Introduction
Gastric cancer is the third major cause of the
cancer-related mortality in Japan. Owing to endoscopic examination
and surgery combined with chemotherapy, the prognosis of patients
with gastric cancer has improved (1,2).
However, there are still cases of gastric cancer which are
diagnosed at an advanced stage, and in these cases, prognosis is
unfavorable. To date, the factors that are involved in the
pathogenesis and the progression of the gastric cancer have not yet
been fully elucidated.
For the elucidation of the pathogenesis of the
disease, the comprehensive profiling of proteins, DNA, RNA and
metabolites has been carried out (3,4).
This profiling would provide invaluable information useful for the
identification of molecules of therapy, diagnosis and tumor
biology. The comprehensive profiling of proteins and mRNAs has been
carried out in gastric cancers (5–9).
These molecular studies have revealed that gastric cancers are
classified into 4 subtypes: The CpG island methylator phenotype,
hypermutated, genomically stable and chromatin instability
(6). These subtypes appear to
represent the pathogenetic differences of gastric cancers. It is
expected that the elucidation of the pathogenesis of gastric
cancers leads to the development of specific treatment (8). Molecular information is required for
the development of individualized therapy (2).
In the present study, the comprehensive protein
profiling of gastric cancers was carried out using protein samples
extracted from formalin-fixed paraffin-embedded (FFPE) specimens.
Bioinformatics analyses suggested the significance of the
expression of proteins in DNA damage response (DDR). DDR is the
consecutive sequence from the detection of damaged DNA, the
aggregation of transducers and modulators at the damaged site, cell
cycle arrest and the repair of the damaged DNA. The rapid and
uncontrolled progression of the cell cycle of carcinoma cells
causes frequent errors in genome duplication, and the defect in DDR
may enhance genetic instability and lead to the progression of
carcinoma (10). In this study,
the expression of molecules of DDR and the phosphorylation state of
proteins were examined in cases of gastric cancer and in cultured
gastric cancer cell lines. The preserved expression and
phosphorylation of DDR proteins appeared to be associated with a
favorable prognosis.
Materials and methods
Cases of human gastric cancer
Cases of gastric cancer were retrieved from the
archives of the pathology records of Nippon Medical School Hospital
(Tokyo, Japan) from 2011 to 2016. In total, 17 cases were used for
the profiling of expressed proteins, and they were selected
randomly from the pathology records. A total of 42 cases were used
for the histological and immunohistochemical analyses. The cases
were randomly selected from the cases of gastric cancer at
pathological stages from I to III. The patients underwent
gastrectomy, but they did not receive chemotherapy or radiation
therapy prior to surgery. The study was conducted according to the
declaration of Helsinki and the Japanese Society of Pathology. This
study was approved by the Ethics Committee of Nippon Medical School
Hospital (#29-06-764). Informed consent was obtained from all
patients.
Comprehensive profiling of protein
expression
FFPE specimens from 17 cases of gastric cancer were
used for the comprehensive profiling of proteins by liquid
chromatography-tandem mass spectrometry (LC-MS/MS). The sections of
gastric cancer (10-µm-thick) were deparaffinized in xylene
and rehydrated through a graded alcohol series. Following staining
with hematoxylin, the cancer tissues were dissected out under a
stereoscopic microscope (AZ-STDM; Nikon Co., Tokyo, Japan) and
lysed in buffer with 100 mM Tris-HCl (pH 9.0)/12 mM deoxycholate/12
mM lauroyl sarcosine. Following the quantification of the protein
concentration by Bradford method using Bio-Rad Protein assay
(Bio-Rad, Laboratories, Inc., Hercules, CA, USA), 10 µg of
extracted protein was reduced in 11.4 mM dithiothreitol (DTT)/1.8
mM Tris (2-carboxyethyl) phosphine hydrochloride and alkylated in
54 mM iodoacetamide. It was further digested with proteomics-grade
trypsin (Agilent Technologies Inc., Santa Clara, CA, USA) at 37°C
for 24 h, and the protein was purified with a PepClean C-18 spin
column (Thermo Fisher Scientific, K.K., Tokyo, Japan). In total, 2
µg of digested protein was injected into a Nano cHiPLC Trap
column (0.2×0.5 mm, ChromXP C18-CL 3 µm) and further
separated through a Eksigent nano LC Ekspert415 system using a
reverse-phase C-18 column (0.075×150 mm, ChromXP C18-CL 3
µm) (all from K.K. Sciex Japan, Tokyo, Japan). The protein
solution was run with the gradient concentration of acetonitrile
from 2 to 32% in 0.1% formic acid in acetonitrile at a flow rate of
300 nl/min for 120 min. Eluted peptides were analyzed by Triple TOF
5600+ mass spectrometer (K.K. Sciex Japan). The data of
10 most intense peaks of each full MS scan were acquired. All MS/MS
spectral data were analyzed by MASCOT 2.4 (Matrix Science K.K.,
Tokyo, Japan) with SwissProt 2015_02. The following parameter
settings were used: Trypsin cleavage; two missed cleavage sites
allowed for cysteine carbamidomethylation (C-terminus) and
methionine oxidation (N-terminus). Peptide mass tolerance was set
to ±50 ppm, and fragment MS/MS tolerance were set to 0.05 Da. The
amounts of identified proteins were expressed as normalized
spectral abundance factor (NSAF).
Bioinformatics analyses
Protein expression was analyzed by hierarchical
clustering, orthogonal partial least squares-discriminant analysis
(OPLS-DA) and Ingenuity Pathway Analysis (IPA). Analyses were done
using NSAF of protein.
Hierarchical clustering was carried out using
Cluster 3.0 and visualized using TreeView software (Howard Hughes
Medical Institutes, University of California at Berkeley, Berkeley,
CA, USA). Analysis with OPLS-DA was carried out using SIMCA version
14 (Umetrics, Umea, Sweden) to identify proteins which have a
significant influence on the magnitude {p[1]} and reliability
{p(corr)[1]} to separate into 2 clusters. The results were
visualized as S-PLOT, a scatter plot with magnitude {p[1]} as the
x-axis and reliability {p(corr)[1]} as the y-axis. The reliability,
which was ≥0.6 and ≤−0.6 was considered significant. Pathways of
expressed proteins were analyzed by IPA (Qiagen, Redwood City, CA,
USA). The scores of networks were calculated by the numbers of
focus proteins. The percentages of the number of reliable proteins
evaluated by OPLS-DA in the number of focus proteins of each
network identified by IPA were then calculated. It was expected
that the combined analysis of OPLS-DA and IPA was useful to
identify the biologically significant proteins from the profiled
proteins.
Histological examination of human gastric
cancers
A total of 42 cases of gastric cancer were used for
the histological and immunohistochemical analyses. The histological
subtypes were classified according to the classification by Lauren
(11). Pathological T-factors,
lymphovascular invasion and lymph node metastasis were also
reviewed. The review of the histology was performed by 3
investigators (H.A., R.W. and Z.N.).
Immunohistochemistry
Immunohistochemistry was performed using the
polymer-based two-step method. Briefly, the paraffin-embedded
sections were deparaffinized and hydrated in phosphate-buffered
saline (PBS). After the blocking of endogenous peroxidase, the
sections were pretreated, if necessary, and then incubated with
primary antibodies listed in Table
I at 4°C overnight. The sections were then incubated with
Simple Stain MAX-PO (R) for rabbit primary antibody or (M) for
mouse primary antibody (Nichirei Inc., Tokyo, Japan). Peroxidase
activity was visualized by diaminobenzidine.
 | Table IThe antibodies used in the present
study. |
Table I
The antibodies used in the present
study.
| Antibody | Clone | Species | Dilution WB | IHC | PT | Company/provider |
|---|
| KU70 | EPR4026 | Rabbit | 1:10,000 | 1:2,000 | – | Abcam K.K., Tokyo,
Japan |
| p-KU70 (Ser5) | ab61783 | Rabbit | 1:10,000 | 1:2,000 | TB | Abcam K.K., Tokyo,
Japan |
| ILF2 | ab28772 | Rabbit | 1:1,000 | – | CB | Abcam K.K., Tokyo,
Japan |
| CHK1 | 2G1D5 | Mouse | 1:1,000 | 1:500 | TB | Cell Signaling
Technology, Japan, K.K. Tokyo, Japan |
| p-CHK1
(Ser317) | D92H3 | Rabbit | 1:1,000 | 1:5,000 | CB | Cell Signaling
Technology, Japan, K.K. Tokyo, Japan |
| CHK2 | D9C6 | Rabbit | 1:1,000 | 1:1,000 | CB | Cell Signaling
Technology, Japan, K.K. Tokyo, Japan |
| p-CHK2 (Thr68) | C13C1 | Rabbit | 1:1,000 | 1:1,000 | TB | Cell Signaling
Technology, Japan, K.K. Tokyo, Japan |
| TP53 | DO7 | Mouse | 1:1,000 | – | CB | Agilent
Technologies Japan, Ltd. Tokyo, Japan |
Culture of gastric cancer cell lines and
exposure to ultraviolet (UV) radiation
Four gastric cancer cell lines, NS-8, MKN-7, NUGC-4
and KATO-III, were used in this study. The cells were cultured in
DMEM with 10% fetal bovine serum and penicillin/streptomycin. The
cells were exposed to UV radiation at a dose of 40 J/m2.
