Introduction
Colorectal cancer (CRC) is one of the most common
types of malignancy and the third leading cause of
cancer-associated mortality worldwide (1). Great progress has been achieved with
regards to CRC therapy, including targeted therapy and immune
therapy; however, the prognosis of patients with advanced CRC
remains poor. Tumor metastasis and progression are the main causes
of cancer-associated mortality in patients with CRC (2); however, the molecular mechanisms
underlying tumor metastasis and progression are complex and
unclear. Therefore, it is of great importance to uncover the
mechanisms underlying CRC progression and metastasis, in order to
develop novel anticancer treatment options.
Sequencing of the human genome revealed that the
vast majority of our genome is transcribed to produce non-coding
RNAs (ncRNAs), including small and long ncRNAs (lncRNAs) (3). lncRNAs are defined as a class of
non-coding RNA transcripts >200 nucleotides long, which have
little or no protein-coding capacity (4). It has previously been reported that
lncRNAs serve important roles in regulating numerous biological
processes, including cell proliferation, cell invasion, cell
differentiation and chromosome inactivation (5). lncRNAs can be classified into various
categories, including transcripts, antisense lncRNAs, long
intergenic ncRNAs and pseudogenes, according to their structure and
function (6,7). They may act as scaffolds, guides,
decoys and tethers of other molecules to regulate the expression of
other genes (4). lncRNAs are
considered novel factors in cancer research, and accumulating
evidence has demonstrated that they may function as oncogenes or
tumor suppressor genes, depending on the circumstance.
The HOX gene family, which was first reported in the
study of homeosis in Drosophila, contains a series of
evolutionarily conserved genes that have important roles in
embryonic development (8). The
lncRNA HOXD cluster antisense RNA 1 (HOXD-AS1), which is located
between the HOXD1 and HOXD3 genes, is encoded by a member of the
same gene family that encodes HOX transcript antisense RNA (HOTAIR)
(9). Similar to HOTAIR, HOXD-AS1
also serves an important role in tumor progression and metastasis
(10,11). A recent study demonstrated that
HOXD-AS1 is upregulated in bladder cancer, and affects the
apoptosis and metastasis of tumor cells (12). However, the role and underlying
molecular mechanisms of HOXD-AS1 in CRC development and metastasis
remain to be elucidated; therefore, the present study aimed to
explore the potential role and molecular mechanisms of HOXD-AS1 in
CRC progression and metastasis.
Materials and methods
Clinical samples
The human CRC tissue and adjacent normal tissue
samples used in the present study were obtained from 136 patients
at the Affiliated 3201 Hospital of Xi'an Jiaotong University
(Hanzhong, China). The present study was approved by the Medical
Ethics Committee of the Institutional Review Board of the
Affiliated 3201 Hospital of Xi'an Jiaotong University. Written
informed consent was obtained from all participants in the present
study. None of the patients received chemotherapy or radiotherapy
prior to surgery. The clinicopathological information of the 136
patients who underwent surgery at the Affiliated 3201 Hospital of
Xi'an Jiaotong University between March 2010 and May 2013,
including sex, age, tumor size, tumor depth, differentiation, T
stage, lymph node invasion and distant metastasis, were
recorded.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA was extracted from tissue and cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA), according to the manufacturer's protocol.
RNA was reverse transcribed into cDNA using Superscript III
transcriptase, according to the manufacturer's protocol
(Invitrogen; Thermo Fisher Scientific, Inc.). qPCR analysis was
used to detect HOXD-AS1 expression, as previously described
(13). qPCR was performed using
the FastStart Universal SYBR-Green Master (#4913914001; Roche
Diagnostics, Basel, Switzerland) on an ABI QuantStudio 6 Flex
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) as
follows: Denaturation at 95°C for 10 min, followed by 40 cycles at
95°C for 5 sec, 60°C for 40 sec and 72°C for 45 sec. GAPDH was used
as an endogenous control to normalize HOXD-AS1 expression levels.
qPCR analysis of microRNA (miRNA/miR)-217 expression was performed
according to a previously described method (14). qPCR analysis for the detection of
Vimentin, N-cadherin, slug, E-cadherin, α-cadherin, β-cadherin,
cluster of differentiation (CD)44, CD133, CD24, CD166,
octamer-binding transcription factor 4 (Oct4), leucine-rich
repeat-containing G-protein coupled receptor 5 (Lgr5), sex
determining region Y-box 2 (Sox2) and Nanog was performed according
to a previously described method (15). All primers for qPCR are listed in
Table I. Relative quantification
of RNA expression was calculated using the 2−ΔΔCq method
(16). Each sample was tested in
triplicate.
 | Table IPrimers used for reverse
transcription-quantitative polymerase chain reaction in the present
study. |
Table I
Primers used for reverse
transcription-quantitative polymerase chain reaction in the present
study.
| Gene | Primer sequences
(5′-3′) |
|---|
| lncRNA | F:
5′-GGCTCTTCCCTAATGTGTGG-3′ |
| HOXD-AS1 | R:
5′-CAGGTCCAGCATGAAACAGA-3′ |
| N-cadherin | F:
5′-GGTGGAGGAGAAGAAGACCAG-3′ |
| R:
5′-GGCATCAGGCTCCACAGTG-3′ |
| Vimentin | F:
5′-GAGAACTTTGCCGTTGAAGC-3′ |
| R:
5′-GCTTCCTGTAGGTGGCAATC-3′ |
| Snail | F:
5′-CCTCCCTGTCAGATGAGGAC-3′ |
| R:
5′-CCAGGCTGAGGTATTCCTTG-3′ |
| Slug | F:
5′-GGGGAGAAGCCTTTTTCTTG-3′ |
| R:
5′-TCCTCATGTTTGTGCAGGAG-3′ |
| E-cadherin | F:
5′-TGCCCAGAAAATGAAAAAGG-3′ |
| R:
5′-GTGTATGTGGCAATGCGTTC-3′ |
| α-catenin | F:
5′-AGCGAATTGTGGCAGAGTGT-3′ |
| R:
5′-GTCTACGCAAGTCCCTGGTC-3′ |
| β-catenin | F:
5′-ACAACTGTTTTGAAAATCCA-3′ |
| R:
5′-CGAGTCATTGCATACTGTCC-3′ |
| CD44 | F:
5′-TTGCAGTCAACAGTCGAAGAAG-3′ |
| R:
5′-CCTTGTTCACCAAATGCACCA-3′ |
| Oct4 | F:
5′-CTTGCTGCAGAAGTGGGTGGAGGAA-3′ |
| R:
5′-CTGCAGTGTGGGTTTCGGGCA-3′ |
| CD133 | F:
5′-TGGATGCAGAACTTGACAACGT-3′ |
| R:
5′-ATACCTGCTACGACAGTCGTGGT-3′ |
| CD24 | F:
5′-TGAAGAACATGTGAGAGGTTTGAC-3′ |
| R:
5′-GAAAACTGAATCTCCATTCCACAA-3′ |
| CD166 | F:
5′-TCCTGCCGTCTGCTCTTCT-3′ |
| R:
5′-TTCTGAGGTACGTCAAGTCGG-3′ |
| Lgr5 | F:
5′-TATCTGGCTCCGAGTGCTTGCC-3′ |
| R:
5′-ATCATAGCCAGAGATGGATACC-3′ |
| Sox2 | F:
5′-GCCGATGTGAAACTTTTGTCG-3′ |
| R:
5′-GGCAGCGTGACTTATCCTTCT-3 |
| Nanog | F:
5′-AATACCTCAGCCTCCAGCAGATG-3′ |
| R:
5′-TGCGTCACACCATTGCTATTCTTC-3′ |
| GAPDH | F:
5′-TGCACCACCAACTGCTTAGC-3′ |
| R:
5′-GGCATGGACTGTGGTCATGAG-3′ |
| U6 | F: 5′-CTCGCTTCGGCA
GCACA-3′ |
| R:
5′-AACGCTTCACGAATT TGCGT-3′ |
| miR-217 | F:
5′-TACTCAACTCACTACTGCATCAGGA-3′ |
| R:
5′-TATGGTTGTTCTGCTCTCTGTGTC-3′ |
Cell culture
CRC cell lines [HCT116, American Type Culture
Collection (ATCC)-CCL-247; SW620, ATCC-CCL-227; SW480,
ATCC-CCL-228; DLD-1, ATCC-CCL-221; HT-29, ATCC-HTB-38; LoVo,
ATCC-CCL-229] and the human normal colonic epithelial cell line
CCD-112CoN (ATCC-CRL-1541) were purchased from the ATCC (Manassas,
VA, USA). Cells were cultured according to the manufacturer's
protocols: HCT116 and HT-29 cells were cultured in McCoy's 5A,
SW620 and SW480 cells were cultured in Leibowitz L-15, LoVo cells
were cultured in F12K, CCD-112CoN cells were cultured in DMEM, and
DLD-1 cells were cultured in RPMI-1640 (all from Gibco; Thermo
Fisher Scientific, Inc.). All of the media were supplemented with
10% fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific,
Inc.).
