Introduction
Human breast cancer is one of the most commonly
diagnosed cancers worldwide (1).
It is estimated that the number of patients with breast cancer
increases annually by ~1.7 million globally; ~245,000 new cases of
breast cancer and >45,000-related are recorded in the USA each
year, as well as ~170,000 new cases and >40,000-related deaths
in China (2,3). Although breast cancer diagnosis and
therapy have both improved, satisfactory therapeutic effects have
not yet been achieved due to disease complexity. Tumorigenesis is a
multifactorial and multistep process involving a range of genetic
alterations, including the activation of oncogenes, the
inactivation of anti-oncogenes and the abnormal expression of
cancer-related genes (4-6). Previous studies have unraveled some
of the pathological mechanisms involved in breast tumorigenesis
(7,8); however, these have yet to be fully
elucidated. Hence, it is important to fully characterize the
molecular mechanisms responsible for breast cancer progression, and
these may then be used to identify novel biomarkers and therapeutic
targets for early diagnosis and therapy.
MicroRNAs (miRNAs or miRs) are an abundant class of
non-coding RNAs, approximately 18-24 nucleotides in length, that
bind to the 3ʹ-untranslated regions (UTRs) of target genes and
regulate expression post-transcriptionally (9-11).
Over the past decade, miRNAs have been demonstrated to actively
participate in various biological effects, including cancer cell
survival, growth, motility, autophagy and apoptosis (12-15).
Numerous miRNAs reportedly function either as oncogenes or
anti-oncogenes. Studies have indicated that miRNAs are often
aberrantly regulated and play prohibitive or oncogenic roles in
breast cancer (16,17). Recently, the clinical significance
of miR-1254 has been verified, along with its role in
tumorigen-esis and progression, including lung cancer, thyroid
cancer and oral squamous cell carcinoma (18-20).
In this study, using bioinformatics software, we
predicted that miR-1254 could target Ras-association domain family
9 (RASSF9). RASSF9 is a RAS-association domain family gene and is
expressed in multiple tissues. The RASSF family includes 10 genes
from RASSF1 to RASSF10 and is subdivided into C-terminally
(RASSF1-6) and N-terminally (RASSF7-10). The N-terminal RASSF genes
are involved in cell growth, survival and apoptosis, among other
processes (21). Evidence suggests
that RASSF7 promotes lung cancer cell growth, migration and
invasion by inhibiting the phosphorylation of mammalian Ste20-like
kinase 1, large tumor suppressor kinase 1 and yes-associated
protein (22). RASSF8 suppresses
cell proliferation, migration and invasion in lung cancer, gastric
cancer and cervical cancer (23-26).
RASSF10 acts as a novel tumor suppressor in liver cancer, breast
cancer, thyroid cancer, gastric cancer, lung cancer, colorectal
cancer, esophageal squamous cell carcinoma and hepatocarcinoma
(27-32). However, the functions of RASSF9 in
many types of cancer, including breast cancer, remain unknown.
In the present study, we investigated the function
and mechanisms of action of miR-1254 in human breast cancer. We
demonstrate that the expression of miR-1254 is markedly upregulated
in human breast cancer tissues and is associated with certain
clinicopathological characteristics. Furthermore, miR-1254 potently
promotes breast cancer cell growth and cycle transition. In
particular, we demonstrate that RASSF9 is a direct target of
miR-1254. Thus, these findings indicate that miR-1254 may be a new
target for breast carcinoma therapy.
Materials and methods
Preparation of tissue samples
Human breast cancer samples (n=79) were collected
from patients who were diagnosed at the Department of Oncology
Surgery (the First Affiliated Hospital, Xi’an Jiaotong University,
Xi’an, China) between January, 2015 and March, 2016. Informed
consent was obtained from each patient prior to specimen
collection, and the tissues were stored at −80˚C.
Clinicopathological data were obtained by reviewing the
pathological records. The study protocol was approved by the Ethics
Committee of Xi’an Jiaotong University. The Cancer Genome Atlas
(TCGA) datasets from 114 normal samples and 1,099 cancer samples
were analyzed for miR-1254 and RASSF9 expression levels.
Cell culture
The human breast carcinoma cell lines, T47D, MCF-7,
ZR-75-30 and MDA-MB-231, and the normal breast epithelial cell
line, HB2, were obtained from the Cell Bank of Genechem (Genechem
Co., Ltd., Shanghai, China). The cells were tested and
authenticated by the Cell Bank of Genechem. The cells
(1×105 cells/ml) were cultivated in Dulbecco’s modified
Eagle’s medium supplemented with 10% fetal bovine serum (both from
Thermo Fisher Scientific, Inc., Waltham, MA, USA) and were cultured
at 37˚C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
RNA was extracted from the human tissues and breast
cancer cell lines using TRIzol reagent (Invitrogen/Thermo Fisher
Scientific, Inc. The SYBR Premix Ex Taq II and PrimeScript RT
Reagent kits (Takara Biotechnology, Inc., Dalian, China) were used
to examine miR-1254 and RASSF9 mRNA expression. RT-qPCR were
performed with the iCycler iQ Multicolor RT-qPCR Detection System
(Bio-Rad Laboratories, Inc., Hercules, CA, USA) under the following
conditions: 95˚C for 10 min, followed by 45 cycles at 95˚C for 10
sec, 60˚C for 20 sec, and 72˚C for 30 sec. All the primer sequences
were as follows: miR-1254 reverse-transcribed primer,
5ʹ-GTCGTATCCAGTGCGTGTCGTGGAGTCGG CAATTGCACTGGATACGACCTGCAGT-3ʹ;
miR-1254 forward, 5ʹ-ATCCAGTGCGTGTCGTG-3ʹ and reverse, 5ʹ-TGC
TAGCCTGGAAGCTGGAGC-3ʹ; U6 reverse-transcribed primer,
5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ; U6 forward,
5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and reverse,
5ʹ-CGCTTCACGAATTTGCGTGT CAT-3ʹ; RASSF9 forward,
5ʹ-ACAACAATCCCGCAGTTCAAA-3ʹ and reverse,
5ʹ-GTGTCTGGATTTCCAGGGTGA-3ʹ; β-actin forward,
5ʹ-TGGCACCCAGCACAATGAA-3ʹ and reverse, 5ʹ-CTA
AGTCATAGTCCGCCTAGAAGCA-3ʹ. The data were normalized to RNU6B (U6)
or β-actin gene expression. The relative expression of genes was
calculated with the 2−ΔΔCq method (33).
Expression vector construction
hsa-miR-1254 precursor expression vector (named
miR-1254) and a control vector (named Control) were constructed
using chemosynthetic oligonucleotides and interpolated into
pcDNA6.2-GW/EmGFPmiR plasmids (both from Genechem Co., Ltd.).
RASSF9 gene DNA was inserted in the pCMV2-GV146 vector (Genechem
Co., Ltd.). Transfection was conducted using Lipofectamine 2000
(Invitrogen/Thermo Fisher Scientific, Inc.). Cells were transfected
and cultured for 48 h before performing the assays.
Dual-luciferase assay
The possible target genes of miR-1254 were predicted
by using bioinformatics software miRanda (www.ma.uni-heidelberg.de/apps/zmf/mirwalk/) and RegRNA
(http://regrna.mbc.nctu.edu.tw/html/prediction.html).
The binding site of miR-1254 in the 3ʹ-UTR of RASSF9 was
constructed and inserted in the pmirGLO Dual-Luciferase expression
vector (named RASSF9-WT; Genechem Co., Ltd.). Mutated sequences of
RASSF9 were also constructed (named RASSF9-MT). miR-1254 plasmids
and reporter plasmids were co-transfected into 293 cells (Genechem
Co., Ltd.) Cells were collected at 24 h post-transfection, and the
reporter activity was detected using a Dual-Luciferase assay system
(Promega Corp., Madison, WI, USA).
Anti-miR-1254/RASSF9 siRNA synthesis and
transfection
Interfering RNA oligonucleotides served as miR-1254
inhibitor (named anti-miR-1254) and were synthesized by GenePharma
(Shanghai, China). The sequence of anti-miR-1254 was
5ʹ-ACUGCAGGCUCCAGCUUCCAGGCU-3ʹ. Scrambled siRNA served as the
control (named anti-miR-Control), and the sequence was
5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ. RNA oligonucleotides were transfected
into breast cancer MCF-7/T47D cells using Lipofectamine 2000. Small
interfering RNA (siRNA) were used to silence the human RASSF9 gene.