At 2 h following exposure, the cells were washed with PBS and lysed
in 50 mM Tris-HCl (pH 7.6). The sample was then sonicated for 10
min.
Western blot analysis
The cell lysate was electrophoresed and blotted onto
a polyvinylidene difluoride membrane. After the blocking in 5% skim
milk in buffer of 0.2 M Tris-HCl (pH 7.6)/150 mM NaCl/0.01%
Tween-20, the membrane was incubated with the primary antibodies
listed in Table I overnight. The
membrane was then incubated with peroxidase-labeled anti-mouse
immunoglobulin antibody (#A106PU) or anti-rabbit immunoglobulin
antibody (#A102PU) (both from American Qualex Scientific Products,
San Clemente, CA, USA), and the peroxidase activity was detected as
chemiluminescence using SuperSignal West Dura Extended Duration
Substrate (Thermo Fisher Scientific, K.K.).
Analysis of apoptotic cell death in
cultured cells
Cell death was examined using the Apoptosis,
Necrosis and Healthy Cell Quantitation kit (Biotium, Inc., Hayward,
CA, USA). Briefly, the cultured cells were washed with PBS, and
incubated with a mixture of FITC-labeled Annexin V and Hoechst
33342 at room temperature for 15 min. Apoptosis was identified when
cells were stained positive for FITC. The frequency of apoptotic
cell death was calculated by dividing the number of apoptotic cells
by the total number of cells. A total of at least total 500 cells
was counted.
Statistical analysis
All quantitative data are expressed as the means ±
SD. The distribution of cases was analyzed by the Chi-square test
with Fisher's correction. The data of 2 groups were analyzed by the
Mann-Whitney U test. P-values <0.05 was considered to indicate
statistically significant differences. All statistical analyses
were carried out using JMP 13.0 software (SAS Institute Japan,
Ltd., Tokyo, Japan).
Results
Comprehensive profiling of protein
expression and bioinformatics analyses
The comprehensive profiling of proteins was
performed with the proteins purified from the FFPE samples of 17
cases of gastric cancer. A total of 5,338 proteins were identified
by LC-MS/MS. NSAFs of housekeeping proteins, such as β-actin and
histones were comparable among the cases (data not shown).
Among the 5,338 proteins, 483 proteins (9%) were
expressed in all 17 cases. The other proteins were not detected at
least one case. Unsupervised hierarchical clustering was performed
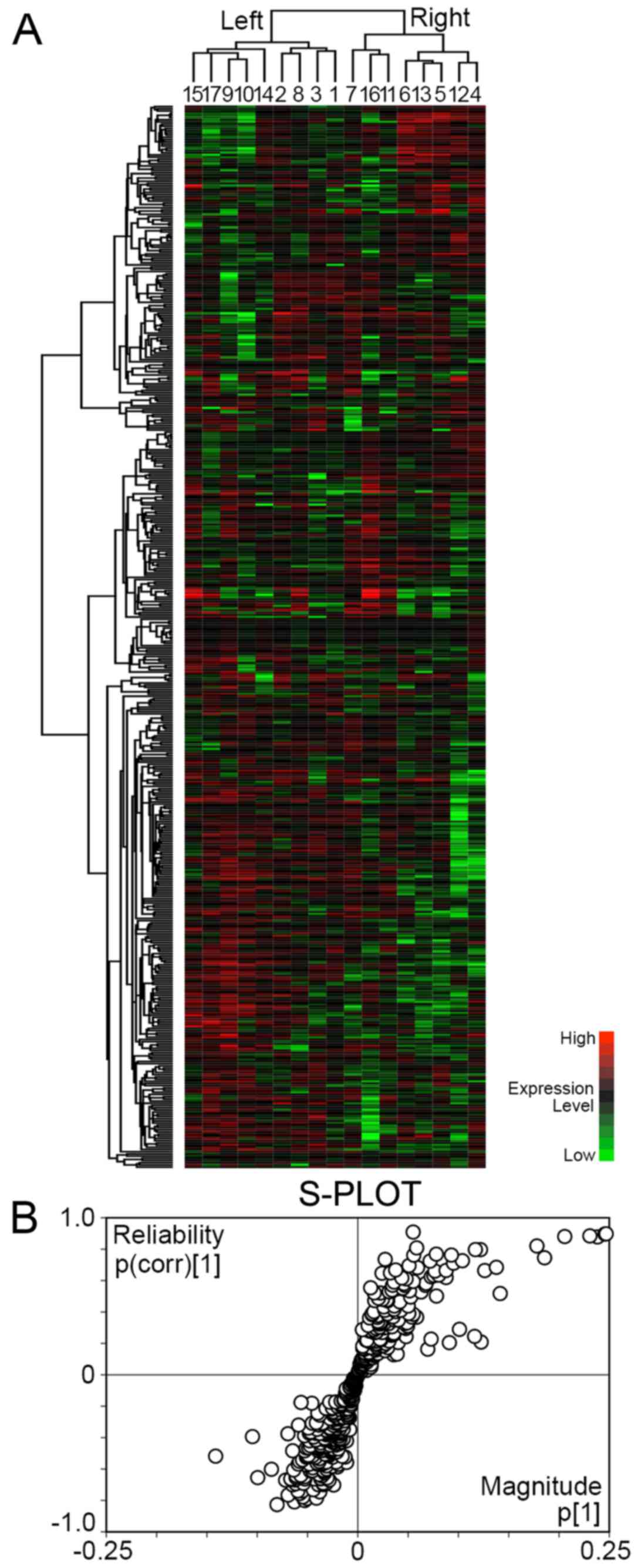
with these 483 proteins (Fig. 1A).
The gastric cancers were separated into 2 major clusters. The left
cluster included 9 cases, and the right cluster included 8 cases.
There was no significant difference as regards age, location,
pathological T factor and pathological stage between the 2 clusters
(Table II). All cases in the
right cluster were male. The intestinal subtype was predominant in
the left cluster, whereas the diffuse subtype was predominant in
the right cluster.
 | Table IIClinicopathological characteristics
of the patients with gastric cancer in the left and right
clusters. |
Table II
Clinicopathological characteristics
of the patients with gastric cancer in the left and right
clusters.
| Left cluster
(n=9) | Right cluster
(n=8) | P-value |
|---|
| Age (years) | 72±12 | 74±7 | ns |
| Sex | | | |
| Male | 4 | 8 | P<0.05 |
| Female | 5 | 0 | |
| Location | | | |
| Upper | 4 | 1 | ns |
| Middle | 1 | 1 | |
| ML | 1 | 2 | |
| Lower | 3 | 4 | |
| Lauren
classification | | | |
| Intestinal | 6 | 2 | P<0.05 |
| Mixed | 3 | 1 | |
| Diffuse | 0 | 5 | |
| pT | | | |
| pT1 | 6 | 5 | ns |
| pT2 | 1 | 3 | |
| pT3 | 2 | 0 | |
| pT4 | 0 | 0 | |
| Stage | | | |
| I | 6 | 5 | ns |
| II | 2 | 3 | |
| III | 1 | 0 | |
| IV | 0 | 0 | |
To identify the proteins reliable to separate the
left and right clusters, OPLS-DA was carried out with NSAF of 483
proteins and visualized by S-PLOT (Fig. 1B). The reliability {p(corr)[1]} of
33 proteins was ≥0.6, and it was considered that they had
significant reliability to discriminate the right cluster (Table III). On the other hand, the
reliability of 46 proteins was ≤−0.6, and they had significant
reliability for the discrimination of the left cluster (Table IV).
 | Table IIIProteins identified by orthogonal
partial least squares-discriminant analysis (the reliability is
≥0.6). |
Table III
Proteins identified by orthogonal
partial least squares-discriminant analysis (the reliability is
≥0.6).