Plasmid construction and cell
transfection
Full-length fragments of HOXD-AS1 were obtained form
HCT116 cells and subcloned into pcDNA3.1(+) vector (Thermo Fisher
Scientific, Inc.) for HOXD-AS1 overexpression (forward,
5′-GGTCGACGTTTGTGCCGCGCGC-3′ and reverse,
5′-CGCGGCCGCTGACACTTTGAA-3′); the vector was subsequently named the
pcDNA-HOXD-AS1 vector. SW480 cells grown to 70–80% confluence were
infected with pcDNA-HOXD-AS1 vector (150 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. In
addition, HCT116 and LoVo cells were grown to 70–80% confluence and
were transfected with small interfering (si)RNA (100 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) to knockdown HOXD-AS1, according to the
manufacturer's protocol. siRNA targeting HOXD-AS1 and relative
negative control (NC) siRNA (5′-GGCCAGAGC AGCAGGGCCC-3′) were
provided by Shanghai GenePharma Co., Ltd. (Shanghai, China). The
target sequences used in the present study were as follow: siRNA1,
5′-GAAAGAAGGACCAAAGTAA-3′; and siRNA2, 5′-GCACAAAGGAACAAGG AAA-3′.
The expression levels of HOXD-AS1 post-transfection were assessed
by RT-qPCR; 48 h post-transfection, the cells underwent further
experimentation.
Cell proliferation assays
Cell proliferation was determined using the MTT kit
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), according to the
manufacturer's protocol. For the colony formation assay, CRC cells
were trypsinized, and single-cell suspensions containing
3×103 cells were plated into 6-well plates and cultured
with RPMI-1640 supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.). The plates were incubated at 37°C in an
atmosphere containing 5% CO2 for 14 days. Images of the
colonies containing ≥50 cells were then captured and colonies were
counted by ImageJ (V.1.8.0) software [National Institutes of Health
(NIH), Bethesda, MD, USA]. Data are presented as the means ±
standard deviation of five randomly scored fields.
Invasion assays
Cell invasion was assessed by Transwell assay
(Corning, Inc., Corning, NY, USA), according to the manufacturer's
protocol. The membrane inserts were coated with diluted Matrigel
(BD Biosciences, San Jose, CA, USA). Cells (1×105) in
100 µl serum-free medium were added to the upper chamber and
cultured for 24 h, and the lower chamber of each well insert was
filled with 600 µl 20% serum-containing medium. Subsequently, the
membrane inserts were removed and stained with 0.1% crystal violet
for 15 min at room temperature; the number of invasive cells was
counted under an inverted microscope and images were captured.
Sphere-forming assays
A sphere-forming assay was performed according to a
previously published method (15)
with minor modifications. Briefly, cell suspensions
(1.0×103 cells/well) were seeded in 6-well ultralow
attachment plates (Corning, Inc.) in serum-free Dulbecco's modified
Eagle's medium/F12 (Gibco; Thermo Fisher Scientific, Inc.)
containing 20 ng/ml basic fibroblast growth factor, 20 ng/ml
epidermal growth factor (both from Miltenyi Biotec, Inc.) and 2 mM
L-glutamine (Mediatech, Inc., Manassas, VA, USA). After culturing
for 7 days, the size and number of tumor spheres were evaluated
using light microscopy (Olympus Corporation, Tokyo, Japan).
Immunofluorescence analyses
Immunofluorescence analyses were conducted according
to a previously described method (17). The following antibodies were used
in the present study: E-cadherin (#3199S) and N-cadherin (#14215S)
(both from Cell Signaling Technology, Inc., Danvers, MA, USA). A
goat anti-rabbit secondary antibody immunoglobulin (Ig)G (A21537)
and a goat anti-mouse IgG (A21538) (both from EMD Millipore,
Bedford, MA, USA) were used. Sections were then visualized under a
confocal laser microscope (Olympus Corporation).
RNA immunoprecipitation (RIP) assay
RIP was performed using a Magna RIP™ RNA-Binding
Protein Immunoprecipitation kit (EMD Millipore), according to the
manufacturer's protocol. Briefly, cell lysates were incubated with
RIP buffer containing magnetic beads conjugated with negative
control normal mouse IgG or human anti-argonaute 2 (Ago2) antibody.
The samples were then incubated with proteinase K to isolate
immunoprecipitated RNA. Finally, the purified RNA was extracted and
analyzed by qPCR to confirm the presence of the binding
targets.
Lentivirus production and infection
Cells were constructed with a stable knockdown of
HOXD-AS1 according to a previously published study (18). Briefly, short hairpin (sh) RNAs
directed against human HOXD-AS1 (target sequence,
5′-GAAAGAAGGACCAAAGTAA-3′) or scrambled oligo-nucleotides were
ligated into the LV-3 (pGLVH1/GFP+Puro) vector (Shanghai GenePharma
Co., Ltd.). Two hundreds and ninety-three cells were cotransfected
with lentiviral vectors (or the control lentiviral vectors) and
Lenti-Pac HIV Expression Packaging Mix using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h, lentiviral particles in the
supernatant were harvested and filtered by centrifugation at 500 ×
g for 10 min. Cells were then infected with lentivirus or negative
control (NC) lentivirus. To select stably transfected cells, the
cells were treated with puromycin (2 µg/ml) for 2 weeks.
Green fluorescent protein-positive cells were named sh-HOXD-AS1 and
sh-NC, and were used for subsequent assays.
Tumorigenesis and metastasis assays
A total of 40 female BABL/c athymic nude mice (age,
5 weeks; weight, 15.4–17.2 g) were obtained from the Beijing Vital
River Laboratory Animal Technology Co., Ltd. (Beijing, China), and
were maintained in the Xi'an Jiaotong University animal facilities,
approved by the China Association for Accreditation of Laboratory
Animal Care. The maintenance conditions for the mice were as
follows: Temperature, 26–28°C; humidity, 40–60%; ventilation, 10–15
times/h; 10-h light/14-h dark cycle; ad libitum access to
food and water. The food and water were obtained from the Beijing
Keao Cooperation Feed Co., Ltd. (Beijing, China), as recommended by
the China Association for Accreditation of Laboratory Animal Care.
For the tumorigenesis analysis, 1×106 sh-HOXD-AS1 and
sh-NC cells were inoculated into the right flanks of each mouse.
Tumor diameter (mm) was measured every week, and the tumor volumes
were calculated using the following formula: Volume = (shortest
diameter)2 × (longest diameter) × 0.5. After 7 weeks,
the mice were sacrificed, and the tumors were excised. For the
metastasis analysis, 2×106 cells were inoculated into
the tail vein of each mouse. After 6 weeks, the mice were
sacrificed, and the lungs and livers were excised, fixed with 4%
paraformaldehyde at room temperature for 30 min and paraffin
embedded. Consecutive tissue sections (4 µm) were made and
were stained with hematoxylin and eosin; micro-metastases in the
lungs and livers were evaluated under a dissecting microscope. All
animal experiments were performed according to the NIH guidelines
on the use of experimental animals. The in vivo experiments
were approved by the Ethics Committee of the Institutional Review
Board of the Affiliated 3201 Hospital of Xi'an Jiaotong University
on Animal Experiments.
Vector construction and luciferase
reporter assay
The 3′-end fragment from HOXD-AS1 containing the
predicted miR-217 binding site was extracted from HCT116 cells and
amplified using PCR and subcloned into a pmirGLO Luciferase Target
Expression Vector (Promega Corporation, Madison, WI, USA) to form
the HOXD-AS1 wild-type (pmirGLO-HOXD-AS1-wt) vector. In addition, a
mutated miR-217 binding sequence was constructed, which was
subcloned into the expression vector to form the
pmirGLO-HOXD-AS1-mt vector. 293T cells were cultured in a 6-well
plate and grown to 70–80% confluence for cell transfection. Cell
were cotransfected with pmirGLO (50 nM), pmirGLO-HOXD-AS1-wt (50
nM) or pmirGLO-HOXD-AS1-mt (50 nM), and miR-217 mimics
(5′-UACUGCAUCAGGAACUGAUUGGA-3′) (100 nM) or NC
(5′-UUGUACUACACAAAAGUACUG-3′) (100 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), and were cultured in a humidified atmosphere
containing 5% CO2 at 37°C for 24 h. Finally, the
relative luciferase activity was measured using the Dual-Luciferase
Reporter Assay kit (Promega Corporation) after 48 h, according to
the manufacturer's protocol. For overexpression or knockdown of
miR-217, HCT116 and LoVo cells grown to 70–80% confluence were
transfected with miR-217 mimics (100 nM) or NC mimics (100 nM), and
miR-217 inhibitor (5′-UACUGCAUCAGGAACUGAUUGGA-3′) (100 nM) or
inhibitor NC (5′-GCCUCCGGCUUCGCACCUCU-3′) (100 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The PCR
primer sequences are listed in Table
I.
