Introduction
Renal cell carcinoma (RCC) with t(6;11)
translocation is a new subtype of RCC, which was first reported by
Argani et al in 2001 (1)
and was officially recognized in the 2013 International Society of
Urological Pathology Vancouver Classification of Renal Neoplasia
(2). In a previous study, we first
reported a Chinese case of RCC with t(6;11) translocation with a
novel Alpha-TFEB fusion point (3).
To the best of our knowledge, approximately 63 cases have been
described in the literature (1,3–29).
The reported age range is from 3 to 77.6 years (mean age, 33.9
years; median age, 34 years), predominantly occurring in young
adults with a slight male predominance. RCC with t(6;11)
translocation has a broad morphological appearance without a
distinctive gross appearance; thus, it can be easily misdiagnosed
as another type of renal neoplasm. Immunohistochemical and
molecular genetic analyses are essential for the accurate diagnosis
of RCC with t(6;11) translocation.
RCC with t(6;11) translocation has been
characterized by the fusion of the Alpha gene on 11q12 or q13 with
the TFEB gene on 6p21, resulting in the overexpression of TFEB
protein. The Alpha gene, also known as MALAT1, encodes a ~7.5 kb or
~8.5 kb transcript (4,30). There are no indications for RNA
splicing in the Alpha gene and open reading frames are shorter than
55 amino acids within the transcript. It is assumed that the Alpha
gene does not encode a functional protein (4). Therefore, from these data, it is
doubtful whether Alpha gene is a strong promoter that upregulates
TFEB expression in RCC with t(6;11) translocation, and whether the
product of fusion gene Alpha-TFEB promotes cell canceration.
In this study, we examined the role of the Alpha
gene in this rare tumor entity and the function of the fusion gene
Alpha-TFEB product in vitro and in vivo. Our results
revealed that the Alpha gene was a strong promoter. The stable
transfection of Alpha-TFEB into HK-2 and CaKi-2 cells promoted the
expression of Alpha-TFEB mRNA and TFEB protein. Furthermore, the
overexpression of TFEB increased cell proliferation and enhanced
the cell invasive ability, and decreased cell apoptosis in
Alpha-TFEB stably transfected cell lines in vitro. The
results of in vivo experiments revealed that the
overexpression of TFEB promoted tumorigenicity in nude mice, which
indicated that the overexpression of TFEB confers a potent
oncogenic signal and may thus be a novel therapeutic target in RCC
with t(6;11) translocation.
Materials and methods
Promoter prediction and primers
synthesis
As our research group previously reported, the break
point of the Alpha gene was at nucleotide 1810 and fell in the 1205
bp breakpoint cluster region (3).
Based on this, the promoter region prediction for the Alpha gene
(GenBank Accession no. AF203815) was performed using online
promoter prediction software (www.genomatix.de), and 5 pairs of primers with
restrictive sites were designed for different lengths of Alpha. In
addition, the promoter sequences of the normal TFEB gene (named
pTFEB) were searched from the eukaryotic promoter database and the
corresponding primers were designed (Table I). The primers were synthesized by
Invitrogen (Shanghai, China).
 | Table ISequences of primers used for PCR
amplification and plasmid construction. |
Table I
Sequences of primers used for PCR
amplification and plasmid construction.
| Groups | Primer pairs | Primer sequence
(5′→3′) | Genomic position
(5′→3′) | Size of PCR
products (bp) |
|---|
| Alpha1 | F1
(SacI) | GAGCTCGATCAGAGTGGGCCACTGCCA | 1–21 | 1,854 |
| R1
(NcoI) | CCATGGGCAGGGGGAGGCCAGAATGA | 1854–1829 | |
| Alpha2 | F2
(HindIII) | AAGCTTTTGTGAGGTGTTTGATGACC | 396–415 | 1,459 |
| R2
(NcoI) | CCATGGGCAGGGGGAGGCCAGAATGA | 1854–1829 | |
| Alpha3 | F3
(KpnI) | GGTACCGCTAAGGGCAAAATGTACAAACT | 1451–1473 | 404 |
| R3
(NcoI) | CCATGGGCAGGGGGAGGCCAGAATGA | 1854–1829 | |
| Alpha4 | F4
(SacI) | GAGCTCAGTAAAGCCCTGAACTATCA | 278–297 | 256 |
| R4
(NcoI) | CCATGGCAGCTTATGGAACTTGAAT | 533–515 | |
| Alpha5 | F5
(SacI) | GAGCTCGTGATCGAATTCCGGTGATGCGAGT | 617–642 | 139 |
| R5
(NcoI) | CCATGGACTTATCTGCGGTTTCCT | 755–738 | |
| pTFEB | F
(SacI) | GAGCTCGACTCTGGACTTTCTCTAATAATAA | | 600 |
| R
(NcoI) | CCATGGCCTGAGCTTGCTGTCATGTT | | |
Ethics approval and consent
The ethics approval and consent for the use of human
tissue was confirmed by the Ethics Committee of Anhui Medical
University.
DNA extraction and PCR
Genomic DNA, extracted from formalin-fixed,
paraffin-embedded tumor tissue samples obtained at surgery from a
patient (26-year-old male) with RCC with t(6;11) translocation, as
previously described (3), served
as templates for PCR according to the manufacturer's instructions
of Phusion High-Fidelity PCR kit (Thermo Fisher Scientific,
Carlsbad, CA, USA). The PCR conditions were as follows:
Pre-denaturation at 95°C for 5 min, 35 cycles of denaturation at
95°C for 30 sec, annealing at 55°C (Alpha1, Alpha2 and Alpha4),
52°C (Alpha3), 58°C (Alpha5) for 30 sec, extension at 72°C for 2
min (Alpha1), 90 sec (Alpha2), 30 sec (Alpha3, Alpha4, Alpha5),
followed by a final extension at 72°C for 10 min. PCR amplification
for pTFEB was performed with the DNA from one healthy individual as
templates. All PCR products were confirmed by 1.5% agarose gel
electrophoresis and purified using the MiniBEST DNA Extraction kit
(Takara, Dalian, China) according to the manufacturer's
recommendations.
Construction of recombinant reporter
plasmids
The purified PCR products were cloned into pGEM-T
vectors (Promega, Madison, WI, USA) with T4 ligase (Takara) at
16°C. The constructs were then transformed into the freshly
prepared competent cells DH5α (Tiangen Biotech Co., Ltd, Beijing,
China). Following cell culture for 16 h at 37°C,
Ampicillin-resistant bacterial colonies were selected randomly and
amplified. The selected cloning plasmids, named Alpha1-T, Alpha2-T,
Alpha3-T, Alpha4-T, Alpha5-T and pTFEB-T were identified by double
enzyme digestion and sequencing. The collected DNA fragments were
incorporated into pGL3-Enhancer vectors to construct recombinant
reporter plasmids named pGL3-Enhancer-Alpha1, pGL3-Enhancer-Alpha2,
pGL3-Enhancer-Alpha3, pGL3-Enhancer-Alpha4, pGL3-Enhancer-Alpha5
and pGL3-Enhancer-pTFEB.
Luciferase assay
The 293T cell lines were purchased from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China), and
cultured in DMEM (Gibco, Carlsbad, CA, USA) with 10% fetal bovine
serum (FBS) (Gibco, Scoresby, VIC, Australia). The recombinant
plasmids, pGL3-Enhancer-Alpha1, 2, 3, 4, 5, pTFEB, were transfected
into 293T cells with the pRL-TK vector (Promega, Madison) using
Lipofectamine 2000 (Life Technologies, Carlsbad, CA, USA) according
to the manufacturer's instructions. The pGL3-Control, pGL3-Basic
and pRL-TK Vector were used as positive, negative and internal
controls, respectively. Luciferase activity was assessed at 48 h
following transfection using the Bright-Glo™ Luciferase Assay
system (Promega). The ratio of Firefly/Renilla luciferase
was calculated. Each plasmid experiment was replicated with 6
duplication wells.
Overlap PCR for Alpha-TFEB fusion gene
amplification
The previously constructed Alpha1-T and TFEB cDNA
plasmids (Generay Biotech Co., Ltd., Shanghai, China) were used as
templates for overlap PCR cloning. Four primers (Alpha-F, Alpha-R,
TFEB-F and TFEB-R) were designed (Table II). The PCR conditions were as
follows: Alpha gene (1,854 bp): 98°C for 3 min, then 35 cycles of
98°C for 5 sec, 54°C for 30 sec, 72°C for 120 sec, followed by a
final extension at 72°C for 10 min; TFEB gene (915 bp): 98°C for 3
min, then 35 cycles of 98°C for 5 sec, 57°C for 30 sec, 72°C for 55
sec, followed by a final extension at 72°C for 10 min. The PCR
products were confirmed by 1% agarose gel electrophoresis and
purified. The Alpha and TFEB products (100 nmol/ml) were then
arranged in annealing buffer solution (0.01 M Tris-HCl pH 7.5,
0.001 M EDTA, 0.1 M NaCl), water-bathed at 95°C for 10 min, and
cooled down for 1–2 h at room temperature. The amplification of the
Alpha-TFEB fusion gene (2,769 bp) was performed with the primers
Alpha-F and TFEB-R according to the instructions provided with the
Phusion High-Fidelity PCR kit (Thermo Fisher Scientific). The
amplification condition was identical with that of Alpha gene.
 | Table IISequences of primers used for
Alpha-TFEB fusion gene amplification. |
Table II
Sequences of primers used for
Alpha-TFEB fusion gene amplification.
| Primer sequence
(5′→3′) | Genome position
(5′→3′) | Size of PCR
products (bp) |
|---|
| Alpha | F: ATCGATGATCAGAGTGGGCCACTGCCA | 1–21 | 1,854 |
| R:
TTTTAGTAGCTTTTTGATGTGATTTTTAACCAACTTCC | 1854–1829 | |
| TFEB | F:
AAAGCTACTAAAAATGGCGTCACGCATAGGGTT | 1–20 | 915 |
| R: GGATCCTCACAGCACATCGCCCTCCTCCATG | 890–915 | |
Construction of recombinant lentiviral
vector
The purified PCR product of Alpha-TFEB was subcloned
into the pGEM-T vector with T4 ligase and then transformed into
DH5α as described above. The cloning plasmids were screened with
Ampicillin, and plasmid DNA was extracted and sequenced. The
selected cloning plasmids named Alpha-TFEB-T and pLVX-Puro vector
(Clontech, Kusatsu, Japan) were digested with BspDI and
BamHI (New England Biolabs, Ipswich, MA, USA). The digested
products were purified, and the target gene and the linearized
pLVX-Puro vector were then ligated by T4 ligase. The constructed
plasmid, named pLVX-Puro-Alpha-TFEB, was transformed into DH5α
competent cells. The constructed vector was screened and purified
as described above. Double enzyme digestion was performed to
confirm the ligation and the products were observed by 1% agarose
gel electrophoresis.