| Protein | Magnitude p[1] | Reliability
p(corr)[1] |
|---|
| INA | 0.055052 | 0.909151 |
| ACTA2 | 0.246902 | 0.897744 |
| ACTC1 | 0.245703 | 0.896767 |
| ACTA1 | 0.230718 | 0.884632 |
| VIM | 0.205729 | 0.880668 |
| ACTB | 0.238275 | 0.878102 |
| ACTBL2 | 0.178069 | 0.820505 |
| PRPH | 0.058522 | 0.808254 |
| TPM1 | 0.116885 | 0.797396 |
| TPM2 | 0.121969 | 0.797050 |
| FLNA | 0.078055 | 0.768442 |
| TPM3 | 0.093025 | 0.762556 |
| CSRP1 | 0.055634 | 0.761816 |
| TAGLN | 0.185655 | 0.745214 |
| FLNC | 0.027218 | 0.735016 |
| FTL | 0.103966 | 0.726628 |
| ANXA6 | 0.071863 | 0.724106 |
| ANXA5 | 0.095132 | 0.710682 |
| MSN | 0.050898 | 0.695238 |
| DES | 0.137757 | 0.684517 |
| S100A4 | 0.059389 | 0.682374 |
| MYH10 | 0.037659 | 0.667677 |
| MYL6 | 0.087813 | 0.664537 |
| POTEKP | 0.126213 | 0.664068 |
| POTEE | 0.076739 | 0.660707 |
| RDX | 0.032470 | 0.649216 |
| PKLR | 0.024108 | 0.646927 |
| COL6A3 | 0.080483 | 0.634928 |
| COL6A2 | 0.075606 | 0.632119 |
| COL6A1 | 0.072951 | 0.624386 |
| LUM | 0.087745 | 0.619709 |
| POTEI | 0.043763 | 0.616854 |
| VCL | 0.048017 | 0.610764 |
 | Table IVProteins identified by orthogonal
partial least squares-discriminant analysis (the reliability is
≤−0.6). |
Table IV
Proteins identified by orthogonal
partial least squares-discriminant analysis (the reliability is
≤−0.6).
| Protein | Magnitude p[1] | Reliability
p(corr)[1] |
|---|
| HYOU1 | −0.031015 | −0.600712 |
| NME1 | −0.085839 | −0.601319 |
| RPS16 | −0.063733 | −0.606282 |
| ATIC | −0.033951 | −0.609055 |
| HNRNPA1L2 | −0.045016 | −0.610854 |
| ETFB | −0.047257 | −0.611190 |
| PDLIM1 | −0.034208 | −0.612325 |
| RNPEP | −0.032576 | −0.612375 |
| CUTA | −0.032975 | −0.624701 |
| RPS5 | −0.038159 | −0.635456 |
| RPSA | −0.056624 | −0.640193 |
| RPS13 | −0.061149 | −0.641738 |
| XRCC5 | −0.035668 | −0.643890 |
| HNRNPC | −0.052836 | −0.649307 |
| RPS19 | −0.061370 | −0.653405 |
| TXN | −0.099468 | −0.654578 |
| SFPQ | −0.030426 | −0.656324 |
| RPL4 | −0.036438 | −0.660385 |
| HNRNPU | −0.036768 | −0.661809 |
| RPS18 | −0.066594 | −0.662418 |
| PSMA6 | −0.052565 | −0.663324 |
| EEF1B2 | −0.043041 | −0.663354 |
| PHB | −0.065979 | −0.667600 |
| C1QBP | −0.062352 | −0.669720 |
| DDX3X | −0.028476 | −0.670196 |
| HSPD1 | −0.071720 | −0.670892 |
| PABPC1 | −0.034400 | −0.690058 |
| HSD17B10 | −0.054045 | −0.691120 |
| NANS | −0.031787 | −0.693264 |
| PDCD6 | −0.061551 | −0.693424 |
| HNRNPM | −0.034036 | −0.699508 |
| SYNCRIP | −0.039143 | −0.699748 |
| AKR1A1 | −0.064921 | −0.700549 |
| CTNND1 | −0.023981 | −0.701687 |
| HNRNPH1 | −0.044206 | −0.731575 |
| XRCC6 | −0.059573 | −0.739444 |
| RPS4X | −0.055168 | −0.739747 |
| RPS23 | −0.037777 | −0.745383 |
| CCT2 | −0.057704 | −0.755868 |
| ETHE1 | −0.069280 | −0.765860 |
| ILF2 | −0.054563 | −0.777686 |
| CIRBP | −0.041115 | −0.785409 |
| HNRNPF | −0.050593 | −0.787704 |
| EEF2 | −0.052731 | −0.800286 |
| VCP | −0.062993 | −0.809144 |
| RPS7 | −0.080280 | −0.826483 |
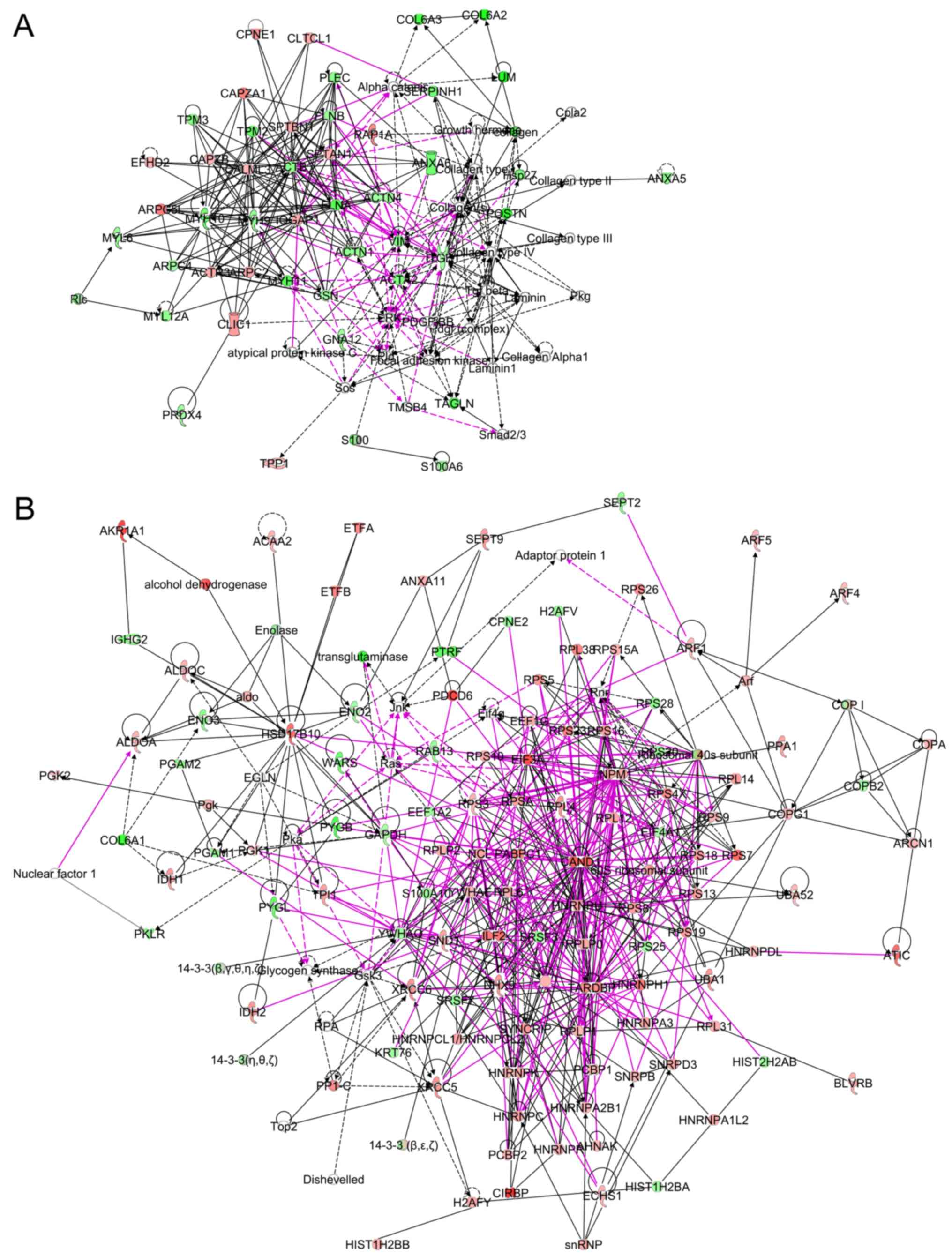
Networks of proteins were analyzed by IPA with NSAF
of 483 proteins, and a total of 24 networks was identified. To
identify proteins which are biologically significant in gastric
cancer, protein expression was further analyzed by combining the
results of OPLS-DA and IPA. The proteins identified as reliable for
the discrimination of right clusters were enriched in networks 2
and 19 (Table V). The percentages
of proteins identified by OPLS-DA (double underlined proteins in
Table V) in the focus molecules of
network 2 and 19 were 23% (7/31) and 54% (7/13), respectively. They
were intermediate filaments, matrix proteins and their binding
proteins. Some were associated with cell death. The combined
networks of 2 and 19 are shown in Fig.