Western blot analysis
For immunoblotting of astrocyte elevated gene-1
(AEG-1), enhancer of zeste homolog 2 (EZH2) and GAPDH, rabbit EZH2
antibody (#5246) was purchased from Cell Signaling Technology,
Inc., and rabbit AEG-1 antibody (abs124058) and mouse GAPDH
antibody (ab127428) were obtained from Absin Bioscience, Inc.
(Shanghai, China). Western blotting was performed as previously
described (19).
Statistical analysis
All statistical analyses were performed using SPSS
version 16.0 (SPSS, Inc., Chicago, IL, USA) or GraphPad 5.0
(GraphPad Software, Inc., La Jolla, CA, USA) software. Data are
expressed as the means ± standard deviation; the experiments were
repeated three times. The significant differences between two
groups were analyzed by Student's t-test, and between multiple
groups were analyzed by one-way analysis of variance and
Newman-Keuls multiple comparison test.. The association between
clinicopathological parameters and HOXD-AS1 expression was analyzed
using χ2 test as appropriate. Overall survival rate was
determined using the Kaplan-Meier method with the log-rank test.
Survival data were evaluated using univariate and multivariate Cox
proportional hazards models. P<0.05 was considered to indicate a
statistically significant difference.
Results
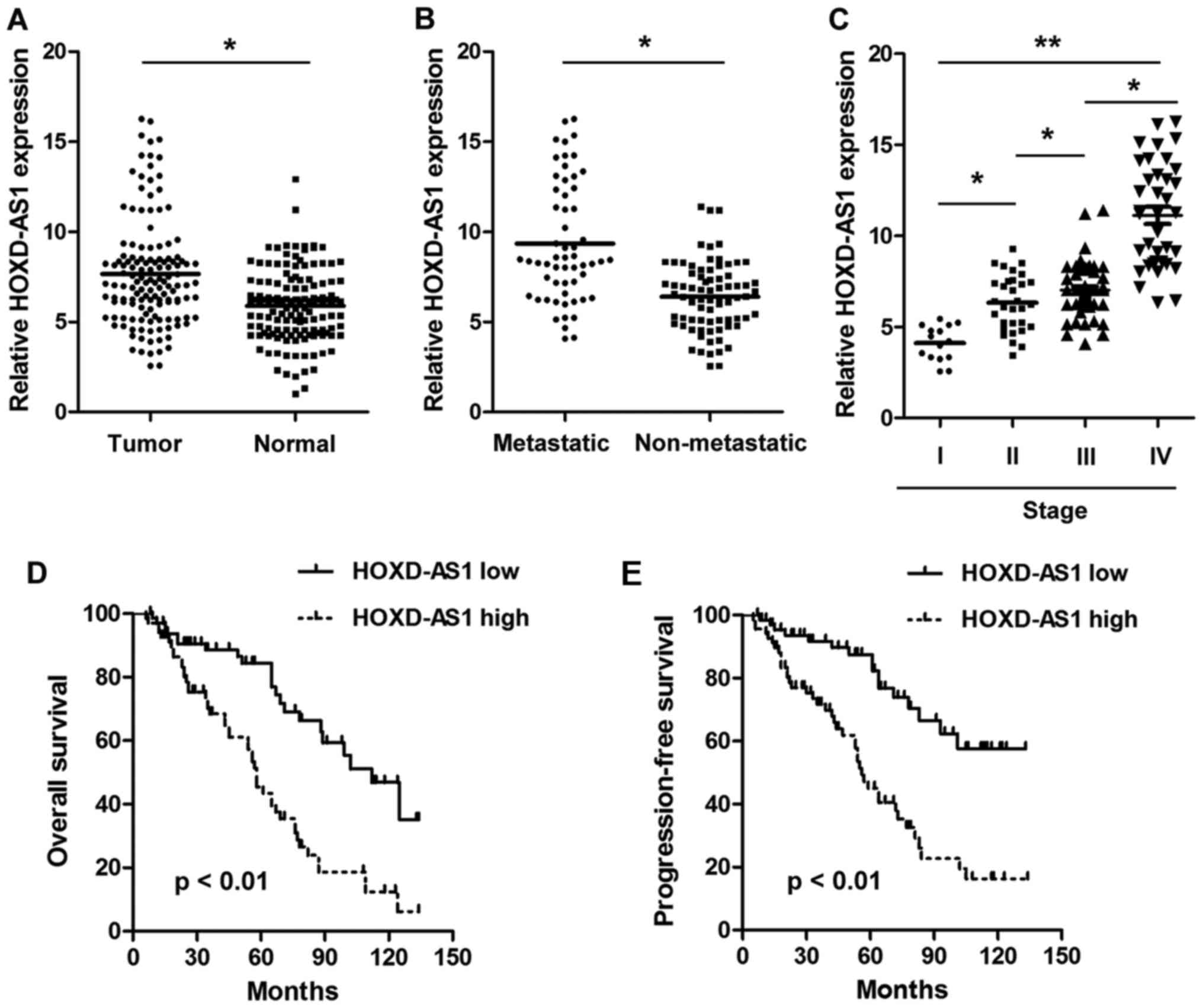
HOXD-AS1 is overexpressed in CRC and is
associated with poor prognosis in patients with CRC
The present study detected the expression levels of
HOXD-AS1 in paired CRC tissues and adjacent normal tissues. As
shown in Fig. 1A, HOXD-AS1 was
significantly upregulated in CRC tissues compared with in adjacent
normal tissues (7.656±0.755 vs. 5.888±0.472; P<0.001).
Furthermore, the expression levels of HOXD-AS1 were significantly
higher in tissues with distant metastasis compared with in those
without distant metastasis (9.350±0.836 vs. 6.396±0.515;
P<0.001) (Fig. 1B). In
addition, HOXD-AS1 expression was significantly increased in
advanced stage patients compared with in earlier stage patients
(Fig. 1C). To explore the
prognostic implications of HOXD-AS1, patients with CRC were divided
into two groups according to the median expression of HOXD-AS1 in
tumor tissues (7.12); namely, a high expression group and a low
expression group. The results demonstrated that patients in the
high HOXD-AS1 expression group presented with significantly worse
overall survival and progression-free survival compared with those
in the low HOXD-AS1 expression group (P<0.01) (Fig. 1D and E). Notably, HOXD-AS1
expression was significantly associated with differentiation
(P<0.001), distant metastasis (P<0.001) and TNM stage
(P=0.009) (Table II). Univariate
analysis demonstrated that HOXD-AS1 expression (P=0.006), distant
metastasis (P=0.005) and TNM stage (P=0.011) were significantly
associated with prognosis. However, multivariate analysis indicated
that only HOXD-AS1 expression (P=0.018) was an independent
prognostic factor in patients with CRC (Table III).
 | Table IIAssociation between
clinicopathological parameters and long non-coding RNA HOXD cluster
antisense RNA 1 expression in 136 patients with colorectal
cancer. |
Table II
Association between
clinicopathological parameters and long non-coding RNA HOXD cluster
antisense RNA 1 expression in 136 patients with colorectal
cancer.
| Characteristic | n | High
expression | Low expression | P-value |
|---|
| Age (years) | | | | 0.492 |
| <60 | 64 | 30 | 34 | |
| ≥60 | 72 | 38 | 34 | |
| Sex | | | | 0.729 |
| Male | 78 | 38 | 40 | |
| Female | 58 | 30 | 28 | |
| Tumor size | | | | 0.122 |
| <4 cm | 71 | 31 | 40 | |
| ≥4 cm | 65 | 37 | 28 | |
|
Differentiation | | | | <0.001 |
| Well | 28 | 9 | 19 | |
| Moderate | 63 | 24 | 39 | |
| Poor | 45 | 35 | 10 | |
| Lymph node
invasion | | | | 0.855 |
| Absent | 45 | 22 | 23 | |
| Present | 91 | 46 | 45 | |
| Distant
metastasis | | | | <0.001 |
| Absent | 84 | 31 | 53 | |
| Present | 52 | 37 | 15 | |
| TNM stage | | | | 0.009 |
| I–II | 42 | 14 | 28 | |
| III–IV | 94 | 54 | 40 | |
 | Table IIIUnivariate and multivariate analysis
of potential prognostic factors in 136 patients with colorectal
cancer. |
Table III
Univariate and multivariate analysis
of potential prognostic factors in 136 patients with colorectal
cancer.
