese Academy of Medical Science. Mice were housed and
maintained in laminar flow cabinets under specific pathogen free
conditions with free access to food and water. When the female
BALB/C athymic nude mice (n=4) were 7–8 weeks of age, each mouse
was inoculated with Saos-2 or U2OS cells transfected with si-SNHG1
or si-Scramble in 0.2 ml of medium subcutaneously in the forelimb.
The tumor cells were allowed to grow for 4 weeks. The tumor growth
was evaluated by the measurement of the length and the width with
electronic calipers and the tumor volume was determined using the
formula: Volume (mm3) = (length x width2)/2.
The mice were sacrificed in 28 days, and then the tumors were
excised and weighed.
Cell migration assay
The wound healing assay was applied to test cell
migration. The Saos-2 and U2OS cells were plated onto 6-well plates
and transfected with si-SNHG1 or si-Scramble. The migration status
was evaluated by measuring the movement of cells into a scraped
area created by a micropipette pipette tip. The spread of wound
closure was observed after 24 h. The wound area was measured and
the percentage of closure of denuded area was calculated using the
ImageJ software (NIH, Bethesda, MD, USA).
Cell invasion assay
Cell invasion assays were conducted by 24-well
Transwell chambers with 8.0 μm pore size polycarbonate
membrane (Corning Inc., Corning, NY, USA). The Saos-2 and U2OS
cells were seeded on the top side of the membrane precoated with
Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) and then
transfected with si-SNHG1 or si-Scramble. After 48 h of incubation,
the non-invading cells were carefully removed from the upper
surface and invading cells were fixed with methanol and stained
with crystal violet (Sigma-Aldrich) on the lower surface of the
membrane. The cell numbers were calculated in each well with six
random fields at ×200. Experiments were performed at least 3
times.
Luciferase assay
To construct reporter vector, one fragment of NOB1
3′-UTR [wild-type (wt) or mutant (mut), respectively] and two
fragments of SNHG1 mRNA containing predicted miR-326 binding site 1
or 2 were separately amplified and fused to a modified pcDNA3.1
vector containing a luciferase gene, which was cloned into upstream
of cloning sites. Mut reporter vectors were performed using the
Mutagenesis kit (Stratagene). The OS cells U2OS were seeded into
24-well plates and transfected with the different reporter plasmid
including pcDNA-wt-SNHG1-1, pcDNA-mut-SNHG1-1, pcDNA-wt-SNHG1-2,
pcDNA-mut-SNHG1-2, pcDNA-wt-NOB1 and pcDNA-mut-NOB1, together with
miR-326 mimics/inhibitor or corresponding mimic/inhibitor NC using
Lipofectamine 2000 (Invitrogen). Forty-eight hours after the
transfection, the luciferase activity was determined by the
Dual-Light luminescent reporter gene assay (Applied Biosystems).
Each experiment was performed at least 3 times in individual
experiments. The ratio of Renilla luciferase to firefly
luciferase was calculated for each well.
Immunohistochemistry
Immunohistochemistry was used to detect NOB1 protein
expression in OS tissues. NOB1 antibody was purchased from Santa
Cruz Biotechnology. Immunohistochemistry was conducted on
paraformaldehyde-fixed paraffin sections. The percentage of
positive tumor cells was graded according to the following
criteria: 0, <10%; 1, 10-30%; 2, 31-50%; 3, >50%. The OS
tissues with different NOB1 expression from patients were divided
into the low-expression group (0 or 1) and the high-expression
group (2 or 3).
Statistical analysis
All statistical analyses were performed using SPSS
14.0 software (SPSS, Inc., Chicago, IL, USA). Experiments were
conducted in triplicate and data are presented as the mean ± SEM.
The results were analyzed with the Student's t-test. The survival
analysis was performed using the Kaplan-Meier method. P<0.05 was
defined as significant and P<0.01 was defined very
significant.
Results
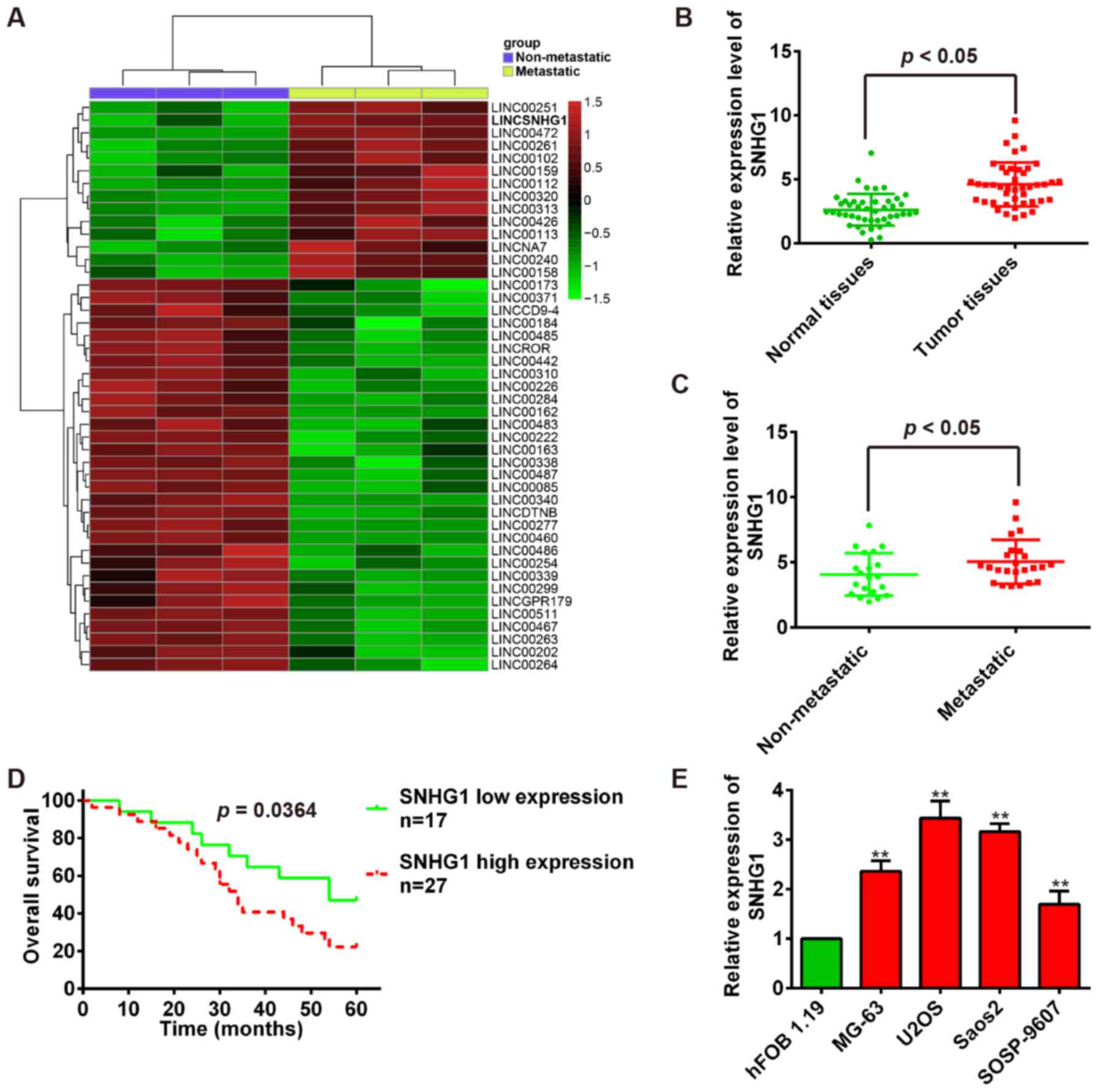
lncRNA SNHG1 is upregulated in OS tissues
and cell lines
To determine lncRNAs involved in OS tumorigenesis,
we screened GEO database and obtained one human lncRNA microarray
datasets (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE85537)
to search differentially expressed lncRNAs between OS bone tissues
and OS lung metastasis tissues. Especially, we observed that SNHG1
is upregulated in OS lung metastasis tissues compared with OS bone
tissues in dataset (Fig. 1A). To
validate this result, the quantitative reverse transcription-PCR
(qRT-PCR) was used to examine SNHG1 expression in 44 pairs of tumor
tissues and adjacent normal tissues. Our results showed that SNHG1
expression is significantly higher in tumor tissues than that in
adjacent normal tissues (P<0.01; Fig. 1B). Then, we measured SNHG1 levels
in OS metastatic tissues (n=24) and no-metastatic tissues (n=20)
using qRT-PCR. Consistent with OS tissue data, the SNHG1 expression
levels were significantly upregulated in OS metastatic tissues
compared with no-metastatic tissues (P<0.01; Fig. 1C). To evaluate the correlation
between SNHG1 expression and OS patients' prognosis, we performed
the Kaplan-Meier survival analysis to plot OS curves and found that
patients with high SNHG1 expression (n=27) had a lower overall
survival percentage than those with low SNHG1 expression (n=17)
(P<0.01; Fig. 1D).