2A. Among the 322 connections, 53 connections (pink-colored
lines in Fig. 2A, 17%) were
interactions between the proteins in 2 networks. On the other hand,
the proteins identified as reliable for the discrimination of the
left cluster by OPLS-DA were enriched in networks 1, 3, 5 and 9
(Table V). The percentages of
proteins identified by OPLS-DA (underlined proteins in Table V) in the focus molecules of
networks 1, 3, 5 and 9 were 27% (9/33), 29% (8/28), 15% (4/27) and
16% (4/25), respectively. Combined networks formed tight
connections (Fig. 2B), and 231
pink-colored connections out of 557 connections (42%) were
interactions among 4 networks. The focus molecules were tightly
linked, suggesting the close functional associations among the
proteins (Fig. 2B). The majority
of these proteins were ribosomal proteins and heterogeneous nuclear
ribonucleoproteins (hnRNPs). The networks also included XRCC5
(KU80), XRCC6 (KU70) and interleukin enhancer binding protein 2
(ILF2).
 | Table VNetworks identified by Ingenuity
Pathway Analysis. |
Table V
Networks identified by Ingenuity
Pathway Analysis.
| ID | Molecules in
network | Score | Focus
molecules | Top diseases and
functions |
|---|
| 1 | 60S ribosomal
subunit, CAND1, CIRBP, EEF1A2, EEF1G,
EIF3A, HNRNPA1L2,
HNRNPCL1/HNRNPCL2, HNRNPDL, HNRNPK,
HNRNPR, HNRNPU, KRT76,
PABPC1,
PCBP1, PCBP2, Ras, RPL4, RPL14,
RPL38, RPLP0, RPS3, RPS8, RPS9,
RPS10, RPS16, RPS19, RPS25,
RPSA,
SND1, SRSF3, SRSF7, SYNCRIP, TARDBP,
UBA52 | 57 | 33 | RNA
post-transcriptional modification, protein synthesis, Cancer |
| 2 | ACTB,
ACTN1, ACTN4, ACTR3, ANXA6, ARPC2,
ARPC4, ARPC5L, CALML3, CAPZA1,
CAPZB, CLIC1, CLTCL1, CPNE1,
EFHD2, ERK, FLNA, FLNB,
GSN, IQGAP1, MYH9, MYH10, MYH11,
MYL6,
MYL12A, PLEC, PRDX4, Rlc, S100, S100A6,
SPTAN1, SPTBN1, TMSB4, TPM2, TPM3 | 52 | 31 | Cellular assembly
and organization, xellular function and maintenance, cell-to-cell
signaling and interaction |
| 3 | ARCN1, Arf,
ARF1, ARF4, ARF5, ATIC, COPI, COPA,
COPB2, COPG1, ECHS1, EIF4A1, Eif4g,
H2AFV, HNRNPA3, HNRNPH1, NPM1,
nuclear factor 1, Pka, PKLR, PPA1,
RAB13, ribosomal 40s subunit, Rnr, RPS5, RPS7, RPS13, RPS18, RPS20,
RPS23,
RPS26, RPS28, RPS15A, RPS4X, UBA1 | 44 | 28 | Cancer, cell death
and survival, organismal injury and abnormalities |
| 5 | ACAA2,
adaptor protein 1, AKR1A1, alcohol dehydrogenase, aldo,
ALDOA, ALDOC, ANXA11, COL6A1, CPNE2, EGLN,
ENO2, ENO3, enolase, ETFA, ETFB, GAPDH,
HSD17B10,
IDH1, IDH2, IGHG2, Jnk, PDCD6, PGAM1,
PGAM2, Pgk, PGK1, PGK2, PTRF,
PYGB, PYGL, SEPT2, SEPT9, TPI1,
transglutaminase | 42 | 27 | Cancer, cell death
and survival, organismal injury and abnormalities |
| 9 | 14-3-3 (β,ε,ζ),
14-3-3 (β,γ,θ,η,ζ), 14-3-3 (η,θ,ζ), AHNAK, H2AFY,
BLVRB, DHX9, dishevelled, glycogen synthase, Gsk3,
HIST1H2BA, HIST1H2BB, HIST2H2AB,
HNRNPA2B1, HNRNPC, ILF2, NCL, PP1-C,
RPA, RPL6, RPL12, RPL31, RPLP1,
RPLP2, S100A10, snRNP, SNRPB, SNRPD3,
SRSF1, Top2, WARS, XRCC5, XRCC6, YWHAE,
YWHAQ | 37 | 25 | RNA
post-transcriptional modification, cell morphology, cellular
function and maintenance |
| 19 | ACTA2, α catenin,
ANXA5,
atypical protein kinase C, COL6A2, COL6A3, collagen, collagen
α1, collagen type I, collagen type II, collagen type III, collagen
type IV, collagen(s), Cpla2, focal adhesion kinase, GNA12,
growth hormone, Hsp27, ITGB1, laminin, laminin1,
LUM, Pdgf,
PDGF BB, Pkg, Pld, POSTN, RAP1A, SERPINH1,
Smad2/3, Sos, TAGLN, Tgfβ, TPP1,
VIM | 14 | 13 | Developmental
disorder, organismal injury and abnormalities, connective tissue
disorders |
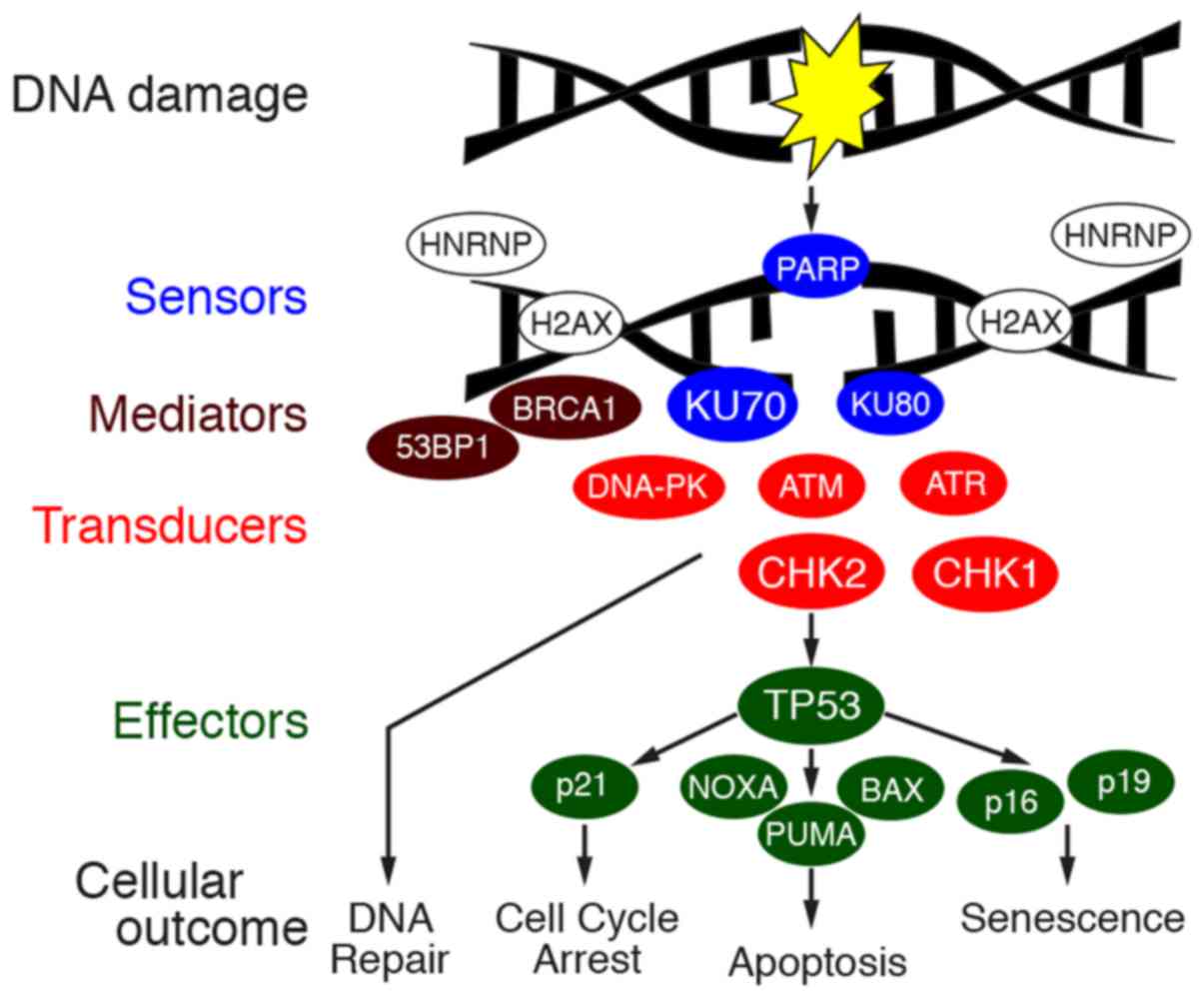
KU70 and KU80 are sensors of damaged DNA (12). ILF2 regulates the splicing of
mRNAs, and it has been shown that ILF2 regulates RNA processing of
genes involved in DDR (13).