Lentiviral packaging and titer
determination
The 293FT cells (a kind gift from Professor Jason
Chen, Columbia University, New York, NY, USA) were cultured in DMEM
containing 10% FBS in a 37°C incubator with 5% CO2. The
pLVX-Puro-Alpha-TFEB plasmid and its packaging plasmid containing
4.5 µg psPAX2 and 4.5 µg pMD2.G (Clontech) were
co-transfected into 293FT cells using Lipofectamine 2000 (Life
Technologies). After 72 h, supernatants were collected from these
cells and passed through a 0.45 µm filter (EMD Millipore,
Billerica, MA, USA). Lentivirus titer determination was performed
using the Lenti-X p24 Rapid Titer kit (Clontech), and the viral
titer of this package was 8.0×106 TU/ml. The ultimate
titer was 8.0×108 TU/ml using Lenti-X Concentrator
(Clontech). The concentrated vector was stored at −80°C until
use.
Cell culture and stable transfection
The CaKi-2 (human papillary renal cell carcinoma
cell line) (31) and HK-2 (normal
human renal epithelial cell line) cells were obtained from Vinhaket
Biological Technology Co., Ltd. (Shanghai, China). The cells were
seeded at a density of 1×105 cells/ml on 24-well plates
in complete medium without antibiotics for 14 h, and the culture
medium was then replaced with 0.3 ml fresh medium with 4
µg/ml retronectin (Takara). The HK-2 and CaKi-2 cells were
infected with pLVX-Puro-Alpha-TFEB at a multiplicity of infection
(MOI) of 20 according to the pre-experimental result. The positive
cell clones containing the Alpha-TFEB fusion gene were screened out
by puromycin (Thermo Fisher Scientific) (1.5 µg/ml for the
HK-2 cell line and 2.0 µg/ml for the CaKi-2 cell line) and
named HK-2-Alpha-TFEB and CaKi-2-Alpha-TFEB.
Reverse transcription PCR (RT-PCR)
The 4 groups of cells, HK-2, CaKi-2, HK-2-Alpha-TFEB
and CaKi-2-Alpha-TFEB, were digested and collected. Primers were
designed using Primer 5.0 software (Table III) and synthesized by
Invitrogen. RT-PCR of the Alpha-TFEB fusion gene was performed with
the PrimeScript™ One Step kit (Takara) following the manufacturer's
instructions. β-actin was used as an internal control. The one-step
RT-PCR conditions were 50°C for 30 min, 94°C for 2 min, followed by
35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec, and
a final extension of 72°C for 10 min. The PCR products were
confirmed by 1.5% agarose gel electrophoresis. The experiments were
run 3 times in independent conditions.
 | Table IIISequences of primers used for
Alpha-TFEB mRNA. |
Table III
Sequences of primers used for
Alpha-TFEB mRNA.
| Groups | Primer sequence
(5′→3′) | Size of PCR
products (bp) |
|---|
| Alpha-TFEB | F:
AGAAGATGAGGGTGTTTACG | 407 |
| R:
TTGTTCCCATAGGTCTCG | |
| β-actin | F:
CTCCATCCTGGCCTCGCTGT | 268 |
| R:
GCTGTCACCTTCACCGTTCC | |
Western blot analysis
Total protein was extracted with lysis buffer (pH
7.4, containing 0.1% SDS, 100 mM NaCl, 1% Triton-X 100, 10 mM Tris,
1 mM EDTA, 0.5% sodium deoxycholate, 1 mM PMSF, 60 µg/ml
aprotinin, 10 µg/ml leupeptin, and 1 µg/ml pepstatin)
at 24 h after transfection and protein concentrations were
determined using a BCA protein assay kit (23227; Pierce
Biotechnology, Rockford, IL, USA). The extracted proteins were
subjected to 10% SDS-PAGE and electrotransferred onto PVDF
membranes (Millipore, Molsheim, France). The membranes were then
incubated with primary antibodies including TFEB (1:500, sc-11005,
Santa Cruz Biotechnology, Santa Cruz, CA, USA) and β-actin
(1:1,000, sc-376421, Santa Cruz Biotechnology) overnight at 4°C
followed by incubation with donkey anti-goat IgG-HRP (1:5,000,
sc-2020, Santa Cruz Biotechnology) and goat anti-mouse IgG-HRP
(1:5,000, sc-2005, Santa Cruz Biotechnology) at room temperature
for 1 h, respectively. Immunoreactive proteins were detected using
the SuperSignal West Femto kit (Pierce Biotechnology) and images
were captured using the Tanon-5500 Multi Image System (Tanon
Science & Technology Ltd., Shanghai, China). β-actin was used
as the internal reference.
Immunofluorescence assay
The HK-2, CaKi-2, HK-2-Alpha-TFEB and
CaKi-2-Alpha-TFEB cells were plated at 1×104 cells/ml in
24-well chamber slides. The cells were washed, fixed, permeated and
blocked as previously described (32). The cells were then incubated with
the primary antibody TFEB (1:250, sc-11005, Santa Cruz
Biotechnology) for 1 h at 37°C. After washing 3 times with PBS, the
cells were incubated with the secondary fluorescence antibody
(donkey anti-goat IgG-FITC, 1:1,000, sc-2024, Santa Cruz
Biotechnology) for 30 min at 37°C. The cells were then mounted with
70% glycerol and observed under an inverted fluorescence microscope
(Olympus IX73002; Olympus, Tokyo, Japan).
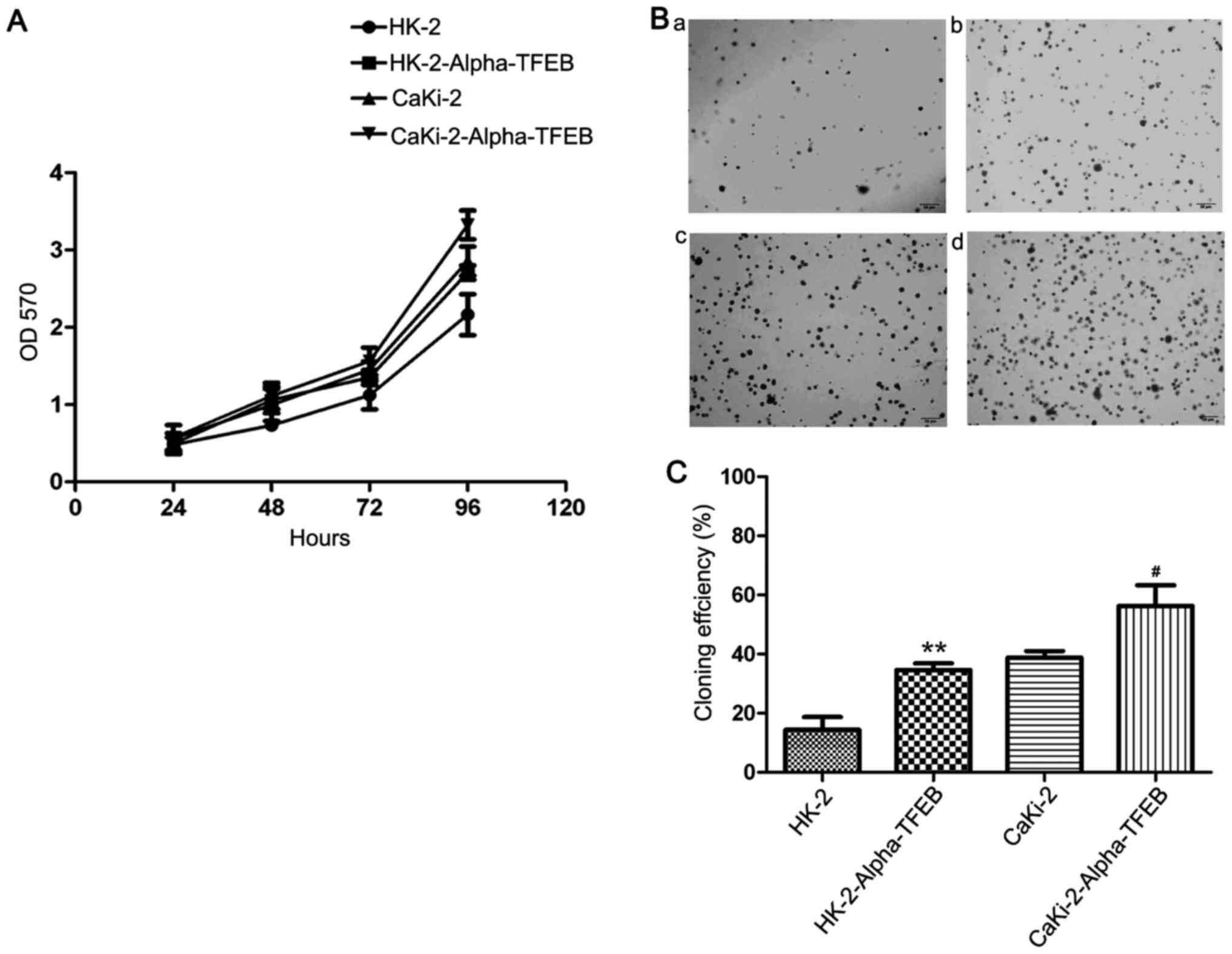
Cell proliferation assay
Cell proliferation was measured by MTT assay. The
cells were plated into 96-well plates (1×103 cells/well)
and each cell line in 8 wells. The blank group contained only
medium without cells. At 24, 48, 72 and 96 h of incubation, 10
µl MTT (5 mg/ml) were added to each well followed by
incubation for an additional 4 h at 37°C. Subsequently, 100
µl DMSO were then added for 10 min to dissolve the formazan
crystals. The absorbance values were measured using a microplate
reader (Bio-Rad, Hercules, CA, USA) at 570 nm. All assays were
repeated 3 times.