Human RASSF9 siRNA (sense, 5ʹ-GAGAAUGAAAGAGCU GGAUTT-3ʹ and
antisense, 5ʹ-AUCCAGCUCUUUCAUUCU CTT-3ʹ), and negative siRNA
(NC-siRNA sense, 5ʹ-UUCUCCG AACGUGUCACGUTT-3ʹ and antisense,
5ʹ-ACGUGACACG UUCGGAGAATT-3ʹ) were synthesized by GenePharma. The
siRNAs were transfected into the cells using Lipofectamine 2000 and
diluted to 70 nM for use in the experiment.
MTT assay
MCF-7/T47D cells (4,000 cells/well in 100 µl
DMEM) were seeded into 96-well plates and cultivated for 24 h,
followed by transfection with the Control, miR-1254,
anti-miR-Control, anti-miR-1254, NC-siRNA (70 nM), RASSF9 siRNA (70
nM), Vector control or RASSF9 expression vector for 24, 48 or 72 h.
Cell viability was then examined by MTT assay and the absorbance
was read at 492 nm using a microplate reader (FLUOstar OPTIMA; BMG
Labtech, Ortenberg, Germany).
Cell counting assay
To measure cell growth, 1×105 cells were
seeded into 60-mm-diameter plates and cultured for 24 h, followed
by transfection with the Control, miR-1254, anti-miR-Control,
anti-miR-1254, NC-siRNA (70 nM), RASSF9 siRNA (70 nM), Vector
control or RASSF9 expression vector. Cell numbers were counted at
24, 48 and 72 h following treatment using a Countess automated cell
counter (Invitrogen, Eugene, OR, USA).
Cell cycle analysis
The MCF-7/T47D cells were incubated at 37˚C for 48
h, prior to harvesting and fixing in 70% ice-cold ethanol. The
cells were then dyed using propidium iodide and RNase A
(DNAse-free) (Sigma, St. Louis, MO, USA). Finally, the cell cycle
distribution was assessed via fluorescence-activated cell sorting
(BD Biosciences, San Jose, CA, USA), and the proportions calculated
with ModFit software (Bio-Rad Laboratories, Hercules, CA, USA).
Cell apoptosis analysis
The MCF-7/T47D cells were seeded into 6-well plates
in triplicate and treated for 48 h with the Control, miR-1254,
anti-miR-Control, anti-miR-1254, NC-siRNA (70 nM), RASSF9 siRNA (70
nM), Vector control or RASSF9 expression vector. An Annexin V FITC
Apoptosis Detection kit (Invitrogen/Thermo Fisher Scientific, Inc.)
was used to measure cell apoptosis. During the early stage of
apoptosis, cells begin to display phosphatidylserine (PS) on the
cell surface membranes where it is readily detectable by staining
the cells with Annexin V-FITC. As the plasma membrane becomes
increasingly permeable during the later stages of apoptosis, PI can
move across the cell membrane and bind to DNA. The stained cells
were detected using a flow cytometer (BD Biosciences), and
apoptosis was analyzed using ModFit software.
Western blot analysis
Human breast cancer cells were lysed with RIPA lysis
buffer (Wolsen, Xi’an, China). Total proteins were measured with
the BCA assay. then separated and transferred onto a nitrocellulose
membrane (Roche Diagnostics, Basel, Switzerland). A total of 40
µg of protein was then isolated by 10% SDS-PAGE and
transferred onto a nitrocellulose membrane (Roche Diagnostics). The
membrane was blocked with 5% non-fat milk in Tris-buffered saline
Tween-20 (TBST) for 1 h at room temperature. The membrane was then
incubated with the following primary antibodies: Rabbit polyclonal
anti-RASSF9 (PA5-58878, 1:1,000; Invitrogen/Thermo Fisher
Scientific, Inc.), rabbit polyclonal anti-p-AKT (sc-135651,
1:1,000), rabbit polyclonal anti-AKT (sc-8312, 1:2,000), mouse
monoclonal anti-Cyclin D1 (sc-70899, 1:1,000), mouse monoclonal
anti-CDK2 (sc-70829, 1:1,000), rabbit polyclonal anti-p53 (sc-6243,
1:1,000) and mouse monoclonal anti-β-actin (sc-58673, 1:3,000) (all
from Santa Cruz Biotechnology, Inc., Danvers, CA, USA). Next, they
were incubated with horseradish peroxidase (HRP)-conjugated
secondary antibodies (sc-2005, goat anti-mouse IgG-HRP, 1:5,000;
sc-2004, goat anti-rabbit IgG-HRP; Santa Cruz Biotechnology, Inc.)
at 37˚C for 2 h. The membrane was subsequently treated with ECL
reagent (Pierce, Rockford, IL, USA) for chemiluminescence
detection, and the blots were analyzed using Quantity One imaging
software.
Statistical analysis
The data are presented as the means ± SEM from at
least 3 experiments. P<0.05 was considered to indicate a
statistically significant difference. Data analysis was conducted
using SPSS v.20.0 software (IBM Corporation, Armonk, NY, USA). A
Student’s t-test was used to analyze the differences between 2
groups. A Chi-square test was employed to analyze the comparisons
between miR-1254 expression and certain clinicopathologic
characteristics. Correlations between miR-1254 and RASSF9
expression in breast cancer tissues were estimated using Pearson’s
correlation analysis.
Results
miR-1254 is frequently upregulated in
human breast cancer tissues and cell lines
To examine the function of miR-1254 in breast
cancer, we performed RT-qPCR to examine its expression in 79
primary breast carcinoma specimens and 79 adjacent normal breast
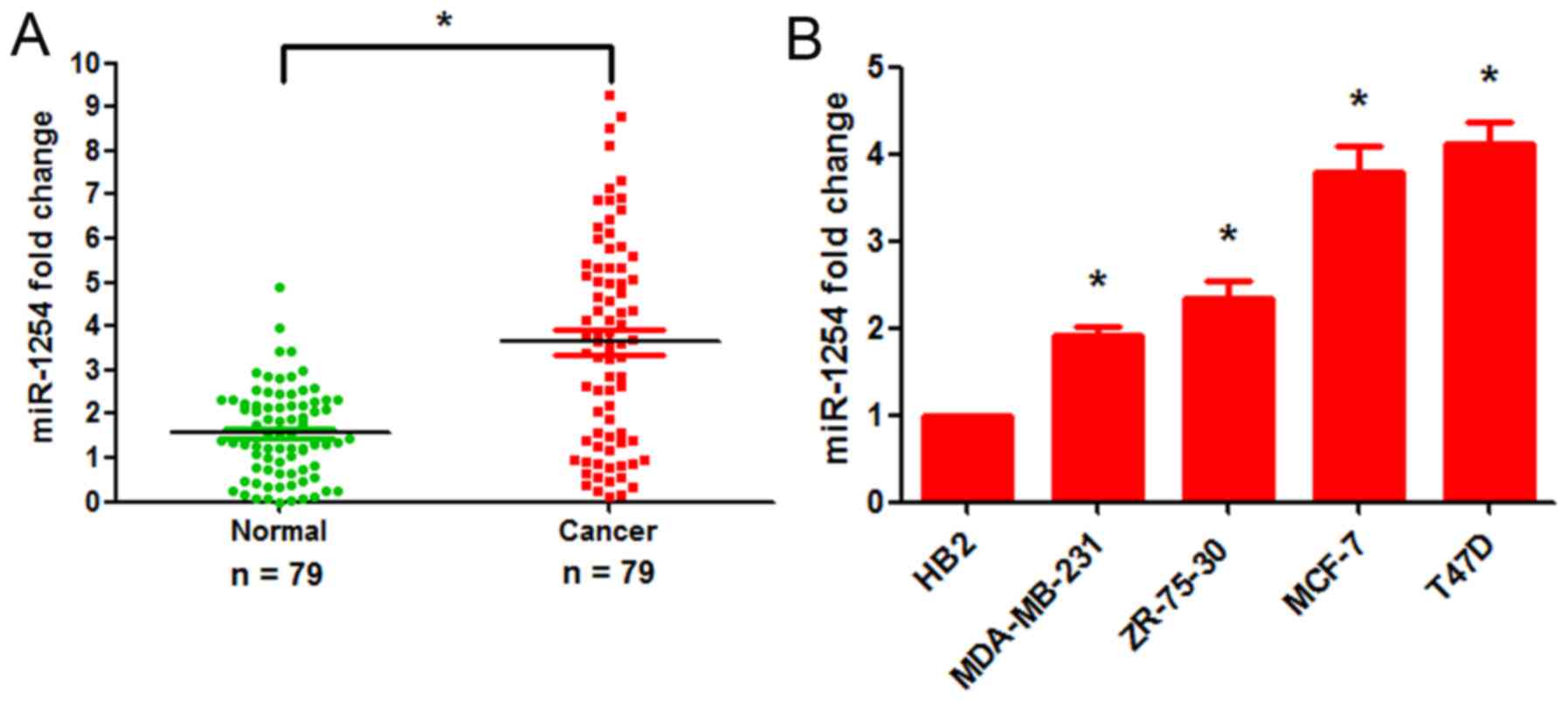
tissues, and in breast tumor cell lines. The results of RT-qPCR
assays revealed that the expression of miR-1254 was markedly
increased in 74.7% (59/79) of the breast cancer tissue samples (a
quantitative ratio of >1.2 was considered as upregulation)
(Fig. 1A and Table I). Further experiments revealed
associations between the miR-1254 levels and certain
clinicopathological characteristics of the patients with breast
cancer. A high miR-1254 expression was associated with tumor size,
T stage, TNM stage and estrogen receptor and progesterone receptor
positivity (Table I). However,
miR-1254 expression was not associated with patient age, lymph node
metastasis, tumor grade and human epidermal growth factor receptor
2 positivity. Additionally, miR-1254 expression was significantly
upregulated in the breast cancer cell lines (T47D, MCF-7, ZR-75-30
and MDA-MB-231) compared with the HB2 normal breast epithelial cell
line (Fig. 1B). These results
indicate that miR-1254 may be a useful biomarker for assessing the
malignant status of breast cancer.