Subsequently, the SNHG1 expression levels were further confirmed in
OS cells, and the SNHG1 expression is also significantly higher in
OS cell lines MG-63, U2OS, Saos-2, SOSP-9607 than that in
osteoblastic cell line hFOB 1.19 (P<0.01; Fig. 1E). Collectively, these data
uncovered that SNHG1 expression is upregulated in OS tissues and
high SNHG1 expression predicts poor prognosis in OS, suggesting
SNHG1 might act as an oncogene in OS tumori-genesis.
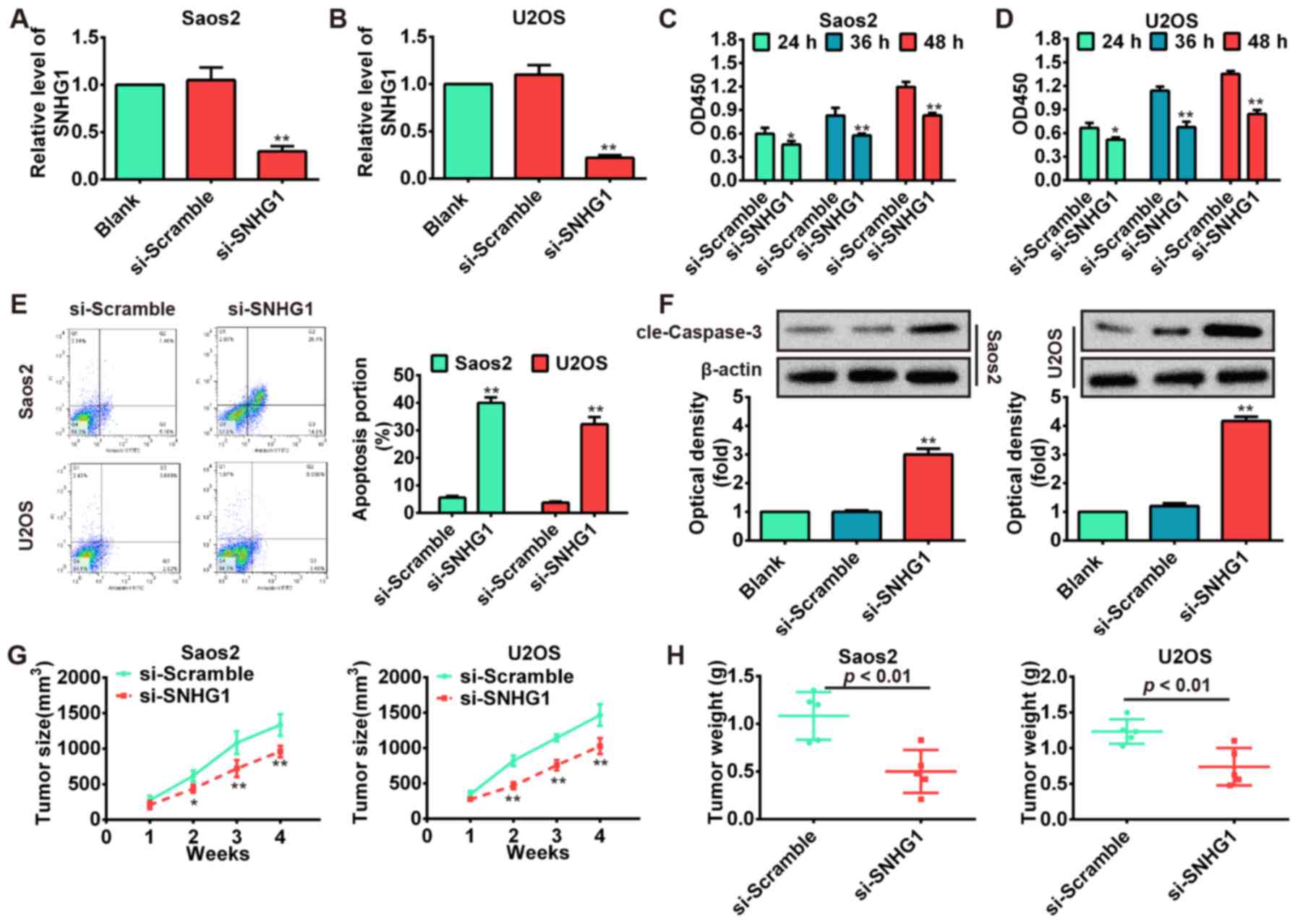
Knockdown of SNHG1 inhibits OS cell
proliferation, induces apoptosis and suppresses tumor growth
To explore the functions of SNHG1 in OS
tumorigenesis, the Saos-2 and U2OS cells were transfected with
si-SNHG1 and scrambled negative control si-Scramble. The results
showed that SNHG1 level was markedly downregulated in Saos-2 and
U2OS cells transfected with si-SNHG1 compared with si-Scramble
(P<0.01; Fig. 2A and B).
Subsequently, we performed the CCK-8 assay to determine cell
proliferation and its result showed that knockdown of SNHG1
significantly inhibited Saos-2 and U2OS cell proliferation compared
with scrambled negative control si-Scramble (P<0.01; Fig. 2C and D). Then, flow cytometric
analysis was used to determine whether the effect of SNHG1 on OS
cell proliferation is regulated by altering cell apoptosis. The
Saos-2 and U2OS cells were stained with Annexin V-FITC and PI
following transfection of cells with si-SNHG1 and si-Scramble. As
shown in Fig. 2E, the apoptosis
rates of Saos-2 and U2OS cells were increased in si-SNHG1 group
compared with si-Scramble group (P<0.01). To further investigate
the induction of apoptotic pathways following knockdown of SNHG1 in
OS cells, the protein level of cleaved-caspase-3 was analyzed by
western blot assays. Our results demonstrated that knockdown of
SNHG1 significantly increased cleaved-caspase-3 expression level in
si-SNHG1 group compared with si-Scramble group (P<0.01; Fig. 2F). To investigate whether knockdown
of SNHG1 affects tumorigenesis in vivo xenograft model was
used, SNHG1-downregulating Saos-2 and U2OS cells and their parallel
si-Scramble carrying cells were subcutaneously injected into the
nude mice. The tumor growth was evaluated by measurement of tumor
size and weight after injection. We observed that
SNHG1-downregulating cells displayed significant decrease in the
tumor size and weight compared with control groups (P<0.01;
Fig. 2G and H). These data
indicate that downregulation of SNHG1 possessed tumor-suppressive
effects that could suppress cell proliferation and induce apoptosis
in vitro and inhibit tumor growth in vivo.
