Introduction
Esophageal carcinoma (EC) is one of the most
frequently occurring malignances worldwide (1). EC is divided into two main
histological types, including esophageal squamous cell carcinoma
(ESCC) and esophageal adenocarcinoma (ECA); ESCC as a main
histological type is more common in Africa, Iran and North China
(2,3). Despite tremendous advances being made
in therapeutic strategies, the 5-year survival rate for patients
with ESCC remains markedly poor (4–8).
Notably, even though chemotherapy is an effective therapeutic
approach for patients with ESCC (9), the development of drug resistance has
become the most severe concern, and is the main cause of treatment
failure in patients with ESCC (10). Therefore, it is imperative to
elucidate the mechanisms responsible for drug resistance in order
to improve the survival rate of patients with ESCC.
MicroRNAs (miRNAs or miRs) are a class of small
non-coding RNAs comprising of 19–25 nucleotides in length, which
regulate gene expression by targeting related genes (11,12).
Ample evidence has revealed that miRNAs are involved in a number of
cellular processes, such as cell apoptosis, cell cycle, cell
invasion and metastasis, the regulation of signaling networks and
drug resistance (13–19). Furthermore, miRNAs are tightly
associated with tumor initiation, development and progression in a
variety of tumors via the modulation of their target gene levels
(20–23); hence, miRNAs may function as either
oncogenes or tumor suppressors in different types of tumors
(20,24–27).
Therefore, it is imperative to interpret the function of miRNAs in
the occurrence and development of a large number of tumors, which
suggests that miRNAs, as novel and promising therapeutic targets,
may exhibit huge clinical value in the future. In addition, miRNAs
may function as molecular markers for early the diagnosis of a
number of tumors (28,29), which may aid in the development of
a number of diagnostic agents for multiple tumor types.
miRNA-125a-5p (miR-125a-5p), a type of newly discovered miRNA
molecule, has been verified to be involved in the development and
progression of a number of tumors, including laryngeal cancer
(30), hepatocellular carcinoma
(31–33), lung cancer (34) and prostate carcinoma (35). The tumor suppressive function of
miR-125a-5p has also been supported by a number of investigations
on a variety of tumor types (32,36,37).
Notably, miR-125a has been shown to enhance the sensitivity of
paclitaxel-resistant colon cancer cells to paclitaxel (38), suggesting that miR-125a may prove
to be a novel ancillary drug for use in chemotherapy for patients
with tumors. These data imply that miR-125a-5p has tremendous
potential for use in the diagnosis, treatment and prognosis of a
wide range of tumors.
In the current study, we examined miRNA-125a-5p
expression in ESCC tissues and cell lines, and verified its role in
the regulation of the proliferation, the cell cycle, apoptosis, and
in the migratory and invasive abilities of ESCC cells. miR-125a-5p
was found to enhance the sensitivity of ESCC cells to cisplatin by
suppressing the activation of the signal transducer and activator
of transcription-3 (STAT3) signaling pathway. Most importantly,
interleukin (IL)-6, a widely reported activator of the STAT3
signaling pathway (39), abrogated
the inactivated status of the STAT3 signaling pathway elicited by
the combined use of miR-125a-5p and cisplatin, which was
accompanied by cell phenotypic recovery. Taken together, the data
from the current study suggest that the manipulation of miR-125a-5p
may be used as a strategy with which to enhance the cytotoxic
effects of cisplatin on ESCC via the suppression of the activation
of the STAT3 signaling pathway.
Materials and methods
Patients and tissue samples
This study was approved by the institutional
Research Ethics Committee of Zhengzhou University. A total of 56
cases of ESCC tissues and paired normal esophageal epithelial
tissues were treated with surgical resection alone from the First
Affiliated Hospital of Zhengzhou University, Zhengzhou, China from
May, 2010 to August, 2012. The clinical characteristics of the
patients with ESCC are summarized in Table I. All tissue samples were confirmed
by a pathologist. All samples were obtained with informal written
and none of the patients had received any treatments prior to
surgery. The tissues were immediately frozen in liquid nitrogen
until RnA extraction.
 | Table IClinical characteristics of the
patients with esophageal squamous cell carcinoma. |
Table I
Clinical characteristics of the
patients with esophageal squamous cell carcinoma.
| Characteristic | n=56 |
|---|
| Age (years) | |
| ≥60 | 33 |
| <60 | 23 |
| Sex | |
| Male | 38 |
| Female | 18 |
| TNM staging | |
| I and II | 26 |
| III and IV | 30 |
| Histological
grade | |
| High
differentiation | 15 |
| Moderate
differentiation | 19 |
| Poor
differentiation | 22 |
| Lymph node
metastasis | |
| Yes | 21 |
| No | 35 |
Cell lines and cell culture
The human ESCC cell lines, including Eca109, EC9706,
EC1, TE1, KYSE450 and KYSE70, as well as normal esophageal
epithelial cells, Het-1A, were maintained in liquid nitrogen in our
laboratory. The cell lines above were cultured in RPMi-1640 medium
supplemented with 10% fetal bovine serum (FBS; Gibco/Thermo Fisher
Scientific, Grand island, ny, USA), 100 U/ml penicillin and 100
µg/ml streptomycin (both from Sigma-Aldrich, St. Louis, MO,
USA) in a humidified 5% CO2 incubator at 37°C.
Cell transfection
The miR-125a-5p mimic, miR-125a-5p inhibitor and the
negative control (nC) (Ribobio, Guangzhou, China) at a final
concentration of 30 nM were transfected into the EC1 and TE1 cells
using Lipofectamine™ 2000 (Invitrogen/Life Technologies, Carlsbad,
CA, USA) according to manufacturer’s instructions. All transfection
experiments were performed in triplicate for each treatment group
at 24, 48, 72 and 96 h for cell proliferation assay and at 48 h for
the other experiments.
Prediction of target genes
The potential target genes of miR-125a-5p were
searched using online webpage TargetScan (http://www.targetscan.org/vert_72/), miRanda
(http://www.microrna.org/microrna/home.do) and miRDB
(http://www.mirdb.org/).
Plasmid construction and luciferase
reporter assay
The human STAT3 3′-UTR-wild-type (STAT3-3′-UTR-WT)
region containing the miR-125a-5p binding sequence was amplified by
PCR, and the STAT3-3′-UTR-mutation (STAT3-3′-UTR-MUT) region with a
substitution of 8 bp in the miR-125a-5p binding region was
generated using a QuikChange Site-Directed Mutagenesis kit
(Stratagene, La Jolla, CA, USA). The STAT3-3′-UTR-WT and
STAT3-3′-UTR-MUT were inserted into the downstream region of the
Firefly luciferase gene, respectively. The EC1 and TE1 cells were
co-transfected using reporter plasmids (400 per 20 ng internal
control Renilla luciferase plasmid pRL-SV40) and miR-125a-5p
mimic or NC by Lipofectamine 2000 (Invitrogen/Life Technologies)
according to the manufacturer’s instructions. Subsequnetly,
luciferase activity was determined using the Dual Luciferase Assay
kit (Promega, Madison, Wi, USA) using a Synergy H1 hybrid reader
(Biotek, Winooski, VT, USA) at 48 h following transfection.
Finally, the luciferase activity was normalized to the
Renilla luciferase activity.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated from the tissues and cells,
and subjected to miRNA First Strand cDNA Synthesis kit (cat. no.
B532453; Sangon Biotech, Shanghai, China) using the specific
miR-125a-5p reverse transcription primer,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCACAGGT-3′ and the U6 gene
reverse transcription primer,
5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAAAATA-3′.
Quantitative PCR (qPCR; Tiangen Biotech, Beijing, China) was used
to determine miR-125a-5p expression using the ABI 7500 Real-time
PCR System (Applied Biosystems, Foster City, CA, USA) by the
addition of miR-125a-5p specific amplification primers as follows:
5′-ACACTCCAGCTGGGTCCCTGAGACCCTTTAAC-3′ (forward) and
5′-TGGTGTCGTGGAGTCG-3′ (reverse).
Western blot analysis
Total proteins were isolated from the ESCC cells
using RiPA lysis buffer (Solarbio, Beijing, China). The protein
concentration was determined using a Micro BCA Protein Assay kit
(cat. no. 23235; Pierce Biotechnology, inc., Rockford, IL, USA).
The proteins (50 µg/lane) were separated by 12% SDS-PAGE,
and then transferred onto PvDF membranes (Millipore Corporation,
Billerica, MA, USA). After blocking with skimmed milk, primary
antibodies against E-cadherin (cat. no. ab1416, 1:50 dilution),
N-cadherin (cat. no. ab98952, 1:500 dilution), Vimentin (cat. no.
ab8978, 1:100 dilution), VEGF (cat. no. ab69479, 1:100 dilution),
β-actin (cat. no. ab8226, 1:500 dilution) (all from Abcam,
Cambridge, MA, USA), total STAT3 (t-STAT3, cat. no. 9139, 1:1,000
dilution) and phosphorylated STAT3 (p-STAT3, cat. no. 4113, 1:1,000
dilution) (all from Cell Signaling Technology, Beverly, MA, USA)
were incubated with the PvDF membranes (Millipore Corporation)
overnight at room temperature. Subsequently, Horseradish peroxidase
(HRP)-labeled secondary antibody (cat. no. SE131, 1:5,000 dilution;
Solarbio) was applied to the PvDF membranes. Finally, the protein
signal was developed using enhanced chemiluminescence reagents
(Beyotime Biotech, Haimen, China). The densitometry of the protein
bands was performed using ImageJ software v1.8.0 (National
Institutes of Health, Bethesda, MD, USA).
Cell counting kit-8 (CCK-8) assay
The EC1 and TE1 cells (2,000 cells/well) were seeded
into a 96-well plate, and these cells were then transfected with
miR-125a-5p mimic, miR-125a-5p inhibitor and NC, and treated with
cisplatin (0, 1, 2, 5, 10 and 15 µg/ml; Hansoh
Pharmaceutical Co. Ltd., Jiangsu, China) or iL-6 (20 µg/ml;
PeproTech inc., Rocky Hill, NJ, USA) in triplicate were applied to
the corresponding wells. Cell viability was determined using the
CCK-8 kit (Beyotime Biotech) according to the manufacturer’s
instructions by measuring the absorbance at 450 nm on a microplate
reader (Thermo Fisher Scientific, Waltham, MA, USA).
Cell cycle detection
Cell cycle assay was conducted as described in a
previous study (40). Briefly, the
EC1 and TE1 cells were harvested at 48 h following transfection
with miR-125a-5p mimic, miR-125a-5p inhibitor or NC, rinsed using
PBS and fixed in 70% ethanol overnight at 4℃. After rinsing thrice,
propidium iodide (Pi; Sigma-Aldrich) was used to treat the cells,
and a flow cytometer (BD Biosciences, San Diego, CA, USA) was used
to detect the DNA contents.
Cell apoptosis assay
Cell apoptosis assay was performed as described in a
previous study (40) using Annexin
v FiTC/Pi (Sigma-Aldrich). In brief, the EC1 and TE1 cells were
collected using trypsinase, and Annexin v/Pi reagents were added to
the cells for 30 min. Finally, a flow cytometer (BD Biosciences)
was utilized to determine cell apoptosis.