 | Table IAssociation of miR-1254 expression
with clinicopathological characteristics of the patients with
breast cancer. |
Table I
Association of miR-1254 expression
with clinicopathological characteristics of the patients with
breast cancer.
| Characteristic | Numberof cases | miR-1254 expression
| P-value |
|---|
High
(n=59) | Low
(n=20) |
|---|
| Age | | | | 0.836 |
| ≥45 years | 62 | 46 | 16 | |
| <45 years | 17 | 13 | 4 | |
| Tumor size | | | | 0.018a |
| <20 mm | 52 | 35 | 17 | |
| ≥20 mm | 27 | 24 | 3 | |
| Lymph node
metastasis | | | | 0.756 |
| Yes | 54 | 40 | 14 | |
| No | 25 | 19 | 6 | |
| Grade | | | | 0.907 |
| 1 | 23 | 18 | 5 | |
| 2 | 41 | 30 | 11 | |
| 3 | 15 | 11 | 4 | |
| T stage | | | | 0.008a |
| T1 | 22 | 12 | 10 | |
| T2 | 41 | 33 | 8 | |
| T3 | 7 | 6 | 1 | |
| T4 | 9 | 8 | 1 | |
| TNM stage | | | | 0.026a |
| I | 24 | 16 | 8 | |
| II | 33 | 24 | 9 | |
| III | 12 | 10 | 2 | |
| IV | 10 | 9 | 1 | |
| ER | | | | 0.003a |
| Positive | 51 | 45 | 6 | |
| Negative | 28 | 14 | 14 | |
| PR | | | | 0.031a |
| Positive | 46 | 38 | | |
| Negative | 33 | 21 | 12 | |
| HER2 | | | | 0.452a |
| Positive | 30 | 23 | 7 | |
| Negative | 49 | 36 | 13 | |
miR-1254 promotes MCF-7/T47D cell growth
and induces G1-S phase transition
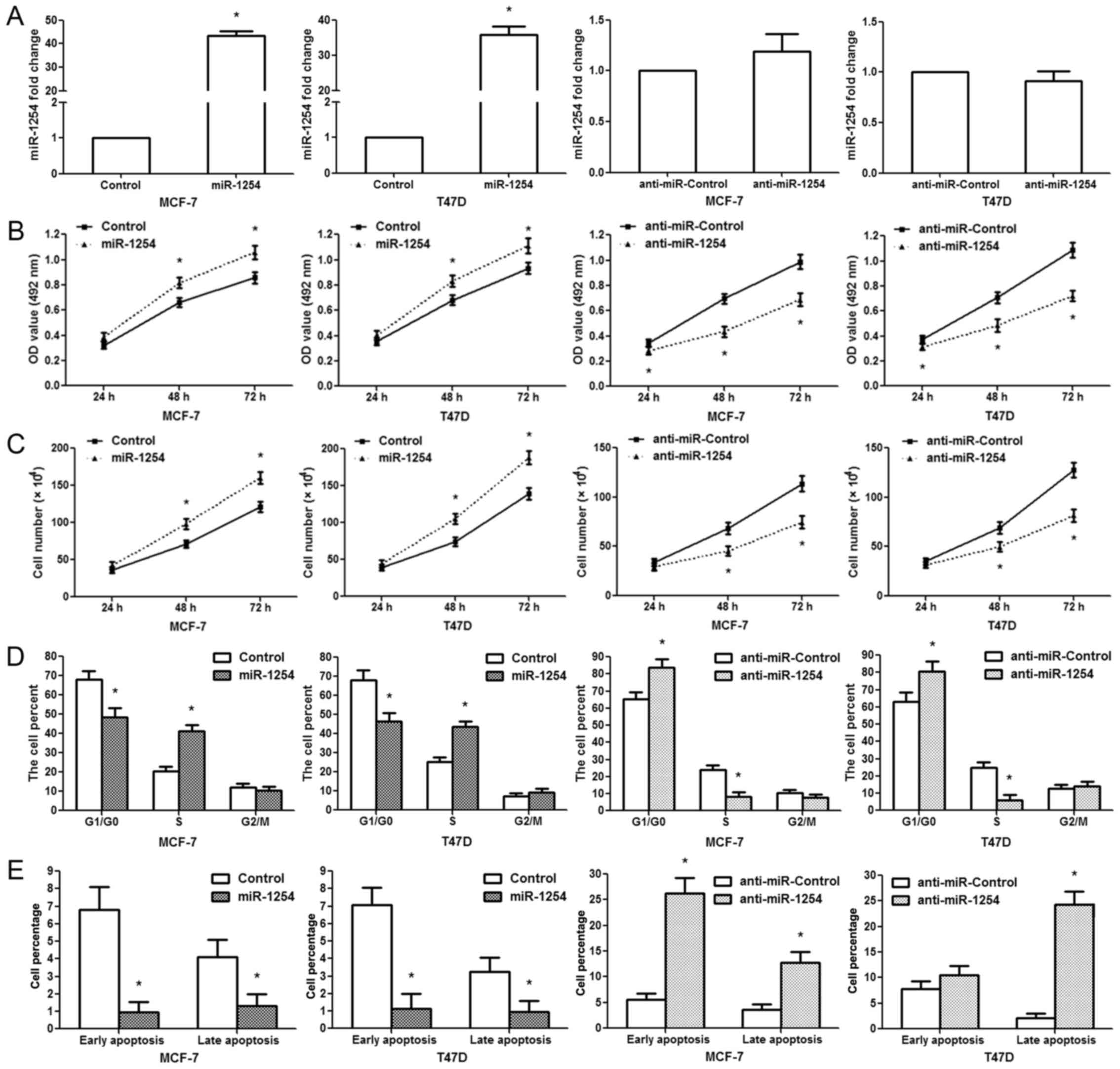
The MCF-7/T47D cells were transfected with the
miR-1254 precursor expression vector, empty vector, miR-1254
antisense oligonucleotides, or negative control. RT-qPCR was
conducted to examine the expression levels of miR-1254 following
treatment. Our data indicated that miR-1254 expression was markedly
upregulated in the cells transfected with the miR-1254 vector
compared with those transfected with the empty vector; however, no
significant difference was observed between the anti-miR-1254 and
anti-miR-Control groups (Fig. 2A).
