Introduction
Prostate adenocarcinoma (PRAD) is one of the most
common malignant tumor types of the male reproductive system and
primarily occurs in elderly individuals (aged >65 years)
(1,2). According to the most recent global
statistical data, the number of newly diagnosed PRAD cases in 2012
has reached 1.1 million (3). The
morbidity and mortality associated with PRAD are higher in
developed than in developing countries. In the US, the predicted
number of new cases of PRAD for 2018 is 164,690 and the number of
associated mortalities is 29,430 (4). The mortality from PRAD is
substantially reduced by early screening for prostate-specific
antigen (PSA); However, the number of individuals who die from PRAD
is second only to the number of patients that die from lung cancer,
and PRAD therefore ranks second among cancer-associated deaths in
males (4,5). According to cancer statistics for
2015, 60,300 individuals were diagnosed with PRAD and 26,600
patients succumbed to the disease in China. While these figures are
lower than those for the US, the incidence of PRAD is rapidly
increasing each year in China (6).
Only a small number of indicators are currently used
for predicting the prognosis of PRAD patients, and each indicator
has its own advantages and disadvantages. PSA is currently the most
commonly used diagnostic screening and prognostic indicator for
PRAD; however, its application has poor specificity (7,8).
Circulating tumor cells provide excellent prognostic evaluation of
PRAD, but their use is limited by high equipment requirements and
costs (9). micro (mi)RNAs have
also been demonstrated to have a high prognostic value for PRAD,
but the technology for the extraction and identification of
relevant miRNAs in limited samples remains to be fully developed
and implemented (8,10). Therefore, novel and effective
predictors for the prognosis of patients with PRAD are urgently
required.
One strategy for identifying prognostic predictors
for PRAD is the assessment of alternative splicing (AS), but this
field has remained largely unexplored. AS is an indispensable
process in prokaryotic gene expression (11) and is able to turn mRNA precursors
into different types of mature mRNAs through different processes to
increase the diversity of mRNA types (12). Increasing evidence indicates that
AS has an important role in the development of PRAD (13-16).
The splice isoforms generated by AS are involved in different
aspects of tumor physiology, including growth, apoptosis,
infiltration, metastasis, angiogenesis and metabolism (14). The study of AS in tumors is helping
to elucidate the mechanisms underlying tumor pathogenicity and may
be a strategy in the search for reliable and effective diagnostic
and prognostic targets. AS is classified into the following 7
types, based on the type of splicing: Alternate acceptor site (AA),
alternate donor site (AD), alternate promoter (AP), alternate
terminator (AT), exon skip (ES), mutually exclusive exons (ME) and
retained intron (RI). The products of AS are termed splicing events
(17,18). In the present study, the
percent-spliced-in (PSI) value (ranging from 0 to 1) was used to
perform an intuitive quantitative analysis and comparison of
splicing events (18,19).
AS involves the use of splicing factors as executive
proteins, so that changes in splicing factor expression directly
cause an abnormal expression of splicing events (20). Numerous studies have confirmed that
mutations in splicing factors are closely associated with the
development and progression of tumors (21,22)
and that the expression levels of splicing factors differ
significantly between tumor cells and juxtacancerous tissue
(20,23). For instance, serine- and
arginine-rich splicing factor 1 was reported to be highly expressed
in various tumor types (24), and
pre-mRNA processing factor 6 was indicated to promote tumor cell
growth (25). Therefore, exploring
the potential regulatory association between splicing factors and
splicing events may aid in identifying the pathogenic mechanisms
underlying tumor development.
In recent years, the analysis of splicing events in
tumor research has become possible with the introduction of deep
sequencing techniques. The Cancer Genome Atlas (TCGA) database
(https://portal.gdc.cancer.gov/) now
provides RNA-sequencing data of various types of tumor and
juxta-cancerous tissues, while no normal tissues were included.
Ryan et al (26) used the
RNA-seq data provided by TCGA to establish a TCGASpliceSeq database
(http://bioinformatics.mdanderson.org/TCGASpliceSeq/index.jsp),
which offers a convenient way for researchers to study splicing
events in tumors. The aim of the present study was to perform a
re-calculation and in-depth analysis to determine the disease-free
survival-associated splicing events (DFS-SEs) in patients with
PRAD. The present study endeavored to construct a prognostic index
(PI) for DFS-SEs and to evaluate the prognostic values of these
indicators in PRAD using time-dependent receiver-operator
characteristic (tROC) and Kaplan-Meier curve analyses. In addition,
a correlation analysis was used to construct a potential regulatory
network to explain the associations between splicing factors and
splicing events.
Material and methods
Data acquirement and organization
The splicing event data for PRAD were downloaded
from the TCGASpliceSeq database. In addition, the clinical data for
500 patients with PRAD were acquired from TCGA database.
In total, 91 patients with a 'Person neoplasm cancer
status' of 'Discrepancy', 'Not Available' or 'Unknown', 3 patients
with a 'patient death reason' of 'Other, non-malignant disease' or
'Unknown' and 9 patients with '<30' in 'days to last followup'
were excluded, leaving 397 patients. After matching the patients
with their TCGASpliceSeq database entries according to their
labeling numbers, 394 patients were included in the present
study.
'With tumor' in 'person neoplasm cancer status' was
regarded as the endpoint. The DFS time in 'days to last followup'
was used to calculate the DFS of patients with PRAD.
Identification and analysis of
DFS-SEs
Univariate Cox regression analysis was performed to
analyze and identify DFS-SEs. Cytoscape software was used to plot
the interactive association between the genes corresponding to
DFS-SEs. The Database for Annotation, Visualization and Integrated
Discovery (DAVID) Functional Annotation Result Summary (https://david.ncifcrf.gov/summary.jsp,
version 6.8) was used for Gene Ontology (GO) and Kyoto Encyclopedia
of Genes and Genomes (KEGG) pathway enrichment analysis of the
genes corresponding to the top 500 most significant DFS-SEs in
PRAD. The top five pathways identified by the GO and KEGG analyses
were displayed.
Construction of the PI model for PRAD
based on splicing events
To construct powerful PIs, multivariate Cox
regression analysis was used to exclude irrelevant splicing events.
Multivariate Cox regression analyses were performed for the top 10
DFS-SEs that had the highest prognostic values for each splicing
type and the top 10 DFS-SEs that had the highest prognostic values
for all splicing types. Splicing events with P<0.05 were
selected to constitute PIs. The calculation formula was as
follows:
where β is the regression coefficient.
Evaluation of the prognostic value of
PIs
The efficacy of the PIs in predicting the cancer
status after five years was evaluated by tROC analysis with the R
package 'survivalROC' (27).
Kaplan-Meier curve analysis was performed for the DFS time and
cancer status of the patients with PRAD to evaluate the prognostic
efficacy of the most significant top 10 DFS-SEs and PIs. Univariate
Cox regression analysis was used to calculate the hazard ratio (HR)
values of the PIs and other clinical parameters for PRAD
recurrence.
Construction of a correlation network of
DFS-SEs in PRAD
Information on the splicing factors was downloaded
from the SpliceAid2 database (http://www.introni.it/splicing.html) (28) and included 66 splicing factors. All
splicing events for the splicing factors were identified based on
the results of the univariate Cox regression analysis. A total of
22 splicing events with P<0.01 were selected. In addition, the
top 50 splicing events with the highest prognostic value according
to the univariate Cox analysis were assessed, and events with
P<1×10−20 were inputted into Cytoscape 3.5.1 (The
Cytoscape Consortium, New York, NY, USA) to construct the
network.
Statistical analysis
UpSet was used to visualize the intersection between
genes and the 7 splicing types (29). SPSS software version 22 (IBM Corp.,
Armonk, NY, USA) was used for the univariate and multivariate Cox
regression, Kaplan-Meier curve and Pearson correlation analyses.
The tROC analysis was performed with R (version 3.4.3). P<0.05
was considered to indicate a statistically significant difference.
GraphPad 7.0 (GraphPad Inc., La Jolla, CA, USA) was used to plot
the ROC and Kaplan-Meier curves.
Results
Comprehensive analysis of splicing events
in the PRAD cohort from TCGA dataset
In the present study, a total of 44,070 splicing
events were identified in the cohort of PRAD patients, including
3,524 of the AA type, 3,101 of the AD type, 9,035 of the AP type,
8,663 of the AT type, 16,772 of the ES type, 228 of the ME type and
2,747 of the RI type (Table I).