Soft agar assay
The cells were plated in 6-well plates and dispersed
by slightly shaking the plates. The cell suspension (0.05 ml) was
mixed with 0.7% agar and 2X DMEM culture medium and plated on top
of 1.2% base agar layer in 60-mm dishes. The mixture was incubated
at 37°C in a 5% CO2 humidified incubator for 10 to 14
days until significant colony formation was observed. The number of
colonies was counted using an inverted microscope (Olympus IX73002;
Olympus). The colony formation efficiency was calculated using the
following formula: Colony forming efficiency (%) = (colonies
formed/cells incubated) ×100%.
Invasion assay
Matrigel Transwell assays were performed to assess
the cell invasive potential (BD Biosciences, Erembodegem, Belgium).
The cells were starved in DMEM without FBS overnight, and the cell
suspension (200 µl) was then loaded to the upper chamber and
DMEM medium containing 10% FBS was added to the lower chamber as a
chemoattractant. After 24 h, the invading cells were methanol-fixed
and stained with 0.1% crystal violet (Sigma, St. Louis, MO, USA).
Five low-magnification areas (×100) were randomly selected and
counted. All assays were performed in triplicate.
Flow cytometric analysis
Cell apoptosis was determined by flow cytometric
analysis using the Annexin V-FITC/PI apoptosis kit (BD Biosciences)
according to the manufacturer's instructions. The HK-2, CaKi-2,
HK-2-Alpha-TFEB and CaKi-2-Alpha-TFEB cells were separately seeded
into 6-well plates and incubated for 24 h at 37°C. The culture
media in 3 wells were removed and the cells were washed twice with
PBS, and then starved in serum-free DMEM. Following starvation for
12, 24 and 48 h, the cells were stained with 10 µl of
Annexin V-FITC and 10 µl of PI, and analyzed using a flow
cytometer (Beckman Coulter, Boulevard Brea, CA, USA).
Establishment of tumor xenografts in nude
mice
The cells were trypsinized and resuspended in DMEM
without FBS. A total of 15 BALB/C-nu/nu 5-week-old female nude mice
with an average weight of 18–20 g were purchased from the National
Rodent Laboratory Animal Resources, Shanghai, China. They were kept
in an environment with a controlled temperature (25±2°C), humidity
(50–70%) with free access to food and water. The mice were randomly
divided into 5 groups of 3 mice in each as follows: The HK-2 group,
CaKi-2 group, HK-2-Alpha-TFEB group, CaKi-2-Alpha-TFEB group and
DMEM medium without FBS group. A total of 100 µl
(1×108) cells were injected into the armpits of the nude
mice. At 4 weeks after the injection, the mice were euthanized, and
the tumors were removed and weighed. Tumor volume was calculated
using the following formula: Tumor volume = 0.5 × length ×
width2. The experiments were approved by the
Institutional Animal Care and Use Committee of Anhui Medical
University (Hefei, China) and were in compliance with all
regulatory guidelines.
Statistical analysis
Data are presented as the means ± standard
deviation. Statistical differences between sample means were
analyzed by an unpaired, two-tailed Student's t-test using SPSS
16.0 software. A two-way ANOVA was used to assess the significance
between different groups. Multiple comparison analysis was
performed using the Tukey method. Significance was set at
P<0.05.
Results
Promoter predictions and PCR
products
According to the results of Alpha gene promoter
predictions with score cut-off value of 0.80 (Fig. 1A), 5 pairs of primers with
restrictive sites were designed for different lengths of Alpha
(Fig. 1B). The Alpha1 and Alpha4
contain the 369–419 promoter prediction region. Alpha1, Alpha2 and
Alpha5 all contain the 643–693 prediction region. Alpha1, Alpha2
and Alpha3 all contain the 1646–1696 prediction region. One pair of
primer for pTFEB was also designed. The size of the PCR products
for Alpha1, 2, 3, 4, 5 was 1,854, 1,459, 404, 256 and 139 bp,
respectively (Fig. 1C). The PCR
product of pTFEB was 600 bp (data not shown).
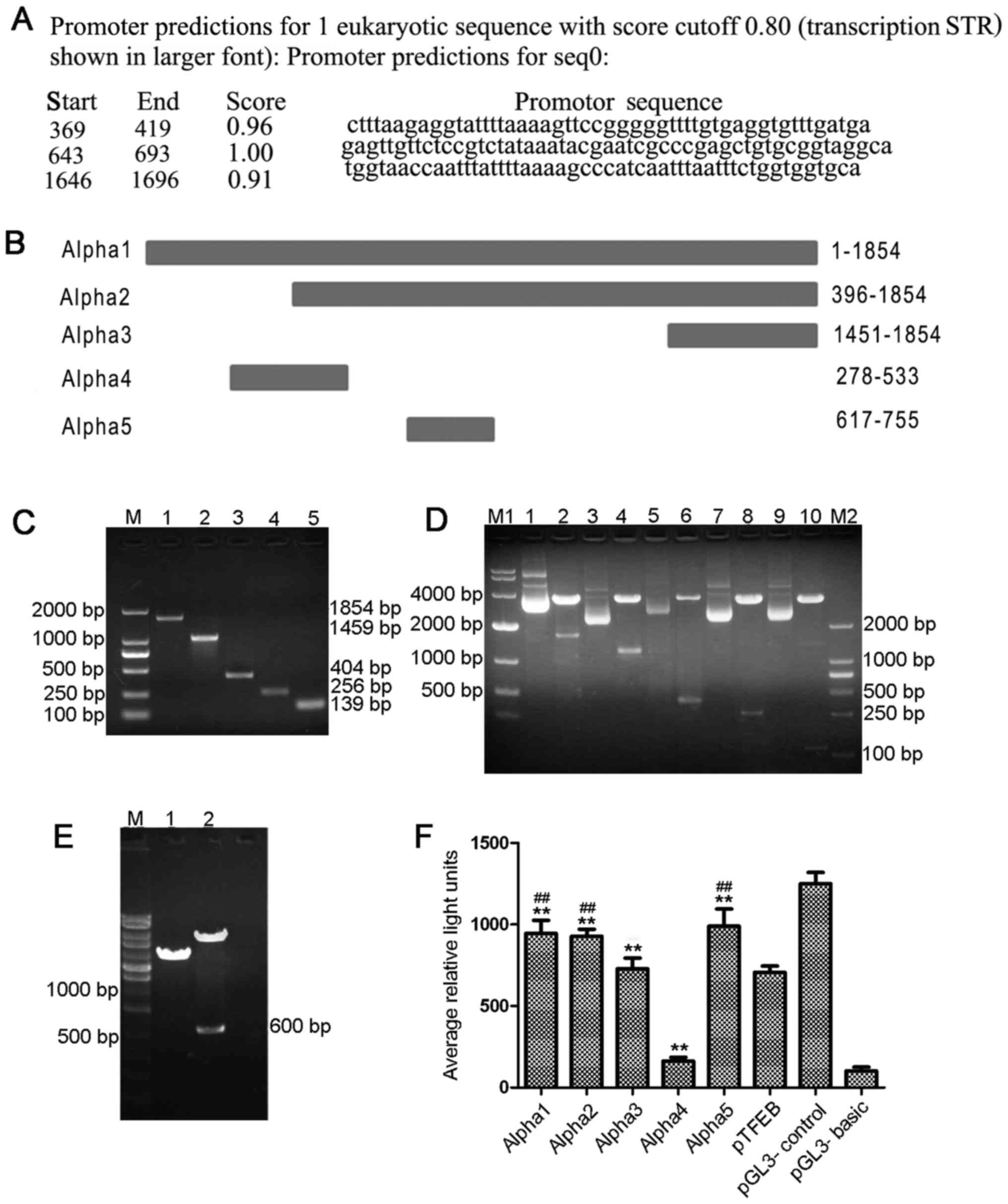 | Figure 1Alpha gene is a strong promoter. (A)
The result of Alpha gene promoter predictions with a score cut-off
value of 0.80. (B) The genetic positions of Alpha1, 2, 3, 4 and 5.