Would healing assay
Cell migration was investigated by wound healing
migration assay as previously described (41). Briefly, a Ibidi Culture-Insert 2
well (Ibidi Company, Martinsried, Germany) was placed in a 24-well
plate, and the EC1 and TE1 cells transfected with miR-125a-5p
mimic, miR-125a-5p inhibitor or NC were digested and seeded into
24-well culture plates at a density of 5×105 cells/well
using RPMi-1640 medium containing 10% FBS. The Culture-Insert 2
well was gently removed 24 h following appropriate cell attachment.
At 0, 12 and 24 h, images were obtained at the same position under
an inverted microscope (Nikon Instruments, Tokyo, Japan),
respectively. The migration distances were quantified by measuring
the distances from the wound edges.
Cell invasion assay
Cell invasion assay was performed using a Transwell
chamber with Matrigel (BD Biosciences). Briefly, the EC1 and TE1
cells at a density of 1×105 were added to the upper
layer of the chamber, and 20% FBS was added to the bottom layer of
the chamber. Invasive cells were fixed using methanol and stained
with crystal violet (Sigma-Aldrich) for 5 min at room temperature
48 h after treatment. Finally, invasive cell numbers were counted
under a field of ×200 magnification under an inverted fluorescence
microscope (Nikon Instruments).
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6.0 software. Data are expressed as the means ± SD, derived
from experiments with at least 3 independently repeats. Comparisons
between two groups were made using the Student’s t-test, and
comparisons between more than two were made using one-way ANOVA
followed by Dunnett’s test. Values of P<0.05 were considered to
indicate statistically significant differences.
Results
Decreased expression of miR-125a-5p in
ESCC tissues and cells
Ample evidence has demonstrated that miRNAs are
widely involved in tumor development and progression (42,43).
In this study, in order to determine whether miR-125a-5p is tightly
associated with the occurrence and development of ESCC, we examined
its expression patterns in ESCC tissues and cells. We found that
relative level of miR-125a-5p in ESCC tissues was significantly
lower than that in normal esophageal epithelial tissues
(P<0.0001) (Fig. 1A). Further
analysis revealed that the relative level of miR-125a-5p in the
patients with I + II stage disease was markedly higher than in
those with iii + iv stage disease (P<0.01) (Fig. 1B), suggesting that miR-125a-5p is
tightly associated with the tumor clinical staging in ESCC. To
further determine the prognostic value of miR-125a-5p, Kaplan-Meier
survival analysis was used to evaluate the association of the
miR-125a-5p expression level with the prognosis of patients with
ESCC. The results revealed that a high expression of miR-125a-5p
contributed to a better overall survival of patients with ESCC
(Fig. 1C). Furthermore, an in
vitro analysis demonstrated that the relative level of
miR-125a-5p in ESCC cells (Eca109, EC9706, EC1, TE1, KYSE450 and
KYSE70) was evidently lower than that in the normal esophageal
epithelial cell line, Het-1A (P<0.01) (Fig. 1D), which further supported the data
obtained from ESCC tissues. These findings suggest that miR-125a-5p
is involved in the development, progression and prognosis of ESCC
and that its upregulation contributes to an improved prognosis of
patients with ESCC. Therefore, it is very imperative to examine the
function of miR-125a-5p in the occurrence and development of
ESCC.
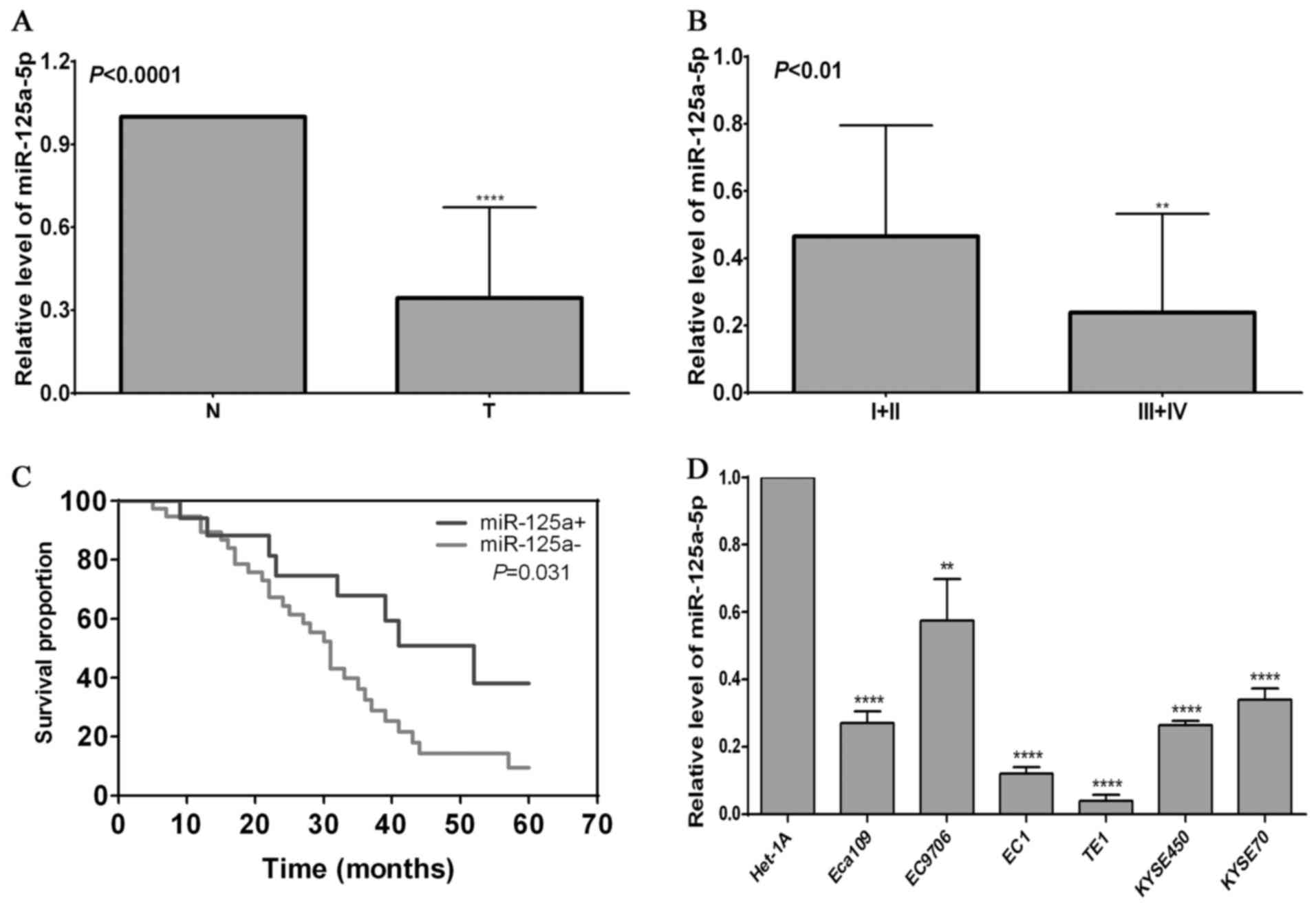 | Figure 1Expression pattern of miR-125a-5p in
esophageal squamous cell carcinoma (ESCC) and its association with
the prognosis of patients with ESCC. (A) miR-125a-5p level in
normal esophageal epithelial tissues (N) and ESCC tissues (T).
Total RNA was isolated from 56 ESCC tisues and paired normal
esophageal epithelial tissues, and subjected to analysis using the
cDnA synthesis kit. RT-qPCR was used to detect the miR-125a-5p
level in ESCC tissues and paired normal esophageal epithelial
tissues; ****P<0.0001, compared with normal tissues.
(B) Expression of miR-125a-5p is tightly associated with tumor TnM
staging. TnM staging in a variety of ESCC tissues was confirmed by
pathology, and RT-qPCR was used to detect the miR-125a-5p level in
ESCC tissues; **P<0.01, compared with ESCC with i and
ii staging. (C) Association of miR-125a-5p level with the prognosis
of the patients with ESCC. Kaplan-Meier analysis was used to
evaluate the association of miR-125a-5p with the overall survival
rate, and the log-rank (Mantel-Cox) test was used to determine the
difference between positive miR-125a-5p and negative miR-125a-5p
expression. (D) Expression of miR-125a-5p in various ESCC cell
lines (Eca109, EC9706, EC1, TE1, KYSE450 and KYSE70 cells) and the
normal esophageal epithelial cell line, Het-1A;
**P<0.01 and ****P<0.0001, compared
with the Het-1A cells. |
Role of miR-125a-5p in the regulation of
the proliferation, cell cycle and apoptosis of ESCC cells
To preliminarily elucidate the underlying function
of miR-125a-5p in ESCC, we transfected NC, miR-125a-5p mimic and
miR-125a-5p inhibitor into EC1 and TE1 ESCC cells, and RT-qPCR was
employed to examine miR-125a-5p expression in the ESCC cells. We
found that relative level of miR-125a-5p in the miR-125a-5p mimic
group was significantly higher than that in the control group and
NC group in the EC1 and TE1 cells (P<0.05), whereas transfection
with the miR-125a-5p yielded opposite results in the EC1 and TE1
cells (Fig. 2A). Further analysis
using CCK-8 assay confirmed that miR-125a-5p overexpression
contributed to the suppression of the proliferation of the EC1 and
TE1 cells at various time points, including 24, 48, 72 and 96 h,
compared with the control group and NC group (P<0.05), and
opposite data were observed in the miR-125a-5p inhibitor group
(Fig. 2B). Cell cycle analysis
revealed that the cell numbers in the G0/G1 phase in the
miR-125a-5p mimic group were markedly higher than those in the
control group and NC group and opposite results were observed with
the cells in the S phase (P<0.05); however, transfection with
miR-125a-5p inhibitor markedly decreased the cell numbers in the
G0/G1 phase and increased the cell numbers in the S phase compared
with the control group and NC group (Fig. 2C and D). Moreover, cell apoptosis
assay revealed that miR-125a-5p upregulation markedly promoted the
apoptosis of the EC1 and TE1 cells, and miR-125a-5p downregulation
evidently suppressed cell apoptosis, compared with the control
group and NC group (Fig. 2E and
F). These data suggest that miR-125a-5p functions as a tumor
suppressor in ESCC.