hnRNPs and ribosomal proteins stabilize mRNAs and regulate the
splicing and translation of mRNAs, and it was recently shown that
hnRNPs and ribosomal proteins stabilize the genome when DNA is
damaged (14). The tight
connections of these proteins raise the possibility of the
involvement of DDR in gastric cancer. DDR is exerted by the
orchestration of molecules, sensors of damaged DNA, transducers,
mediators and effectors to repair the damaged DNA and to determine
the fate of the cell (Fig. 3). The
transduction pathway is exerted by the phosphorylation of proteins.
Thus, in this study, the expression and phosphorylation of key
proteins of DDR were examined by immunostaining of 42 cases of
gastric cancer.
Clinicopathological characteristics and
DNA damage response proteins in gastric cancers
The clinicopathological characteristics of 42 cases
of gastric cancer are summarized in Table VI. The age of the patients varied
from 53 to 86 years. In total, 35 cases were male, and 7 cases were
female. Histological subtypes were evaluated by Lauren
classification, and the number of cases of intestinal, diffuse and
mixed types were 20 (48%), 12 (29%) and 10 (23%) cases,
respectively (Table VI).
 | Table VIClinicopathological characteristics
and immunohistochemical results of the cases of gastric cancer. |
Table VI
Clinicopathological characteristics
and immunohistochemical results of the cases of gastric cancer.
| No | Age/sex | Lauren | pT | pN | Stage | KU70 | pKU70 | CHK1 | pCHK1 | CHK2 | pCHK2 | Rec | Prognosis |
|---|
| 1 | 53/M | Intestinal | 1a | 0 | IA | + | + | + | + | + | + | | |
| 2 | 55/M | Intestinal | 3 | 1 | IIB | + | + | + | + | + | + | | |
| 3 | 56/M | Intestinal | 1b1 | 0 | IIA | + | + | + | + | + | + | | |
| 4 | 59/M | Diffuse | 4a | 0 | IIB | + | + | + | + | + | + | | |
| 5 | 62/M | Intestinal | 4a | 3b | IIIB | + | + | + | + | + | + | +, 3 mo | DOD, 18 mo |
| 6 | 62/M | Intestinal | 4a | 2 | IIIB | + | + | + | + | + | + | | |
| 7 | 63/M | Intestinal | 3 | 2 | IIIA | + | + | + | + | + | + | +, 10 mo | DOD, 33 mo |
| 8 | 64/M | Diffuse | 3 | 3b | IIIC | + | + | + | + | + | + | | |
| 9 | 65/M | Mixed | 4a | 1 | IIIA | + | + | + | + | + | + | | |
| 10 | 65/M | Intestinal | 1b2 | 1 | IB | + | + | + | + | + | + | | |
| 11 | 65/M | Intestinal | 1a | 0 | IA | + | + | + | + | + | + | | |
| 12 | 66/M | Diffuse | 2 | 0 | IB | + | + | + | + | + | + | | |
| 13 | 68/M | Mixed | 1b2 | 0 | IA | + | + | + | + | + | + | | |
| 14 | 68/M | Intestinal | 1a | 0 | IA | + | + | + | + | + | + | | |
| 15 | 70/M | Mixed | 2 | 3a | IIIA | + | + | + | + | + | + | | |
| 16 | 70/M | Mixed | 4a | 3a | IIIC | + | + | + | + | + | + | | |
| 17 | 70/M | Intestinal | 3 | 1 | IIB | + | + | + | + | + | + | | |
| 18 | 70/M | Mixed | 4a | 0 | IIB | + | + | + | + | + | + | | |
| 19 | 70/M | Intestinal | 2 | 1 | IIA | + | + | + | + | + | + | | |
| 20 | 70/M | Intestinal | 2 | 0 | IB | + | + | + | + | + | + | | |
| 21 | 70/F | Intestinal | 1a | 0 | IA | + | + | + | + | + | + | | |
| 22 | 71/M | Intestinal | 1b1 | 0 | IA | + | + | + | + | + | + | | |
| 23 | 71/M | Diffuse | 4a | 3b | IIIC | + | + | + | + | − | − | +, 4 mo | DOD, 5 mo |