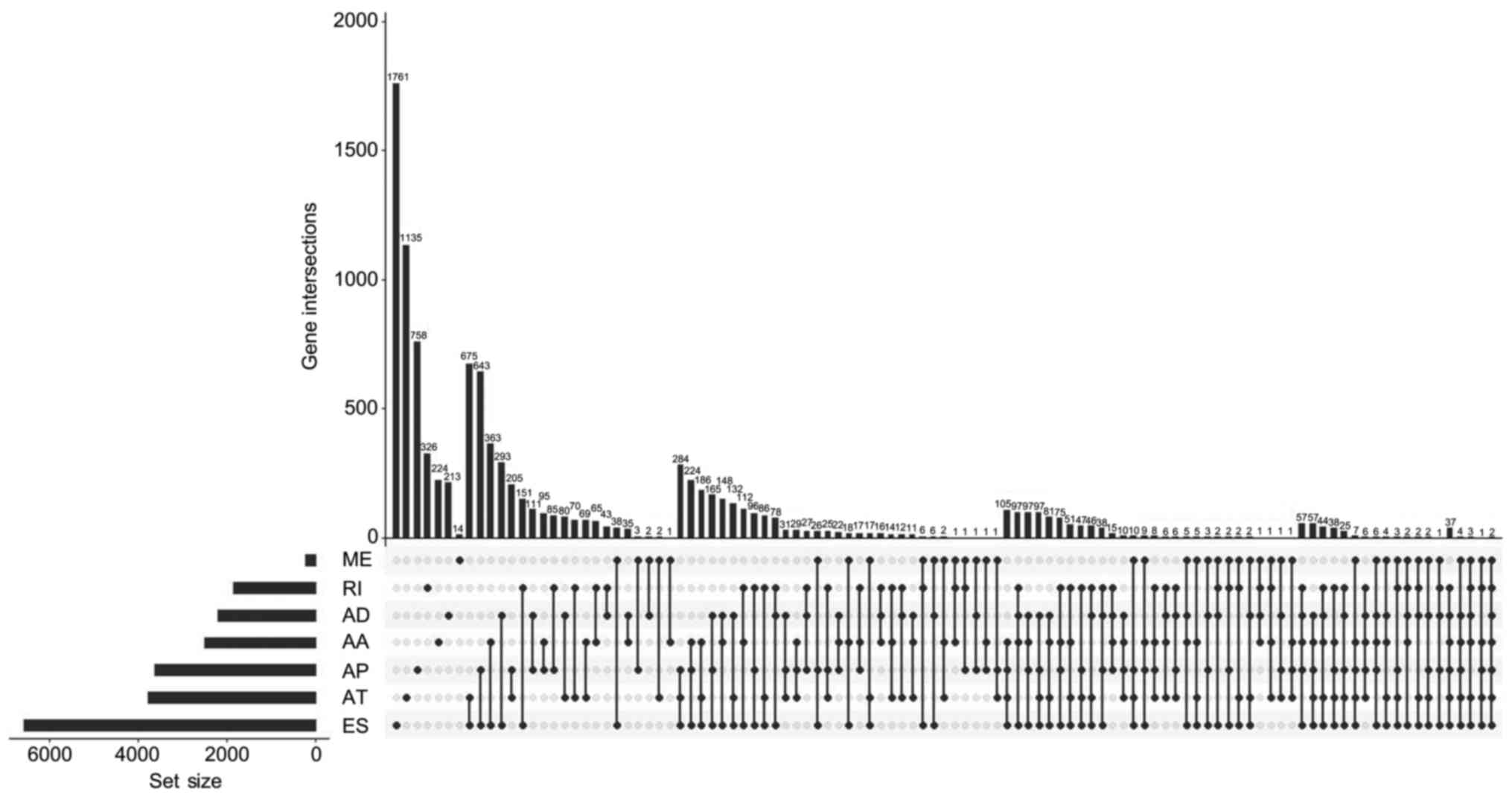
UpSet, which is similar to a Venn diagram, was used to illustrate
the splicing events in PRAD (Fig.
1).
 | Table IOverview of the splicing events in
prostate adenocar-cinoma (n). |
Table I
Overview of the splicing events in
prostate adenocar-cinoma (n).
| Total splicing
events
| DFS-SEs
|
|---|
| Type | Splicing
events | Genes | Splicing
events | Genes |
|---|
| AA | 3524 | 2488 | 373 | 341 |
| AD | 3101 | 2185 | 364 | 325 |
| AP | 9035 | 3621 | 1474 | 826 |
| AT | 8663 | 3781 | 2275 | 1171 |
| ES | 16772 | 6577 | 1956 | 1466 |
| ME | 228 | 221 | 42 | 42 |
| RI | 2747 | 1849 | 425 | 375 |
| Total | 44070 | 10380 | 6909 | 3645 |
Single splicing events were accurately described
using the labelled name of each splicing event, which contained the
gene name, splicing type and AS ID. For instance, for pleckstrin
homology domain containing N1 (PLEKHN1)-ES-1, the gene name is
PLEKHN1, the splicing type is ES and the AS ID is 1.
DFS-SEs in PRAD identified from TCGA
In total, 6,909 splicing events were identified as
DFS-SEs by univariate Cox proportional hazards regression analysis;
these included 373 of the AA type, 364 of the AD type, 1,474 of the
AP type, 2,275 of the AT type, 1,956 of the ES type, 42 of the ME
type and 425 of the RI type (Table
I). The HRs of the top 10 splicing events for PRAD recurrence,
ranked by P-value for each splicing type, are presented in Fig. 2. Kaplan-Meier curve analyses were
performed for the 10 most significant splicing events among all the
DFS-SEs. Patients with PRAD were divided into 'high' and 'low'
groups based on the median PSI value of a certain splicing event.
The patients with PRAD who had a high PSI value for elongin
B-ES-33303, FK506 binding protein 2 (FKBP2)-AP-16603, NHL repeat
containing 3-ES-25701 and thioredoxin-ES-87183 also had a
relatively high chance of DFS, whereas patients with high PSI
values for abhydrolase domain containing 17A-AD-46558,
FKBP2-AP-16602, yippee like 3-AD-36074, high mobility group AT-hook
2-AT-22879 and prostaglandin D2 synthase-AT-88235 had a reduced
probability of DFS (Fig. 3).
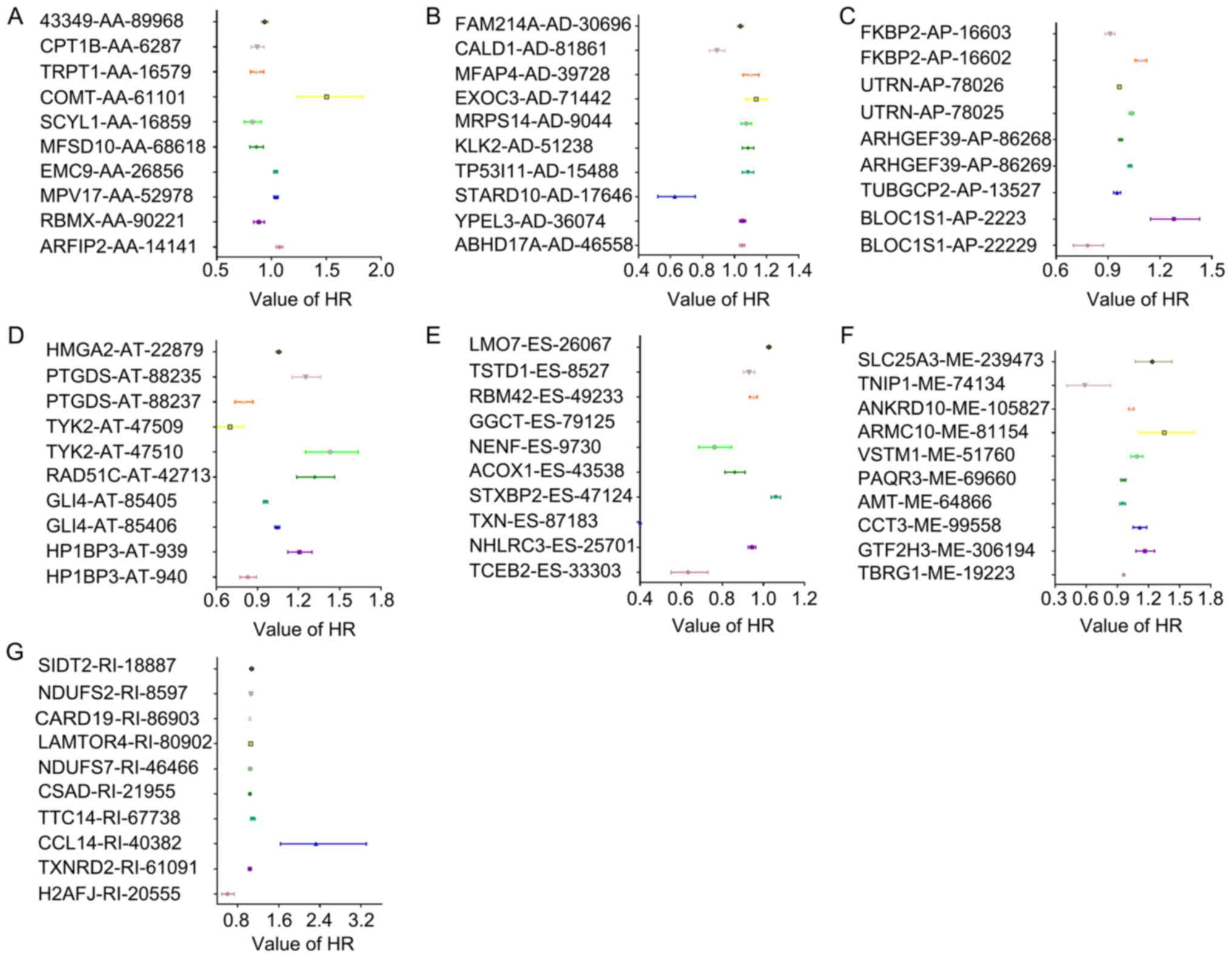 | Figure 2HRs of DFS-SEs for tumor recurrence
in prostate adenocarcinoma. HRs of the top 10 DFS-SEs of (A) AA;
(B) AD; (C) AP; (D) AT; (E) ES; (F) ME; and (G) RI. The horizontal
lines and data-points indicate the HR with 95% confidence interval
for PRAD recurrence. HR, hazard ratio; DFS-SEs, disease-free
survival-associated splicing events; AA, alternate acceptor site;
AD, alternate donor site; AP, alternate promoter; AT, alternate
terminator; ES, exon skip; ME, mutually exclusive exons; RI,
retained intron; ARFIP2, ADP ribosylation factor interacting
protein 2; RBMX, RNA binding motif protein X-linked; MPV17,
mitochondrial inner membrane protein; EMC9, endoplasmic riticulum
membrane protein complex subunit 9; MFSD10, major facilitator
superfamily domain containing 10; SCYL1, SCY1 like pseudokinase 1;
COMT, catechol-O-methyltransferase; TRPT1, tRNA phosphotransferase
1; CPT1B, carnitine palmitoyltransferase 1B; ABHD17A, abhydrolase
domain containing 17A; YPEL3, yippee like 3; STARD10, StAR related
lipid transfer domain containing 10; TP53I11, tumor protein p53
inducible protein 11; KLK2, kallikrein related peptidase 2; MRPS14,
mitochondrial ribosomal protein S14; EXOC3, exocyst complex