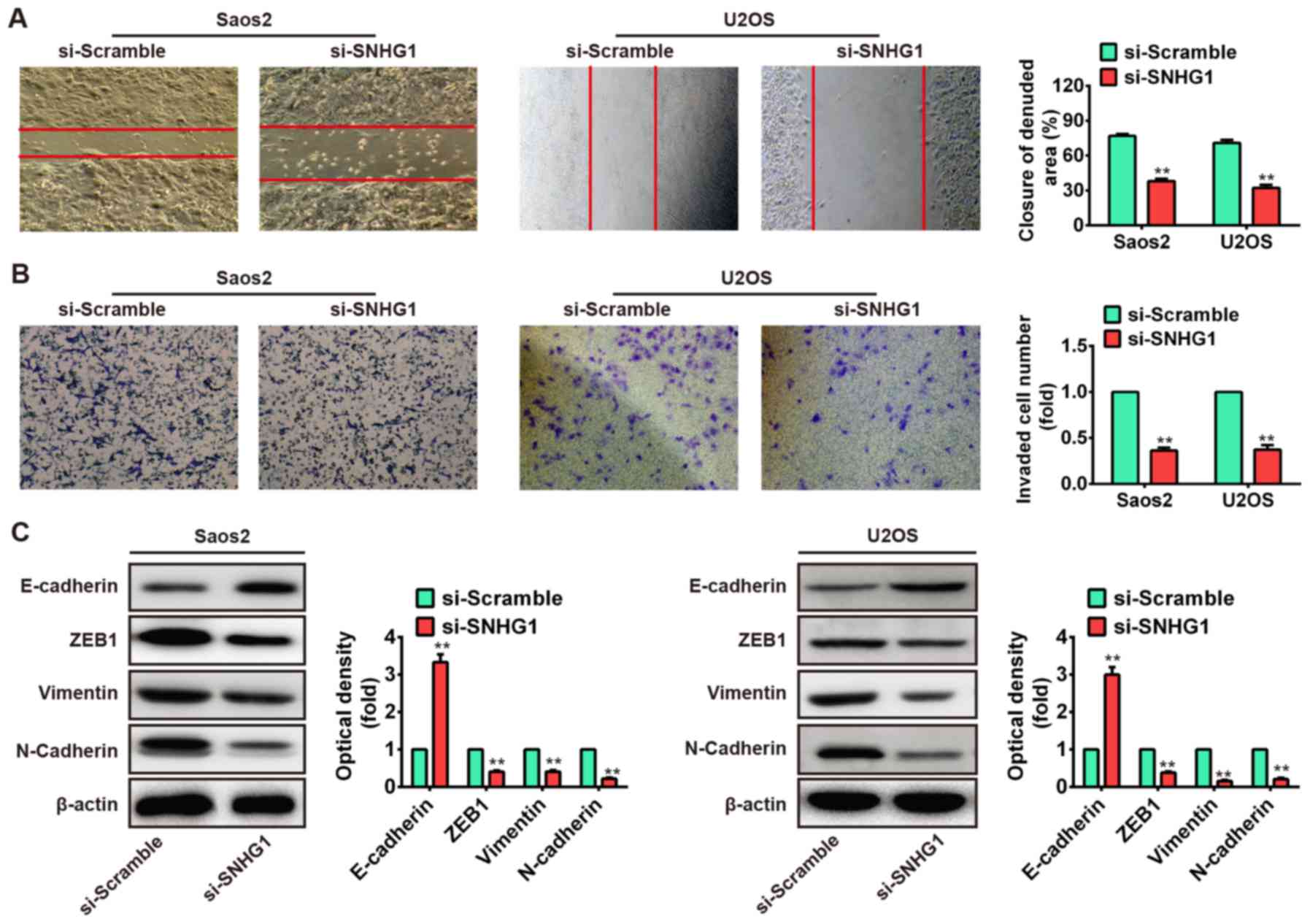
Knockdown of SNHG1 inhibits cell
migration and invasion in vitro
We further assessed the effects of SNHG1
downregulation on cell migration and invasion, which are key
determinants of malignant progression and metastasis. We used wound
healing assay to assess cell migration after transfection with
si-SNHG1 or si-Scramble. As shown in Fig. 3A, the migration of Saos-2 and U2OS
cells that knockdown of SNHG1 were significantly suppressed
compared to the si-Scramble group (P<0.01). Subsequently, the
transwell invasion assay was used to evaluate the cell invasion. We
observed that knockdown of SNHG1 also markedly inhibited cell
invasion in Saos-2 and U2OS cells after transfection with si-SNHG1
compared with that of si-Scramble (P<0.01; Fig. 3B). These data demonstrated that
knockdown of SNHG1 inhibits cell migration and invasion in
vitro, but the possible molecular mechanism needs further
research for deep understanding.
Emerging evidence revealed that cancer progression
is tightly correlated with epithelial-to-mesenchymal transition
(EMT), which induces the cancer cells to acquire mesenchymal
phenotype and metastasize towards distant sites (30–32).
Cancer cells could mislay the expression of cellular adhesion
proteins (E-cadherin) and gain expression of mesenchymal markers
(N-cadherin and vimentin) in the EMT cascade (33,34).
The transcription factor zinc finger E-box-binding homeobox (ZEB)
has an important role in initiation of EMT process. ZEB family
factors (ZEB1 and ZEB2) were identified to act as transcriptional
repressors that include two widely separated clusters of C2H2-type
zinc fingers which bind to paired CAGGTA/G E-box-like promoter
elements. These factors could trigger EMT through suppressing
E-cadherin expression (35–37).
EMT is modulated by a variety of signaling pathways and also by
post-transcriptional mechanisms, including lncRNA (38). Therefore, to further investigate
the molecular mechanism which knockdown of SNHG1 suppresses cell
migration and invasion in vitro, the Saos-2 and U2OS cells
were transfected with si-SNHG1 and si-Scramble, and the expression
levels of ZEB1, E-cadherin, vimentin and N-cadherin were measured
by western blot assays. Our results showed that knockdown of SNHG1
increased E-cadherin protein level but decreased ZEB1, vimentin and
N-cadherin protein levels in both Saos-2 and U2OS cells (Fig. 3C). Taken together, these results
indicated that knockdown of SNHG1 suppresses cell migration and
invasion via mediating EMT.