MTT assays revealed that miR-1254 overexpression facilitated the
proliferation of MCF-7/T47D cells at 48 and 72 h following
transfection; however, anti-miR-1254 inhibited MCF-7/T47D cell
growth at 48 and 72 h following transfection (Fig. 2B).
Similar results were observed for cell counting
assay. miR-1254 overexpression promoted MCF-7/T47D cell
proliferation, while anti-miR-1254 suppressed cell proliferation
(Fig. 2C). Since the cell cycle
involves the regulation of proliferation, we analyzed it following
transfection. We found that miR-1254 overexpression decreased the
G1/G0 phase population and increased the S phase population in the
MCF-7/T47D cells; however, the inhibition of miR-1254 led to a
marked accumulation of cells in the G1/G0 phase and a reduction of
the S phase cell population (Fig.
2D). Apoptosis analysis revealed that the proportion of cells
undergoing early and late stage apoptosis decreased markedly in the
miR-1254 overexpression group, while it increased significantly in
the anti-miR-1254 group (Fig. 2E).
These data suggest that miR-1254 promotes breast cancer cell
proliferation and G1-S transition, and inhibits cell apoptosis.
RASSF9 is a direct target of
miR-1254
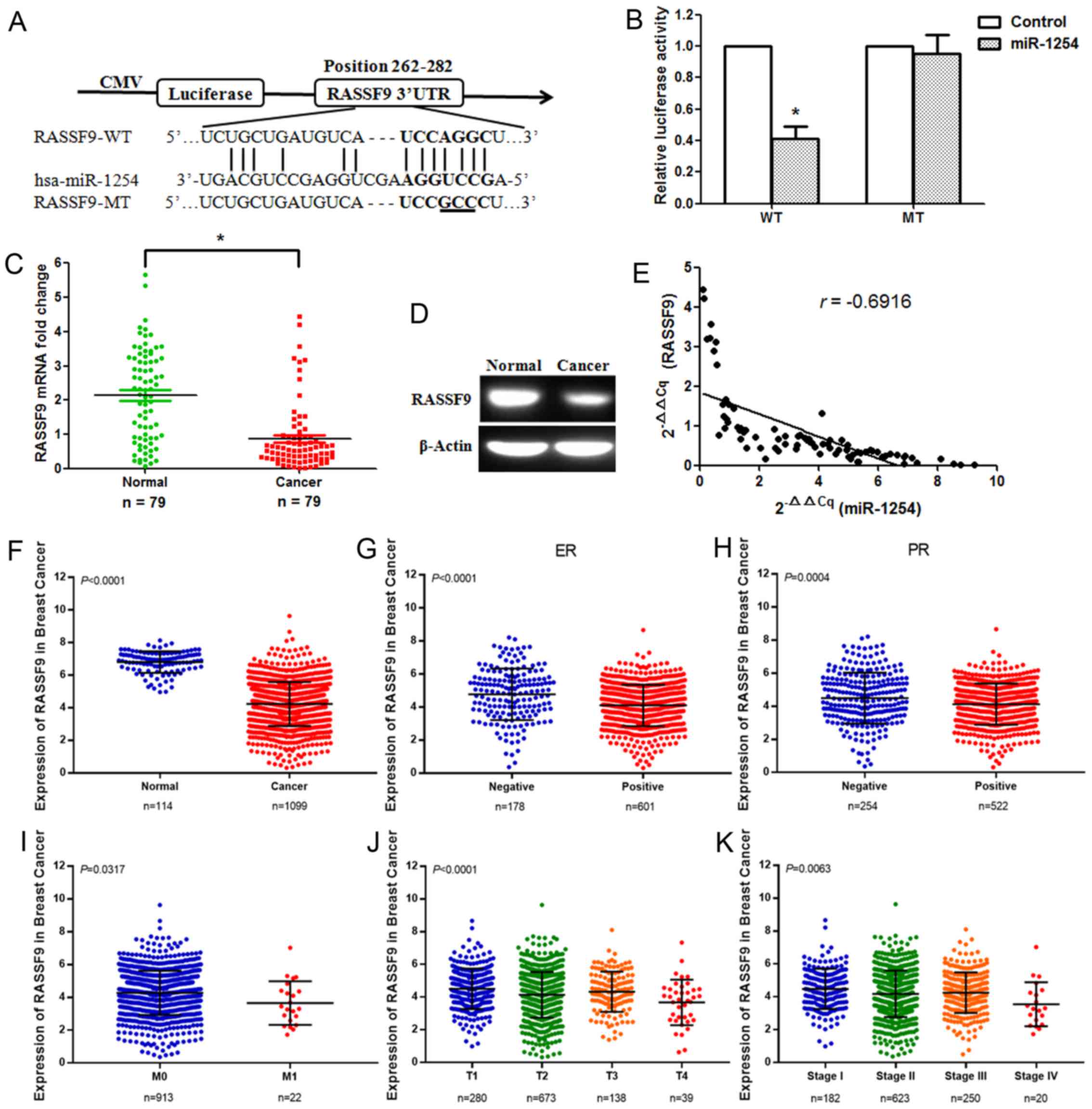
miRanda and RegRNA were used to predict many
possible target genes of miR-1254. Among the candidates, RASSF9 was
selected for further research. We discovered a binding site for
miR-1254 in the 3ʹ-UTR of RASSF9 mRNA ranging from 262-282 bp
(Fig. 3A). To confirm whether
miR-1254 directly targets RASSF9, a dual-luciferase reporter assay,
including the WT and MT 3ʹ-UTRs of RASSF9, was performed. Reporter
plasmids and pre-miR-1254 or pmirGLO control vector (Control) were
co-transfected into 293T cells. The luciferase activity of the
pre-miR-1254/WT-RASSF9-UTR-transfected cells decreased
significantly, but that of the
pre-miR-1254/MT-RASSF9-UTR-transfected cells failed to suppress the
relative luciferase activity (Fig.
3B), suggesting that miR-1254 can bind to the 3ʹ-UTR of RASSF9.
Subsequently, the RASSF9 mRNA and protein expression levels were
measured. The results revealed that RASSF9 expression was
significantly downregulated in the breast cancer tissues (Fig. 3C and D). The effect of miR-1254 on
RASSF9 expression was assessed based on the results of RT-qPCR. A
significantly negative correlation was identified between RASSF9
and miR-1254 (Fig. 3E; n=79,
r=−0.6916, P<0.001, Pearson’s correlation analysis). The TCGA
data revealed that RASSF9 expression was markedly downregulated in
breast cancer tissues. A low RASSF9 expression was associated with
estrogen receptor positivity, progesterone receptor positivity, M
stage, T stage and TNM stage (Fig.
3F-K). In addition, to verify the changes in RASSF9 expression
in breast cancer tissues, its mRNA levels were detected in the 79
breast cancer tissues by RT-qPCR. A low RASSF9 expression was found
to be associated with tumor size, lymph node metastasis, T stage,
TNM stage, estrogen receptor positivity and progesterone receptor
positivity (Table II).
 | Table IIAssociation of RASSF9 mRNA expression
with the clinicopathological characteristics of patients with
breast cancer. |
Table II
Association of RASSF9 mRNA expression
with the clinicopathological characteristics of patients with
breast cancer.