component 3; MFAP4, microfibril associated protein 4; CALD1,
caldesmon 1; FAM214A, family with sequence similarity 214 member A;
FKBP2, FK506 binding protein 2; UTRN, utrophin; ARHGEF39, Rho
guanine nucleotide exchange factor 39; TMUB1, transmembrane and
ubiquitin like domain containing 1; TUBGCP2, tubulin γ complex
associated protein 2; BLOC1S1, biogenesis of lysosomal organelles
complex 1 subunit 1; HMGA2, high mobility group AT-hook 2; PTGDS,
prostaglandin D2 synthase; TYK2, tyrosine kinase 2; RAD51C, RAD51
paralog C; GLI4, GLI family zinc finger 4; HP1BP3, heterochromatin
protein 1 binding protein 3; TCEB2, elongin B; NHLRC3, NHL repeat
containing 3; TXN, thioredoxin; STXBP2, syntaxin binding protein 2;
ACOX1, acyl-CoA oxidase 1; NENF, neudesin neurotrophic factor;
GGCT, γ-glutamylcyclotransferase; RBM42, RNA binding motif protein
42; TSTD1, thiosulfate sulfurtransferase like domain containing 1;
LMO7, LIM domain 7; TBRG1, transforming growth factor β regulator
1; GTF2H3, general transcription factor IIH subunit 3; CCT3,
chaperonin containing TCP1 subunit 3; AMT, aminomethyltransferase;
PAQR3, progestin and adipoQ receptor family member 3; VSTM1, V-set
and transmembrane domain containing 1; ARMC10, armadillo repeat
containing 10; ANKRD10, ankyrin repeat domain 10; TNIP1, tumor
necrosis factor α-induced protein 3 interacting protein 1; SLC25A3,
solute carrier family 25 member 3; H2AFJ, H2A histone family member
J; TXNRD2, TXN reductase 2; CCL14, C-C motif chemokine ligand 14;
TTC14, tetratricopeptide repeat domain 14; CSAD, cysteine sulfinic
acid decarboxylase; NDUFS7, NADH:ubiquinone oxidoreductase core
subunit S7; LAMTOR4, late endosomal/lysosomal adaptor,
mitogen-activated protein kinase and mammalian target of rapamycin
activator 4; CARD19, caspase recruitment domain family member 19;
SIDT2, SID1 transmembrane family member 2. |
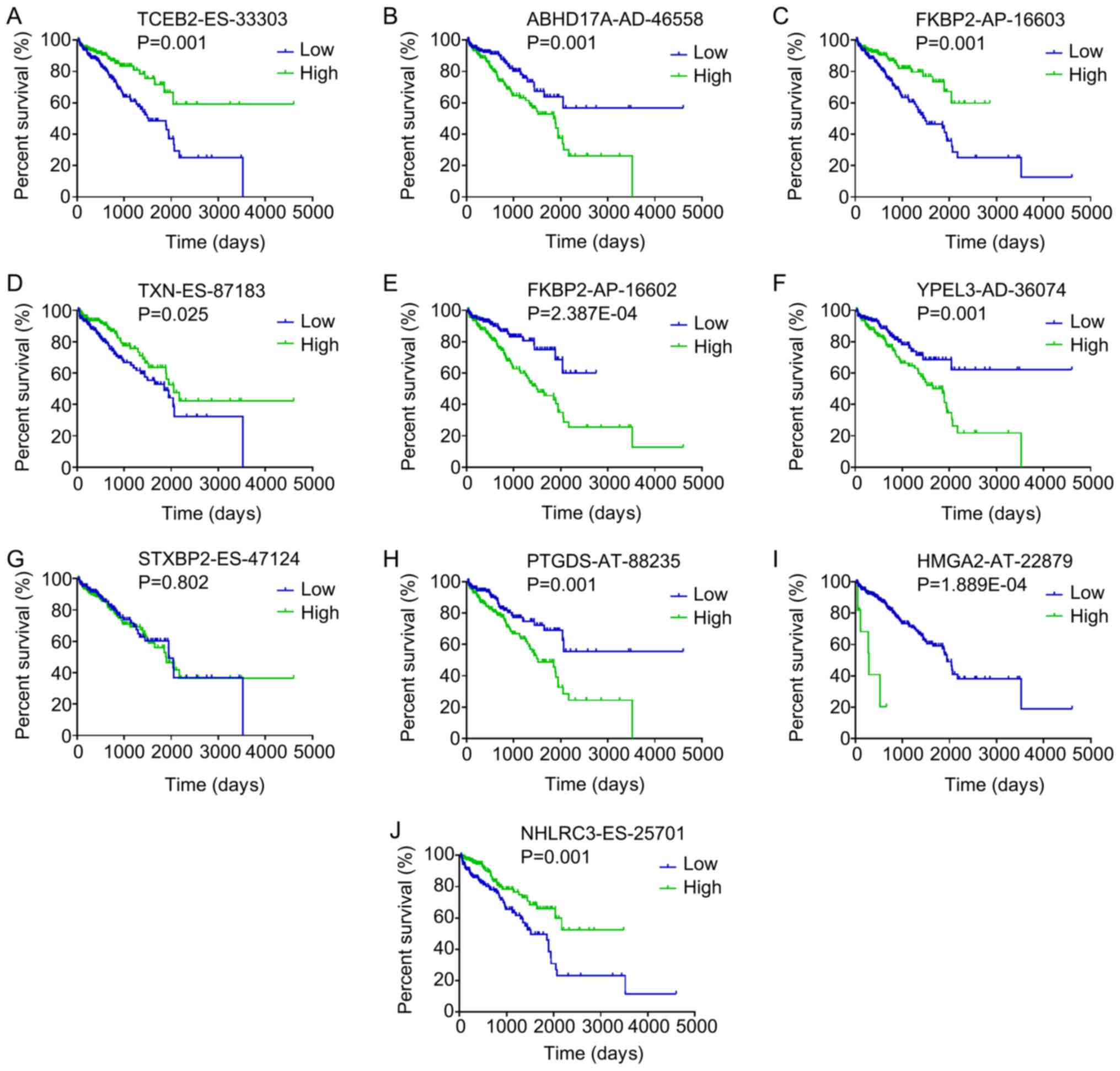 | Figure 3Kaplan-Meier curves for the top 10
most significant disease-free survival-associated splicing events
in PRAD identified by univariate Cox proportional hazards
regression analysis. Patients with PRAD were divided into 'high'
and 'low' groups based on the median percent-spliced-in value of a
certain splicing event. Kaplan-Meier curves for (A) TCEB2-ES-33303,
(B) ABHD17A-AD-46558, (C) FKBP2-AP-16603, (D) TXN-ES-87183, (E)
FKBP2-AP-16602, (F) YPEL3-AD-36074, (G) STXBP2-ES-47124, (H)
PTGDS-AT-88235, (I) HMGA2-AT-22879 and (J) NHLRC3-ES-25701. PRAD,
prostate adenocarcinoma; AA, alternate acceptor; AD, alternate
donor; AP, alternate promoter; AT, alternate terminator; ES, exon
skip; ME, mutually exclusive exons; RI, retained intron; TCEB2,
elongin B; ABHD17A, abhydrolase domain containing 17A; FKBP2, FK506
binding protein 2; TXN, thioredoxin; YPEL3, yippee like 3; STXBP2,
syntaxin binding protein 2; PTGDS, prostaglandin D2 synthase;
HMGA2, high mobility group AT-hook 2; NHLRC3, NHL repeat containing
3. |
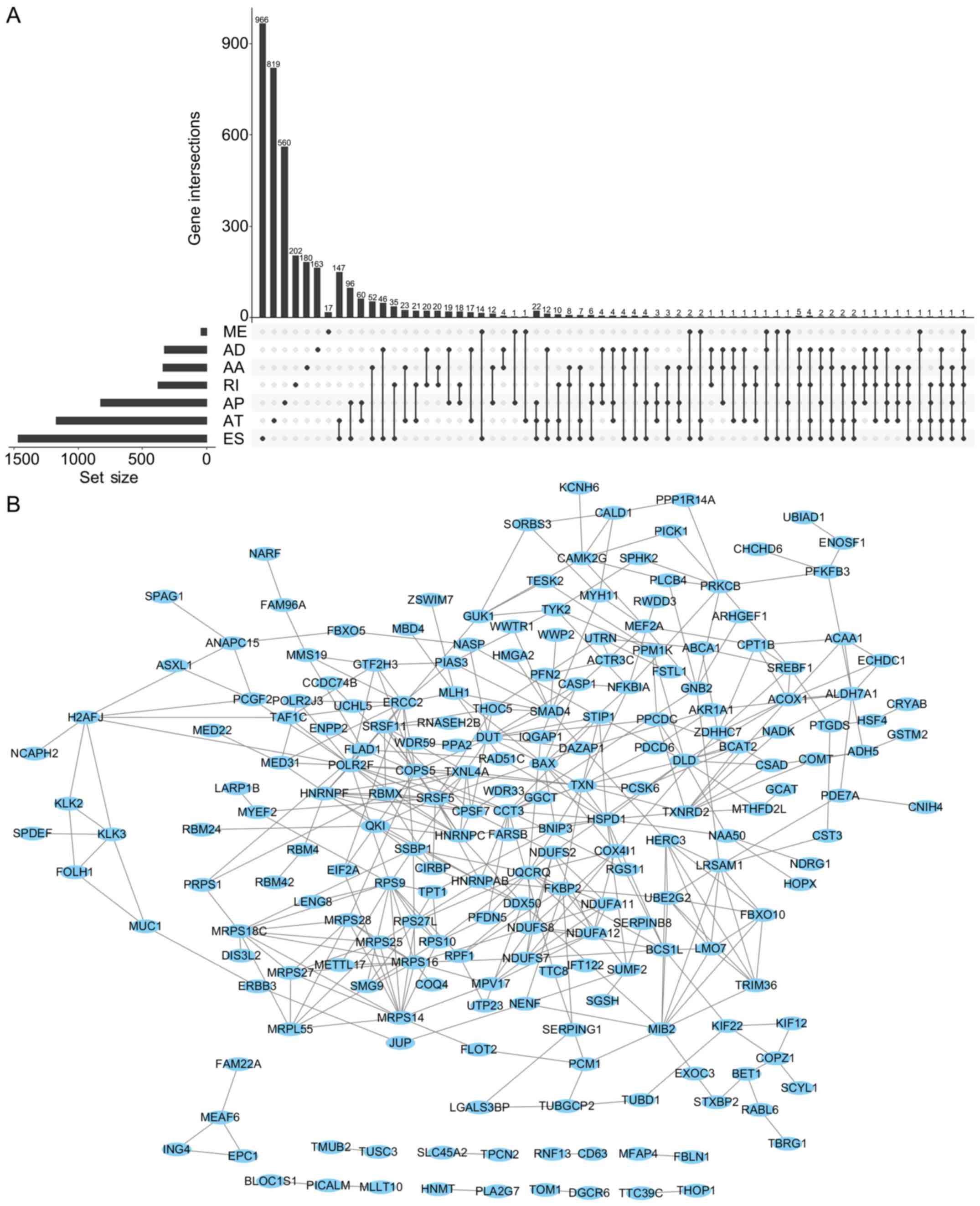
The UpSet was also used to quantitatively display
the intersection of genes and splicing types of the DFS-SEs in PRAD
(Fig. 4A). Numerous splicing
events of certain genes, including inositol hexakisphosphate kinase
2, mammalian target of rapamycin-associated protein, LST8 homolog
and autophagy related 16 like 2, were associated with DFS.