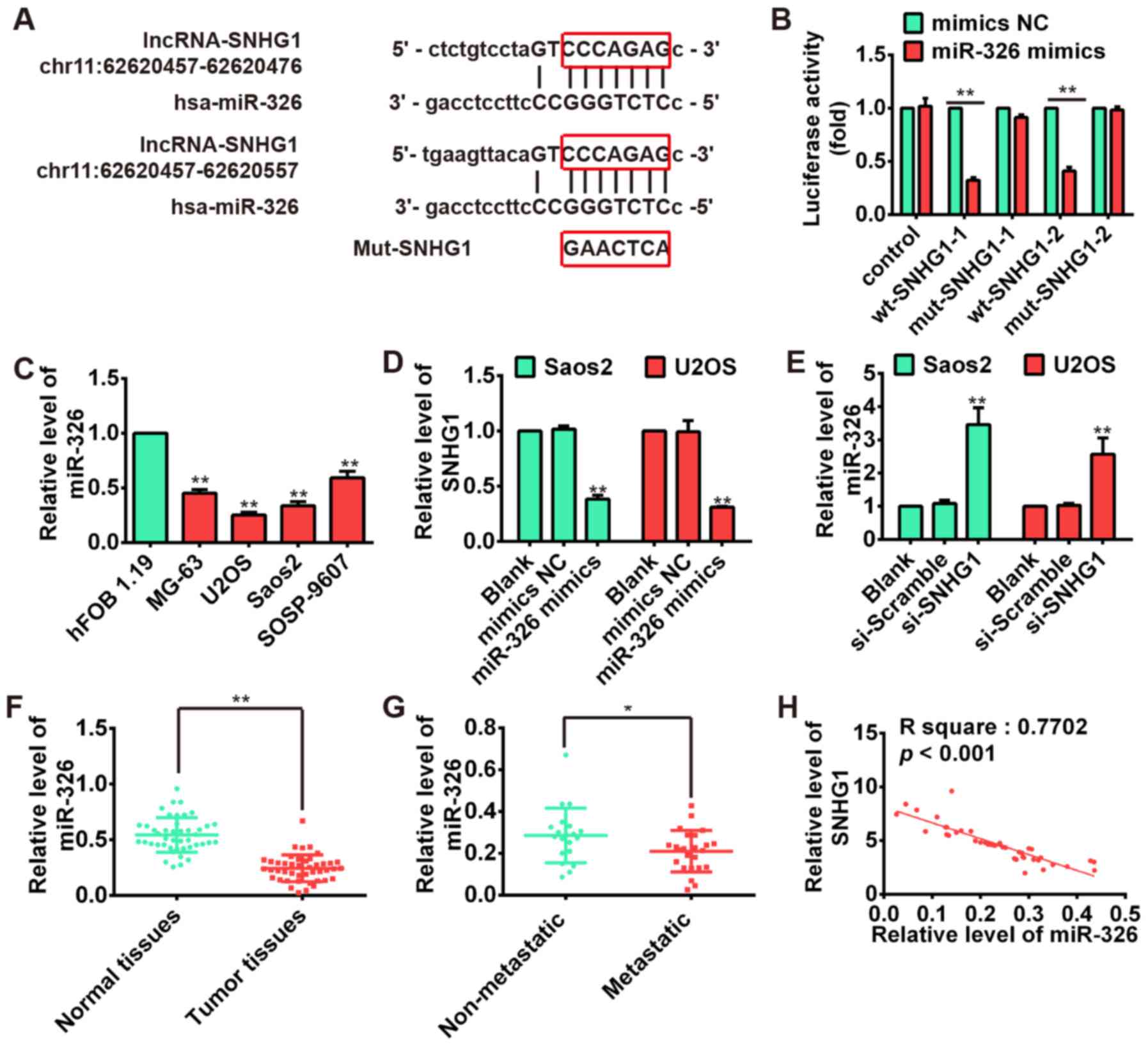
SNHG1 and miR-326 interact with and
repress each other
Recently, lncRNA has been identified to be a sponge
for modulating the expression and activity of miRNA (21,22).
To investigate if the expression of SNHG1 was regulated by miRNA,
two bioinformatic databases including TargetScan and microRNA.org were used to predict potential miRNAs,
and found several potential miRNA-binding sites including miR-326.
Previous studies documented that miR-326 may act as a tumor
suppressor in a variety of cancers including glioma,
medulloblastoma, cholangiocarcinoma and chronic lymphocytic
leukemia (39–41). Our previous study also reported
that miR-326 could repress the proliferation, migration and
invasion of OS cells (42). Thus,
the miR-326 was selected for further bioinformatic predication, we
constructed the 3′-UTR reporter vectors coupled with full length of
SNHG1 3′-UTR with wt or mut miR-326 binding site 1 or 2 (Fig. 4A). Luciferase assay demonstrated
that miR-326 could suppress the expression of reporter gene
containing wt 3′-UTR but not that containing mut 3′-UTR (Fig. 4B), suggesting SNHG1 is a direct
target of miR-326. Moreover, we found that miR-326 is significantly
upregulated in OS cell lines MG-63, U2OS, Saos-2, SOSP-9607
compared with osteoblastic cell line hFOB 1.19, exhibiting an
inverse relationship with the SNHG1 expressions in OS cell lines
(P<0.01; Fig. 4C). Furthermore,
the results of qRT-PCR showed that the SNHG1 level could be
repressed by miR-326 overexpression (Fig. 4D), whereas miR-326 expression was
also upregulated after knockdown of SNHG1 in both Saos-2 and U2OS
cells (Fig. 4E). Subsequently, we
performed the qRT-PCR to determine the miR-326 level in tumor
tissues. The result showed that the expression of miR-326 in tumor
tissues was much lower than that in corresponding normal tissues
(P<0.01; Fig. 4F), whereas the
miR-326 expression in metastatic tissues was also much lower than
non-metastatic tissues (P<0.01; Fig. 4G). Moreover, correlation analysis
presented a strong negative relationship between SNHG1 and miR-326
expression in OS tissues (R2=0.7702, P<0.001;
Fig. 4H). These data suggested
that there was reciprocal repression between SNHG1 and miR-326.
miR-326 inhibits cell proliferation,
induces apoptosis and suppresses migration and invasion
To further verify the tumor-suppressive effects of
miR-326 on OS cells, the Saos-2 and U2OS cells were transfected
with miR-326 mimics and mimics NC, and the cell proliferation,
apoptosis, migration and invasion were investigated using CCK-8
assay, flow cytometric analysis, wound healing and Transwell
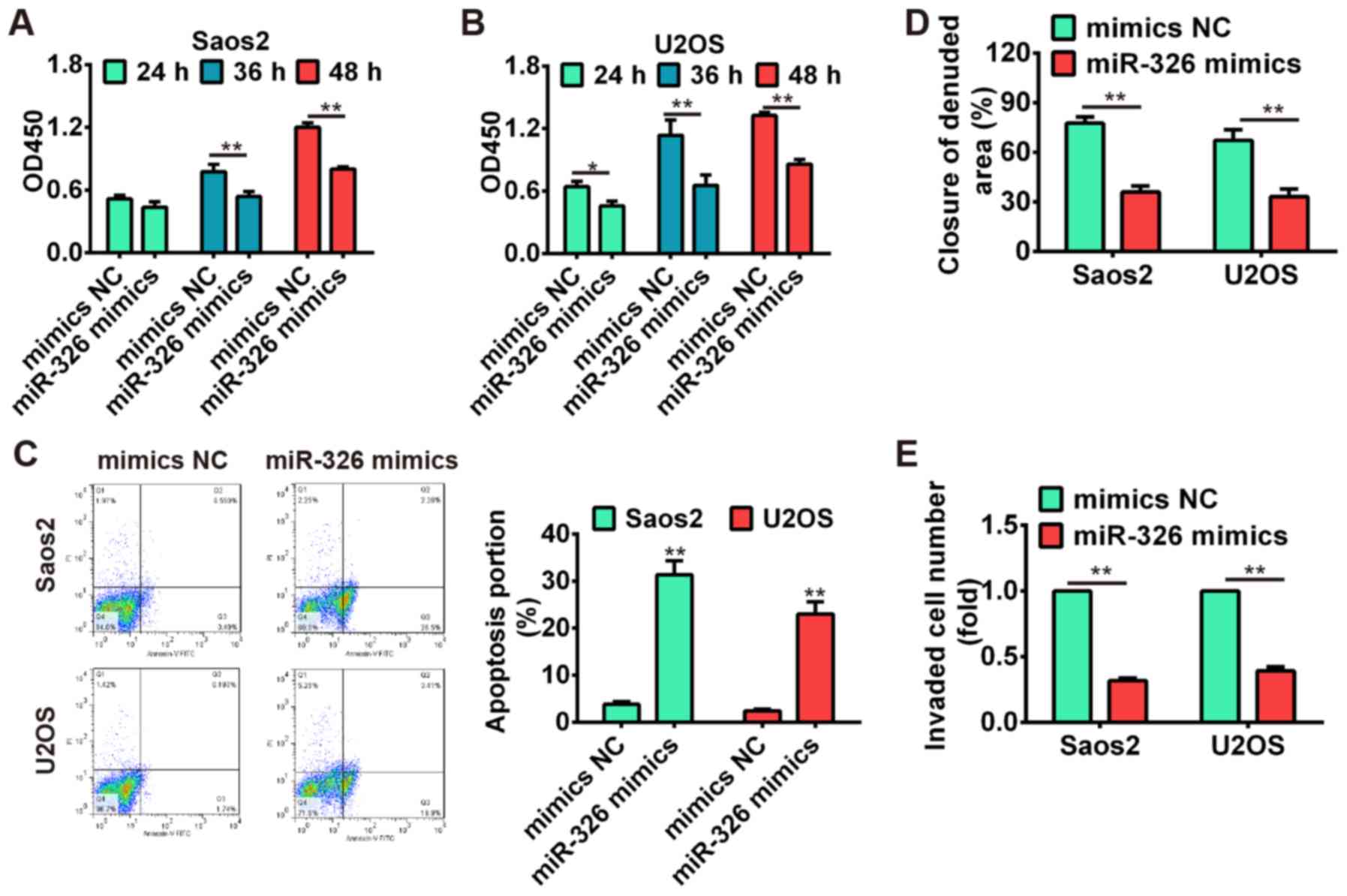
invasion assay, respectively. Our results showed that
overexpression of miR-326 significantly suppressed cell
proliferation compared with mimics NC group (P<0.01; Fig. 5A and B), whereas miR-326
upregulation also induced apoptosis in both Saos-2 and U2OS cells
(Fig. 5C). Additionally, the wound
healing and transwell invasion assay demonstrated that cell
migration and invasion were significantly inhibited after
overexpres-sion of miR-326 compared with mimics NC group
(P<0.01; Fig. 5D and E). These
data indicated that miR-326 harbored the tumor-suppressive effects
via suppressing cell growth, migration and invasion.