(C) The size of PCR products for Alpha1, 2, 3, 4, 5 were 1,854,
1,459, 404, 256 and 139 bp, respectively. Lane M1, DNA marker 2000;
lane 1, Alpha1; lane 2, Alpha2; lane 3, Alpha3; lane 4, Alpha4;
lane 5, Alpha5. (D) Enzyme identification of recombinant reporter
plasmids containing different Alpha gene segments. Lane M1, DNA
marker 10000; lane 1, pGL3-Enhancer-Alpha1; lane 2, pGL3-Enhancer
and Alpha1 by enzyme cutting; lane 3, pGL3-Enhancer-Alpha2; lane 4,
pGL3-Enhancer and Alpha2 by enzyme cutting; lane 5,
pGL3-Enhancer-Alpha3; lane 6, pGL3-Enhancer and Alpha3 by enzyme
cutting; lane 7, pGL3-Enhancer-Alpha4; lane 8, pGL3-Enhancer and
Alpha4 by enzyme cutting; lane 9, pGL3-Enhancer-Alpha5; lane 10,
pGL3-Enhancer and Alpha5 by enzyme cutting; lane M2, DNA marker
2000. (E) Enzyme identification of recombinant reporter plasmids
containing pTFEB. Lane M, DNA marker 10000; lane 1,
pGL3-Enhancer-pTFEB; lane 2, pGL3-Enhancer and pTFEB by enzyme
digestion. (F) Luciferase activity analysis of Alpha1, Alpha2,
Alpha3, Alpha4, Alpha5 and pTFEB. **P<0.01, compared
with the pGL3-Basic group. ##P<0.01, compared with
the pTFEB group. |
Identification of recombinant reporter
plasmids
Through TA cloning, the cloning plasmids Alpha1-T,
Alpha2-T, Alpha3-T, Alpha4-T, Alpha5-T and pTFEB-T were identified
by double enzyme digestion and confirmed by sequencing. The
recombinant reporter plasmids, pGL3-Enhancer-Alpha1,
pGL3-Enhancer-Alpha2, pGL3-Enhancer-Alpha3, pGL3-Enhancer-Alpha4,
pGL3-Enhancer-Alpha5 and pGL3-Enhancer-pTFEB, were identified by
double enzyme digestion, and target fragments (1,854, 1,459, 404,
256, 139 and 600 bp) were shown by electrophoresis (Fig. 1D and E). The results indicated that
recombinant reporter plasmids were successfully constructed
containing the target segments.
Alpha gene is a strong promoter
Compared with the pGL3-Basic group, the luciferase
activity of the Alpha1, Alpha2, Alpha3, Alpha4 and Alpha5 groups
significantly increased (P<0.01). The luciferase activity was
also markedly increased in the Alpha1, Alpha2 and Alpha5 groups
compared with that of the pTFEB group, and the luciferase activity
of Alpha 5 was the strongest (P<0.01) (Fig. 1F and Table IV). Alpha1, Alpha2 and Alpha5 all
contain the 643–693 base sequence. The experimental data showed
that the Alpha gene in RCC with t(6;11) translocation has a
promoter activity, and the strongest active region is located in
the 643–693 base sequence.
 | Table IVPromoter activity analysis of
different Alpha gene segments. |
Table IV
Promoter activity analysis of
different Alpha gene segments.
| Groups | Luciferase
activity | t-value | P-valuea |
|---|
| pTFEB | 706.61±103.31 | | |
| pGL3-Basic
Vector | 113.00±53.05 | 21.68 | <0.001 |
| pGL3-Control
Vector |
1,206.28±162.96 | 10.99 | <0.001 |
| Alpha1 | 945.89±170.79 | 19.76 | <0.001 |
| Alpha2 | 966.56±117.35 | 28.12 | <0.001 |
| Alpha3 | 752.89±147.22 | 17.35 | 0.283 |
| Alpha4 | 189.94±50.67 | 4.45 | <0.001 |
| Alpha5 |
1,042.28±166.54 | 22.56 | <0.001 |
Successful construction of Alpha-TFEB
lentiviral expression vector
The Alpha-TFEB fusion gene was amplified by overlap
PCR and the expected product of Alpha-TFEB fusion, 2,769 bp, was
detected by 1% agarose gel eletrophoresis (Fig. 2A). Sequencing analysis confirmed
that the inserted Alpha-TFEB fragment in the pGEM-T vector was
consistent with the target sequence (Fig. 2B). The cloning plasmid Alpha-TFEB-T
was identified by double enzyme digestion and shown by 1% agarose
eletrophoresis (Fig. 2C). The
Alpha-TFEB lentiviral expression vector, pLVX-Puro-Alpha-TFEB, was
digested by restriction enzyme and distinguished by eletrophoresis
(Fig. 2D). The data indicated that
the lentiviral expression vector containing Alpha-TFEB fusion was
successfully constructed.
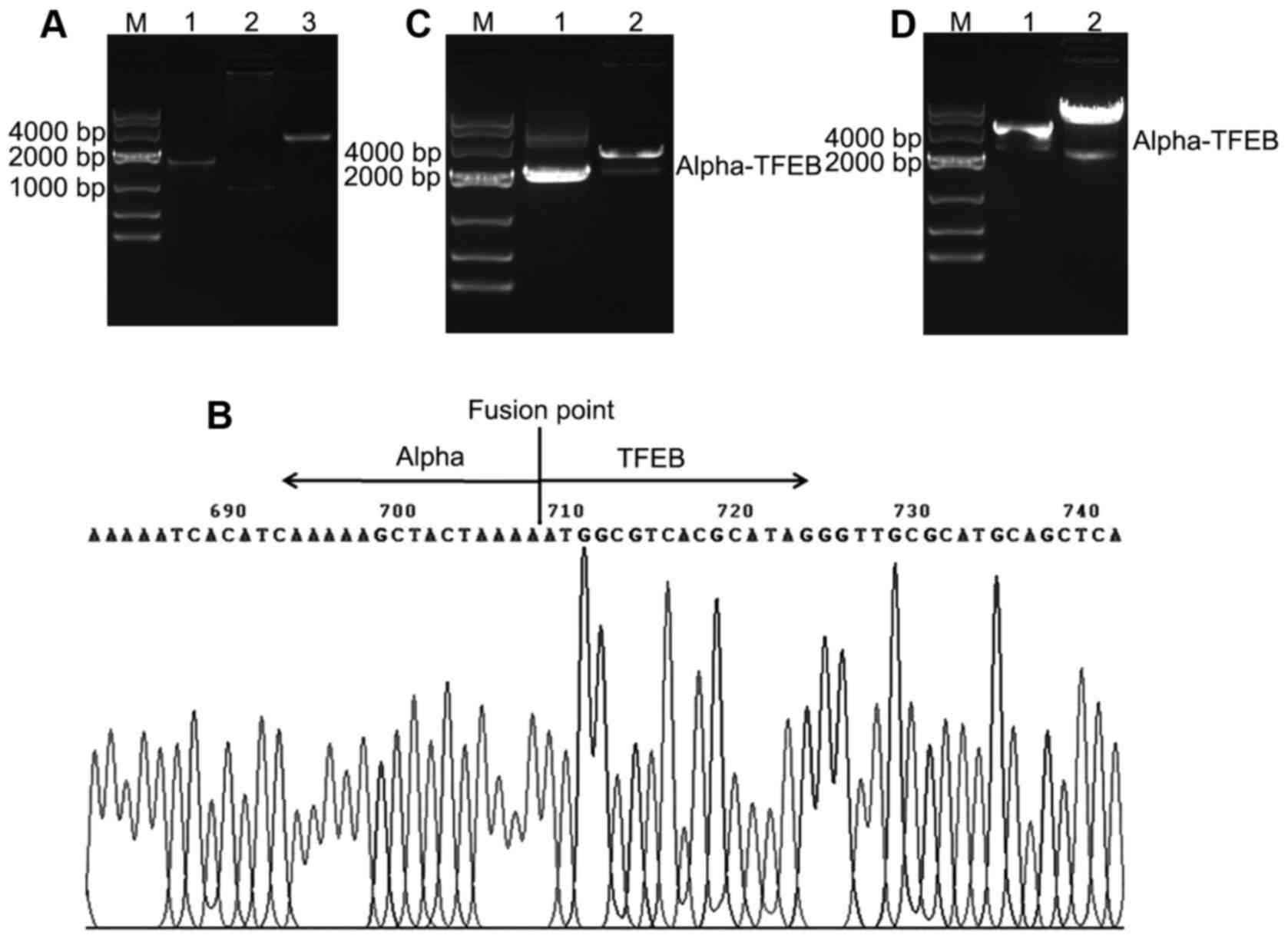 | Figure 2Construction of Alpha-TFEB lentiviral
expression vector. (A) Alpha-TFEB fusion gene was amplified by
overlap PCR. Lane M, DNA marker 10000; lane 1, amplified Alpha gene
(1,854 bp); lane 2, amplified TFEB gene (915 bp); lane 3, amplified
Alpha-TFEB fusion gene (2,769 bp). (B) The cloning plasmids
containing Alpha-TFEB were sequenced. (C) Double enzyme digestion
of cloning plasmid Alpha-TFEB-T. Lane M, DNA marker 10000; lane 1,
Alpha-TFEB-T; lane 2, pGEM-T vector and Alpha-TFEB by enzyme
cutting. (D) Enzyme identification of recombinant lentiviral vector
pLVX-Puro-Alpha-TFEB. Lane M, DNA marker 10000; lane 1,
pLVX-Puro-Alpha-TFEB; lane 2, pLVX-Puro vector and Alpha-TFEB by
enzyme cutting |
Stable transfection of Alpha-TFEB
upregulates the expression of Alpha-TFEB mRNA and TFEB protein
The results of RT-PCR revealed that the expression
of Alpha-TFEB mRNA could be detected in the HK-2-Alpha-TFEB and
CaKi-2-Alpha-TFEB (407 bp) cells (lanes 1 and 2), while there was
no mRNA expression of the target gene in untransfected cells HK-2
and CaKi-2 (Fig. 3A; lanes 3 and
4). The RT-PCR product of β-actin (268 bp) is shown in Fig. 3B. Sequencing analysis for the
RT-PCR product of Alpha-TFEB mRNA confirmed that it was consistent
with the target sequence. The results of western blot analysis
revealed that the relative expression levels of TFEB protein (53
kDa) were significantly higher in the HK-2-Alpha-TFEB and
CaKi-2-Alpha-TFEB groups as compared to the HK-2 and CaKi-2 groups
(Fig. 3C). Immunofluorescence
assay revealed that green fluorescence was also evident in the
HK-2-Alpha-TFEB and CaKi-2-Alpha-TFEB cells, while there was very
little fluorescence observed in the HK-2 and CaKi-2 cells (Fig. 3D).