Selection of the top 500 DFS-SEs and input of their corresponding
356 genes into Cytoscape for analysis provided the gene interaction
network (Fig. 4B). Of note, a
total of 356 counterpart genes of the 500 most significant DFS-SEs
were linked to mitochondria and associated pathways according to GO
annotation, including 'mitochondrial electron trans-port, NADH to
ubiquinone', 'mitochondrial translational elongation' and
'mitochondrial translational termination' in the category
Biological Process (BP) and 'mitochondrion', 'mitochondrial inner
membrane' in the category Cellular Component (CC) (Fig. 5).
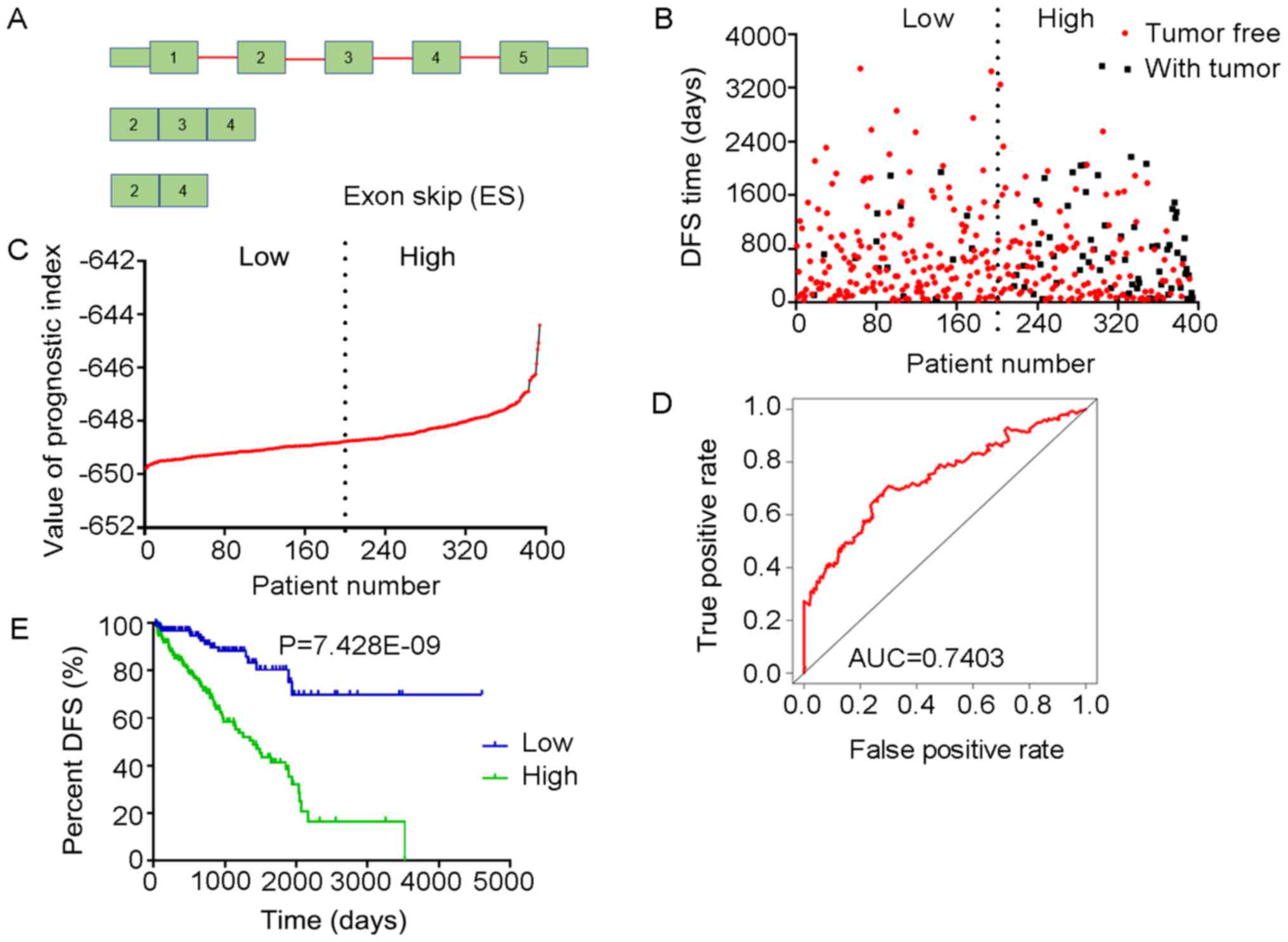
PI models based on the DFS-SEs in
patients with PRAD
Multivariate Cox regression analyses were performed
based on the top 10 significant DFS-SEs for each of the seven
splicing types and for all splicing types. PIs constructed based on
splicing events of the AA, AD, AP, AT, ES, ME, RI and all splicing
types, and their associated data are presented in Figs. 6-13, respectively. For simplification,
these eight PIs were abbreviated as PI-AA, PI-AD, PI-AP, PI-AT,
PI-ES, PI-ME, PI-RI and PI-ALL, respectively. The calculation of
PIs was performed according to the formula mentioned above.