| 24 | 73/F | Mixed | 3 | 0 | IIA | + | + | + | + | + | − | | |
| 25 | 74/M | Mixed | 3 | 2 | IIB | + | + | + | + | + | − | | |
| 26 | 74/M | Intestinal | 2 | 0 | IB | + | + | + | + | + | + | | |
| 27 | 75/M | Diffuse | 2 | 1 | IIB | + | + | + | + | + | + | | |
| 28 | 75/F | Diffuse | 3 | 3a | IIIB | + | + | + | + | + | + | | |
| 29 | 76/M | Intestinal | 1b1 | 0 | IA | + | + | + | + | + | + | | |
| 30 | 76/M | Diffuse | 1b2 | 0 | IA | + | + | + | + | + | + | | |
| 31 | 76/M | Diffuse | 2 | 0 | IB | + | + | + | + | + | + | | |
| 32 | 76/F | Intestinal | 2 | 0 | IB | + | + | + | + | + | + | | |
| 33 | 77/M | Mixed | 4a | 3a | IIIC | + | + | + | + | − | − | | DOOD |
| 34 | 78/M | Diffuse | 1a | 0 | IA | + | − | + | + | + | + | | |
| 35 | 81/M | Intestinal | 1b2 | 0 | IA | + | + | + | + | + | + | | DOOD |
| 36 | 81/M | Diffuse | 3 | 1 | IIB | + | + | + | + | + | + | | |
| 37 | 81/M | Diffuse | 1b2 | 2 | IIA | + | + | + | + | + | + | | |
| 38 | 82/F | Mixed | 1b2 | 0 | IA | + | + | + | + | + | + | | |
| 39 | 83/F | Mixed | 3 | 2 | IIIB | + | + | + | + | + | − | +, 6 mo | DOD, 12 mo |
| 40 | 85/M | Intestinal | 1b1 | 0 | IA | + | + | + | + | + | + | | |
| 41 | 86/M | Diffuse | 2 | 1 | IIA | + | + | + | + | + | − | | |
| 42 | 86/F | Intestinal | 1a | 0 | IA | + | + | + | + | + | + | | |
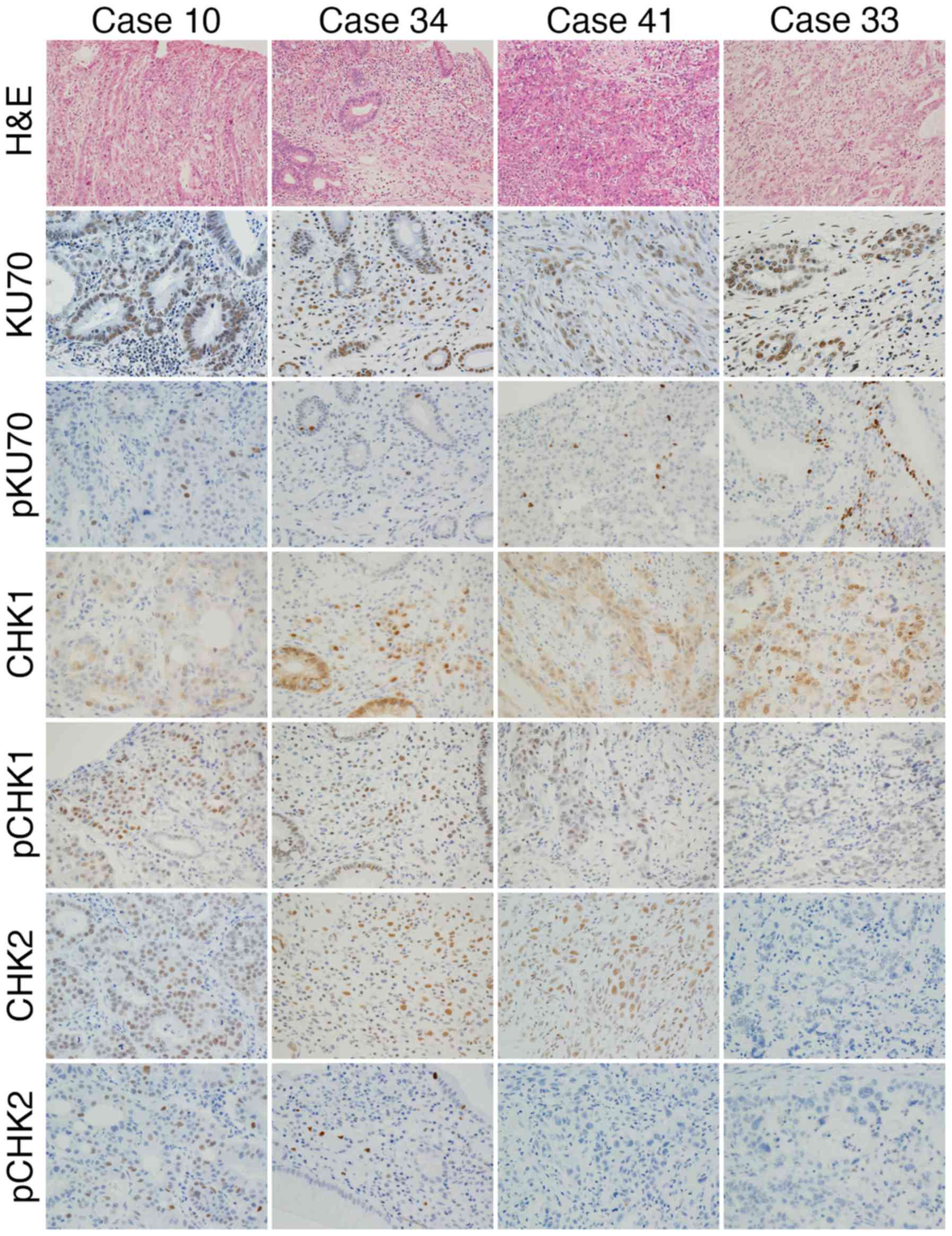
Immunohistochemistry was performed using antibodies
against KU70, pKU70, CHK1, pCHK1, CHK2 and pCHK2 (Fig. 4). The expression of KU70 was found
in all cases, however, the phosphorylation was not apparent in one
case (case 34) (Fig. 4 and
Table VI). The expression of CHK1
and protein phosphorylation was observed in all cases. In 4 cases
(cases 24, 25, 39 and 41) (Fig. 4
and Table VI), the expression of
CHK2 was found in tumor cells; however, the positive reaction of
pCHK2 was not apparent in tumor cells. The expression of CHK2 was
not found in 2 cases (cases 23 and 33) (Fig. 4 and Table VI), and pCHK2 was also absent. The
expression and phosphorylation of proteins of the DDR pathway
appeared to be impaired in 7 (cases 23, 24, 25, 33, 34, 39 and 41)
out of the 42 cases (17%) (Table
VI).
Recurrence was noted in 2 out of 7 cases (29%), in
which expression and phosphorylation were impaired (cases 23 and
39) (Table VI). One patient
succumbed to the disease. On the other hand, out of remaining 35
cases, in which the DDR pathway was intact, recurrence was noted in
2 cases (6%, cases 5 and 7) (Table
VI).
DNA damage response proteins and
apoptotic cell death in gastric cancer cell lines
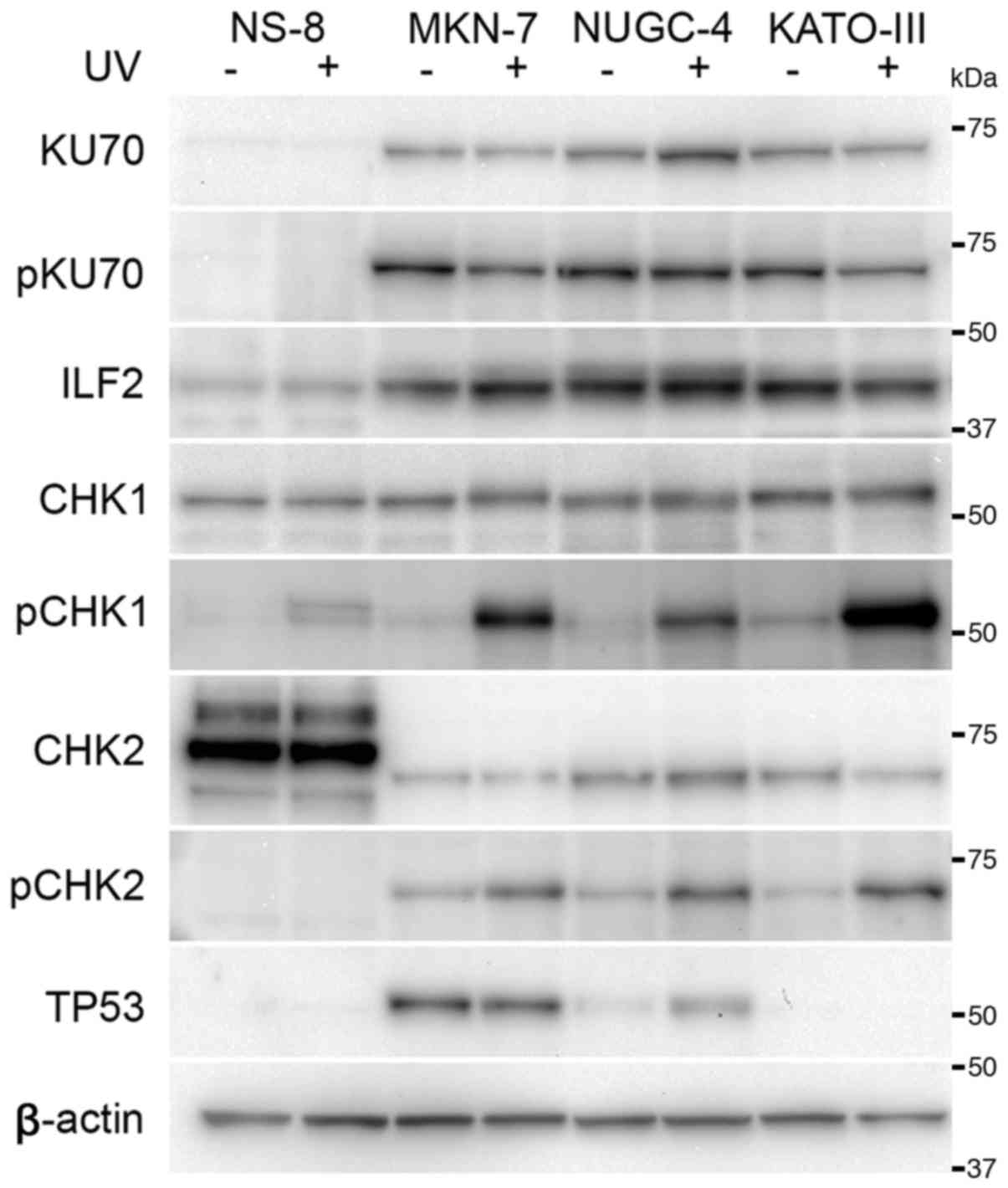
KU70 was expressed in all the cell lines apart from
the NS-8 cells (Fig. 5). The
differences in the expression levels of KU70 were not evident among
the other cells. The expression of ILF2 was observed in all cell
lines; however, the expression appeared to be decreased in the NS-8
cells. The expression level of CHK1 appeared similar among the
cultured cells. CHK2 was overexpressed in the NS-8 cell; however,
its molecular size differed from that observed in the other cell
lines. The expression of TP53 was not detected in the NS-8 and
KATO-III cells. TP53 was overexpressed in the MKN-7 cells, in which
the TP53 gene is mutated (15).