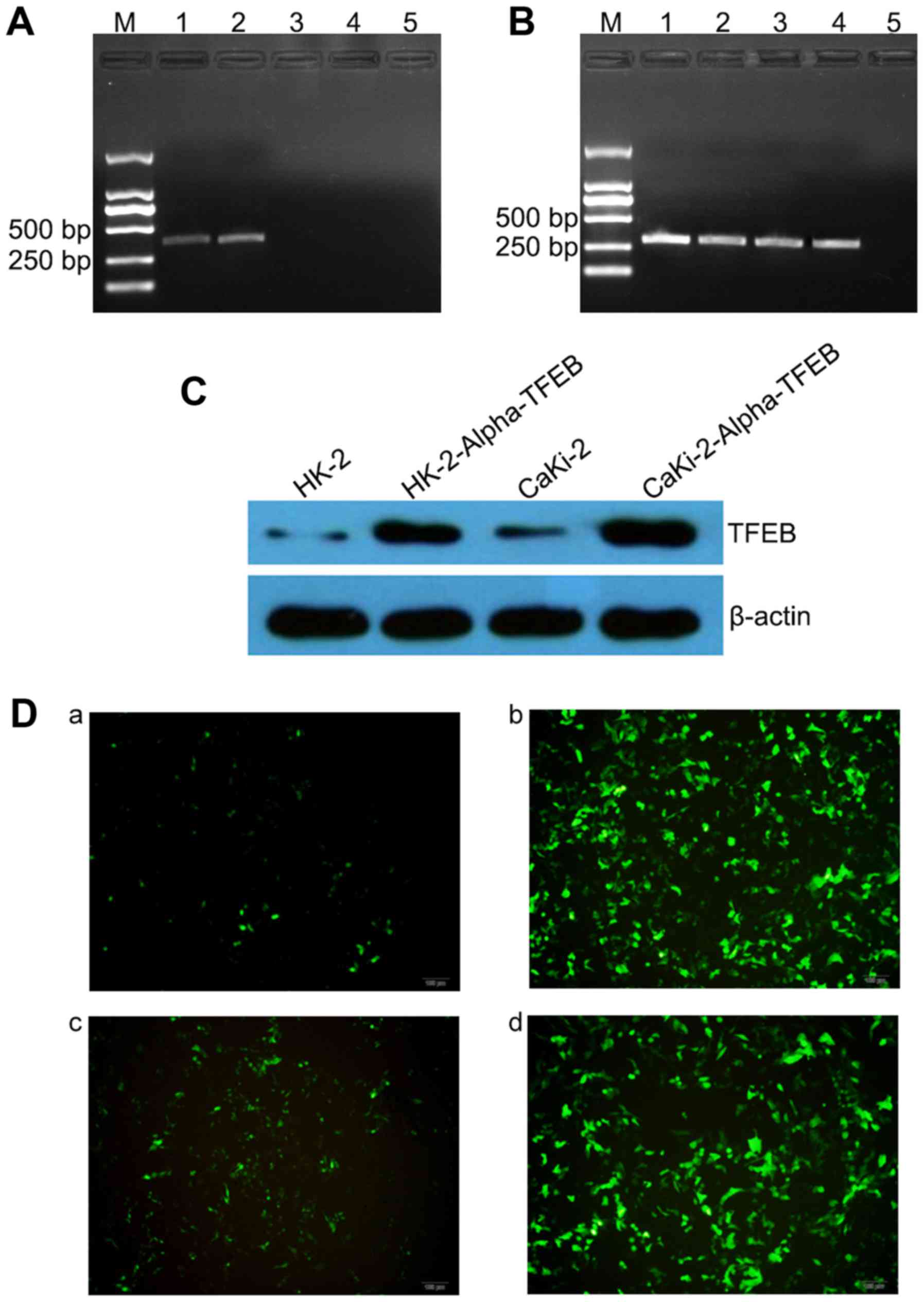 | Figure 3Overexpression of Alpha-TFEB mRNA and
TFEB protein in stably transfected Alpha-TFEB cells. (A) Expression
of Alpha-TFEB mRNA was analyzed by RT-PCR (407 bp). Lane M, DNA
marker 2000; lane 1, HK-2-Alpha-TFEB cells; lane 2,
CaKi-2-Alpha-TFEB cells; lane 3, HK-2 cells; lane 4, CaKi-2 cells;
lane 5, blank control (without templates). (B) β-actin (268 bp) was
used as an internal control. Lane M, DNA marker 2000; lane 1,
HK-2-Alpha-TFEB cells; lane 2, CaKi-2-Alpha-TFEB cells; lane 3,
HK-2 cells; lane 4, CaKi-2 cells; lane 5, blank control (without
templates). (C) TFEB protein levels were examined by western blot
analysis. β-actin was used as an internal control. (D) TFEB protein
levels were analyzed by indirect immunofluorescence assay. Panel a,
HK-2 cells; panel b, HK-2-Alpha-TFEB cells; panel c, CaKi-2 cells;
panel d, CaKi-2-Alpha-TFEB cells. |
Overexpression of TFEB promotes cell
proliferation, colony formation and invasion, and suppresses cell
apoptosis
The stable transfection of Alpha-TFEB upregulated
the expression of Alpha-TFEB mRNA and TFEB protein in the
HK-2-Alpha-TFEB and CaKi-2-Alpha-TFEB cells. To assess the function
of stably transfected cells overexpressing TFEB, an MTT assay, soft
agar assay, Matrigel Transwell assay, and flow cytometric analysis
were performed. The results of MTT assay revealed that the
proliferation rate of the HK-2-Alpha-TFEB group following
transfection at 24, 48, 72 and 96 h (0.4893±0.1135, 1.0505±0.1103,
1.3591±0.1490 and 2.7889±0.1670, respectively) was markedly higher
than that of the HK-2 group (0.4885±0.0602, 0.7163±0.0620,
1.1188±0.1512 and 2.2284±0.2819, respectively). Similarly, the
proliferation rate of the CaKi-2-Alpha-TFEB group (0.5718±0.0954,
1.1873±0.1231, 1.5718±0.1499, 3.6584±0.1597, respectively) was
significantly higher than that of the CaKi-2 group (0.5513±0.0654,
0.9993±0.1223, 1.4022±0.1574, 3.0461±0.1969, respectively)
(Fig. 4A). These effects were
time-dependent. A similar result was observed in the soft agar
assay: There was a significant difference in the colony-forming
efficiency between the HK-2-Alpha-TFEB group and HK-2 group
(t=14.20, P=0.0049), and between the CaKi-2-Alpha-TFEB group and
the Caki-2 group (t=6.145, P=0.0255) (Fig. 4B and C). Matrigel Transwell assay
also demonstrated that the overexpression of TFEB significantly
increased the migratory and invasive capacity of the HK-2 and
CaKi-2 cells (Fig. 5A and B). Flow
cytometric analysis indicated that the apoptotic ratio of the
HK-2-Alpha-TFEB group following transfection at 12, 24, and 48 h
(4.80±1.15, 27.03±1.10 and 46.43±1.97%, respectively) was
significantly lower than that of the HK-2 group (18.87±0.35,
37.67±1.91 and 64.17±2.22%, respectively). Likewise, the apoptotic
ratio of the CaKi-2-Alpha-TFEB group (4.37±1.11, 23.60±1.85 and
41.00±0.61%, respectively) was significantly lower than that of the
CaKi-2 group (9.77±0.55, 26.77±2.41 and 45.20±2.61%, respectively)
(Fig. 5C). These data indicated
that the over-expression of TFEB promoted cell proliferation,
colony formation and invasion, and inhibited cell apoptosis.