Prognostic value of PIs in patients with
PRAD
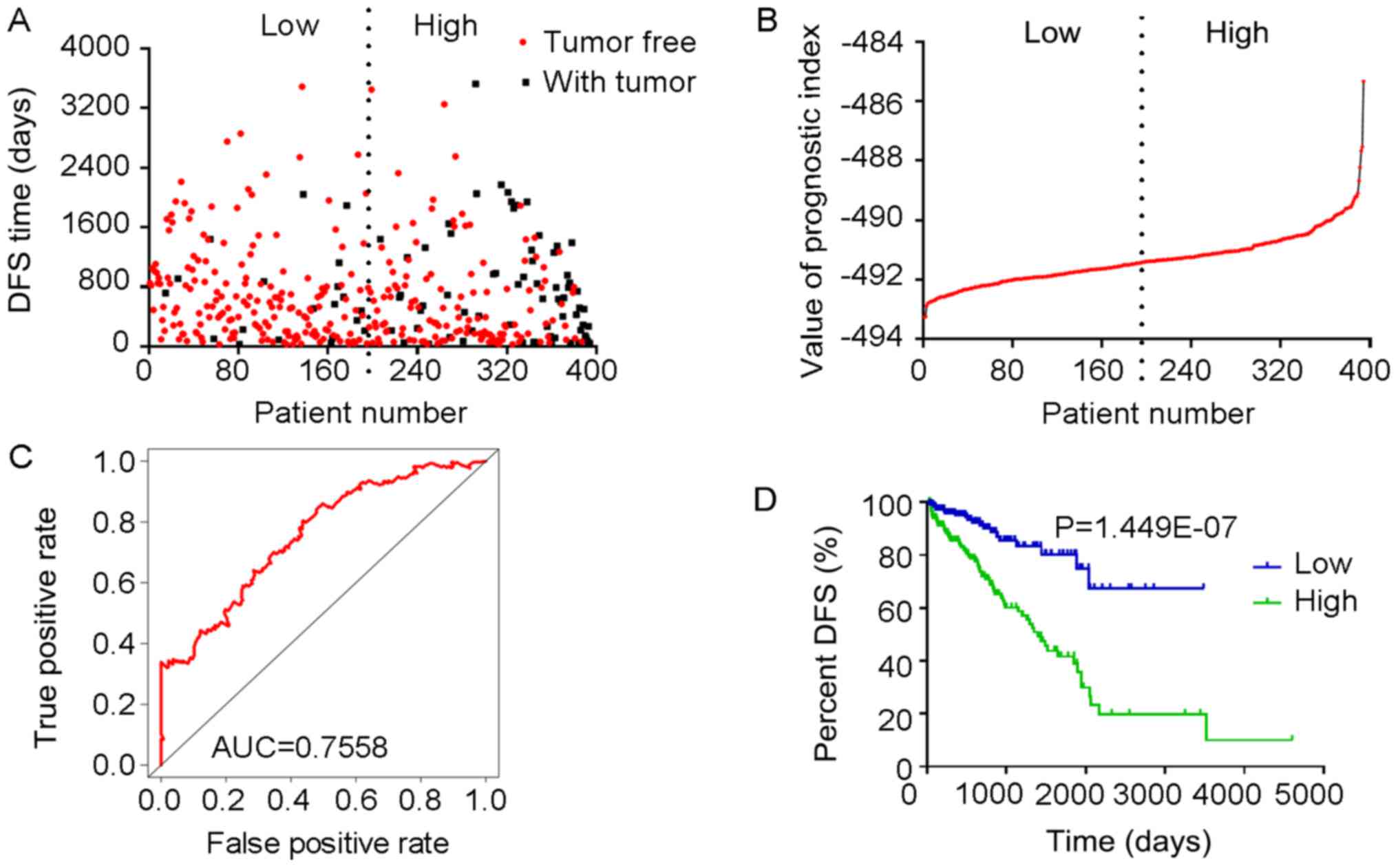
The tROC curve analyses indicated that PI-ME was the
most effective PI at predicting the cancer status after five years,
with an AUC value of 0.7606 (Fig.
13D). This was followed by PI-ALL and PI-ES, which exhibited
AUC values of 0.7558 (Fig. 10E)
and 0.7403 (Fig. 6E),
respectively. The median values of the eight PIs were then used to
classify the patients with PRAD into low- and high-level groups for
the Kaplan-Meier curve analyses. The low and high groups into which
the patients with PRAD were stratified based on the median value of
PI-ES exhibited the most significant difference in DFS (low vs.
high, 3,588.45±250.51 vs. 1,531.08±136.50 days;
P=7.43×10−9). After this, the most significant
differences between low and high groups were those with
stratification based on PI-ME and PI-AD (Table II).
 | Table IIAnalysis of Kaplan-Meier curves of PI
for evaluating the DFS time of patients with prostate
adenocarcinoma stratified by the PI value (cutoff at median). |
Table II
Analysis of Kaplan-Meier curves of PI
for evaluating the DFS time of patients with prostate
adenocarcinoma stratified by the PI value (cutoff at median).
| Type/group | DFS time
(days) | P-value |
|---|
| PI-AA | | |
| Low
(<−5.88) |
2583.742±174.316 |
4.13×10−5 |
| High (≥−5.88) |
1778.112±200.024 | |
| PI-AD | | |
| Low
(<−8.67) |
3306.887±246.458 |
5.99×10−5 |
| High (≥−8.67) |
1704.737±145.898 | |
| PI-AP | | |
| Low
(<4.89) |
1721.248±135.488 |
1.95×10−2 |
| High (≥4.89) |
2741.437±305.572 | |
| PI-AT | | |
| Low
(<−34.84) |
3223.958±272.204 |
6.80×10−6 |
| High
(≥−34.84) |
1659.270±149.443 | |
| PI-ES | | |
| Low
(<−648.78) |
3588.446±250.513 |
7.43×10−9 |
| High
(≥−648.78) |
1531.083±136.504 | |
| PI-ME | | |
| Low
(<−1.56) |
3412.259±231.568 |
2.95×10−7 |
| High (≥−1.56) |
1450.951±129.526 | |
| PI-RI | | |
| Low
(<0.87) |
2395.445±160.811 |
4.59×10−3 |
| High (≥0.87) |
1784.987±247.797 | |
| PI-ALL | | |
| Low
(<−491.40) |
2751.999±171.559 |
1.45×10−7 |
| High
(≥−491.40) |
1700.247±184.410 | |
The prognostic value of the PIs and other clinical
parameters, including age, Gleason score, PSA value, pathologic T
stage and pathologic N stage, were then analyzed using univariate
Cox analysis. Except for age, the other clinical parameters and the
eight PIs all had a high predictive value regarding the DFS of
patients with PRAD (Table III).
In addition, the multivariate analysis revealed that only the
pathologic T stage, PI-AT, PI-ME and PI-ALL had an independent
significant prognostic value for predicting the DFS of patients
with PRAD (Table III).
 | Table IIILogistic regression analysis of the
association of clinical parameters and the PIs with the risk of
prostate adenocarcinoma recurrence. |
Table III
Logistic regression analysis of the
association of clinical parameters and the PIs with the risk of
prostate adenocarcinoma recurrence.
| Univariate Cox
analysis
| Multivariate Cox
analysis
|
|---|
| Clinical
feature | HR (95% CI) | P-value | HR (95% CI) | P-value |
|---|
| Age (≥61 vs. <61
years) | 1.463
(0.952-2.246) |
8.20×10−2 | 1.103
(0.663-1.834) |
7.06×10−1 |
| Gleason score (≥8
vs. <8) | 4.515
(2.738-7.444) |
3.47×10−9 | 1.429
(0.689-2.961) |
3.38×10−1 |
| PSA value (≥0.1 vs.
<0.1) | 5.587
(3.451-9.043) |
2.54×10−12 | 2.946
(1.702-5.099) |
1.14×10−4 |
| Pathologic T stage
(T3/T4 vs. T1/T2) | 7.010
(3.233-15.199) |
8.17×10−7 | 4.180
(1.403-12.450) |
1.00×10−1 |
| Pathologic N stage
(N1 vs. N0) | 2.635
(1.666-4.166) |
3.42×10−5 | 1.024
(0.589-1.780) |
9.34×10−1 |
| PI-AA (≥−5.88 vs.
<−5.88) | 2.531
(1.598-4.008) | 7.57
×10−8 | 1.588
(0.847-2.980) |
1.50×10−1 |
| PI-AD (≥−8.67 vs.
<−8.67) | 2.479
(1.567-3.922) | 1.05
×10−4 | 0.942
(0.502-1.769) |
8.53×10−1 |
| PI-AP (≥4.89 vs.
<4.89) | 0.578
(0.370-0.902) | 1.60
×10−2 | 1.080
(0.600-1.946) |
7.97×10−1 |
| PI-AT (≥−34.84 vs.
<−34.84) | 2.604
(1.668-4.065) | 2.54
×10−5 | 1.173
(0.635-2.167) |
6.11×10−1 |
| PI-ES (≥−648.78 vs.
<−648.78) | 4.097
(2.439-6.881) | 9.77
×10–8 | 2.599
(1.329-5.084) |
5.00×10−3 |
| PI-ME (≥−1.56 vs.
<−1.56) | 3.152
(1.985-5.004) | 1.12
×10−6 | 1.775
(0.999-3.155) |
5.00×10−2 |
| PI-RI (≥0.87 vs.
<0.87) | 1.836
(1.199-2.811) | 5.00
×10−3 | 0.624
(0.337-1.157) |
1.34×10−1 |
| PI-ALL (≥−491.40
vs. <−491.40) | 3.512
(2.131-5.789) | 8.28
×10−7 | 1.273
(0.641-2.528) |
4.91×10−1 |
Potential correlation network of DFS-SEs
in PRAD
Splicing factors are key genes for the regulation of
the development of splicing events (30). Splicing factors also have their own
corresponding splicing events. A correlation analysis was used to
examine the potential regulatory association between the splicing
events of splicing factors and non-splicing factors and to
construct a regulatory network.
A total of 66 splicing factors were obtained from
the SpliceAid2 database. Based on the univariate Cox analysis, 22
splicing events of the 66 splicing factors with P<0.01 were
selected, as well as the most significant top 50 splicing events of
non-splicing factors with P<2.5×10−06. The
correlations between the PSI values of the 22 splicing events of
the splicing factors and the PSI values of the top 50 splicing
events of non-splicing factors were calculated. Splicing events
with P<1×10−20 according to the correlation analysis
were then introduced into Cytoscape to generate the network
(Fig. 14A). The red nodes in the
graph indicate the splicing events of the splicing factors (n=17)
and the light blue nodes indicate the splicing events of
non-splicing factors (n=25). The triangular nodes indicate the
splicing events associated with good prognosis and the oval nodes
indicate the splicing events associated with poor prognosis. The
blue lines indicate negative correlations between two splicing
events and the green lines indicate positive correlations between
two splicing events.