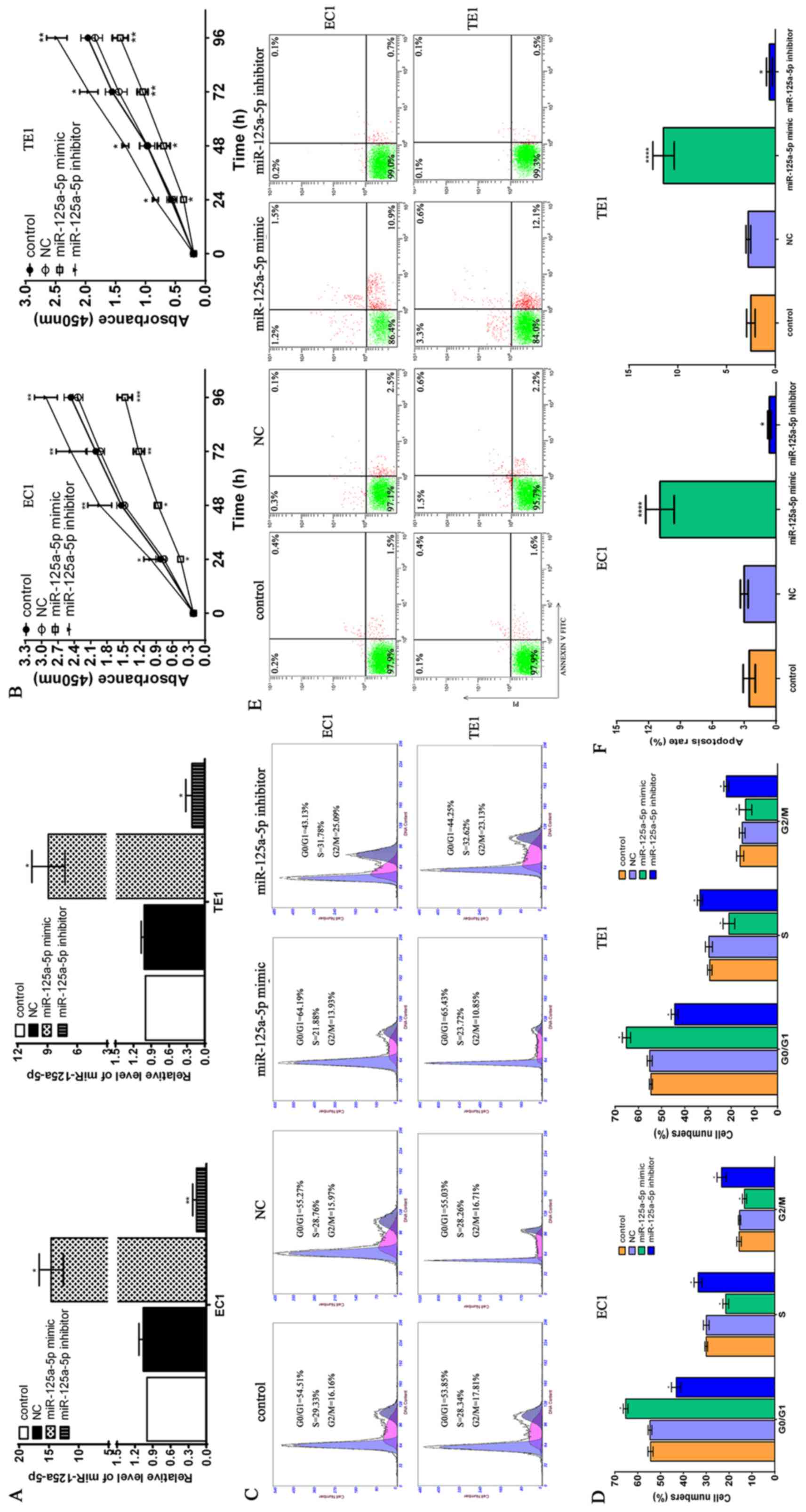 | Figure 2The potential role of miR-125a-5p in
the proliferation, cell cycle progression and apoptosis of
esophageal squamous cell carcinoma (ESCC) cells. (A) miR-125a-5p
mimic or inhibitor triggered the marked upregulation or
downregulation miR-125a-5p of ESCC EC1 and TE1 cells, respectively.
NC, miR-125a-5p mimic and inhibitor were transfected into EC1 and
TE1 ESCC cells, and total RNA was extracted from the EC1 and TE1
cells using TRizol reagent at 48 h after transfection.
Subsequently, cDnA was synthesized using the cDnA synthesis kit and
RT-qPCR was used to determine the miR-125a-5p level in the EC1 and
TE1 cells; *P<0.05, compared with the control group
and NC group. (B) Effects of miR-125a-5p upregulation or
downregulation on the proliferation of ESCC EC1 and TE1 cells. The
CCK-8 kit was used to determine cell proliferation according to the
standard protocol, and the absorbance value at 450 nm was obtained
using a microplate reader; *P<0.05,
**P<0.01 and ***P<0.001, compared with
the control group and NC group. (C) miR-125a-5p upregulation or
downregulation altered the cell cycle distribution in the G0/G1 and
S phase in the ESCC EC1 and TE1 cells. (D) Statistical analysis of
cell cycle distribution in ESCC cells; *P<0.05,
compared with the control group and NC group. (E) Effects of
miR-125a-5p upregulation or downregulation on the apoptosis of ESCC
cells. (F) Statistical analysis of the number of apoptotic ESCC
cell; *P<0.05 and ****P<0.0001,
compared with the control group and NC group. |
miR-125a-5p suppresses the migratory and
invasive abilities of ESCC cells via the regulation of the
epithelial-mesenchymal transition (EMT) process
Accumulating evidence has demonstrated that miRNAs
are widely involved in the migration and invasion in a wide range
of tumors (44,45). However, whether miR-125a-5p is
tightly associated with the invasion and metastasis of ESCC remains
undetermined. Thus, in this study, we examined the effects of
miR-125a-5p mimic or inhibitor on the migratory and invasive
abilities of EC1 and TE1 cells. The results from wound healing
assay and Transwell chamber assay revealed that miR-125a-5p
overexpression significantly suppressed the cell migratory and
invasive abilities; conversely, miR-125a-5p downregulation markedly
promoted the migratory and invasive abilities of the EC1 and TE1
cells, compared with the control group and NC group (P<0.01)
(Fig. 3A–D). A number of studies
have revealed that EMT plays an important role in invasion,
metastasis, and carcinogenesis in a variety of tumors (46,47).
Thus, in this study, to elucidate the underlying mechanisms
responsible for the suppressive effects of miR-125a-5p on the
invasive ability of ESCC cells, we detected the expression levels
of EMT-related molecular markers in ESCC cells. We found that
miR-125a-5p overexpression led to E-cadherin upregulation, and the
downregulation of N-cadherin and Vimentin in the EC1 and TE1 cells,
whereas miR-125a-5p downregulation decreased the E-cadherin level,
and enhanced the levels of N-cadherin and Vimentin, compared with
the control group and NC group (Fig.
3E and F), suggesting miR-125a-5p inhibits the EMT process in
ESCC. These findings indicate that miR-125a-5p mediates the
suppression of the migration and invasion of ESCC cells via the
inhibition of the EMT process.
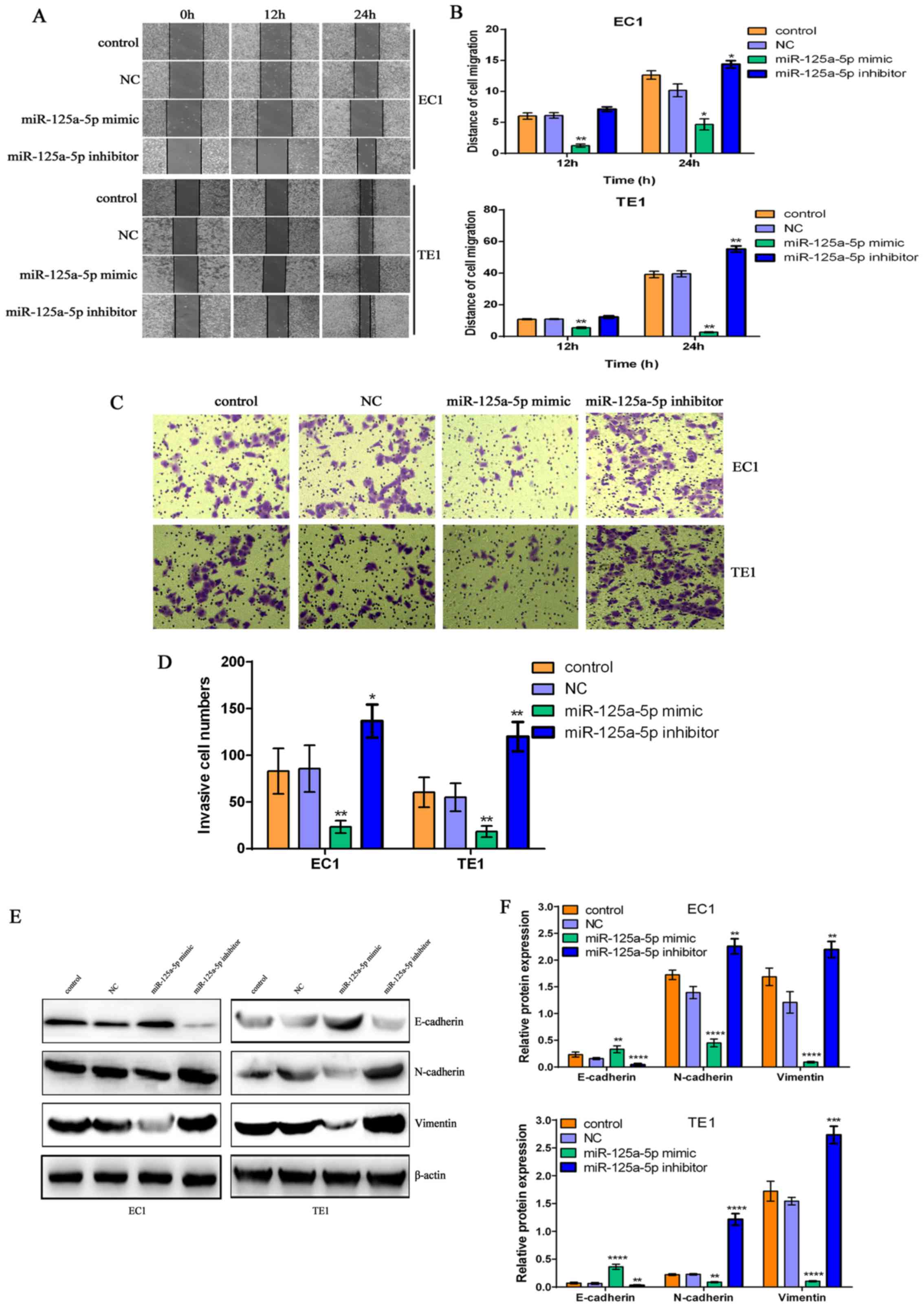 | Figure 3The miR-125a-5p-mediated changes in
the migratory and invasive abilities of esophageal squamous cell
carcinoma (ESCC) cells are tightly associated with the
epithelial-mesenchymal transition (EMT) process. (A) miR-125a-5p
overexpression or downregulation evidently reduced or promoted the
migratory ability of EC1 and TE1 ESCC cells, respectively. (B)
Migration distance was counted to evaluate the migratory ability of
EC1 and TE1 cells; *P<0.05 and
**P<0.01, compared with the control group and NC
group. (C) miR-125a-5p overexpression or downregulation markedly
suppressed or enhanced the invasive ability of EC1 and TE1 ESCC
cells. (D) Statistical analysis of the invasive numbers of EC1 and
TE1 ESCC cells; *P<0.05 and **P<0.01,
compared with the control group and NC group. (E) miR-125a-5p
overexpression suppressed the protein expression levels of
N-cadherin and Vimentin, and promoted E-cadherin protein
expression; miR-125a-5p downregulation promoted the expression
levels of N-cadherin and Vimentin proteins, and suppressed
E-cadherin protein expression in EC1 and TE1 ESCC cells. Western
blot analysis was utilized to investigate the expression levels of
the EMT-related molecular markers, E-cadherin, N-cadherin and
Vimentin, and β-actin was used as a loading control. (F) Relative
levels of E-cadherin, N-cadherin and Vimentin in ESCC EC1 and TE1
cells; **P<0.01, ***P<0.001 and
****P<0.0001, compared with the control group and NC
group. |
miR-125a-5p upregulation enhances the
cytotoxic effects of cisplatin, induces cell apoptosis and reduces
the migratory and invasive abilities of ESCC cells
Chemoresistance is the main cause of tumor treatment
failure, and aberrant miRNA levels are closely linked to
chemosensitivity and chemoresistance in a wide range of tumors. To
further investigate whether miR-125a-5p is tightly implicated in
chemosensitivity and chemoresistance in ESCC cells, in this study,
we investigated the effects of various concentrations of cisplatin
combined with NC or miR-125a-5p transfection on the proliferation,
apoptosis, migration and invasion of ESCC cells. We found that
miR-125a-5p significantly enhanced the cytotoxic effects of
cisplatin on EC1 and TE1 cells, compared with NC group treated with
cisplatin (Fig. 4A). Further
analysis revealed that transfection with miR-125a-5p mimic alone or
treatment with cisplatin alone markedly induced cell apoptosis and
reduced the migratory and invasive abilities of the EC1 and TE1
cells, compared with the NC group (P<0.05) (Fig. 4B–G). However, miR-125a-5p in
combination with cisplatin was the most effective in the induction
of cell apoptosis and the decrease in the migratory and invasive
abilities of the EC1 and TE1 cells (Fig. 4B–G). To further elucidate the
underlying mechanisms of the combined effects of miR-125a-5p and
cisplatin, we further examined the levels of the EMT-related
proteins, E-cadherin, N-cadherin and Vimentin. We found that
transfection with miR-125a-5p alone, and treatment with cisplatin
alone or their combination significantly promoted E-cadherin
expression, and suppressed the expression levels of N-cadherin and
vimentin, compared with the NC group (P<0.05) in the EC1 and TE1
cells. However, the combined use of miR-125a-5p and cisplatin was
the most effective in suppressing the EMT process (Fig. 4H and I). These data indicate that
miR-125a-5p plays a pivotal role in enhancing the
cisplatin-mediated chemo-sensitivity of ESCC cells via suppressing
the EMT process.