At 2 h following exposure to UV radiation, the
expression level of KU70 appeared to be unaltered, and its
phosphorylation was not evident after 2 h (Fig. 5). The phosphorylation of CHK1 was
increased following exposure to UV radiation in cell lines;
however, this increase was not evident in the NS-8 cells. The
expression level of CHK2 remained unaltered following exposure to
UV radiation, and CHK2 was phosphorylated in the cultured cells
apart from the NS-8 cells. Overexpressed TP53 in the MKN-7 remained
unaltered following exposure to UV radiation. In the NUGC-4 cells,
the expression of TP53 was upregulated at 2 h following exposure to
UV radiation.
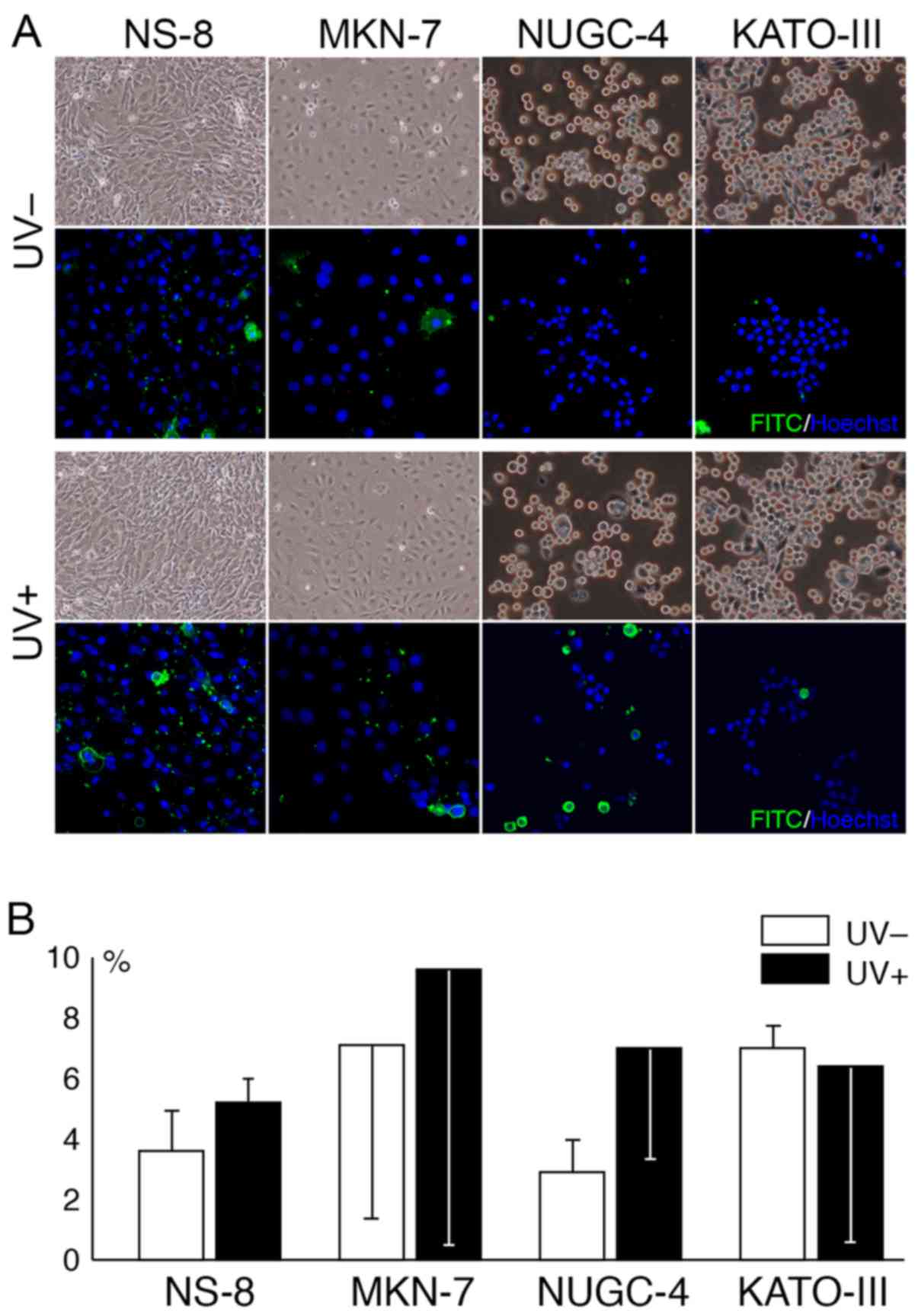
Morphologically, some cultured cells appeared to be
swollen at 2 h following exposure to UV radiation (Fig. 6A). The amount of apoptotic cells
was increased following exposure to UV radiation in all cell lines
apart from the KATO-III cells. The increase in apoptotic cell death
following exposure to UV radiation appeared to be pronounced in the
NUGC-4 cells, although the difference did not reach statistical
significance (Fig. 6B).
Discussion
The present study revealed the biological and
clinical significance of DDR in gastric cancer. The comprehensive
protein profiling and bioinformatics analyses suggested the
possible involvement of DDR in gastric cancer. The expression and
phosphorylation of the proteins of DDR were verified and examined
in 42 cases of gastric cancer. Among these cases, the expression
and phosphorylation of proteins of DDR appeared abnormal in 7 out
of 42 cases (17%). Although the number of cases was limited, the
recurrence in the cases with an impaired expression and the
phosphorylation of DDR proteins was more frequent than that in
cases with a preserved expression and phosphorylation of the
proteins. In the cultured gastric cancer cells, the effective
induction of apoptotic cell death following exposure to UV
radiation was observed in one cell line with a preserved expression
and phosphorylation of DDR proteins.
The comprehensive profiling of molecules in gastric
cancers has been reported in the literature (5,6,16).
It was indicated that gastric cancers are categorized into 4
subtypes, CpG island methylator phenotype, hypermutated,
genomically stable and chromatin instability (6). However, definitive molecular
alterations, which are involved in pathogenesis and progression of
gastric cancer, have not been identified. In the present study, for
the identification of molecules or pathway commonly involved in the
pathobiology of gastric cancer, the proteins that were expressed in
all 17 cases gastric cancer were analyzed. The number of these
proteins constituted 9% (483/5,338) of the total proteins
identified by LC-MS/MS, and the proteins with a subtle expression,
as well as proteins expressed specifically in certain subtypes of
gastric cancer may have been excluded from the analyses. This
analytical strategy may pronounce the alterations of proteins
commonly expressed in gastric cancer.
DDR has been implicated in the tumorigenesis and
progression of human cancers (17–19).
The impairment of DDR causes the genetic instability of the cell.
DDR may serve not only as a barrier to neoplastic cells, but also
as a barrier of the progression of neoplastic cells to highly
malignant cells (17). The
biological significance of DDR in gastric cancer has not yet been
fully elucidated. It was previously reported that the expression
level of CHK2 was increased in cases with a mutation of the TP53
gene. The expression level of CHK1 was also increased in gastric
cancers; however, there was no significant correlation between the
expression level of CHK1 and the mutational state of TP53 (20). It has also been demonstrated that
the loss of ATM and CHK2 is 17.3 and 12.2% in gastric cancers
(21). The percentage of the
absence of the expression and/or phosphorylation of CHK2 in gastric
cancer in the present study was in agreement with these findings,
and its expression and/or phosphorylation were not noted in 6 cases
out of the 42 cases (14.3%). Furthermore, the phosphorylation of
KU70 appeared to be impaired in one case by immunostaining in the
present study.
The impairment of DDR may affect the prognosis of
gastric cancer. It has been reported that patients with a positive
expression of ATM, CHK2 and TP53 present with a favorable
prognosis, and the loss of CHK2 is associated with the reduction of
ATM, and/or TP53 is an independent factor for a poor prognosis
(21). In the present study,
recurrence was noted in 2 out of 7 cases (29%), in which the
expression and phosphorylation of DDR-related proteins were
impaired, whereas recurrence was noted in 2 out of 35 cases (6%),
in which the expression and phosphorylation DDR were intact. The
impairment of DDR appeared to be associated with an unfavorable
prognosis. The follow-up duration, and the number of cases was
limited in the present study. Further studies are warranted in
order to clarify the prognostic significance of DDR.