|
Characteristics | Number of
cases | RASSF9 mRNA
expression
| P-value |
|---|
High
(n=16) | Low
(n=63) |
|---|
| Age | | | | 0.803 |
| ≥45 years | 62 | 12 | 50 | |
| <45 years | 17 | 4 | 13 | |
| Tumor size | | | | 0.009a |
| <20 mm | 52 | 14 | 38 | |
| ≥20 mm | 27 | 2 | 25 | |
| Lymph node | | | 0.015a | |
| metastasis | | | | |
| Yes | 54 | 5 | 49 | |
| No | 25 | 11 | 14 | |
| Grade | | | | 0.637 |
| 1 | 23 | 5 | 18 | |
| 2 | 41 | 8 | 33 | |
| 3 | 15 | 3 | 12 | |
| T stage | | | | 0.023a |
| T1 | 22 | 7 | 15 | |
| T2 | 41 | 8 | 33 | |
| T3 | 7 | 1 | 6 | |
| T4 | 9 | 0 | 9 | |
| TNM stage | | | | 0.013a |
| I | 24 | 5 | 19 | |
| II | 33 | 8 | 25 | |
| III | 12 | 2 | 10 | |
| IV | 10 | 1 | 9 | |
| ER | | | | 0.001a |
| Positive | 51 | 3 | 48 | |
| Negative | 28 | 13 | 15 | |
| PR | | | | 0.045a |
| Positive | 46 | 6 | 40 | |
| Negative | 33 | 10 | 23 | |
| HER2 | | | | 0.118 |
| Positive | 30 | 4 | 26 | |
| Negative | 49 | 12 | 37 | |
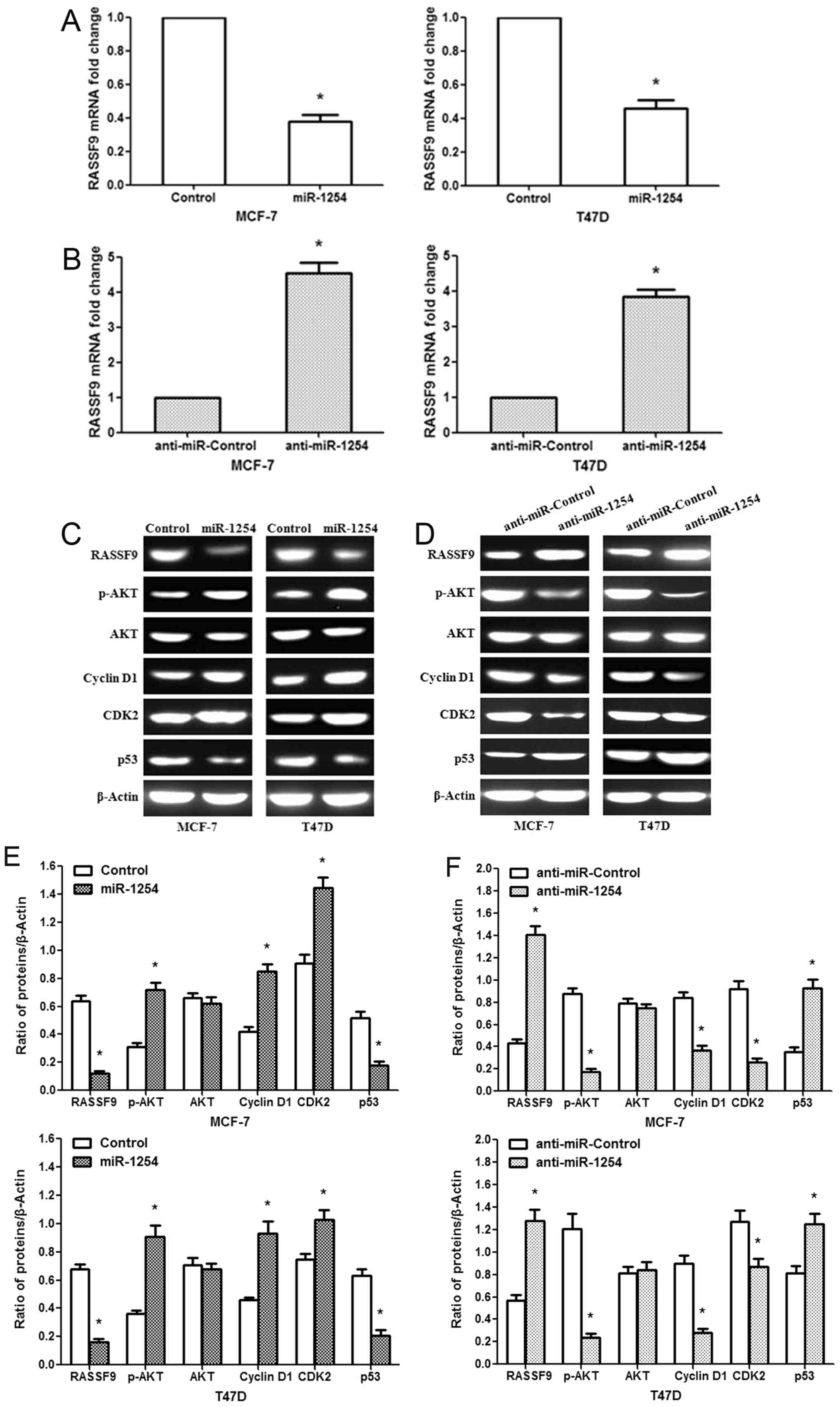
In addition, we found that miR-1254 overexpression
markedly downregulated the mRNA expression of RASSF9 in MCF-7/T47D
cells, while anti-miR-1254 increased RASSF9 mRNA expression
(Fig. 4A and B). Similar results
were observed for the protein levels (Fig. 4C and D). To further explore the
potential mechanisms of the miR-1254-regulated cell proliferation,
cell cycle transition and apoptosis, we measured protein expression
in the AKT signaling pathway, and that of G1 phase regulators and
p53. The data revealed that miR-1254 overexpression increased
p-AKT, Cyclin D1 and CDK2 protein expression, and decreased the p53
protein levels in the MCF-7/T47D cells (Fig. 4C). By contrast, anti-miR-1254
inhibited p-AKT, Cyclin D1 and CDK2 protein expression, and
promoted p53 expression (Fig. 4D).
These results indicate that miR-1254 may modulate breast cancer
cell proliferation and apoptosis through the regulation of the AKT
and p53 signaling pathways by targeting RASSF9.
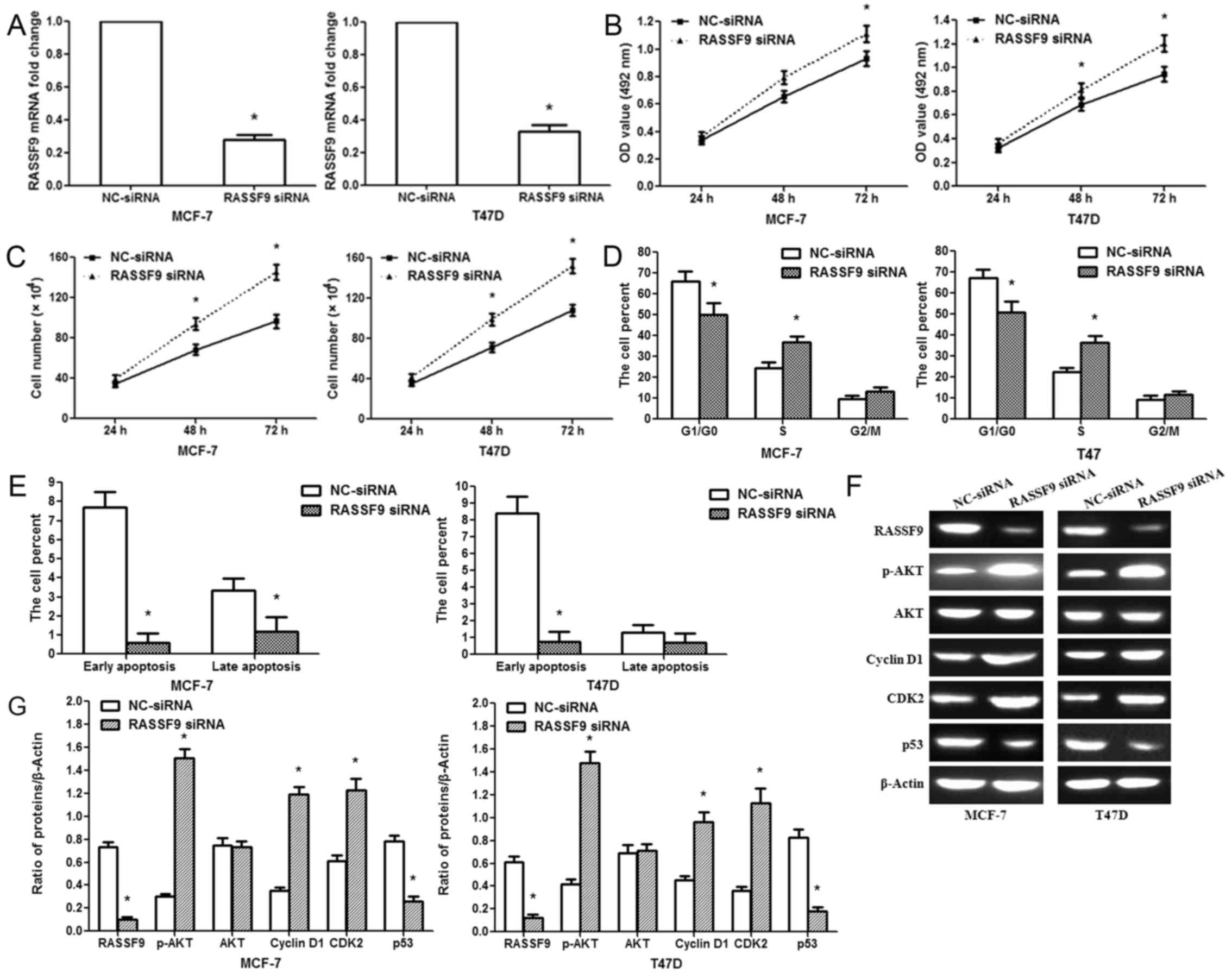
Knockdown of RASSF9 facilitates
MCF-7/T47D cell proliferation
We knocked down RASSF9 expression in the MCF-7/T47D
cells via RNA interference to affirm its involvement in the
pro-tumor functions of miR-1254. RASSF9 mRNA expression was
specifically knocked down in the MCF-7/T47D cells using siRNA
(Fig. 5A). The silencing of RASSF9
significantly increased cell activity at 48 and 72 h following
treatment (Fig. 5B). A cell
counting assay revealed that the silencing of RASSF9 promoted
MCF-7/T47D cell proliferation (Fig.