| Characteristic | Univariate analysis
| Multivariate
analysis
|
|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (years) | 0.85
(0.65–1.23) | 0.157 | – | – |
| Sex | 1.13
(0.73–1.51) | 0.541 | – | – |
|
Differentiation | 1.09
(0.88–1.49) | 0.159 | – | – |
| Tumor size | 1.07
(0.82–1.34) | 0.657 | – | – |
| Lymph node
invasion | 1.23
(1.13–1.90) | 0.134 | – | – |
| Distant
metastasis | 1.45
(1.04–2.13) | 0.005a | 1.12
(1.04–1.99) | 0.103 |
| TNM stage | 1.16
(1.07–2.15) | 0.011a | 1.18
(1.03–1.72) | 0.151 |
| HOXD-AS1 level | 1.67
(1.55–2.61) | 0.006a | 1.51
(1.21–2.53) | 0.018a |
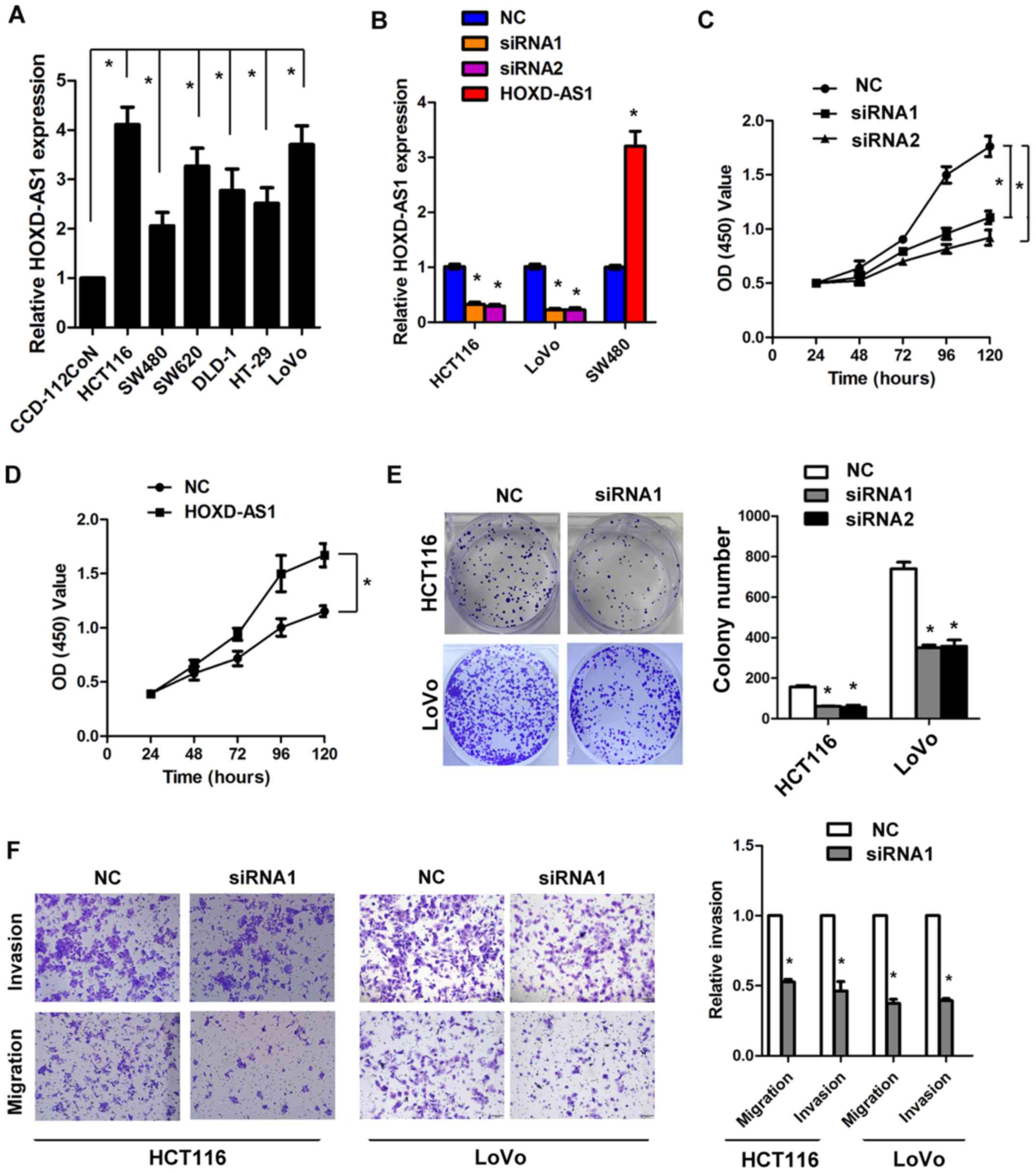
Knockdown of HOXD-AS1 significantly
inhibits cell viability, proliferation and invasion in vitro
Since HOXD-AS1 is significantly overexpressed in
CRC, the present study aimed to determine whether HOXD-AS1 is
involved in the regulation of CRC cell biology. The expression
levels of HOXD-AS1 were detected in CRC cell lines. The results
demonstrated that HOXD-AS1 was markedly increased in CRC cell lines
compared with in the normal colonic cell line CCD-112CoN
(P<0.05; Fig. 2A). Knockdown of
HOXD-AS1 reduced the expression of HOXD-AS1 in HCT116 and LoVo
cells by >50%, whereas overexpression of HOXD-AS1 increased the
expression of HOXD-AS1 in SW480 cells by 3.2-fold (P<0.05;
Fig. 2B). The MTT assay
demonstrated that knockdown of HOXD-AS1 inhibited cell viability in
HCT116 cells, whereas ectopic overexpression of HOXD-AS1 promoted
cell viability in SW480 cells (P<0.05; Fig. 2C and D). In addition, knockdown of
HOXD-AS1 significantly inhibited colony formation in HCT116 and
LoVo cells (P<0.05; Fig. 2E).
Similarly, knockdown of HOXD-AS1 inhibited cell migration and
invasion in HCT116 and LoVo cells by >50% (P<0.05; Fig. 2F).
Knockdown of HOXD-AS1 significantly
inhibits EMT and stem cell potential in CRC cells
It has previously been reported that EMT and cancer
stem cells are involved in tumor progression (20,21);
the present study conducted experiments to investigate this. As
shown in Fig. 3A, knockdown of
HOXD-AS1 increased E-cadherin expression and reduced Vimentin
expression in HCT116 cells. Furthermore, RT-qPCR analysis indicated
that knockdown of HOXD-AS1 decreased the expression levels of
Vimentin, N-cadherin, Slug and Snail, and increased the expression
levels of E-cadherin, α-catenin and β-catenin (P<0.05; Fig. 3B). In addition, knockdown of
HOXD-AS1 inhibited stem cell formation in HCT116 and LoVo cells
(P<0.05; Fig. 3C). RT-qPCR
analysis also demonstrated that knockdown of HOXD-AS1 decreased the
expression levels of stem cell markers (CD44, CD133, CD24, CD166,
Oct4, Lgr5, Sox2 and Nanog) in HCT116 and LoVo cells (P<0.05;
Fig. 3D).