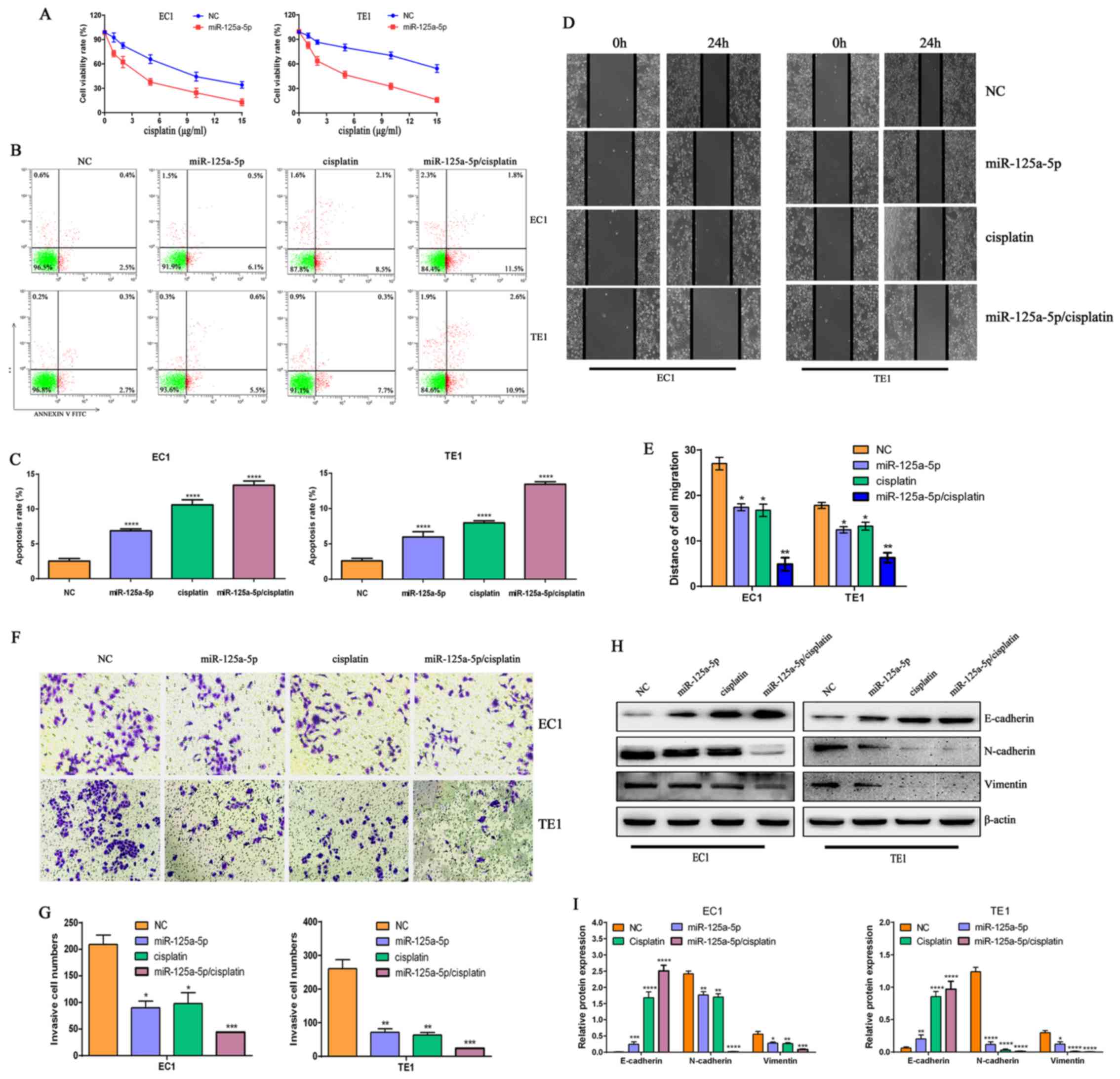 | Figure 4miR-125a-5p enhances the sensitivity
of esophageal squamous cell carcinoma (ESCC) cells to cisplatin.
(A) miR-125a-5p increased the cell-killing effects of cisplatin on
EC1 and TE1 ESCC cells. (B) miR-125a-5p combined with cisplatin
significantly promoted the apoptosis of EC1 and TE1 ESCC cells. (C)
Statistical analysis of the numbers of apoptotic EC1 and TE1 ESCC
cells subjected to the different treatments;
****P<0.0001, compared with NC group. (D) miR-125a-5p
in combination with cisplatin significantly inhibited the migratory
ability of EC1 and TE1 ESCC cells. (E) Migration distance was
counted to evaluate the migratory ability of EC1 and TE1 cells;
*P<0.05 and **P<0.01, compared with the
NC group. (F) miR-125a-5p in combination with cisplatin
significantly inhibited the invasive ability of EC1 and TE1 ESCC
cells. (G) Statistical analysis of the invasive numbers of EC1 and
TE1 ESCC cells subjected to various treatments;
*P<0.05, **P<0.01 and
***P<0.001, compared with the NC group. (H)
miR-125a-5p in combination with cisplatin promoted E-cadherin
expression, and suppressed the expression levels of n-cadherin and
vimentin in EC1 and TE1 ESCC cells. Western blot analysis was
utilized to examine the expression levels of the EMT-related
molecular markers, E-cadherin, N-cadherin and Vimentin, and β-actin
was used as a loading control. (I) Relative levels of E-cadherin,
N-cadherin and Vimentin in EC1 and TE1 ESCC cells,
*P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001, compared
with the NC group. |
STAT3 is a direct target of miR-125a-5p
in ESCC
To clarify the possible molecular mechanisms of the
chemosensitivity triggered by miR-125a-5p in ESCC, we performed a
search for the potential target genes of miR-125a-5p using
TargetScan, miRanda and miRDB. We found that STAT3 was a potential
target gene of miR-125a-5p (Fig.
5A), and corresponding STAT3-3′-UTR-WT and STAT3-3′-UTR-MUT
plasmids were constructed (Fig.
5A). Subsequently, these vectors, along with NC or miR-125a-5p
were co-transfected into the EC1 and TE1 cells, and the luciferase
activity was determined by measuring the relative luciferase
intensity. We found that miR-125a-5p significantly decreased the
luciferase activity in the STAT3-3′UTR-WT group, but it did not
affect the luciferase activity in the cells in the STAT3-3′UTR-MUT
group (Fig. 5B and C), suggesting
that miR-125a-5p can directly bind to the 3′UTR region of STAT3. To
validate the results mentioned above, we further performed western
blot analysis to detect the expression levels of t-STAT3, p-STAT3
and its downstream target gene, VEGF, in the EC1 and TE1 cells. The
results demonstrated that miR-125a-5p overexpression significantly
decreased the protein levels of t-STAT3, p-STAT3 and VEGF in the
EC1 and TE1 cells, compared with the control and NC group
(P<0.01) (Fig. 5D–G). These
findings suggest that STAT3 is a direct target gene of miR-125a-5p,
and that miR-125a-5p suppresses the invasive ability of ESCC cells,
and enhances chemosensitivity and that these effects may be
mediated via the suppression of the activation of the STAT3
signaling pathway in ESCC.