5C). The silencing of RASSF9 decreased the G1/G0 phase
population and increased the S phase population of the MCF-7/T47D
cells (Fig. 5D). Furthermore, the
silencing of RASSF9 inhibited the apoptosis of MCF-7/T47D cells
(Fig. 5E). These effects were
similar to those caused by miR-1254 overexpression, suggesting a
similar effect caused by both RASSF9 knockdown and miR-1254
overexpression. In addition, we examined the knockdown efficiency
of RASSF9 siRNA. RASSF9 protein expression decreased in the siRNA
group; p53 protein expression was also decreased. However, p-AKT,
Cyclin D1 and CDK2 protein expression increased in the siRNA group
(Fig. 5F).
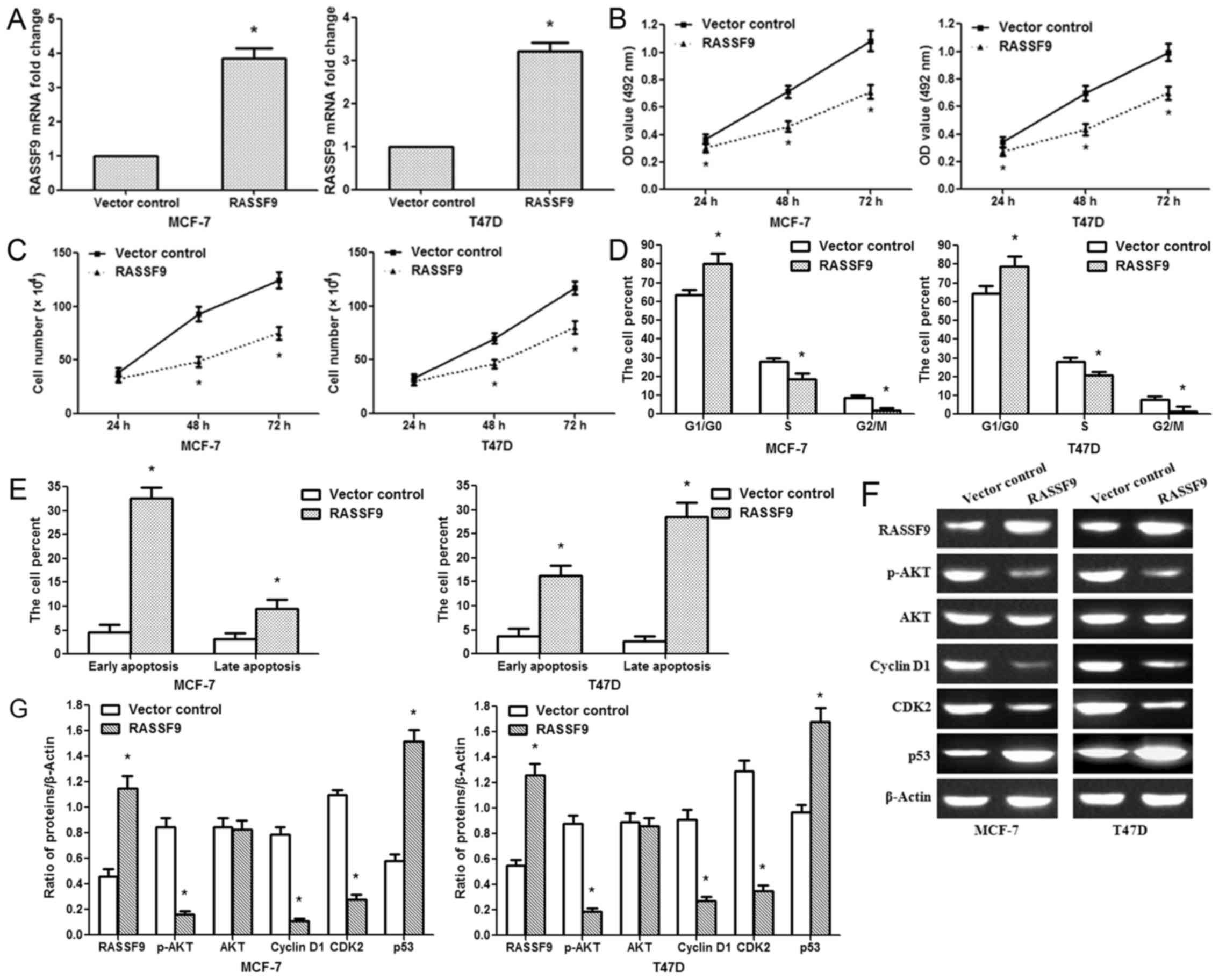
Overexpression of RASSF9 inhibits
MCF-7/T47D cell proliferation
We then constructed a RASSF9 overexpression plasmid.
In the MCF-7/T47D cells, the plasmid efficiently upregulated the
RASSF9 mRNA levels (Fig. 6A).
Based on the results of cell viability and cell counting assays,
the upregulation of RASSF9 inhibited MCF-7/T47D cell growth
(Fig. 6B and C). The impact of
RASSF9 expression on the cell cycle was measured by flow cytometry,
and it was found that RASSF9 overexpression increased the G1 phase
cell proportion and decreased the S and G2/M phase cell proportion
(Fig. 6D). As for the effect of
RASSF9 expression on cell apoptosis, RASSF9 overexpression
significantly induced early and late apoptosis (Fig. 6E). Further analysis revealed that
RASSF9 overexpression inhibited G1-S phase transition by
suppressing p-AKT, Cyclin D1 and CDK2 expression. Additionally, it
promoted the expression of key apoptosis regulators, such as p53
(Fig. 6F). Therefore, these data
indicate that miR-1254 regulates breast cancer cell progression by
targeting RASSF9 through the AKT and p53 signaling pathways.
Discussion
Previous studies have demonstrated that miRNAs play
crucial roles in regulating tumor cell survival, proliferation,
apoptosis, differentiation, migration and invasion (9,34).
It has been found that abnormally expressed miRNAs are associated
with breast carcinogenesis and progression (35,36).
Although miRNA profiles have been well-characterized in breast
cancer, the roles and mechanisms of action of dysregulated miRNAs
remain largely unknown. Authenticating miRNAs and elucidating their
biological effects in breast cancer is crucial for identifying
novel targets for diagnosis and treatment. It has been reported
that miR-1254 expression is increased in lung cancer and promotes
lung cancer cell proliferation by targeting SFRP1 (18). However, miR-1254 has been shown to
inhibit oral squamous cell carcinoma metastasis (20). Nevertheless, the clinical
implication and function of miR-1254 in breast cancer remains
unclear. In the present study, we found that miR-1254 expression
was markedly upregulated in breast cancer tissues and cell lines.
The results revealed that a high miR-1254 expression was associated
with tumor size, T stage, TNM stage, estrogen receptor positivity
and progesterone receptor positivity in breast cancer. The results
demonstrated that miR-1254 overexpression promoted breast cancer
cell growth by inducing G1-S phase transition and inhibiting cell
apoptosis in vitro. Anti-miR-1254 inhibited breast cancer
cell growth and induced cell apoptosis, whereas there was no
significant difference between the anti-miR-1254 and
anti-miR-Control groups. This is due to the fact that the inhibitor
may suppress the function of miR-1254 by binding to it, but does
not affect the expression of miR-1254. These findings suggest that
miR-1254 plays a crucial role in breast cancer development and
progression.