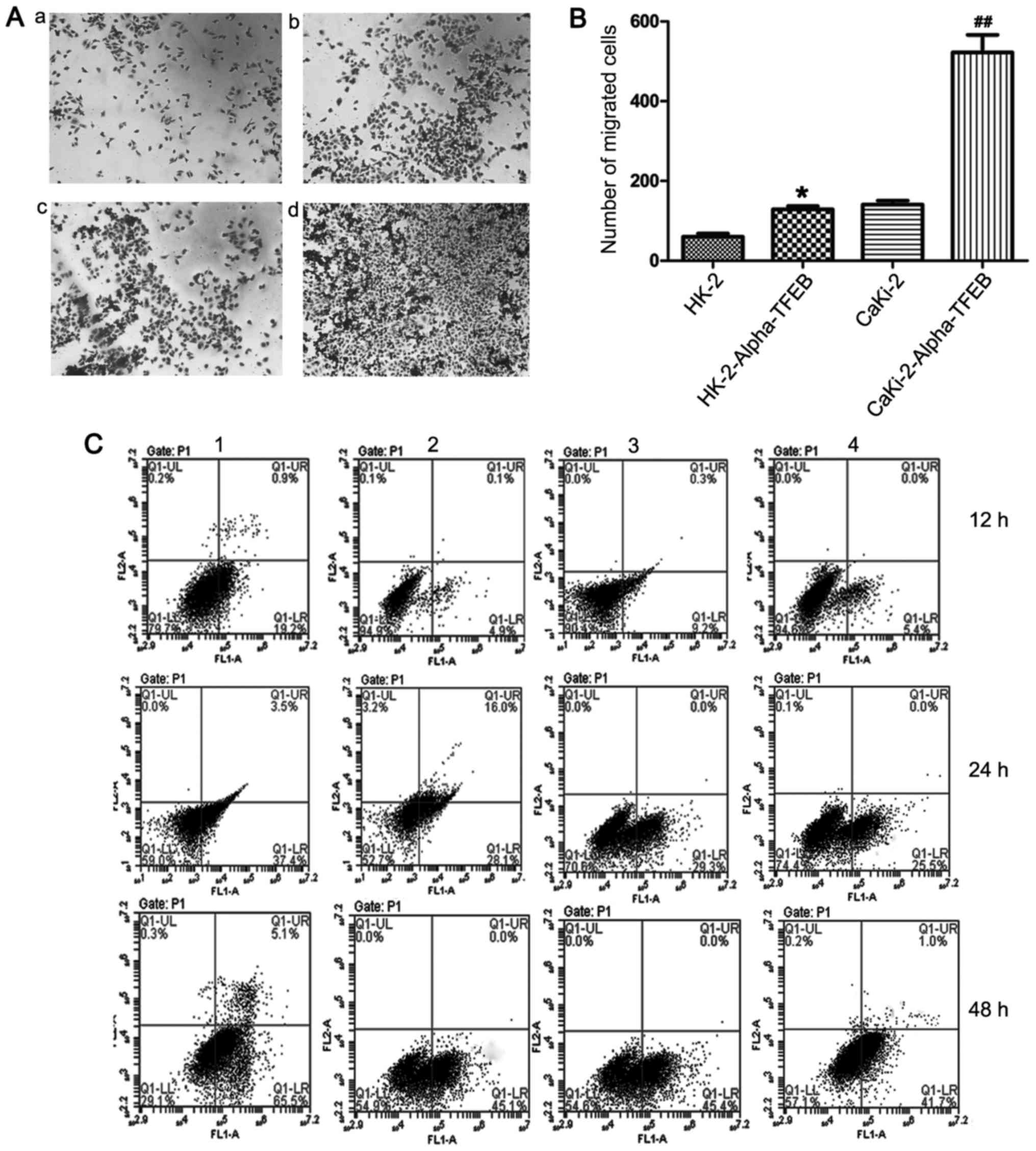 | Figure 5Overexpression of TFEB promotes the
cell migration and suppresses the cell apoptosis. (A and B) The
migration and invasion capacity was detected by Matrigel Transwell
assay. Panel a, HK-2 cells; panel b, HK-2-Alpha-TFEB cells; panel
c, CaKi-2 cells; panel d, CaKi-2-Alpha-TFEB cells.
*P<0.05, compared with the HK-2 group.
##P<0.01, compared with the CaKi-2 group. (C) The
apoptotic ratio of stably transfected and untransfected cells was
analyzed by flow cytometry. Upper left quadrant, necrotic cells;
bottom left quadrant, live cells; upper right quadrant, late
apoptotic cells; lower right quadrant, early apoptotic cells.
Panels 1, HK-2 cells; panels 2, HK-2-Alpha-TFEB cells; panels 3,
CaKi-2 cells; panels 4, CaKi-2-Alpha-TFEB cells. |
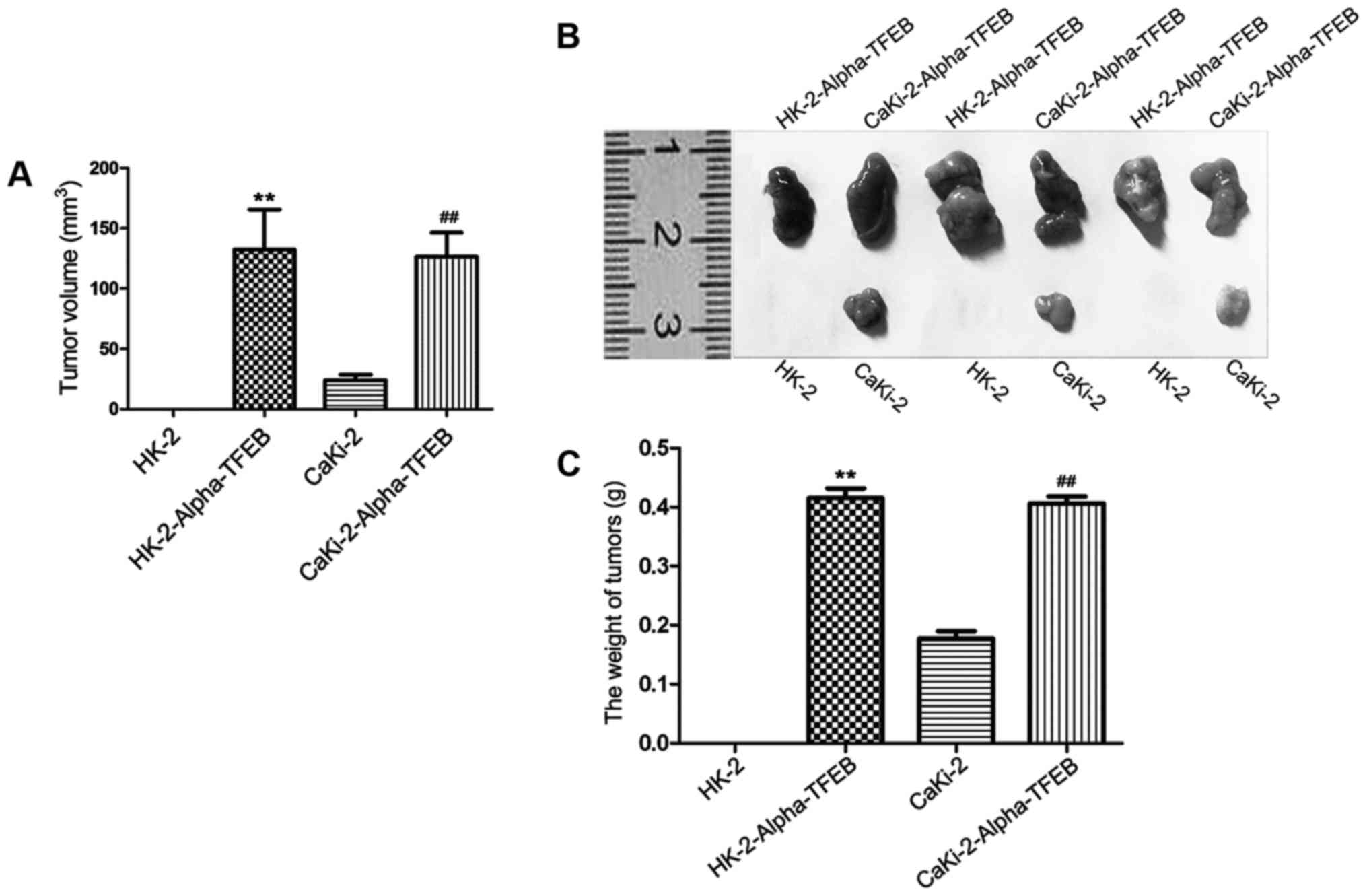
Overexpression of TFEB promotes
tumorigenicity in nude mice
To examine the effects on tumorigenesis of the
overexpression of TFEB in nude mice, in vivo tumor formation
assay was carried out. The results of the in vivo
experiments revealed that a tumor mass could be observed in the
CaKi-2 group, HK-2-Alpha-TFEB group and CaKi-2-Alpha-TFEB group,
while no mass was observed in the HK-2 group and DMEM medium
without FBS group. In the HK-2-Alpha-TFEB group and
CaKi-2-Alpha-TFEB group, a significant increase in tumor volume and
weight, compared to the HK-2 group and CaKi-2 group was observed
(Fig. 6A and B). The maximum
diameter of the largest tumor was 11.0 mm and the maximum tumor
volume was 198.0 mm3. The weight of the mice upon
sacrifice was 24.87±0.74, 21.17±1.27, 20.27±1.11, 19.90±1.31 and
25.33±0.85 g in the HK-2 group, CaKi-2 group, HK-2-Alpha-TFEB
group, CaKi-2-Alpha-TFEB group and DMEM medium without FBS group,
respectively (data not shown). No mice developed multiple tumors.
The average tumor weight in the HK-2-Alpha-TFEB group (0.42±0.07 g)
was significantly higher than that in the HK-2 group (0.00±0.00 g)
(P<0.01). The average tumor weight in the CaKi-2-Alpha-TFEB
group (0.41±0.03 g) was also higher than that in the CaKi-2 group
(0.17±0.02 g; P<0.01) (Fig.
6C).
Discussion
RCCs constitute multiple heterogeneous cancer types
that account for approximately 90% of all adult renal malignancies.
The most common types are clear cell, papillary and chromophobe,
each with a different histology and clinical course (18). RCC with translocation is a distinct
subtype of RCCs harboring recurrent gene rearrangements (33). The majority of RCCs with
translocation are with the Xp11.2 translocation which usually
occurs in children and young adults, resulting in various types of
gene fusions involving the TFE3 gene located on chromosome Xp11.2
(33,34). Another less common translocation is
the t(6;11) translocation in RCC, involving the fusion between the
Alpha gene and TFEB gene (1). The
majority of cases of RCC with t(6;11) translocation seem to be
indolent (3,11,14),
although recurrence occurs in 17% of patients (15) and some adult patients present with
metastasis or succumb to the disease (9,13,15).
It has been almost 2 decades since the first case of RCC with
t(6;11) translocation reported in 2001 (1). However, its molecular biology remains
greatly uncharacterized and effective targeted therapy has yet to
be identified (35).