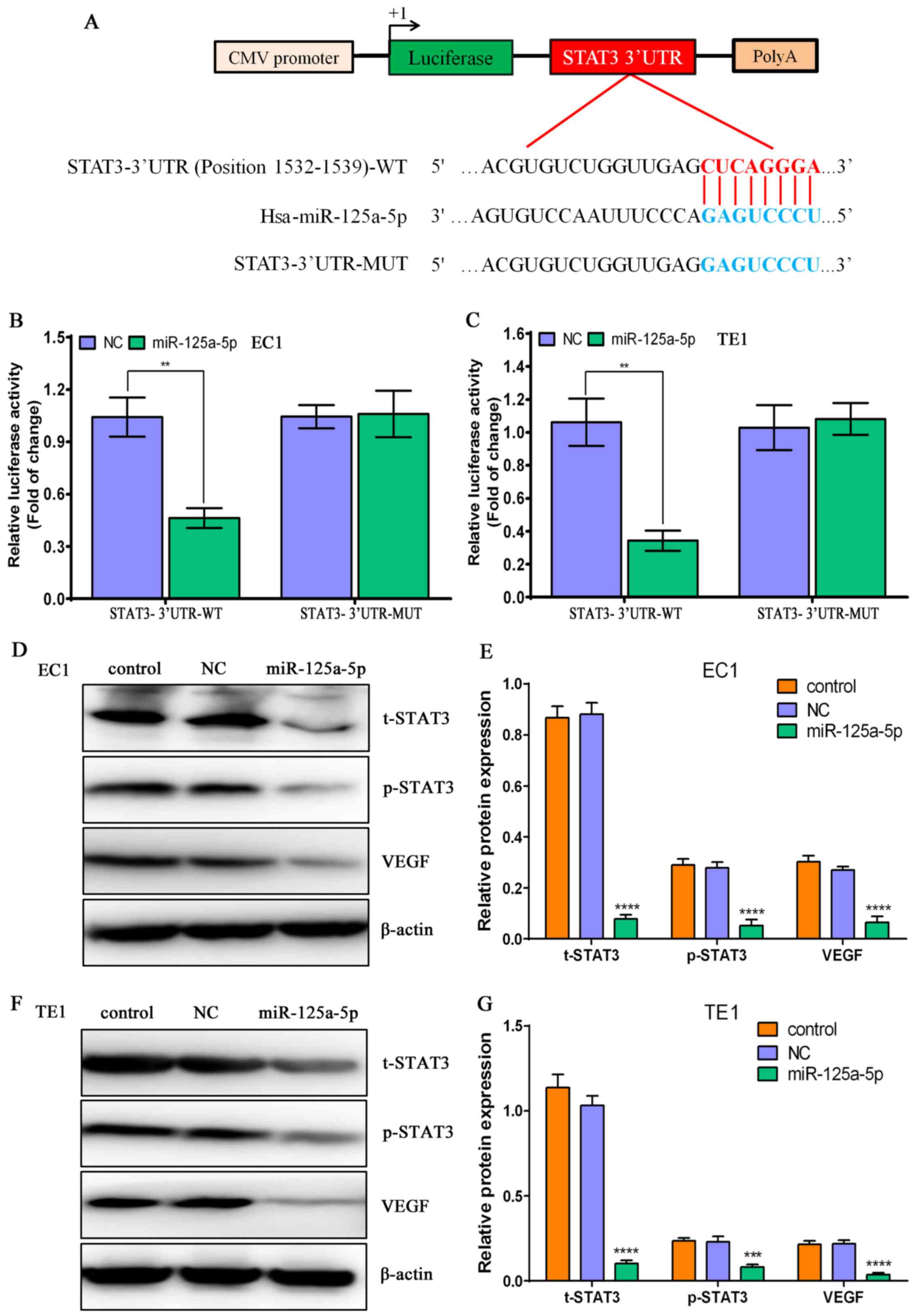 | Figure 5STAT3 is a direct target of
miR-125a-5p in esophageal squamous cell carcinoma (ESCC) cells. (A)
The signal transducer and activator of transcription-3 (STAT3)
3′-UTR sequences including wild-type (WT) or mutant (MUT) were
inserted into the downstream of luciferase reporter vector
according to diagrammatic presentation. (B) The luciferase activity
was determined by co-transfecting the vectors (STAT3 3′-UTR-WT and
MUT) combined with NC or miR-125a-5p mimic into EC1 ESCC cells;
**P<0.01, compared with the NC group. (C) The
luciferase activity was determined by co-transfecting the vectors
(STAT3 3′-UTR-WT and MUT) combined with NC or miR-125a-5p mimic
into TE1 ESCC cells; **P<0.01, compared with the NC
group. (D) Western blot analysis of the protein expression levels
of t-STAT3, p-STAT3 and vascular endothelial growth factor (VEGF)
in the EC1 ESCC cells subjected to various treatments, and β-actin
was used as a loading control. (E) Relative protein levels of
t-STAT3, p-STAT3 and VEGF in EC1 ESCC cells subjected to different
treatments; ****P<0.0001, compared with the control
group and NC group. (F) Western blot analysis of the protein
expression levels of t-STAT3, p-STAT3 and VEGF in TE1 ESCC cells
subjected to various treatments, and β-actin was used as a loading
control. (G) Relative protein levels of t-STAT3, p-STAT3 and VEGF
in TE1 ESCC cells subjected to various treatments;
***P<0.001 and ****P<0.0001, compared
with the control group and NC group. |
Combination of miR-125a-5p with cisplatin
inactivates the STAT3 signaling pathway in ESCC cells
To further explore the underlying mechanisms of the
antitumor effects mediated by miR-125a-5p in combination with
cisplatin in ESCC, we examined the protein expression levels of
t-STAT3, p-STAT3 and VEGF in ESCC cells subjected to different
treatments. We found that miR-125a-5p alone or cisplatin alone
significantly downregulated the protein levels of t-STAT3, p-STAT3
and vEGF, compared with the NC group (P<0.01) (Fig. 6A and B). However, the combination
of miR-125a-5p with cisplatin exerted the most prominent inhibitory
effect on the expression levels of these proteins (Fig. 6A and B). These findings suggest
that miR-125a-5p and cisplatin play a synergistic antitumor role in
ESCC via suppressing the activation of the STAT3 signaling
pathway.
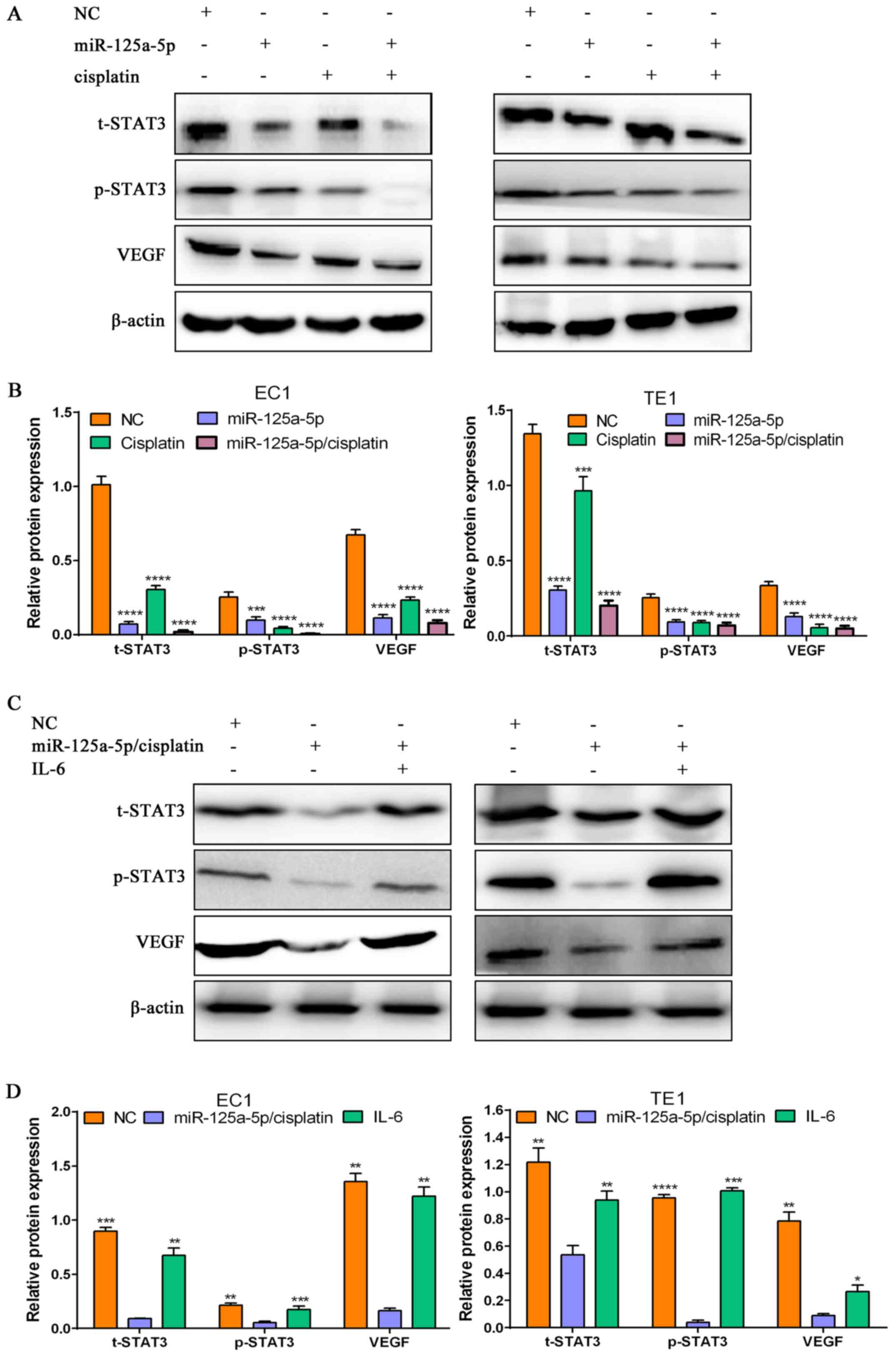 | Figure 6miR-125a-5p/cisplatin suppresses
signal transducer and activator of transcription-3 (STAT3)
activation and vascular endothelial growth factor (VEGF)
expression, and the STAT3 activator, interleukin (IL)-6
re-activates the STAT3 signaling pathway in esophageal squamous
cell carcinoma (ESCC) cells treated with miR-125a/cisplatin. (A)
Western blot analysis of the protein expression levels of t-STAT3,
p-STAT3 and VEGF in the NC group, miR-125a-5p alone group,
cisplatin alone group and miR-125a-5p/cisplatin combination group,
and β-actin was employed as a loading control. (B) The relative
protein levels of t-STAT3, p-STAT3 and VEGF in the NC group,
miR-125a-5p alone group, cisplatin alone group and
miR-125a-5p/cisplatin combination group; ***P<0.001
and ****P<0.0001, compared with the NC group. (C)
Western blot analysis of the protein expression levels of t-STAT3,
p-STAT3 and vEGF in the NC group, miR-125a-5p/cisplatin combination
group and miR-125a-5p/cisplatin/IL-6 combination group, and β-actin
was employed as a loading control. (D) The relative protein levels
of t-STAT3, p-STAT3 and VEGF in the NC group, miR-125a-5p/cisplatin
combination group and miR-125a-5p/cisplatin/IL-6 combination group;
*P<0.05, **P<0.01,
***P<0.001 and ****P<0.0001, compared
with the miR-125a-5p/cisplatin combination group. |
IL-6 attenuates the inhibitory effects of
miR-125a-5p combined with cisplatin on the activation of the STAT3
signaling pathway in ESCC cells
To determine whether the re-activation of the STAT3
signaling pathway evoked by IL-6 (a widely reported activator of
the STAT3 signaling pathway) can abolish the inactivation of the
STAT3 signaling pathway induced by miR-125a-5p in combination with
cisplatin in ESCC cells, western blot analysis was employed to
examine the activation status of the STAT3 signaling pathway in the
EC1 and TE1 cells. The results indicated that IL-6 markedly
increased the protein expression levels of t-STAT3, p-STAT3 and
VEGF, compared with the miR-125a-5p/cisplatin group (P<0.01) in
the EC1 and TE1 cells (Fig. 6C and
D), suggesting that IL-6 can re-activate the STAT3 signaling
pathway in ESCC cells treated with a combination of miR-125a-5p and
cisplatin.
STAT3 activation evoked by IL-6
attenuates the suppressive effects mediated by
miR-125a-5p/cisplatin on the proliferation and invasion of ESCC
cells and also attenuates the pro-apoptotic effects
To further determine whether STAT3 activation can
partially recover the phenotype of ESCC cells, which had been
altered by miR-125a-5p/cisplatin, IL-6 was used to treat the ESCC
cells, and CCK-8, flow cytometry and Transwell chamber assay were
used to examine the status of proliferation, apoptosis and invasion
of EC1 and TE1 cells, respectively. We found that
miR-125a-5p/cisplatin/iL-6 treatment significantly increased the
viability of the EC1 and TE1 cells at 24, 48 and 72 h, compared
with the miR-125a-5p/cisplatin group (Fig. 7A). Cell apoptosis assay revealed
that miR-125a-5p/cisplatin/IL-6 treatment markedly decreased the
apoptosis of the EC1 and TE1 cells, compared with the
miR-125a-5p/cisplatin group (P<0.01) (Fig. 7B and C). Notably, we found that
miR-125a-5p/cisplatin/IL-6 treatment evidently restored the
invasive ability of the EC1 and TE1 cells, compared with the
miR-125a-5p/cisplatin group (P<0.05) (Fig. 7D and E). These data suggest that
miR-125a-5p/cisplatin exerts antitumor effects by inhibiting the
activation of the STAT3 signaling pathway.