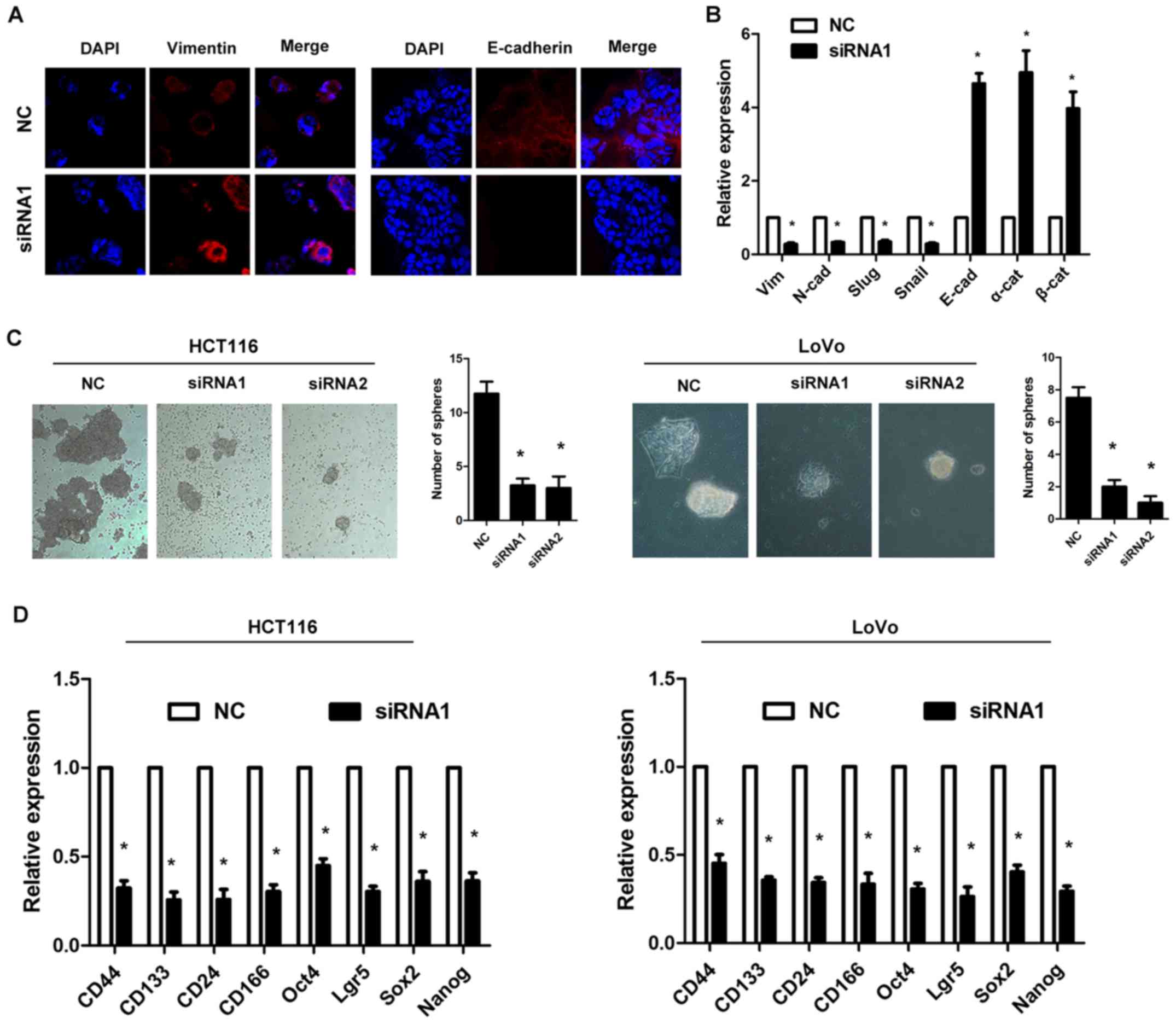 | Figure 3Long non-coding RNA HOXD-AS1
regulates epithelial-mesenchymal transition and stem cell formation
in colorectal cancer cells. (A) Knockdown of HOXD-AS1 increased
E-cadherin expression and reduced Vimentin expression in HCT116
cell. Magnification, ×200. (B) RT-qPCR analysis demonstrated that
knockdown of HOXD-AS1 decreased the expression levels of Vimentin,
N-cadherin, Slug and Snail, and increased the expression of
E-cadherin, α-catenin and β-catenin in HCT116 cells
(*P<0.05 vs. NC). (C) Knockdown of HOXD-AS1 inhibited
stem cell formation in HCT116 and LoVo cells (*P<0.05
vs. NC). Magnification, ×200. (D) RT-qPCR analysis demonstrated
that knockdown of HOXD-AS1 decreased the expression levels of stem
cell markers (CD44, CD133, CD24, CD166, Oct4, Lgr5, Sox2 and Nanog)
in HCT116 and LoVo cells (*P<0.05 vs. NC). CD,
cluster of differentiation; HOXD-AS1, HOXD cluster antisense RNA 1;
Lgr5, leucine-rich repeat-containing G-protein coupled receptor 5;
NC, negative control; Oct4, octamer-binding transcription factor 4;
siRNA, small interfering RNA; Sox2, sex determining region Y-box
2. |
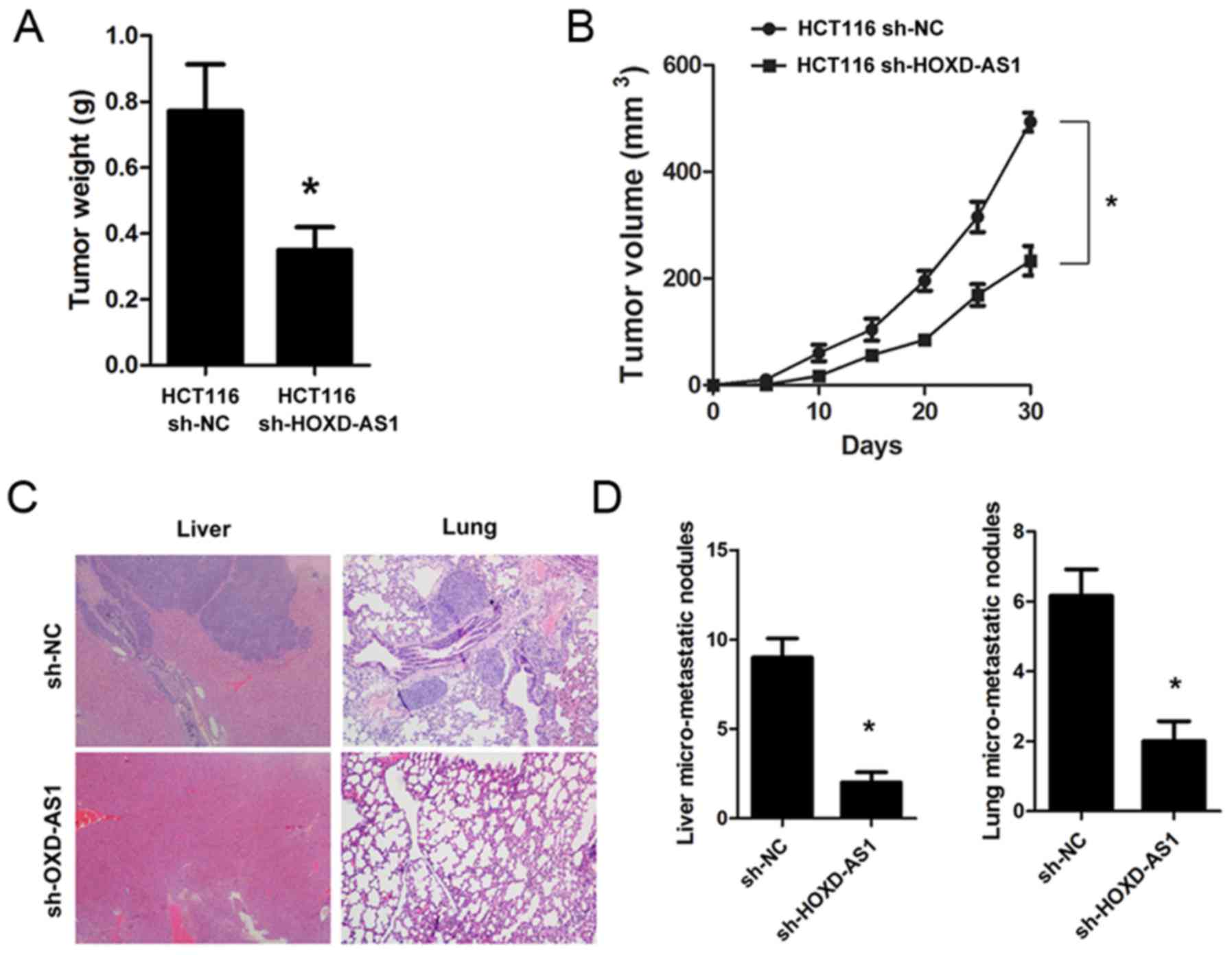
Knockdown of HOXD-AS1 suppresses
tumorigenesis and metastasis in vivo
To explore the biological role of HOXD-AS1 in
vivo, sh-HOXD-AS1 and sh-NC cells were constructed, and were
implanted into mice. The results demonstrated that knockdown of
HOXD-AS1 significantly inhibited tumorigenesis; the tumor weight
(0.771±0.46 vs. 0.349±0.23; P<0.001) and tumor volume
(P<0.001) were significantly reduced in mice implanted with
sh-HOXD-AS1 HCT116 cells (Fig. 4A
and B). Furthermore, knockdown of HOXD-AS1 significantly suppressed
liver (9.00±1.76 vs. 2.00±0.57; P<0.001) and lung metastasis
(6.16±0.74 vs. 2.00±0.54; P<0.001) in nude mice (Fig. 4C and D).