miR-326 suppresses NOB1 expression by
directly targeting its 3′-UTR
Increasing evidence revealed that the human nin one
binding protein (NOB1) acts as an oncogene in different cancers
(43–45), and miR-326 functions as a tumor
suppressor by targeting the NOB1 in some cancers including
colorectal carcinoma (46) and
glioma (39). Therefore, we
speculated whether NOB1 is a target of miR-326 in OS cells, and
performed bioinformatic analysis to predict the putative targets of
miR-326, and found that NOB1 might be a target gene of miR-326 and
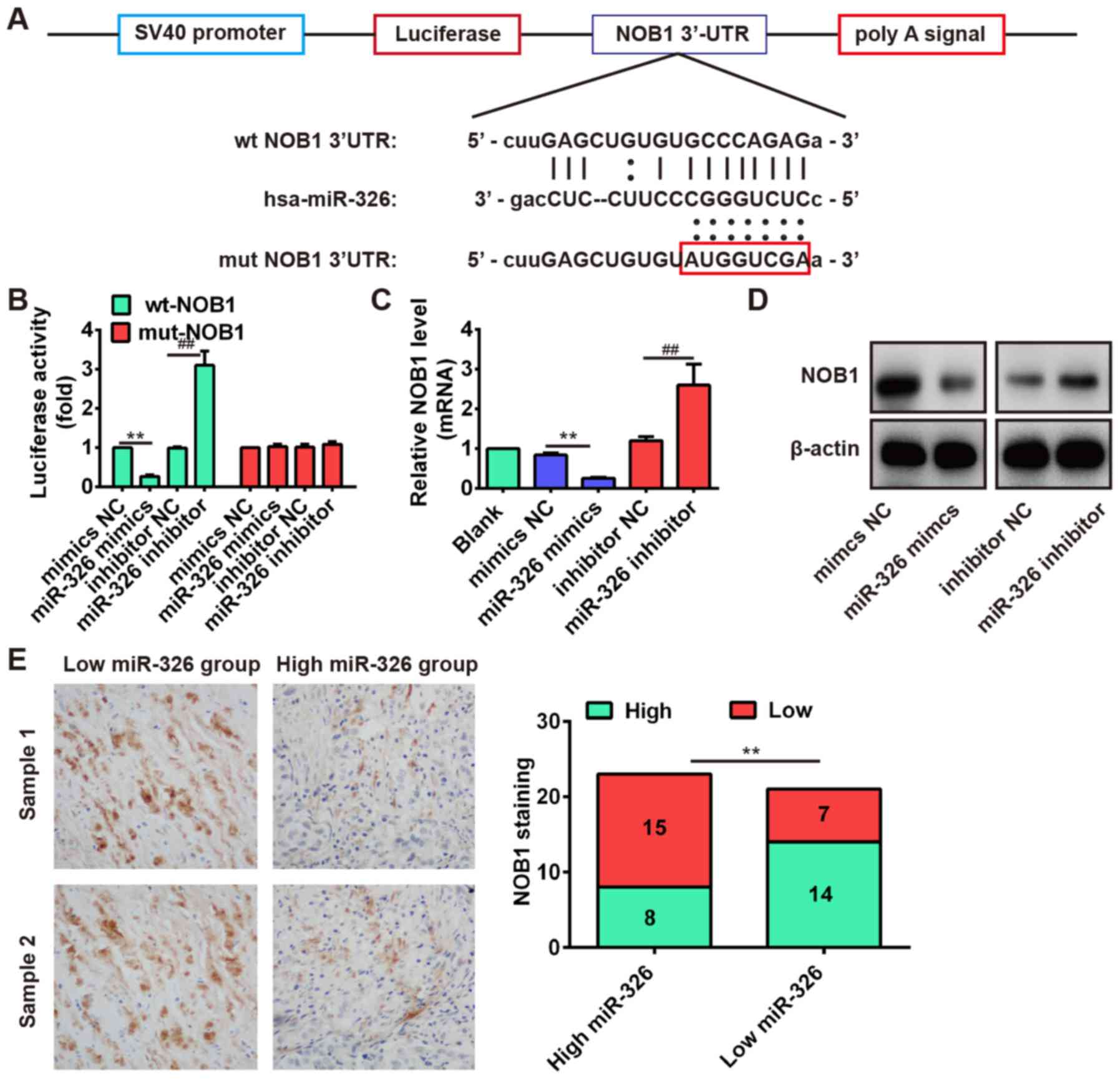
the target site located in the 3′-UTR (Fig. 6A). To verify this bioinformatic
prediction, we constructed luciferase-reporter vectors containing
the wt or mut 3′-UTR segments of NOB1 (Fig. 6A). The wt or mut reporter plasmid
was cotransfected into Saos-2 cells along with miR-326
mimics/inhibitor or NC, and measured the luciferase activity. We
observed that miR-326 mimic significantly inhibited the luciferase
activity compared with the mimic NC, but miR-326 inhibitor
significantly enhanced the luciferase activity compared with the
inhibitor NC in the presence of the wt 3′-UTR (P<0.01; Fig. 6B). Additionally, miR-326 did not
repress the luciferase activity of the reporter vector containing
3′-UTR of NOB1 with mutations in the miR-326-binding site (Fig. 6B). To further determine that the
expression of NOB1 is controlled by miR-326, we performed the
western blot analysis and qRT-PCR analysis to detect the protein
and mRNA level for NOB1, respectively. Our results showed that the
protein and mRNA level of NOB1 were significantly inhibited after
miR-326 overexpression, but markedly increased after knockdown of
miR-326 compared with NC (Fig. 6C and
D). These results demonstrated that NOB1 was a direct target
for miR-326 in OS cells.
In addition, we performed the immunohistochemistry
assay to measure the expression levels of NOB1 in OS tissues. Based
on NOB1 expression level, the OS tissues were divided into
low-expression group (scores 0 and 1) and high expression group
(scores 2 and 3). In the high miR-326 group (n=23), there were 15
cases with low expression of NOB1 and 8 cases with high expression
of NOB1. In the low miR-326 group (n=21), there were 7 cases with
low expression of NOB1 and 14 cases with high expression of NOB1
(Fig. 6E). Furthermore,
correlation analysis showed that the miR-326 expression was
inversely correlated with NOB1 expression in OS tissues (Fig. 6E, r=−0.5723, P=0.0064). These data
indicated that NOB1 might be a target of miR-326 in OS tissues as
well.