The translocation of t(6;11) (p21;q12 or q13) in RCC
causes the fusion of the Alpha gene with the TFEB gene (6). To date, all Alpha-TFEB fusions were
shown to fall in the 1,205 bp breakpoint cluster region of the
Alpha gene, and in the 289 bp breakpoint cluster region numbered
from the 5′ end of exon3 of TFEB gene (35). We have previously detected a novel
fusion point of Alpha-TFEB which the breakpoint was at nucleotide
1810 in the Alpha and −94 in the TFEB (3). Argani et al (6) demonstrated that DNA-PCR and RT-PCR
products of the Alpha-TFEB fusion gene were identical due to the
lack of splice signals in an intronless gene Alpha. The fusion of
Alpha-TFEB results in the overexpression of a full-length wild-type
TFEB protein as opposed to a chimeric protein (4). To date, no TFEB fusion partners other
than Alpha have been detected in RCC with t(6;11) translocation
(35). Therefore, from these data,
it is doubtful as to whether the Alpha gene is a strong promoter
that upregulates TFEB expression. In this study, in order to
clarify this issue, luciferase assay for 5 different lengths of
Alpha gene and normal TFEB gene was performed, and our results
suggested that the Alpha gene has a strong promoter activity and
the strongest promoter activity region is located in the 643–693
base sequence.
Based on the above-mentioned results, we constructed
an Alpha-TFEB lentiviral expression vector and transfected this
vector into HK-2 cells and CaKi-2 cells. Following the successful
transfection of Alpha-TFEB, the expression of Alpha-TFEB mRNA could
be detected in the HK-2-Alpha-TFEB and CaKi-2-Alpha-TFEB cells,
while there was no mRNA expression of the target gene in
untransfected HK-2 and CaKi-2 cells. This result was identical to
that obtained from the tissue test of RCC with t(6;11)
translocation and normal kidney (6). Kuiper et al (4) reported that Alpha-TFEB fusion
transcripts were expressed in t(6;11)(p21;q13)-positive cells, but
not in control human genomic DNA by northern blotting, and
Alpha-TFEB mRNA was detected in tissue from RCC with t(6;11)
translocation, but not in normal kidney tissue by RT-PCR analysis.
Real-time RT-RCR analysis, reported by Kuiper et al
(4), revealed that the Alpha-TFEB
mRNA levels were up to 60-fold upregulated in RCC with t(6;11)
translocation cells as compared to wild-type TFEB mRNA levels in
normal kidney samples. A native TFEB protein, but not a chimeric
protein, was overexpressed in RCC with t(6;11) translocation.
Argani et al (6) detected
strong nuclear TFEB labeling in all 7 cases of RCC with t(6;11)
translocation, whereas 1,089 other unrelated neoplasms and normal
tissues did not show nuclear TFEB labeling. We have previously
reported a high Alpha-TFEB DNA level and strong nuclear TFEB
labeling in tissue from RCC with t(6;11) translocation (3). In this study, the results of western
blot analysis revealed that the expression levels of TFEB protein
were significantly higher in the positively transfected cells as
compared to the untransfected cells, which suggested that the
fusion of Alpha-TFEB led to a high expression of TFEB protein. The
role of Alpha may not be specific in RCC. However, the fusion of
Alpha-TFEB is specific to RCC with t(6;11) translocation.
The TFEB gene is encoded by 51,083 bp on chromosome
6p21.1 and produces a 2,364 bp mRNA transcript consisting of a 302
bp 5′ untranslated region followed by a start codon in exon 3, and
a stop codon in exon 10 followed by a 621 bp 3′ untranslated region
(35). TFEB mRNA encodes a 476 AA
protein with a 54 AA basic region and helix-loop-helix domain
including a putative nuclear localization signal (35). TFEB is one of 4 members of the
microphthalmia transcription factor (MiT) family, which also
includes TFE3, TFEC and MiTF (33). RCCs with Xp11.2 and t(6;11) are now
grouped as MiT family translocation RCC in the light of clinical,
morphologic and molecular mimics. MiT members share similar
DNA-binding and activation domains. Their functions are diverse,
and are related to cell growth and differentiation (36). The oncogenic activity of MiT
members has been reported in clear cell sarcoma (37). Giatromanolaki et al
(38) demonstrated that the
overexpression of TFEB was associated with autophagy, the migratory
phenotype and a poor prognosis in non-small cell lung cancer. TFEB
may promote the growth of pancreatic ductal adenocarcinoma by
autophagy regulation (39). The
role of TFEB in RCC with t(6;11) translocation has yet to be
determined.
No cell line has yet to be derived from patients
with RCC with t(6;11) translocation (35). Lentiviral vectors are useful
experimental instruments for stable gene delivery. In this study,
we constructed an Alpha-TFEB lentiviral expression vector.
Following the successful transfection of Alpha-TFEB into HK-2 cells
and CaKi-2 cells, the expression levels of Alpha-TFEB products were
significantly higher in positively transfected cells as compared to
untransfected cells. We also explored the role of Alpha-TFEB
product by in vitro experiments. An MTT assay, soft agar
assay, Matrigel Transwell assay, and flow cytometric analysis were
performed. Our results revealed that the overexpression of TFEB
promoted cell proliferation, colony formation and invasion, and
inhibited cell apoptosis. Furthermore, to validate the
effectiveness of the overexpression of TFEB, an in vivo
tumor formation assay was performed. The results of the in
vivo experiments revealed that a tumor mass was observed in the
CaKi-2 group, HK-2-Alpha-TFEB group and CaKi-2-Alpha-TFEB group,
but not in the HK-2 group and culture medium group. The
HK-2-Alpha-TFEB group and CaKi-2-Alpha-TFEB group had a
significantly increased tumor volume and weight, compared to the
HK-2 group and CaKi-2 group. These findings were consistent with
those of our in vitro experiments, which indicated that the
significant transcriptional and translational overexpression of
TFEB may ultimately drive renal tumorigenesis by controlling cell
growth, differentiation and metabolism. Chronic myeloid leukemia
(CML) is driven by BCR-ABL fusion and BCR-ABL inhibitors are
effective therapies in CML (40).
The anaplastic lymphoma kinase (ALK) inhibitor, crizotinib, was
first approved by the US Food and Drug Administration for the
treatment of ALK-rearranged non-small cell lung cancer in 2011
(41). Whether TFEB inhibitor is a
potential target for the therapy of patients with RCC with t(6;11)
translocation warrants further investigation.
In conclusion, Alpha is a strong promoter and the
strongest promoter activity region is located in the 643-693 base
sequence. The stable transfection of Alpha-TFEB into HK-2 cells and
CaKi-2 cells promoted the expression of Alpha-TFEB mRNA and TFEB
protein. Furthermore, the overexpression of TFEB increased cell
proliferation and invasion, and decreased cell apoptosis in cells
stably transfected with Alpha-TFEB expression vector in
vitro. The in vivo experiments using nude mice indicated
that the overexpression of TFEB promoted tumorigenicity in nude
mice, suggesting that TFEB may function as a cancer oncogene. These
data suggest that the effects of Alpha-TFEB are specific in this
type of tumor, which results in the overexpression of a native TFEB
protein and then promotes cell canceration. The overexpression of
TFEB confers a potent oncogenic signal and may be a novel
therapeutic target in RCC with t(6;11) translocation.
Acknowledgments
This study was supported by the National Natural
Science Foundation of China (Grant no. 81202006), the Anhui
Provincial Natural Science Foundation, China (Grant no.
1208085MH175) and Grants for Scientific Research of BSKY (no.
XJ201101) from Anhui Medical University.
References
|
1
|
Argani P, Hawkins A, Griffin CA, Goldstein
JD, Haas M, Beckwith JB, Mankinen CB and Perlman EJ: A distinctive
pediatric renal neoplasm characterized by epithelioid morphology,
basement membrane production, focal HMB45 immunoreactivity, and
t(6;11)(p21.1;q12) chromosome translocation. Am J Pathol.
158:2089–2096. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Srigley JR, Delahunt B, Eble JN, Egevad L,
Epstein JI, Grignon D, Hes O, Moch H, Montironi R, Tickoo SK, et
al: ISUP Renal Tumor Panel: The International Society of Urological
Pathology (ISUP) Vancouver Classification of Renal Neoplasia. Am J
Surg Pathol. 37:1469–1489. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhan HQ, Wang CF, Zhu XZ and Xu XL: Renal
cell carcinoma with t(6;11) translocation: A patient case with a
novel Alpha-TFEB fusion point. J Clin Oncol. 28:e709–e713. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kuiper RP, Schepens M, Thijssen J, van
Asseldonk M, van den Berg E, Bridge J, Schuuring E, Schoenmakers EF
and van Kessel AG: Upregulation of the transcription factor TFEB in
t(6;11)(p21;;q13)-positive renal cell carcinomas due to promoter
substitution. Hum Mol Genet. 12:1661–1669. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Davis IJ, Hsi BL, Arroyo JD, Vargas SO,
Yeh YA, Motyckova G, Valencia P, Perez-Atayde AR, Argani P, Ladanyi
M, et al: Cloning of an Alpha-TFEB fusion in renal tumors harboring
the t(6;11)(p21;q13) chromosome translocation. Proc Natl Acad Sci
USA. 100:6051–6056. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Argani P, Laé M, Hutchinson B, Reuter VE,
Collins MH, Perentesis J, Tomaszewski JE, Brooks JS, Acs G, Bridge
JA, et al: Renal carcinomas with the t(6;11)(p21;;q12):
Clinicopathologic features and demonstration of the specific
alpha-TFEB gene fusion by immunohistochemistry, RT-PCR, and DNA
PCR. Am J Surg Pathol. 29:230–240. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Argani P, Laé M, Ballard ET, Amin M,
Manivel C, Hutchinson B, Reuter VE and Ladanyi M: Translocation
carcinomas of the kidney after chemotherapy in childhood. J Clin
Oncol. 24:1529–1534. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pecciarini L, Cangi MG, Lo Cunsolo C,
Macri' E, Dal Cin E, Martignoni G and Doglioni C: Characterization
of t(6;11) (p21;q12) in a renal-cell carcinoma of an adult patient.
Genes Chromosomes Cancer. 46:419–426. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Camparo P, Vasiliu V, Molinie V, Couturier
J, Dykema KJ, Petillo D, Furge KA, Comperat EM, Lae M, Bouvier R,
et al: Renal translocation carcinomas: Clinicopathologic,
immunohistochemical, and gene expression profiling analysis of 31
cases with a review of the literature. Am J Surg Pathol.