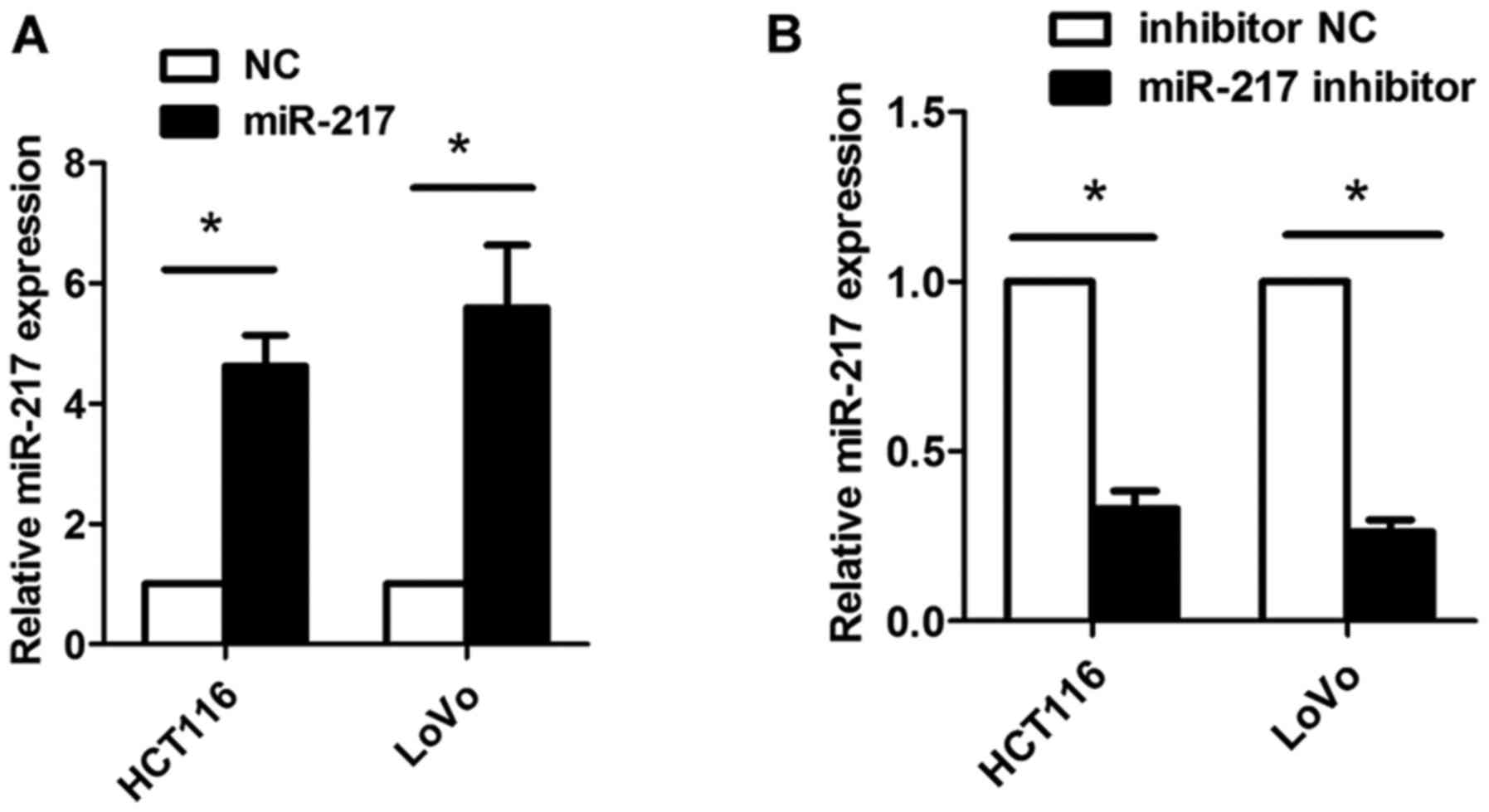
HOXD-AS1 acts as a sponge of miR-217 in
CRC cells
It has previously been reported that some lncRNAs
can act as competing endogenous (ce)RNAs for miRNAs. In the present
study, using bioinformatics tools (miRcode, http://www.mircode.org/), it was identified that
HOXD-AS1 contains some miRNA-binding sites. However, further
analysis indicated that only miR-217 could regulate the expression
of HOXD-AS1 in CRC cells (data not shown); therefore, the present
study focused on miR-217 (Figs. 5
and 6). miR-217-binding sites were
detected in HOXD-AS1 at chr2-q31.1 (Fig. 5A). Subsequently, miR-217 was
overexpressed or knocked down in HCT116 and LoVo cells following
transfection with miR-217 mimics or a miR-217 inhibitor; successful
transfection was confirmed by RT-qPCR (Fig. 6). In addition, HOXD-AS1 expression
was significantly inhibited in HCT116 and LoVo cells transfected
with miR-217 mimics (P<0.05; Fig.
5B); however, knockdown of HOXD-AS1 did not affect the
expression levels of miR-217 (Fig.
5C). Inhibition of miR-217 led to increased expression of
HOXD-AS1; however, increased expression of HOXD-AS1 caused by
miR-217 inhibition could be reduced by the suppression of HOXD-AS1
(P<0.05; Fig. 5D). Furthermore,
overexpression of miR-217 significantly reduced luciferase activity
in cells transfected with pmirGLO-HOXD-AS1-wt, but not in cells
transfected with pmirGLO-HOXD-AS1-mt (Fig. 5E). In addition, RIP analysis
indicated that miR-217 and HOXD-AS1 could be significantly enriched
in HCT116 and LoVo cells, as Ago2 is the vital component of the
RNA-induced silencing complex (RISC), thus indicating that miR-217
and HOXD-AS1 are in the same RISC (P<0.05; Fig. 5F and G). Furthermore, the present
study performed western blotting, the results of which confirmed
that knockdown of HOXD-AS1 decreased the protein expression levels
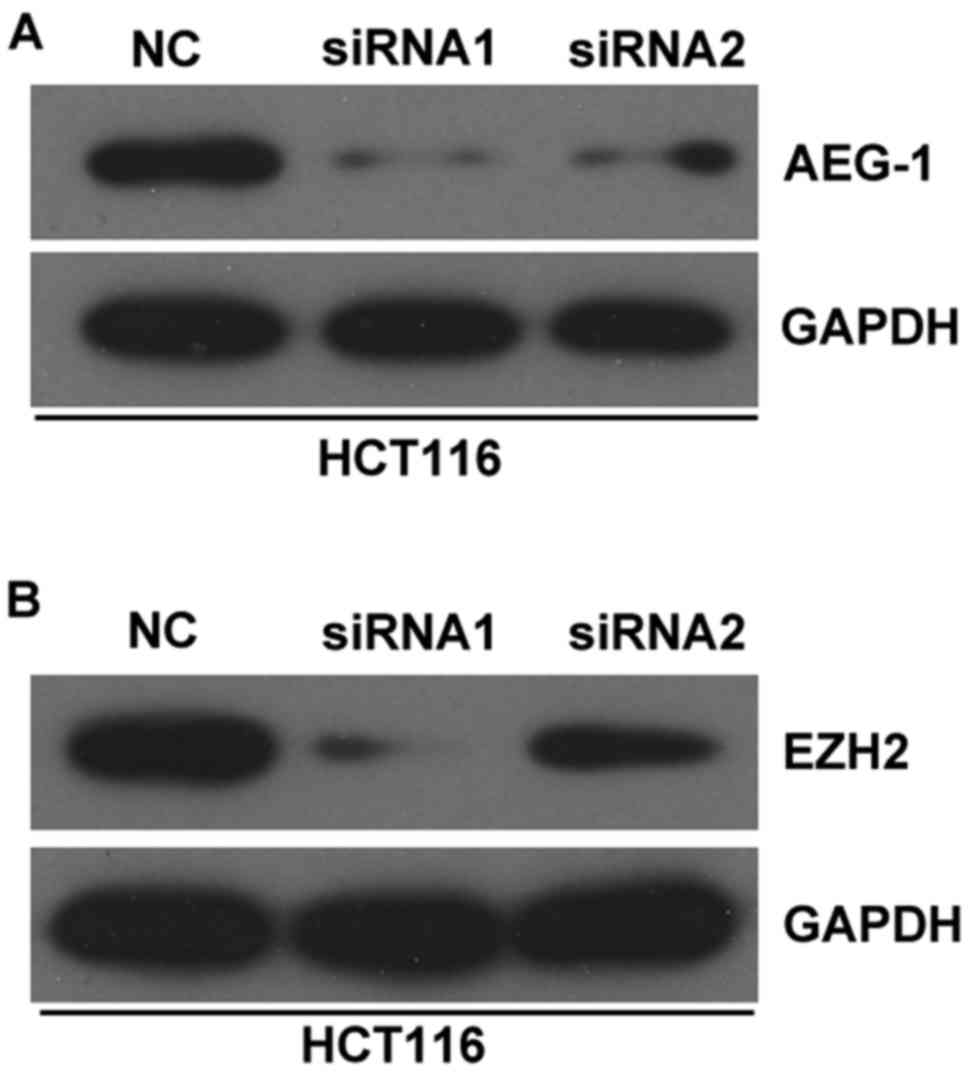
of AEG-1 and EZH2 in CRC cells (Fig.
7).