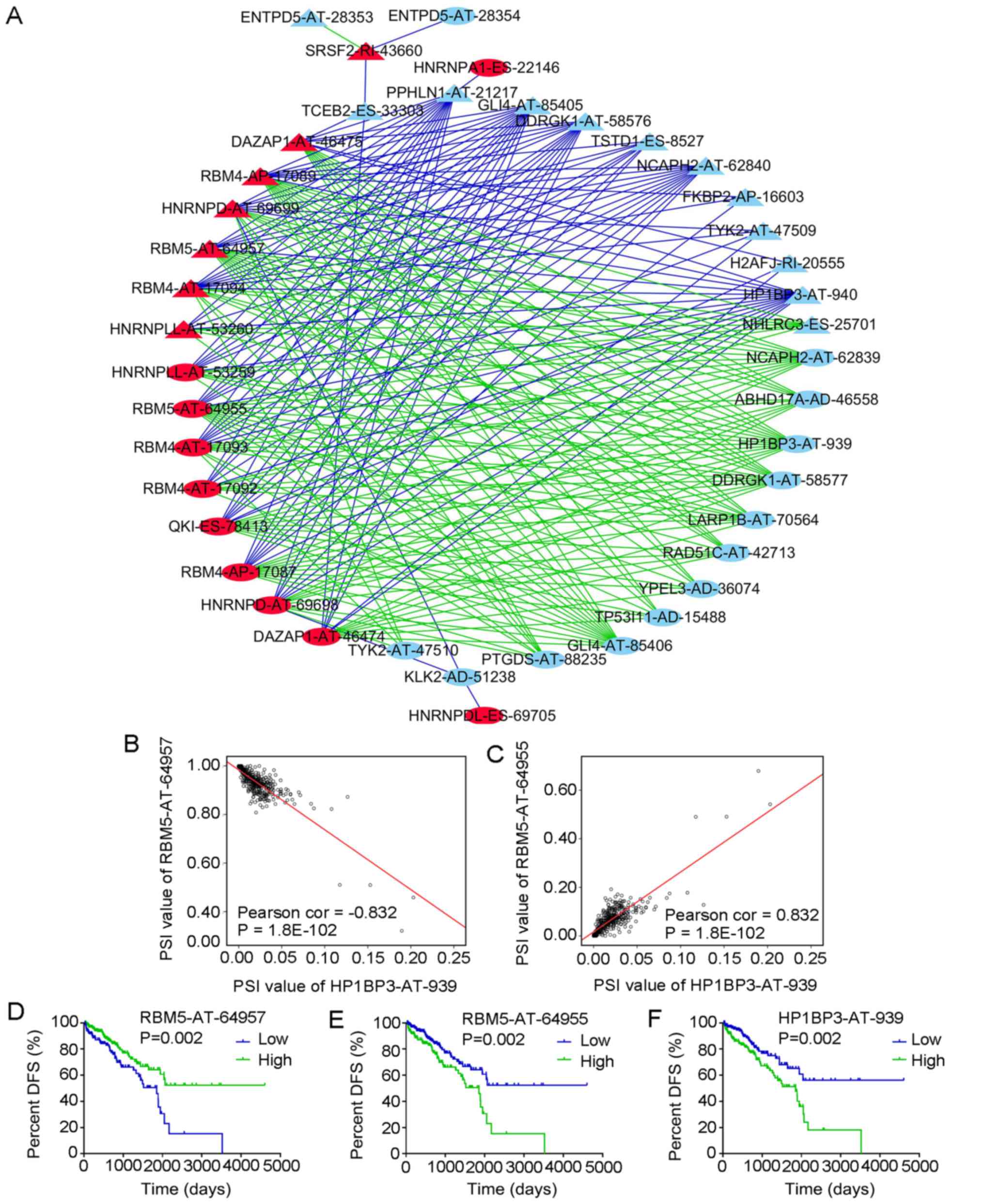 | Figure 14Correlation analysis between splicing
events of splicing factors and DFS-associated splicing events. (A)
Correlation network of splicing events in PRAD. Red nodes represent
splicing events of splicing factors, light blue nodes represent
splicing events of non-splicing factors, triangle nodes represent
splicing events positively associated with DFS, oval nodes
represent splicing events negatively associated with DFS, blue
lines represent negative correlations and green lines represent
positive correlations. (B) Correlation of the PSI value between
RBM5-AT-64957 and HP1BP3-AT-939. RBM5-AT-64957 is one of the
splicing events of the splicing factor RBM5. (C) Correlation of the
PSI value between RBM5-AT-64955 and HP1BP3-AT-939. RBM5-AT-64955 is
one of the splicing events of the splicing factor RBM5. (D-F)
Kaplan-Meier curves for evaluating the DFS time of PRAD patients
stratified by the median PSI value of (D) RBM5-AT-64957, (E)
RBM5-AT-64955 and (F) HP1BP3-AT-939 (high/low). PRAD, prostate
adenocarcinoma; DFS, disease-free survival; AT, Alternate
Terminator type of splicing events; PSI, percent-spliced-in; RBM4,
RNA binding motif protein 4; DAZAP1, DAZ-associated protein 1; QKI,
QKI, KH domain containing RNA binding; HNRNPDL, heterogeneous
nuclear ribonucleoprotein D like; SRSF2, serine and arginine rich
splicing factor 2; TSTD1, thiosulfate sulfurtransferase like domain
containing 1; HP1BP3, heterochromatin protein 1 binding protein 3;
DDRGK1, DDRGK domain containing 1; GLI4, GLI family zinc finger 4;
PPHLN1, periphilin 1; YPEL3, yippee like 3; NCAPH2, non-SMC
condensin II complex subunit H2; LARP1B, La ribonucleoprotein
domain family member 1B; TYK2, tyrosine kinase 2; ABHD17A,
abhydrolase domain containing 17A; H2AFJ, H2A histone family member
J; TP53I11, tumor protein p53 inducible protein 11; RAD51C, RAD51
paralog C; KLK2, kallikrein related peptidase 2; PTGDS,
prostaglandin D2 synthase; NHLRC3, NHL repeat containing 3; TCEB2,
elongin B; FKBP2, FK506 binding protein 2; ENTPD5, ectonucleoside
triphosphate diphosphohydrolase 5. |
The correlation network revealed a complex
association between the splicing events of splicing and
non-splicing factors. For instance, the splicing event of splicing
factor serine and arginine-rich splicing factor 2-RI-43660 was
positively correlated with ectonucleoside triphosphate
diphosphohydrolase 5 (ENTPD5)-AT-28353 and was negatively
correlated with ENTPD5-AT-28354. Of note, the splicing events with
good DFS were predominantly negatively correlated with the splicing
events of the splicing factors, whereas the splicing events with
poor DFS were predominantly positively correlated with the splicing
events of the splicing factors (Fig.
14A). Fig. 14B and C also
present two pairs of splicing events of splicing and non-splicing
factors with the most significant positive and negative
correlations among all combinations and their Kaplan-Meier curve
analyses. The PSI values of RNA binding motif protein 5
(RBM5)-AT-64957 and hetero-chromatin protein 1 binding protein 3
(HP1BP3)-AT-939 were negatively correlated (Fig. 14B), while the PSI values of
RBM5-AT-64955 and HP1BP3-AT-939 were positively correlated
(Fig. 14C). Patients with PRAD
were then divided into low and high groups based on the median PSI
value of certain splicing events. PRAD patients in the low group of
RBM5-AT-64957 had a significantly higher DFS than those in the high
group (Fig. 14D), while PRAD
patients in the high RBM5-AT-64955 or HP1BP3-AT-939 group had a
markedly higher DFS than those in the respective low group
(Fig. 14E and F).
Discussion
AS is an important regulatory mechanism of gene
expression, and abnormalities of this mechanism may cause the
development of various diseases (14), including the entirety of the
processes of cancer development and progression. Certain products
of AS may therefore be used as diagnostic and prognostic
indicators, as well as therapeutic targets for cancer (14,20).
A small number of studies have been published on the use AS in
PRAD, but these have primarily focused on splicing variants of the
androgen receptor (14,15,31-33),
proprotein convertase subtilisin/kexin type 6 (34), CD44 (35) and staphylococcal nuclease and tudor
domain containing 1 (36).
However, studies providing a comprehensive evaluation of splicing
events in PRAD are scarce.
In the present study, splicing events in PRAD were
comprehensively evaluated based on data from the TCGASpliceSeq
database. In those analyses, the total number of splicing events
greatly exceeded the number of genes, indicating that AS markedly
increases the diversity of gene expression products in PRAD, as has
been observed in other diseases (14,20,37).
One of the highlights of the present study was the comprehensive
and systematic presentation of all splicing events and DFS-SEs in
PRAD, which may provide clues for studying the functions of AS and
its products in PRAD.