32:656–670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Geller JI, Argani P, Adeniran A, Hampton
E, De Marzo A, Hicks J and Collins MH: Translocation renal cell
carcinoma: Lack of negative impact due to lymph node spread.
Cancer. 112:1607–1616. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hora M, Hes O, Urge T, Eret V, Klecka J
and Michal M: A distinctive translocation carcinoma of the kidney
['rosette-like forming,' t(6;11), HMB45-positive renal tumor]. Int
Urol Nephrol. 41:553–557. 2009. View Article : Google Scholar
|
|
12
|
Suárez-Vilela D, Izquierdo-García F,
Méndez-Álvarez JR, Miguélez-García E and Domínguez-Iglesias F:
Renal translocation carcinoma with expression of TFEB: Presentation
of a case with distinctive histological and immunohistochemical
features. Int J Surg Pathol. 19:506–509. 2011. View Article : Google Scholar
|
|
13
|
Ishihara A, Yamashita Y, Takamori H and
Kuroda N: Renal carcinoma with (6;11)(p21;q12) translocation:
Report of an adult case. Pathol Int. 61:539–545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Petersson F, Vaněček T, Michal M,
Martignoni G, Brunelli M, Halbhuber Z, Spagnolo D, Kuroda N, Yang
X, Cabrero IA, et al: A distinctive translocation carcinoma of the
kidney; 'rosette forming,' t(6;11), HMB45-positive renal tumor: A
histomorphologic, immunohistochemical, ultrastructural, and
molecular genetic study of 4 cases. Hum Pathol. 43:726–736. 2012.
View Article : Google Scholar
|
|
15
|
Inamura K, Fujiwara M, Togashi Y, Nomura
K, Mukai H, Fujii Y, Yamamoto S, Yonese J, Fukui I and Ishikawa Y:
Diverse fusion patterns and heterogeneous clinicopathologic
features of renal cell carcinoma with t(6;11) translocation. Am J
Surg Pathol. 36:35–42. 2012. View Article : Google Scholar
|
|
16
|
Argani P, Yonescu R, Morsberger L, Morris
K, Netto GJ, Smith N, Gonzalez N, Illei PB, Ladanyi M and Griffin
CA: Molecular confirmation of t(6;11)(p21;q12) renal cell carcinoma
in archival paraffin-embedded material using a break-apart TFEB
FISH assay expands its clinicopathologic spectrum. Am J Surg
Pathol. 36:1516–1526. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rao Q, Liu B, Cheng L, Zhu Y, Shi QL, Wu
B, Jiang SJ, Wang Y, Wang X, Yu B, et al: Renal cell carcinomas
with t(6;11)(p21;q12): A clinicopathologic study emphasizing
unusual morphology, novel alpha-TFEB gene fusion point,
immunobiomarkers, and ultrastructural features, as well as
detection of the gene fusion by fluorescence in situ hybridization.
Am J Surg Pathol. 36:1327–1338. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhong M, De Angelo P, Osborne L,
Paniz-Mondolfi AE, Geller M, Yang Y, Linehan WM, Merino MJ,
Cordon-Cardo C and Cai D: Translocation renal cell carcinomas in
adults: A single-institution experience. Am J Surg Pathol.
36:654–662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bambury RM, Battley JE, McCarthy A, Brady
C, O'Reilly S, Kelly PJ, O'Brien F, Sweeney P, Fleming S, Mayer NJ,
et al: Translocation renal cell carcinomas: An evolving entity and
a member of the microphthalmia transcription factor-associated
family of tumors. Clin Genitourin Cancer. 11:357–361. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rao Q, Zhang XM, Tu P, Xia QY, Shen Q,
Zhou XJ and Shi QL: Renal cell carcinomas with t(6;11)(p21;;q12)
presenting with tubulocystic renal cell carcinoma-like features.
Int J Clin Exp Pathol. 6:1452–1457. 2013.
|
|
21
|
Chaste D, Vian E, Verhoest G and Blanchet
P: Translocation renal cell carcinoma t(6;11)(p21;;q12) and sickle
cell anemia: First report and review of the literature. Korean J
Urol. 55:145–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Smith NE, Illei PB, Allaf M, Gonzalez N,
Morris K, Hicks J, Demarzo A, Reuter VE, Amin MB, Epstein JI, et
al: t(6;11) renal cell carcinoma (RCC): Expanded
immunohistochemical profile emphasizing novel RCC markers and
report of 10 new genetically confirmed cases. Am J Surg Pathol.
38:604–614. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hora M, Urge T, Trávníček I, Ferda J,
Chudáček Z, Vaněček T, Michal M, Petersson F, Kuroda N and Hes O:
MiT translocation renal cell carcinomas: Two subgroups of tumours
with translocations involving 6p21 [t (6; 11)] and Xp11.2 [t (X;1
or X or 17)]. Springerplus. 3:2452014. View Article : Google Scholar
|
|
24
|
Peckova K, Vanecek T, Martinek P, Spagnolo
D, Kuroda N, Brunelli M, Vranic S, Djuricic S, Rotterova P, Daum O,
et al: Aggressive and nonaggressive translocation t(6;11) renal
cell carcinoma: Comparative study of 6 cases and review of the
literature. Ann Diagn Pathol. 18:351–357. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhao Y, Yao J, Chen N, Zeng H and Zhang W:
Renal cell carcinomas with t(6;11) (p21;q12): Presentation of two
cases with computed tomography findings. Jpn J Radiol. 33:380–383.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arneja SK and Gujar N: Renal cell
carcinoma with t(6:11) (p21;q12). A case report highlighting
distinctive immunohistologic features of this rare tumor. Int J
Surg Case Rep. 7C:16–19. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lilleby W, Vlatkovic L, Meza-Zepeda LA,
Revheim ME and Hovig E: Translocational renal cell carcinoma
(t(6;11)(p21;q12) with transcription factor EB (TFEB) amplification
and an integrated precision approach: A case report. J Med Case
Reports. 9:2812015. View Article : Google Scholar
|
|
28
|
Xia Q, Shi S, Shen Q, Wei X, Wang X, Ma H,
Lu Z, Zhou X and Rao Q: Renal cell carcinoma with
t(6;11)(p21;2;q13)/MALAT1-TFEB fusion: A clinical and pathological
analysis. Zhonghua Bing Li Xue Za Zhi. 44:895–899. 2015.In
Chinese.
|
|
29
|
Williamson SR, Eble JN and Palanisamy N:
Sclerosing TFEB-rearrangement renal cell carcinoma: A recurring
histologic pattern. Hum Pathol. 62:175–179. 2017. View Article : Google Scholar
|
|
30
|
Guru SC, Agarwal SK, Manickam P, Olufemi
SE, Crabtree JS, Weisemann JM, Kester MB, Kim YS, Wang Y,
Emmert-Buck MR, et al: A transcript map for the 2.8-Mb region
containing the multiple endocrine neoplasia type 1 locus. Genome
Res. 7:725–735. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brodaczewska KK, Szczylik C, Fiedorowicz
M, Porta C and Czarnecka AM: Choosing the right cell line for renal
cell cancer research. Mol Cancer. 15:832016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lee M, Dworkin AM, Gildea D and Trivedi
NS; NISC Comparative Sequencing Program, Moorhead GB and Crawford
NP: RRP1B is a metastasis modifier that regulates the expression of
alternative mRNA isoforms through interactions with SRSF1.
Oncogene. 33:1818–1827. 2014. View Article : Google Scholar :
|
|
33
|
Magers MJ, Udager AM and Mehra R: MiT
family translocation-associated renal cell carcinoma: A
contemporary update with emphasis on morphologic, immunophenotypic,
and molecular mimics. Arch Pathol Lab Med. 139:1224–1233. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhan HQ, Chen H, Wang CF and Zhu XZ: A
case of PSF-TFE3 gene fusion in Xp11.2 renal cell carcinoma with
melanotic features. Hum Pathol. 46:476–481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kauffman EC, Ricketts CJ, Rais-Bahrami S,
Yang Y, Merino MJ, Bottaro DP, Srinivasan R and Linehan WM:
Molecular genetics and cellular features of TFE3 and TFEB fusion
kidney cancers. Nat Rev Urol. 11:465–475. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Argani P: MiT family translocation renal
cell carcinoma. Semin Diagn Pathol. 32:103–113. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Davis IJ, Kim JJ, Ozsolak F, Widlund HR,
Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT,
Lessnick SL, et al: Oncogenic MITF dysregulation in clear cell
sarcoma: Defining the MiT family of human cancers. Cancer Cell.
9:473–484. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giatromanolaki A, Kalamida D, Sivridis E,
Karagounis IV, Gatter KC, Harris AL and Koukourakis MI: Increased
expression of transcription factor EB (TFEB) is associated with
autophagy, migratory phenotype and poor prognosis in non-small cell
lung cancer. Lung Cancer. 90:98–105. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Klein K, Werner K, Teske C, Schenk M,
Giese T, Weitz J and Welsch T: Role of TFEB-driven autophagy
regulation in pancreatic cancer treatment. Int J Oncol. 49:164–172.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Pinilla-Ibarz J, Sweet K, Emole J and
Fradley M: Long-term BCR-ABL1 tyrosine kinase inhibitor therapy in
chronic myeloid leukemia. Anticancer Res. 35:6355–6364.
2015.PubMed/NCBI
|
|
41
|
Ou SH: Crizotinib: A novel and
first-in-class multitargeted tyrosine kinase inhibitor for the
treatment of anaplastic lymphoma kinase rearranged non-small cell
lung cancer and beyond. Drug Des Devel Ther. 5:471–485. 2011.
View Article : Google Scholar : PubMed/NCBI
|