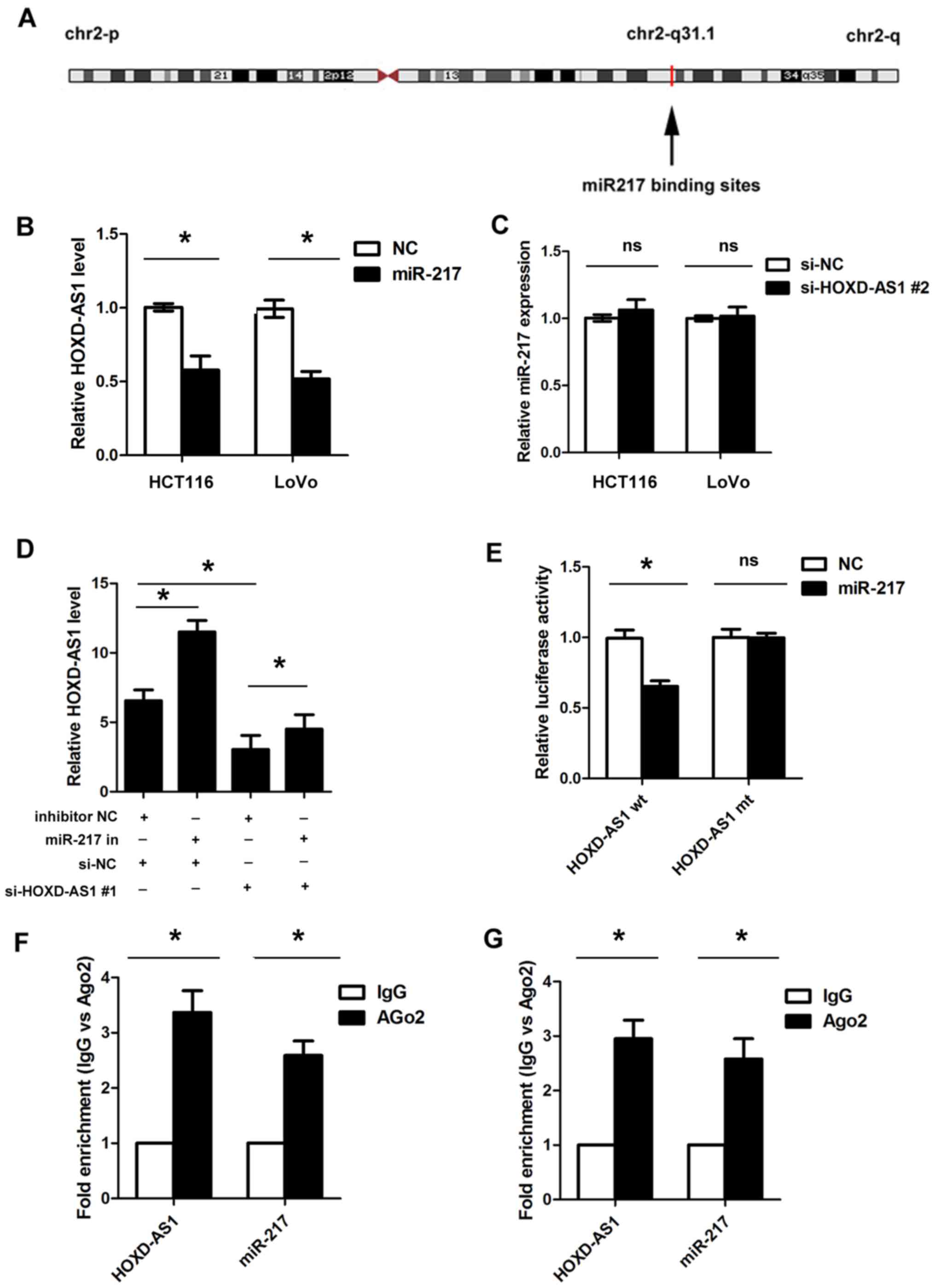 | Figure 5HOXD-AS1 acts as a sponge of miR-217
in CRC cells. (A) Binding sites between HOXD-AS1 and miR-217 at
chr2-q31.1. (B) Overexpression of miR-217 significantly inhibited
the expression of HOXD-AS1 in HCT116 and LoVo cells
(*P<0.05). (C) Knockdown of HOXD-AS1 did not affect
the expression levels of miR-217. (D) Inhibition of miR-217 led to
increased expression of HOXD-AS1; however, the increased expression
of HOXD-AS1 caused by miR-217 inhibition could be reduced by
suppression of HOXD-AS1 (*P<0.05). (E) Ectopic
overexpression of miR-217 significantly reduced the luciferase
activity in cells transfected with the HOXD-AS1 wt vector, but not
in those transfected with the HOXD-AS1 mt vector
(*P<0.05). RNA immunoprecipitation analysis indicated
that miR-217 and HOXD-AS1 could be significantly enriched in (F)
HCT116 cells and (G) LoVo cells (*P<0.05). Ago2,
argonaute 2; HOXD-AS1, HOXD cluster antisense RNA 1; IgG,
immunoglobulin G; miR-217, microRNA-217; mt, mutant; NC, negative
control; ns, not significant; si, small interfering; wt,
wild-type. |
Discussion
Numerous studies have indicated that lncRNAs are
frequently dysregulated in tumors, including CRC (22,23).
Some lncRNAs are considered biomarkers for the diagnosis, prognosis
and treatment of various tumors. For example, it has been reported
that colorectal cancer-associated lncRNA promotes CRC progression
through activating the Wnt/β-catenin signaling pathway via
inhibition of activator protein 2α (24). In addition, Chen et al
revealed that the lncRNA X-inactive specific transcript (XIST)
promotes CRC progression and metastasis by regulating EMT (25). The present study demonstrated that
the lncRNA HOXD-AS1 was significantly upregulated in CRC tissues
and cell lines. Overexpression of HOXD-AS1 was associated with an
aggressive tumor phenotype and poor prognosis in CRC. Furthermore,
HOXD-AS1 was identified as an independent prognostic indicator in
patients with CRC. These results demonstrated that HOXD-AS1 may be
an important biomarker in CRC.
The results of present in vitro and in
vivo experiments indicated that HOXD-AS1 may regulate cell
proliferation, cell migration and cell invasion, thus resulting in
increased tumorigenesis and metastasis in CRC. In agreement with
the present results, previous studies have reported that HOXD-AS1
is involved in the progression and metastasis of bladder cancer,
gastric cancer and hepatocellular carcinoma (12,13,26).
These results highlighted the potential of targeting HOXD-AS1 as a
novel therapeutic strategy in CRC.
EMT is an important process associated with tumor
metastasis and cancer stem cells are the major source of tumor
progression; therefore, the present study investigated the effects
of HOXD-AS1 on EMT and stem cells in CRC. The results demonstrated
that knockdown of HOXD-AS1 inhibited EMT and stem cell properties
in CRC. This finding is in agreement with previous reports that
demonstrated that cancer stem cells are the key source of tumor
progression and metastasis (20).
Notably, a direct relationship has been observed between EMT and
enhanced stem cell properties (21). Taken together, these data suggested
that HOXD-AS1 may promote EMT and stem cell properties, thus
resulting in enhanced proliferation, migration and invasion,
finally accelerating tumor progression and metastasis.
An increasing number of studies have indicated that
a widespread interaction network exists involving ceRNAs, in which
lncRNAs can sequester miRNAs, titrating them off their binding
sites on mRNAs (27). For example,
lncRNA highly upregulated in liver cancer is overexpressed in liver
carcinoma and can promote tumor progression via the suppression of
miR-372 (28). In addition, it has
been reported that the lncRNA H19 acts as a ceRNA to regulate let-7
expression, thus promoting breast cancer progression (29). Recently, Chen et al reported
that the lncRNA XIST may promote gastric cancer by acting as a
ceRNA for miR-101 to regulate EZH2 expression (18). The present study used a
bioinformatics database and identified several miRNAs that may
interact with HOXD-AS1. Notably, miR-217 was confirmed to be
regulated by HOXD-AS1 in CRC cells according to the following
evidence: i) Ectopic overexpression of miR-217 could decrease the
expression of HOXD-AS1, whereas inhibition of miR-217 increased the
expression of HOXD-AS1; ii) luciferase activity assay confirmed
that miR-217 could bind to the 3′-untranslated region of HOXD-AS1;
iii) RIP analysis indicated that miR-217 and HOXD-AS1 could be
significantly enriched in HCT116 and LoVo cells by Ago2. A previous
study demonstrated that lncRNAs can regulate the expression of
miRNAs (18); however, the present
study indicated that HOXD-AS1 did not affect the expression of
miR-217; the underlying mechanisms require further exploration.
Similar to these results, it has been reported that HOXD-AS1 acts
as a ceRNA for miR-19a in hepatocellular carcinoma (13). Previous studies indicated that
miR-217 may inhibit cell proliferation and induce cell apoptosis in
CRC cells (30,31). Notably, it has been revealed that
miR-217 may suppress EMT in other malignancies, such as gastric
cancer (32), lung cancer
(33) and pancreatic cancer
(34). The present study confirmed
that HOXD-AS1 could act as a ceRNA for miR-217; this finding may
partly explain why HOXD-AS1 regulates proliferation and EMT in CRC
cells. To investigate whether HOXD-AS1 regulates the target genes
of miR-217, western blotting was conducted and the results
confirmed that knockdown of HOXD-AS1 was able to decrease the
expression of AEG-1 and EZH2 in CRC cells. AEG-1 and EZH2 have been
reported to be target genes of miR-217 in previous studies
(30,35,36).