RASSF9-null mice exhibit a marked variety in
epithelial organization, including cell proliferation and
differentiation (37). In this
study, to the best of our knowledge, we present the first evidence
that miR-1254 promotes breast cancer cell proliferation through the
miR-1254 target, RASSF9. RASSF9 was predicted to be a target of
miR-1254 by two databases. The results of reporter assays revealed
that miR-1254 overexpression inhibited RASSF9 3ʹ-UTR luciferase
reporter activity, which was abrogated by the mutation of the
miR-1254 binding site; we also found a negative correlation between
miR-1254 and RASSF9 expression in breast carcinoma tissues;
miR-1254 overexpression inhibited RASSF9 mRNA and protein
expression in breast cancer cells. Our findings also demonstrated
that the knockdown of RASSF9 promoted breast cancer cell
proliferation and suppressed apoptosis, and that the overexpression
of RASSF9 inhibited cell proliferation and induced apoptosis. These
results suggest that miR-1254 acts as a positive regulator or
oncogene for cell proliferation, partially mediated by inhibiting
RASSF9 expression.
PI3K/AKT is one of the most effective growth
signaling pathways in cancer (38). Aberrations in the AKT signaling
pathway are involved in tumorigenesis and progression, particularly
in breast, liver, lung, prostate, colorectal and renal cancer
(39). AKT activity is reportedly
associated with various clinicopathological characteristics of
cancer (40). AKT regulates the
roles of certain substrates related to the cell cycle by directly
phosphorylating target proteins or indirectly controlling protein
expression (39). The downstream
regulated genes of AKT, Cyclin D1 and CDK2, are crucial
transcriptional factors in the G0/G1 phase (41). Cyclin A-CDK2 and Cyclin D-CDK4/6
protein kinase complexes, important cell cycle regulators, regulate
progression from the G1/G0 to the S phase (42). D-Cyclins drive entry into the S
phase by releasing E2F transcription factors following
extracellular mitogenic stimulation. Cyclin A-CDK2 protein kinase
complexes regulate proliferation and the cell cycle in lung and
renal cancer (43,44). The findings of the present study
demonstrate that miR-1254 overexpression and RASSF9 siRNA can
promote Cyclin D1 expression and drive more cells into the S phase
by activating the AKT signaling pathway, while anti-miR-1254 and
RASSF9 overexpression may inhibit Cyclin D1 expression and induce
G1-phase arrest by suppressing the AKT signaling pathway. These
results suggest that miR-1254 may promote the expression of Cyclin
D1 and induce G1-S phase transition by activating the AKT signaling
pathway via targeting RASSF9.
p53 is a transcription factor able to regulate the
expression of numerous downstream target genes responsible for
directly or indirectly controlling senescence, apoptosis, DNA
repair and genetic stability in response to various cellular
stresses (45). The results of
this study indicated that miR-1254 overexpression or RASSF9 siRNA
inhibited breast cancer cell apoptosis by suppressing p53
expression; anti-miR-1254 and RASSF9 overexpression induced
apoptosis by promoting p53 expression. These results indicate that
miR-1254 may suppress breast cancer cell apoptosis by regulating
p53 expression via RASSF9.
In conclusion, this study demonstrated that miR-1254
functions as an oncogene in breast cancer. We found that miR-1254
was both upregulated and associated with certain
clinicopathological characteristics of patients with breast cancer.
miR-1254 promoted breast cancer cell proliferation by activating
the AKT signaling pathway and suppressed apoptosis through the
regulation of p53 expression via RASSF9. These findings indicate
that miR-1254 plays a crucial role in breast cancer progression and
may thus represent a latent novel target for breast cancer
therapy.
Acknowledgments
Not applicable.
Funding
This study was supported by the Shaanxi Province
Natural Science Foundation (grant no. 2016JM4263) and the China
Postdoctoral Science Foundation (grant no. 2016M546537).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
BL and JH designed the experiments. PC and JW
collected the clinical data and samples. BL, PC, JW, LW, MR and RZ
performed the experiments. BL and JW performed the statistical
analysis. BL, LW and JH wrote and edited the manuscript. JH
supervised the study. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
For the use of patient samples, informed consent was
obtained from each patient prior to specimen collection. The study
protocol was approved by the Ethics Committee of Xi’an Jiaotong
University (Xi’an, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Obayashi S, Horiguchi J, Higuchi T,
Katayama A, Handa T, Altan B, Bai T, Bao P, Bao H, Yokobori T, et
al: Stathmin1 expression is associated with aggressive phenotypes
and cancer stem cell marker expression in breast cancer patients.
Int J Oncol. 51:781–790. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan L, Strasser-Weippl K, Li JJ, St Louis
J, Finkelstein DM, Yu KD, Chen WQ, Shao ZM and Goss PE: Breast
cancer in China. Lancet Oncol. 15:e279–e289. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Espinoza-Sánchez NA, Vadillo E, Balandrán
JC, Monroy-García A, Pelayo R and Fuentes-Pananá EM: Evidence of
lateral transmission of aggressive features between different types
of breast cancer cells. Int J Oncol. 51:1482–1496. 2017. View Article : Google Scholar :
|
|
5
|
Zhao L, Liu Y, Tong D, Qin Y, Yang J, Xue
M, Du N, Liu L, Guo B, Hou N, et al: MeCP2 promotes gastric cancer
progression through regulating FOXF1/Wnt5a/β-Catenin and MYOD1/
Caspase-3 signaling pathways. EBioMedicine. 16:87–100. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Raman V, Fuentes Lorenzo JL, Stashenko EE,
Levy M, Levy MM and Camarillo IG: Lippia origanoides extract
induces cell cycle arrest and apoptosis and suppresses NF-κB
signaling in triple-negative breast cancer cells. Int J Oncol.
51:1801–1808. 2017. View Article : Google Scholar :
|
|
7
|
Bhardwaj A, Rosen D, Liu M, Liu Y, Hao Q,
Ganesan N, Etzel CJ, Gullett A, Albarracin CT and Bedrosian I:
Suppression of Akt-mTOR pathway-a novel component of oncogene
induced DNA damage response barrier in breast tumorigenesis. PLoS
One. 9:e970762014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Byler S, Goldgar S, Heerboth S, Leary M,
Housman G, Moulton K and Sarkar S: Genetic and epigenetic aspects
of breast cancer progression and therapy. Anticancer Res.
34:1071–1077. 2014.PubMed/NCBI
|
|
9
|
Zhao LY, Tong DD, Xue M, Ma HL, Liu SY,
Yang J, Liu YX, Guo B, Ni L, Liu LY, et al: MeCP2, a target of
miR-638, facilitates gastric cancer cell proliferation through
activation of the MEK1/2-ERK1/2 signaling pathway by upregulating
GIT1. Oncogenesis. 6:e3682017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hyrina A, Olmstead AD, Steven P, Krajden
M, Tam E and Jean F: Treatment-induced viral cure of hepatitis c
virus-infected patients involves a dynamic interplay among three
important molecular players in lipid homeostasis: Circulating
microRNA (miR)-24, miR-223, and proprotein convertase
subtilisin/kexin type 9. EBioMedicine. 23:68–78. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhao LY, Yao Y, Han J, Yang J, Wang XF,
Tong DD, Song TS, Huang C and Shao Y: miR-638 suppresses cell
proliferation in gastric cancer by targeting Sp2. Dig Dis Sci.
59:1743–1753. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xu J, Lin H, Li G, Sun Y, Chen J, Shi L,
Cai X and Chang C: The miR-367-3p increases sorafenib chemotherapy
efficacy to suppress hepatocellular carcinoma metastasis through
altering the androgen receptor signals. EBioMedicine. 12:55–67.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Selbach M, Schwanhäusser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shi M, Liu D, Shen B and Guo N: Helpers of
the cellular gatekeeper-miRNAs dance in P53 network. Biochim
Biophys Acta. 1805:218–225. 2010.
|
|
15
|
Sahu A, Jha PK, Prabhakar A, Singh HD,
Gupta N, Chatterjee T, Tyagi T, Sharma S, Kumari B, Singh S, et al:
MicroRNA-145 impedes thrombus formation via targeting tissue factor
in Venous thrombosis. EBioMedicine. 26:175–186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng Y, Lv X, Wang X, Wang B, Shao X,
Huang Y, Shi L, Chen Z, Huang J and Huang P: MiR-181b promotes
chemoresistance in breast cancer by regulating Bim expression.