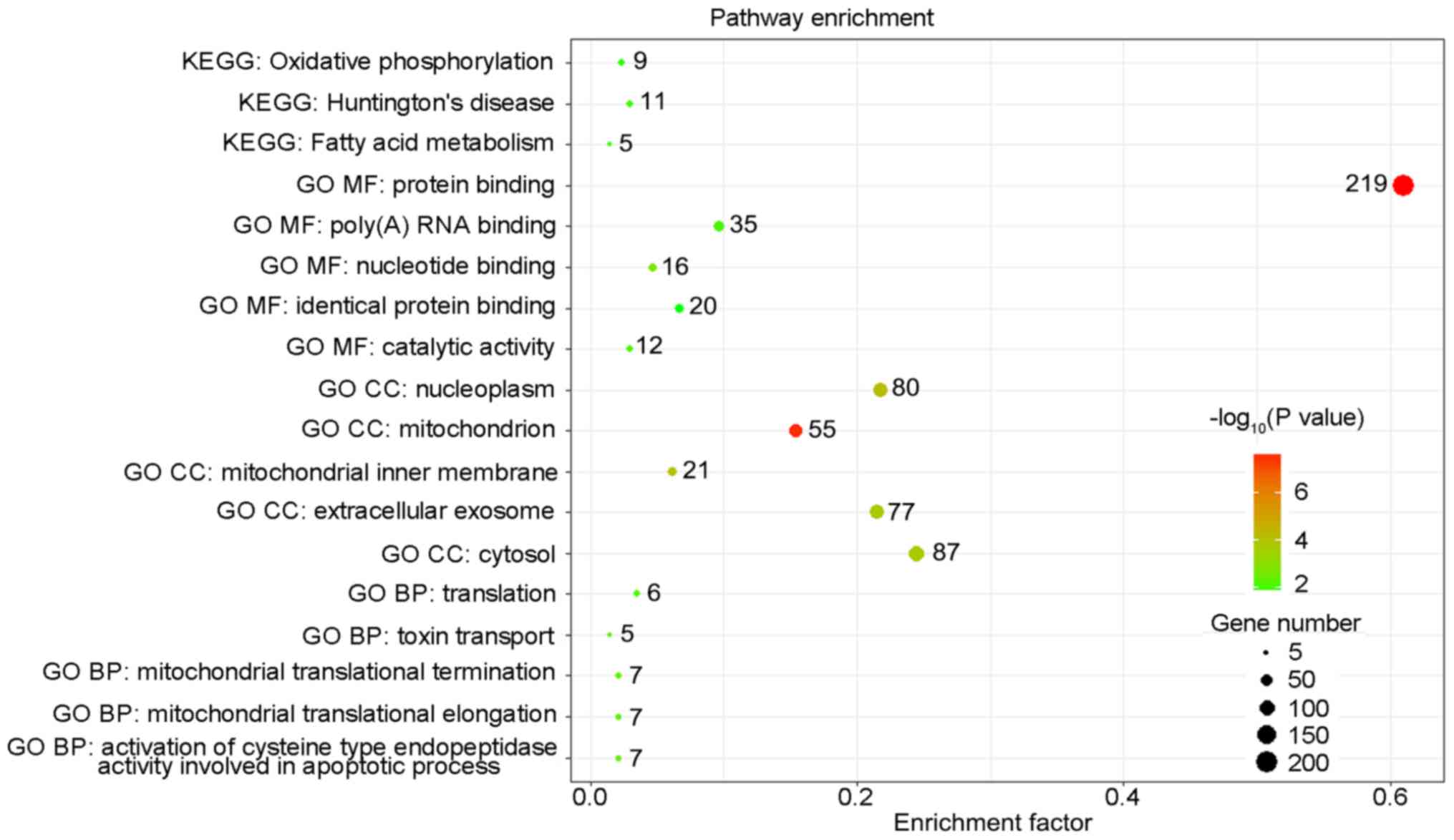
The present study also analyzed the functions of
genes corresponding to DFS-SEs in PRAD, indicating that these genes
were mainly enriched in the fatty acid (FA) metabolism and
oxidative phosphorylation pathways. An increasing number of cohort
studies and experiments have confirmed a close association between
FA metabolism and PRAD. For instance, a cohort study by Brasky
et al (38) indicated an
increased risk for the development of PRAD in males who had high
blood concentrations of long-chain ω-3 polyunsaturated (PU)FAs
(20:5ω3; 22:5ω3; 22:6ω3). The risk of PRAD was also positively
correlated with the percentage of plasma phospholipids and
saturated FAs. In addition, dietary n-6 PU fats, primarily linoleic
acids, were significantly associated with an increased risk of PRAD
(39). Among low-risk PRAD
patients, the ω-3 PUFAs, particularly eicosapentaenoic acid, in
prostate tissues may be a protective factor against PRAD (40).
In addition to risk prediction, FA metabolism also
appears to have an important role in the survival and prognosis of
patients with PRAD. De novo FA synthesis is important for the
survival and progression of PRAD cells. FA synthase, a key enzyme
in FA synthesis, is frequently highly expressed in human PRAD, and
its expression is closely associated with poor prognosis and low
survival rates (41). In
vitro, treatment with phenethyl isothiocyanate inhibited the
oxidative phosphorylation activity in LNCaP and PC-3 human prostate
cancer cells and promoted the production of reactive oxygen species
to induce cancer cell death (42).
Of note, the GO analysis of the present study
indicated that the genes involved in CC and BP pathways were mainly
enriched in the mitochondria and their associated pathways. The
mitochondrion is the major location of cellular FA metabolism and
oxidative phosphorylation (43).
These results were consistent with those of the KEGG analysis,
which indicated enrichment of the DFS-SEs in these two pathways and
further supported the accuracy and reliability of the present
bioinformatics analyses.
In the present study, splicing variants were
indicated to have prognostic value in patients with PRAD. The eight
PIs were constructed based on DFS-SEs without taking the
therapeutic strategies, including targeted molecular therapy and
radiation therapy, into account, which were excluded by a
multivariate Cox analysis (data not shown). tROC analysis indicated
that in patients with PRAD, the AUC of PI-ES was 0.7403 for
predicting the cancer status after five years. In addition, the HR
of PI-ES for tumor recurrence was 4.097 (95% CI: 2.439-6.881,
P=9.77×10−8) according to univariate Cox analysis and
2.599 (95% CI: 1.329-5.084, P=0.005) according to multivariate Cox
analysis, which were better than previously reported values
focusing on only a single indicator, including programmed death-1
receptor methylation (44),
pituitary homeobox 3 methylation (45) and cysteine dioxygenase 1 promoter
methylation (46).
The functions of different transcripts of the same
gene may differ. Therefore, an increasing number of studies have
been assessing the functions of specific mature mRNAs or
transcripts of genes generated by AS (47-50).
In addition, as the major executors of AS, splicing factors have
important roles in the production of the splicing variants of genes
(51,52). Therefore, a regulatory network of
splicing factors and the AS products of regulated genes was
constructed in the present study to elucidate pathogenic mechanisms
of PRAD. The DFS-SEs identified by univariate Cox analysis were
subjected to correlation analyses for the construction of a more
accurate correlation network between splicing events of splicing
factors and other genes. The splicing events of non-splicing
factors associated with a relatively long DFS exhibited a
significant negative correlation with the splicing events of the
splicing factors, while the opposite trend was identified for
splicing events of non-splicing factors associated with poor DFS.
At present, no relevant literature is available to confirm the
potential regulatory associations for the splicing events
discovered in the present study.
The major data of the present study comprehensively
assessed the prognostic value of splicing events in PRAD. Of note,
the present study had certain limitations. The lack of
PRAD-associated mortalities in the TCGA database meant that only
'with tumor' was available as the endpoint in the prognostic
analysis, rather than 'death'. Validation in a larger cohort in a
future study is also required. The present study also identified
numerous splicing events that may theoretically influence the
biological behavior of PRAD, so these splicing events require
verification in experimental biological studies.
In conclusion, the present study comprehensively
assessed AS events in PRAD. PIs generated from DFS-SEs had a high
prognostic value. In addition, a more accurate regulatory network
for AS in PRAD was provided based on a correlation analysis.
Acknowledgments
Not applicable.
Funding
This study was supported by the Innovation Project
of Guangxi Graduate Education (grant no. YCBZ2017044), the
Promoting Project of Basic Capacity for Young and Middle-aged
University Teachers in Guangxi (grant no. KY2016YB090) and the
International Communication of Guangxi Medical University Graduate
Education (2017) from the Education Department of Guangxi.
Availability of data and materials
The PSI value of splicing events and clinical
information in PRAD analyzed in this study may be acquired from the
TCGASpliceSeq and TCGA.
Author's contributions
ZNM provided the study design. ZGH and RQH performed
the analyses and calculations. ZGH, RQH and ZNM all contributed to
the writing of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Salinas CA, Tsodikov A, Ishak-Howard M and
Cooney KA: Prostate cancer in young men: An important clinical
entity. Nat Rev Urol. 11:317–323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tabayoyong W and Abouassaly R: Prostate
Cancer Screening and the Associated Controversy. Surg Clin North
Am. 95:1023–1039. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Etzioni R, Tsodikov A, Mariotto A, Szabo
A, Falcon S, Wegelin J, DiTommaso D, Karnofski K, Gulati R, Penson
DF, et al: Quantifying the role of PSA screening in the US prostate
cancer mortality decline. Cancer Causes Control. 19:175–181. 2008.
View Article : Google Scholar
|
|
6
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kontos CK, Adamopoulos PG and Scorilas A:
Prognostic and predictive biomarkers in prostate cancer. Expert Rev
Mol Diagn. 15:1567–1576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dimakakos A, Armakolas A and Koutsilieris
M: Novel tools for prostate cancer prognosis, diagnosis, and
follow-up. BioMed Res Int. 2014:8906972014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thalgott M, Rack B, Maurer T, Souvatzoglou
M, Eiber M, Kress V, Heck MM, Andergassen U, Nawroth R, Gschwend
JE, et al: Detection of circulating tumor cells in different stages
of prostate cancer. J Cancer Res Clin Oncol. 139:755–763. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hessels D and Schalken JA: Urinary
biomarkers for prostate cancer: A review. Asian J Androl.
15:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wagner SD and Berglund JA: Alternative
pre-mRNA splicing. Methods Mol Biol. 1126:45–54. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kelemen O, Convertini P, Zhang Z, Wen Y,
Shen M, Falaleeva M and Stamm S: Function of alternative splicing.