SNHG1 induces NOB1 upregulation via
inhibiting miR-326 expression
Above results showed that SNHG1 could suppress the
miR-326 which acts as a tumor suppressor by targeting the NOB1 in
OS cells. Therefore, we speculated whether SNHG1 could indirectly
mediate NOB1 expression via inhibiting the miR-326 level. To
demonstrate this speculation, SNHG1 expression was dysregulated by
SNHG1 overexpression plasmid pcDNA-SNHG1 or pcDNA-mut-SNHG1
transfection in Saos-2 cells, or cotransfection with miR-326 mimic,
and the NOB1 expression was detected using western blot analysis.
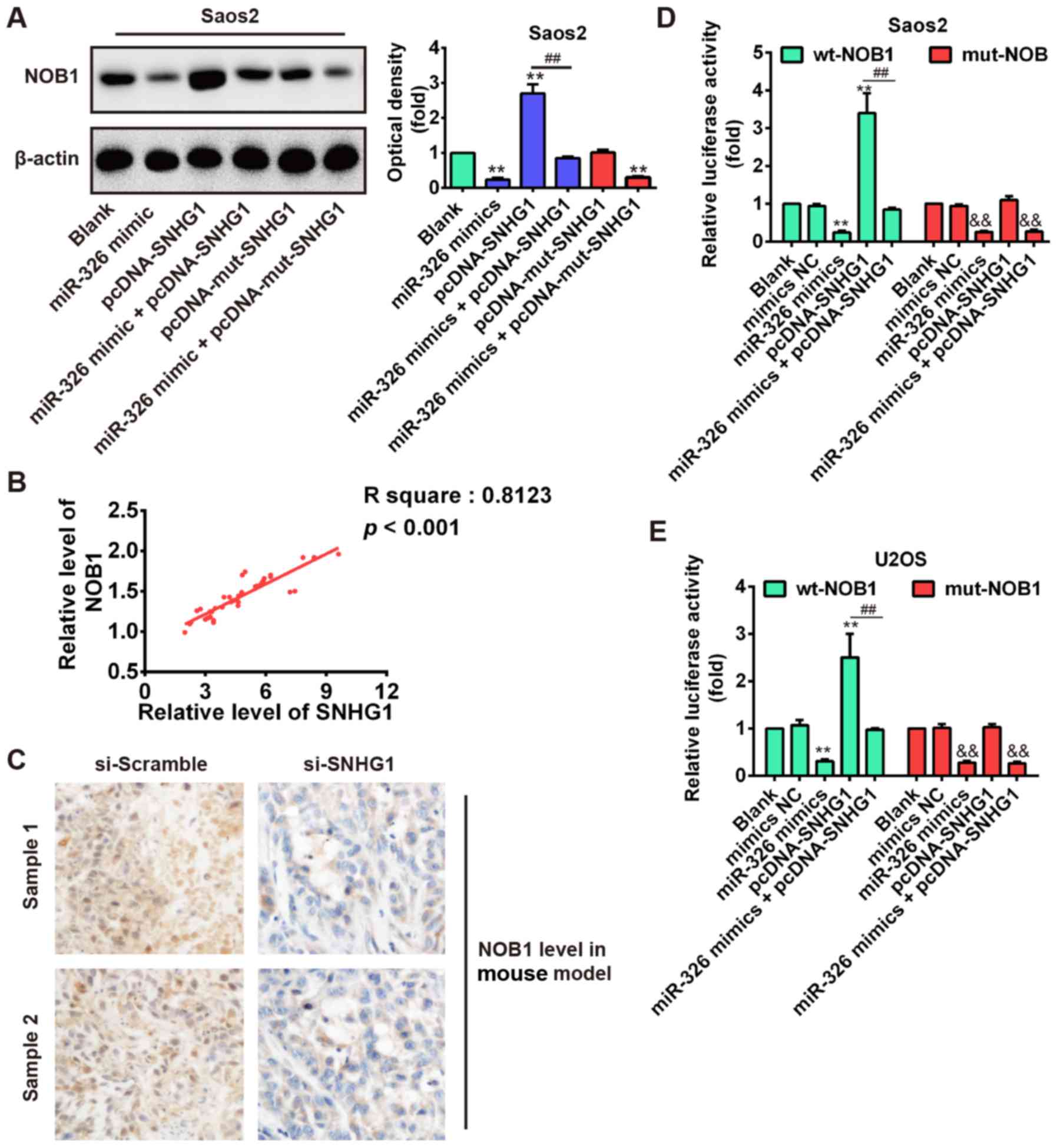
As shown in Fig. 7A, SNHG1
overexpression significantly increased NOB1 expression compared
with blank, but SNHG1-induced upregulation of NOB1 was reversed by
miR-326 overexpression in Saos-2 cells after co-transfection with
miR-326 mimic and pcDNA-SNHG1. Furthermore, we observed that
mut-SNHG1 overexpression did not affect the NOB1 level, whereas
miR-326 upregulation also significantly reduced NOB1 expression in
Saos-2 cells after co-transfection with miR-326 mimic and
pcDNA-mut-SNHG1 compared with blank (P<0.01; Fig. 7A). Additionally, correlation
analysis further verified a strong positive relationship between
SNHG1 and NOB1 expression in OS tissues (R2=0.8123,
P<0.01; Fig. 7B). Furthermore,
immunohistochemical results also demonstrated that knockdown of
SNHG1 increased the NOB1 expression level in xenograft tumor
tissues (Fig. 7C). These results
suggested that SNHG1 may indirectly increase NOB1 level through
regulating miR-326 expression. To further explore this mechanism,
the wt or mut reporter plasmid that contained the wt or mut 3′-UTR
segments of NOB1 was cotransfected into Saos-2 and U2OS cells along
with miR-326 mimics, pcDNA-SNHG1, mimics NC, or miR-326 mimics +
pcDNA-SNHG1 and measured the luciferase activity. Our results
showed that miR-326 mimics or pcDNA-SNHG1 significantly inhibited
or enhanced the luciferase activity compared with the NC (blank and
mimic NC) in the presence of the wt 3′-UTR, but the
pcDNA-SNHG1-enhanced effect was abolished by miR-326 mimics in
Saos-2 and U2OS cells after cotransfection with miR-326 mimics and
pcDNA-SNHG1 (Fig. 7D and E). In
addition, pcDNA-SNHG1 did not enhance the luciferase activity
compared with the NC (blank and mimic NC) in the presence of the
mut 3′-UTR. These data indicated that the tumorigenic SNHG1
upregulated NOB1 expression via suppressing miR-326 level in
OS.
Discussion
Accumulating evidence demonstrated that ectopic
expression of lncRNAs could act as oncogenes or tumor suppressors
in various cancer progression and development (13). However, only few reports have
discussed their contribution in OS. In the present study, we found
that SNHG1 was upregulated in OS tissues and cell lines. Knockdown
of SNHG1 inhibited cell proliferation, migration and invasion, and
induced apoptosis via activating cleaved-caspase-3. Meanwhile, the
xenograft model showed that SNHG1 downregulation suppressed tumor
growth in vivo. Furthermore, our data confirmed that there
was reciprocal repression between SNHG1 and miR-326 which
functioned as a tumor suppressor through suppressing NOB1
expression. Additionally, we verified that tumorigenic SNHG1
modulated NOB1 expression via suppressing miR-326 level in OS,
suggesting that SNHG1 might function as an oncogene in OS through
upregulating NOB1 level.