The aberrant expression of DDR proteins was noted in
cultured gastric cancer cell lines. The expression of KU70, ILF2
and CHK2 was aberrant in one cell line, the NS-8 cells, which has
characteristic features of the amplification of the N-ras
gene and the production of α-fetoprotein (22). The consecutive alteration of the
expression and phosphorylation of DDR protein following exposure to
UV radiation were further examined, and the phosphorylation of CHK1
and CHK2 was decreased in NS-8. The upregulation of TP53 following
exposure to UV radiation was noted in only one cell line, NUGC-4,
and the increase in apoptotic cell death was evident only in this
cell line. It is thus considered that the preservation of DDR and
the TP53 response are necessary for the determination of cell fate
and the induction of apoptosis. This may in part account for the
favorable outcome in patients with gastric cancer with a preserved
expression of DDR proteins (21).
DDR may affect the susceptibility to chemotherapy in
gastric cancer. The expression of XRCC1 is increased in
cisplatin-resistant cancer cells (23). In cases of gastric cancer, the
elevated expression of γH2AX and pATM is adverse for
progression-free and overall survival (24). In addition, the targeting of CHK2
enhances cell death by treatment with cisplatin and paclitaxel in
cultured cell lines (25). It thus
appears that the expression of DDR proteins attenuates the
susceptibility to chemotherapy, although this may be affected by
other genetic factors, such as ARID1A (24). The association of DDR with the
efficacy of chemotherapy needs to be elucidated.
Glossary
Abbreviations
Abbreviations:
|
DDR
|
DNA damage response
|
|
ILF2
|
interleukin binding factor 2
|
|
hnRNPs
|
heterogeneous nuclear
ribonucleoproteins
|
|
FFPE
|
formalin-fixed paraffin-embedded
|
|
IPA
|
Ingenuity Pathway Analysis
|
|
LC-MS/MS
|
liquid chromatography-tandem mass
spectrometry
|
|
NSAF
|
normalized spectral abundance
factor
|
|
OPLS-DA
|
orthogonal partial least squares
discriminant analysis
|
|
PBS
|
phosphate-buffered saline
|
|
TBS
|
Tris-buffered saline
|
|
TUNEL
|
terminal deoxynucleotidyl
transferase-mediated dUTP nick-end labeling
|
|
UV
|
ultraviolet
|
|
XRCC
|
X-ray repair cross complementing
|
Acknowledgments
The authors would like to thank Ms. Kiyoko Kawahara,
Mr. Takenori Fujii, Mr. Kiyoshi Teduka, Ms. Yoko Kawamoto and Ms.
Taeko Kitamura for their assistance with this manuscript.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Kodera Y: The current state of stomach
cancer surgery in the world. Jpn J Clin Oncol. 46:1062–1071. 2016.
View Article : Google Scholar
|
|
2
|
Shitara K: Chemotherapy for advanced
gastric cancer: Future perspective in Japan. Gastric Cancer.
20(Suppl 1): 102–110. 2017. View Article : Google Scholar
|
|
3
|
Eke I, Makinde AY, Aryankalayil MJ, Ahmed
MM and Coleman CN: Comprehensive molecular tumor profiling in
radiation oncology: How it could be used for precision medicine.
Cancer Lett. 382:118–126. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pan L, Aguilar HA, Wang L, Iliuk A and Tao
WA: Three-dimensionally functionalized reverse phase glycoprotein
array for cancer biomarker discovery and validation. J Am Chem Soc.
138:15311–15314. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Boussioutas A, Li H, Liu J, Waring P, Lade
S, Holloway AJ, Taupin D, Gorringe K, Haviv I, Desmond PV, et al:
Distinctive patterns of gene expression in premalignant gastric
mucosa and gastric cancer. Cancer Res. 63:2569–2577.
2003.PubMed/NCBI
|
|
6
|
Bass AJ, Thorsson V, Shmulevich I,
Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C,
Shen H, et al Cancer Genome Atlas Research Network: Comprehensive
molecular characterization of gastric adenocarcinoma. Nature.
513:202–209. 2014. View Article : Google Scholar :
|
|
7
|
Sousa JF, Ham AJ, Whitwell C, Nam KT, Lee
HJ, Yang HK, Kim WH, Zhang B, Li M, LaFleur B, et al: Proteomic
profiling of paraffin-embedded samples identifies
metaplasia-specific and early-stage gastric cancer biomarkers. Am J
Pathol. 181:1560–1572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sunakawa Y and Lenz HJ: Molecular
classification of gastric adenocarcinoma: Translating new insights
from the cancer genome atlas research network. Curr Treat Options
Oncol. 16:172015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tan IB, Ivanova T, Lim KH, Ong CW, Deng N,
Lee J, Tan SH, Wu J, Lee MH, Ooi CH, et al: Intrinsic subtypes of
gastric cancer, based on gene expression pattern, predict survival
and respond differently to chemotherapy. Gastroenterology.
141:476–485. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jackson SP and Bartek J: The DNA-damage
response in human biology and disease. Nature. 461:1071–1078. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lauren P: The two histological main types
of gastric carcinoma: Diffuse and so-called intestinal-type
carcinoma. An attempt at a histo-clinical classification. Acta
Pathol Microbiol Scand. 64:31–49. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Blanpain C, Mohrin M, Sotiropoulou PA and
Passegué E: DNA-damage response in tissue-specific and cancer stem
cells. Cell Stem Cell. 8:16–29. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Marchesini M, Ogoti Y, Fiorini E, Aktas
Samur A, Nezi L, D'Anca M, Storti P, Samur MK, Ganan-Gomez I,
Fulciniti MT, et al: ILF2 is a regulator of RNA splicing and DNA
damage response in 1q21-amplified multiple myeloma. Cancer Cell.
32:88–100. e1062017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kai M: Roles of RNA-binding proteins in
DNA damage response. Int J Mol Sci. 17:3102016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yokozaki H: Molecular characteristics of
eight gastric cancer cell lines established in Japan. Pathol Int.
50:767–777. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Kan J, Yuen ST, Shi ST, Chu KM,
Law S, Chan TL, Kan Z, Chan AS, Tsui WY, et al: Exome sequencing
identifies frequent mutation of ARID1A in molecular subtypes of
gastric cancer. Nat Genet. 43:1219–1223. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartkova J, Horejsí Z, Koed K, Krämer A,
Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et
al: DNA damage response as a candidate anti-cancer barrier in early
human tumorigenesis. Nature. 434:864–870. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Halazonetis TD, Gorgoulis VG and Bartek J:
An oncogene-induced DNA damage model for cancer development.
Science. 319:1352–1355. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Poehlmann A and Roessner A: Importance of
DNA damage checkpoints in the pathogenesis of human cancers. Pathol
Res Pract. 206:591–601. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Shigeishi H, Yokozaki H, Oue N, Kuniyasu
H, Kondo T, Ishikawa T and Yasui W: Increased expression of CHK2 in
human gastric carcinomas harboring p53 mutations. Int J Cancer.
99:58–62. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee HE, Han N, Kim MA, Lee HS, Yang HK,
Lee BL and Kim WH: DNA damage response-related proteins in gastric
cancer: ATM, Chk2 and p53 expression and their prognostic value.
Pathobiology. 81:25–35. 2014. View Article : Google Scholar
|
|
22
|
Matsunobu T, Ishiwata T, Yoshino M,
Watanabe M, Kudo M, Matsumoto K, Tokunaga A, Tajiri T and Naito Z:
Expression of keratinocyte growth factor receptor correlates with
expansive growth and early stage of gastric cancer. Int J Oncol.
28:307–314. 2006.PubMed/NCBI
|
|
23
|
Xu W, Wang S, Chen Q, Zhang Y, Ni P, Wu X,
Zhang J, Qiang F, Li A, Røe OD, et al: TXNL1-XRCC1 pathway
regulates cisplatin-induced cell death and contributes to
resistance in human gastric cancer. Cell Death Dis. 5:e10552014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ronchetti L, Melucci E, De Nicola F,
Goeman F, Casini B, Sperati F, Pallocca M, Terrenato I, Pizzuti L,
Vici P, et al: DNA damage repair and survival outcomes in advanced
gastric cancer patients treated with first-line chemotherapy. Int J
Cancer. 140:2587–2595. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Gutiérrez-González A, Belda-Iniesta C,
Bargiela-Iparraguirre J, Dominguez G, García Alfonso P, Perona R
and Sanchez-Perez I: Targeting Chk2 improves gastric cancer
chemotherapy by impairing DNA damage repair. Apoptosis. 18:347–360.
2013. View Article : Google Scholar
|