Gene. 514:1–30. 2013. View Article : Google Scholar
|
|
13
|
Krause WC, Shafi AA, Nakka M and Weigel
NL: Androgen receptor and its splice variant, AR-V7, differentially
regulate FOXA1 sensitive genes in LNCaP prostate cancer cells. Int
J Biochem Cell Biol. 54:49–59. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lapuk AV, Volik SV, Wang Y and Collins CC:
The role of mRNA splicing in prostate cancer. Asian J Androl.
16:515–521. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu LL, Xie N, Sun S, Plymate S, Mostaghel
E and Dong X: Mechanisms of the androgen receptor splicing in
prostate cancer cells. Oncogene. 33:3140–3150. 2014. View Article : Google Scholar
|
|
16
|
Sprenger CC and Plymate SR: The link
between androgen receptor splice variants and castration-resistant
prostate cancer. Horm Cancer. 5:207–217. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Omenn GS, Menon R and Zhang Y: Innovations
in proteomic profiling of cancers: Alternative splice variants as a
new class of cancer biomarker candidates and bridging of proteomics
with structural biology. J Proteomics. 90:28–37. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Y, Sun N, Lu Z, Sun S, Huang J, Chen Z
and He J: Prognostic alternative mRNA splicing signature in
non-small cell lung cancer. Cancer Lett. 393:40–51. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ryan MC, Cleland J, Kim R, Wong WC and
Weinstein JN: SpliceSeq: A resource for analysis and visualization
of RNA-Seq data on alternative splicing and its functional impacts.
Bioinformatics. 28:2385–2387. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sveen A, Kilpinen S, Ruusulehto A, Lothe
RA and Skotheim RI: Aberrant RNA splicing in cancer; expression
changes and driver mutations of splicing factor genes. Oncogene.
35:2413–2427. 2016. View Article : Google Scholar
|
|
21
|
Yoshida K and Ogawa S: Splicing factor
mutations and cancer. Wiley Interdiscip Rev RNA. 5:445–459. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ogawa S: Splicing factor mutations in
myelodysplasia. Int J Hematol. 96:438–442. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Silipo M, Gautrey H and Tyson-Capper A:
Deregulation of splicing factors and breast cancer development. J
Mol Cell Biol. 7:388–401. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karni R, de Stanchina E, Lowe SW, Sinha R,
Mu D and Krainer AR: The gene encoding the splicing factor SF2/ASF
is a proto-oncogene. Nat Struct Mol Biol. 14:185–193. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Adler AS, McCleland ML, Yee S, Yaylaoglu
M, Hussain S, Cosino E, Quinones G, Modrusan Z, Seshagiri S, Torres
E, et al: An integrative analysis of colon cancer identifies an
essential function for PRPF6 in tumor growth. Genes Dev.
28:1068–1084. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res.
44:D1018–D1022. 2016. View Article : Google Scholar :
|
|
27
|
Heagerty PJ, Lumley T and Pepe MS:
Time-dependent ROC curves for censored survival data and a
diagnostic marker. Biometrics. 56:337–344. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Piva F, Giulietti M, Burini AB and
Principato G: SpliceAid 2: A database of human splicing factors
expression data and RNA target motifs. Hum Mutat. 33:81–85. 2012.
View Article : Google Scholar
|
|
29
|
Lex A, Gehlenborg N, Strobelt H, Vuillemot
R and Pfister H: UpSet: Visualization of intersecting sets. IEEE
Trans Vis Comput Graph. 20:1983–1992. 2014. View Article : Google Scholar
|
|
30
|
Fredericks AM, Cygan KJ, Brown BA and
Fairbrother WG: RNA-Binding Proteins: Splicing Factors and Disease.
Biomolecules. 5:893–909. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
He Y, Lu J, Ye Z, Hao S, Wang L, Kohli M,
Tindall DJ, Lim B, Zhu R, Wang L, et al: Androgen receptor splice
variants bind to constitutively open chromatin and promote
abiraterone-resistant growth of prostate cancer. Nucleic Acids Res.
46:1895–1911. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Munkley J, Livermore K, Rajan P and
Elliott DJ: RNA splicing and splicing regulator changes in prostate
cancer pathology. Hum Genet. 136:1143–1154. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ciccarese C, Santoni M, Brunelli M, Buti
S, Modena A, Nabissi M, Artibani W, Martignoni G, Montironi R,
Tortora G, et al: AR-V7 and prostate cancer: The watershed for
treatment selection? Cancer Treat Rev. 43:27–35. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Couture F, Sabbagh R, Kwiatkowska A,
Desjardins R, Guay SP, Bouchard L and Day R: PACE4 Undergoes an
Oncogenic Alternative Splicing Switch in Cancer. Cancer Res.
77:6863–6879. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hernandez JR, Kim JJ, Verdone JE, Liu X,
Torga G, Pienta KJ and Mooney SM: Alternative CD44 splicing
identifies epithelial prostate cancer cells from the mesenchymal
counterparts. Med Oncol. 32:1592015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cappellari M, Bielli P, Paronetto MP,
Ciccosanti F, Fimia GM, Saarikettu J, Silvennoinen O and Sette C:
The transcriptional co-activator SND1 is a novel regulator of
alternative splicing in prostate cancer cells. Oncogene.
33:3794–3802. 2014. View Article : Google Scholar
|
|
37
|
Robert C and Watson M: The incredible
complexity of RNA splicing. Genome Biol. 17:2652016. View Article : Google Scholar
|
|
38
|
Brasky TM, Darke AK, Song X, Tangen CM,
Goodman PJ, Thompson IM, Meyskens FL Jr, Goodman GE, Minasian LM,
Parnes HL, et al: Plasma phospholipid fatty acids and prostate
cancer risk in the SELECT trial. J Natl Cancer Inst. 105:1132–1141.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bassett JK, Severi G, Hodge AM, MacInnis
RJ, Gibson RA, Hopper JL, English DR and Giles GG: Plasma
phospholipid fatty acids, dietary fatty acids and prostate cancer
risk. Int J Cancer. 133:1882–1891. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Moreel X, Allaire J, Léger C, Caron A,
Labonté MÈ, Lamarche B, Julien P, Desmeules P, Têtu B and Fradet V:
Prostatic and dietary omega-3 fatty acids and prostate cancer
progression during active surveillance. Cancer Prev Res (Phila).
7:766–776. 2014. View Article : Google Scholar
|
|
41
|
Gang X, Yang Y, Zhong J, Jiang K, Pan Y,
Karnes RJ, Zhang J, Xu W, Wang G and Huang H: P300
acetyltransferase regulates fatty acid synthase expression, lipid
metabolism and prostate cancer growth. Oncotarget. 7:15135–15149.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Xiao D, Powolny AA, Moura MB, Kelley EE,
Bommareddy A, Kim SH, Hahm ER, Normolle D, Van Houten B and Singh
SV: Phenethyl isothiocyanate inhibits oxidative phosphorylation to
trigger reactive oxygen species-mediated death of human prostate
cancer cells. J Biol Chem. 285:26558–26569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen ZP, Li M, Zhang LJ, He JY, Wu L, Xiao
YY, Duan JA, Cai T and Li WD: Mitochondria-targeted drug delivery
system for cancer treatment. J Drug Target. 24:492–502. 2016.
View Article : Google Scholar
|
|
44
|
Goltz D, Gevensleben H, Dietrich J,
Ellinger J, Landsberg J, Kristiansen G and Dietrich D: Promoter
methylation of the immune checkpoint receptor PD-1 (PDCD1) is an
independent prognostic biomarker for biochemical recurrence-free
survival in prostate cancer patients following radical
prostatectomy. OncoImmunology. 5:e12215552016. View Article : Google Scholar :
|
|
45
|
Holmes EE, Goltz D, Sailer V, Jung M,
Meller S, Uhl B, Dietrich J, Röhler M, Ellinger J, Kristiansen G,
et al: PITX3 promoter methylation is a prognostic biomarker for
biochemical recurrence-free survival in prostate cancer patients
after radical prostatectomy. Clin Epigenetics. 8:1042016.
View Article : Google Scholar :
|
|
46
|
Meller S, Zipfel L, Gevensleben H,
Dietrich J, Ellinger J, Majores M, Stein J, Sailer V, Jung M,
Kristiansen G, et al: CDO1 promoter methylation is associated with
gene silencing and is a prognostic biomarker for biochemical
recurrence-free survival in prostate cancer patients. Epigenetics.
11:871–880. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gonzàlez-Porta M, Frankish A, Rung J,
Harrow J and Brazma A: Transcriptome analysis of human tissues and
cell lines reveals one dominant transcript per gene. Genome Biol.
14:R702013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Soussi T, Leroy B and Taschner PE:
Recommendations for analyzing and reporting TP53 gene variants in
the high-throughput sequencing era. Hum Mutat. 35:766–778. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Thadani-Mulero M, Portella L, Sun S, Sung
M, Matov A, Vessella RL, Corey E, Nanus DM, Plymate SR and
Giannakakou P: Androgen receptor splice variants determine taxane
sensitivity in prostate cancer. Cancer Res. 74:2270–2282. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Carstens RP, Eaton JV, Krigman HR, Walther
PJ and Garcia-Blanco MA: Alternative splicing of fibroblast growth
factor receptor 2 (FGF-R2) in human prostate cancer. Oncogene.
15:3059–3065. 1997. View Article : Google Scholar
|
|
51
|
Anczuków O and Krainer AR: Splicing-factor
alterations in cancers. RNA. 22:1285–1301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lee Y and Rio DC: Mechanisms and
regulation of alternative Pre-mRNA splicing. Annu Rev Biochem.
84:291–323. 2015. View Article : Google Scholar : PubMed/NCBI
|