Many studies confirmed that SNHG1 was upregulated in
different cancers, such as lung, breast cancer and hepatocellular
carcinoma (24–26). SNHG1 overexpression is associated
with poor prognosis of hepatocellular carcinoma patients (24). However, the potential roles of
SNHG1 in OS remain unclear. Consistent with previous studies, our
results also showed that SNHG1 was upregulated in OS tissues and
cell lines, and Kaplan-Meier survival analysis demonstrated that
high SNHG1 expression predicts poor overall survival of OS
patients. These data suggested that SNHG1 was dysregulated in OS,
and might function as an oncogene in OS tumorigenesis. To further
explore this mechanism, the SNHG1 function was investigated using
siRNA-mediated knockdown experiments. We observed that knockdown of
SNHG1 suppressed cell proliferation, migration and invasion and
prompted apoptosis via activating cleaved-caspase-3 pathway.
Xenograft tumor model showed that knockdown of SNHG1 inhibited
tumor growth in vivo. Increasing evidence identified that
cancer is tightly involved with EMT, which induces the cancer cells
to acquire mesenchymal phenotype and metastasize towards distant
sites (30–32). Therefore, to further explore the
molecular mechanism which knockdown of SNHG1 suppresses cell
migration and invasion in vitro, we performed western blot
analysis to determine EMT markers (E-cadherin, vimentin, N-cadherin
and ZEB1) in OS cells after SNHG1 downregulation. Our data showed
that knockdown of SNHG1 increased E-cadherin protein level but
decreased vimentin, N-cadherin and ZEB1 which promoted EMT by
repressing expression of E-cadherin (35–37).
These results suggested that knockdown of SNHG1 suppresses cell
migration and invasion via modulating EMT.
In recent years, ceRNA hypothesis has been widely
reported and some studies have verified the interaction between
lncRNA and miRNA in cancers (47,48),
we hypothesized that SNHG1 may act as a ceRNA in OS. To confirm our
hypothesis, we used two bioinformatic databases including
TargetScan and microRNA.org to predict potential
miRNAs, and found several potential miRNA-binding sites including
miR-326. Recent studies, and our previous demonstration, indicated
that miR-326 may act as a tumor suppressor in many cancers
including OS (39–41). Thus, the miR-326 was selected and
further investigated by bioinformatics. Our results showed that
SNHG1 is a direct target of miR-326, and overexpression of miR-326
abolished SNHG1 upregulation in OS cells, conversely, SNHG1
downregulation increased miR-326 expression levels. Furthermore, we
found that miR-326 was downregulated in OS tissues especially
metastatic tissues and cell lines, and exhibited an inverse
correlation with SNHG1 expression in OS tissues. These results
suggested that the reciprocal regulation of SNHG1 and miR-326 might
have an important role in OS progression.
Increasing evidence identified that miR-326
functions as a tumor suppressor by targeting the NOB1 which acts as
an oncogene in different cancers (39,43–46).
Consistent with previous studies, our data further confirmed that
miR-326 repressed cell proliferation, migration and invasion, and
induced apoptosis via directly targeting NOB1 3′-UTR. Moreover,
miR-326 expression was inversely correlated with NOB1 expression in
OS tissues. Based on these results, we speculated whether SNHG1
could indirectly modulate NOB1 expression as ceRNA to inhibit the
activity of miR-326. Our results showed that upregulation of SNHG1
increased NOB1 expression, but SNHG1-induced upregulation of NOB1
was reversed by miR-326 overexpression. Furthermore, further
experiments verified a strong negative relationship between SNHG1
and miR-326 expression in OS tissues, and the miR-326 suppressive
effect on NOB1 was abolished by SNHG1 overexpression in OS cells.
Taken together, these results indicated tumorigenic SNHG1 as a
sponge to suppress miR-326 activity by sequestering it away from
its target NOB1 in OS.
Recently, lncRNAs have been demonstrated to be
involved in differentiation, proliferation and development, as well
as cell cycle regulation and programmed cell death (49,50).
Previous studies reported that lncRNAs were identified to act as
oncogenes or tumor suppressor genes to regulate OS progression,
such as H19, MALAT1, ANCR, HOTAIR, TUG1, MEG3, HIF2PUT and TUSC7
(16). According to the ceRNA
hypothesis, lncRNAs interacted with other RNA transcripts via miRNA
response elements as the letters of a novel RNA language (6). Liang et al (51) revealed that H19 promotes epithelial
to mesenchymal transition and acts as a ceRNA for miR-138 and
miR-200a in colorectal cancer. Therefore, we speculated that more
lncRNAs may be involved in the progression of OS, whether SNHG1
functions as a ceRNA for more miRNAs need to be further
investigated in subsequent studies.
In summary, our results demonstrated that SNHG1 was
upregulated in OS tissues and cell lines. Knockdown of SNHG1
suppressed OS cell growth and metastasis in vitro and in
vivo. More importantly, we verified that SNHG1 inhibits the
activity of miR-326 by acting as an endogenous sponge, through
increasing the expression of its target gene, NOB1. The present
study not only uncovered the important role of SNHG1/miR-326/NOB1
signaling pathway in OS pathogenesis but also implicated the
potential role of both SNHG1 and miR-326 in the OS clinical
diagnosis and treatment.
Acknowledgments
The present study was supported by the National
Natural Science Foundation of China (no. 81301588).
References
|
1
|
Link MP, Goorin AM, Miser AW, Green AA,
Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick
JA, et al: The effect of adjuvant chemotherapy on relapse-free
survival in patients with osteosarcoma of the extremity. N Engl J
Med. 314:1600–1606. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: Data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marina N, Gebhardt M, Teot L and Gorlick
R: Biology and therapeutic advances for pediatric osteosarcoma.
Oncologist. 9:422–441. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song QC, Shi ZB, Zhang YT, Ji L, Wang KZ,
Duan DP and Dang XQ: Downregulation of microRNA-26a is associated
with metastatic potential and the poor prognosis of osteosarcoma
patients. Oncol Rep. 31:1263–1270. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cho WCS: MicroRNAs: Potential biomarkers
for cancer diagnosis, prognosis and targets for therapy. Int J
Biochem Cell Biol. 42:1273–1281. 2010. View Article : Google Scholar
|
|
6
|
Khoury S: Circulating microRNAs :
potential biomarkers for common malignancies. Biomark Med.
9:131–151. 2015. View Article : Google Scholar
|
|
7
|
Liang WC, Wang Y, Xiao LJ, Wang YB, Fu WM,
Wang WM, Jiang HQ, Qi W, Wan DC, Zhang JF, et al: Identification of
miRNAs that specifically target tumor suppressive KLF6-FL rather
than oncogenic KLF6-SV1 isoform. RNA Biol. 11:845–854. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lipovich L, Johnson R and Lin CY: MacroRNA
underdogs in a microRNA world: Evolutionary, regulatory, and
biomedical significance of mammalian long non-protein-coding RNA.
Biochim Biophys Acta. 1799:597–615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peng W and Fan H: Long non-coding RNA
PANDAR correlates with poor prognosis and promotes tumorigenesis in
hepatocellular carcinoma. Biomed Pharmacother. 72:113–118. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Deng J, Liang Y, Liu C, He S and Wang S:
The up-regulation of long non-coding RNA AFAP1-AS1 is associated
with the poor prognosis of NSCLC patients. Biomed Pharmacother.
75:8–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li L, Zhang L, Zhang Y and Zhou F:
Increased expression of LncRNA BANCR is associated with clinical
progression and poor prognosis in gastric cancer. Biomed
Pharmacother. 72:109–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen J, Wang Y and Wang D: Noise
perturbation improves supervised speech separation. Latent Variable
Analysis and Signal Separation: Proc of 12th International
Conference, LVA/ICA 2015; Liberec, Czech Rep. August 25–28, 2015;
Vincent E, Yeredor A, Koldovský Z and Tichavský P: Springer
International Publishing, Cham; pp. 83–90. 2015
|
|
16
|
Yang Z, Li X, Yang Y, He Z, Qu X and Zhang
Y: Long noncoding RNAs in the progression, metastasis, and
prognosis of osteosarcoma. Cell Death Dis. 7:e23892016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cong M, Li J, Jing R and Li Z: Long
non-coding RNA tumor suppressor candidate 7 functions as a tumor
suppressor and inhibits proliferation in osteosarcoma. Tumour Biol.