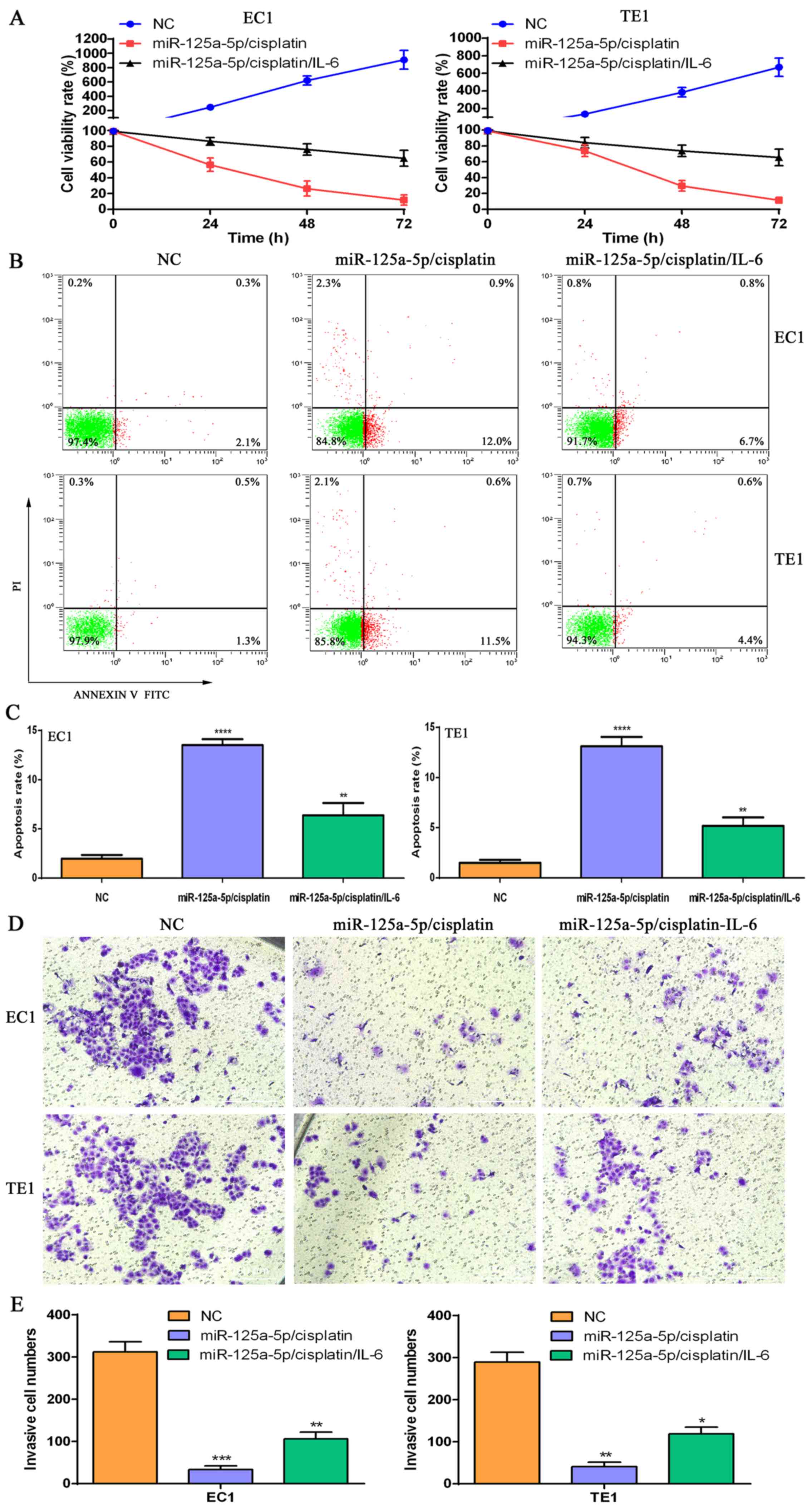 | Figure 7The signal transducer and activator
of transcription-3 (STAT3) activator, interleukin (IL)-6 reverses
the alteration in the cell phenotype mediated by
miR-125a-5p/cisplatin in esophageal squamous cell carcinoma (ESCC)
cells. (A) CCK-8 kit assay for cell proliferation in the NC group,
miR-125a-5p/cisplatin combination group and
miR-125a-5p/cisplatin/IL-6 combination group. (B) Flow cytometry of
cell apoptosis in the NC group, miR-125a-5p/cisplatin combination
group and miR-125a-5p/cisplatin/IL-6 combination group. (C)
Statistical analysis of the apoptotic cell numbers in the NC group,
miR-125a-5p/cisplatin combination group and
miR-125a-5p/cisplatin/IL-6 combination group;
**P<0.01 and ****P<0.0001, compared
with the NC group. (D) Transwell chamber assay for cell invasive
ability in the NC group, miR-125a-5p/cisplatin combination group
and miR-125a-5p/cisplatin/IL-6 combination group. (E) Statistical
analysis of the invasive cell numbers in the NC group,
miR-125a-5p/cisplatin combination group and
miR-125a-5p/cisplatin/IL-6 combination group,
*P<0.05, **P<0.01 and
***P<0.001, compared with the NC group. |
Discussion
Mounting evidence has demonstrated that miRNAs
function as either oncogenes or tumor suppressor genes in various
type tumors, which may bring forth new challenges or may open up
novel opportunities for the use of miRNAs as novel molecular
targets for a myriad of tumors (48,49).
Moreover, miRNAs have been verified as a therapeutic tool in the
management of pancreatic adenocarcinoma in clinical studies
(50,51), suggesting that miRNAs have
important clinical value in many different types of tumors.
Therefore, the identification of key miRNA molecules implicated in
the development and progression of ESCC may provide new diagnostic
and prognostic markers, and may aid in the development of more
effective treatment strategies for patients with ESCC. In the
current study, we found that miR-125a-5p was downregulated in ESCC
tissues and cells, implying that miR-125a-5p may function as a
tumor suppressor in ESCC. Further analysis revealed that a
decreased miR-125a-5p expression was tightly associated with a
higher tumor staging and a lower survival rate of patients with
ESCC, suggesting that miR-125a-5p participates in tumor development
and progression; thus, miR-125a-5p may be a molecular marker for
the malignant degree and prognosis of patients with ESCC.
There is strong evidence that miRNAs are tightly
implicated in a mass of complex regulatory networks of crucial
genes in a variety of tumors (52-54).
It has been indicated that target genes of miRNAs are direct
regulators of the hallmarks of cancer, including proliferation,
cell cycle and cell apoptosis (52). Tao et al found that a
decreased miR-125a expression in osteosarcoma tissues, and its
overexpression contributed to growth suppression in osteosarcoma
cells by downregulating E2F2 expression (55). Moreover, miR-125a-5p overexpression
has been shown to markedly inhibit cell proliferation and tumor
formation in retinoblastoma, exerting antitumor effects by
suppressing the transcriptional co-activator with PDZ binding motif
(TAZ) (36), a critical downstream
component of the Hippo signaling pathway; TAZ overexpression has
been shown to markedly accelerate tumor initiation and progression
(56,57). Qin et al confirmed that
miR-125a-5p overexpression significantly suppressed the
proliferation and migratory ability of cervical cancer cells
through the direct targeted inhibition of ABL2 expression (37). Converse results from leukemia have
revealed that miR-125a overexpression induces daunorubicin
resistance by suppressing the apoptosis of HL-60, K562 and THP-1
cells, which was further verified to be directly correlated with
the downregulation of GRK2 and Puma (58). These findings fully highlighted the
essential role of miR-125a-5p in the regulation of proliferation
and apoptosis in a plethora of tumors. In this study, we found that
miR-125a-5p overexpression significantly inhibited the
proliferation, arrested the cell cycle in the G0/G1 phase and
induced the apoptosis of ESCC cells, whereas miR-125a-5p
downregulation markedly promoted the proliferation and cell cycle
progression, and inhibited the apoptosis of ESCC cells. These
findings suggest that miR-125a-5p may be an important regulator of
cell proliferation, cell cycle and apoptosis in ESCC, and thus the
manipulation of miR-125a-5p may be a novel molecular target for
ESCC.
Understanding the possible mechanisms of tumor
invasion and metastasis remains a formidable challenge for a large
number of tumors. Several studies have focused on the involvement
of miRNAs in tumor invasion and metastasis (59–62).
The EMT process has been reported to be involved in tumor
progression and metastasis, which may be a pivotal mechanism of
tumor invasion and metastasis (63,64).
Recently, increasing evidence has demonstrated that miRNAs
implicated in tumor invasion and metastasis are tightly associated
with the EMT process (65–68). To further interpret the possible
role of miR-125a-5p in invasion and metastasis of ESCC, we further
examined the alterations in the migratory and invasive abilities of
ESCC cells triggered by miRNA-125a-5p. We found that miR-125a-5p
overexpression significantly reduced the migratory and invasive
abilities of ESCC cells, whereas its downregulation promoted the
migratory and invasive abilities of ESCC cells. Further analysis
revealed that transfection with miR-125a-5p mimic markedly
upregulated the protein expression levels of N-cadherin and
Vimentin, and downregulated the E-cadherin protein level in ESCC,
and converse results were observed in the miR-125a-5p inhibitor
group, implying that the involvement of miR-125a-5p in tumor
invasion and metastasis may be partly achieved through the
modulation of EMT-related signaling pathways. These findings
potentiate miR-125a-5p as a potential predictor for the invasion
and metastasis of ESCC.
Chemoresistance is a major hurdle in the treatment
of many tumor patients. Recent studies have revealed that aberrant
miRNA levels are tightly implicated in chemoresistance or
chemosensitivity in a host of tumors (69–74),
suggesting that targeting miRNAs to eradicate chemoresistance or
improve chemosensitivity may be a novel therapeutic strategy for
the therapy of tumor patients. Nishida et al found that
miR-125a-5p evidently suppressed cell proliferation via targeting
ERBB2, and miR-125a-5p markedly enhanced the sensitivity of gastric
carcinoma cells to trastuzumab (75). Another study revealed that miR-200a
increased the chemoresistance of breast cancer cells to
chemotherapeutic agents; by contrast, miR-200a down-regulation
promoted the sensitivity of resistant cancer cells to gemcitabine
(70). Furthermore, the
re-introduction of miR-31 has been shown to significantly promote
clonogenic resistance to cisplatin and carboplatin in NCI-H2452
cells without miR-31 expression (76). Another study revealed that miR-125a
overexpression enhanced the sensitivity of paclitaxel on
paclitaxel-resistant colon cancer cells (38). These findings highlight the crucial
role of miRNAs in potentiating chemosensitivity in the process of
tumor therapy. However, whether miR-125a-5p is implicated in the
chemosensitivity of ESCC cells remains unknown. In this study, as
expected, miR-125a-5p significantly elevated the killing efficacy
of cisplatin on ESCC cells, and miR-125a-5p/cisplatin significantly
promoted the apoptosis and reduced the migratory and invasive
abilities of ESCC cells. Further analysis revealed that
co-treatment with miR-125a-5p and cisplatin markedly increased the
E-cadherin level and reduced the levels of N-cadherin and Vimentin,
suggesting that the miR-125a-5p-mediated enhancement of the
cisplatin sensitivity of ESCC cells may be tightly associated with
the suppression of the EMT process. The data presented herein
suggest that miR-125a-5p markedly improved the therapeutic efficacy
of cisplatin in ESCC, and thus miR-125a-5p may be a novel ancillary
drug of cisplatin for use in the treatment of patients with ESCC in
the future.
Several studies have demonstrated that the STAT3
signaling pathway is implicated in drug resistance in a variety of
tumors (77,78), and its activation not only promotes
tumor growth rapidly, but also imparts therapeutic resistance in
cancer cells (77,79–81).