These findings further confirmed that HOXD-AS1 may function as a
ceRNA for miR-217.
In conclusion, to the best of our knowledge, the
present study is the first to indicate that HOXD-AS1 is
significantly upregulated in CRC tissues and cell lines. HOXD-AS1
expression was also significantly associated with aggressive tumor
phenotype and poor prognosis in patients with CRC. Knockdown of
HOXD-AS1 inhibited CRC cell proliferation, migration, EMT and stem
cell formation in vitro, as well as tumor growth and
metastasis in vivo. Therefore, HOXD-AS1 may be considered a
useful tumor biomarker in CRC, and targeting HOXD-AS1 may be a
novel therapeutic strategy for patients with CRC.
Abbreviations:
|
CRC
|
colorectal cancer
|
|
EMT
|
epithelial-mesenchymal transition
|
|
lncRNA
|
long non-coding RNA
|
|
HOXD-AS1
|
HOXD cluster antisense RNA 1
|
|
siRNA
|
small interfering RNA
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81372533).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
XL, YL, and LP performed the molecular studies and
drafted the manuscript. XZ and ZF performed the in vivo
assays. BY and TL collected the clinical data and tissue samples.
WM and ZhL performed the statistical analysis. ZeL designed the
study and helped to draft the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Affiliated 3201 Hospital of Xi'an Jiaotong
University (Hanzhong, China), and written informed consent was
obtained from all patients. All of the animal experiments were
performed according to the National Institutes of Health (Bethesda,
MD, USA) guidelines on the use of experimental animals, and were
approved by the Ethics Committee of the Institutional Review Board
of the Affiliated 3201 Hospital of Xi'an Jiaotong University on
Animal Experiments.
Consent for publication
Consent for the publication of the clinical and
pathological data was obtained from all patients who were involved
in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Goldstein DA, Zeichner SB, Bartnik CM,
Neustadter E and Flowers CR: Metastatic Colorectal Cancer: A
Systematic Review of the Value of Current Therapies. Clin
Colorectal Cancer. 15:1–6. 2016. View Article : Google Scholar :
|
|
3
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rinn JL and Chang HY: Genome regulation by
long noncoding RNAs. Annu Rev Biochem. 81:145–166. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Graham A, Papalopulu N and Krumlauf R: The
murine and Drosophila homeobox gene complexes have common features
of organization and expression. Cell. 57:367–378. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Huo X, Yang XR, He J, Cheng L,
Wang N, Deng X, Jin H, Wang N, Wang C, et al: STAT3-mediated
upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer
metastasis by regulating SOX4. Mol Cancer. 16:1362017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gu P, Chen X, Xie R, Han J, Xie W, Wang B,
Dong W, Chen C, Yang M, Jiang J, et al: lncRNA HOXD-AS1 regulates
proliferation and chemo-resistance of castration-resistant prostate
cancer via recruiting WDR5. Mol Ther. 25:1959–1973. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Li J, Zhuang C, Liu Y, Chen M, Chen Y,
Chen Z, He A, Lin J, Zhan Y, Liu L, et al: Synthetic
tetracycline-controllable shRNA targeting long non-coding RNA
HOXD-AS1 inhibits the progression of bladder cancer. J Exp Clin
Cancer Res. 35:992016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lu S, Zhou J, Sun Y, Li N, Miao M, Jiao B
and Chen H: The noncoding RNA HOXD-AS1 is a critical regulator of
the metastasis and apoptosis phenotype in human hepatocellular
carcinoma. Mol Cancer. 16:1252017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM and
Chen J: The miR-217 microRNA functions as a potential tumor
suppressor in pancreatic ductal adenocarcinoma by targeting KRAS.
Carcinogenesis. 31:1726–1733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang JX, Chen ZH, Xu Y, Chen JW, Weng HW,
Yun M, Zheng ZS, Chen C, Wu BL and Li EM: Downregulation of
MicroRNA-644a promotes esophageal squamous cell carcinoma
aggressiveness and stem cell-like phenotype via dysregulation of
ITX2. Clin Cancer Res. 23:298–310. 2017. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Lu YX, Ju HQ, Wang F, Chen LZ, Wu QN,
Sheng H, Mo HY, Pan ZZ, Xie D, Kang TB, et al: Inhibition of the
NF-κB pathway by nafamostat mesilate suppresses colorectal cancer
growth and metastasis. Cancer Lett. 380:87–97. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chen DL, Ju HQ, Lu YX, Chen LZ, Zeng ZL,
Zhang DS, Luo HY, Wang F, Qiu MZ, Wang DS, et al: Long non-coding
RNA XIST regulates gastric cancer progression by acting as a
molecular sponge of miR-101 to modulate EZH2 expression. J Exp Clin
Cancer Res. 35:1422016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Teng KY, Qiu MZ, Li ZH, Luo HY, Zeng ZL,
Luo RZ, Zhang HZ, Wang ZQ, Li YH and Xu RH: DNA polymerase η
protein expression predicts treatment response and survival of
metastatic gastric adenocarcinoma patients treated with
oxaliplatin-based chemotherapy. J Transl Med. 8:1262010. View Article : Google Scholar
|
|
20
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Jiang C, Li X, Zhao H and Liu H: Long
non-coding RNAs: Potential new biomarkers for predicting tumor
invasion and metastasis. Mol Cancer. 15:622016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ma Y, Yang Y, Wang F, Moyer MP, Wei Q,
Zhang P, Yang Z, Liu W, Zhang H, Chen N, et al: Long non-coding RNA
CCAL regulates colorectal cancer progression by activating
Wnt/β-catenin signalling pathway via suppression of activator
protein 2α. Gut. 65:1494–1504. 2016. View Article : Google Scholar
|
|
25
|
Chen DL, Chen LZ, Lu YX, Zhang DS, Zeng
ZL, Pan ZZ, Huang P, Wang FH, Li YH, Ju HQ, et al: Long noncoding
RNA XIST expedites metastasis and modulates epithelial-mesenchymal
transition in colorectal cancer. Cell Death Dis. 8:e30112017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zheng L, Chen J, Zhou Z and He Z:
Knockdown of long non-coding RNA HOXD-AS1 inhibits gastric cancer
cell growth via inactivating the JAK2/STAT3 pathway. Tumour Biol.
39:10104283177053352017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:102017. View Article : Google Scholar
|
|
30
|
Wang B, Shen ZL, Jiang KW, Zhao G, Wang
CY, Yan YC, Yang Y, Zhang JZ, Shen C, Gao ZD, et al: MicroRNA-217
functions as a prognosis predictor and inhibits colorectal cancer
cell proliferation and invasion via an AEG-1 dependent mechanism.
BMC Cancer. 15:4372015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Flum M, Kleemann M, Schneider H, Weis B,
Fischer S, Handrick R and Otte K: miR-217-5p induces apoptosis by
directly targeting PRKCI, BAG3, ITGAV and MAPK1 in colorectal
cancer cells. J Cell Commun Signal. Sep 14–2017.Epub ahead of
print. PubMed/NCBI
|
|
32
|
Liu YP, Sun XH, Cao XL, Jiang WW, Wang XX,
Zhang YF and Wang JL: MicroRNA-217 suppressed
epithelial-to-mesenchymal transition in gastric cancer metastasis
through targeting PTPN14. Eur Rev Med Pharmacol Sci. 21:1759–1767.
2017.PubMed/NCBI
|
|
33
|
Lu L, Luo F, Liu Y, Liu X, Shi L, Lu X and
Liu Q: Posttranscriptional silencing of the lncRNA MALAT1 by
miR-217 inhibits the epithelial-mesenchymal transition via enhancer
of zeste homolog 2 in the malignant transformation of HBE cells
induced by cigarette smoke extract. Toxicol Appl Pharmacol.
289:276–285. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Deng S, Zhu S, Wang B, Li X, Liu Y, Qin Q,
Gong Q, Niu Y, Xiang C, Chen J, et al: Chronic pancreatitis and
pancreatic cancer demonstrate active epithelial-mesenchymal
transition profile, regulated by miR-217-SIRT1 pathway. Cancer
Lett. 355:184–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Miao S, Mao X, Zhao S, Song K, Xiang C, Lv
Y, Jiang H, Wang L, Li B, Yang X, et al: miR-217 inhibits laryngeal
cancer metastasis by repressing AEG-1 and PD-L1 expression.
Oncotarget. 8:62143–62153. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen DL, Zhang DS, Lu YX, Chen LZ, Zeng
ZL, He MM, Wang FH, Li YH, Zhang HZ, Pelicano H, et al:
microRNA-217 inhibits tumor progression and metastasis by
downregulating EZH2 and predicts favorable prognosis in gastric
cancer. Oncotarget. 6:10868–10879. 2015.PubMed/NCBI
|