37:9441–9450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li F, Cao L, Hang D, Wang F and Wang Q:
Long non-coding RNA HOTTIP is up-regulated and associated with poor
prognosis in patients with osteosarcoma. Int J Clin Exp Pathol.
8:11414–11420. 2015.PubMed/NCBI
|
|
19
|
Zhuang LK, Xu GP, Pan XR, Lou YJ, Zou QP,
Xia D, Yan WW, Zhang YT, Jia PM and Tong JH: MicroRNA-181a-mediated
downregulation of AC9 protein decreases intracellular cAMP level
and inhibits ATRA-induced APL cell differentiation. Cell Death Dis.
5:e11612014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
21
|
Ma MZ, Chu BF, Zhang Y, Weng MZ, Qin YY,
Gong W and Quan ZW: Long non-coding RNA CCAT1 promotes gallbladder
cancer development via negative modulation of miRNA-218-5p. Cell
Death Dis. 6:e15832015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cui M, Xiao Z, Wang Y, Zheng M, Song T,
Cai X, Sun B, Ye L and Zhang X: Long noncoding RNA HULC modulates
abnormal lipid metabolism in hepatoma cells through an
miR-9-mediated RXRA signaling pathway. Cancer Res. 75:846–857.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding
F, Li D and Yang T: Long noncoding RNA SNHG1 predicts a poor
prognosis and promotes hepatocellular carcinoma tumorigenesis.
Biomed Pharmacother. 80:73–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
You J, Fang N, Gu J, Zhang Y, Li X, Zu L
and Zhou Q: Noncoding RNA small nucleolar RNA host gene 1 promote
cell proliferation in nonsmall cell lung cancer. Indian J Cancer.
51:e99–e102. 2014. View Article : Google Scholar
|
|
26
|
Yu F, Bracken CP, Pillman KA, Lawrence DM,
Goodall GJ, Callen DF and Neilsen PM: p53 represses the oncogenic
Sno-MiR-28 derived from a SnoRNA. PLoS One. 10:e01291902015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Zhang Z, Xiong L, Guo C, Jiang T,
Zeng L, Li G and Wang J: SNHG1 lncRNA negatively regulates
miR-199a-3p to enhance CDK7 expression and promote cell
proliferation in prostate cancer. Biochem Biophys Res Commun.
487:146–152. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang H, Zhou D, Ying M, Chen M, Chen P,
Chen Z and Zhang F: Expression of long non-coding RNA (lncRNA)
small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular
carcinoma through suppressing miR-195. Med Sci Monit. 22:4820–4829.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou L, Rui JA, Ye DX, Wang SB, Chen SG
and Qu Q: Edmondson-Steiner grading increases the predictive
efficiency of TNM staging for long-term survival of patients with
hepatocellular carcinoma after curative resection. World J Surg.
32:1748–1756. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hollier BG, Evans K and Mani SA: The
epithelial-to-mesenchymal transition and cancer stem cells: A
coalition against cancer therapies. J Mammary Gland Biol Neoplasia.
14:29–43. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ahmed N, Abubaker K, Findlay J and Quinn
M: Epithelial mesenchymal transition and cancer stem cell-like
phenotypes facilitate chemoresistance in recurrent ovarian cancer.
Curr Cancer Drug Targets. 10:268–278. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Latifi A, Abubaker K, Castrechini N, Ward
AC, Liongue C, Dobill F, Kumar J, Thompson EW, Quinn MA, Findlay
JK, et al: Cisplatin treatment of primary and metastatic epithelial
ovarian carcinomas generates residual cells with mesenchymal stem
cell-like profile. J Cell Biochem. 112:2850–2864. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar :
|
|
34
|
Cesi V, Casciati A, Sesti F, Tanno B,
Calabretta B and Raschellà G: TGFβ-induced c-Myb affects the
expression of EMT-associated genes and promotes invasion of
ER+ breast cancer cells. Cell Cycle. 10:4149–4161. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Derynck R and Akhurst RJ: Differentiation
plasticity regulated by TGF-beta family proteins in development and
disease. Nat Cell Biol. 9:1000–1004. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Vandewalle C, Van Roy F and Berx G: The
role of the ZEB family of transcription factors in development and
disease. Cell Mol Life Sci. 66:773–787. 2009. View Article : Google Scholar
|
|
38
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7's suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhou J, Xu T, Yan Y, Qin R, Wang H, Zhang
X, Huang Y, Wang Y, Lu Y, Fu D, et al: MicroRNA-326 functions as a
tumor suppressor in glioma by targeting the Nin one binding protein
(NOB1). PLoS One. 8:e684692013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Meng F, Henson R, Lang M, Wehbe H,
Maheshwari S, Mendell JT, Jiang J, Schmittgen TD and Patel T:
Involvement of human micro-RNA in growth and response to
chemotherapy in human cholangiocarcinoma cell lines.
Gastroenterology. 130:2113–2129. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ferretti E, De Smaele E, Miele E, Laneve
P, Po A, Pelloni M, Paganelli A, Di Marcotullio L, Caffarelli E,
Screpanti I, et al: Concerted microRNA control of Hedgehog
signalling in cerebellar neuronal progenitor and tumour cells. EMBO
J. 27:2616–2627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cao L, Wang J and Wang PQ: MiR-326 is a
diagnostic biomarker and regulates cell survival and apoptosis by
targeting Bcl-2 in osteosarcoma. Biomed Pharmacother. 84:828–835.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He XW, Feng T, Yin QL, Jian YW and Liu T:
NOB1 is essential for the survival of RKO colorectal cancer cells.
World J Gastroenterol. 21:868–877. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu K, Chen HL, Gu MM and You QS: NOB1
expression predicts early response to cisplatin-based chemotherapy
in patients with advanced non-small cell lung cancer. J Chemother.
28:225–229. 2016. View Article : Google Scholar
|
|
45
|
Huang P, Xi J and Liu S: MiR-139-3p
induces cell apoptosis and inhibits metastasis of cervical cancer
by targeting NOB1. Biomed Pharmacother. 83:850–856. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu L, Hui H, Wang LJ, Wang H, Liu QF and
Han SX: MicroRNA-326 functions as a tumor suppressor in colorectal
cancer by targeting the nin one binding protein. Oncol Rep.
33:2309–2318. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C,
Xu M, Wu F and Mo YY: Negative regulation of lncRNA GAS5 by miR-21.
Cell Death Differ. 20:1558–1568. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Liu XH, Sun M, Nie FQ, Ge YB, Zhang EB,
Yin DD, Kong R, Xia R, Lu KH, Li JH, et al: Lnc RNA HOTAIR
functions as a competing endogenous RNA to regulate HER2 expression
by sponging miR-331-3p in gastric cancer. Mol Cancer. 13:922014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wu L, Jin L, Zhang W and Zhang L: Roles of
long non-coding RNA CCAT2 in cervical cancer cell growth and
apoptosis. Med Sci Monit. 22:875–879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ballantyne MD, Pinel K, Dakin R, Vesey AT,
Diver L, Mackenzie R, Garcia R, Welsh P, Sattar N, Hamilton G, et
al: Smooth muscle enriched long noncoding RNA (SMILR) regulates
cell proliferation. Circulation. 133:2050–2065. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Liang WC, Fu WM, Wong CW, Wang Y, Wang WM,
Hu GX, Zhang L, Xiao LJ, Wan DC, Zhang JF, et al: The lncRNA H19
promotes epithelial to mesenchymal transition by functioning as
miRNA sponges in colorectal cancer. Oncotarget. 6:22513–22525.
2015. View Article : Google Scholar : PubMed/NCBI
|