In this study, we determined whether the involvement of
miRNA-125a-5p in the sensitivity of ESCC cells to cisplatin may be
tightly associated with the activated status of the STAT3 signaling
pathway. Therefore, we performed a search for the potential target
genes of miR-125a-5p using TargetScan, miRanda and miRDB, and found
that STAT3 was the direct target gene of miR-125a-5p by luciferase
reporter assay. Further analysis demonstrated that miR-125a-5p
over-expression significantly reduced the protein levels of
t-STAT3, p-STAT3 and its downstream target gene, VEGF, ESCC cells.
These findings suggest that the miR-125a-5p-mediated
chemo-sensitivity may be tightly associated with the inactivation
of the STAT3 signaling pathway in ESCC. Correspondingly, whether
the re-activation of the STAT3 signaling pathway can reverse the
chemosensitivity mediated by miR-125a-5p overexpression in ESCC,
was examined. IL-6 [an activator of the STAT3 signaling pathway
(39)] was used to re-activate the
the STAT3 signaling pathway in ESCC cells. We found that iL-6
significantly recovered the t-STAT3 and p-STAT3 levels which were
suppressed by miR-125a-5p/cisplatin in the ESCC cells, and further
analysis revealed that the re-activation of the STAT3 signaling
pathway evoked by iL-6 significantly recovered cell viability,
decreased cell apoptosis and promoted the invasion of ECSS cells
co-treated with miR-125a-5p and cisplatin. The data presented
herein suggest that miR-125a-5p enhances the sensitivity of ESCC
cells to cisplatin via suppressing the activation of the STAT3
signaling pathway, which may be an underlying molecular mechanism
through which miR-125a-5p exerts antitumor effects on ESCC.
In conclusion, the data from the present study
suggest that a low level of miR-125a-5p is tightly associated with
a higher tumor staging and a poor prognosis of patients with ESCC.
The overexpression of miR-125a-5p significantly suppressed the
proliferation, arrested the cell cycle in G0/G1 phase, induced the
apoptosis and reduced the migratory and invasive abilities of ESCC
cells, and converse results were observed in miR-125a-5p inhibitor
group, which may be closely associated with the EMT process.
Further analysis indicated that miR-125a-5p enhanced the
sensitivity of ESCC cells to cispl-atin, which may be achieved by
the inactivation of the STAT3 signaling pathway. Most importantly,
the re-activation of the STAT3 signaling pathway triggered by IL-6
prominently led to a recovery of the viability, decreased cell
apoptosis and an increased cell invasive ability of ESCC cells. The
data presented herein may provide a novel therapeutic strategy for
the therapy of patients with ESCC by the combined use of
miR-125a-5p with cisplatin.
Funding
The present study was supported by the National
Natural Science Foundation of China (no. 81272691) and the Key
Scientific Research Projects of Henan Higher Education institutions
(no. 17A180016).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
QF supervised the whole project. QF, YZ and KM
designed the study. YZ, KM and SY performed the majority of the
experiments; XZ contributed to plasmid construction and luciferase
reporter assay; FW performed the CCK-8 experiment; XZ and HL
participated in the design and interpretation of some of the
experiments; YZ and QF interpreted all the results and wrote the
manuscript.
Ethics approval and consent to
participate
This study was approved by the Institutional
Research Ethics Committee of Zhengzhou University. All samples were
obtained with informal written and none of the patients had
received any treatments prior to surgery.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar
|
|
2
|
Conteduca V, Sansonno D, Ingravallo G,
Marangi S, Russi S, Lauletta G and Dammacco F: Barrett’s esophagus
and esophageal cancer: An overview. Int J Oncol. 41:414–424. 2012.
View Article : Google Scholar
|
|
3
|
Arnold M, Soerjomataram I, Ferlay J and
Forman D: Global incidence of oesophageal cancer by histological
subtype in 2012. Gut. 64:381–387. 2015. View Article : Google Scholar
|
|
4
|
Berger AC, Farma J, Scott WJ, Freedman G,
Weiner L, Cheng JD, Wang H and Goldberg M: Complete response to
neoadjuvant chemoradiotherapy in esophageal carcinoma is associated
with significantly improved survival. J Clin Oncol. 23:4330–4337.
2005. View Article : Google Scholar
|
|
5
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
6
|
Law S, Kwong DL, Kwok KF, Wong KH, Chu KM,
Sham JS and Wong J: Improvement in treatment results and long-term
survival of patients with esophageal cancer: Impact of
chemoradiation and change in treatment strategy. Ann Surg.
238:339–3488. 2003.
|
|
7
|
Sjoquist KM, Burmeister BH, Smithers BM,
Zalcberg JR, Simes RJ, Barbour A and Gebski V; Australasian
Gastro-Intestinal Trials Group: Survival after neoadjuvant
chemotherapy or chemoradiotherapy for resectable oesophageal
carcinoma: An updated meta-analysis. Lancet Oncol. 12:681–692.
2011. View Article : Google Scholar
|
|
8
|
Pennathur A, Gibson MK, Jobe BA and
Luketich JD: Oesophageal carcinoma. Lancet. 381:400–412. 2013.
View Article : Google Scholar
|
|
9
|
Hong L, Han Y, Zhang H and Fan D:
Prognostic markers in esophageal cancer: From basic research to
clinical use. Expert Rev Gastroenterol Hepatol. 9:887–889. 2015.
View Article : Google Scholar
|
|
10
|
Haenisch S and Cascorbi I: miRNAs as
mediators of drug resistance. Epigenomics. 4:369–381. 2012.
View Article : Google Scholar
|
|
11
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar
|
|
12
|
Bartel DP: MicroRnAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar
|
|
13
|
Gurtan AM and Sharp PA: The role of miRnAs
in regulating gene expression networks. J Mol Biol. 425:3582–3600.
2013. View Article : Google Scholar
|
|
14
|
Ameres SL and Zamore PD: Diversifying
microRNA sequence and function. Nat Rev Mol Cell Biol. 14:475–488.
2013. View
Article : Google Scholar
|
|
15
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar
|
|
16
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar
|
|
17
|
Tiscornia G and Izpisúa Belmonte JC:
MicroRNAs in embryonic stem cell function and fate. Genes Dev.
24:2732–2741. 2010. View Article : Google Scholar
|
|
18
|
Inui M, Martello G and Piccolo S: MicroRNA
control of signal transduction. Nat Rev Mol Cell Biol. 11:252–263.
2010. View
Article : Google Scholar
|
|
19
|
Ping W, Gao Y, Fan X, Li W, Deng Y and Fu
X: MiR-181a contributes gefitinib resistance in non-small cell lung
cancer cells by targeting GAS7. Biochem Biophys Res Commun.
495:2482–2489. 2018. View Article : Google Scholar
|
|
20
|
Gartel AL and Kandel ES: miRNAs: Little
known mediators of oncogenesis. Semin Cancer Biol. 18:103–110.
2008. View Article : Google Scholar
|
|
21
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar
|
|
22
|
Negrini M, Nicoloso MS and Calin GA:
MicroRNAs and cancer–new paradigms in molecular oncology. Curr Opin
Cell Biol. 21:470–479. 2009. View Article : Google Scholar
|
|
23
|
Spizzo R, Nicoloso MS, Croce CM and Calin
GA: SnapShot: microRNAs in cancer. Cell. 137:586–586.e581. 2009.
View Article : Google Scholar
|
|
24
|
Voorhoeve PM: MicroRNAs: Oncogenes, tumor
suppressors or master regulators of cancer heterogeneity. Biochim
Biophys Acta. 1805.72–86. 2010.
|
|
25
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar
|
|
26
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
27
|
Hammond SM: MicroRNAs as tumor
suppressors. Nat Genet. 39:582–583. 2007. View Article : Google Scholar
|
|
28
|
Baraniskin A, Chomiak M, Ahle G, Gress T,
Buchholz M, Turewicz M, Eisenacher M, Margold M, Schlegel U and
Schmiegel W: MicroRNA-30c as a novel diagnostic biomarker for
primary and secondary B-cell lymphoma of the CNS. J Neurooncol.
137:463–468. 2018. View Article : Google Scholar
|
|
29
|
Li C, Zheng X, Li W, Bai F, Lyu J and Meng
QH: Serum miR-486-5p as a diagnostic marker in cervical cancer:
With investigation of potential mechanisms. BMC Cancer. 18:612018.
View Article : Google Scholar
|
|
30
|
Yao XD, Li P and Wang JS: MicroRNA
differential expression spectrum and microRNA-125a-5p inhibition of
laryngeal cancer cell proliferation. Exp Ther Med. 14:1699–1705.
2017. View Article : Google Scholar
|
|
31
|
Li G, Zhang W, Gong L and Huang X:
MicroRNA-125a-5p inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma by
downregulation of ErbB3. Oncol Res. 2017.
|
|
32
|
Coppola N, de Stefano G, Panella M,
Onorato L, Iodice V, Minichini C, Mosca N, Desiato L, Farella N,
Starace M, et al: Lowered expression of microRNA-125a-5p in human
hepatocellular carcinoma and up-regulation of its oncogenic targets
sirtuin-7, matrix metalloproteinase-11, and c-Raf. Oncotarget.
8:25289–25299. 2017. View Article : Google Scholar
|
|
33
|
Potenza N, Mosca N, Zappavigna S,
Castiello F, Panella M, Ferri C, Vanacore D, Giordano A, Stiuso P,
Caraglia M, et al: MicroRNA-125a-5p is a downstream effector of
sorafenib in its antiproliferative activity toward human
hepatocellular carcinoma cells. J Cell Physiol. 232:1907–1913.
2017. View Article : Google Scholar
|
|
34
|
Zhong L, Sun S, Shi J, Cao F, Han X and
Chen Z: MicroRNA-125a-5p plays a role as a tumor suppressor in lung
carcinoma cells by directly targeting STAT3. Tumour Biol.
39:10104283176975792017. View Article : Google Scholar
|
|
35
|
Fu Y and Cao F: MicroRNA-125a-5p regulates
cancer cell proliferation and migration through NAIF1 in prostate
carcinoma. OncoTargets Ther. 8:3827–3835. 2015. View Article : Google Scholar
|
|
36
|
Zhang Y, Xue C, Zhu X, Zhu X, Xian H and
Huang Z: Suppression of microRNA-125a-5p upregulates the TAZ-EGFR
signaling pathway and promotes retinoblastoma proliferation. Cell
Signal. 28:850–860. 2016. View Article : Google Scholar
|
|
37
|
Qin X, Wan Y, Wang S and Xue M:
MicroRNA-125a-5p modulates human cervical carcinoma proliferation
and migration by targeting ABL2. Drug Des Devel Ther. 10:71–79.
2015.
|
|
38
|
Chen J, Chen Y and Chen Z: MiR-125a/b
regulates the activation of cancer stem cells in
paclitaxel-resistant colon cancer. Cancer Invest. 31:17–23. 2013.
View Article : Google Scholar
|
|
39
|
Kishimoto T: IL-6: From its discovery to
clinical applications. Int Immunol. 22:347–352. 2010. View Article : Google Scholar
|
|
40
|
Lu Z, Liu H, Xue L, Xu P, Gong T and Hou
G: An activated Notch1 signaling pathway inhibits cell
proliferation and induces apoptosis in human esophageal squamous
cell carcinoma cell line EC9706. Int J Oncol. 32:643–651. 2008.
|
|
41
|
Zafar S, Coates DE, Cullinan MP, Drummond
BK, Milne T and Seymour GJ: Effects of zoledronic acid and
geranylgeraniol on the cellular behaviour and gene expression of
primary human alveolar osteoblasts. Clin Oral Investig.