Oncol Rep. 35:683–690. 2016. View Article : Google Scholar
|
|
17
|
Ji Y, Han Z, Shao L and Zhao Y: Evaluation
of in vivo antitumor effects of low-frequency ultrasound-mediated
miRNA-133a microbubble delivery in breast cancer. Cancer Med.
5:2534–2543. 2016. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li H, Yang T, Shang D and Sun Z: miR-1254
promotes lung cancer cell proliferation by targeting SFRP1. Biomed
Pharmacother. 92:913–918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Q, Shen W, Li X, Zhang L and Jin X: The
lncRNA n340790 accelerates carcinogenesis of thyroid cancer by
regulating miR-1254. Am J Transl Res. 9:2181–2194. 2017.PubMed/NCBI
|
|
20
|
Lu M, Chen WH, Wang CY, Mao CQ and Wang J:
Reciprocal regulation of miR-1254 and c-Myc in oral squamous cell
carcinoma suppresses EMT-mediated metastasis and tumorinitiating
properties through MAPK signaling. Biochem Biophys Res Commun.
484:801–807. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sherwood V, Recino A, Jeffries A, Ward A
and Chalmers AD: The N-terminal RASSF family: A new group of
Ras-association-domain-containing proteins, with emerging links to
cancer formation. Biochem J. 425:303–311. 2010. View Article : Google Scholar
|
|
22
|
Zheng X, Dong Q, Zhang X, Han Q, Han X,
Han Y, Wu J, Rong X and Wang E: The coiled-coil domain of oncogene
RASSF 7 inhibits hippo signaling and promotes non-small cell lung
cancer. Oncotarget. 8:78734–78748. 2017.PubMed/NCBI
|
|
23
|
Lock FE, Underhill-Day N, Dunwell T,
Matallanas D, Cooper W, Hesson L, Recino A, Ward A, Pavlova T,
Zabarovsky E, et al: The RASSF8 candidate tumor suppressor inhibits
cell growth and regulates the Wnt and NF-κB signaling pathways.
Oncogene. 29:4307–4316. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Liu W, Zhang YP and Huang XR: The
miR-224 promotes non-small cell lung cancer cell proliferation by
directly targeting RASSF8. Eur Rev Med Pharmacol Sci. 21:3223–3231.
2017.PubMed/NCBI
|
|
25
|
He C, Wang L, Zhang J and Xu H:
Hypoxia-inducible microRNA-224 promotes the cell growth, migration
and invasion by directly targeting RASSF8 in gastric cancer. Mol
Cancer. 16:352017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Huang Y, Li Y, Wang FF, Lv W, Xie X and
Cheng X: Over-expressed miR-224 promotes the progression of
cervical cancer via targeting RASSF8. PLoS One. 11:e01623782016.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hesson LB, Dunwell TL, Cooper WN,
Catchpoole D, Brini AT, Chiaramonte R, Griffiths M, Chalmers AD,
Maher ER and Latif F: The novel RASSF6 and RASSF10 candidate tumour
suppressor genes are frequently epigenetically inactivated in
childhood leukaemias. Mol Cancer. 8:422009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schagdarsurengin U, Richter AM, Wohler C
and Dammann RH: Frequent epigenetic inactivation of RASSF10 in
thyroid cancer. Epigenetics. 4:571–576. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu W, Wang J, Wang L, Qian C, Qian Y,
Xuan H, Zhuo W, Li X, Yu J and Si J: Ras-association domain family
10 acts as a novel tumor suppressor through modulating MMP2 in
hepatocarcinoma. Oncogenesis. 5:e2372016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Richter AM, Walesch SK and Dammann RH:
aberrant promoter methylation of the tumour suppressor RASSF10 and
its growth inhibitory function in breast cancer. Cancers (Basel).
8:262016. View Article : Google Scholar
|
|
31
|
Li X, Liang Q, Liu W, Zhang N, Xu L, Zhang
X, Zhang J, Sung JJ and Yu J: Ras association domain family member
10 suppresses gastric cancer growth by cooperating with GSTP1 to
regulate JNK/c-Jun/AP-1 pathway. Oncogene. 35:2453–2464. 2016.
View Article : Google Scholar
|
|
32
|
Lu D, Ma J, Zhan Q, Li Y, Qin J and Guo M:
Epigenetic silencing of RASSF10 promotes tumor growth in esophageal
squamous cell carcinoma. Discov Med. 17:169–178. 2014.PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) methods. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
34
|
Padi SK, Zhang Q, Rustum YM, Morrison C
and Guo B: MicroRNA-627 mediates the epigenetic mechanisms of
vitamin D to suppress proliferation of human colorectal cancer
cells and growth of xenograft tumors in mice. Gastroenterology.
145:437–446. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gong C, Tan W, Chen K, You N, Zhu S, Liang
G, Xie X, Li Q, Zeng Y, Ouyang N, et al: Prognostic value of a
BCSC-associated microRNA signature in hormone receptor-positive
HER2-negative breast cancer. EBioMedicine. 11:199–209. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng Q, Cui X, Zhang D, Yang Y, Yan X,
Liu M, Niang B, Aziz F, Liu S, Yan Q, et al: miR-200b inhibits
proliferation and metastasis of breast cancer by targeting
fucosyltransferase IV and α1, 3-fucosylated glycans. Oncogenesis.
6:e3582017. View Article : Google Scholar
|
|
37
|
Lee CM, Yang P, Chen LC, Chen CC, Wu SC,
Cheng HY and Chang YS: A novel role of RASSF9 in maintaining
epidermal homeostasis. PLoS One. 6:e178672011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ejaz A, Mitterberger MC, Lu Z, Mattesich
M, Zwierzina ME, Hörl S, Kaiser A, Viertler HP, Rostek U, Meryk A,
et al: Weight loss upregulates the small gtpase diras3 in human
white adipose progenitor cells, which negatively regulates
adipogenesis and activates autophagy via akt-mtor inhibition.
EBioMedicine. 6:149–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Xu N, Lao Y, Zhang Y and Gillespie DA:
AKT: A double-edged sword in cell proliferation and genome
stability. J Oncol. 2012:9517242012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang J, Tong DD, Xue M, Jiang QY, Wang
XF, Yang PB, Ni L, Zhao LY and Huang C: FAM196B acts as oncogene
and promotes proliferation of gastric cancer cells through AKT
signaling pathway. Cell Mol Biol. 63:18–23. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maddika S, Ande SR, Wiechec E, Hansen LL,
Wesselborg S and Los M: Akt-mediated phosphorylation of CDK2
regulates its dual role in cell cycle progression and apoptosis. J
Cell Sci. 121:979–988. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhao LY, Zhang J, Guo B, Yang J, Han J,
Zhao XG, Wang XF, Liu LY, Li ZF, Song TS, et al: MECP2 promotes
cell proliferation by activation ERK1/2 and inhibiting p38 activity
in human hepatocellular carcinoma HEPG2 cells. Cell Mol Biol.
59:OL1876–OL1881. 2013.
|
|
43
|
Juengel E, Euler S, Maxeiner S, Rutz J,
Justin S, Roos F, Khoder W, Nelson K, Bechstein WO and Blaheta RA:
Sulforaphane as an adjunctive to everolimus counteracts everolimus
resistance in renal cancer cell lines. Phytomedicine. 27:1–7. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chu X, Zhang T, Wang J, Li M, Zhang X, Tu
J, Sun S, Chen X and Lu F: Alternative splicing variants of human
Fbx4 disturb cyclin D1 proteolysis in human cancer. Biochem Biophys
Res Commun. 447:158–164. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li T, Kon N, Jiang L, Tan M, Ludwig T,
Zhao Y, Baer R and Gu W: Tumor suppression in the absence of
p53-mediated cell cycle arrest, apoptosis, and senescence. Cell.
149:1269–1283. 2012. View Article : Google Scholar : PubMed/NCBI
|