20:2023–2035. 2016. View Article : Google Scholar
|
|
42
|
Nana-Sinkam SP and Croce CM: MicroRNA
regulation of tumorigenesis, cancer progression and interpatient
heterogeneity: Towards clinical use. Genome Biol. 15:4452014.
View Article : Google Scholar
|
|
43
|
Wang X, Ivan M and Hawkins SM: The role of
MicroRNA molecules and MicroRNA-regulating machinery in the
pathogenesis and progression of epithelial ovarian cancer. Gynecol
Oncol. 147:481–487. 2017. View Article : Google Scholar
|
|
44
|
Kim J, Yao F, Xiao Z, Sun Y and Ma L:
MicroRNAs and metastasis: Small RNAs play big roles. Cancer
Metastasis Rev. 37:5–15. 2018. View Article : Google Scholar
|
|
45
|
Pastorkova Z, Skarda J and Andel J: The
role of microRNA in metastatic processes of non-small cell lung
carcinoma. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub.
160:343–357. 2016. View Article : Google Scholar
|
|
46
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar
|
|
47
|
Giannelli G, Koudelkova P, Dituri F and
Mikulits W: Role of epithelial to mesenchymal transition in
hepatocellular carcinoma. J Hepatol. 65:798–808. 2016. View Article : Google Scholar
|
|
48
|
Dorrance AM, Neviani P, Ferenchak GJ,
Huang X, Nicolet D, Maharry KS, Ozer HG, Hoellarbauer P, Khalife J
and Hill EB: Targeting leukemia stem cells in vivo with
antagomiR-126 nanoparticles in acute myeloid leukemia. Leukemia.
29:2143–2153. 2015. View Article : Google Scholar
|
|
49
|
Jiang X, Bugno J, Hu C, yang Y, Herold T,
Qi J, Chen P, Gurbuxani S, Arnovitz S, Strong J, et al: Eradication
of acute myeloid leukemia with FLT3 ligand-targeted miR-150
nanoparticles. Cancer Res. 76:4470–4480. 2016. View Article : Google Scholar
|
|
50
|
Keklikoglou I, Hosaka K, Bender C, Bott A,
Koerner C, Mitra D, Will R, Woerner A, Muenstermann E and Wilhelm
H: MicroRNA-206 functions as a pleiotropic modulator of cell
proliferation, invasion and lymphangiogenesis in pancreatic
adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene.
34:4867–4878. 2015. View Article : Google Scholar
|
|
51
|
Papaconstantinou IG, Lykoudis PM, Gazouli
M, Manta A, Polymeneas G and voros D: A review on the role of
microRNA in biology, diagnosis, and treatment of pancreatic
adenocarcinoma. Pancreas. 41:671–677. 2012. View Article : Google Scholar
|
|
52
|
Robb T, Reid G and Blenkiron C: Exploiting
microRNAs as cancer therapeutics. Target Oncol. 12:163–178. 2017.
View Article : Google Scholar
|
|
53
|
Li Z, Peng Z, Gu S, Zheng J, Feng D, Qin Q
and He J: Global analysis of miRNA-mRNA interaction network in
breast cancer with brain metastasis. Anticancer Res. 37:4455–4468.
2017.
|
|
54
|
Cora’ D, Re A, Caselle M and Bussolino F:
MicroRNA-mediated regulatory circuits: Outlook and perspectives.
Phys Biol. 14:0450012017. View Article : Google Scholar
|
|
55
|
Tao T, Shen Q, Luo J, Xu Y and Liang W:
MicroRNA-125a regulates cell proliferation via directly targeting
E2F2 in osteosarcoma. Cell Physiol Biochem. 43:768–774. 2017.
View Article : Google Scholar
|
|
56
|
Hsu YL, Hung JY, Chou SH, Huang MS, Tsai
MJ, Lin YS, Chiang Sy, Ho YW, Wu CY and Kuo PL: Angiomotin
decreases lung cancer progression by sequestering oncogenic yAP/TAZ
and decreasing Cyr61 expression. Oncogene. 34:4056–4068. 2015.
View Article : Google Scholar
|
|
57
|
Brusgard JL, Choe M, Chumsri S, Renoud K,
MacKerell AD Jr, Sudol M and Passaniti A: RUnX2 and TAZ-dependent
signaling pathways regulate soluble E-Cadherin levels and
tumorsphere formation in breast cancer cells. Oncotarget.
6:28132–28150. 2015. View Article : Google Scholar
|
|
58
|
Bai H, Zhou L, Wang C, Xu X, Jiang J, Qin
Y, Wang X, Zhao C and Shao S: Involvement of miR-125a in resistance
to daunorubicin by inhibiting apoptosis in leukemia cell lines.
Tumour Biol. 39:10104283176959642017. View Article : Google Scholar
|
|
59
|
Palma Flores C, García-Vázquez R, Gallardo
Rincón D, Ruiz-García E, Astudillo de la Vega H, Marchat LA,
Salinas Vera YM and López-Camarillo C: MicroRnAs driving invasion
and metastasis in ovarian cancer: Opportunities for translational
medicine (Review). Int J Oncol. 50:1461–1476. 2017. View Article : Google Scholar
|
|
60
|
Verma V and Lautenschlaeger T: MicroRNAs
in non-small cell lung cancer invasion and metastasis: From the
perspective of the radiation oncologist. Expert Rev Anticancer
Ther. 16:767–774. 2016. View Article : Google Scholar
|
|
61
|
Chan SH and Wang LH: Regulation of cancer
metastasis by microRNAs. J Biomed Sci. 22:92015. View Article : Google Scholar
|
|
62
|
Zhao X, Li X and Yuan H: microRNAs in
gastric cancer invasion and metastasis. Front Biosci. 18:803–810.
2013. View Article : Google Scholar
|
|
63
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar
|
|
64
|
Kalluri R and Weinberg RA: The basics of
epithelial-mesenchymal transition. J Clin Invest. 119:1420–1428.
2009. View Article : Google Scholar
|
|
65
|
Chen W, Kong KK, Xu XK, Chen C, Li H, Wang
FY, Peng XF, Zhang Z, Li P and Li JL: Downregulation of miR-205 is
associated with glioblastoma cell migration, invasion, and the
epithelial-mesenchymal transition, by targeting ZEB1 via the
Akt/mTOR signaling pathway. Int J Oncol. 52:485–495. 2018.
|
|
66
|
Chen C, Yang Q, Wang D, Luo F, Liu X, Xue
J, yang P, Xu H, Lu J, Zhang A, et al: MicroRNA-191, regulated by
HIF-2α, is involved in EMT and acquisition of a stem cell-like
phenotype in arsenite-transformed human liver epithelial cells.
Toxicol In Vitro. 48:128–136. 2018. View Article : Google Scholar
|
|
67
|
Huang J, He Y, Mcleod HL, Xie Y, Xiao D,
Hu H, Chen P, Shen L, Zeng S and Yin X: miR-302b inhibits
tumorigenesis by targeting EphA2 via Wnt/β-catenin/EMT signaling
cascade in gastric cancer. BMC Cancer. 17:8862017. View Article : Google Scholar
|
|
68
|
Xu X, Cao L, Zhang Y, Lian H, Sun Z and
Cui Y: MicroRNA-1246 inhibits cell invasion and epithelial
mesenchymal transition process by targeting CXCR4 in lung cancer
cells. Cancer Biomark. 21:251–260. 2018. View Article : Google Scholar
|
|
69
|
Berman M, Mattheolabakis G, Suresh M and
Amiji M: Reversing epigenetic mechanisms of drug resistance in
solid tumors using targeted microRNA delivery. Expert Opin Drug
Deliv. 13:987–998. 2016. View Article : Google Scholar
|
|
70
|
Yu SJ, Yang L, Hong Q, Kuang XY, Di GH and
Shao ZM: MicroRnA-200a confers chemoresistance by antagonizing
TP53inP1 and yAP1 in human breast cancer. BMC Cancer. 18:742018.
View Article : Google Scholar
|
|
71
|
Xiong J, Wang D, Wei A, Ke N, Wang Y, Tang
J, He S, Hu W and Liu X: MicroRNA-410-3p attenuates gemcitabine
resistance in pancreatic ductal adenocarcinoma by inhibiting
HMGB1-mediated autophagy. Oncotarget. 8:107500–107512. 2017.
View Article : Google Scholar
|
|
72
|
Leivonen SK, Icay K, Jäntti K, Siren I,
Liu C, Alkodsi A, Cervera A, Ludvigsen M, Hamilton-Dutoit SJ and
d’Amore F: MicroRNAs regulate key cell survival pathways and
mediate chemosensitivity during progression of diffuse large B-cell
lymphoma. Blood Cancer J. 7:6542017. View Article : Google Scholar
|
|
73
|
Gabra MM and Salmena L: microRNAs and
acute myeloid leukemia chemoresistance: A mechanistic overview.
Front Oncol. 7:2552017. View Article : Google Scholar
|
|
74
|
Yang RM, Zhan M, Xu SW, Long MM, Yang LH,
Chen W, Huang S, Liu Q, Zhou J, Zhu J, et al: miR-3656 expression
enhances the chemosensitivity of pancreatic cancer to gemcitabine
through modulation of the RHOF/EMT axis. Cell Death Dis.
8:e31292017. View Article : Google Scholar
|
|
75
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar
|
|
76
|
Moody HL, Lind MJ and Maher SG:
MicroRNA-31 regulates chemosensitivity in malignant pleural
mesothelioma. Mol Ther Nucleic Acids. 8:317–329. 2017. View Article : Google Scholar
|
|
77
|
Ara T, Nakata R, Sheard MA, Shimada H,
Buettner R, Groshen SG, Ji L, Yu H, Jove R and Seeger RC: Critical
role of STAT3 in IL-6-mediated drug resistance in human
neuroblastoma. Cancer Res. 73:3852–3864. 2013. View Article : Google Scholar
|
|
78
|
Suh YA, Jo SY, Lee HY and Lee C:
Inhibition of IL-6/STAT3 axis and targeting Axl and Tyro3 receptor
tyrosine kinases by apigenin circumvent taxol resistance in ovarian
cancer cells. Int J Oncol. 46:1405–1411. 2015. View Article : Google Scholar
|
|
79
|
Garcia R, Bowman TL, Niu G, Yu H, Minton
S, Muro-Cacho CA, Cox CE, Falcone R, Fairclough R and Parsons S:
Constitutive activation of Stat3 by the Src and JAK tyrosine
kinases participates in growth regulation of human breast carcinoma
cells. Oncogene. 20:2499–2513. 2001. View Article : Google Scholar
|
|
80
|
Epling-Burnette PK, Liu JH,
Catlett-Falcone R, Turkson J, Oshiro M, Kothapalli R, Li Y, Wang
JM, Yang-Yen HF and Karras J: Inhibition of STAT3 signaling leads
to apoptosis of leukemic large granular lymphocytes and decreased
Mcl-1 expression. J Clin Invest. 107:351–362. 2001. View Article : Google Scholar
|
|
81
|
Huang W, Dong Z, Chen Y, Wang F, Wang CJ,
Peng H, He Y, Hangoc G, Pollok K and Sandusky G: Small-molecule
inhibitors targeting the DNA-binding domain of STAT3 suppress tumor
growth, metastasis and STAT3 target gene expression in vivo.
Oncogene. 35:8022016. View Article : Google Scholar
|





















